Abstract
Tumor protein phosphorylation analysis may provide insight into intracellular signaling networks underlying tumor behavior, revealing diagnostic, prognostic or therapeutic information. Human tumors collected by The Cancer Genome Atlas (TCGA) program potentially offer the opportunity to characterize activated networks driving tumor progression, in parallel with the genetic and transcriptional landscape already documented for these tumors. However, a critical question is whether cellular signaling networks can be reliably analyzed in surgical specimens, where freezing delays and spatial sampling disparities may potentially obscure physiological signaling. To quantify the extent of these effects, we analyzed the stability of phosphotyrosine (pTyr) sites in ovarian and colon tumors collected under conditions of controlled ischemia and in the context of defined intratumoral sampling. Cold-ischemia produced a rapid, unpredictable, and widespread impact on tumor pTyr networks within 5 minutes of resection, altering up to 50% of pTyr sites by more than 2-fold. Effects on adhesion and migration, inflammatory response, proliferation, and stress response pathways were recapitulated in both ovarian and colon tumors. Additionally, sampling of spatially distinct colon tumor biopsies revealed pTyr differences as dramatic as those associated with ischemic times, despite uniform protein expression profiles. Moreover, intra-tumoral spatial heterogeneity and pTyr dynamic response to ischemia varied dramatically between tumors collected from different patients. Overall, these findings reveal unforeseen phosphorylation complexity, thereby increasing the difficulty of extracting physiologically relevant pTyr signaling networks from archived tissue specimens.In light of this data, prospective tumor pTyr analysis will require appropriate sampling and collection protocols to preserve in vivo signaling features.
Keywords: Phosphorylation, heterogeneity, ischemia, signaling, tumor
Introduction
Protein post-translational modifications (PTMs), such as phosphorylation, regulate the stability, localization, and activity of cellular components (1-3). Accordingly, dynamic phosphorylation plays a vital role in coordinating information flow within the cell and regulating emergent tumor responses ranging from proliferation to invasion and angiogenesis. Phosphorylation occurs predominantly on serine and threonine residues with tyrosine phosphorylation accounting for only ∼0.05% of all phosphorylation events in eukaryotic cells (4). Although tyrosine phosphorylation is rare and tyrosine kinases represent only 0.3% of the genome, these enzymes represent close to 30% of the known oncoproteins such as SRC, EGFR, and BCR-ABL (5). Their disproportionate role in oncology, combined with their structural druggability, make tyrosine kinases highly desirable therapeutic targets. Quantitative analysis of protein tyrosine phosphorylation in human tumor tissue specimens can provide insight into intracellular signaling networks underlying tumor behavior while identifying activated kinases and their substrates, signaling components that may represent druggable targets.
Human tumors collected by The Cancer Genome Atlas (TCGA) program potentially offer the opportunity to characterize activated signaling networks driving tumor progression, in parallel with the genetic and transcriptional landscape already documented for these tumors (6-8). The National Cancer Institute's Clinical Proteomic Tumor Analysis Consortium (NCI-CPTAC) seeks to provide proteomic characterization of tumors genomically annotated by TCGA programs (9). State-of-the-art mass spectrometry (MS)-based proteomics provides quantitative, systematic analysis of protein phosphorylation profiles (10-12) with the potential to directly identify activated signaling networks in tumors; information that is difficult to derive from genetic based studies. However, tumor specimen analysis is complicated by possible temporal delays in tissue acquisition and processing along with spatial sampling differences due to the heterogeneous nature of human tumors. Freezing delays following sample resection and processing subject the specimen to ischemia; unfortunately the exact time to freezing for archived tissue specimens is often undocumented. To date, studies on the effects of ischemia on tissue phosphorylation have been reported, yet the sample collection and analysis approaches used did not allow assessment of short periods of ischemia (13) or permit comprehensive analysis of pTyr signaling (14).
The heterogeneous nature of human tumors has been widely documented (15,16). Inter-patient and intra-tumoral variations are evident at a macroscopic histological level and at molecular, genetic, and epigenetic levels through clonal evolution and tumor microenvironment influences (17,18). A previous report investigated phosphorylation patterns across distinct anatomic metastatic lesions of prostate cancer. However, the number of phosphorylation sites profiled was limited and intra-lesion comparison was not performed (19).The influence of proximal spatial heterogeneity within a tumor specimen on pTyr signaling networks has yet to be determined. Collectively, these pre-analytical factors may influence protein phosphorylation measurements, thereby obscuring physiological signaling networks (20). Therefore, we sought to investigate how pre-analytical variations in sample collection and processing can ultimately affect downstream pTyr signaling analysis of human tumors.
Materials and Methods
Human ovarian and colon tumor collection
High-grade serous ovarian carcinoma tissue from five patients was collected as previously reported (21). Colon adeno carcinoma biopsy tissue was collected from five patients at the Cooperative Human Tissue Network at Vanderbilt University Medical Centre (CHTN-VUMC), in accordance with IRB-approved protocols. Following vessel ligation, surgical specimen removal was performed and the first core biopsy was taken immediately thereafter (t=0), transferred to prechilled cryovials and snap frozen in liquid nitrogen. Further core biopsies were collected and frozen after 10, 30, and 60 minutes of cold ischemia. Only specimens meeting pathology quality inclusion criteria of left sided colon adenocarcinoma cancers in which clamp time can easily be determined, minimal tumor diameter of 4 cm and no prior chemotherapy and/or radiation were released for analysis.
Protein extraction, digestion, and iTRAQ labeling of peptides from ovarian and colon tumors
Approximately 50-100 mg (total wet weight) of each of the timepoint samples was homogenized separately for protein extraction and digestion as described previously (22). Desalted peptides were labeled with multiplex iTRAQ (Isobaric Tags for Relative and Absolute Quantification) reagents as reported previously (22). Briefly, 800 μg peptide per sample for each of the ovarian and colon tumor timepoints was labeled with two tubes of iTRAQ reagent (according to the labeling scheme shown in Supplementary Fig. 2 and 5).
Phosphotyrosine peptide enrichment
Phosphotyrosine peptides were enriched prior to mass spectrometry analyses using a cocktail of anti-phosphotyrosine antibodies followed by immobilized metal affinity chromatography (IMAC) as previously described (22).
Mass-spectrometry based phosphotyrosine analysis
Peptides were chromatographically separated and subsequently analyzed by Orbitrap Elite mass spectrometer (Thermo Scientific), database-searched, validated and normalized as previously reported (22).
Phosphotyrosine data analysis
The total list of peptides and proteins identified and quantified can be found in Supplementary Table 1, 2 and 4. All mass spectra, in the original instrument vendor format, contributing to this study may be downloaded from: https://cptac-data-portal.georgetown.edu/cptacPublic/
Mass-spectrometry based protein expression analysis
Approximately 10% (∼300 μg) of iTRAQ labeled peptides from each colorectal tumor pTyr IP supernatant was separated off-line on C18 column. A total of 80 fractions were collected, non-contiguously pooled into 20 final fractions, and each subjected to an independent LC-MS/MS analysis. Each fraction was separated by reverse phase UHPLC (Easy-nLC 1000, Thermo Scientific) before nanoelectrospray directly into a Q-Exactive mass spectrometer (Thermo Scientific).
Protein expression data analysis
Peptide and protein identification were performed with the Proteome Discoverer software (version 1.4; Thermo Scientific) using Mascot search engine (version 2.4.1, Matrix Science). MS/MS spectra were searched against a human protein sequence database (NCBInr, 2012 release, 35,586 sequences). For each protein expression experiment, one combined database search was performed where the data files from all 20 independent MS fraction analyses were searched collectively as a single input dataset. The total list of peptides and proteins identified and quantified can be found in Supplementary Table 5.
Affinity propagation clustering analysis
Quantitative temporal profiles of all pTyr sites within an individual patient dataset were clustered using the affinity propagation algorithm proposed by Frey and Dueck (23). Individual patient datasets were subjected to independent analysis (see Supplementary Fig. 4) as described previously (22).
Statistical Analysis and Annotation
Hierarchical clustering analysis and heatmap construction was performed using the built-in Bioinformatics Toolbox function ‘Clustergram’ in Matlab (R2013b, The Mathworks Inc.) with Euclidean pairwise distance metric. Statistical analysis was conducted using GraphPad Prism 5.0a software. Pearson correlation analysis (two-tailed), and pairwise Student t tests (two-tailed) were used for calculating the significance of the differences and significance was accepted when P < 0.05. Kinase enrichment analysis was performed using the webtool at http://amp.pharm.mssm.edu/lib/kea.jsp and conducted as described previously (24). The input dataset was compiled from the union of ischemia-regulated pTyr sites found in ≥4 ovarian tumor samples and ≥4colon tumor samples. Predicted kinases were plotted onto a dendrogram of the human kinome (25) using the webtool at http://web.cecs.pdx.edu/∼josephl/kinome-cluster/. Panther Gene ontology (GO) annotations were identified by uploading UniProt ID lists to the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org/).
Results
Post-excision ischemia induces rapid, widespread, patient-dependent alterations to pTyr networks
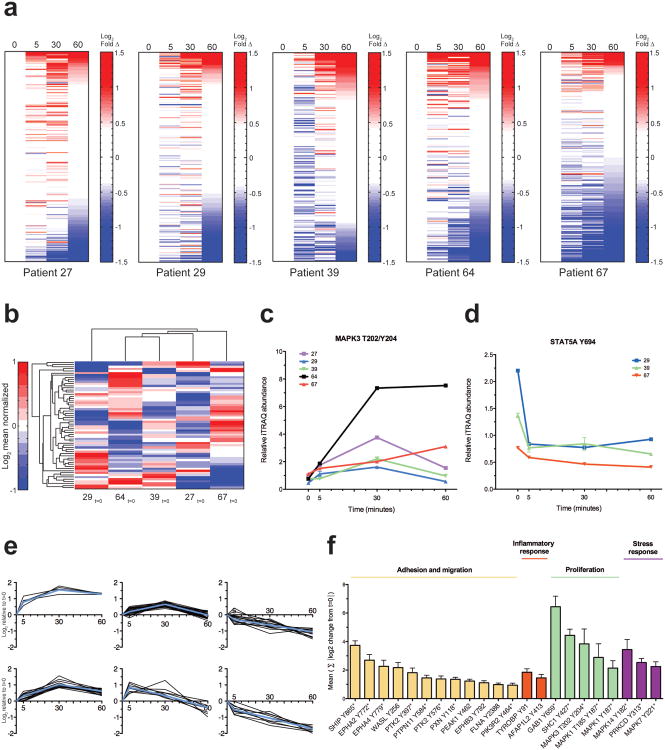
Post-resection ischemia time is generally undocumented in TCGA samples but may range from minutes to an hour during processing and pathological inspection. To understand the effect of cold-ischemia on protein tyrosine phosphorylation, we collected patient-derived ovarian tumor samples from five individuals undergoing debulking surgery and performed a controlled ischemia 4-point time-course (Supplementary Fig. 2). Ischemia time course sets from each patient were analyzed separately, providing detection and quantitation of several hundred pTyr sites per patient (Supplementary Table 1). Within 5 minutes of cold-ischemia, the fraction of pTyr sites within a specimen that showed quantitative fluctuations ranged from 29-55%, depending on the patient (Fig. 1a and Supplementary Table 1). Intriguingly, the temporal response to ischemia was patient specific. Two patients, 39 and 67, showed a predominant decrease across many pTyr sites within the first five minutes, while the other patients had a more mixed response, with comparable proportions of increasing and decreasing sites. We hypothesized that some of the inter-patient differences in response to ischemia could be attributed to patient-specific baseline phosphorylation profiles. To investigate this possibility, the ‘0 min’ samples from each ovarian patient were analyzed simultaneously by quantitative multiplex MS, allowing the relative levels of each pTyr site at resection to be directly compared between patients. Euclidean distance hierarchical clustering indicated variation in the basal pTyr profiles of each patient, suggestive of distinct phosphorylation network states at excision (Fig. 1b, Supplementary Table 2).This result agrees with previous reports noting that tumors with similar driver mutations can demonstrate distinct signaling network profiles (22,26). Integration of the ischemia time-course datasets with the inter-patient basal resection profile dataset revealed dependencies of ischemic response dynamics on initial baseline phosphorylation (Supplementary Table 2). For example, in the ERK1 activation loop the initial basal values were comparable, but the magnitude of change with ischemia time was distinct for different patients (Fig. 1c and Supplementary Fig. 3). For other sites, such as the activating site on STAT5A, the extent of ischemic regulation was dependent on the pTyr abundance at resection (Fig. 1d and Supplementary Fig. 3). These data suggest that documentation of the temporal delay between resection and freezing might not be sufficient to back-calculate the basal phosphorylation status prior to ischemia, since the dynamics for some phosphosites are patient specific.
Figure 1.
Post-excision ischemia induces rapid, widespread, patient-dependent alterations to pTyr networks. (a) Heatmaps of all quantified peptides from individual patient-derived ovarian tumor 4-timepoint ischemia study. Rows indicate mass spectrometry-derived quantitative levels of pTyr sites in log2 scale relative to t=0 min, rank ordered by value at t=60 min. Onset of color in the heatmaps corresponds to changes ≥3 s.d. from value at t=0 (red, increasing; blue, decreasing). See Supplementary Table 1 for details. (b)Hierarchical-clustered heatmap of quantitative pTyr profiles of individual ovarian tumors at resection. Quantitative levels of pTyr sites grouped in rows, in log2 mean normalized scale. (c, d) Select examples of distinct inter-patient ischemia regulated temporal dynamics. Colored lines represent relative inter-patient phosphorylation levels, mean ± s.d. of technical replicates is shown. Temporal data values shown are derived from independent patient datasets and normalized with t=0 value measured in b (See Supplementary Fig. 3 for additional examples). (e) Affinity propagation-derived clusters for pTyr sites detected in patient 27 (See Supplementary Fig. 4 for affinity propagation clusters of other patients). Subplots signify groups of pTyr sites with similar quantitative trends across time (x-axis). Solid lines denote trends of individual pTyr sites. Mean log2-temporal values relative to t=0 min are shown. Exemplar pTyr site of each cluster shown in blue. (f) Ischemia-regulated phosphorylation sites common to all patients with functional categorization. Ischemia index (y-axis) for each site represents absolute sum of the log2 changes relative to t=0 at5, 30, 60 minutes. Mean value ± SEM from all patients is plotted. Asterisks denote sites known to be implicated in protein activity or function.
To identify systemic response to ischemia, affinity propagation analysis (23) was used to classify distinct clusters of pTyr sites with comparable temporal trends. Interestingly, several clusters behaved in a similar manner across patients, suggesting a set of co-regulated signaling responses to ischemia (Supplementary Fig. 4 and Supplemental Table 3). One cluster characterized by rapid increase in phosphorylation within 5 minutes of cold-ischemia included MAPK12, MAPK13, MAPK14, SHC1, GAB1, and SHIP (Fig. 1e, left). These acute responders to ischemia agree with the known roles of these proteins in stress responses and MAPK signaling (27-30). A separate cluster of pTyr sites whose phosphorylation decreased with increasing ischemia time included ephrin receptors, tensins, and plakophilins. These phosphoproteins are suggestive of modulation of cell adhesion, cytoskeletal, and migration related processes, all consistent with perfusion stress (Fig. 1e, right)(31-33). Other clusters include pTyr sites with minimal quantitative fluctuations, as well as sites with bi-directional regulation during the course of 60 minute cold-ischemia (Fig. 1e, middle). A‘core’ ischemia-regulated phosphorylation signature containing twenty-two phosphorylation sites was detected across all ovarian tumor specimens. We devised a quantitative ‘ischemia-index’ to compare the relative severity of ischemia response among this consensus set. This signature represents four broad functional categories: adhesion and migration, inflammatory response, proliferation, and stress response (Fig. 1f). Given the primary importance of these pathways in tumor biology, delayed-freezing artifacts will likely create erroneous biological interpretations if tumor specimens intended for pTyr analysis are not immediately frozen.
Spatial phosphorylation heterogeneity is apparent in human tumor samples
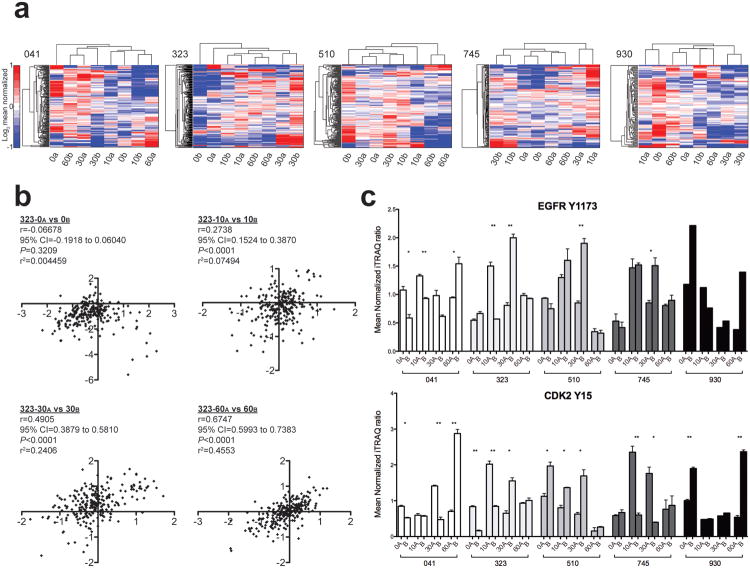
The heterogeneous nature of human tumors has been described (15-18). However, the effects of inter-tumor spatial heterogeneity on cellular signaling networks are unknown. To understand the impact of spatially resolved samples on protein phosphorylation profiles, we analyzed the pTyr profiles of paired spatially distinct primary colorectal tumor samples from five individuals undergoing resection (Supplementary Fig. 5, and Supplementary Table 4). As with the ovarian tumors, a controlled ischemia time-course enabled analysis of pTyr dynamics and the generality of ischemia effects across tumors. Unsupervised hierarchical clustering of intratumoral paired-timepoint samples indicated the presence of pTyr heterogeneity in spatially proximal samples (Fig. 2a), as indicated by the greater difference in the paired time-point samples (i.e., 0a and 0b) compared to the non-paired points (i.e., 0b and 10a in patient 041). In patients 323 and 745, where the 0 min paired-timepoints clustered together, significant differences were still apparent. Technical measurement error did not contribute significantly to the observed variation, and was not the cause of the observed heterogeneity (Supplementary Fig. 6).Pearson correlation analysis of the quantitative pTyr data for each pair of time points (Supplementary Fig. 7) indicated weak correlations and minimal shared covariance between spatially proximal samples, even in tumor 323, the most ‘homogeneous’ tumor, where paired samples appear to cluster (Fig. 2b). Spatial heterogeneity has the potential to impact clinical decisions, as clinically diagnostic pTyr markers were statistically different in proximal regions at resection (Fig. 2c, 0 min pairs), in some cases by more than 2-fold. Freezing delays can further alter phosphorylation heterogeneity, as paired timepoints often provided different signaling profiles (Fig. 2c; 10, 30, 60 min pairs).
Figure 2.
Spatial phosphorylation heterogeneity is apparent in human colon tumors. (a) Separate unsupervised hierarchical-clustered heatmaps represent the spatial and temporal sample set of each patient-derived colon tumor. Patient identifier shown to top left of each heatmap. Rows indicate mass spectrometry-derived quantitative levels of pTyr sites in log2 mean normalized scale. Onset of saturated color in the heatmaps corresponds to changes ≥2-fold from the mean (red, increasing; blue, decreasing). (b) Pearson correlation analysis was used to quantify the direction and magnitude of correlation among the spatially distinct colorectal tumor samples. Data points presented in each plot are the log2mean-centered value for a given pTyr site between paired spatially-distinct samples at the indicated timepoints in patient 323. r=Pearson correlation coefficient; 95% CI= 95% confidence interval of r; P=two-tailed p-value. See Supplementary Fig. 7 for correlation analysis of other patients. (c) Phosphorylation levels of clinically useful pTyr sites are plotted for each patient spatial and temporal sample set. Samples are shaded according to patient and columns grouped to indicate identical timepoint samples that are spatially distinct. Mean value ± SEM is plotted. Statistically significant differences are indicated. Note: ** indicates P=0.001 to 0.01, and * indicates P=0.01 to 0.05.
Spatial and temporal protein expression differences are minimal in tumor samples
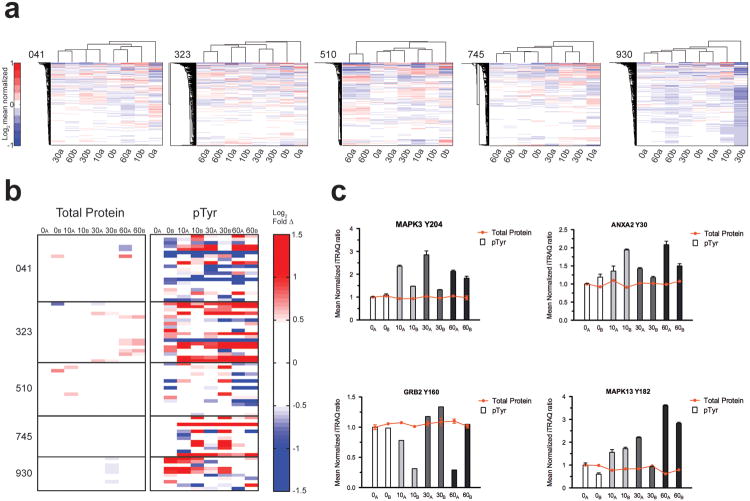
To evaluate whether intra-tumor protein expression heterogeneity contributed to the observed spatial and temporal pTyr differences, we performed protein expression analysis of the colorectal specimens (Supplementary Fig. 8, Supplementary Table 5). Unsupervised hierarchical clustering indicated that protein expression is mostly uniform temporally and spatially (Fig. 3a).Where quantitative protein expression differences were observed between paired timepoints, they were muted compared to the large differences identified in pTyr profiles. To systematically evaluate the relationship between protein expression heterogeneity and pTyr levels, we identified 76 pTyr sites where corresponding protein levels were also available (Fig. 3b, Supplementary Table 6). Whereas protein expression was invariant with both spatial location and ischemia, pTyr levels showed dramatic spatial and ischemic time-dependent variation. These results suggest that differential ischemic responses could occur in adjacent parts of a tumor. For example, selected pTyr sites in patient 041 exhibited spatially distinct changes during ischemia, despite uniform expression of the corresponding proteins (Fig. 3c).
Figure 3.
Spatial and temporal protein expression differences are minimal in colorectal tumors. (a)Separate unsupervised hierarchical-clustered heatmaps represent the spatial and temporal sample set of each patient-derived colon tumor. Patient identifier shown to top left of each heatmap. Rows indicate mass spectrometry-derived quantitative levels of protein expression in log2 mean normalized scale. Onset of saturated color in the heatmaps corresponds to changes ≥2-fold from the mean (red, increasing; blue, decreasing). (b) Quantitative levels of protein (from a) and matching pTyr sites (from 2a) on the same protein. Rows represent values in log2 scale relative to t=0a min. Onset of color in the heatmaps represents changes greater than ∼1.4-fold from the mean (red, increasing; blue, decreasing) to emphasize uniformity in protein expression values compared to pTyr values. Plotted pTyr values are also quantitatively normalized to the protein expression levels. See Supplementary Table 6 for details. (c) Selected examples of corresponding protein and pTyr levels from b. Columns are shaded to group identical timepoint samples that are spatially distinct. Mean value ± SEM is plotted.
Ischemia alterations occur across tumor types impacting a core set of functional classes yet has the potential to affect biologically diverse pTyr pathways
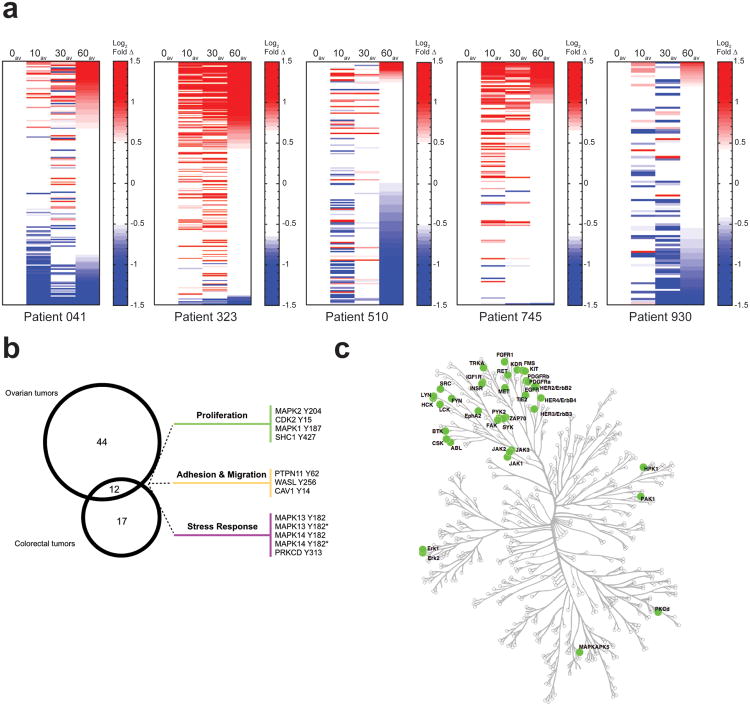
To reduce the impact of spatial heterogeneity on phosphorylation and identify consistent ischemia-related changes, we averaged the pTyr measurements from paired-timepoint samples. As with the ovarian tumors, a large proportion of pTyr sites showed significant quantitative changes following delayed freezing (Fig. 4a, Supplementary Table 4). These results demonstrate that delayed-freezing effects can alter tyrosine phosphorylation in multiple tumor types. As with the ovarian cancer samples, inter-patient differences were observed for temporal dynamics (Supplementary Table 4). From these data, we extracted a minimal ischemia-regulated signature of 12 pTyr sites common to both ovarian and colorectal samples; adhesion and migration, proliferation, and stress response pathways are represented within this signature (Fig. 4b). Due to the inter-patient biological variation, many additional specimen-specific ischemia-driven pTyr perturbations were documented. Accordingly, when all tumor variation is considered it becomes apparent that extensive ischemia-dependent alterations affect most tyrosine kinases (Fig. 4c, Supplemental Table 7). The pathways impacted by ischemia encompass diverse kinases and downstream effectors across broad biological functions and processes (Supplementary Fig. 9). Therefore, ischemia, particularly of unknown duration, may distort pTyr network profiles to an extent that cannot be reliably corrected.
Figure 4.
Ischemia alterations occur across tumor types impacting a core set of functional classes yet has the potential to affect biologically diverse pTyr pathways. (a) Heatmaps of individual patient-derived colorectal tumor 4-timepoint ischemia study. Rows indicate mass spectrometry-derived quantitative levels of pTyr sites in log2 scale relative to t=0 min, rank ordered by value at t=60av min. Average value of paired timepoints is plotted. Onset of color in the heatmaps corresponds to changes ≥3 s.d. from value at t=0av (red, increasing; blue, decreasing). See Supplementary Table 4 for details. (b) Venn diagram indicating ischemia-phosphorylation signature observed in multiple tumors. 56 ischemia-regulated sites were detected in ≥4 ovarian tumor samples and 29 ischemia-regulated sites were detected in ≥4 colorectal tumor samples. (c) Kinase enrichment analysis (from union of data in b) was performed to identify kinases with high-confidence enrichment and plotted onto a map of the human kinome using Kinome Cluster to highlight the diverse set of kinases affected by delayed freezing. Green circle=kinases significantly altered in ≥4 colon tumors and/or ≥4 ovarian tumors.
Discussion
There is a strong impetus to comprehensively integrate genetic profiles of tumors with corresponding proteomic expression and protein phosphorylation datasets (9).The salient findings of this study indicate that measurement of pTyr signaling nodes in human tumors is i) susceptible to extensive post-resection ischemia effects creating rapid and systemic changes that alter the initial in situ tumor phosphorylation profile, and ii) distinct intra-tumor phosphorylation profiles are apparent indicating spatial micro-heterogeneity, and presumably signaling differences, within specimens. Our findings suggest that the tumor specimens are actively regulating signaling events despite their loss of blood supply and attachment to the surrounding tissue. Hypoxia, hypoglycemia, acidosis, hypothermia, and osmotic disturbances are perturbations to which the tumor acutely responds until cryopreservation. The immediate implication is that insight regarding tumor pTyr signaling in human tumor specimens may be greatly misleading or incorrect depending on the nature and duration of tumor harvesting/processing before analysis. Unfortunately, retrospective extrapolation of an accurate in vivo phosphorylation state appears to be prohibitively difficult due to unique dynamics on each pTyr site coupled with unpredictable patient specific responses to ischemia (Fig. 1c, d and Supplementary Fig. 3).
The temporal trends and directionality of pTyr fluctuation observed are consistent with the physiological effects of tumor resection and a step-wise signaling response. In fact, the pTyr sites within the identified temporal clusters appear to correlate with progressive stages of ischemic stress (Fig. 1e, Supplementary Fig. 4, and Supplementary Table 3). For example, physical stresses from wounding, hypoxia and osmotic shock result in the immediate activation of response pathways to promote tissue repair and regeneration. This activity is consistent with the observed rapid hyperphoshorylation of p38 MAPKs (ie. MAPK12, MAPK13, MAPK14) through oxidative stress-sensing ASK1 and osmosensing OSM (34,35). Activation of these pathways can in turn trigger signaling programs necessary for the production of pro-inflammatory cytokines and tissue repair (36). This immediate signaling cluster also included increased phosphorylation of mitogenic nodes on SHC1, GAB1, and MAPK1 and MAPK3 sites to potentially initiate proliferative regeneration. Sustained ischemia times were accompanied by more gradual cellular responses. Increased phosphorylation of PRKCD, a known substrate of a caspase-3 during apoptosis, was observed across all ovarian tumors. Lack of perfusion leads to cellular dehydration, shrinkage and distortion. Accordingly, phosphorylation decreases on EPHA2, EPHA4, EPHB2, PARD3, PKP4, and TNS3 agree with physiologic changes in loss of cell-cell and ECM contacts (31-33). Collectively, the directionality and timing of several phosphorylation sites are consistent with the physiologic stages of severe ischemia.
Of note, the core pTyr ischemia signature described overlaps with functional pathways known to be relevant in the signaling of cancer cells such as adhesion, migration, and proliferation (37,38). Importantly, in many cases the pTyr sites annotated are directly implicated in regulating protein function—for example 17 of 22 pTyr sites in the core ischemia signature (Fig. 1f) have been shown experimentally to directly modulate protein kinase activity and function. As such, these are not uncharacterized phosphorylation events but rather functionally relevant pTyr sites with probable signaling consequences. Moreover, while phosphorylation changes of greater than two-fold were detected during the ischemia timecourse (20-28% and 25-48% of peptides in ovarian and colon tumors respectively; Supplemental Table 2 and 4) and often used to prioritize biologically significant signaling changes, the magnitude of change does not always correlate with signaling significance. In fact, modest changes to phosphorylation levels may correspond to meaningful biological results as demonstrated in the context of MEK, K-RasG12D, or EGFRvIII, where slight changes to the signaling activity have profound effects on viability and oncogenicity (39-41). As such, the subset of pTyr sites with seemingly insignificant variations may still in fact push signaling networks away from a fine-tuned steady state (42).
It is import to emphasize that the extensive list of ischemia-regulated pTyr sites in ovarian and colon tumors (Fig. 4c and Supplementary Fig. 9) spans a multitude of pathways and processes and are not limited to obvious stress response pathway proteins (ie. p38 MAPKs). While it is tempting to speculate that tumors with strong driver signaling (ie. HER2 overexpression) could still generate pTyr signatures which overshadow ischemia induced signatures, this is conceptually unlikely for at least two reasons. First, oncogenic signaling mutations do not always exhibit enhanced levels of phosphorylation—but can instead present persistent, minor increases to achieve oncogenic network states(40,41). Secondly, strong pTyr signaling would likely impinge on the nodes and pathways affected by ischemia (ie. proliferation, migration, adhesion, etc.) preventing definite attribution of the source. Thus, it will not be feasible for future pTyr experiments to simply exclude a set of ‘ischemia susceptible’ proteins and derive a quantitatively reliable dataset since the boundaries of susceptible and stable pTyr sites are not entirely clear, appear to be patient specific and cannot be predicted a priori based on our current knowledge.
The use of surgically excised tumors in this study was paramount to recapitulate a typical biospecimen collection scenario, however an unexpected caveat of this analysis was the realization that pTyr tumor heterogeneity exists even on a relatively proximal scale. Spatial heterogeneity of tyrosine phosphorylation may have significant impact on clinical decisions. As alluded to in Figure 2c, monitoring of therapeutic efficacy in pre-clinical or clinical trials is often examined by measuring phosphorylation levels on kinases and other signaling targets. However, using pTyr as a diagnostic proxy in human specimens could pose challenging, as inadequate assessment of clinically relevant pTyr sites is possible depending on the extent of tumor spatial heterogeneity and breadth of sampling. Ideally, this variation should be acknowledged and accounted for in current and future studies to allow appropriate interpretation of pTyr levels in human tumor specimens (e.g. multiple, spatially distinct pre- and post-therapy biopsies when evaluating drug efficacy.) A concurrent CPTAC study of the ischemia-driven effects on serine/threonine phosphorylation in tumor samples revealed complementary insights (21). Although examination of the same ovarian tumor specimens used here suggested that serine/threonine phosphorylation might be less susceptible to ischemia, perturbations were observed in approximately 6% of the ∼9,000 pSer/pThr sites measured (based on those sites overlapping in at least 3 samples), with the majority of these sites concentrated in stress signaling pathways. This result is in contrast to this study, where perturbations were observed in 62% of 217 sites measured in ovarian tumors (based on those sites overlapping in at least 3 samples) and ∼44% of 57 sites measured across both ovarian and colon tumors (overlap in at least 3 samples). These differences are likely attributable to the highly regulated nature of pTyr sites compared to pSer/pThr sites, and may be indicative of the biological relevance of the regulated phosphorylation sites measured in both studies.
In summary, immediate freezing of human tumor specimens is necessary to minimize post-resection artifacts that could confound identification of physiological pTyr signaling networks. Intra-tumoral phosphorylation heterogeneity suggests that performing single biopsies of primary tumors or metastases, as is usual clinical practice, may provide erroneous or incomplete profiles of signaling systems in tumors. These data suggest that multiple biopsies, immediately flash-frozen, may be necessary to accurately assess the signaling characteristics of human tumors.
Supplementary Material
Acknowledgments
We thank Henry Rodriguez, Christopher Kinsinger, and Robert Rivers at the NCI as well as members of the White lab for their helpful comments and critical review of this manuscript.
Grant Support: This work was supported by NIH grant U24 CA159988.
Financial Support: This work was supported by NIH grant U24 CA159988.
Footnotes
Conflicts of Interest: The authors disclose no potential conflicts of interest.
Author Contributions: A.S.G, H.J., D.C.L. and F.M.W. designed the research, performed experiments, analyzed data and wrote the manuscript; R.J.C.S., K.S., K.W., M.K.W., A.J.H., D.A.L. and D.C.L. provided tumor specimens.
References
- 1.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–36. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 2.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 3.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–72. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77:1311–5. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Futreal PA, Kasprzyk A, Birney E, Mullikin JC, Wooster R, Stratton MR. Cancer and genomics. Nature. 2001;409:850–2. doi: 10.1038/35057046. [DOI] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, et al. Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 2013;3:1108–12. doi: 10.1158/2159-8290.CD-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends Mol Med. 2012;18:43–51. doi: 10.1016/j.molmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Johnson H, White FM. Seminars in Cell and Developmental Biology. Elsevier Ltd; 2012. Toward quantitative phosphotyrosine profiling in vivo; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altelaar AFM, Munoz J, Heck AJR. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 13.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Molecular & Cellular Proteomics. 2008;7:1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giindisch S, Grundner-Culemann K, Wolff C, Schott C, Reischauer B, Machatti M, et al. Delayed times to tissue fixation result in unpredictable global phosphoproteome changes. J Proteome Res. 2013;12:4424–34. doi: 10.1021/pr400451z. [DOI] [PubMed] [Google Scholar]
- 15.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–5. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 16.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 17.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 19.Drake JM, Graham NA, Lee JK, Stoyanova T, Faltermeier CM, Sud S, et al. Metastatic castration-resistant prostate cancer reveals intrapatient similarity and interpatient heterogeneity of therapeutic kinase targets. Proc Natl Acad Sci USA. 2013;110:E4762–9. doi: 10.1073/pnas.1319948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker AF, Dragovich T, Ihle NT, Williams R, Fenoglio-Preiser C, Powis G. Stability of phosphoprotein as a biological marker of tumor signaling. Clin Cancer Res. 2005;11:4338–40. doi: 10.1158/1078-0432.CCR-05-0422. [DOI] [PubMed] [Google Scholar]
- 21.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Molecular & Cellular Proteomics. 2014 doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson H, del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Molecular & Cellular Proteomics. 2012;11:1724–40. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–6. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 24.Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–6. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 26.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–17. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 28.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, et al. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 29.Holgado-Madruga M, Emlet DR, Moscatello DK, Godwin AK, Wong AJ. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–4. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 30.Kavanaugh WM, Pot DA, Chin SM, Deuter-Reinhard M, Jefferson AB, Norris FA, et al. Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr Biol. 1996;6:438–45. doi: 10.1016/s0960-9822(02)00511-0. [DOI] [PubMed] [Google Scholar]
- 31.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui Y, Liao YC, Lo SH. Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3. Mol Cancer Res. 2004;2:225–32. [PubMed] [Google Scholar]
- 33.Bass-Zubek AE, Godsel LM, Delmar M, Green KJ. Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr Opin Cell Biol. 2009;21:708–16. doi: 10.1016/j.ceb.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsukawa J, Matsuzawa A, Takeda K, Ichijo H. The ASK 1-MAP kinase cascades in mammalian stress response. J Biochem. 2004;136:261–5. doi: 10.1093/jb/mvh134. [DOI] [PubMed] [Google Scholar]
- 35.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5:1104–10. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 36.Moens U, Kostenko S, Sveinbjørnsson B. The Role of Mitogen-Activated Protein Kinase-Activated Protein Kinases (MAPKAPKs) in Inflammation. Genes (Basel) 2013;4:101–33. doi: 10.3390/genes4020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Huang PH, Miraldi ER, Xu AM, Kundukulam VA, del Rosario AM, Flynn RA, et al. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol Biosyst. 2010;6:1227–37. doi: 10.1039/c001196g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman IN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–35. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 42.Creixell P, Schoof EM, Erler JT, Linding R. Navigating cancer network attractors for tumor-specific therapy. Nat Biotechnol. 2012;30:842–8. doi: 10.1038/nbt.2345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.