Abstract
Purpose
DNA methyltransferase 3A ( DNMT3A) is one of the commonly mutated genes in acute myelogenous leukemia (AML). Reports on the prognostic significance of DNMT3A mutations have been inconsistent, and most of the data is available only for patients 60 years of age or younger. We hypothesized that this inconsistency is due to an interaction between the dose of anthracycline used in induction therapy and DNMT3A status. We studied whether patients with DNMT3A-mutated AML treated with standard dose anthracyclines had an inferior survival compared to patients with other mutation profiles or those who received high dose therapy.
Experimental design
152 patients in this retrospective cohort study (median age, 54 years) with de-novo AML underwent induction therapy and next-generation sequencing of 33 commonly mutated genes in hematologic malignancies, including DNMT3A, FLT3-ITD, NPM1, and IDH1/2. Cox regression was used to if those with DNMT3A mutations who were treated with standard dose anthracycline had inferior survival.
Results
DNMT3A mutations, found in 32% of patients, were not associated with an inferior survival. Dose escalation of anthracycline in the induction regimen was associated with improved survival in those with DNMT3A mutations but not those with wild-type DNMT3A. Patients with DNMT3A mutations who received standard dose induction had shorter survival time than other patient groups (10.1 months vs. 19.8 months, p=0.0129). This relationship remained significant (HR: 1.90, p=0.006) controlling for multiple variables.
Conclusions
Patients with DNMT3A-mutated AML have an inferior survival when treated with standard-dose anthracycline induction therapy. This group should be considered for high-dose induction therapy.
Keywords: Acute myeloid leukemia, DNMT3A, prognosis, induction chemotherapy
Introduction
The choice of induction and post-remission therapy in AML is guided by certain prognostic factors. Karyotype has historically been the largest determinant of prognosis 2,3, but this inadequately predicts outcome in a large proportion of patients, particularly those with no karyotypic abnormalities. Recurrent gene mutations in NPM1 and CEBPA, and internal tandem duplications (ITD) in FLT3 have been recognized as important in AML pathogenesis and prognosis. 4 More recently, an additional class of genes recurrently mutated in AML genomes has been identified that normally function in the epigenetic regulation of transcription. These include DNMT3A, TET2, IDH1/IDH2, and ASXL1. 5,6,7,8,9,10,1 A growing body of evidence supports a pathogenic role for these mutations in AML. 11
DNMT3A is one of the most commonly mutated genes in AML genomes and has been the topic of significant analysis since it was first noted by Ley et al. 12 It encodes one of the DNA methyltransferases, and along with DNMT3B, is responsible for adding a methyl group to cytosine/guanine residues. The prevalence of mutations in DNMT3A ranges from 18–36% and is enriched in normal karyotype AML. 13,12,14,15,8,16,17,18 The most frequently mutated residue of the DNMT3A gene occurs in the methyltransferase domain at Arginine 882, leading to decreased methylation activity in vitro 15 as well as decreased methylation levels in select genomic regions. 15,12 Additional mutations seen throughout the gene have also been described and are thought to also disrupt normal methylation activity. However, it has not been consistently associated with an altered gene expression pattern. 12
Despite an incomplete understanding of the functional changes induced by DNMT3A mutations, the initial studies of this gene mutation consistently showed that it conferred a poor prognosis. 12,14,8 However, more recent studies have contradicted this finding, and have shown no difference in overall survival based on DNMT3A mutational status in large, homogenously treated patient cohorts. 1,19,18,13 While the differences in prognostic significance in these studies may be due to a number of causes, including both patient factors and the location of the mutation, one interesting possibility that could account for these differences may be the intensity of therapy in these patient cohorts.
Patel et al recently noted that DNMT3A status affected the response to high-dose induction therapy in patients under age 60. 1 In patients with wildtype DNMT3A, NPM1 and MLL, there was no effect of dose escalation of daunorubicin from 45mg/m2 to 90mg/m2 on overall survival, whereas those with DNMT3A mutations did experience a survival benefit from a higher dose of daunorubicin. Since this observation may offer insight into the biologic characteristics of DNMT3A mutations and affect the choice of induction therapy, we further explored this relationship in a unique patient cohort. This cohort included many patients over age 60 in whom the value of high dose therapy is unclear.
Methods
Patient samples and treatment
Between January 2001 and August 2011, 172 patients with newly diagnosed AML consented to donation of their bone marrow or peripheral blood samples to the tissue bank at our institution. All patients consented to genetic analysis and clinical assessment on the basis of an institutional review board approved protocol with accompanying HIPAA authorization, and 167 underwent next-generation sequencing on the basis of available leukemia cell DNA.
Of these 167 patients, 152 underwent induction therapy and all analyses were restricted to this group (Supplemental Figure S1). The regimen selected for each patient was based on treating physician preference, but generally included 3 days of an anthracycline and 7 days of cytarabine. Patients without adequate cytoreduction at the day 14-marrow assessment were retreated at their nadir with the same drugs unless they were felt to have failed therapy. For the purposes of this study, we defined induction therapy as high-dose for those who received a cumulative dose of ≥270mg/m2 of daunorubicin as either a single induction of 90mg/m2/day or a double-induction with 45mg/m2/day-60mg/m2/day daunorubicin or 72mg/m2 of idarubicin, given as 12mg/m2/day on initiated on day 1 and again day 14. All other regimens were classified as standard dose therapy.
Cytogenetic analysis
All patients underwent cytogenetic analysis. Karyotype results were classified as good, intermediate, or poor risk according to the Medical Research Council criteria. 20 Patients with missing cytogenetic data, including those with failed cytogenetic testing, were classified as unknown.
Next-generation sequencing
Mutational analysis was performed using a targeted next generation sequencing panel (ASXL1, ATM, BRAF, CBL, CDKN2A, DDX3X, DNMT3A, ETV6, EZH2, FBXW7, FLT3 (ITD and TKD) GNAS, IDH1, IDH2, JAK2, KIT, KLHL6, KRAS, MAPK1, MYD88, NOTCH1, NPM1, NRAS, PTEN, PTPN11, PHF6, RUNX1, SF3B1, TET2, TP53, WT1, XPO1, ZMYM3). In short, DNA was quantified using a fluorescent based measurement (Qubit, Life Technologies, Ca) and 20–250 ng of DNA was used for custom target enrichment. Following library preparation with the TruSeq Amplicon assay (Ilumina, Ca) libraries were pooled and sequenced on the Miseq to an average depth of coverage greater than 1000x. This mean depth allowed for the most challenging amplicon to reach a minimum depth of coverage of 250 reads at all copy neutral loci. Data was then processed using a custom analysis pipeline composed of commercial, publically available and in house developed tools. 21
Statistical analysis
All hypothesis tests were 2-sided with statistical significance set as p<0.05. All analyses were performed in STATA Version 12.0 (StataCorp, College Station, TX). Baseline characteristics were compared between the mutated and wildtype DNMT3A status using the chi-squared test for categorical variable and the Wilcoxon rank sum test for continuous variables.
Survival distributions for overall survival (OS) and relapse free survival (RFS) were computed using the Kaplan-Meier method and compared using the log-rank test to determine statistical differences in the distributions for the exposure groups. A Cox regression model was used to adjust for covariates including age over 60, cytogenetic risk group, sex, allogeneic transplant, and FLT3-ITD, NPM1, IDH1 and IDH2 mutations. A backwards elimination procedure was used to create the final multivariate model. Because an interaction between high-dose therapy and DNMT3A status was noted, an interaction term defined as DNMT3A mutated treated with standard-dose therapy compared to all other groups (DNMT3A wild type or DNMT3A mutated treated with high dose therapy) was retained in the multivariate model. We anticipated a sample size of 175 patients, with 22 (12.5%) in the DNMT3A-mutated/standard-dose group. A post-hoc calculation using bootstrap methods was used to estimate the power of the log-rank test used to test the hypothesis that that there was a difference in survival among patients with a DNMT3A mutation who received standard dose anthracycline (n=33, 3-year survival rate=13.1%) compared those without a DNMT3A mutation and those with a DNMT3A mutation who received high dose anthracycline(n=119, 3-year survival=33.9%). The estimate of the power of the test was 73% (95% CI = 70%–76%).
Results
Patient cohort
This patient cohort included all patients with a diagnosis of AML seen at the Hospital of the University of Pennsylvania between January 2001 and August 2011 who provided adequate tissue and gave informed consent for these studies (Supplementary Figure S1). Patient, disease, and treatment information is detailed in Table 1. Of note, the age range for this study was 19–86 years with a median age of 55, and 44% were ≥60 years. All cytogenetic risk groups are represented, with the intermediate risk group representing the largest fraction at 62%. High-dose induction therapy (as defined above) was given to 32% of all patients. The median follow-up time was 12.6 months.
Table 1.
Patient, Disease, and Treatment Characteristics
| Full cohort (n=152) | DNMT3A mutant (n=49, 32%) | DNMT3A wild type (n=103, 68%) | Significance | |
|---|---|---|---|---|
|
| ||||
| Age at diagnosis, median, yrs (range) | 54 (19–79) | 54.4 (26–78) | 54.1 (19–79) | 0.6759 |
|
| ||||
| Age ≥ 60 | 39% | 29% | 44% | 0.074 |
|
| ||||
| Male | 57% | 45% | 63% | 0.034 |
|
| ||||
| WBC at diagnosis (mean) | 58,232 | 76,569 | 49,509 | 0.021 |
|
| ||||
| WBC ≥100,000 | 21% | 31% | 17% | 0.057 |
|
| ||||
| Cytogenetics risk groups | 0.003 | |||
| Favorable | 13% | 0% | 19% | |
| Intermediate | 64% | 82% | 56% | |
| Poor | 15% | 10% | 17% | |
| Unknown | 7% | 8% | 7% | |
|
| ||||
| FLT3-ITD mutant | 32% | 43% | 26% | 0.039 |
|
| ||||
| NPM1 mutant | 33% | 65% | 25% | <0.001 |
|
| ||||
| IDH1 mutant | 8% | 16% | 4% | 0.008 |
|
| ||||
| IDH2 mutant | 14% | 14% | 14% | 0.908 |
|
| ||||
| High-dose therapy | 32% | 33% | 32% | 0.844 |
|
| ||||
| Double-induction | 15% | 12% | 17% | 0.493 |
Abbreviations: n, number of patients; yrs, years
Frequency and spectrum of DNMT3A mutations
Of the 152 patients assessed for mutation status, 52 (31.1%) harbored mutations in the DNMT3A gene (Supplementary Table S1). As expected, missense mutations in the R882 codon were the most common change, found in 57.7% (30/52) of those with DNMT3A mutations. Of those 30 patients, one also had a concurrent non-R882 mutation. An additional 21 patients had single non-R882-DNMT3A mutations and one additional patient had with 2 non-R882 mutations. For subsequent analyses, DNMT3A mutated included both missense mutations in the R882 codon as well as the non-R882 mutations.
Association of DNMT3A with patient, disease, and treatment characteristics
The association of DNMT3A mutations with patient, disease, and treatment characteristics is detailed in Table 1 and Supplemental Figure S1. At diagnosis, patients with DNMT3A mutations were younger and less likely to be male compared to DNMT3A wild-type (33% vs. 50% were 60 years or older and 46% vs. 63% were male). More patients with DNMT3A mutations were in the intermediate cytogenetic risk group (79% vs 54%). The mean WBC count at diagnosis was also higher in those with DNMT3A mutations (74,700 vs 51,500).
DNMT3A mutations occurred concomitantly with FLT3-ITD, NPM1, and IDH1 mutations more frequently than with wild-type DNMT3A, as seen in Table 1. When analysis was restricted to those with intermediate-risk cytogenetics, only NPM1 remained associated with DNMT3A (70.9% vs 33.9%, p= <0.001). Concomitant mutations in DNMT3A, NPM1 and FLT3-ITD occurred 18/167 patients as compared to the 6/167 expected by chance alone (p= 0.011). This triple-mutant genotype was initially noted by the Cancer Atlas Genome Study for AML and suggests biologic cooperation among these genes. 22
Since we were interested in the interaction between the dose of anthracycline and DNMT3A status, we looked at the differences in induction chemotherapy dose in those with mutated or wild-type (wt) DNMT3A. The percentage of patients who received high dose induction therapy or double induction did not differ based on DNMT3A (Table 1).
Association of DNMT3A mutations with clinical outcomes
There was no difference in OS or RFS based on DNMT3A status alone, with median survival of 17.3 months and RFS of 13.8 months for DNMT3A-mutant compared to 16 and 13.1 for DNMT3A-wt (p= 0.3297 and p=0.222, respectively) (Figure 1).
Figure 1. Survival by DNMT3A status.
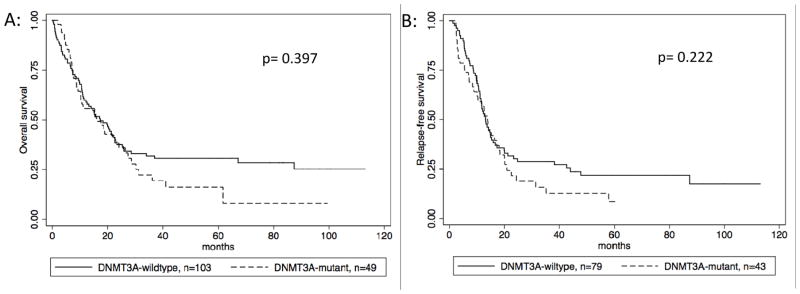
Figure 1A: Overall survival stratified by DNMT3A status, 1B: Relapse free survival stratified by DNMT3A status.
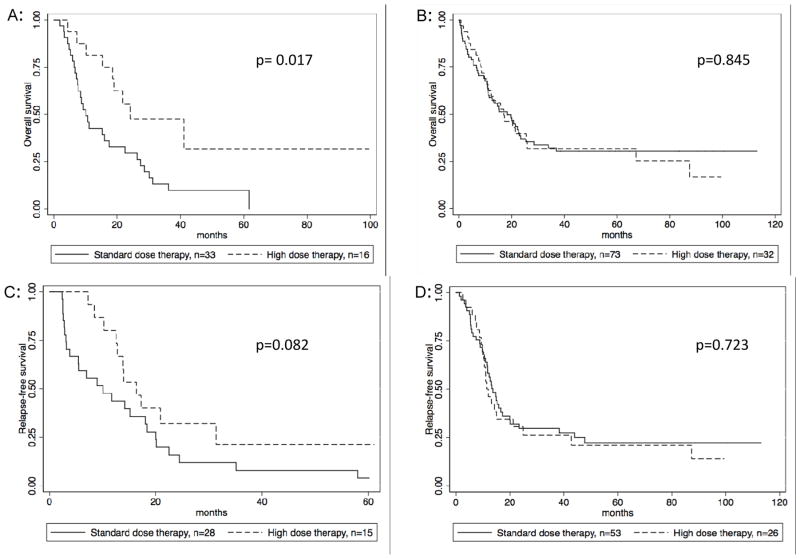
A mutational analysis of the ECOG 1900 trial patients demonstrated that the benefit of anthracycline-intensified induction was seen only in those with a particular mutation profile, including DNMT3A mutations. 1 We found a similar pattern in our institution’s cohort. Patients with mutated DNMT3A had an improved overall survival with high-dose therapy (p=0.017) as compared to those with DNMT3A-wt, who did not benefit from intensified therapy (Figure 2). Those with a mutated DNMT3A also had improved RFS with high-dose therapy, although this did not meet statistical significance (p=0.082).
Figure 2. Effect of dose escalation depends on DNMT3A status.
Figure 2 A: OS in DNMT3A-mutant, stratified by anthracycline dose B: OS in DNMT3A-wildtype, stratified anthracycline dose C: RFS in DNMT3A-mutant, stratified by anthracycline dose D: RFS in DNMT3A-wildtype, stratified by anthracycline dose
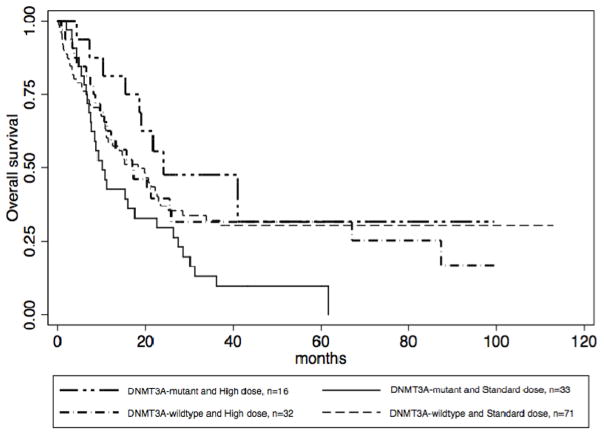
Of the 152 patients who received induction therapy, 33 (21.7%) had both a DNMT3A mutation and received standard-dose induction. We found that patients with this profile had worse prognosis, with a median survival of 10.1 months compared to 19.8 months for all other patients (p=0.0129) (Figure 3a). Of note, there was no survival difference between the 3 patient subsets (DNMT3A-wildtype/standard dose; DNMT3A-wildtype/high dose; DNMT3A-mutant/high dose) that make up the comparator group (Figure 4) (p=0.845, 0.2637, 0.2767).
Figure 3. Survival stratified by anthracycline dose and DNMT3A status.
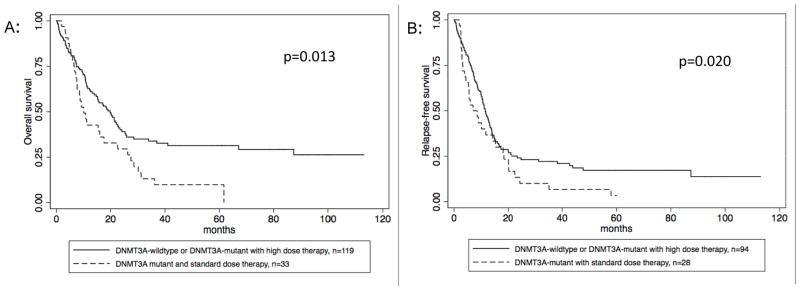
Figure 3 A: OS, comparing the DNMT3A mutant patients who received standard dose therapy to all other patients (DNMT3A-wildtype and DNMT3A mutant who received high dose therapy) B: RFS, comparing the DNMT3A mutant patients who received standard dose therapy to all other patients (DNMT3A-wildtype and DNMT3A-mutant who received high dose therapy)
Figure 4. Overall survival by DNMT3A status and anthracycline dose.
Figure 4: Overall survival, as stratified by the presence or absence of a DNMT3A mutation and the anthracycline dose received.
This relationship of poorer survival in the DNMT3A mutant/standard dose group persisted on multivariate analysis after adjustment for other known prognostic factors, including age >60 years, karyotype, FLT3-ITD, and NPM1 mutations (HR: 1.89, p=0.006). With the exception of FLT3-ITD, all known prognostic factors were significantly associated with survival in the univariate analyses (Table 2). A similar effect was seen for RFS, with a median RFS of 10.1 months for those patients with a DNMT3A mutation treated with standard dose therapy and 13.6 months for all others (p=0.020)(Figure 3b). This relationship was also significant in the multivariate analysis (Table 2).
Table 2.
Multivariate Analysis
| Covariate | Overall Survival | Relapse Free Survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate HR | p-value | Multivariate HR | p-value | Univariate HR | p-value | Multivariate HR | p-value | |
| DNMT3A mut and stnd dose (vs. DNMT3A wt or DNMT3A mut and high dose) | 1.71 | 0.014 | 1.90 | 0.006 | 1.47 | 0.070 | 1.69 | 0.017 |
| DNMT3A mut (vs. DNMT3A wt) | 1.21 | 0.331 | ------ | ------ | 1.02 | 0.911 | ------ | ------ |
| High dose (vs. stnd dose) | 0.83 | 0.355 | ------ | ------ | 0.83 | 0.339 | ------ | ------ |
| Age ≥60 (vs. Age <60) | 2.16 | <0.001 | 2.62 | <0.001 | 2.25 | <0.001 | 2.10 | <0.001 |
| Intermediate Cytogenetics (vs. Favorable) | 2.59 | 0.016 | 2.71 | 0.016 | 1.27 | 0.388 | 1.37 | 0.317 |
| Poor Cytogenetics (vs. Favorable) | 3.29 | 0.006 | 2.66 | 1.10 | 1.68 | 0.096 | 1.19 | 0.616 |
| Unknown Cytogenetics (vs. Favorable) | 2.55 | 0.0800 | 3.62 | 0.022 | 1.42 | 0.391 | 1.68 | 0.268 |
| Female sex (vs. Male sex) | 0.92 | 0.645 | ------ | ------ | 0.93 | 0.656 | ------ | ------ |
| FLT3-ITD (vs. no FLT3-ITD) | 1.45 | 0.068 | 1.87 | 0.006 | 1.15 | 0.455 | 1.56 | 0.037 |
| NPM1 mut (vs. NPM1 wt) | 0.84 | 0.390 | 0.50 | 0.002 | 0.79 | 0.174 | 0.55 | 0.005 |
| IDH1 mutated (vs. IDH1 wt) | 0.91 | 0.792 | ------ | ------ | 0.75 | 0.375 | ------ | ------ |
| IDH2 mutated (vs. IDH2 wt) | 0.93 | 0.782 | ------ | ------ | 0.87 | 0.585 | ------ | ------ |
| Allo tx (vs. no allo-tx) | 0.46 | <0.001 | ------ | ------ | 0.73 | 0.072 | ------ | ------ |
Abbreviations: mut, mutant; vs, versus; wt, wildtype; stnd, standard; Allo tx, Allogeneic transplant; HR, hazard ratio
Discussion
Mutational analysis in AML is being used to supplement traditional cytogenetic analysis in order to better understand prognosis and guide post-remission therapy. It has emerging implications for targeted therapy. The results of this study suggest it may also help to determine induction chemotherapy. Although DNMT3A mutations have a controversial impact on survival, this appears to be at least partially explained by an interaction with the dose of induction chemotherapy. Our study of a large single institutional cohort of AML patients confirms that patients with mutated, but not wildtype, DNMT3A have an inferior prognosis if treated with standard doses of anthracycline chemotherapy during induction therapy.
This finding was consistent throughout our analysis, including our multivariate analysis that adjusted for age, cytogenetics, FLT3-ITD and NPM1. As this was a retrospective study performed on patients who were not randomized to different doses of anthracyclines, it is possible that the inferior survival seen in DNMT3A mutant patients who received standard dose anthracycline was due to selection bias. Indeed, we were not able to collect and adjust for performance status. However, this finding was seen only in patients with DNMT3A mutations, the minority of the cohort, suggesting that the inferior survival of this group is due to a true biologic effect, not simply selection bias for those with a poor performance status who were unable to tolerate higher doses of anthracycline chemotherapy. Furthermore, even patients with mutated DNMT3A who achieved a complete response with standard dose anthracyclines had an inferior RFS, demonstrating that the improved survival seen in the patients who received high dose therapy was not the results of failure to provide a second induction to unfit patients with residual disease.
The biology driving this relationship is not certain. DNMT3A mutations have been linked to changes in methylation patterns in affected genomes 15,12 and extensive methylation loss when occurring with FLT3-ITD and NPM1 mutations together. 22 It is possible that the pattern of changes in methylation through DNMT3A mutations could affect response to anthracyclines. Alternatively, DNMT3A mRNA and protein have been shown to be upregulated in response to increasing doses of doxorubicin in human colorectal cell lines, and silencing of DNMT3A increased the percentage of senescent cells in response to treatment with doxorubicin. 23 DNMT3A mutations, particularly the single amino acid mutation, R882, has been shown to result in decreased function of the methyltransferase enzyme in in vitro studies. 15 It is plausible that the decreased function of DNMT3A allows for a better response to high dose anthracycline chemotherapy.
We defined high dose anthracycline for this study as either a cumulative dose of ≥270 mg/m2 of daunorubicin (single induction of 90mg/m2/day or double induction with 45–60mg/m2) or 72mg/m2 of idarubicin (double induction of 12mg/m2 of idarubicin). 49 patients received this high dose therapy; this included 24 patients who received “double-induction” with 2 rounds of standard induction. This was generally performed at the treating physician’s discretion in response to an inadequately ablated day 14 bone marrow biopsy. As such, we considered this a single high-dose regimen. We feel this is a reasonable and physiologic approach, and previous studies of anthracycline induction have been performed in a “response-adapted” method using double-induction as needed, then using the total dose of anthracycine received to guide subsequent trials. 24,25,26
Our findings of inferior survival in patients with DNMT3A mutations who receive standard doses of anthracyclines support those of Patel et al in the ECOG 1900 cohort.1 With 2 studies now revealing this interaction, it seems reasonable to use the findings to guide therapy. Anthracycline escalation led to an improved survival without an increase in toxicity in patients under age 60 in ECOG 1900. 27 Thus, we would not recommend a return to standard dose induction for those without DNMT3A mutations without further studies. However, in patients over age 60, the role of dose escalation is uncertain. A cooperative group study published by Lowenberg et al compared 45mg/m2 to 90mg/m2 of daunorubicin in patients over age 60 with newly diagnosed AML and found that while there was an improved CR rate in those who received high dose anthracycline, there was no difference in overall survival or in toxicity profile. 28 Similarly, the Acute Leukemia French Association (ALFA)-9801 study found no difference in CR, OS, or EFS for dose-escalated therapy compared to standard dose idarubicin in patients age 50–70 with AML.29 As such, high dose therapy has not been routinely adopted for this age group, although the lack of excess toxicity in these trials suggests that anthracycline induction may not need to be dose-attenuated either. 30
Patients with DNMT3A mutations who are ≥60 years old may be a select group for whom higher dose anthracycline is reasonable. However, DNMT3A mutational analysis is often not feasible prior to the initiation of induction therapy. For example, current processing time for this test at our center is 7–10 days. Our cohort of patients receiving induction therapy ranged from age 19 to 79 including 59 patients age ≥60. Fourteen patients ≥ 60 years old received high dose therapy; 10 of whom received it as a double-induction. Therefore, one strategy in older patients may be to give standard dose induction with 45mg/m2, and, if subsequently found to have a DNMT3A mutation, they could receive a second dose of daunorubicin at 45mg/m2 on day 14 regardless of bone marrow biopsy results at that time to ensure that they receive full high dose anthracycline induction.
Mutational analysis of leukemic cells in patients with newly diagnosed AML is becoming more feasible and the number of clinical applications is growing. These results suggest that DNMT3A mutations alter the response to anthracycline chemotherapy, and define a group for whom high-dose therapy is particularly useful. Furthermore, it suggests that chemotherapy dose should be considered in the algorithm when incorporating comprehensive gene mutation signatures into risk-adapted post-remission therapy plans. Future studies are necessary to determine the biology that guides this relationship to allow further personalization of treatment plans in this AML subtype. Significantly larger patient cohorts are necessary to define the behavior of rare subtypes—such as the triple DNMT3A, FLT3-ITD, and NPM1 mutant—and their response to chemotherapy.
Supplementary Material
Statement of Translational Relevance.
The emergence of comprehensive mutational testing in AML has led to significant excitement in the leukemia community but also some concern for how to use the wide array of new genetic tests. One gene of interest is the DNMT3A gene. Previous work by Patel and colleagues1 suggested that patients <60 years old with AML that express mutant DNMT3A require higher doses of anthracycline in their induction regimen in order to obtain equivalent results as DNMT3A wild type patients. However, this observation has not previously been reproduced. Here we describe our retrospective cohort study which confirms that mutated DNMT3A can predict for both overall and relapse-free survival with standard doses of anthracycline induction therapy, including patients ≥60 years. This decreased prognosis can be overcome by treating patients with higher doses of anthracycline. This confirms that patients with DNMT3A mutated AML should be treated with higher doses of anthracycline.
Acknowledgments
Financial Support: MC is supported by NIH grant 1R01CA149566, Veterans Administration Merit Award 1I01BX000918-01 and by the Leukemia and Lymphoma Society”
Footnotes
The authors declare no conflicts of interest.
References
- 1.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. The New England journal of medicine. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 3.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–83. [PubMed] [Google Scholar]
- 4.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. The New England journal of medicine. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 5.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:475–86. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 9.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–9. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nature reviews Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 12.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaidzik VI, Schlenk RF, Paschka P, Stolzle A, Spath D, Kuendgen A, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–77. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 14.Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2889–96. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 15.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 16.Fried I, Bodner C, Pichler MM, Lind K, Beham-Schmid C, Quehenberger F, et al. Frequency, onset and clinical impact of somatic DNMT3A mutations in therapy-related and secondary acute myeloid leukemia. Haematologica. 2012;97:246–50. doi: 10.3324/haematol.2011.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renneville A, Boissel N, Nibourel O, Berthon C, Helevaut N, Gardin C, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26:1247–54. doi: 10.1038/leu.2011.382. [DOI] [PubMed] [Google Scholar]
- 18.Roller A, Grossmann V, Bacher U, Poetzinger F, Weissmann S, Nadarajah N, et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia. 2013;27:1573–8. doi: 10.1038/leu.2013.65. [DOI] [PubMed] [Google Scholar]
- 19.Markova J, Michkova P, Burckova K, Brezinova J, Michalova K, Dohnalova A, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. European journal of haematology. 2012;88:128–35. doi: 10.1111/j.1600-0609.2011.01716.x. [DOI] [PubMed] [Google Scholar]
- 20.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 21.Daber RD, Sukhadia S, Morrissette JJD. Understanding the limitations of NGS informatics, an approach to clinical pipeline validation using artificial data sets. Cancer genetics. doi: 10.1016/j.cancergen.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Gao Y, Zhang G, Huang S, Dong Z, Kong C, et al. DNMT3a plays a role in switches between doxorubicin-induced senescence and apoptosis of colorectal cancer cells. International journal of cancer Journal international du cancer. 2011;128:551–61. doi: 10.1002/ijc.25365. [DOI] [PubMed] [Google Scholar]
- 24.Ohno R, Kobayashi T, Tanimoto M, Hiraoka A, Imai K, Asou N, et al. Randomized study of individualized induction therapy with or without vincristine, and of maintenance-intensification therapy between 4 or 12 courses in adult acute myeloid leukemia. AML-87 Study of the Japan Adult Leukemia Study Group. Cancer. 1993;71:3888–95. doi: 10.1002/1097-0142(19930615)71:12<3888::aid-cncr2820711216>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T, Miyawaki S, Tanimoto M, Kuriyama K, Murakami H, Yoshida M, et al. Randomized trials between behenoyl cytarabine and cytarabine in combination induction and consolidation therapy, and with or without ubenimex after maintenance/intensification therapy in adult acute myeloid leukemia. The Japan Leukemia Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1996;14:204–13. doi: 10.1200/JCO.1996.14.1.204. [DOI] [PubMed] [Google Scholar]
- 26.Miyawaki S, Tanimoto M, Kobayashi T, Minami S, Tamura J, Omoto E, et al. No beneficial effect from addition of etoposide to daunorubicin, cytarabine, and 6-mercaptopurine in individualized induction therapy of adult acute myeloid leukemia: the JALSG-AML92 study. Japan Adult Leukemia Study Group. International journal of hematology. 1999;70:97–104. [PubMed] [Google Scholar]
- 27.Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, et al. Anthracycline dose intensification in acute myeloid leukemia. The New England journal of medicine. 2009;361:1249–59. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, et al. High-dose daunorubicin in older patients with acute myeloid leukemia. The New England journal of medicine. 2009;361:1235–48. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- 29.Pautas C, Merabet F, Thomas X, Raffoux E, Gardin C, Corm S, et al. Randomized study of intensified anthracycline doses for induction and recombinant interleukin-2 for maintenance in patients with acute myeloid leukemia age 50 to 70 years: results of the ALFA-9801 study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:808–14. doi: 10.1200/JCO.2009.23.2652. [DOI] [PubMed] [Google Scholar]
- 30.Luger SM. Treating the elderly patient with acute myelogenous leukemia. Hematology/the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2010;2010:62–9. doi: 10.1182/asheducation-2010.1.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.