Abstract
High-resolution digitalizing of histology slides facilitates the development of computational alternatives to manual quantitation of features of interest. We developed a MATLAB-based quantitative histological analysis tool (QuHAnT) for the high-throughput assessment of distinguishable histological features. QuHAnT validation was demonstrated by comparison with manual quantitation using liver sections from mice orally gavaged with sesame oil vehicle or 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; 0.001–30 µg/kg) every 4 days for 28 days, which elicits hepatic steatosis with mild fibrosis. A quality control module of QuHAnT reduced the number of quantifiable Oil Red O (ORO)-stained images from 3,123 to 2,756. Increased ORO staining was measured at 10 and 30 µg/kg TCDD with a high correlation between manual and computational volume densities (Vv), although the dynamic range of QuHAnT was 10-fold greater. Additionally, QuHAnT determined the size of each ORO vacuole, which could not be accurately quantitated by visual examination or manual point counting. PicroSirius Red quantitation demonstrated superior collagen deposition detection due to the ability to consider all images within each section. QuHAnT dramatically reduced analysis time and facilitated the comprehensive assessment of features improving accuracy and sensitivity and represents a complementary tool for tissue/cellular features that are difficult and tedious to assess via subjective or semiquantitative methods.
Keywords: bioinformatics, histopathology, morphometry, hepatic, TCDD, quantitative, high throughput
Introduction
Digital pathology and whole slide imaging (WSI) are becoming increasingly popular in research, clinical, and toxicological settings (Pantanowitz et al. 2011; Romero Lauro et al. 2013; Garrido et al. 2013; Ying and Monticello 2006). Digitized images from tissue sections facilitate storage and sharing of histological slides and allow quantitative analyses in a rapid and unbiased manner (Pantanowitz et al. 2011; Romero Lauro et al. 2013; Garrido et al. 2013; Ying and Monticello 2006). Furthermore, sharing of WSI is an effective strategy to minimize interlaboratory variation in histological assessments influenced by tissue quality and internal immunohistochemical protocols (Mengel et al. 2002; Oyama et al. 2007), by reducing inter- and intraobserver variation in the quantitation of histological features (Oyama et al. 2007; Mengel et al. 2002; Lawrie et al. 2012; Huang et al. 2013).
Quantitation of features of interest in clinical and toxicological studies largely consists of subjective or semiquantitative scoring systems used by trained pathologists (Ge et al. 2010; Gurcan et al. 2009). For example, the nonalcoholic steatohepatitis Clinical Research Network developed the nonalcoholic fatty liver disease (NAFLD) activity score to standardize NAFLD severity assessments using H&E-stained tissue sections based on a scale of 0 to 8 that considers lipid accumulation, inflammation, and hepatocellular ballooning (Levene et al. 2012; Nishida et al. 2013). Similarly, the Ishak system assesses liver fibrosis on a 0 to 6 scale based on observed characteristics (Standish et al. 2006; Ishak et al. 1995). Comparable scoring systems are commonly used in toxicity and carcinogenicity studies to determine exposure limits (Crissman et al. 2004; Boorman et al. 2002). However, these systems can be onerous and involve several pathologists or working groups for large studies (Boorman et al. 2002) that may not be entirely free of bias and subjectivity.
Alternatively, digital image analyses provide ratio scale (e.g., 0–100%) as opposed to ordinal scale measurements (e.g., severity scale of 0–8), thus improving the quantitative characterization of the histopathological response (Garrido et al. 2013; Huang et al. 2013). Manual point counting and computational feature extraction have been used to analyze digital images. Morphometry is commonly used for quantitation of histological features (Bringhenti et al. 2013; Frantz et al. 2013) aided by programs such as STEPanizer and Image-Pro Plus (Tschanz et al. 2011; Bringhenti et al. 2013; Frantz et al. 2013). Although visual-based assessment is the current standard for quantitation of histological features, it is time consuming, tedious, and vulnerable to inter- and intraobserver variability. To address these limitations, automated feature extraction approaches have been developed (Ge et al. 2010; Rexhepaj et al. 2008). Commercial tools are available for automated detection and quantitation of features of interest but typically require purchasing a license and rarely take advantage of advanced computational resources such as high performance or cloud computing that facilitate high-throughput analysis and the storage of memory-intensive digital data. In-house developed tools have also been created to examine specific histological features of interest. For example, a MATLAB algorithm was developed for the quantitation of estrogen and progesterone receptor staining in breast cancer tumor sections (Rexhepaj et al. 2008) but is not extendible to other types of features.
This study describes the development of the quantitative histological analysis tool (QuHAnT), a MATLAB-based automated image analysis tool developed for the high-throughput quantitative assessment of pathologist identified and characterized histopathological features of interest. Unlike other approaches, QuHAnT uses a modular framework for (1) threshold determination, (2) quality control, (3) feature extraction and quantitation, and (4) result output. Comparison to manual point counting using the dose-dependent increase in Oil Red O (ORO) and PicroSirius Red (PSR) staining in livers of mice following treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; Boverhof et al. 2005; Pierre et al. 2013; Lu et al. 2011; Kopec et al. 2010) demonstrated that QuHAnT outperforms manual point counting and provides additional complementary quantitative data. In summary, QuHAnT is an accurate and extensible high-throughput tool that can be used to quantitate distinguishable histological features that are difficult to assess using subjective or semiquantitative methods such as the modest increase in collagen deposition induced by TCDD and the dose-dependent increase in micro- versus macrovesicular steatosis.
Materials and Methods
Animal Husbandry and Treatment
Female C57BL/6 mice received on postnatal day (PND) 25 were obtained from Charles Rivers Laboratories (Portage, MI). Mice were housed in polycarbonate cages with cellulose fiber chips (Aspen Chip Laboratory Bedding, Northeastern Products, Warrensberg, NY) and maintained at 30 to 40% humidity and a 12-hr light/dark cycle. Mice were fed ad libitum with Harlan Teklad 22/5 Rodent Diet 8940 (Madison, WI) and had free access to deionized water. On PND28 (day 0) and every following 4th day (days 4, 8, 12, 16, 20, and 24), animals were orally gavaged with 0.1 ml sesame oil vehicle control or 0.001, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, or 30 µg/kg of TCDD (Dow Chemical Company, Midland, MI) for a total of 7 doses over 28 days. On day 28, mice were killed, and the right liver lobe was fixed in 10% neutral-buffered formalin (Sigma-Aldrich, MO) for collagen staining or frozen in Tissue-Tek O.C.T. compound (Sakura, CA) for lipid staining. All animal procedures were approved by the Michigan State University Institutional Animal Care and Use Committee (AUF08/12-149-00).
Histological Processing and WSI
All histological processing was performed at the Michigan State University Investigative HistoPathology Laboratory (humanpathology.msu.edu/histology). Staining of frozen liver sections for lipids using ORO (Sigma-Aldrich) was performed as previously described (Kopec et al. 2011). Briefly, livers were sectioned at 6 µm, stained with an ORO solution, and counterstained with Gill 2 hematoxylin (Thermo Fisher Scientific, MA). For collagen staining by PSR, paraffin-embedded livers were sectioned at 4 to 5 µm and stained with H&E and 0.1% PSR. Slides were digitized using the Olympus Virtual Slide System VS110 (Olympus, PA) at 20× magnification. For quantitative analyses of ORO and PSR staining, images were randomly sampled using Visiomorph Microimager (Visiopharm, Denmark) across 2 adjacent sections on the same slide (N = 6 individual livers) in each treatment group. The tif format was used to obtain the highest quality images, although any standard image format (e.g., jpeg, png) can be used.
Quantitative Analyses
Manual point counting was performed using STEPanizer (Tschanz et al. 2011). Briefly, a 4 × 4 or 16 × 16 test system grid was superimposed on each image and the number of points overlapping a feature of interest (e.g., ORO, PSR) were counted. Volume density was calculated as the sum of positive hits (Ppositive staining) divided by the total number of tissue hits (Ptissue) for each section (Vv = (Ppositive staining/Ptissue) × 100).
For QuHAnT analysis, hue, saturation, and value (HSV) thresholds were determined using the ImageJ color threshold tool (http://rsb.info.nih.gov/ij/). Prior to quantitation using QuHAnT, visual quality control was performed using the incorporated quality control module to remove images containing false positives or debris. Volume density was estimated as the sum of the area of positive staining (Apositive staining) divided by the sum of tissue area (Atissue) for each liver section (Vv = (Apositive staining/Atissue) × 100). Only features larger than 6.3 µm2 (4 pixels) were considered to minimize noise. For the assessment of the variability associated with slide coverage, 100% of slides from 3 animals (vehicle, 0.01, and 30 µg/kg) were sampled and quantitated and randomly selected a posteriori using an in-house written python script (www.python.org). QuHAnT analyses were performed using the Michigan State University High Performance Computing Center (HPCC). Statistical analyses were performed using SAS 9.2 (SAS Institute Inc., NC).
Results
Implementation of QuHAnT
Figure 1 provides an overview of the implementation of QuHAnT compared to manual point counting using the dose-dependent increase in ORO staining of hepatic vacuoles in mice following oral gavage with TCDD (Boverhof et al. 2005; Kopec et al. 2010; Lu et al. 2011; Pierre et al. 2013). For the assessment of QuHAnT performance, 42 to 55 images were randomly sampled using Visiomorph Microimager across 2 adjacent sections on the same slide for each animal in sesame oil vehicle control, 0.001-, 0.01-, 0.03-, 0.1-, 0.3-, 1-, 3-, 10-, or 30-µg/kg TCDD treatment groups (n = 6 animals) resulting in a total of 3,123 images across all the treatment groups.
Figure 1.
Overview of comparative histological feature quantitation analysis approaches for dose-dependent 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) elicited steatosis by manual point counting using STEPanizer (Tschanz et al. 2011) and the developed quantitative histological analysis tool (QuHAnT).
For QuHAnT analysis, HSV thresholds used for image segmentation (herein referred to as feature extraction) were determined using ImageJ (http://rsb.info.nih.gov/ij/) from a random subset of images (~1%of images) containing positive staining, background tissue, and/or blank background. For ORO, optimal HSV thresholds used for feature extraction were determined to be 0 to 50 and 225 to 255 (hue), 125 to 255 (saturation), and 0 to 255 (value), while optimal total tissue feature extraction thresholds were 0 to 255 (hue), 20 to 255 (saturation), and 0 to 255 (value). Of the 3,123 captured images, 367 were eliminated from further analysis due to the absence of any tissue or the presence of false positives (e.g., ORO droplets present in absence of tissue) or the presence of debris (e.g., bubbles) as identified using the quality control module within QuHAnT. Although the tool provides the option to perform image correction using external editors, this was not performed due to the availability of additional images to compensate for eliminated images. QuHAnT quantitated the remaining 2,756 images in less than 1 hr using the HPCC at Michigan State University. Data output consisted of 3, delimited files containing information of each individual feature within an image and background tissue area. The output summary provided feature count, mean size, and total area as well as background tissue information for each image. The data can then be imported into any common analysis software including SAS (SAS Institute Inc.) and R (http://www.r-project.org/). QuHAnT MATLAB code is freely available for download at dbzach.fst.msu.edu.
For validation, the same initial 3,123 images were also assessed using manual point counting. A 4 × 4 grid was superimposed on each image by STEPanizer (Tschanz et al. 2011), and the number of points overlapping an ORO-stained feature or unstained area were manually counted. In cases where errors were observed, overlapping points were ignored (not counted) as no initial quality screening was performed for manual point counting. Elimination of the same 367 images omitted from QuHAnT analysis for manual point counting did not result in significantly different Vv estimates (data now shown).
Comparing QuHAnT to Manual Point Counting Vv Estimates
Both manual point counting and QuHAnT analyses identified a dose-dependent ~5% and ~15% increase in hepatic ORO Vv estimates at 10- and 30-µg/kg TCDD, respectively (Table 1). Although high-dose Vvs were similar across both methods, estimates using manual point counting ranged from 0.4 to 14.6% while QuHAnT estimates ranged from 0.04 to 14%, representing a 10-fold increase in the dynamic range. Dose-dependent manual point counting and QuHAnT Vv estimates were consistent with visual assessment by a board-certified veterinary pathologist (J.H.) of whole liver slides, which reported mild to moderate hepatic lipid accumulation at 10-µg/kg TCDD and widespread microvesicular and macrovesicular lipid accumulation in the centriacinar, mid-zonal, and periportal regions of the liver at 30-µg/kg TCDD. Correlation analysis between manual point counting and QuHAnT Vv estimates revealed a high concordance between both measures (Pearson’s correlation coefficient of 0.82; Figure 2). QuHAnT estimates, although highly correlated to manual estimates overall, are ~10-fold lower than the corresponding manual point counting estimates at low Vv values while high Vv values are comparable (slope > 1). Similar high concordance between manual point counting and QuHAnT Vv estimates were found in ORO-stained liver sections processed and quantitated in an independent study using the same feature extraction HSV threshold values (data not shown).
Table 1.
Comparison of manual point counting and QuHAnT analysis of Oil Red O and PicroSirius Red volume density (Vv) estimates in ~50 images per slide of TCDD-treated mouse liver sections.
| Vv (% tissue area) | ||
|---|---|---|
| Dose (µg/kg TCDD) | Manual point counting | QuHAnT |
| Oil Red O | ||
| 0 | 0.36 ± 0.13 | 0.11 ± 0.03 |
| 0.001 | 0.60 ± 0.25 | 0.05 ± 0.01 |
| 0.01 | 0.48 ± 0.19 | 0.07 ± 0.03 |
| 0.03 | 0.65 ± 0.22 | 0.20 ± 0.10 |
| 0.1 | 0.41 ± 0.14 | 0.08 ± 0.04 |
| 0.3 | 0.85 ± 0.10 | 0.1 ± 0.04 |
| 1 | 2.43 ± 0.68 | 0.17 ± 0.06 |
| 3 | 3.28 ± 1.72 | 0.48 ± 0.17 |
| 10 | 5.63a ± 1.85 | 4.07a ± 1.26 |
| 30 | 14.61a ± 1.94 | 13.99a ± 3.56 |
| PicroSirius Red | ||
| 0b | 0.96 ± 0.26 | 0.80 ± 0.10 |
| 30b | 1.60 ± 0.43 | 1.34a ± 0.20 |
Note. ANOVA= analysis of variance; PSR= PicroSirius Red; QuHAnT= quantitative histological analysis tool; TCDD = 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Significant differences (p ≤ .05) compared to vehicle control determined by ANOVA followed by Dunnett’s post hoc test.
Manual point counting estimates for PSR was performed on a feasible ~50 images per slide and compared to 100% sampling for QuHAnT analysis.
Figure 2.
Correlation analysis of manual point counting and quantitative histological analysis tool (QuHAnT) estimated (A) Oil Red O and (B) PicroSirius Red volume densities (Vvs). Data points represent the mean Vv (percentage of tissue area) estimated for each liver section using 42 to 55 images at 20× magnification per liver section. A Pearson’s correlation coefficient of 0.82 (p ≤ .05) was calculated for Oil Red O Vv estimates. No significant correlation was observed for PicroSirius Red manual point counting and QuHAnT Vv estimates.
Superior QuHAnT Performance
Manual point counting estimates improve with increased grid density (i.e., 16 × 16) since more points are counted at the expense of analysis time. A sample image was quantitated using 4 × 4 and 16 × 16 grid densities in order to determine the tradeoff between accuracy and quantitation time, and for accuracy comparison to QuHAnT, which represents a theoretical ~1.3 million points (each pixel represents 1 point) per image (Table 2). The QuHAnT Vv estimate of 28% was found to be much closer to the 33% manual point counting estimate determined using a 16 × 16 grid compared to the 6% estimate on the 4 × 4 grid. Vv estimates of 28 to 33% for the sample image are also more consistent with visual assessment of the images. However, improved accuracy with increased grid density in manual point counting came a ≥5-fold increase in assessment time while QuHAnT analysis was ~5% of the time required for manual point counting using a 4 × 4 grid while simultaneously providing a more comprehensive assessment (i.e., considered each pixel).
Table 2.
Accuracy and time comparison for manual point counting and QuHAnT.
| Quantitation method |
Grid densitya (W × H) |
Volume density (%) |
Analysis time (image−1) |
Study analysis (hr)b |
|---|---|---|---|---|
| Point counting | 4 × 4 | 6 | 1–2 min | 45 |
| 16 × 16 | 33 | 5–6 min | 225 | |
| QuHAnT | 1,280 × 1,024 | 28 | 1 secb | 1c |
Note. QuHAnT = quantitative histological analysis tool.
Image used for comparison across grid sizes is shown in Figure 4 (top).
Manual point counting was performed on 3,123 images, and QuHAnT analysis was performed on 2,756 images.
Analysis time was determined based on the use of the Intel Xeon E5620 2.4 GHz core processor with TurboBoost within the High Performance Computing Center at Michigan State University.
Unlike manual point counting, computational approaches can also provide estimates for the number of individual features per tissue area, and the size distribution of features of interest at little to no cost in accuracy, effort, or time. Testing of this advantage was demonstrated using a previously established 15-µm diameter (~176 µm2 area) threshold to distinguish microvesicular from macrovesicular lipid droplets (Zaitoun et al. 2001). Applying this threshold, QuHAnT not only identified independent dose-dependent increases in microvesicular and macrovesicular lipid droplets but also distinguished the emergence of macrovesicular lipid droplets at lower doses (10-µg/kg TCDD, Table 3) compared to visual assessment by a pathologist (J.H.), who only reported macrovesicular lipid droplets at 30-µg/kg TCDD. However, it should be noted that this approach assumes that each distinct feature represents a single spherical droplet which, by visual assessment, appears to be largely true (droplets appear as circles in the images and are not expected to have any specific orientation).
Table 3.
Total count of lipid vacuoles representing microvesicular or macrovesicular steatosis determined using QuHAnT for ~50 images per slide of TCDD-treated mouse liver sections.
| Vacuolization type ([count/tissue area] × 10−6) | ||
|---|---|---|
| Dose (µg/kg TCDD) | Microvesicular (≤176 µm2)a |
Macrovesicular (≥176 µm2)a |
| 0 | 28.70 ± 8.55 | 1.42 ± 0.28 |
| 0.001 | 12.40 ± 3.32 | 0.62 ± 0.08 |
| 0.01 | 19.00 ± 11.2 | 0.76 ± 0.23 |
| 0.03 | 70.30 ± 38.6 | 1.14 ± 0.36 |
| 0.1 | 22.10 ± 12.2 | 1.09 ± 0.35 |
| 0.3 | 28.20 ± 12.8 | 1.03 ± 0.20 |
| 1 | 100.08 ± 71.5829 | 3.42 ± 2.61 |
| 3 | 117.26 ± 41.3956 | 6.47 ± 2.43 |
| 10 | 437.40b ± 71.298 | 97.30b ± 35.10 |
| 30 | 555.13b ± 65.1883 | 251.47b ± 56.02 |
Note. ANOVA = analysis of variance; QuHAnT = quantitative histological analysis tool; TCDD = 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Area of 176 µm2 was estimated based on a diameter of 15 µm previously reported to distinguish between microvesicular steatosis from macrovesicular steatosis (Zaitoun et al. 2001).
Significant differences (p ≤ .05) compared to vehicle control determined by ANOVA followed by Dunnett’s post hoc test.
Influence of Random Tissue Sampling on Vv Estimates
To quantitate a feasible number of images by manual point counting in a reasonable amount of time, ~50 images were randomly sampled for each pair of liver sections per slide representing only~40% of the tissue section. However, the high-throughput capability of QuHAnT allows whole slide analysis (quantitation of the whole liver section) with little additional effort and time. To investigate optimal sampling coverage, images from 3 liver sections were randomly sampled from ~1 to 100% a total of 50 times at each percentage (e.g., 20 images were randomly sampled 50 times), and the standard error of the mean was calculated to determine the variability in Vv estimates (Figure 3). Random sampling was associated with large variability when considering only 40% of the tissue section particularly for sections with a large Vv (30-µg/kg TCDD sample) or features with nonhomogenous distribution (e.g., perivenular fibrosis). In general, Vv estimates exhibited higher variability when <95% of the tissue section was considered. The effect of sampling coverage on Vv estimate variability was less in vehicle and in liver sections for lower TCDD doses with much lower ORO Vv estimates (Figure 4).
Figure 3.
Variability of (A) Oil Red O and (B) PicroSirius Red volume density (Vv) estimates using quantitative histological analysis tool (QuHAnT) with increasing slide coverage and random sampling of liver sections. Data points represent standard error of the mean for 50 individual randomly selected images of liver sections from mice dosed every 4 days for 28 days with sesame oil vehicle, 0.01-µg/kg 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; Oil Red O only), or 30-µg/kg TCDD; 40% slide coverage representing the coverage used for quantitation approach comparisons is represented by a dashed vertical line (left panel). The right panel (expanded inset from left panel [gray background]) represents 95% slide coverage where variability is minimized.
Figure 4.
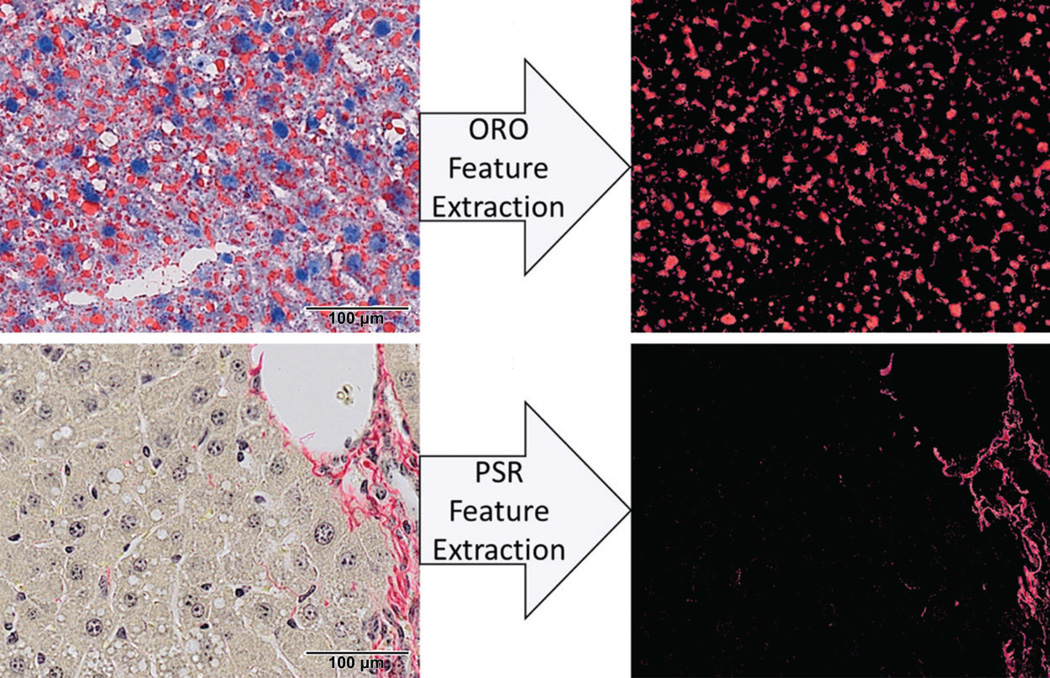
Examples of Oil Red O (top) and PicroSirius Red (bottom) images (left) and extracted features (right). Images represent a 20× liver section from mice dosed every 4 days for 28 days with 30-µg/kg 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). ImageJ was used to establish independent hue, saturation, and value (HSV) thresholds. In addition to quantitation of features of interest by quantitative histological analysis tool (QuHAnT), the digital images provide a digital copy that can be shared upon request for further quality control or independent visual assessment.
Extensibility of QuHAnT
In addition to hepatic fat accumulation, TCDD induces immune cell infiltration and collagen deposition in mice (Lu et al. 2011; Pierre et al. 2013; Boverhof et al. 2005), which was also observed in this study following visual assessment by a board-certified veterinary pathologist (J.H.). The extensibility of QuHAnT was investigated by examining PSR- (Table 1; Figures 2 and 3) and F4/80 (data now shown)-stained liver sections from the same animals. No significant increase in PSR staining was detected by manual point counting or by QuHAnt when only ~40% of randomly sampled images were analyzed. In contrast, a significant increase in PSR staining was detected at 30 µg/kg TCDD when 100% of the stained liver sections were sampled and quantified using QuHAnT. Although Vv values for PSR staining were similar for both approaches, there was no correlation between QuHAnT and manual point counting (Figure 2B). QuHAnT was able to detect perivenular and sinusoidal fibrosis missed by manual point counting following higher percentage of sampling of liver sections. Large variability in Vv estimates were found at low percentage of tissue sampling levels, while very little variation was observed at ≥95% tissue sampling, similar to ORO estimates (Figure 3, right panel). These results suggest manual counting could be improved by increasing the percentage of tissue sampling or the use of a mixed size point count grid with a coarse and fine mesh for the reference area and the structure on interest area, respectively, but at the expense of increased analysis time, effort, and labor. Unlike lipid vacuoles that can be assumed to be spherical with no specific orientation, collagen deposition form continuous fibers confound accurate counting of individual features or determination of feature size. Therefore, these parameters were not assessed for PSR staining, although QuHAnT could be used to calculate these values using standard morphometric concepts.
Discussion
Complex diseases and adverse effects induced by chemical or biological agents are rarely the result of disruption of a single gene or pathway. Histopathology can be used to identify apical responses and/or key events within a mode of action. It also places complex biological responses and potential interactions reflected in transcriptomic, proteomic, and metabolomic studies into cellular and morphological, as well as spatial and temporal, context within a tissue or specimen. Histopathology has historically been a qualitative technique with lesion severity scoring largely based on visual assessment, semiquantitative scoring, and/or manual quantitation (Ge et al. 2010; Gurcan et al. 2009; Hadi et al. 2010; Crissman et al. 2004; Boorman et al. 2002). Whole slide digital imaging represents an emerging approach that facilitates automated quantitative measures of histopathological features in a rapid and reproducible manner reducing human workload and minimizing inter- and intraobserver biases and variability (Pantanowitz et al. 2011; Romero Lauro et al. 2013; Garrido et al. 2013; Ying and Monticello 2006). Interoperability, and standards for image collection, analysis and reporting are in development to ensure rigorous and reproducible quantitative results between independent laboratories (Chieco et al. 2013; Eliceiri et al. 2012). In this study, we describe the development and testing of QuHAnT, a flexible high-throughput computational tool that reduces biases and variability for the quantitation of pathologist characterized features while dramatically reducing human workload.
ORO- and PSR-stained liver sections from mice dosed every 4 days for 28 days with TCDD were used to compare QuHAnT with manual point counting. TCDD elicits dose-dependent hepatic lipid accumulation and collagen deposition (Boverhof et al. 2005; Kopec et al. 2010; Lu et al. 2011; Pierre et al. 2013) and provides a relevant model for QuHAnT performance testing. Comparative analysis indicates QuHAnT accurately quantitated increases in lipid vacuolization compared to manual point counting as demonstrated by the high Vv value concordance between these approaches. These results confirm that QuHAnT is a valuable complement to pathologist-directed assessments and suggest that it is a more comprehensive approach for the quantitation of histological features compared to manual point counting (Hadi et al. 2010; Levene et al. 2012; Garrido et al. 2013).
Unlike ORO, pathologist observed that increases in PSR staining were detectable only using QuHAnT due to the ability to consider all the images comprising the section. PSR staining of collagen deposition was dispersed throughout the section as marbled veins highly localized to perivenular areas. Consequently, the manual quantitation of collagen deposition would require a more comprehensive and labor-intensive assessment. For example, PSR quantitation is highly correlated to automated analysis when using a 200-point grid for manual quantitation (Hadi et al. 2010). However, using a 200-point grid density or alternative manual approach is not practical when examining thousands of images further emphasizing the value of a high-throughput image analysis tool.
In addition to showing high concordance with point counting, QuHAnT provided feature size characteristics that cannot be easily acquired using manual approaches (Day and James 1998; Deutsch et al. 2014). More specifically, QuHAnT was used to provide the absolute number of vacuoles and determine their size distribution, another challenging manual quantitation task. These data were used to identify the dose-dependent transition from microvesicular to macrovesicular steatosis, which is associated with increasing disease severity (Day and James 1998; Deutsch et al. 2014).
The greatest advantage is QuHAnT’s ability to comprehensively assess feature characteristics, such as profile counts, area, length, and size, for thousands of images in a fraction of the time compared to manual morphometry. This high-throughput capability and the use of the HPCC at Michigan State University reduced analysis time to the extent that entire tissue sections could be quantitated rapidly and consistently. This refinement was particularly evident for features that exhibit random localization such as hepatic lipid accumulation or collagen deposition. Furthermore, our studies show that minimal variability in Vv estimates was achieved when ≥95% of the images for the entire specimen were considered for both high TCDD dose ORO and PSR staining. These results provide compelling evidence that whole slide analysis improves detection and quantitation of histological features compared to random sampling of images. Although high-performance computing was a significant factor in reducing analysis time, QuHAnT can also be implemented on any personal computer and maintain assessments in a fraction of time compared to manual point counting.
Although our study was limited to ORO and PSR, QuHAnT is extensible to a variety of features of interest. It provides a framework that includes quality control, feature extraction, quantitation, and result output that simplifies the extension of QuHAnT to other features of interest. In theory, QuHAnT could be used for any in situ hybridization and/or immunocyto-chemically distinguishable feature either by finding appropriate thresholds for feature extraction or by creating custom modules to meet the research needs. Image analysis data have also been used to investigate associations between feature staining and related quantitative data such as hepatocellular hypertrophy, circulating hormone levels, and differential gene expression (Kind 2000; Garrido et al. 2013). Moreover, QuHAnT data obtained using standard morphometry concepts facilitate quantitative dose–response modeling to estimate points of departure to determine acceptable levels of exposure (U.S. Environmental Protection Agency 2012; Davis et al. 2011). In contrast, semiquantitative scoring of histological features by manual morphometry is suboptimal for modeling, limiting the quantitative potential of large scale toxicological studies such as those submitted to, or contracted by, regulatory agencies.
Imaging technologies have been incorporated into various drug development stages including target discovery, candidate screening, and early safety evaluation (Lang et al. 2006). QuHAnT, as well as other similar approaches, extends image analysis into preclinical development (Garrido et al. 2013) and risk assessment. QuHAnT is a high-throughput image analysis tool that aids in subjective or semiquantitative feature analysis by maximizing the extraction of quantitative information from data-rich histopathology images while minimizing bias by reducing interobserver variability. Particularly important for the reduction in interobserver reliability is the transparency in setting feature detection thresholds which is not typically reported. Our study has shown that QuHAnT outperforms manual morphometry in accuracy, reproducibility, and analysis time as well as providing additional complementary information not easily obtained using manual approaches. It is also easily extensible to other distinguishable histological features of interest. The ability to phenotypically anchor transcriptomic, proteomic, and metabolomic responses to quantitated key histopathological effects facilitates a more statistically robust interpretation of the data and facilitates the differentiation of adverse effects from adaptive responses. QuHAnT also supports the identification and refinement of mechanistically based biomarkers of adverse reactions and advances the assessment of the potential toxicological relevance of the mode of action to humans.
Acknowledgments
The authors would like to thank Drs. Michelle Angrish, Agnes Forgacs, and Anna Kopec as well as Kelly Fader for assistance with animal experiments and Dr. Katryn Allen for technical assistance with histological slide scanning.
The author(s) disclosed receipt of the following support for the research, authorship, and/or publication of this article: The presented work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (NIEHS SBRP P42ES04911). TRZ is partially supported by AgBioResearch at Michigan State University.
Abbreviations
- DIA
digital image analysis
- HSV
hue, saturation, and value
- NAFLD
nonalcoholic fatty liver disease
- ORO
Oil Red O
- PND
postnatal day
- PSR
PicroSirius Red
- QuHAnT
quantitative histological analysis tool
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- Vv
volume density
- WSI
whole slide imaging.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Boorman GA, Haseman JK, Waters MD, Hardisty JF, Sills RC. Quality review procedures necessary for rodent pathology databases and toxicogenomic studies: The National Toxicology Program experience. Toxicol Pathol. 2002;30:88–92. doi: 10.1080/01926230252824752. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-mediated hepatotoxicity. Toxicol Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Bringhenti I, Moraes-Teixeira JA, Cunha MR, Ornellas F, Mandarimde-Lacerda CA, Aguila MB. Maternal obesity during the preconception and early life periods alters pancreatic development in early and adult life in male mouse offspring. PLoS One. 2013;8:e55711. doi: 10.1371/journal.pone.0055711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieco P, Jonker A, De Boer BA, Ruijter JM, Van Noorden CJ. Image cytometry: Protocols for 2D and 3D quantification in microscopic images. Prog Histochem Cytochem. 2013;47:211–333. doi: 10.1016/j.proghi.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Crissman JW, Goodman DG, Hildebrandt PK, Maronpot RR, Prater DA, Riley JH, Seaman WJ, Thake DC. Best practices guideline: Toxicologic histopathology. Toxicol Pathol. 2004;32:126–131. doi: 10.1080/01926230490268756. [DOI] [PubMed] [Google Scholar]
- Davis JA, Gift JS, Zhao QJ. Introduction to benchmark dose methods and U.S. EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol Appl Pharmacol. 2011;254:181–191. doi: 10.1016/j.taap.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Day CP, James OF. Hepatic steatosis: Innocent bystander or guilty party? Hepatology. 1998;27:1463–1466. doi: 10.1002/hep.510270601. [DOI] [PubMed] [Google Scholar]
- Deutsch MJ, Schriever SC, Roscher AA, Ensenauer R. Digital image analysis approach for lipid droplet size quantitation of Oil Red O-stained cultured cells. Anal Biochem. 2014;445:87–89. doi: 10.1016/j.ab.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Eliceiri KW, Berthold MR, Goldberg IG, Ibanez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, Stuurman N, Swedlow JR, Tomancak P, Carpenter AE. Biological imaging software tools. Nat Meth. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz ED, Crespo-Mascarenhas C, Barreto-Vianna AR, Aguila MB, Mandarim-de-Lacerda CA. Renin–angiotensin system blockers protect pancreatic islets against diet-induced obesity and insulin resistance in mice. PLoS One. 2013;8:e67192. doi: 10.1371/journal.pone.0067192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido R, Zabka TS, Tao J, Fielden M, Fretland A, Albassam M. Quantitative histological assessment of xenobiotic-induced liver enzyme induction and pituitary–thyroid axis stimulation in rats using whole-slide automated image analysis. J Histochem Cytochem. 2013;61:362–371. doi: 10.1369/0022155413482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F, Lobdell HT, Zhou S, Hu C, Berk PD. Digital analysis of hepatic sections in mice accurately quantitates triglycerides and selected properties of lipid droplets. Exp Biol Med. 2010;235:1282–1286. doi: 10.1258/ebm.2010.010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: A review. IEEE Rev Biomed Eng. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi AM, Mouchaers KT, Schalij I, Grunberg K, Meijer GA, Vonk-Noordegraaf A, van der Laarse WJ, Belien JA. Rapid quantification of myocardial fibrosis: A new macro-based automated analysis. Analytical Cell Pathol. 2010;33:257–269. doi: 10.3233/ACP-CLO-2010-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, de Boer WB, Adams LA, MacQuillan G, Rossi E, Rigby P, Raftopoulos SC, Bulsara M, Jeffrey GP. Image analysis of liver collagen using sirius red is more accurate and correlates better with serum fibrosis markers than trichrome. Liver Int. 2013;33:1249–1256. doi: 10.1111/liv.12184. [DOI] [PubMed] [Google Scholar]
- Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RNM, Phillips MJ, Portmann BG, Poulsen H, Scheuer PJ, Schmid M, Thaler H. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- Kind CN. The application of in-situ hybridisation and immunocytochemistry to problem resolution in drug development. Toxicol Lett. 2000;112–113:487–492. doi: 10.1016/s0378-4274(99)00204-0. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Mets BD, Tashiro C, Potter D, Sharratt B, Harkema JR, Zacharewski TR. PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: Comparative toxicogenomic effects of dioxin and nondioxin-like ligands. Toxicol Appl Pharmacol. 2010;243:359–371. doi: 10.1016/j.taap.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AK, D’souza ML, Mets BD, Burgoon LD, Reese SE, Archer KJ, Potter D, Tashiro C, Sharratt B, Harkema JR, Zacharewski TR. Non-additive hepatic gene expression elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) co-treatment in C57BL/6 mice. Toxicol Appl Pharmacol. 2011;256:154–167. doi: 10.1016/j.taap.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nature reviews. Drug Discovery. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Ballabio E, Soilleux E, Sington J, Hatton CS, Dirnhofer S, Tzankov A. Inter- and intra-observational variability in immunohistochemistry: A multicentre analysis of diffuse large B-cell lymphoma staining. Histopathology. 2012;61:18–25. doi: 10.1111/j.1365-2559.2012.04179.x. [DOI] [PubMed] [Google Scholar]
- Levene AP, Kudo H, Armstrong MJ, Thursz MR, Gedroyc WM, Anstee QM, Goldin RD. Quantifying hepatic steatosis—More than meets the eye. Histopathology. 2012;60:971–981. doi: 10.1111/j.1365-2559.2012.04193.x. [DOI] [PubMed] [Google Scholar]
- Lu H, Cui W, Klaassen CD. Nrf2 protects against 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced oxidative injury and steatohepatitis. Toxicol Appl Pharmacol. 2011;256:122–135. doi: 10.1016/j.taap.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel M, von Wasielewski R, Wiese B, Rudiger T, Muller-Hermelink HK, Kreipe H. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J Pathol. 2002;198:292–299. doi: 10.1002/path.1218. [DOI] [PubMed] [Google Scholar]
- Nishida T, Tsuneyama K, Fujimoto M, Nomoto K, Hayashi S, Miwa S, Nakajima T, Nakanishi Y, Sasaki Y, Suzuki W, Iizuka S, Nagata M, Shimada T, Aburada M, Shimada Y, Imura J. Spontaneous onset of nonalcoholic steatohepatitis and hepatocellular carcinoma in a mouse model of metabolic syndrome. Lab Invest. 2013;93:230–241. doi: 10.1038/labinvest.2012.155. [DOI] [PubMed] [Google Scholar]
- Oyama T, Ishikawa Y, Hayashi M, Arihiro K, Horiguchi J. The effects of fixation, processing and evaluation criteria on immunohistochemical detection of hormone receptors in breast cancer. Breast Cancer. 2007;14:182–188. doi: 10.2325/jbcs.976. [DOI] [PubMed] [Google Scholar]
- Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, Collins LC, Colgan TJ. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S, Chevallier A, Teixeira-Clerc F, Ambolet-Camoit A, Bui LC, Bats AS, Fournet JC, Fernandez-Salguero P, Aggerbeck M, Lotersztajn S, Barouki R, Coumoul X. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol Sci. 2013;137:111–124. doi: 10.1093/toxsci/kft236. [DOI] [PubMed] [Google Scholar]
- Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, Duffy MJ, Jirstrom K, Gallagher WM. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: Application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10:R89. doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Lauro G, Cable W, Lesniak A, Tseytlin E, McHugh J, Parwani A, Pantanowitz L. Digital pathology consultations—a new era in digital imaging, challenges and practical applications. J Digital Imaging. 2013;26:668–677. doi: 10.1007/s10278-013-9572-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standish RA, Cholongitas E, Dhillon A, Burroughs AK, Dhillon AP. An appraisal of the histopathological assessment of liver fibrosis. Gut. 2006;55:569–578. doi: 10.1136/gut.2005.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschanz SA, Burri PH, Weibel ER. A simple tool for stereological assessment of digital images: The STEPanizer. J Microscopy. 2011;243:47–59. doi: 10.1111/j.1365-2818.2010.03481.x. [DOI] [PubMed] [Google Scholar]
- USEPA (U.S. Environmental Protection Agency) Benchmark Dose Technical Guidance, 40CFR Part 261, US Government Printing Office. Washington, DC: USEPA; 2012. [Google Scholar]
- Ying X, Monticello TM. Modern imaging technologies in toxicologic pathology: An overview. Toxicol Pathol. 2006;34:815–826. doi: 10.1080/01926230600918983. [DOI] [PubMed] [Google Scholar]
- Zaitoun AM, Al Mardini H, Awad S, Ukabam S, Makadisi S, Record CO. Quantitative assessment of fibrosis and steatosis in liver biopsies from patients with chronic hepatitis C. J Clin Pathol. 2001;54:461–465. doi: 10.1136/jcp.54.6.461. [DOI] [PMC free article] [PubMed] [Google Scholar]