Abstract
In arid ecosystems environmental factors such as geoclimatic conditions and agricultural practices are of major importance in shaping the diversity and functionality of plant-associated bacterial communities. Assessing the influence of such factors is a key to understand (i) the driving forces determining the shape of root-associated bacterial communities and (ii) the plant growth promoting (PGP) services they provide. Desert oasis environment was chosen as model ecosystem where agriculture is possible by the microclimate determined by the date palm cultivation. The bacterial communities in the soil fractions associated with the root system of date palms cultivated in seven oases in Tunisia were assessed by culture-independent and dependent approaches. According to 16S rRNA gene PCR-DGGE fingerprinting, the shapes of the date palm rhizosphere bacterial communities correlate with geoclimatic features along a north-south aridity transect. Despite the fact that the date palm root bacterial community structure was strongly influenced by macroecological factors, the potential rhizosphere services reflected in the PGP traits of isolates screened in vitro were conserved among the different oases. Such services were exerted by the 83% of the screened isolates. The comparable numbers and types of PGP traits indicate their importance in maintaining the plant functional homeostasis despite the different environmental selection pressures.
1. Introduction
The southern regions of Tunisia are very arid and the date palm (Phoenix dactylifera L.) is a key plant determining in the oasis agroecosystem a microclimate that favours agriculture [1]. The palms protection provides many ecosystem services, including ameliorating oasis temperature, changing floodwater dynamics and facilitating wildlife, and making agriculture possible under harsh environmental conditions [2]. In the world, oases cover about 800 000 ha and support the living of 10 million people. In Tunisia more than four millions of date palm trees are spread onto 32 000 ha of oasis in the southern part of the country [3, 4]. As a result of the oases overexploitation and strong anthropogenic pressures, these ecosystems are becoming increasingly fragile. Furthermore, despite the oasis potential to tolerate several abiotic stresses typical of arid environment, the ongoing climate change is enhancing the environmental pressure on the date palm affecting growth and development, especially in the Middle East [5].
Besides the well-known plant growth promoting properties typical of rhizospheres in temperate soils in nonarid ecosystems, rhizosphere bacteria in arid soils contribute in counteracting drought and salinity stresses, by providing services such as, among others, physical protection of the root from mechanical stress against the dry soil particles, induction of plant physiological responses against water losses [6], or productions of metabolites contributing to the maintenance of the plant hormone and nutrient homeostasis, [7]. In particular PGP (plant growth promoting) bacteria, naturally associated with plants, have been shown to be essential partners for improving plant tolerance to stressful conditions [8]. The exploration of plants naturally adapted to extreme condition may allow a reservoir of biodiversity exploitable to understand the ecological service enclosed in these ecosystems [8, 9]. In this context, ecological niche presented in the oasis ecosystem could provide a new model to study and dissect the key factors driving the stability of this ecosystem [10]. Little information is available about the microbiological functionality of both oasis and date palm. For instance the potential PGP services provided by the root-associated bacteria appear to be invariant with respect to geoclimatic factors despite provided by different bacterial communities, according to observations across a north to south aridity transect that included Tunisia [11].
Since plants contribute to shape soil microbial diversity [12, 13], the aim of this work was to assess bacterial communities associated with the date palm rhizosphere soil, the root surrounding soil and the bulk soil fractions in seven Tunisian oases, in order to evaluate if along a north-south transect (i) the assemblage of bacterial communities in the palm root soil fractions was driven by the geoclimatic factors and (ii) the ecological services were preserved in the soil fractions of the root system. The structure of the bacterial communities associated with the soil fractions of date palm in the seven oases was dissected by 16S rRNA gene-based PCR-DGGE (denaturing gradient gel electrophoresis) analysis. The results were analysed in function of geoclimatic factor and oasis origin, and compared with the diversity of the cultivable bacteria and their PGP potential.
2. Materials and Methods
2.1. Site Description and Sampling
The sampling was carried out from seven oases in different geographic locations in Tunisia, along a latitude/longitude gradient, respectively from 32° to 34°N and from 7° to 9°E (Figure 1(a) and Supplementary Table 1 in the Supplementary Material available online at http://dx.doi.org/10.1155/2015/153851). A traditional crop management was used in all the oases, including groundwater-based flooding irrigation and fertilization with organic fertilizers. In each oasis, the roots of three date palm trees of similar age, lying in the distance of less than 15 meters and growing in the same soil were separately collected at 20–30 cm depth in order to obtain the adhering rhizosphere soil (R) tightly attached to roots. After removing the roots, the root surrounding soil (S) was collected. Bulk soil samples (B) not influenced by date palm root system were also sampled as control. All soil samples were collected under sterile conditions using sterile tools. Recovered samples were stored at −20°C for molecular analysis or at 4°C for isolation.
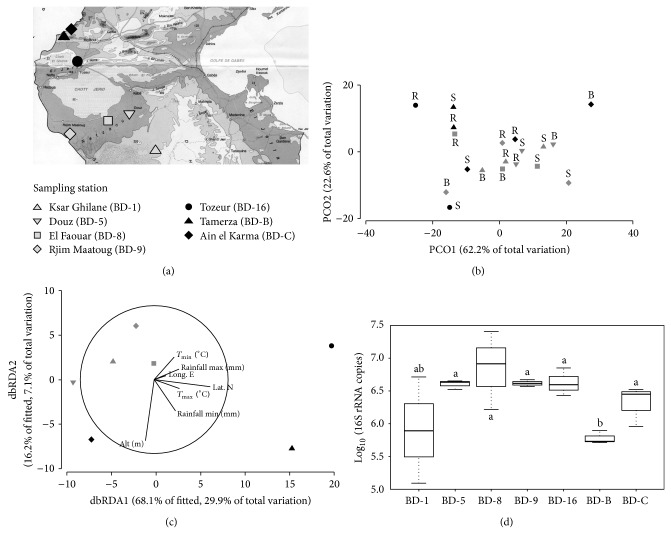
Figure 1.
Station location and analysis of bacterial community structure associated with soil fraction of date root system. (a) The location of the studied oases is indicated on the map of Tunisia. (b) A principal coordinate analysis (PCO), deduced from the 16S rRNA gene-based PCR-DGGE profiles, resumes the diversity of the bacterial communities associated with the date palm root soil fractions. 84.8% of total variation is explained in the presented PCO. The soil fractions analysed are R: rhizosphere; S: root surrounding soil; and B: bulk soil. (c) Dist LM analysis to evaluate which are the main geoclimatic variables influencing the structure of the bacterial communities associated with date palm root soil fractions. Lat. N: latitude north; Long. E: longitude east; Alt.: altitude; T min: minimum temperature; T max: maximum temperature; Rainfall min: minimum rainfall; Rainfall max: maximum rainfall. (d) Box-plot graph represents the quantification of 16S rRNA gene by qPCR. The number of copies is expressed in Log10. Statistical analysis (pairwise test) of bacterial assemblage across locations was indicated by the letter.
2.2. Total DNA Extraction, PCR-DGGE, and Profile Analysis
Total DNA from soil samples was extracted by commercial kit FASTDNA SPIN KIT for soil (Qbiogene, Carlsbad, USA) according to the manufacturer's procedure. PCR amplification was performed in a final volume of 50 μL using primers 907R and 357F, adding a GC-clamp to the forward primer [14, 15]. The reaction mixture was prepared with 1X PCR buffer, 2.5 mM MgCl2, 0.12 mM deoxynucleoside triphosphate, 0.3 mM of each primer and 1U Taq DNA polymerase and 10 ng of pooled DNA obtained from the three plant replicates were added as template. PCR products were resolved on 7% (w/v) polyacrylamide gel in 1X TAE pH 7.4 with a 40–60% denaturing gradient. Gels were run at 90 V for 17 h at 60°C in DCode apparatus (Bio-Rad, Italy). After electrophoresis, gels were stained with ethidium bromide solution for 30 min, washed with sterile distilled water, and photographed on a UV transillumination table. The DGGE band profiles were converted into numerical values using Image J (version 1.46) and XLSTAT software.
2.3. Real Time PCR
Quantitative real time PCR (q-PCR) was performed on a Chromo4 real time PCR machine (Bio-Rad) to measure the presence and concentration of bacterial 16S rRNA gene associated with the rhizosphere fractions. The reactions were performed with IQ SYBR Green Supermix (Bio-Rad), using primers targeting the 16S rRNA gene (Bac357-F and Bac907-R) [16]. PCR SYBR green reactions were prepared by using the “Brilliant SYBR Green QPCR Master Mix” kit (Stratagene) in 96-well plates. The reaction mix (25 mL) contained 1X Brilliant SYBR Green (2.5 mM MgCl2), 0.12 mM of each primers, and approximately 100 ng of extracted DNsA. The DNA obtained from the three plants sampled in the same station was pooled and used as template to carry out the real time assay in triplicate. At the end of each real time PCR, a melting curve analysis was performed for verifying the specificity of PCR products. To construct standard curves, the 16S rRNA gene of Asaia sp. was amplified by PCR and cloned using the pGEM T-easy Vector Cloning Kit (Promega). q-PCR data relative to the 16S rRNA gene concentration were log-transformed.
2.4. Isolation of Cultivable Bacteria
One gram of rhizosphere soil (R) from each sample was suspended in 9 mL of sterile physiological solution (9 g/L NaCl) and shaken for 15 min at 200 rpm at room temperature. Suspensions were diluted in tenfold series and plated in triplicate onto TSA (Tryptic Soy Agar), YEM (Yest Extract Mannitol), and KB (King'B agar) culture media. After three days at 30°C colonies were randomly selected and spread on the original medium for three times to avoid contamination risks. Pure strains were frozen in 25% glycerol at −80°C. A total of 440 isolates were collected. The isolates were named based on the station and the medium from which they were isolated.
2.5. DNA Extraction, Dereplication, and Identification of Isolates
Genomic DNA was recovered from the isolates using a boiling lysis. Bacterial cells were suspended in 50 μL of sterile TE (10 mM Tris/HCl, pH 8, 1 mM EDTA) and incubated at 100°C for 8 min. After centrifugation (13000 g, 10 min), the supernatant containing the released DNA was stored at −20°C and used as template for PCR. Amplification of 16S–23S internal transcribed spacer region (ITS) was performed using the universal primers S-DBact-0008-a-S-20 (5′-CTA CGG CTA CCT TGT TAC GA-3′) and S-D-Bact-1495-a-S-20 (5′-AGA GTT TGA TCC TGG CTC AG-3′) according to the procedure described previously by Fhoula et al. [17]. Two μL of the PCR products were checked by electrophoresis in 1.5% agarose gel and stained with ethidium bromide. Gel images were captured using Gel Doc 2000 system (Bio-Rad, Tunis, Tunisia), and bacteria redundancy was reduced by evaluating the different ITS profiles. One strain per each ITS haplotype was used in the phylogenetic analysis and for further experiments. A total of 98 strains were characterized by 16S rRNA gene sequencing using the primers S-D-Bact-1494-a-20 (5′-GTC GTA ACA AGG TAG CCG TA-3′) and L-D-Bact-0035-a-15 (5′-CAA GGC ATC CAC CGT-3′). PCR amplification was carried out as described by Fhoula et al. [17]. The 16S rRNA PCR amplicons were purified with Exonuclease-I and Shrimp Alkaline Phosphatase (Exo-Sap, Fermentas, Life Sciences) following the manufacturer's standard protocol. Sequencing of the purified amplicons was performed using a Big Dye Terminator cycle sequencing kit V3.1 (Applied Biosystems) and an Applied Biosystems 3130XL Capillary DNA Sequencer machine. The obtained sequences, with an average length of 750 bp, were compared with those available at the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) using the basic local alignment search tool (BLAST) algorithm [18]. The 16S rDNA sequences were submitted to the NCBI nucleotide database under the accession number KJ956590 to KJ956687.
Phylogenetic analysis of the 16S rRNA gene sequences was conducted with molecular evolutionary genetics analysis (MEGA) software, version 6 [19]. Trees were constructed by using neighbor-joining method [20].
2.6. Characterization of Plant Growth Promoting Activity and Abiotic Stress Resistance
The 98 bacterial strains identified were screened for production of indole acetic acid (IAA), siderophores and ammonia, mineral phosphate solubilization, protease and cellulose activity, and tolerance to several abiotic stresses. Quantitative production of IAA was determined as described by Ouzari et al. [21]. Briefly, after incubation in minimal medium supplemented with glucose (100 g/L) and L-tryptophane (10 μg/mL), using Salkowski's reagent, the colour absorbance was read, after 20–25 min, at 535 nm. Concentration of IAA produced was measured by comparison with a standard graph of IAA. Ability of bacteria to solubilize inorganic phosphate was evaluated as described by Nautiyal [22], by the observation of clear halo around the bacterial colony grown in Pikovskaya medium. To demonstrate the production of siderophore, the tested strains were spotted on nutrient agar plates. After incubation for 48–72 h at 30°C, the grown strains were overlaid with CAS medium supplemented with agarose (0.9% w/v). Positive test was noted when colour modification around colonies from blue to orange was observed [23]. Ammonia production was assayed by inoculation of bacterial strains in 10 mL of peptone water and using Nessler's reagent (0.5 mL). Ammonia producing strains were identified when brown to yellow colour was developed [24]. Protease (casein degradation) and cellulase activities were determined by spot inoculation of the strains on Skimmed milk and CMC agar media, respectively. A clear halo around the colonies indicates the ability of the strains to produce the degrading enzymes [25].
Tolerance to osmotic stress was evaluated by adding to tryptic soy broth (TSB) medium 30% of polyethylene glycol (PEG 8000). Resistance to salt was assessed by adding 5, 10, 15, 20, 25, and 30% NaCl to the culture media and incubating the plates at 30°C for 5 days. The ability to growth at 45, 50, and 55°C was checked in TSA by incubation at the indicated temperatures for 5 days. Tolerance to acid (3 and 4) and basic (10 and 12) pH was assessed by adjusting the medium with concentrated HCl (12 N) and NaOH (3 M), respectively.
2.7. Statistical Analysis
Significant differences in soil bacterial community structure were investigated by permutational analysis of variance (PERMANOVA, [26]). Distance-based multivariate analysis for a linear model (DistLM, [27]) was used to determine which significant environmental variables explain the observed similarity among the samples. The Akaike information criterion (AIC) was used to select the predictor variables [28]. The contribution of each environmental variable was assessed: firstly the “marginal test” is used to assess the statistical significance and percentage contribution of each variable by itself and secondly the “sequential test” was employed to explain the biotic similarity considering all the variable contributions. All the statistical tests were performed by PRIMER v. 6.1 [29], PERMANOVA+for PRIMER routines [30].
3. Results and Discussion
3.1. Environment Parameters Directly Influence Bacterial Communities Associated with Palm Rhizosphere
The diversity of bacterial communities associated with the date palm root system from each of the seven studied oasis was investigated through the analysis of the diversity of the 16S rRNA gene in the rhizosphere (R) and root surrounding soil (S) fractions. Bulk soil (B) was also included as a comparative fraction not directly influenced by the plant root. Separation among palm bacterial communities located in north (BD-16, BD-B, and BD-C) and south (BD-1, BD-5, BD-8, and BD-9) oases was supported by a principal coordinate analysis (PCO) (Figure 1(b)), suggesting that geoclimatic conditions influence the bacterial community structure. Statistical analysis confirmed the grouping observed in the PCO analysis with a significant difference between north and south oases (PERMANOVA, df = 1.55; F = 8.06; P = 0.0017) but not a significant separation mediated by the aquifer used to irrigate the oases (PERMANOVA, df = 1.55; F = 1.45; P = 0.21).
Within the two macroregions, the north and south groups of oases, we observed a significant difference among oases (Supplementary Table 2) indicating the presence of oasis-specific bacterial community supporting a concept of “ecological island.” Pairwise analysis showed that such differences observed among the oases predominantly occurred in the north regions (Supplementary Table 3), possibly because the south region (closest to desert) presents harsher conditions that select a more restricted type of bacteria. These ecological islands represent specific cluster of biological diversity that may contribute to the overall regional bacterial community functionality and furthermore increase the level of resilience to environmental change of the entire system [30].
Along the transect, the soil fraction communities were significantly different (PERMANOVA, df = 2.55; F = 2.70; P = 0.03). In particular the rhizosphere community, that resides in the first millimeter of soil adhering to the root, appeared completely different from the root surrounding soil (PERMANOVA, t = 2.04; P = 0.017, p-pht) and bulk soil (PERMANOVA, t = 2.05; P = 0.019, p-pht), suggesting the influence of palm root exudates in shaping the bacterial community. Generally, the rhizosphere is the transition zone between the root surface and soil where the released exudates and the rhizodeposition favour microbial proliferation, inducing changes in the structure and in the chemical-physical properties of the soil [31]. Indeed, the analysis of bacterial abundance in the rhizosphere showed a numbers of 16S rRNA copies ranging from 5.88 ± 0.78 to 6.63 ± 0.15 (Figure 1(d)). Despite similar values observed in the rhizosphere community, a statistical difference among the stations was identified (PERMANOVA, df = 6.20; F = 2.93; P = 0.041), mainly influenced by environmental factor directly linked to location, such as altitude and temperature maximum (DistLM, P = 0.03).
Despite the rhizosphere effect observed along the transect, in each oasis considered separately from the others, rhizobacterial community appeared directly connected to that present in the root surrounding soil and the bulk soil, since no statistically significant differences in the bacterial diversity were observed among the different soil fractions within each station (R, S e B: PERMANOVA, df = 12.55; F = 1.62; P = 0.057). The rhizosphere effect is particularly noticeable in nutrient-poor soils and under severe abiotic stresses, as previously observed for herbaceous and arboreal plants grown in arid lands [7, 8, 32, 33]. In the oasis model the selection mediated by “oasis ecosystem” appeared stronger than the one exerted by the plant root system (Supplementary Table 2). Naturally, most of the desert microbial communities seem to be structured solely by abiotic processes [34, 35]. However, desert farming may strongly affect the sand/soil microbial diversity reshaping the structure of the resident microbial communities [8, 9, 36, 37]. During long-term desert farming land management, such as that occurring in the studied oases, the structure of rhizosphere bacterial community is strongly influenced by the plant and the desert farming practices that determine drastic shifts in the composition of the original desert soil communities [7, 8].
Dist LM multivariate analysis was performed in order to correlate the differences in the structure of bacterial communities in the different oases with environmental parameters. The selection of soil microorganisms by the rhizosphere is a complex process controlled by several factors, often not easily correlated to the environmental settings [38]. Nevertheless, Dist LM analysis showed that geoclimatic parameters contributed to drive the assemblage of the bacterial communities. In particular, marginal test showed latitude, longitude, altitude, minimum temperature, and minimum rainfall as significant variables singularly involved in the selection of bacterial assemblages (Table 1(a)). Sequential test confirmed latitude, altitude, and temperature as variables involved in the bacterial community shaping (Table 1(b)). We can assume that a concurrence of environmental factors, including a hot and dry climate, may influence the differences among the bacterial communities of the soil fractions (R, S, and B) associated with the root system of date palm cultivated in the oases in the north and south macroregions examined (Figure 1(c)).
Table 1.
Environmental factors associated to the structure of the date palm soil bacterial community. Relationships between bacterial assemblages and climate features using nonparametric multivariate multiple regression analysis (DISTLM). (a) Marginal test considers each single geographical variables and their contribution to explain the total variability. (b) Sequential test explaining the total variation with the contribution of all the variables accounted together. Lat. N: latitude north; Long. E: longitude east; Alt.: altitude; T min: minimum temperature; T max: maximum temperature; Rainfall min: minimum rainfall; Rainfall max: maximum rainfall; F: statistic F; P: probability (in bold the variables statistically significant; P < 0.05); Prop.: proportion of total variation explained; Cumul.: cumulative variation explained by the variables listed; Res df: residual degrees of freedom.
(a) Marginal test
| Variable | SS (trace) | F | P |
|---|---|---|---|
| Lat. N | 1855.5 | 6.4211 | 0.0028 |
| Long. E | 1444.3 | 4.8698 | 0.0097 |
| Alt (m) | 980.15 | 3.2116 | 0.0376 |
| T min (°C) | 1170.8 | 3.8811 | 0.0193 |
| T max (°C) | 363.14 | 1.147 | 0.3034 |
| Rainfall min (mm) | 1187.3 | 3.9401 | 0.0198 |
| Rainfall max (mm) | 588.06 | 1.8821 | 0.136 |
(b) Sequential test
| Variable | AIC | SS | F | P | Cumul. | Res. df |
|---|---|---|---|---|---|---|
| (+) Lat. N | 319.28 | 1855.5 | 6.4211 | 0.0022 | 0.10627 | 54 |
| (+) Long. E | 320.07 | 331.89 | 1.1517 | 0.2998 | 0.12528 | 53 |
| (+) Alt (m) | 317.33 | 1240.6 | 4.5974 | 0.0122 | 0.19633 | 52 |
| (+) T min (°C) | 316.7 | 644.65 | 2.4558 | 0.084 | 0.23325 | 51 |
| (+) T max (°C) | 309.31 | 2066 | 9.1244 | 0.0006 | 0.35158 | 50 |
| (+) Rainfall min (mm) | 303.19 | 1528.3 | 7.6468 | 0.001 | 0.43911 | 49 |
| (+) Rainfall max (mm) | 303.19 | < 0.01 | 0 | 1 | 0.43911 | 49 |
3.2. Cultivable Bacterial Communities Associated with Date Palm Soil Fractions
The isolation of native bacterial species associated with date palm root soil was performed using nonspecific media, in order to select a wide range of genera of possible plant growth promoters [39–41].
A total of 440 isolates were retrieved from the seven analyzed stations. To manage such a large set of isolates, total DNA was extracted from each isolate and 16S–23S rRNA gene internal transcribed spacers (ITS) were amplified. ITS characterization represents a useful molecular tool for the discrimination of bacterial isolates up to the subspecies level [42–45]. Within the whole bacterial collection, ITS-PCR fingerprinting revealed 30 distinct haplotypes (H1-H30). Haplotypes H4 and H20 were the most frequent and were represented by 46 and 26 isolates, respectively. Representative isolates (from one to four strains for each haplotype, summing up a total of 98 isolates) were subjected to species identification using partial 16S rRNA gene sequencing (Supplementary Figure 1).
A wide diversity was detected into date palm rhizosphere bacterial community along the studied aridity transect in Tunisia. Significant differences were observed in the structure of the bacterial communities in the rhizosphere of the analyzed oases, in particular for the differential distribution pattern of the major bacterial genera (Figure 2(a)). According to the cluster analysis at the genus level performed on the entire strain collection, the composition of the cultivable rhizobacterial communities associated with date palm in the seven oases shared about the 65% similarity.
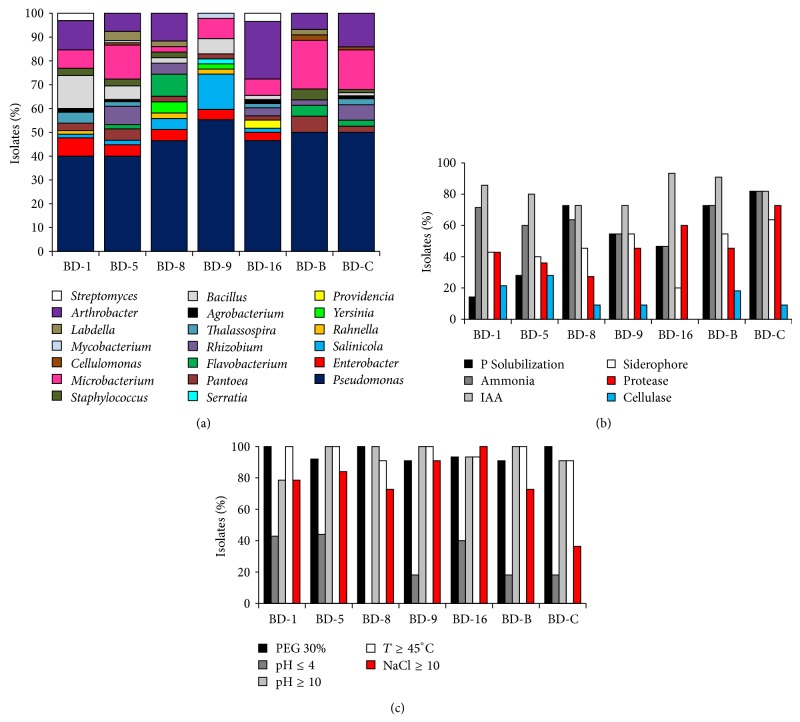
Figure 2.
Diversity and functionality of cultivable bacteria islated from date palm rhizosphere. (a) Phylogenetic identification at the genus level of culturable bacteria associated with date palm rhizosphere. (b) Percentage of date palm rhizosphere-associated bacteria showing PGP activity. (c) Percentage of isolates displaying the assayed abiotic stress tolerance in the bacterial collection of strains associated with date palm cultivated in the seven oases analysed.
The phylogenetic identification of cultivable bacteria highlighted a predominance of gram-negative bacteria (66%), belonging to the Gammaproteobacteria (57%), Alphaproteobacteria (7%), and Betaproteobacteria (1%) subclasses. The remaining isolates were affiliated to the Firmicutes (7%), Actinobacteria (26%), and Bacteroidetes (2%) classes. Members of these taxa are frequently associated with different plant species and types, confirming that soil is the main reservoir of plant-associated bacteria [46]. The strains were allocated into 20 different genera of variable occurrence (Figure 2(a)), showing a high genetic diversity in the date palm rhizosphere presumably influenced by the combined effects of root exudates and agricultural management practices, particularly important under the arid pedoclimatic conditions [47]. The rhizobacterial communities were dominated by Pseudomonas, as previously described in herbaceous plants, arboreal and plant adapted to arid climates [11, 48–50].
Together with Pseudomonas, Pantoea and Microbacterium genera were observed in all stations followed by Bacillus and Arthrobacter, which were reported in six out of seven stations. As well, Enterobacter, Salinicola, Rhizobium, and Staphylococcus were recorded among 5 stations, suggesting the adaptation of these genera to the oasis environment. Except Labedella, the genera found in association with date palm rhizosphere have been previously recognized as being capable of colonizing plant root systems in arid environment [11, 38, 48, 50–52].
3.3. Characterization of Rhizobacteria PGP Potential
The plant microbiome is a key determinant of plant health and productivity. Plant-associated microbes can help plants stimulate growth, promote biotic and abiotic stress resistance and influence crop yield and quality by nutrient mobilization and transport [6]. While the possibility to contribute to control biotic stresses by plant-associated microorganisms is well characterized, less is known for abiotic stress. However, several promising examples of stress protecting bacteria are already reported in the literature [7, 38, 53]. Recent works demonstrated that drought-exposed plants cultivated under desert farming are colonized by bacterial communities with high PGP potential [7, 8]. Such a PGP potential can promote increased tolerance to water shortage, mediated by the induction of a larger root system (up to 40%) that enhances water uptake [7]. To assess if the oasis date palm PGP potential was conserved in the rhizosphere soil, 98 isolates were evaluated for a series of PGP traits. The majority (85%) of isolates showed multiple PGP activities, which may promote plant growth directly, indirectly, or synergistically. Only 15% of the rhizobacteria showed one or no activity, while no strains displayed all the screened PGP activities. The most common PGP trait was auxin production (83%), followed by ammonia synthesis (63%) and biofertilization activities such as solubilization of phosphates (48%) and siderophore production (44%). In our rhizobacterial collection, the IAA production was equally distributed among the seven oases selected along the aridity transect (Figure 2(b)), similarly to previous observations in other arboreal plants cultivated along a latitude transect [11], confirming that IAA synthesis is a widespread PGP trait. The IAA production ranged from 2.5 to 85 μg/mL with 49% of the strains producing an amount ranging from 10 to 20 μg/mL and the 38% showing higher levels of IAA (more than 20 μg/mL). As already described in the literature, Pseudomonas, Bacillus, Pantoea, Staphylococcus, and Microbacterium were the most abundant taxa implicated in IAA production [48, 54, 55]. The high frequency of IAA producing strains suggests a role of PGP bacteria in contributing to regulate the root surface extension and consequently the potential of water and nutrient uptake [56].
Phosphorus, together with iron and nitrogen, is a key nutrient for plant, particularly in oasis soil where the availability of nutrient sources of animal origin is scarce [57]. The ability of rhizobacteria to solubilize phosphate (48%) through the production of organic acids or phytases can be very important in arid ecosystems [58, 59]. Strains of Pantoea, Enterobacter, Pseudomonas, Streptomyces, and Rhizobium genera were the most efficient solubilizers, as previously showed in other arid contexts such as Tunisian grapevine [48] and different crops in Bolivia [60]. Several siderophore-producing bacteria were observed in the rhizosphere (44%) probably because this PGP trait confers competitive colonization ability in iron-limiting soils. Iron is made available for the plant host and consequently exerts a biocontrol role reducing iron-dependent spore germination of fungi [61]. The siderophore-producing bacteria belonged mainly to the Pseudomonas genus (67%), followed by Bacillus (7%) and Pantoea (7%). Predominance of siderophore release by Pseudomonas bacteria was already reported in the rhizosphere of other plants [62, 63]. In addition to siderophore production, cell wall degrading enzymes implicated in fungal inhibition and the organic matter turnover [51] were investigated. The 49% and 15% of the examined isolates were able to produce proteases and cellulases, respectively, with the most active strains belonging to Serratia marcescens and Sinorhizobium meliloti, respectively [64, 65]. Ammonia production can indirectly affect plant growth through nitrogen supply [66]. This trait was represented in 64% of the isolates, confirming its spread in the palm-bacteria association.
Further analyses were performed to evaluate the adaptability of isolates to abiotic stresses (Figure 2(c)). Drought stress resistance was presented by 95% of the strains that could grow in presence of increasing concentrations of PEG. Most of the strains (98%) were able to grow at 45°C, while only 39% at 50°C. The capacity to tolerate high temperature drastically decreased (5%) at 55°C and only Bacillus and Pseudomonas strains showed this ability [67, 68]. Moderate halotolerance was presented by 75% of the isolates, while 50% tolerated up to 15% NaCl, 20% actively grew in presence of 20% NaCl, and only the 6% were extremely halotolerant (25% NaCl), indicating salinity as a major selective factor for the bacterial microbiomes in the Tunisian date palm oases. The formulation of halotolerant PGPR could be an interesting alternative for agriculture productivity in the oasis [69]. The tested rhizobacteria could grow in a wide pH range. Within the bacterial collection 96% and the 75% of the strains were facultative alkalophiles able to grow in basic media (up to pH = 12), while 34% of them could grow in acidic media (pH = 4) and only 6% was facultative acidophiles growing down to pH = 3.
4. Conclusion
Date palm represents the key plant species in desert oases being essential in determining the oasis microclimate that can allow agriculture. Palm exerts both physical and functional services involved in the creation of ideal condition for desert farming. Palm root system and rhizosphere soil showed a complex diversity that enclosed a reservoir of PGP bacteria involved in the regulation of plant homeostasis. Future work is needed to perform experiment about the ability of selected bacterial isolates in promoting plant growth under greenhouse and field conditions. In this context, the selection of autochthonous bacteria, together with the desert farming practices, could have promising perspectives for sustainable agriculture in oasis ecosystem.
Supplementary Material
Supplementary material contains additional information about the geo-climatic data and biodiversity of bacterial community in the different studied oases.
Acknowledgments
This work was supported by the project BIODESERT GA-245746 “Biotechnology from desert microbial extremophiles for supporting agriculture research potential in Tunisia and Southern Europe” (European Union), Fondazione Project BIOGESTECA n° 15083/RCC “Fondo per la promozione di accordi istituzionali” (Regione Lombardia, Italy) through a fellowship to RM. ER was supported by Università degli Studi di Milano, DeFENS, European Social Fund (FSE), and Regione Lombardia (contract “Dote Ricerca”). Thanks are due to Marco Fusi for invaluable help in statistical analysis. Research reported in this publication was supported by the King Abdullah University of Science and Technology (KAUST).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Raoudha Ferjani, Ramona Marasco, and Eleonora Rolli contributed equally to the work.
References
- 1.Chao C. T., Krueger R. R. The date palm (Phoenix dactylifera L.): overview of biology, uses, and cultivation. HortScience. 2007;42(5):1077–1082. [Google Scholar]
- 2.de Haas H. Migration and Agricultural Transformations in the Oases of Morocco and Tunisia. Utrecht, The Netherlands: KNAG; 2001. [Google Scholar]
- 3.Elhoumaizi M. A., Saaidi M., Oihabi A., Cilas C. Phenotypic diversity of date-palm cultivars (Phoenix dactylifera L.) from Morocco. Genetic Resources and Crop Evolution. 2002;49(5):483–490. doi: 10.1023/A:1020968513494. [DOI] [Google Scholar]
- 4.Hammadi H., Mokhtar R., Mokhtar E., Ali F. New approach for the morphological identification of date palm (Phoenix dactylifera L.) cultivars from Tunisia. Pakistan Journal of Botany. 2009;41(6):2671–2681. [Google Scholar]
- 5.Sen Z., Al Alsheikh A., Al-Dakheel A. M., et al. Climate change and Water Harvesting possibilities in arid regions. International Journal of Global Warming. 2011;3(4):355–371. doi: 10.1504/11.44399. [DOI] [Google Scholar]
- 6.Ryu C. M., Farag M. A., Hu C. H., et al. Bacterial volatiles promote growth in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America. 2013;100(8):4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Zelicourt A., Al-Yousif M., Hirt H. Rhizosphere microbes as essential partners for plant stress tolerance. Molecular Plant. 2013;6(2):242–245. doi: 10.1093/mp/sst028. [DOI] [PubMed] [Google Scholar]
- 8.Marasco R., Rolli E., Ettoumi B., et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0048479.e48479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köberl M., Müller H., Ramadan E. M., Berg G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0024452.e24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar K., Amaresan N., Bhagat S., Madhuri K., Srivastava R. C. Isolation and characterization of rhizobacteria associated with coastal agricultural ecosystem of rhizosphere soils of cultivated vegetable crops. World Journal of Microbiology and Biotechnology. 2011;27(7):1625–1632. doi: 10.1007/s11274-010-0616-z. [DOI] [Google Scholar]
- 11.Marasco R., Rolli E., Fusi M., et al. Plant growth promotion potential is equally represented in diverse grapevine root-associated bacterial communities from different biopedoclimatic environments. BioMed Research International. 2013;2013:17. doi: 10.1155/2013/491091.491091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Othman A. A., Amer W. M., Fayez M., Hegazi N. A. Rhizosheath of sinai desert plants is a potential repository for associative diazotrophs. Microbiological Research. 2004;159(3):285–293. doi: 10.1016/j.micres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Saul-Tcherkas V., Unc A., Steinberger Y. Soil microbial diversity in the vicinity of desert shrubs. Microbial Ecology. 2013;65(3):689–699. doi: 10.1007/s00248-012-0141-8. [DOI] [PubMed] [Google Scholar]
- 14.Muyzer G., de Waal E. C., Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merlino G., Rizzi A., Schievano A., et al. Microbial community structure and dynamics in two-stage vs single-stage thermophilic anaerobic digestion of mixed swine slurry and market bio-waste. Water Research. 2013;47(6):1983–1995. doi: 10.1016/j.watres.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Fhoula I., Najjari A., Turki Y., Jaballah S., Boudabous A., Ouzari H. Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of tunisia. BioMed Research International. 2013;2013:14. doi: 10.1155/2013/405708.405708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Ouzari H., Khsairi A., Raddadi N., et al. Diversity of auxin-producing bacteria associated to Pseudomonas savastanoi-induced olive knots. Journal of Basic Microbiology. 2008;48(5):370–377. doi: 10.1002/jobm.200800036. [DOI] [PubMed] [Google Scholar]
- 22.Nautiyal C. S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiology Letters. 1999;170(1):265–270. doi: 10.1016/S0378-1097(98)00555-2. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Miranda S., Cabirol N., George-Téllez R., Zamudio-Rivera L. S., Fernández F. J. O-CAS, a fast and universal method for siderophore detection. Journal of Microbiological Methods. 2007;70(1):127–131. doi: 10.1016/j.mimet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Cappuccino J. C., Sherman N. Microbiology: A Laboratory Manual. 3rd. New York, NY, USA: Benjamin Cummings; 1992. [Google Scholar]
- 25.Cattelan M. E., Hartel P. G., Fuhrmann J. J. Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Science Society of America Journal. 1999;63(6):1670–1680. doi: 10.2136/sssaj1999.6361670x. [DOI] [Google Scholar]
- 26.Anderson M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26(1):32–46. [Google Scholar]
- 27.Anderson M. J. DISTLM v.2: A FORTRAN Computer Program to Calculate a Distance-Based Multivariate Analysis for a Linear Model. Department of Statistics, University of Auckland; 2002. [Google Scholar]
- 28.Bozdogan H. Model selection and Akaike's Information Criterion (AIC): the general theory and its analytical extensions. Psychometrika. A Journal of Quantitative Psychology. 1987;52(3):345–370. doi: 10.1007/BF02294361. [DOI] [Google Scholar]
- 29.Clarke K., Gorley R. “PRIMER v6.” User Manual/Tutorial, Plymouth Routine in Multivariate Ecological Research. Plymouth Marine Laboratory; 2006. [Google Scholar]
- 30.Anderson M. J., Gorley R. N., Clarke K. R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth, UK: PRIMER-E; 2008. [Google Scholar]
- 31.Cook W. M., Lane K. T., Foster B. L., Holt R. D. Island theory, matrix effects and species richness patterns in habitat fragments. Ecology Letters. 2002;5(5):619–623. doi: 10.1046/j.1461-0248.2002.00366.x. [DOI] [Google Scholar]
- 32.Walker T. S., Bais H. P., Grotewold E., Vivanco J. M. Root exudation and rhizosphere biology. Plant Physiology. 2003;132(1):44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angel R., Soares M. I. M., Ungar E. D., Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME Journal. 2010;4(4):553–563. doi: 10.1038/ismej.2009.136. [DOI] [PubMed] [Google Scholar]
- 34.Bachar A., Soares M. I. M., Gillor O. The effect of resource islands on abundance and diversity of bacteria in arid soils. Microbial Ecology. 2012;63(3):694–700. doi: 10.1007/s00248-011-9957-x. [DOI] [PubMed] [Google Scholar]
- 35.Cary S. C., McDonald I. R., Barrett J. E., Cowan D. A. On the rocks: The microbiology of Antarctic Dry Valley soils. Nature Reviews Microbiology. 2010;8(2):129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 36.Caruso T., Chan Y., Lacap D. C., Lau M. C. Y., McKay C. P., Pointing S. B. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME Journal. 2011;5(9):1406–1413. doi: 10.1038/ismej.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köberl M., Ramadan E. M., Adam M., et al. Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in egypt. FEMS Microbiology Letters. 2013;342(2):168–178. doi: 10.1111/1574-6968.12089. [DOI] [PubMed] [Google Scholar]
- 38.Armada E., Roldán A., Azcon R. Differential activity of autochthonous bacteria in controlling drought stress in native lavandula and salvia plants species under drought conditions in natural arid soil. Microbial Ecology. 2014;67(2):410–420. doi: 10.1007/s00248-013-0326-9. [DOI] [PubMed] [Google Scholar]
- 39.Bachar A., Al-Ashhab A., Soares M. I. M., et al. Soil microbial abundance and diversity along a low precipitation gradient. Microbial Ecology. 2010;60(2):453–461. doi: 10.1007/s00248-010-9727-1. [DOI] [PubMed] [Google Scholar]
- 40.Bertrand H., Nalin R., Bally R., Cleyet-Marel J.-C. Isolation and identification of the most efficient plant growth-promoting bacteria associated with canola (Brassica napus) Biology and Fertility of Soils. 2001;33(2):152–156. doi: 10.1007/s003740000305. [DOI] [Google Scholar]
- 41.Siddikee M. A., Chauhan P. S., Anandham R., Han G.-H., Sa T. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. Journal of Microbiology and Biotechnology. 2010;20(11):1577–1584. doi: 10.4014/jmb.1007.07011. [DOI] [PubMed] [Google Scholar]
- 42.Mapelli F., Marasco R., Rolli E., et al. Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. BioMed Research International. 2013;2013:13. doi: 10.1155/2013/248078.248078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daffonchio D., De Biase A., Rizzi A., Sorlini C. Interspecific, intraspecific and interoperonic variability in the 16S rRNA gene of methanogens revealed by length and single-strand conformation polymorphism analysis. FEMS Microbiology Letters. 1998;164(2):403–410. doi: 10.1016/S0378-1097(98)00246-8. [DOI] [PubMed] [Google Scholar]
- 44.Daffonchio D., Borin S., Frova G., Manachini P. L., Sorlini C. PCR fingerprinting of whole genomes: the spacers between the 16s and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis . International Journal of Systematic Bacteriology. 1998;48(1):107–116. doi: 10.1099/00207713-48-1-107. [DOI] [PubMed] [Google Scholar]
- 45.Daffonchio D., Cherif A., Borin S. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group”. Applied and Environmental Microbiology. 2000;66(12):5460–5468. doi: 10.1128/AEM.66.12.5460-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gürtler V., Stanisich V. A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142(1):3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 47.Berg G., Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology. 2009;68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Pichel F., Johnson S. L., Youngkin D., Belnap J. Small-scale vertical distribution of bacterial biomass and diversity in biological soil crusts from arid lands in the Colorado Plateau. Microbial Ecology. 2003;46(3):312–321. doi: 10.1007/s00248-003-1004-0. [DOI] [PubMed] [Google Scholar]
- 49.Joshi P., Bhatt A. B. Diversity and function of plant growth promoting Rhizobacteria associated with wheat Rhizosphere in North Himalayan Region. International Journal of Environmental Sciences. 2011;1(6):p. 1135. [Google Scholar]
- 50.Muleta D., Assefa F., Hjort K., Roos S., Granhall U. Characterization of rhizobacteria isolated from wild Coffea arabica L. Engineering in Life Sciences. 2009;9(2):100–108. doi: 10.1002/elsc.200700031. [DOI] [Google Scholar]
- 51.Kumar G., Kanaujia N., Bafana A. Functional and phylogenetic diversity of root-associated bacteria of Ajuga bracteosa in Kangra valley. Microbiological Research. 2012;167(4):220–225. doi: 10.1016/j.micres.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 52.El Hidri D., Guesmi A., Najjari A., et al. Cultivation-dependant assessment, diversity, and ecology of haloalkaliphilic bacteria in arid saline systems of southern Tunisia. BioMed Research International. 2013;2013:15. doi: 10.1155/2013/648141.648141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alavi P., Starcher M. R., Zachow C., Müller H., Berg G. Root-microbe systems: the effect and mode of interaction of stress protecting agent (SPA) Stenotrophomonas rhizophila DSM14405T. Frontiers in Plant Science. 2013;4:p. 141. doi: 10.3389/fpls.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barazani O., Friedman J. Is IAA the major root growth factor secreted from plant-growth-mediating bacteria? Journal of Chemical Ecology. 1999;25(10):2397–2406. doi: 10.1023/A:1020890311499. [DOI] [Google Scholar]
- 55.Trivedi P., Spann T., Wang N. Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microbial Ecology. 2011;62(2):324–336. doi: 10.1007/s00248-011-9822-y. [DOI] [PubMed] [Google Scholar]
- 56.Marasco R., Rolli E., Vigani G., et al. Are drought-resistance promoting bacteria cross-compatible with different plant models? Plant Signaling and Behavior. 2013;8(10) doi: 10.4161/psb.26741.e26741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schachtman D. P., Reid R. J., Ayling S. M. Phosphorus uptake by plants: from soil to cell. Plant Physiology. 1998;116(2):447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vyas P., Rahi P., Gulati A. Stress tolerance and genetic variability of phosphate-solubilizing fluorescent Pseudomonas from the cold deserts of the trans-himalayas. Microbial Ecology. 2009;58(2):425–434. doi: 10.1007/s00248-009-9511-2. [DOI] [PubMed] [Google Scholar]
- 59.Ndung'u-Magiroi K. W., Herrmann L., Okalebo J. R., Othieno C. O., Pypers P., Lesueur D. Occurrence and genetic diversity of phosphate-solubilizing bacteria in soils of differing chemical characteristics in Kenya. Annals of Microbiology. 2012;62(3):897–904. doi: 10.1007/s13213-011-0326-2. [DOI] [Google Scholar]
- 60.Fürnkranz M., Müller H., Berg G. Characterization of plant growth promoting bacteria from crops in Bolivia. Journal of Plant Diseases and Protection. 2009;116(4):149–155. [Google Scholar]
- 61.Ribeiro C. M., Cardoso E. J. B. N. Isolation, selection and characterization of root-associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia) Microbiological Research. 2012;167(2):69–78. doi: 10.1016/j.micres.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Tian F., Ding Y., Zhu H., Yao L., Du B. Genetic diversity of siderophore-producing Bacteria of tobacco rhizosphere. Brazilian Journal of Microbiology. 2009;40(2):276–284. doi: 10.1590/S1517-83822009000200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma A., Johri B. N. Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiological Research. 2003;158(3):243–248. doi: 10.1078/0944-5013-00197. [DOI] [PubMed] [Google Scholar]
- 64.Kumar D. J. M., Lawrence L., Rajan R., Priyadarshini S., Yachittybabu S., Kalaichelvan P. T. Characterization of lipase and protease from Serratia marcescens DEPTK21 and its destaining capability. Asian Journal of Experimental Biological Sciences. 2012;3(3):621–628. [Google Scholar]
- 65.Michaud P., Belaich A., Courtois B., Courtois J. Cloning, sequencing and overexpression of a Sinorhizobium meliloti M5N1CS carboxymethyl-cellulase gene. Applied Microbiology and Biotechnology. 2002;58(6):767–771. doi: 10.1007/s00253-002-0953-4. [DOI] [PubMed] [Google Scholar]
- 66.Wani P. A., Khan M. S., Zaidi A. Synergistic effects of the inoculation with nitrogen-fixing and phosphate-solubilizing rhizobacteria on the performance of field-grown chickpea. Journal of Plant Nutrition and Soil Science. 2007;170(2):283–287. doi: 10.1002/jpln.200620602. [DOI] [Google Scholar]
- 67.Scheldeman P., Herman L., Foster S., Heyndrickx M. Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. Journal of Applied Microbiology. 2006;101(3):542–555. doi: 10.1111/j.1365-2672.2006.02964.x. [DOI] [PubMed] [Google Scholar]
- 68.Kanwar S. S., Gupta M., Gupta R., Kaushal R. K., Chimni S. S. Properties of hydrogel-entrapped lipase of thermophilic Pseudomonas aeruginosa BTS-2. Indian Journal of Biotechnology. 2006;5(3):292–297. [Google Scholar]
- 69.Diatta I. L. D., Kane A., Agbangba C. E., et al. Inoculation with arbuscular mycorrhizal fungi improves seedlings growth of two sahelian date palm cultivars (Phoenix dactylifera L., cv. Nakhla hamra and cv. Tijib) under salinity stresses. Advances in Bioscience and Biotechnology. 2014;5(1):64–72. doi: 10.4236/abb.2014.51010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material contains additional information about the geo-climatic data and biodiversity of bacterial community in the different studied oases.