Abstract
Purpose
To elucidate additional substrate specificities of ALDH1B1 and determine the effect that human ALDH1B1 polymorphisms will have on substrate specificity.
Methods
Computational-based molecular modeling was used to predict the binding of the substrates propionaldehyde, 4-hydroxynonenal, nitroglycerin, and all-trans retinaldehyde to ALDH1B1. Based on positive in silico results, the capacity of purified human recombinant ALDH1B1 to metabolize nitroglycerin and all-trans retinaldehyde was explored. Additionally, metabolism of 4-HNE by ALDH1B1 was revisited. Databases queried to find human polymorphisms of ALDH1B1 identified three major variants: ALDH1B1*2 (A86V), ALDH1B1*3 (L107R), and ALDH1B1*5 (M253V). Computational modeling was used to predict the binding of substrates and of cofactor (NAD+) to the variants. These human polymorphisms were created and expressed in a bacterial system and specific activity was determined.
Results
ALDH1B1 metabolizes (and appears to be inhibited by) nitroglycerin and has favorable kinetics for the metabolism of all-trans retinaldehyde. ALDH1B1 metabolizes 4-HNE with higher apparent affinity than previously described, but with low throughput. Recombinant ALDH1B1*2 is catalytically inactive, whereas both ALDH1B1*3 and ALDH1B1*5 are catalytically active. Modeling indicated that the lack of activity in ALDH1B1*2 is likely due to poor NAD+ binding. Modeling also suggests that ALDH1B1*3 may be less able to metabolize all-trans retinaldehyde and that ALDH1B1*5 may bind NAD+ poorly.
Conclusions
ALDH1B1 metabolizes nitroglycerin and all-trans-retinaldehyde. One of the three human polymorphisms, ALDH1B1*2, is catalytically inactive, likely due to poor NAD+ binding. Expression of this variant may affect ALDH1B1-dependent metabolic functions in stem cells and ethanol metabolism.
Keywords: ALDH1B1, aldehyde dehydrogenase, substrate specificity, polymorphism, computational modeling
Introduction
The aldehyde dehydrogenase superfamily (ALDH) is a group of enzymes responsible for the metabolism of a diversity of exogenous and endogenous aldehydes, ranging from the developmentally crucial retinaldehyde to acetaldehyde, a major toxic byproduct of ethanol consumption (1). ALDH1B1, first described in 1991 (2), is a NAD+-dependent mitochondrial enzyme capable of metabolizing short chain aldehydes, including acetaldehyde (3). Studies in humans and mice have revealed ALDH1B1 to be expressed at high levels in the liver, intestinal tract, cornea, lens, testes and, to a lesser extent, heart and lung (2, 4, 5).
Enzymes with high amino acid similarity often share similar substrate specificities. ALDH1B1 shares 72 percent amino acid identity with ALDH2, and 64 percent amino acid identity with ALDH1A1. ALDH2 has been shown to possess three types of catalytic activity, namely aldehyde dehydrogenase, esterase, and nitroglycerin reductase (6). ALDH1B1 has previously been reported to catalyze two of these activities: aldehyde dehydrogenase and esterase (4). It is not known whether ALDH1B1 has nitroglycerin reductase activity. As noted, initial reports of ALDH1B1 substrate specificity indicated a preference for short chain aldehydes, including acetaldehyde and propionaldehyde (3). More recently, Stagos and colleagues described a broader range of substrates for ALDH1B1, including acetaldehyde (Km = 55 μM), benzaldehyde (Km = 50 μM), and p-nitrophenyl acetate (Km = 288 μM). Unlike ALDH2, ALDH1B1 metabolizes 4-HNE very poorly (Km = 3,383 μM) but had some activity towards malondialdehyde (Km = 466 μM) (5), making it unlikely that ALDH1B1 plays a large role in detoxifying these products of lipid peroxidation. Retinoic acid signaling plays a role in the development and homeostasis of many human tissues (7). The oxidation of retinaldehyde to the biologically active retinoic acid represents another important ALDH family function and there is some evidence that ALDH1B1 may play a role in retinoic acid signaling. For example, ALDH1B1 activity may be downregulated by retinoic acid and retinaldehyde (8), and a role for ALDH1B1 in granulocytic development of human hematopoietic stem cells has been proposed through a mechanism involving retinoic acid signaling (9). In addition, ALDH1B1 has been shown to be a stem cell / progenitor marker in the development of the pancreas (10). Finally, the amino acid sequence similarity between ALDH1B1 and traditional retinaldehyde-metabolizing enzymes, such as the ALDH1A subfamily, lends further support to the possibility that ALDH1B1 may play a role in these pathways.
ALDH2 plays an important role in the metabolic activation of nitroglycerin (11), an anti-anginal drug that has been used for more than a century. Individuals lacking ALDH2 activity (e.g., those possessing the ALDH2*2 polymorphism) retain some responsiveness to nitroglycerin, suggesting the existence of alternate, ALDH2-independent pathways of activation (12). Nitroglycerin acts as a potent inhibitor of ALDH2. Through such an action, nitroglycerin inhibits its own bioactivation (leading to its diminished efficacy with continued administration, a process called tolerance) and can cause ALDH2 dysfunction (13).
Given the range of substrates metabolized by ALDH1B1, it is important to understand mutations that could affect its activity. Early studies found ALDH1B1 to be polymorphic (2, 14). A search of current databases revealed three polymorphisms which are non-synonymous and present at a frequency of at least 1% - ALDH1B1*2 (Ala86Val), ALDH1B1*3 (Leu107Arg), and ALDH1B1*5 (Met253Val) (Table 1). ALDH1B1*4 has been named, but is a silent (synonymous) mutation. ALDH polymorphisms can have significant pathophysiological sequelae. For example, a polymorphism of ALDH2, ALDH2*2, causes marked reductions in acetaldehyde metabolism and consequent flushing syndrome and ethanol avoidance in hetero- and homozygotes. Early studies in a limited number of subjects found no significant association between the ALDH1B1 genotypes and alcoholism or alcohol aversion (14, 15). A more recent, larger study examined associations between polymorphisms in several ALDHs (including ALDH1B1) and alcoholism and cardiovascular risk factors including weekly alcohol intake, cholesterol, triglycerides, and systolic and diastolic blood pressure. Individuals with ALDH1B1*2 exhibited increased non-drinking behaviors (average of less than one drink per week), as well as increased systolic blood pressure (16). No associations were found in ALDH1B1*3 individuals. The same research group performed a follow-up study that expanded the population used by Hussemoen et al. (2008) to include an additional 6,784 adults (17). It examined associations between ALDH genotypes and a variety of behavioral and physiological factors including ethanol consumption behaviors and ethanol hypersensitivity reactions, such as itchy runny nose, sneezing, shortness of breath, rash, itching or swelling. An increase in the number of alcohol hypersensitivity reactions was observed in ALDH1B1*2 individuals, suggesting increased acetaldehyde toxicity, which is consistent with poorer metabolism of acetetaldehyde by ALDH1B1. As was the case in the previous study, ALDH1B1*3 polymorphisms did not correlate with any change in epidemiological parameters.
Table 1.
Computational modeling of interactions between ALDH isozymes and substrates.
| ALDH1A1 | ALDH2 | ALDH1B1*1 | ALDH1B1*2 | ALDH1B1*3 | ALDH1B1*5 | |
|---|---|---|---|---|---|---|
| Propionaldehyde | ||||||
| Km (μM) | 21.0 | 2.4 | 14.0 | ND | ND | ND |
| CYS H bond | 2.48 (150 | 151) | 1.97 (161 | 144) | 1.88 (165 | 145) | 1.90 (164 | 162) | 1.92 (161 | 105) | 1.89 (157 | 156) |
| ASN H bond | 1.91 (164 | 90) | 2.42 (131 | 94) | 2.07 (142 | 114) | 3.21 (106 | 116) | 2.38 (145 | 119) | 3.42 (105 | 93) |
| Interaction Energy (kcal / mol) | ||||||
| Total | −21.26 | −22.80 | −22.28 | −23.52 | −18.49 | −18.79 |
| Electrical | −10.17 | −13.56 | −14.43 | −15.49 | −9.55 | −9.59 |
| Van der Waals | −11.09 | −9.24 | −7.85 | −8.04 | −8.93 | −9.20 |
| 4-HNE | ||||||
| Km (μM) | 1.7 | 0.9 | 18.5 | ND | ND | ND |
| CYS H bond | 2.04 (161 | 117) | 1.92 (166 | 146) | 1.85 (174 | 152) | 1.82 (160 | 156) | 2.11 (149 | 93) | 1.86 (177 | 126) |
| ASN H bond | 1.90 (158 | 107) | 1.93 (147 | 93) | 1.90 (156 | 104) | 2.92 (105 | 136) | 3.10 (152 | 155) | 2.14 (134 | 114) |
| Interaction Energy (kcal / mol) | ||||||
| Total | −43.92 | −36.62 | −45.40 | −42.74 | −40.25 | −40.23 |
| Electrical | −25.41 | −14.27 | −23.90 | −24.48 | −15.57 | −16.30 |
| Van der Waals | −18.51 | −22.35 | −21.50 | −18.26 | −24.67 | −23.93 |
| All-trans retinaldehyde | ||||||
| Km (μM) | 26.8 | NS | 24.9 | ND | ND | ND |
| CYS H bond | 2.08 (153 | 134) | NP | 1.78 (176 | 137) | 1.84 (177 | 166) | NP | 1.82 (164 | 140) |
| ASN H bond | 1.82 (165 | 92) | NP | 2.32 (134 | 115) | 2.48 (113 | 100) | NP | 2.99 (109 | 114) |
| Interaction Energy (kcal / mol) | ||||||
| Total | −58.82 | NP | −55.50 | −63.18 | NP | −56.95 |
| Electrical | −13.22 | NP | −17.92 | −25.52 | NP | −17.05 |
| Van der Waals | −45.60 | NP | −37.59 | −37.66 | NP | −39.89 |
| Nitroglycerin | ||||||
| Km (μM) | ND | 11.3 | ND | ND | ND | ND |
| CYS H bond | 2.33 (143 | 118) | 2.26 (143 | 116) | 2.01 (176 | 140) | 2.05 (168 | 131) | 2.43 (170 | 90) | 2.13 (167 | 105) |
| ASN H bond | 2.25 (144 | 133) | 2.52 (133 | 125) | 2.90 (111 | 140) | 2.96 (105 | 142) | 2.00 (154 | 138) | 293 (98 | 169) |
| Interaction Energy (kcal / mol) | ||||||
| Total | −39.35 | −38.65 | −44.25 | −48.47 | −36.58 | −44.27 |
| Electrical | −10.46 | −12.09 | −17.89 | −23.48 | −10.23 | −16.47 |
| Van der Waals | −28.88 | −26.56 | −26.36 | −25.00 | −26.35 | −27.80 |
H bonds are in the format - length Å (θ AHD | θ HAY)
ND - no data is available or this substrate was not examined
NS - not a substrate for this enzyme
NP - no binding pose found
Given the number of known and proposed roles for ALDH1B1 in vitro and in vivo, and the effects that have been shown in population association studies, it is important to understand the substrate specificity of ALDH1B1 and the impact polymorphisms have on the function of ALDH1B1. Computational modeling of the binding of substrates to ALDH1B1 can be used to predict the substrate specificity of ALDH1B1 and the impact mutations may have. This modeling is facilitated and underpinned by an understanding of the well-studied and highly conserved catalytic mechanism of the aldehyde dehydrogenase activity of the ALDH superfamily. A catalytic cysteine (CYS319 in ALDH1B1) makes a nucleophilic attack on the carbonyl carbon of the aldehyde, removing a hydride ion which reduces NAD+ to NADH. A glutamate (GLU285 in ALDH1B1) serves as a general base (or activates a water molecule to do so), attacking the carbonyl carbon with sulfur as a leaving group. The side chain amide nitrogen of an asparagine (ASN186 in ALDH1B1) and the peptide nitrogen of the catalytic cysteine stabilize the oxyanion in the thiochemical transition state and orient the thiohemiacetal for hydride transfer to NAD+ (18, 19).
We have used computational methods to investigate the binding of known and previously unreported substrates to ALDH1B1. Using recombinant human ALDH1B1, we have characterized the enzyme kinetics of two additional substrates of ALDH1B1: nitroglycerin and all-trans retinaldehyde. Based on the results of the computational docking, we also revisited enzyme kinetics of another substrate, 4-HNE. In addition, we have created computational models of ALDH1B1 and its polymorphic variants, and docked them against known substrates in order to: 1) provide a physicochemical basis for observed epidemiological differences, and 2) predict differences in substrate specificities that might arise from polymorphic variants. We then expressed ALDH1B1 and its polymorphic variants in vitro in order to confirm the results of our in silico docking studies. Finally, we have investigated the computational models of ALDH1B1 variants to provide a mechanism for the results seen in vitro.
Methods
Computational modeling
Modeling was performed for a total of six proteins: ALDH1A1, ALDH2, ALDH1B1 and the variants, ALDH1B1*2, ALDH1B1*3 and ALDH1B1*5. Wild-type ALDH1B1 is sometimes referred to as ALDH1B1*1 where it might be otherwise confused with one of its variants. ALDH1A1 and ALDH2 were included in experiments as positive controls since they are established metabolizers of all-trans retinaldehyde (ALDH1A1), acetaldehyde (ALDH2), 4-HNE (ALDH2) and nitroglycerin (ALDH2). Crystal structures were downloaded from the Protein Data Bank (20). The B subunit of human ALDH2 (PDB ID: 1O01, (21)) was used directly for docking. Homology models were created for human ALDH1B1 (from ALDH2 – PDB ID: 1O01) and for human ALDH1A1 (from sheep ALDH1A1 – PDB ID: 1BXS, (22)). Once mitochondrial leader sequences are removed, ALDH1B1 is the same length as ALDH2 (i.e., 517 AA), and these sequences align with no gaps, making ALDH2 an ideal template for creating a homology model of ALDH1B1. An alignment of ALDH1B1 and ALDH2 is provided in supplemental Figure S1. ALDH1B1*2, ALDH1B1*3 and ALDH1B1*5 were created as mutations of the ALDH1B1 homology model. Homology models were created in MODELLER 9.12 (23). 100 models were created using random seeds and the best model was picked by DOPE score. The best model was then minimized using NAMD 2.9 (24). Briefly, the protein was solvated with explicit water (TIP3P) molecules (30Å minimum padding in each direction) and 20 mM MgCl2 was added as a buffer. All molecules were typed with the CHARMM force field (CHARMM22 for proteins and CGenFF for small molecules). Energy minimization was calculated using periodic boundary conditions until the average step size was less than 0.001 kcal (approximately 50,000 steps). Minimization was performed twice, first with the protein held rigid to minimize the solvent only, and then with the entire system allowed to move.
Substrates were prepared using MGLTools (v1.5.6) and then docked into homology models using AutoDock Vina (v1.1.2) (25) 100 times using random seeds. Ligands were treated as flexible, but isomerization was not allowed, e.g. from all-trans retinaldehyde to 9- or 13-cis retinaldehyde. Hydrogen bond lengths and angles were measured between substrates and critical amino acids (with a cutoff of 3.5Å) using the BINANA python script (26), with minor modifications to output values that were calculated internally but not reported in the published script. Among the multiple poses found for each protein-substrate interaction, the best pose was selected from those poses that made two critical interactions: hydrogen-bonding of the side-chain amide nitrogen of asparagine (ASN186) and the peptide nitrogen of the catalytic cysteine (CYS319) to the carbonyl oxygen of the substrate. Where multiple poses were found that met this requirement, the pose that had the minimum hydrogen bond distances and maximum AHD and HAY bond angles for these interactions was chosen. The best poses (approximately 10) for each substrate/protein interaction were chosen and subjected to energy minimization as described above. The highest ranked final pose was selected from the minimized poses using the same criteria described above.
Interaction energy (i.e., the sum of pairwise Van der Waals and electrostatic energy between the ligand and protein) was calculated for each final minimized docking pose using NAMD.
The cofactor NAD+ was also docked into ALDH1B1 and variant homology models as described above. Hydrogen bonds between cofactor (NAD+) and ALDH protein were measured up to 3.5Å in length. The best pose was selected based on interactions and similarity to positioning reported in the literature for other ALDHs (18). As a crude measure of the position of NAD+ relative to the substrate, the distance between the carbonyl oxygen of the docked pose of propionaldehyde and the center of the nicotinamide ring of the docked pose for NAD+ was measured. As a measure of hydrogen bond conservation, the number of amino acids making hydrogen bonds to NAD+ were counted as described previously for ALDH2 by Steinmetz and colleagues (18).
The root mean square distance (RMSD) between the α-carbon of amino acid residues of wild-type vs. variant proteins was measured for individual amino acids, for each element of secondary structure, and for the entire protein using MODELLER 9.12. Overlays of wild-type vs. variant proteins were created using structural alignments performed by Discovery Studio Visualizer (Accelrys, San Diego, CA).
ALDH substrate metabolism in vitro
Recombinant human wild-type ALDH1A1, ALDH1B1, and ALDH2 proteins were expressed in SF9 cells and purified by FPLC as described previously (4). ALDH1B1 and ALDH2 were activated by incubation in 50 mM β-mercaptoethanol for 1 h in quantities to yield 1 mM β-mercaptoethanol in the final reaction. ALDH1A1 does not require pre-activation, so β-mercaptoethanol was added with reaction buffers to achieve a final concentration of 1mM.
To monitor the metabolism of nitroglycerin over time, 20 μM nitroglycerin (5mg/mL, ethanol 30% v/v, propylene glycol 30% v/v - American Reagent Inc., Shirley, NY) was added to 25 μg ALDH protein in a buffer containing 1mM NAD+, 1mM glutathione and 1mM dithiothreitol. Reactions were performed in triplicate. Aliquots of reaction mixtures were taken at 0, 1, 2, 5, 10, 30, 60, and 120 min. Reactions in aliquots were quenched by adding 50:50% ice-cold acetonitrile / water, centrifuged at 15,000 RPM for 15 min in a microcentrifuge, and then analyzed by ultra performance liquid chromatography (UPLC). Negative controls (buffered system with no ALDH protein) were included. UPLC analysis (Acquity UPLC, Waters, Milford, MA) of nitroglycerin metabolites was performed using a BEH C18 column (1.7 μm). The UPLC reverse phase consisted of a linear gradient of 0% to 95% B using the solvents - A: 100% acetonitrile and B: 95% water / 5% acetonitrile. Quantitation was performed by comparing peak areas against standard curves of nitroglycerin, 1,2 DNG and 1,3 DNG.
To determine kinetic parameters of all-trans retinaldehyde metabolism, 2.2 μg of activated ALDH protein (or no-enzyme controls) were added to a solution of all-trans retinaldehyde (3.9 - 62.5 μM) in a sodium-pyrophosphate buffer containing 1 mM NAD+ and 1 mM pyrazole (N=7). These experiments were performed under minimal light conditions. After incubation for 30 min, reactions were quenched by adding 50/50% ice-cold acetonitrile / 1-butanol containing the internal standard retinyl acetate, and then extracted and analyzed as described previously (27). Samples were analyzed by UPLC using a mobile phase of 79.5% acetonitrile, 0.5% acetic acid, and 20% water. Quantitation was performed by comparing peak areas against standard curves of retinoic acid, retinaldehyde and retinyl acetate. Kinetic parameters were calculated using SigmaPlot 12 (Systat Software, Inc., San Jose, CA).
To determine the kinetic properties of 4-HNE, 10 μg activated ALDH protein was added to a solution of 4-HNE (2-128 μM) in a sodium-pyrophosphate buffer containing 2 mM NAD+ and 1mM pyrazole (N=4). Production of NADH, used to measure ALDH catalytic activity, was monitored spectrophotofluorometrically at 450 nm (excitation 340 nm; SpectraMax Gemini EM) and normalized to a NADH standard curve. The reaction was monitored and provided a linear increase in NADH from 5 min post substrate addition to 30 min.
ALDH1B1 polymorphisms
ALDH1B1 polymorphic variants were retrieved from listings in the Uniprot, NCBI's refSNP, and GeneCards databases (28-30). Three variants were found that were non-synonymous and with a population frequency exceeding 1%. Sequences were downloaded from the Uniprot database. In this study, amino acids are numbered from the beginning of the translated protein sequence, which include the 17 amino acid mitochondrial leader sequence in amino acid numbering. In some databases, amino acids are numbered without the leader sequence, e.g. ALDH1B1*2 A86V may be listed in some references as A69V. Frequency data were obtained from NCBI's refSNP database. Variant frequency data by race were obtained from NCBI's refSNP database by combining HapMap3 data into the major racial groups African (AFR), Asian (ASN), European (EUR), Indian (IND), and Mexican (MEX).
Generation of ALDH1B1 variant proteins in vitro
Human ALDH1B1 cDNA (NM_000692.3) was purchased from Origene (Rockville, MD) (5). To remove the mitochondrial leader sequence from ALDH1B1, Y19 was mutated to MET, creating an NdeI restriction site. The modified cDNA sequence was then cloned into the pET-15b vector using NdeI and BamHI restriction sites. The expressed protein is HIS-tagged and has the modified sequence MGSSHHHHHHSSGLVPRGSHMSSA... where the underlined MET replaces Y19 and begins the native human ALDH1B1 sequence. This modified plasmid was created and generously provided by the laboratory of Dr. Tom Hurley (Indiana University, Indianapolis, IN). The pET-15b hALDH1B1 plasmid was transformed into E. coli BL21-DE3 Tuner cells.
Cells were expressed in 6 l batches by seeding 15 ml of LB broth (all media was supplemented with 100 μg/ml carbenicillin) with culture from glycerol stocks and growing overnight at 37° C (all growth periods were performed with shaking). This 15 ml culture was centrifuged to remove media then resuspended in 90 ml fresh LB broth and grown for 3 h at 37° C. 15 ml of this culture was added to each of 4 flasks containing 1.5 l media and then grown at 37° C to 0.8 OD600. Flasks were cooled to 16° C and then induced with 0.1 mM IPTG and grown at 16° C for 24 h. Cells were harvested by centrifugation at 5,000 RPM for 15 min and frozen overnight at −80 C. Pellets were thawed and resuspended in a lysis buffer (20 mM Hepes pH 8.0, 0.5 M NaCl, 2 mM β-mercaptoethanol) containing protease inhibitors (cOmplete protease inhibitor cocktail; Roche, Indianapolis, IN) and 1 mg/ml lysozyme (from chicken egg white; Sigma-Aldrich, St. Louis, MO) by gentle shaking at room temperature for 30 min. The cell suspension was subjected to 4 freeze-thaw cycles, i.e., complete freezing in liquid nitrogen followed by thawing in a shaking water bath at 37° C. The suspension was drawn through an 18G needle 10x to shear genomic DNA followed by brief sonication to complete shearing. This solution was ultracentrifuged at 35,000 RPM for 1 h. The cleared lysate was then purified using a Ni-NTA column by applying the lysate to the column and washing with 10 column volumes of lysis buffer containing 10mM imidazole followed by 5 column volumes lysis buffer containing 60 mM imidazole. Protein was eluted with 5 column volumes of lysis buffer containing 250 mM imidazole and concentrated / desalted using centrifugal filter units (Amicon Ultra; Sigma-Aldrich, St. Louis, MO) in 10 mM Tris buffer (pH 7.8). Human ALDH1B1 was verified by denaturing gel electrophoresis (SDS-PAGE) followed by either Coomassie Blue staining or immunoblotting with human ALDH1B1 antibodies (data not shown).
Variant plasmids for ALDH1B1*2, ALDH1B1*3 and ALDH1B1*5 were created by Custom DNA Constructs (University Heights, OH) via site-directed mutagenesis and verified by nucleotide sequencing. Variant proteins were expressed and purified as above.
To determine the specific activity of bacterially expressed ALDH1B1 and variants, ≈ 20 μg activated ALDH protein was added to a solution of propionaldehyde (10 mM) in a sodium-pyrophosphate buffer containing 2 mM NAD+ and 1mM pyrazole (N=4). Production of NADH, used to measure ALDH catalytic activity, was monitored spectrophotofluorometrically at 450 nm (excitation 340 nm; SpectraMax Gemini EM) and normalized to a NADH standard curve. The reaction was monitored and provided a linear increase in NADH from 5 min post substrate addition to 30 min.
Results
Molecular modeling of ALDH1B1 substrate binding
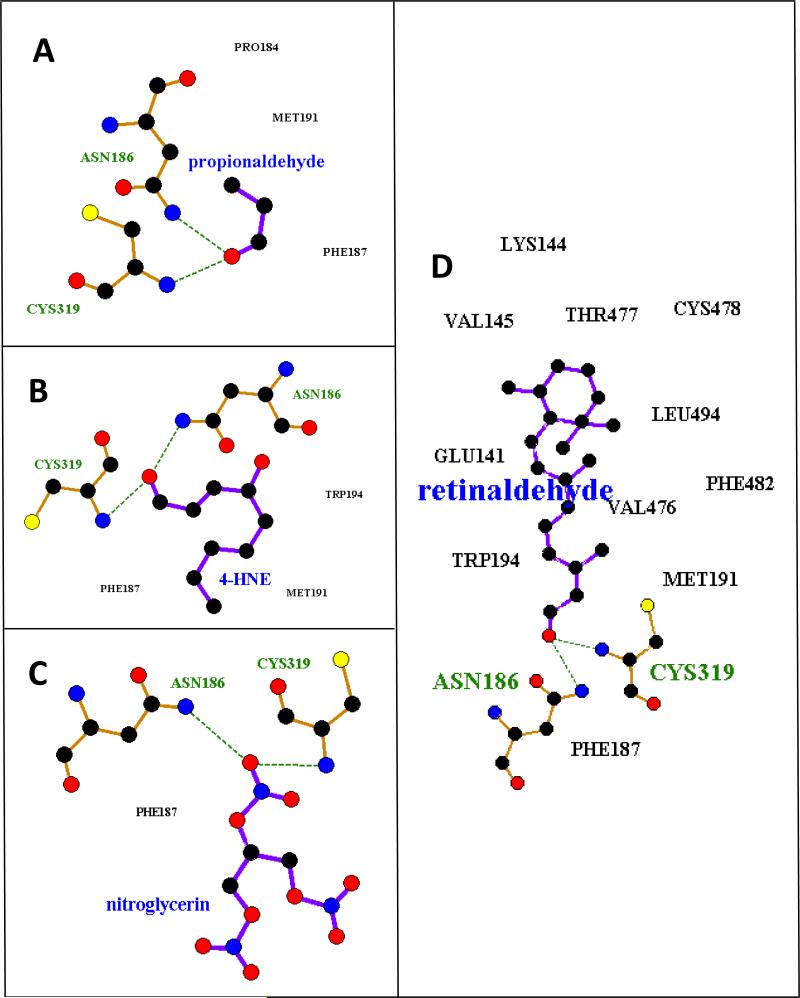
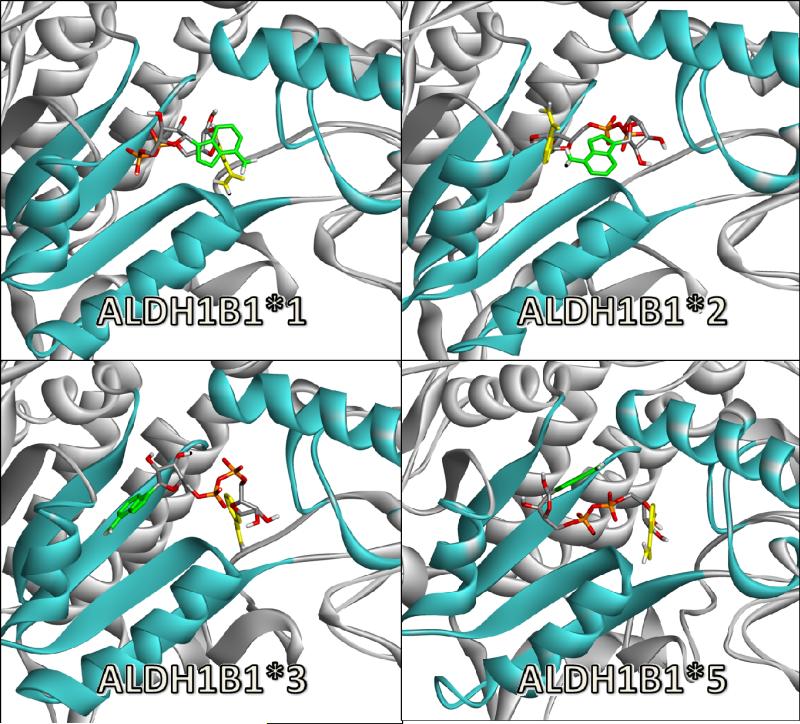
As noted, ALDH2 was used as a positive control because it is known to efficiently metabolize propionaldehyde (Km 2.4 μM (31)), 4-HNE (Km 0.9 μM (32)) and nitroglycerin (Km 11.3 μM (33)). ALDH1A1 was used as a positive control for all-trans retinaldehyde, but is also known to metabolize propionaldehyde (Km 21.0 μM (34)), 4-HNE (Km 1.7 μM (32)) and nitroglycerin. ALDH1B1 has previously been shown to metabolize propionaldehyde efficiently (Km 14.0 μM (4)). Poses containing the two critical hydrogen bonds (ASN, CYS, described above) were identified. Poses without these interactions were scored as non-interacting. Table 1 lists hydrogen bond lengths found (up to 3.5 Å), as well as calculated interaction energies between the substrate and protein. Apparent binding affinity (Km) is also listed if they have been experimentally determined. Although there is no direct correlation between interaction energies or poses and Km, each of the positive control substrates had Km values in the lower micromolar range, indicative of relatively strong binding. Each of the four substrates bound to ALDH1A1 with appropriate docking poses. Similarly, for ALDH2, poses were found for propionaldehyde, 4-HNE, and nitroglycerin. No appropriate docking pose was found for all-trans retinaldehyde with ALDH2, which was expected since ALDH2 has a much narrower substrate binding pocket than ALDH1A1, making it less likely to accommodate larger substrates. Each of the four substrates correctly bound to ALDH1B1 (ALDH1B1*1). Figure 1 shows two dimensional representations of the binding poses for each substrate with ALDH1B1. Additional three dimensional representations of these poses are presented in supplemental Figure S2. Multiple hydrophobic interactions were found for all-trans retinaldehyde, and to a somewhat lesser extent 4-HNE and propionaldehyde. ALDH1B1 has been previously shown to metabolize propionaldehyde, but was reported to have a poor affinity for 4-HNE, which was inconsistent with the appropriate docking poses that were consistently found. Additionally, good docking poses were found for the untested substrates all-trans retinaldehyde and nitroglycerin. To verify the functional implications of these in silico results, we examined the metabolism of 4-HNE, all-trans retinaldehyde and nitroglycerin by ALDH1B1 in vitro.
Figure 1.
Representative docking poses for substrates of ALDH1B1. Amino acids of ALDH1B1 that make hydrogen bonds to the substrate are displayed and labeled in green (hydrogen atoms and bond order are not shown). Amino acids that make hydrophobic interactions with the substrate are labeled in black. Key: Carbon – black, Oxygen – red, Nitrogen – blue, Sulfur – yellow. This figure was created in LigPlot+ (v1.4.5) (55).
Metabolism of all-trans retinaldehyde by ALDH1B1 in vitro
All-trans retinaldehyde is an established substrate for ALDH1A1. In the present study, ALDH1A1 metabolized all-trans retinaldehyde with a Km of 26.8 ± 7.1 μM and a Vmax of 74.2 ± 23.6 nMol/min/mg protein. ALDH1B1 had a similar Km of 24.9 ± 10.7 μM and a lower Vmax of 20.0 ± 7.6 nMol/min/mg protein for all-trans retinaldehyde (Table 2).
Table 2.
Kinetic values for the metabolism of select substrates by ALDH isozymes.
| Substrate | Protein | Km (μM) | Vmax (nmol / min / mg protein ) | Vmax / Km |
|---|---|---|---|---|
| All-trans retinaldehyde | ALDH1A1 | 26.8 ± 7.1 | 74.2 ± 23.6 | 2.8 |
| ALDH1B1 | 24.9 ± 10.7 | 20.0 ± 7.6 | 0.8 | |
| 4 - HNE | ALDH1B1 | 18.5 ± 4.1 | 10.3 ± 0.4 | 0.6 |
Metabolism of 4-HNE by ALDH1B1 in vitro
In this study, ALDH1B1 metabolized 4-HNE with a Km of 18.5 ± 4.1 μM and Vmax of 10.3 ± 0.4 nmol/min/mg protein (Table 2).
Metabolism of nitroglycerin by ALDH1B1 in vitro
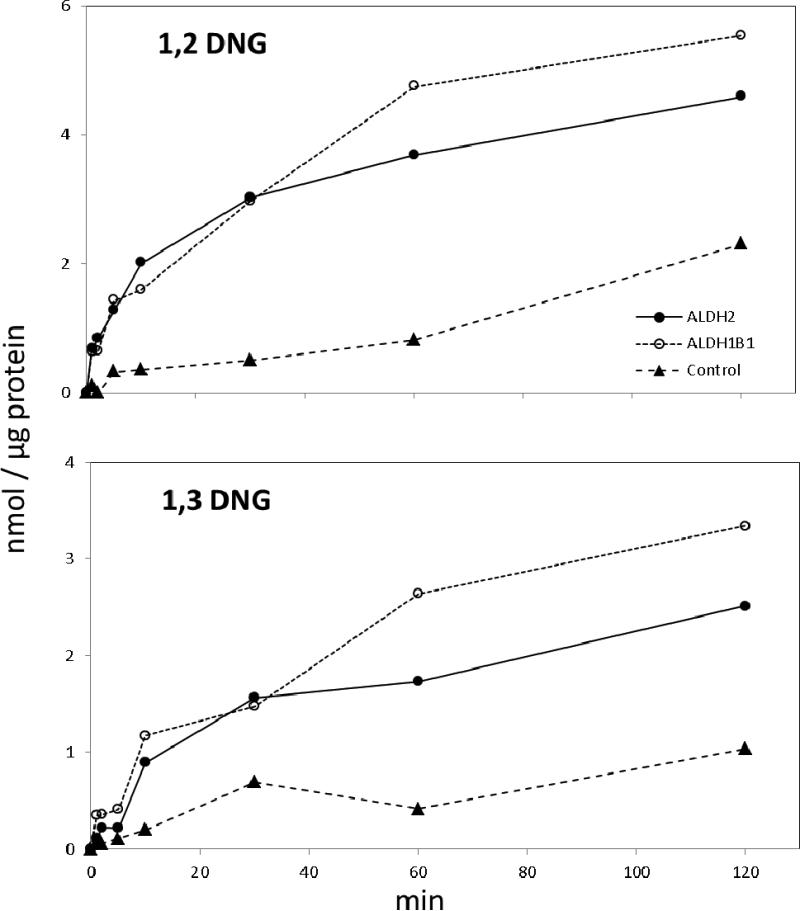
At a 20 μM substrate concentration, ALDH1B1 metabolized nitroglycerin to 1,2-DNG and 1,3-DNG at rates comparable to ALDH2 (Figure 2). Rates of 1,2 DNG production by both enzymes declined sharply after 10 min without depletion of nitroglycerin (data not shown), which is consistent with inhibition of these enzymes by nitroglycerin. Initial rates of catalysis were calculated for both enzymes at 10 minutes. ALDH2 produced 0.20 ± 0.02 nmol 1,2-DNG/min/μg protein, and 0.09 ± 0.02 nmol 1,3-DNG/min/μg protein. ALDH1B1 produced 0.16 ± 0.06 nmol 1,2-DNG/min/μg protein, and 0.12 ± 0.03 nmol 1,3-DNG/min/μg protein. For both enzymes, rates of 1,2-DNG production was higher than rates of 1,3-DNG production (ratios of 1,2-DNG/1,3-DNG for ALDH2 and ALDH1B1 were 2.3, and 1.4, respectively).
Figure 2.
Metabolism of nitroglycerin by recombinant ALDH2 and ALDH1B1. ALDH2 (closed circles), ALDH1B1 (open circles) or buffer solution containing no ALDH protein (closed triangles) were incubated with 20 μM nitroglycerin in a buffer containing NAD+, glutathione and DTT. The rate of production of 1,2 dinitroglycerin (1,2 DNG) (upper panel) and 1,3 dinitroglycerin (1,3 DNG) (lower panel) after nitroglycerin addition was studied by UPLC analysis and normalized to the amount of ALDH protein. For consistency, the negative controls were divided by 25 μg protein, the same as each experimental group.
Human polymorphisms of ALDH1B1
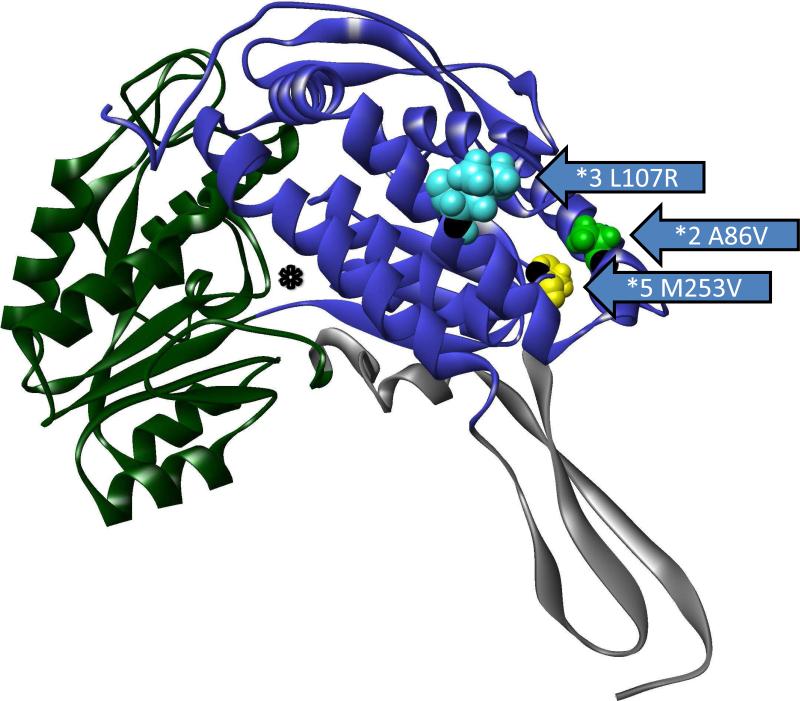
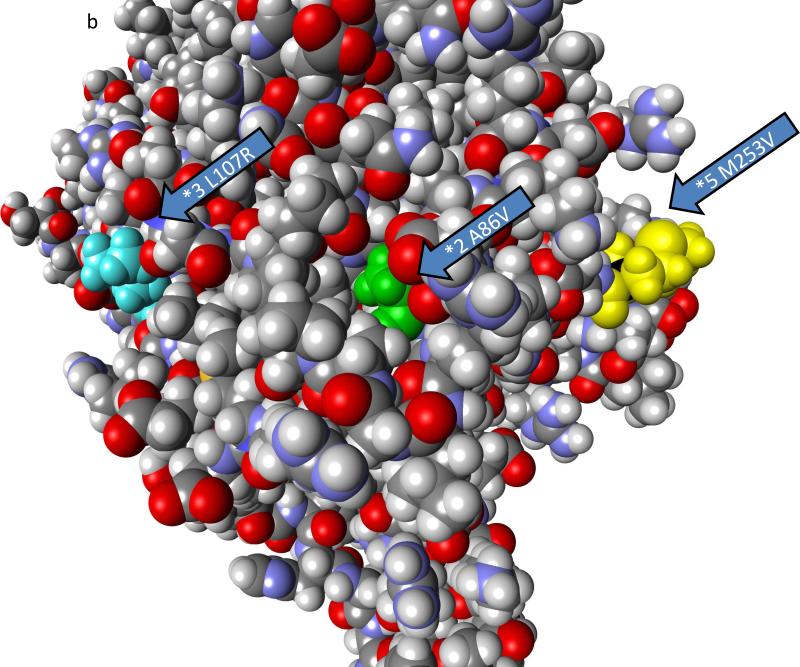
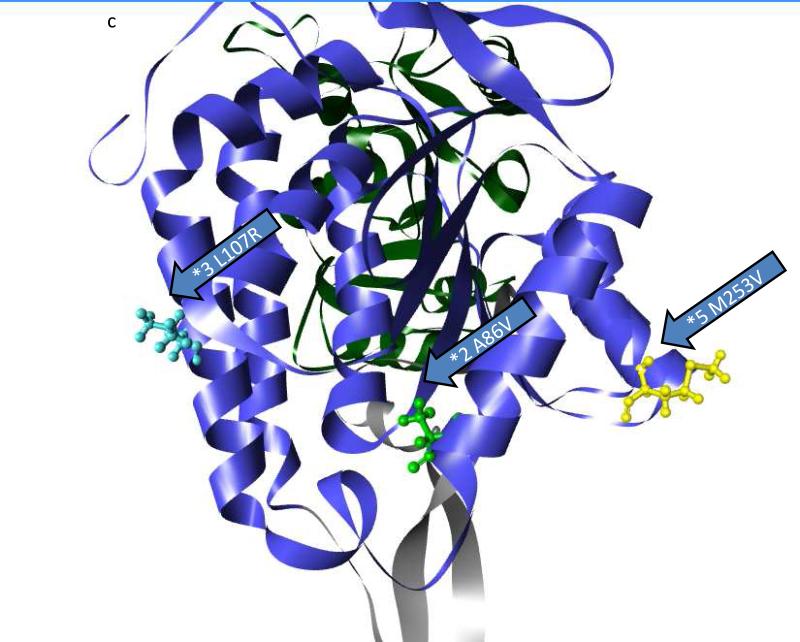
Three polymorphic variants of ALDH1B1 were found in sequence databases that caused amino acid changes and were present at frequencies of greater than 1%. These include ALDH1B1*2 (A86V – dbSNP: rs2228093), ALDH1B1*3 (L107R – dbSNP: rs2073478), and ALDH1B1*5 (M253V – dbSNP: rs4878199) (Table 3). Population frequencies found in the 1000 genomes project and the HapMap3 project and frequency by race are provided in Table 3. The frequency of mutations varied between the races such that the ALDH1B1*2 variant are the most common in Asian and Mexican populations and the ALDH1B1*3 variant was most common in African, European and Indian populations. The ALDH1B1*5 was least frequent in the Asian, European, Indian and Mexican populations (Table 3). The mutations in all three polymorphic variants are present in the NAD+ binding domain of ALDH1B1 (Figure 3a, Figure S1). By homology to ALDH2 (18), ALDH1B1*2 is located in the αA helix, and the amino acid side chain faces inward toward the core of the protein (Figure 3b, 3c, Figure S1). ALDH1B1*3 is located on the αB helix and faces outward at the surface of the protein (Figure 3b, 3c, Figure S1). ALDH1B1*5 is located on the loop between the αF helix and β10 sheet and faces outward at the surface of the protein (Figure 3b, 3c, Figure S1). None of these substitutions are at positions that are involved in the monomer-monomer (dimer-forming) or dimer-dimer (tetramer-forming) interfaces. It should be noted that while ALDH1B1*3 (L107R) does not participate in the interfaces directly, ASN106 forms a hydrogen bond across the dimer axis. This residue is part of an α-helix secondary structure, and thus the residue at position 107 is rotated away from this interface and faces outwards in a tetramer homology model (data not shown), and is unlikely to interact directly across this interface.
Table 3.
Polymorphisms of human ALDH1B1, and variant frequency by race.
| mRNA | Protein | Frequency | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant | Position | Wild-Type | Variant | Position | Wild-Type | Variant | 1000 Genomes | HapMap3 | dbSNP ref |
| ALDH1B1*2 | 369 | C (GCC) | T (GTC) | 86 | A [Ala] | V [Val] | 0.22 | 0.25 | rs2228093 |
| ALDH1B1*3 | 432 | T (CTC) | G (CGC) | 107 | L [Leu] | R [Arg] | 0.42 | 0.39 | rs2073478 |
| ALDH1B1*5 | 869 | A (ATG) | G (GTG) | 253 | M [Met] | V [Val] | 0.07 | 0.10 | rs4878199 |
| HapMap3 Frequency | |||||
|---|---|---|---|---|---|
| Variant | AFR | ASN | EUR | IND | MEX |
| ALDH1B1*2 | 0.11 | 0.41 | 0.14 | 0.27 | 0.39 |
| ALDH1B1*3 | 0.30 | 0.29 | 0.63 | 0.51 | 0.32 |
| ALDH1B1*5 | 0.18 | 0.00 | 0.05 | 0.01 | 0.02 |
Figure 3.
Location of polymorphisms of ALDH1B1. The location of the polymorphic amino acids are shown for ALDH1B1*2 (A86V; light green), ALDH1B1*3 (*3 L107R; light blue) and ALDH1B1*5 (*5 M253V; yellow). a) Homology model of an ALDH1B1 monomer. The protein structure is colored by domain as follows: substrate binding domain (green), cofactor (NAD+) binding domain (blue) and polymerization domain (grey). The asterisk (*) shows the substrate binding tunnel (as shown) which connects to the cofactor binding cavity (if viewed from behind the page). b) Space filling model of the ALDH1B1 protein showing the predicted relative exposure or burial of the mutated amino acids (shown are wild-type residues). c) The position of the polymorphic amino acids relative to secondary structures on the cofactor binding domain. Domains are colored as in (a). Figures were created in Discovery Studio Visualizer.
Computational modeling of ALDH1B1 polymorphisms
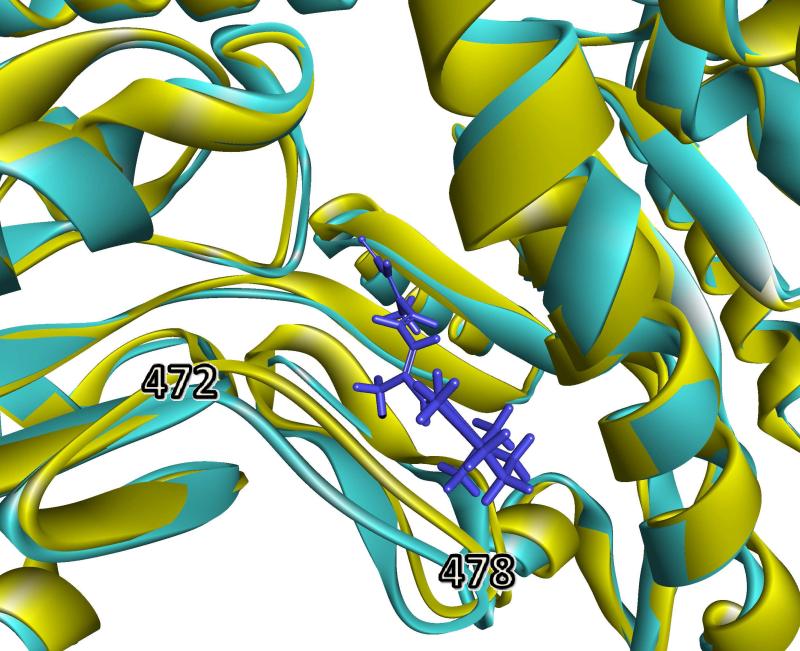
The substrates propionaldehyde, 4-HNE, all-trans retinaldehyde, and nitroglycerin were docked into homology models of ALDH1B1*2, ALDH1B1*3, and ALDH1B1*5 as described above. Poses similar to ALDH1B1*1 (wild-type) were found for all polymorphism / substrate combinations with the single exception that no appropriate docking pose was found for ALDH1B1*3 with all-trans retinaldehyde (Table 1). Figure 4 shows the homology model of ALDH1B1*3 superimposed upon the docked pose of all-trans retinaldehyde into ALDH1B1*1. It shows that the likely reason that no docking poses for all-trans retinaldehyde were found with ALDH1B1*3 was that in the homology model of ALDH1B1*3, a loop comprising amino acids 472-478 was shifted 2.4 Å towards the substrate binding pocket compared to wild-type, leaving insufficient room for docking of the bulky substrate. However, it should be noted that this loop is part of the dimer-forming interface. In the current study, ALDH1B1 is modeled as a monomer, and this allows extra flexibility in loops that would normally be stabilized by interactions between subunits. Thus, without further information, it is likely that this shift represents an artifact of modeling rather than a consequence of the polymorphic variant.
Figure 4.
Comparison of substrate-binding domain in ALDH1B1*1 and ALDH1B1*3. An overlay of ALDH1B1*3 (yellow) over ALDH1B1 wild type (light-blue) with all-trans retinaldehyde bound (dark blue stick representation). The loop comprising amino acids 472-478 was shifted 2.4 Å towards the substrate, blocking that binding position in ALDH1B1*3. This figure was created in Discovery Studio Visualizer.
Given that the human polymorphisms were all located in the cofactor binding domain, NAD+ was also docked against ALDH2, ALDH1B1, and each variant of ALDH1B1. ALDH2 was used as a positive control since it has a known crystal structure with cofactor (NAD+) bound. The binding poses for ALDH1B1 and its polymorphic variants were compared with hydrogen bond interactions reported by Steinmetz and colleagues (18) (Table 4). No individual docking experiment in silico was able to reproduce the exact binding pose reported for the crystal structure of ALDH2. However, each of the docking experiments reproduced five of the seven known hydrogen bonding interactions, with the exception of ALDH1B1*2 which only reproduced four. Notably, ALDH1B1*2 was the only protein that did not reproduce any of the three hydrogen bonding interactions nearest the substrate, i.e., LEU283, GLU416 and TRP185. As a crude measure of the position of NAD+ relative to the substrate, the distance between the carbonyl oxygen of propionaldehyde and the center of the nicotinamide ring of NAD+ was measured. For reference, in the literature, the measured distance for the hydride transfer pose for NAD+ for ALDH2 is 4.7 Å, and the hydrolysis pose for NAD+ has a distance of 7.9 Å (PDB ID: 1O00B and A, respectively, (21)). The distances calculated for ALDH1B1 and variants are presented in Table 4. The docked pose of ALDH1B1 and ALDH1B1*3 had calculated distances similar to that of the known hydride transfer pose. However, ALDH1B1*2 and ALDH1B1*5 both had distances more than double the distance of ALDH1B1, indicating that the docked pose placed the nicotinamide ring far away from the substrate. Qualitative representations of each cofactor binding pose are provided in Figure 5. Most docking poses were able to correctly place the backbone of NAD+ in the correct orientation (also shown in the hydrogen bond data in Table 4), but were differentiated by their placement of the nicotinamide ring and the adenine base. The pose that most resembled known interactions was ALDH1B1*3 which correctly oriented the nicotinamide ring towards the substrate and the adenine base in the cleft between the αF and αG helices (Figure 5). ALDH1B1 was positioned similarly but had the adenine base projecting out of the binding cleft towards the exterior of the protein. For ALDH1B1*5, both the nicotinamide ring and the adenine base projected outwards away from their binding clefts. Finally, the binding pose for ALDH1B1*2 was completely unsuitable, and reversed in overall orientation. The RMSD between Cα for each variant was calculated for each residue and overall for each variant protein compared to the wild-type protein (Table 5). The overall RMSD for each protein was between 0.72 and 0.97 Å. indicating that major deformations of the proteins due to the mutations are unlikely. This is also supported by similar overall minimization energies for the proteins (Figure S3). The RMSD for individual secondary structures was also low, similar to that seen between whole proteins (data not shown). When comparing the individual amino acids involved in binding NAD+, ALDH1B1*3 is most similar to ALDH1B1*1, with only one amino acid (SER263) with an RMSD greater than 1 Å, which is also shifted in each of the other variants. Both ALDH1B1*2 and ALDH1B1*5 have shifts in the LEU286 amino acid residue compared to wild-type. In both cases, the shift is away from the cofactor making interaction less likely. ALDH1B1*2 also has a shift in GLU416 towards the cofactor, likely disrupting the binding pocket further. In terms of these binding metrics, an overall binding suitability of ALDH1B1*3 > ALDH1B1*1 >> ALDH1B1*5 > ALDH1B1*2 is proposed. However, it is important to recognize that, due to the multitude of possible interactions and the flexibility of NAD+, multiple configurations are likely to exist in vivo.
Table 4.
Summary of docking poses for NAD+ binding to ALDH isozymes.
| ALDH2 | ALDH1B1*1 | ALDH1B1*2 | ALDH1B1*3 | ALDH1B1*5 | |
|---|---|---|---|---|---|
| Interaction Energy (kcal / mol) | |||||
| Total | −201.0 | −189.9 | −182.5 | −210.7 | −234.2 |
| Electrical | −156.1 | −132.5 | −129.7 | −160.2 | −189.8 |
| Van der Waals | −44.9 | −57.4 | −52.8 | −50.5 | −44.4 |
| Dist to substrate1 (Å) | 5.4 | 4.4 | 16.9 | 4.1 | 12.0 |
| H-bonds2 | 5(8) | 5(5) | 4(4) | 5(6) | 5(6) |
| LEU286 | LEU286 | LEU286 | |||
| GLU416 | GLU416x3 | GLU416 | GLU416 | GLU416 | |
| TRP185 | TRP185 | TRP185 | TRP185 | ||
| SER263 | SER263x2 | SER263 | SER263x2 | SER263 | |
| GLU212 | GLU212 | GLU212 | GLU212 | GLU212 | GLU212x2 |
| LYS209 | LYS209 | LYS209 | LYS209 | ||
| ILE183 | ILE183 | ILE183 | ILE183 | ||
Distance between the carbonyl oxygen of the docked propionaldehyde and the nicotinamide ring of NAD+
Number of unique hydrogen bonds to each amino acid with total hydrogen bonds in parentheses. Only interactions described for ALDH2 in Steinmetz et al. 1997 are shown. Where multiple interactions to the same amino acid was measured, ×2 or ×3 is indicated.
Figure 5.
Docking poses for NAD+ bound to ALDH1B1 and human variants. The elements comprising the cofactor binding cleft are colored blue and other elements of the protein are colored grey. The cofactor is shown in stick representation with the nicotinamide ring of NAD+ (highlighted in yellow) and the adenine base (highlighted in green).
Table 5.
Root mean square (RMSD) distances between ALDH1B1 variants and wild-type.
| RMSD (Å) | ALDH1B1*2 | ALDH1B1*3 | ALDH1B1*5 |
|---|---|---|---|
| Overall | 0.79 | 0.72 | 0.97 |
| LEU286 | 1.00 | 0.59 | 1.38 |
| GLU416 | 2.10 | 0.37 | 0.48 |
| TRP185 | 0.53 | 0.30 | 0.66 |
| SER263 | 1.44 | 1.03 | 1.32 |
| GLU212 | 0.43 | 0.36 | 1.06 |
| LYS209 | 0.16 | 0.17 | 0.43 |
| ILE183 | 0.72 | 0.53 | 0.66 |
RMSD values 1.0Å or greater are highlighted.
Recombinant expression of human ALDH1B1 and variants
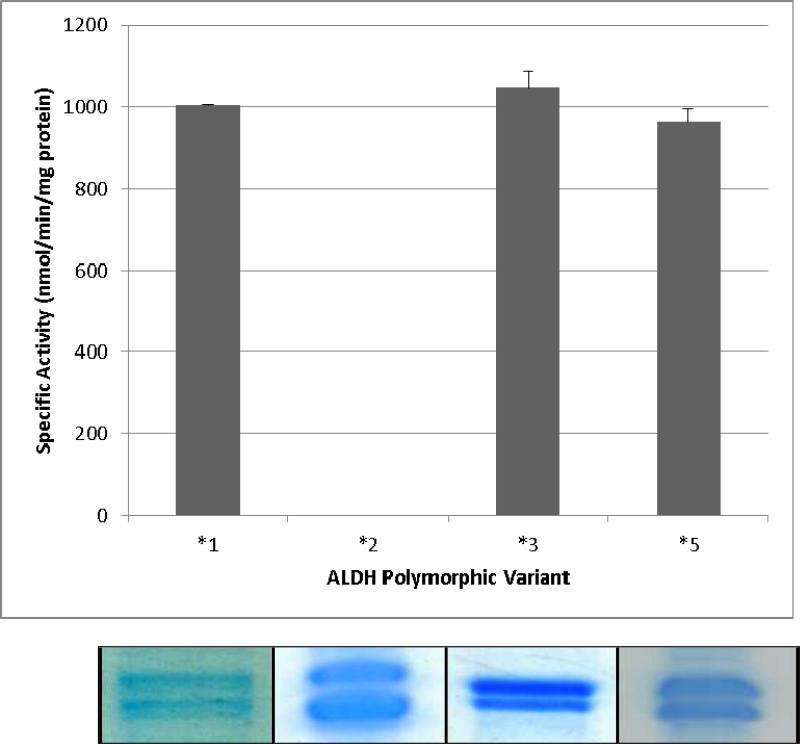
The approximate yield of ALDH1B1 and variants were similar at approximately 75 μg protein/l culture. Attempts to enhance the yield were unsuccessful as increased protein expression inevitably increased the insoluble fraction of the protein. This is consistent with the suggestion that expression of similar ALDH isozymes in this family is chaperone-dependent (35). ALDH1B1 and all variant proteins appeared as a double band between 55-58 kDa on SDS-PAGE (Figure 6). This has been previously observed when ALDH1B1 is expressed in a eukaryotic system (4). Immunobloting of ALDH1B1*1 protein using antibodies against human ALDH1B1 were successful and specific (data not shown). The specific activity of ALDH1B1 using propionaldehyde as a substrate and NAD+ as a cofactor under saturating conditions was 1,004 ± 2 nmol/min/mg protein. The specific activities for the variants were 0 nmol/min/mg protein for ALDH1B1*2, 1,048 ± 39 nmol/min/mg protein for ALDH1B1*3, and 962 ± 32 nmol/min/mg protein for ALDH1B1*5. There was no significant difference between the specific activities of ALDH1B1*1, ALDH1B1*3 and ALDH1B1*5 using an ANOVA and n = 3 (Figure 6).
Figure 6.
Expression and activity of ALDH1B1 variants. (top) The specific activity of ALDH1B1 polymorphic variants was estimated by measuring NADH production from NAD+ using propionaldehyde as a substrate. No significant difference (P > 0.05, ANOVA) was found between the wild-type (ALDH1B1*1) and ALDH1B1*3 or ALDH1B1*5. Data represent the mean ± SE from 3 experiments. (bottom) Coomassie-stained SDS-PAGE of recombinant ALDH1B1 proteins.
Discussion
ALDH1B1 substrate specificity
Computational modeling was used to investigate the substrate specificity of ALDH1B1. In these studies, previously examined (propionaldehyde and 4-HNE) and untested (nitroglycerin and all-trans retinaldehyde) substrates were all found to have favorable docking poses for ALDH1B1 in silico. Based on these results, additional enzyme kinetics studies were performed in vitro to verify the predicted metabolism of all-trans retinaldehyde and nitroglycerin. Moreover, 4-HNE, which was included in the in silico studies as a poor binder based on the previously reported apparent Km of (3,383 μM) (4), made favorable docking interactions in silico, so the kinetics of 4-HNE were revisited as well.
These studies revealed ALDH1B1 to be capable of metabolizing two previously untested substrates, nitroglycerin and all-trans retinaldehyde. Nitroglycerin is metabolized to 1,2 DNG and 1,3 DNG by both ALDH2 and ALDH1B1. A sharp decline in DNG formation occurred after the first 10 minutes for both ALDH isozymes, suggesting that, like ALDH2, ALDH1B1 is subject to rapid inhibition by nitroglycerin (36). This has potential therapeutic implications. First, nitroglycerin is bioactivated through metabolism by ALDHs and 1,2 DNG is thought to be the pharmacologically-active metabolite (37). Inactivation of ALDH2 is thought to underlie the diminishing vasodilator activity of nitroglycerin observed with maintained nitroglycerin exposure or therapy (36). The present results suggest that this may also apply to ALDH1B1. Second, by inhibiting ALDH2 and ALDH1B1, nitroglycerin treatment may adversely affect other physiological processes reliant upon the catalytic activity of these enzymes, such as the development and differentiation of cells due to retinoic acid signaling and the detoxification of exogenous and endogenous aldehydes. In the present study, 1,2 DNG was formed preferentially compared to 1,3 DNG by both ALDH1B1 and ALDH2, but not by a very large extent. This may indicate that, under the experimental conditions utilized, the enzymes were saturated, which has been shown to reduce product specificity in ALDH2. Although the present results are valuable in providing the first demonstration of the capacity of ALDH1B1 to metabolize nitroglycerin, limitations in the sensitivity of UPLC methods utilized in the present study prevent the determination of kinetic properties (i.e., Km and Vmax) for the metabolism of nitroglycerin by ALDH1B1. Future studies using LC/MS will be performed to better define the kinetic properties and the ratio of metabolites created by ALDH1B1 at lower, sub-saturating nitroglycerin concentrations. The inactivating ALDH2 polymorphism, ALDH2*2, has been shown to have 7-10 fold lower activity against nitroglycerin than the wild-type ALDH2, similar to the reduced activity seen for this isozyme for aldehyde substrates (33, 38). Nevertheless, a study in individuals with ALDH2*2 genotypes found that sublingual nitroglycerin retained efficacy in 36.1% of individuals (compared with 81.1% in wild-type ALDH2 individuals) (12). This suggests the presence other enzymes capable of catalytically-activating nitroglycerin. The results of the present study support the hypothesis that ALDH1B1 may be one such enzyme and may serve as an important contributor to the efficacy of nitroglycerin in vivo.
All-trans retinaldehyde is known to be metabolized by ALDH1A1. The present study found the kinetic properties of ALDH1A1 for catalyzing all-trans retinaldehyde to have a Km of 26.8 μM and a Vmax of 74.2 nmol/min/mg protein, a result somewhat higher, but similar to previous reports (e.g. Km = 8.1 μM (39)). Consistent with the computational data, ALDH1B1 showed favorable kinetics for retinaldehyde metabolism in vitro. ALDH1B1 was found to have a similar Km of 24.9 μM but a lower Vmax of 20.0 nmol/min/mg protein than ALDH1A1 for all-trans retinaldehyde. Given the role of retinoic acid signaling in cell development and differentiation, these results, when combined with ALDH1B1 associations with development of hematopoietic stem cells (9) and recent reports that ALDH1B1 may be a cancer stem cell marker (40), suggest that ALDH1B1 may play a role in development and differentiation. Should this indeed be the case, disruption of such a function by inactivating mutations would be predicted to have physiological and pathophysiological consequences.
As noted, computational analyses in the present study showed 4-HNE to have a favorable docking pose with ALDH1B1. This seems inconsistent with the previous observation that 4-HNE is a poor substrate for ALDH1B1 with an apparent Km of 3,383 μM and a Vmax of 2,043 nmol/min/mg protein (4). In the present study, ALDH1B1 was shown to metabolize 4-HNE with higher affinity (Km = 18.5 μM) but lower throughput (10.3 nmol/min/mg protein) than the previous study. In spite of these differing parameter values, our observed Vmax/Km of 0.56 is very similar to that previously reported (0.60) by Stagos and colleagues. The discrepancy is likely due to longer kinetic runs and the more sensitive measurements (fluorescent vs. visible absorbance spectrometry), which were better able to precisely measure the low activity in vitro. Thus, while the previously reported conclusion that 4-HNE is a poor substrate for ALDH1B1 remains valid, this new information is of particular relevance to studies which attempt to model or generalize the binding of substrates to ALDH1B1.
ALDH1B1 polymorphisms
Three human ALDH1B1 variants were discovered that met the criteria of polymorphism (i.e., > 1% frequency and non-synonymous) at of the time of database query. There is currently an explosion of sequence data becoming available as sequencing shifts from “the human genome” to projects like HapMap and the 1000 human genomes project. As more data becomes available, the “frequency in humans” will change, even assuming even coverage of human populations. It has been long known that genetic polymorphisms often have strong racial biases, and this was evident in ALDH variants as well (Table 3). ALDH1B1*2, the inactive variant, is especially prevalent (≈ 40%) in Mexican and Asian populations, and has significant representation (11-27%) in the other racial populations sampled. This is intermediate between ALDH1B1*3 which appears to be widely distributed and even dominant in some populations, possibly because it has little to no effect on enzyme activity, and ALDH1B1*5 which is almost exclusively found in African populations. Recently, another human polymorphism of ALDH1B1 has been reported (V176I - rs113083991), which is also found in the coenzyme binding domain (41). The prediction software PolyPhen-2, which classifies how likely polymorphisms are to affect protein function, correctly assigns ALDH1B1*2 as probably damaging and ALDH1B1*3 and ALDH1B1*5 as benign (42). This software also classifies the mutation V176I as benign. Given the large number of mutations being discovered, predictive software and computational modeling will continue to play an important role in screening mutations and prioritizing experimental work, especially in cases where the recombinant protein is either difficult or time-consuming to obtain.
Computational-based molecular modeling of ALDH1B1 and its polymorphic variants
In all cases except one, docking analyses in the present study suggested that ALDH1B1 polymorphic variants would be able to metabolize the same substrates as the wild-type enzyme. The one exception was that no docking pose was found for all-trans retinaldehyde binding to ALDH1B1*3. Computational modeling indicated that in silico, this was due to a shift in a loop that resulted in narrowing of the substrate binding pocket. As the bulkiest substrate in this study, and one likely to be physiologically important, this means that all-trans retinaldehyde may be a good substrate to test mutations in which the substrate binding cavity may be narrowed. Additional experiments should be carried out to test whether all-trans retinaldehyde metabolism by ALDH1B1*3 is affected in vitro.
Altered cofactor binding plays a role in the changes in catalytic activity of many enzyme variants. An example of this is the ALDH2 polymorphism, ALDH2*2, in which the change in NAD+ binding renders this enzyme catalytically inactive (43, 44). In the present study, ALDH2 and ALDH1B1 were shown to have conserved cofactor binding modalities, with many shared hydrogen interactions, which placed NAD+ in similar positions relative to the substrate, i.e., ≈ 5 Å distance. ALDH1B1*3 had the best NAD+ binding profile with the most conserved interactions and a location near the substrate, followed closely by ALDH1B1*1. These proteins were both fully active in vitro as well. ALDH1B1*2 had a poor binding profile, characterized by few conserved interactions and a location far from the substrate. Lack of cofactor binding is the most likely explanation for the complete lack of enzyme activity seen for ALDH1B1*2 in vitro, documented in the present study. ALDH1B1*5 had a relatively poor binding profile where, despite a number of favorable conserved hydrogen bond interactions, the best docking pose showed a nicotinamide ring that was not appropriately bound to the binding cleft, leaving it far away from the substrate. Similar to ALDH1B1*2, LEU286 was shifted away from the binding site, decreasing the likelihood of necessary interactions. However, this enzyme was fully active in vitro. There are several possibilities which could explain these apparently disparate findings. First, it is possible that ALDH1B1*5 does, in fact, bind NAD+ more poorly than wild-type enzyme, but this had no functional impact on the in vitro experiments because weak binding was overcome by high concentrations of cofactor. Second, the in silico results may simply reflect an artifact or error in the homology model or docking process which would not occur in vivo. Although protein expression was low, making it difficult to perform extensive kinetic studies for each substrate/cofactor, the apparent binding affinity of NAD+ with ALDH1B1*5 should be determined in the future to determine which of these possibilities is occurring.
The pathophysiological implications of ALDH1B1 mutations remain to be established. Known mutations in other ALDH family members have been shown to play a role in a number of disease states (1). Some of these include: increased risk for certain cancers and myocardial infarction with polymorphisms of ALDH2 (45-48); increased risk of spina bifida with polymorphisms of ALDH1A2 (49); γ-hydroxybutyric aciduria with polymorphisms of ALDH5A1 (50); developmental and metabolic abnormalities with polymorphisms of ALDH6A1 (51); and Sjögren-Larsson syndrome with polymorphisms of ALDH3A2 (52). Finally, a linkage analysis of Finnish families identified two chromosomal regions associated with bipolar disorder, 9p13.1, which contains ALDH1B1, among other candidate enzymes, and 7q31 (53). Given the proposed roles for ALDH1B1, it is no surprise that polymorphisms with diminished catalytic activity could have significant pathophysiological consequences.
Present in the liver and intestinal tract and possessing a favorable Km, ALDH1B1 is likely to contribute to both first-pass and systemic acetaldehyde detoxification. Preliminary observations show that ALDH1B1 knockout mice clear acetaldehyde more slowly than wild-type mice, providing additional evidence for a role of ALDH1B1 in ethanol metabolism (Singh and Vasiliou, manuscript in preparation). Previously ALDH1B1 has been shown to metabolize acetaldehyde, and all ALDH1B1 variants are predicted to be capable of binding acetaldehyde, as reflected in appropriate docking poses (data not shown). However, poor cofactor binding in ALDH1B1*2 may prevent this variant from being catalytically active. This would help explain why ALDH1B1*2 was associated with changes in acetaldehyde toxicity in population association studies and ALDH1B1*3 was not (16, 17).
In addition to the factors we have discussed here, other interactions that may affect the metabolic activity of ALDH1B1 variants warrant future experimental consideration. As one example, it will be important to know whether inactivating mutations are dominant or recessive. Similar to ALDH2, ALDH1B1 likely forms homotetramers. The ALDH2*2 variant is dominant negative, meaning that ALDH2*1/*2 heterotetramers are inactive and degraded (8, 54). Work by Linneberg and colleagues suggests that this may not be the case with ALDH1B1*2 because the prevalence of ethanol hypersensitivity reactions increased in a trend-wise fashion (15% in ALDH1B1*1/*1, 19% in ALDH1B1*1/*2, and 31% in ALDH1B1*2/*2) (17). Although genotypes were not statistically tested individually in that study, this single result is not consistent with a dominant negative interaction. Additionally, in the present study, substrate/cofactor interactions have been modeled as monomers. This is a logical initial approach due to (i) the greatly increased computational cost of modeling a full tetrameric protein, and (ii) none of the mutations appear to reside in protein-protein interfaces. While state-of-the-art molecular dynamics software was used, the analyses in this study can be performed on a single modern PC over the course of days to weeks. Modeling of larger systems, such as a tetramer, requires many months to compute and may require more advanced computational resources. These considerations notwithstanding, future modeling should examine the effect that tetramers may have on either (i) restraining the shifts caused by the mutations or (ii) propagating amino acid shifts to dimer or tetramer partners.
Conclusions
Computational-based molecular modeling studies allow prediction of enzyme catalytic activities and may provide a mechanistic explanation of experimental results. The results of the present study offer a possible physicochemical explanation for the differences in ethanol sensitivity between ALDH1B1*2 and ALDH1B1*3. We demonstrated that ALDH1B1 metabolizes nitroglycerin and all-trans retinaldehyde. Computational modeling predicts that some ALDH1B1 polymorphic variants will be catalytically inactive due to poor substrate binding, and/or poor cofactor binding. Clearly, the diminished catalytic activity of the variants may adversely impact physiological processes in which ALDH1B1 has a functional role. As the in vivo functions of ALDH1B1 become more clearly defined, it will be important for investigators to consider the impact polymorphic variants may have in the manifestation of diseases or in variations in the efficacy of therapeutic interventions.
Supplementary Material
Figure S1. Amino acid alignment of ALDH1B1 and ALDH2. An alignment was generated by T-COFFEE (56). Amino acids are classified as identical (*), highly similar (:), somewhat similar (.), or dissimilar ( ). Secondary structures are labeled by homology with ALDH2 with beta pleated sheets tinted blue and alpha helices tinted green. ALDH1B1 polymorphic variants are indicated by arrows and labeled as *2, *3, and *5.
Figure S2. Representative three dimensional docking poses for substrates of ALDH1B1. Amino acids of ALDH1B1 that make hydrogen bonds to the substrate are displayed as ball-and-stick figures and are labeled. Substrates are shown as stick figures (non-polar hydrogen atoms and bond order are not shown). Hydrogen bonds are shown as dashed green lines.
Figure S3. Scoring functions for ALDH1B1 wild type and variant homology models. Initial models were ranked by DOPE score, an unnormalized score with an arbitrary scale designed for choosing the most native-like model from multiple homology models (57). Also shown is the Z-score, a normalized DOPE score in which scores > 0 indicate relatively poor models, and scores < −1 are considered more ‘native-like’. The minimization energy of the final minimized homology model is also shown. Positive numbers or large differences in this score between models might indicate some major defect in protein structure.
Acknowledgements
The authors wish to thank the Computational Chemistry and Biology Core Facility at the University of Colorado Anschutz Medical Campus for their contributions to these studies. The authors wish to thank the laboratory of Dr. Tom Hurley (Indiana University, Indianapolis, IN) for providing the modified ALDH1B1 plasmid and for a critical reading of the manuscript. The Protein Production / Tissue Culture / MoAB Shared Resource at the University of Colorado Cancer Center provided expression of ALDH1B1 in eukaryotic cells, and is supported by the Cancer Center Support Grant (P30CA046934). This work was supported, in part, by the following NIH Grants – EY11490, AA021724, and AA022057. Fellowship support to B.C.J. (F31 AA020728) is also acknowledged.
Abbreviations
- 4-HNE
4-hydroxynonenal
- 1,3 DNG
1,3 dinitroglycerin
- 1,2 DNG
1,2 dinitroglycerin
References
- 1.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu LC, Chang WC. Cloning and characterization of a new functional human aldehyde dehydrogenase gene. J Biol Chem. 1991;266(19):12257–65. [PubMed] [Google Scholar]
- 3.Stewart MJ, Malek K, Xiao Q, Dipple KM, Crabb DW. The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun. 1995;211(1):144–51. doi: 10.1006/bbrc.1995.1789. [DOI] [PubMed] [Google Scholar]
- 4.Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, et al. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38(10):1679–87. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stagos D, Chen Y, Cantore M, Jester JV, Vasiliou V. Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res Bull. 2010;81(2-3):211–8. doi: 10.1016/j.brainresbull.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daiber A, Wenzel P, Oelze M, Schuhmacher S, Jansen T, Munzel T. Mitochondrial aldehyde dehydrogenase (ALDH-2)--maker of and marker for nitrate tolerance in response to nitroglycerin treatment. Chem Biol Interact. 2009;178(1-3):40–7. doi: 10.1016/j.cbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Theodosiou M, Laudet V, Schubert M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci. 2010;67(9):1423–45. doi: 10.1007/s00018-010-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabb DW, Stewart MJ, Xiao Q. Hormonal and chemical influences on the expression of class 2 aldehyde dehydrogenases in rat H4IIEC3 and human HuH7 hepatoma cells. Alcohol Clin Exp Res. 1995;19(6):1414–9. doi: 10.1111/j.1530-0277.1995.tb01000.x. [DOI] [PubMed] [Google Scholar]
- 9.Luo P, Wang A, Payne KJ, Peng H, Wang JG, Parrish YK, et al. Intrinsic retinoic acid receptor alpha-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells. 2007;25(10):2628–37. doi: 10.1634/stemcells.2007-0264. [DOI] [PubMed] [Google Scholar]
- 10.Ioannou M, Serafimidis I, Arnes L, Sussel L, Singh S, Vasiliou V, et al. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev Biol. 2013;374(1):153–63. doi: 10.1016/j.ydbio.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Foster MW, Zhang J, Mao L, Rockman HA, Kawamoto T, et al. An essential role for mitochondrial aldehyde dehydrogenase in nitroglycerin bioactivation. Proc Natl Acad Sci U S A. 2005;102(34):12159–64. doi: 10.1073/pnas.0503723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Chen YG, Xu F, Xue L, Jiang CX, Zhang Y. [The relationship between aldehyde dehydrogenase-2 gene polymorphisms and efficacy of nitroglycerin]. Zhonghua Nei Ke Za Zhi. 2007;46(8):629–32. [PubMed] [Google Scholar]
- 13.Beretta M, Sottler A, Schmidt K, Mayer B, Gorren AC. Partially irreversible inactivation of mitochondrial aldehyde dehydrogenase by nitroglycerin. J Biol Chem. 2008;283(45):30735–44. doi: 10.1074/jbc.M804001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman D, Dave V, Hsu LC, Peters TJ, Yoshida A. Diverse polymorphism within a short coding region of the human aldehyde dehydrogenase-5 (ALDH5) gene. Hum Genet. 1993;92(5):477–80. doi: 10.1007/BF00216454. [DOI] [PubMed] [Google Scholar]
- 15.Sherman DI, Ward RJ, Yoshida A, Peters TJ. Alcohol and acetaldehyde dehydrogenase gene polymorphism and alcoholism. EXS. 1994;71:291–300. doi: 10.1007/978-3-0348-7330-7_29. [DOI] [PubMed] [Google Scholar]
- 16.Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol Clin Exp Res. 2008;32(11):1984–91. doi: 10.1111/j.1530-0277.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 17.Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, et al. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy. 2010;40(1):123–30. doi: 10.1111/j.1365-2222.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- 18.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5(5):701–11. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 19.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4(4):317–26. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42(23):7100–9. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- 22.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6(12):1541–51. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 23.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 24.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durrant JD, McCammon JA. BINANA: a novel algorithm for ligand-binding characterization. J Mol Graph Model. 2011;29(6):888–93. doi: 10.1016/j.jmgm.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClean SW, Ruddel ME, Gross EG, DeGiovanna JJ, Peck GL. Liquid-chromatographic assay for retinol (vitamin A) and retinol analogs in therapeutic trials. Clin Chem. 1982;28(4 Pt 1):693–6. [PubMed] [Google Scholar]
- 28.Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39(Database issue):D214–9. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V. Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug Metab Dispos. 2005;33(5):649–56. doi: 10.1124/dmd.104.002964. [DOI] [PubMed] [Google Scholar]
- 32.Yoval-Sanchez B, Rodriguez-Zavala JS. Differences in susceptibility to inactivation of human aldehyde dehydrogenases by lipid peroxidation byproducts. Chem Res Toxicol. 2012;25(3):722–9. doi: 10.1021/tx2005184. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, et al. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116(2):506–11. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang MF, Han CL, Yin SJ. Substrate specificity of human and yeast aldehyde dehydrogenases. Chem Biol Interact. 2009;178(1-3):36–9. doi: 10.1016/j.cbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, Kim HS, Jeong HS, Lee YS. Chaperonin GroESL mediates the protein folding of human liver mitochondrial aldehyde dehydrogenase in Escherichia coli. Biochem Biophys Res Commun. 2002;298(2):216–24. doi: 10.1016/s0006-291x(02)02423-3. [DOI] [PubMed] [Google Scholar]
- 36.Sydow K, Daiber A, Oelze M, Chen Z, August M, Wendt M, et al. Central role of mitochondrial aldehyde dehydrogenase and reactive oxygen species in nitroglycerin tolerance and cross-tolerance. J Clin Invest. 2004;113(3):482–9. doi: 10.1172/JCI19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Stamler JS. Bioactivation of nitroglycerin by the mitochondrial aldehyde dehydrogenase. Trends Cardiovasc Med. 2006;16(8):259–65. doi: 10.1016/j.tcm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Beretta M, Gorren AC, Wenzl MV, Weis R, Russwurm M, Koesling D, et al. Characterization of the East Asian variant of aldehyde dehydrogenase-2: bioactivation of nitroglycerin and effects of Alda-1. J Biol Chem. 2010;285(2):943–52. doi: 10.1074/jbc.M109.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bchini R, Vasiliou V, Branlant G, Talfournier F, Rahuel-Clermont S. Retinoic acid biosynthesis catalyzed by retinal dehydrogenases relies on a rate-limiting conformational transition associated with substrate recognition. Chem Biol Interact. 2013;202(1-3):78–84. doi: 10.1016/j.cbi.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405(2):173–9. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Way MJ. Computational modelling of ALDH1B1 tetramer formation and the effect of coding variants. Chem Biol Interact. 2014;207:23. doi: 10.1016/j.cbi.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larson HN, Weiner H, Hurley TD. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem. 2005;280(34):30550–6. doi: 10.1074/jbc.M502345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J Biol Chem. 2007;282(17):12940–50. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oze I, Matsuo K, Hosono S, Ito H, Kawase T, Watanabe M, et al. Comparison between self-reported facial flushing after alcohol consumption and ALDH2 Glu504Lys polymorphism for risk of upper aerodigestive tract cancer in a Japanese population. Cancer Sci. 2010;101(8):1875–80. doi: 10.1111/j.1349-7006.2010.01599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoyama A, Muramatsu T, Omori T, Yokoyama T, Matsushita S, Higuchi S, et al. Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis. 2001;22(3):433–9. doi: 10.1093/carcin/22.3.433. [DOI] [PubMed] [Google Scholar]
- 47.Muto M, Hitomi Y, Ohtsu A, Ebihara S, Yoshida S, Esumi H. Association of aldehyde dehydrogenase 2 gene polymorphism with multiple oesophageal dysplasia in head and neck cancer patients. Gut. 2000;47(2):256–61. doi: 10.1136/gut.47.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jo SA, Kim EK, Park MH, Han C, Park HY, Jang Y, et al. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin Chim Acta. 2007;382(1-2):43–7. doi: 10.1016/j.cca.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Deak KL, Dickerson ME, Linney E, Enterline DS, George TM, Melvin EC, et al. Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2. Birth Defects Res A Clin Mol Teratol. 2005;73(11):868–75. doi: 10.1002/bdra.20183. [DOI] [PubMed] [Google Scholar]
- 50.Akaboshi S, Hogema BM, Novelletto A, Malaspina P, Salomons GS, Maropoulos GD, et al. Mutational spectrum of the succinate semialdehyde dehydrogenase (ALDH5A1) gene and functional analysis of 27 novel disease-causing mutations in patients with SSADH deficiency. Hum Mutat. 2003;22(6):442–50. doi: 10.1002/humu.10288. [DOI] [PubMed] [Google Scholar]
- 51.Chambliss KL, Gray RG, Rylance G, Pollitt RJ, Gibson KM. Molecular characterization of methylmalonate semialdehyde dehydrogenase deficiency. J Inherit Metab Dis. 2000;23(5):497–504. doi: 10.1023/a:1005616315087. [DOI] [PubMed] [Google Scholar]
- 52.Rizzo WB, Carney G. Sjogren-Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2). Hum Mutat. 2005;26(1):1–10. doi: 10.1002/humu.20181. [DOI] [PubMed] [Google Scholar]
- 53.Palo OM, Soronen P, Silander K, Varilo T, Tuononen K, Kieseppa T, et al. Identification of susceptibility loci at 7q31 and 9p13 for bipolar disorder in an isolated population. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(3):723–35. doi: 10.1002/ajmg.b.31039. [DOI] [PubMed] [Google Scholar]
- 54.Xiao Q, Weiner H, Crabb DW. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest. 1996;98(9):2027–32. doi: 10.1172/JCI119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–86. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 56.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 57.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Amino acid alignment of ALDH1B1 and ALDH2. An alignment was generated by T-COFFEE (56). Amino acids are classified as identical (*), highly similar (:), somewhat similar (.), or dissimilar ( ). Secondary structures are labeled by homology with ALDH2 with beta pleated sheets tinted blue and alpha helices tinted green. ALDH1B1 polymorphic variants are indicated by arrows and labeled as *2, *3, and *5.
Figure S2. Representative three dimensional docking poses for substrates of ALDH1B1. Amino acids of ALDH1B1 that make hydrogen bonds to the substrate are displayed as ball-and-stick figures and are labeled. Substrates are shown as stick figures (non-polar hydrogen atoms and bond order are not shown). Hydrogen bonds are shown as dashed green lines.
Figure S3. Scoring functions for ALDH1B1 wild type and variant homology models. Initial models were ranked by DOPE score, an unnormalized score with an arbitrary scale designed for choosing the most native-like model from multiple homology models (57). Also shown is the Z-score, a normalized DOPE score in which scores > 0 indicate relatively poor models, and scores < −1 are considered more ‘native-like’. The minimization energy of the final minimized homology model is also shown. Positive numbers or large differences in this score between models might indicate some major defect in protein structure.