Abstract
Determination of variant pathogenicity represents a major challenge in the era of high-throughput sequencing. Erroneous categorization may result if variants affect genes that are in fact dispensable. We demonstrate that this also applies to rare, apparently unambiguous truncating mutations of an established disease gene. By whole-exome sequencing (WES) in a consanguineous family with congenital non-syndromic deafness, we unexpectedly identified a homozygous nonsense variant, p.Arg1066*, in AHI1, a gene associated with Joubert syndrome (JBTS), a severe recessive ciliopathy. None of four homozygotes expressed any signs of JBTS, and one of them had normal hearing, which also ruled out p.Arg1066* as the cause of deafness. Homozygosity mapping and WES in the only other reported JBTS family with a homozygous C-terminal truncation (p.Trp1088Leufs*16) confirmed AHI1 as disease gene, but based on a more N-terminal missense mutation impairing WD40-repeat formation. Morpholinos against N-terminal zebrafish Ahi1, orthologous to where human mutations cluster, produced a ciliopathy, but targeting near human p.Arg1066 and p.Trp1088 did not. Most AHI1 mutations in JBTS patients result in truncated protein lacking WD40-repeats and the SH3 domain; disease was hitherto attributed to loss of these protein interaction modules. Our findings indicate that normal development does not require the C-terminal SH3 domain. This has far-reaching implications, considering that variants like p.Glu984* identified by preconception screening (‘Kingsmore panel’) do not necessarily indicate JBTS carriership. Genomes of individuals with consanguineous background are enriched for homozygous variants that may unmask dispensable regions of disease genes and unrecognized false positives in diagnostic large-scale sequencing and preconception carrier screening.
Introduction
Large-scale sequencing projects have shown that individuals carry ∼100 putative loss-of-function (LoF) alleles. LoF alleles fall into a range of categories, from mutations in non-essential genes to truly deleterious mutations in established recessive disease-causing genes (1). Homozygous LoF variants that do not elicit overt clinical consequences are strong indicators for LoF tolerance (and thus non-essentiality) of the affected genes. Biallelic and, in particular, truncating mutations of genes for severe congenital recessive disorders in healthy individuals are therefore very unusual. Such a finding should prompt reassessment of the functional requirement of the affected gene region or transcript.
AHI1 (Abelson-helper integration site 1; MIM 608894) is such a gene, associated with a severe recessive congenital disorder, Joubert syndrome type 3 (JBTS3; MIM 608629). JBTS3 is characterized by psychomotor delay, cerebellar hypoplasia, consecutive ataxia and an altered respiratory pattern during neonatal development. Retinal degeneration is frequently found. Renal disease, liver defects or skeletal involvement, symptoms that may be present in other genetic subtypes of JBTS, are not typical for JBTS3. The hallmark of JBTS is a midbrain–hindbrain malformation, named ‘molar tooth sign’ (MTS) because of its appearance on axial magnetic resonance imaging.
Here, we report that two homozygous truncating variants in AHI1 are non-penetrant in humans. A zebrafish model also supports non-penetrance of AHI1 LoF variations in the respective gene region. In contrast to obvious non-pathogenic LoFs or gene losses that provide selective advantages, both AHI1 variants are extremely rare in the general population. Our findings draw attention to an underestimated category of major pitfalls in the interpretation of highly parallel sequencing for diagnostics in patients and for preconception carrier screening. Large-scale sequencing data from consanguineous families provide a rich but still largely untapped resource that can be used to minimize erroneous categorization of variants: seemingly unambiguous pathogenic variants are unmasked as benign when they are repeatedly found in healthy probands or patients with unrelated conditions.
Results
Homozygosity mapping and whole-exome sequencing
Family 1
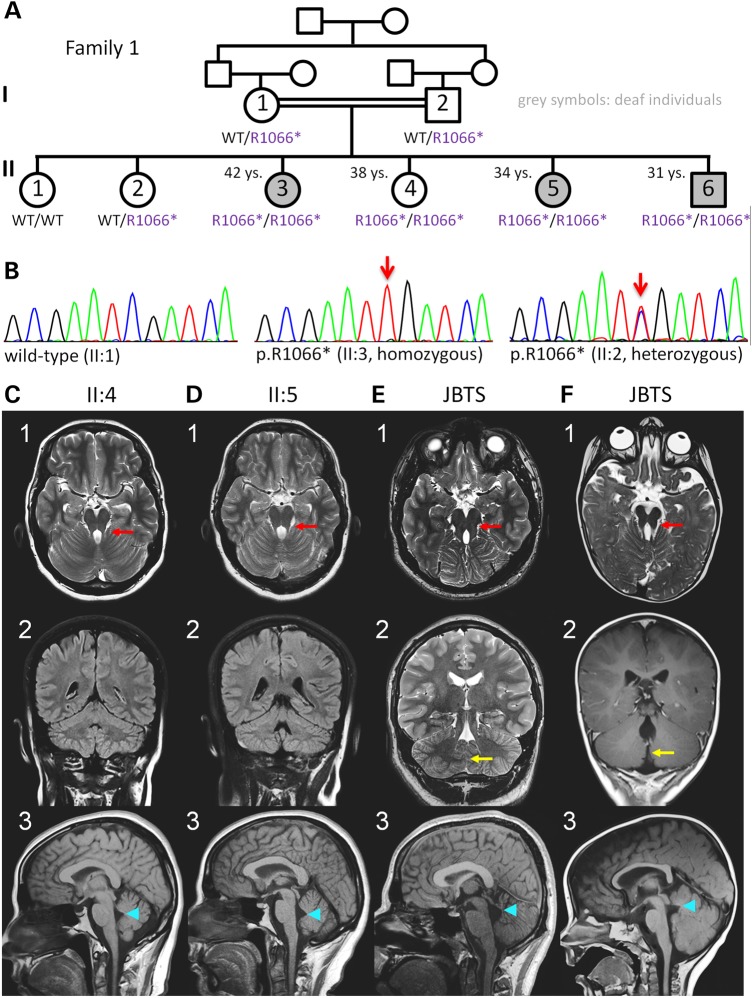
Homozygosity mapping to identify the gene underlying congenital non-syndromic deafness in this consanguineous Palestinian family (Fig. 1A) identified three regions with homozygosity by descent (HBD) in the three affected siblings: A 7.9 Mb interval on chromosome 6q24.1–q24.3, a 14.5 Mb interval on chromosome 7q21.3–q31.1 and a 22.2 Mb region on chromosome 11 (Supplementary Material, Fig. S1A). Sanger sequencing of known deafness genes within these intervals, SLC26A4 and SLC26A5 (both on chromosome 7), RDX and TECTA (both on chromosome 11), and of several functional candidate genes did not reveal any mutations. Subsequent whole-exome sequencing (WES) of one index patient (II:3) revealed homozygous missense variants in three genes located in the mapped HBD regions, namely PTCD1 and MUC17 (both on chromosome 7), and DDX10 (chromosome 11). Segregation analysis excluded causality of the PTCD1 variant, which was homozygous not only in the three hearing-impaired siblings, but also in a healthy sister (II:4). The variants in MUC17 and DDX10 have both been annotated as rare single-nucleotide polymorphisms (SNPs) (rs74974199 and rs138955971, respectively). Because HBD intervals can escape detection when the interval is very small and based on only very few homozygous SNPs, we filtered the entire exome for homozygous mutations irrespective of their chromosomal localization. Although this did not reveal a strong candidate gene for sensory deafness, it exposed a homozygous nonsense mutation, p.Arg1066* (c.3196C>T), in exon 25 of AHI1. Segregation analysis of the p.Arg1066* variant showed that it was heterozygous in both parents and homozygous in the three deaf siblings (II:3, II:5, II:6) and a healthy sister (II:4) (Fig. 1A). These results were confirmed by re-drawn blood samples of Family 1 and by using an alternative primer pair. In II:4, the AHI1 gene is located within a 37.5 Mb region of homozygous SNPs (shared by the three deaf individuals II:3, II:5 and II:6; Supplementary Material, Fig. S2), compatible with true homozygosity of p.Arg1066*AHI1. Furthermore, qPCR with primers amplifying the binding regions of the primers used for PCR and Sanger sequencing did not indicate any deletion of those binding sites that could have led to allelic dropout in PCR amplification (data not shown). The variant was not present in the ESP and the TGP databases, or our extended in-house database of 1629 exomes (regularly updated database comprising not all, but only pre-filtered, rare variants of these exomes), but it was annotated once in heterozygous state (total MAF = 8.6E−6; African MAF = 1.1E−5) in the ExAC database.
Figure 1.
Family 1 with autosomal recessive non-syndromic deafness and a non-manifesting homozygous AHI1 nonsense variant. (A) Pedigree (grey symbols: deaf individuals). Whole-exome sequencing of a sample from the index patient, II:3, did not identify a mutation either in any known deafness gene or in any strong candidate gene in any of the mapped HBD regions shared by II:3, II:5 and II:6. Four siblings were homozygous for the AHI1 nonsense variant p.Arg1066* (in purple = considered non-pathogenic). For clarity, the one-letter code (R1066*) was applied for variant designation. (B) Electropherograms depicting wild type (left), homozygosity (middle) and heterozygosity (right) for p.Arg1066*. (C–F) Cranial MRI of individuals II:4 (at the age of 34 years) and II:5 (at the age of 32 years) in comparison to MRI of two children with JBTS. (C1–F1) Axial sections at the level of the mesencephalon/superior cerebellar peduncles (scp): normal dimensions and orientation of the scp in C1 and D1, ‘molar tooth sign’ with thickened and elongated scp in E1 and F1 (red arrows). (C2–F2) Coronal sections, showing normal appearance of intervening vermis between the cerebellar hemispheres in C2 and D2, but ‘vermal cleft’ due to partial absence of the vermis in E2 and F2 (yellow arrows). (C3–F3) Mid-sagittal views illustrate normal brain stem morphology and normal tent-like triangular shape of the fourth ventricle in C3 and D3; the ‘tip’ of the fourth ventricle roof (fastigium) is at the level of mid-pons. In E3 and F3, the shape of the fourth ventricle is distorted, with rostral displacement of the fastigium, now at the level of mesencephalic-pontine boundary (blue arrowheads point to fastigium).
Family 2
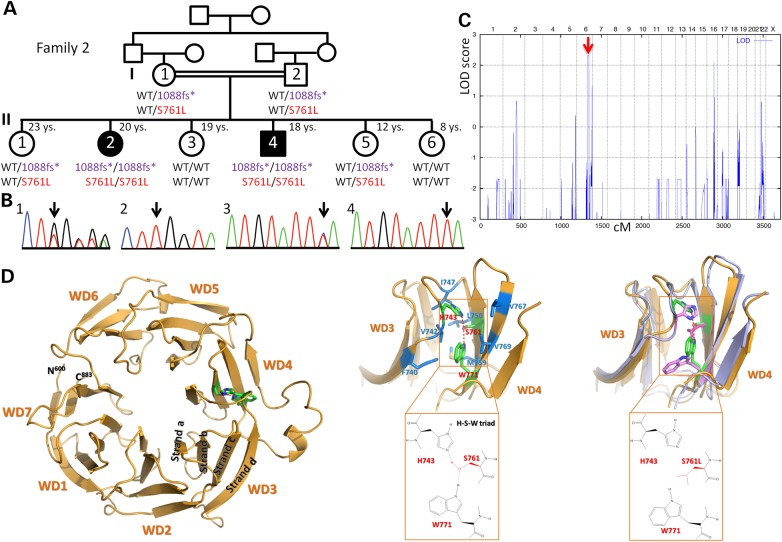
Targeted Sanger sequencing of AHI1 in a cohort with clinically proven JBTS had previously identified the homozygous frameshift AHI1 variant, c.3263_3264delGG (p.Trp1088Leufs*16) in both affected siblings (II:2 and II:4) from this consanguineous Egyptian family (Fig. 2A and B). In the original publication, this AHI1 variant was considered to be causative (2). We now carried out genome-wide homozygosity mapping including additional samples from four healthy sisters and the parents (first cousins), which identified a single HBD region with a combined maximum parametric LOD score of 2.31 in both affected siblings, on chromosome 6q23.2–q24.3 (Fig. 2C). This region contains ∼90 genes, including AHI1. WES of II:2 did not identify homozygous candidate variants in any other gene from that locus, but it revealed the presence of another homozygous AHI1 variant, p.Ser761Leu (Fig. 2A and B). As can be expected from linkage data, both AHI1 variants co-segregate with JBTS in Family 2. Neither p.Trp1088Leufs*16 nor p.Ser761Leu was present in the ESP, the TGP or the ExAC database, or in 1629 in-house exomes. However, p.Trp1088Leufs*16 has been documented in dbSNP (rs387906269), but no MAF is available.
Figure 2.
Family 2 with JBTS and two homozygous AHI1 variants. (A) Pedigree (black symbols: JBTS). The sample of II:2 was subjected to WES. A deceased sister and a healthy brother (whose samples were not available for genotyping) are not shown due to space constraints. For clarity, abbreviated codes (1088fs*, S761L) were used for the AHI1 variants p.Trp1088Leufs*16 and p.Ser761Leu. Both homozygous variants co-segregate with the JBTS phenotype. S761L is considered to be the disease-causing mutation and therefore depicted in red. The 1088fs* variant is considered non-pathogenic and therefore depicted in purple. (B) Electropherograms depicting heterozygosity (1, I:1) and homozygosity (2, II:2) for 1088fs*, as well as heterozygosity (3, I:1) and homozygosity (4, II:2) for S761L. (C) Genome-wide homozygosity mapping implicating the parents and all siblings shown in (A) identified a single HBD region with a combined maximum parametric LOD score of 2.31 in both affected siblings, for a region on chromosome 6q23.2–q24.3 that contains AHI1. (D) Left: Structure of AHI1 protein as predicted by the WDSP algorithm. Middle: More detailed structure of the WD3- and WD4-repeats (with some loops deleted for clarity) that show the hydrogen bond network formed by residues H743-S761-W771 (red/green) and its hydrophobic environment (corresponding residues in blue). Right: The superposition of simulated WD3 and WD4 structures for S761 (wild type, green) and S761L (mutant, pink) shows the overall conformational changes caused by the S761L mutation.
Although compound heterozygous mutations are less likely to be the cause of JBTS in Family 2, we also filtered for genes carrying at least two rare variants in II:2. This revealed heterozygous missense variants in two other JBTS genes, CSPP1 (c.3281A>G/p.Glu1094Gly and c.3478C>T/p.Pro1160Ser) and CEP164 (c.100C>T/p.Arg34Trp and c.1480C>A/p.Pro494Thr). Haplotypes from genome-wide SNP genotyping were not compatible with a causative role of compound heterozygous CSPP1 mutations. For the CEP164 locus, haplotypes would be compatible with JBTS resulting from compound heterozygous mutations in this gene (data not shown). Because both CEP164 variants are categorized as benign polymorphisms by MutationTaster (3), and p.Pro494Thr represents an annotated SNP with a documented global MAF of 0.006 in the general population, CEP164 is very unlikely to be the causative gene for JBTS in Family 2.
Evolutionary conservation and prediction of WD40 structure in wild-type and in mutant AHI1 protein
AHI1 has seven WD40-repeats (Supplementary Material, Table S1). p.Ser761Leu in Family 2 affects an evolutionarily highly conserved residue of AHI1 in the 4th WD40-repeat (WD4) (Supplementary Material, Fig. S3A). In contrast to wild-type AHI1 (Supplementary Material, Fig. S3B), ScanProsite (4) fails to predict the WD-repeat in AHI1p.Ser761Leu (Supplementary Material, Fig. S3C). We therefore modeled the structure of AHI1 (Fig. 2D) as recently described for POC1B, another WD40 protein that we found to be mutated in a family with severe retinal dystrophy and JBTS (5). The p.Ser761 residue is predicted to form a hydrogen bond network with p.His743 and p.Trp771 (Fig. 2D) (6), surrounded by hydrophobic residues p.Phe740, p.Val742, p.Ile747, p.Leu750, p.Met759, p.Val767 and p.Val769. Due to this hydrophobic environment, the hydrogen bonds are very strong. Our earlier studies suggest that each hydrogen bond may cause ∼15 kJ/mol stabilization (7). The p.Ser761Leu mutation is predicted to eliminate the two hydrogen bonds, causing a significant destabilization of the protein. To study the effect of the p.Ser761Leu mutation on structure, we performed preliminary molecular dynamics simulations of wild-type and mutant AHI1. The initial stereo-structures were generated by the WDSP. The simulations were carried out using our recently developed protein force field RSFF1 (see Methods for details). Because of the steric effect of p.Leu761, both p.His743 and p.Trp771 are rotated (Fig. 2D, right). Conformational changes are found in the Loop_da that connect WD3 and WD4 and in the long Loop_cd of WD4 (Supplementary Material, Table S1). We postulate that the structural changes caused by p.Ser761Leu affect the interaction of AHI1 with its binding partners. Dasen et al. (8) found that the p.Ser715Pro mutation of the TLE1 protein abolished its interaction with Hesx1.5. p.Ser715 is part of the TLE1 hydrogen bond network p.Asp719-p.His691-p.Ser715-p.Trp725 (Supplementary Material, Table S2). As predicted for the AHI1 mutation p.Ser761Leu, the TLE1 mutation also disrupts the hydrogen bond network. Because p.Asp-p.His-p.Ser/p.Thr-p.Trp and p.His-p.Ser/p.Thr-p.Trp hydrogen bonds are widely present in WD40 proteins, we expect that mutations of these residues, especially His and Ser/Thr, are deleterious.
Clinical investigations
Family 1
None of the four homozygous AHI1 mutation carriers in this non-syndromic deafness family presented any symptoms or any medical history compatible with JBTS. Cranial MRI was available from II:4 and II:5 and ruled out an MTS (Fig. 1C and D). Detailed ophthalmological investigation of II:4 showed no retinal abnormalities, and abdominal ultrasound failed to detect renal abnormalities.
Family 2
Two siblings from this family have JBTS with retinitis pigmentosa (RP), compatible with JBTS3, the subtype caused by AHI1 mutations (9), and have been reported previously (2).
AHI1 protein analysis
Western blotting with two antibodies against the N-terminal half of AHI1 showed three isoforms in fibroblast lysates from II:4, a homozygous carrier of the AHI1 variant of Family 1 (p.Arg1066*), and from a wild-type control proband (Supplementary Material, Fig. S1B). The observed bands correspond to isoform 2 (ca. 120 kDa), to isoform 3 (ca. 65 kDa) and to a fourth isoform (ca. 50 kDa) of AHI1 (10).
Zebrafish model of human mutations
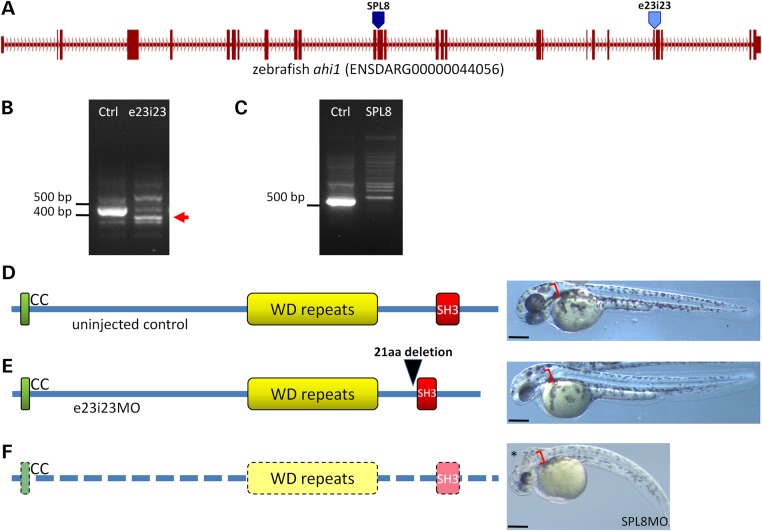
We used zebrafish to investigate the pathogenicity of disruptions in the 3′ region of zebrafish ahi1, corresponding to the locations of the truncating human variants p.Arg1066* and p.Trp1088Leufs*16. Ahi1 protein structure is conserved between zebrafish and human, so we designed a morpholino, e23i23, to alter splicing of a 3′ exon in the region of the gene that encodes the SH3 domain (Fig. 3A). RT–PCR results show an in-frame deletion that abolishes a large portion of the SH3 domain and a concomitant depletion in the normal transcript (Fig. 3B and E). When we analyzed embryos injected with this morpholino for any resulting phenotype, we found that body axis, brain and ear morphologies were comparable to those of uninjected controls (Fig. 3D and E). As a positive control, we injected a previously reported splice-blocking morpholino (11), SPL8, that targets a more 5′ region of the ahi1 transcript (Fig. 3A). In our hands, this morpholino resulted in nearly total knockdown of the normal transcript and we observed strong ciliopathy defects consistent with the previous study (Fig. 3F).
Figure 3.
Zebrafish studies. (A) The predominant ahi1 transcript in zebrafish embryos consists of 27 exons. The position of the previously published splice-blocking MO, SPL8, is indicated with a dark blue arrowhead at the splice acceptor site of exon 13. The e23i23 morpholino used in this study is indicated with a light blue arrowhead at the splice donor site of exon 23. (B) A reduction in the normal splice form, visualized as a 438 bp fragment in the control lane, is observed in e23i23MO-injected embryos at 2 dpf. A smaller running band containing an in-frame 63 bp deletion of the 23rd exon is observed in the e23i23 samples (red arrow). (C) The SPL8 morpholino strongly reduces the normal splice product of 517 bp at 2 dpf. No mis-spliced product was observed in our experiments. (D) The structure of the Ahi1 protein is conserved between human and zebrafish, with intact coiled-coil, WD-repeat and SH3 domains. Uninjected control shows normally elongated body axis, normal brain formation and an otic vesicle with two distinct otoliths (red brackets). (E) The predominant splice form in e23i23 morpholino-injected embryos encodes a protein lacking 21 amino acids of the N-terminal region of the SH3 domain (black arrowhead). e23i23MO-injected embryo is morphologically indistinguishable from controls. (F) Embryos injected with the previously described SPL8 morpholino (11) are depleted of the normal ahi1 transcript and exhibit a phenotype consistent with a severe ciliopathy, including hydrocephaly (asterisk), curved body axis and otic vesicle abnormalities. Scale bars: 250 µm.
Discussion
The criteria for classifying a genetic variant of a known disease gene as pathogenic must be stringent and include the variant's allele frequency and cosegregation with the phenotype in the patient's family and in additional families. The precise classification of variants is required for the reliability of online mutation repositories such as HGMD, a major database accessed by molecular biologists, clinicians and genetic counselors (12).
Heterozygous variants in recessive disease genes that are incidentally identified in healthy individuals (e.g. in preconceptional carrier screening with unremarkable family history) or in a patient whose phenotype is unrelated to the respective gene (e.g. whole-exome or whole-genome sequencing) may be difficult to interpret in terms of their pathogenicity. In contrast, homozygous inactivation of severe recessive disease genes usually results in obvious clinical phenotypes. Of all mutations, truncating and frameshift mutations in known disease genes are therefore most likely to be pathogenic (13). For genes not known to be implicated in monogenic disease, homozygous LoF variants in healthy individuals are strong indicators of LoF tolerance of the affected genes. Genes involved in embryonic development and cellular metabolism, such as AHI1, are rarely LoF tolerant. AHI1 encodes a highly conserved ciliary protein, Jouberin, with a crucial role in embryonic, in particular cerebellar and cortical, development. The longest AHI1 isoform is a 1196-residue (137 kDa) protein with a coiled-coil region, seven WD40-repeats and a Sarcoma homology 3 (SH3) domain. These motifs are known to participate in cell signaling and intracellular trafficking. AHI1 mutations, which are mostly truncating, cluster in the N-terminal half of AHI1 that contains the WD-repeats, but not the SH3 domain, and cause the severe autosomal recessive ciliopathy, JBTS3 (OMIM 608629).
The lack of an overt clinical effect (including an MTS in cranial MRI) of the p.Arg1066* mutation in AHI1 (Fig. 1C and D) is surprising, because this mutation does not have the features typical of a neutral LoF variant: (i) There are no close homologs, hence compensation for AHI1 inactivation by functional redundancy is unlikely. (ii) The mutation is predicted to remove a significant part (10%) of the full-length AHI1 protein, including the SH3 domain. (iii) The p.Arg1066* variant is present at very low frequency in the ExAC database (one out of 116 180 alleles; last accessed on 20 January 2015), and absent from the ESP and TGP databases, and from our extended in-house database of 1629 exomes. Rather, the truncating nature of the p.Arg1066* variant is comparable to the majority of previously described AHI1 mutations in JBTS3 patients, and in heterozygous state, it would have been classified as a pathogenic allele (2). The only indication of the benign character of the p.Arg1066* variant was its homozygous occurrence in three siblings with an unrelated condition (non-syndromic deafness) (14) and in their completely healthy sister. Three rare missense variants affecting AHI1 codon 1066 are currently listed in the ExAC database (last accessed on 20 January 2015). Two of them [p.Arg1066Pro and p.Arg1066Gln (rs544992761)] have been found four times in 114848 alleles, and one missense variant, p.Arg1066Gly, has been found only once. In contrast to the homozygous nonsense mutation described herein, these variants were all heterozygous, thus not adding any information about the essentiality of this AHI1 region.
Of note, one of the three main AHI1 isoforms (transcript variant/isoform 2, RefSeq NM_017651.4) consists of only 21 coding exons (compared with 26 exons in the full-length protein, isoform 1; RefSeq NM_001134831.1) that encode a protein of 1053 residues (120 kDa) lacking the SH3 domain (15). In contrast to the AHI1 full-length isoform 1, isoform 2, running at ∼120 kDa, was more strongly expressed in culture fibroblasts from a homozygous mutation carrier (II:4) than from a control individual. The apparently predominant shorter isoforms (10) are therefore not affected by the p.Arg1066* mutation except that their relative levels of expression are switched around between homozygous mutation carrier and control proband. The reason for this may be a culture-induced shift of expression ratios or interindividual variations.
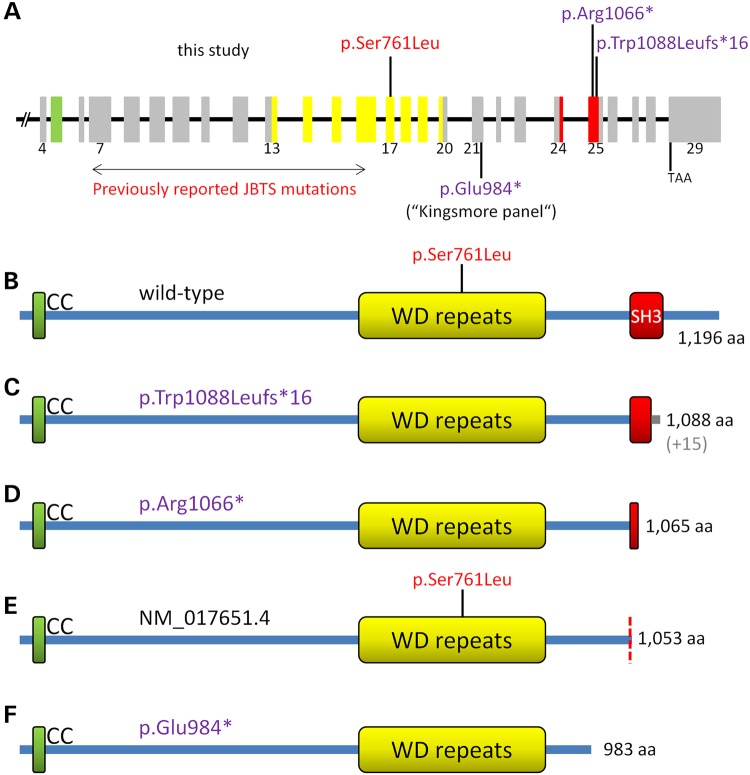
To investigate whether pathogenicity of truncating AHI1 variants could depend upon their location within the gene, we generated zebrafish models. In contrast to zebrafish treated with a morpholino affecting the N-terminal region where human mutations cluster (Fig. 4A), we observed no ciliopathy phenotype after deleting a significant portion of the SH3 domain near the sequence corresponding to the localization of the human p.Arg1066* variant. Hence, the SH3 domain (and downstream AHI1 peptide sequence) may not be essential for AHI1 function. Transcript 2 (Fig. 4E) may define the functionally indispensable part of the gene, and mutations located more 3′, such as p.Arg1066* (Fig. 4C), remain clinically silent. Consistent with this interpretation, all JBTS3-associated AHI1 mutations cluster between the N-terminus and the WD-repeats. The only exception is a homozygous truncating mutation (c.3263_3264delGG/p.Trp1088Leufs*16; Fig. 4A and C) in the same exon as p.Arg1066* (exon 25). This frameshift mutation was identified by targeted AHI1 mutation screening in two siblings with a JBTS3 phenotype who were born to consanguineous parents (2) (here termed ‘Family 2’). p.Trp1088Leufs*16 has been documented in dbSNP as rs387906269, but no MAF is available, indicating that this is a very rare variant. Our genome-wide homozygosity mapping using samples of additional siblings and both parents of family 2, combined with WES of one index patient, confirmed AHI1 as the gene responsible for JBTS. Remarkably, the unimpaired phenotype of the zebrafish model for C-terminal AHI1 truncations and intensive structural modeling strongly indicate that the causative mutation in this family is not the C-terminal frameshift variant p.Trp1088Leufs*, but rather is a missense mutation, p.Ser761Leu, residing in the JBTS3 mutation cluster region (Fig. 4A, B and E). We therefore consider p.Trp1088Leufs*16, like p.Arg1066*, a very rare neutral LoF variant. The p.Trp1088Leufs*16 mutation is particularly misleading not only because of the variant type (frameshift), but also because mapping and WES indicate that AHI1 undoubtedly is the disease gene in this family.
Figure 4.
Schemes of the AHI1 gene and the encoded AHI1/Jouberin protein. (A) AHI1 gene with numbers for coding exons (4–29). The three AHI variants investigated herein are indicated above the gene scheme. Known JBTS-causing AHI1 mutations clustering within exons 7–16 and the p.Glu984* variant [identified in the pilot study for the preconception carrier screen for 448 severe recessive childhood diseases (16)] are indicated below the scheme. Variants considered to be non-pathogenic or of uncertain pathogenicity (p.Glu984*) are depicted in purple. Variants considered to represent disease-causing mutations are in red. Exon colors refer to the protein regions they encode (B–F). (B) AHI1 wild-type protein (the position of the presumably pathogenic p.Ser761Leu mutation is indicated), (C) truncated AHI1 protein predicted to result from the p.Trp1088Leufs*16 variant (in grey: 15 unrelated residues before truncation), (D) truncated AHI1 protein predicted to result from the p.Arg1066* variant, (E) AHI1 isoform encoded by NM_017651.4 (‘transcript variant 2’). The position of p.Ser761Leu mutation in this isoform is indicated. (F) AHI protein truncation predicted to result from the p.Glu984* variant that had been identified in a carrier screen (16).
The pilot study for a preconception carrier screen of 448 severe recessive childhood diseases identified a healthy heterozygous carrier of a novel C-terminal AHI1 nonsense variant, p.Glu984* (Fig. 4A and F), located between the WD-repeat region and the two apparently non-pathogenic LoF variants discussed in our study (16). A conclusive assessment of the p.Glu984* variant is currently not possible. Instead, further studies are needed to define the extent of the proposed C-terminal LoF-tolerant region of AHI1. This region might be limited to the transcript 3-specific sequence or it may extend to the transcript 2 sequence downstream of the WD-repeats. Even if both parents carried the heterozygous p.Glu984* mutation, they might have no elevated risk for JBTS in their offspring, rather than the 25% risk of deleterious recessive mutations. This would have major implications for reproductive decision making.
Many previously reported mutations have recently been reclassified as likely benign because of their high frequency in datasets from large-scale exome and genome sequencing studies (17–20). In some cases, this reclassification questions the categorization of the respective genes as disease causing. For example, our findings from large-scale sequencing of known deafness genes recently disqualified MYO1A as a dominant deafness gene: most MYO1A ‘mutations’, including truncating variants, had MAFs above the threshold for dominant mutations and were present in both heterozygous and homozygous state in healthy individuals (21). Here, we propose that even homozygosity for rare truncating variants in undoubted disease genes must be assessed more rigorously before assignment of pathogenicity. In addition to the nature of the variant, the location of the sequence change within the gene is very important, and more research is needed to define those regions of disease genes that are vulnerable to pathogenic mutations and those that tolerate LoF variants. LoF-tolerant regions of disease genes, as we have identified for AHI1, can be unmasked by systematic analysis of large-scale genome sequencing studies for biallelic mutations in healthy individuals (22–24). In their survey, MacArthur et al. (1) considered novel LoF variants as likely disease causing only when they affect ‘all known transcripts of genes in which other null mutations have been convincingly associated with disease’. We strongly agree with this strict definition; although AHI1 mutations undoubtedly cause severe congenital disease, this gene seems partially dispensable. We propose that LoF variants in transcripts previously not associated with disease-causing mutations require careful evaluation. Such efforts are required to avoid erroneous interpretation, particularly in preconception carrier screening when couples make reproductive decisions. It will also, as in Family 2, help avoid misclassification of non-causative truncating mutations in known disease genes in patients with phenotypes expected for the affected genes.
Materials and Methods
Patients
All samples in this study (patients and healthy relatives) were obtained with written informed consent for the genetic analyses performed in this study. All clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the institutional review board of the Ethics Committee of the University Hospital of Cologne. Family 1 is a consanguineous Palestinian family (the parents of the patients are first-degree cousins) with three siblings affected by congenital deafness. To exclude any mix-up of samples, new samples were taken from all family members, and AHI1 (RefSeq NM_001134831.1) genotyping was repeated with the previously used primers, but also with an alternative pair of primers. Family 2 is a previously reported consanguineous Egyptian family (2) with two siblings affected by JBTS and RP.
Homozygosity mapping
We performed genome-wide linkage analysis of the parents and the six siblings (Family 1) and of the parents and six siblings (Family 2, as displayed in Fig. 2A) as previously described (25). We used 10 K SNP arrays (Family 1; Affymetrix GeneChip Human Mapping 10 K Array, version 2.0; Affymetrix, Santa Clara, CA, USA) and the Illumina HumanCoreExome-12v1-1 BeadChip (Family 2; Illumina Inc., San Diego, CA, USA), respectively, according to the manufacturer's protocol. Linkage analysis was performed assuming autosomal recessive inheritance, full penetrance, consanguinity and a disease gene frequency of 0.0001. Multipoint LOD scores were calculated using the program ALLEGRO (26). All data handling was done using the graphical user interface ALOHOMORA (27).
Whole-exome sequencing
Samples of patient II:3 (Family 1) and of patient II:2 (Family 2) were subjected to WES. We fragmented 1 μg of DNA by sonication (Covaris, Woburn, MA, USA). The fragments were end-repaired and adaptor-ligated including incorporation of sample index barcodes. After size selection, the library was subjected to the enrichment process. For this, we chose the SeqCap EZ Human Exome Library v2.0 kit from NimbleGen (Roche NimbleGen, Madison, WI, USA). The enriched library was subsequently sequenced on an Illumina HiSeq 2000 sequencing instrument using a paired end 2 × 100 bp protocol.
This resulted in 7.8 Gb of mapped sequences, a mean coverage of 77 and 30× coverage of 85% of target sequences. For the data analysis, a local pipeline and interface were used (Varbank v.2.3; https://varbank.ccg.uni-koeln.de). Primary data were filtered according to signal purity by the Illumina Realtime Analysis (RTA) software v1.8. Subsequently, the reads were mapped to the human genome reference build hg19 using the BWA (28) alignment algorithm. GATK v1.6 (29) was used to mark duplicated reads, for local realignment around short insertion and deletions, to recalibrate the base quality scores and to call SNPs and short indels.
Scripts developed in-house at the Cologne Center for Genomics were used to detect protein changes, affected donor and acceptor splice sites, and overlaps with known variants. Acceptor and donor splice site mutations were analyzed with a Maximum Entropy model (30) and filtered for effect changes. In particular, we filtered for high-quality (coverage >15; quality >25) rare (MAF < 0.005) homozygous variants [dbSNP build 135, the database of the 1000 Genomes Project build 20110521, TGP (31)], and the Exome Variant Server, NHLBI Exome Sequencing Project, Seattle, build ESP6500 (32). We also filtered against an in-house database containing all variants from 511 exomes from epilepsy patients to exclude pipeline-related artifacts/false positives (MAF < 0.004). Because the patients came from a consanguineous background, we filtered for variants overlapping with runs of homozygosity (ROH). To detect ROH, we extracted high-quality SNP genotypes from the exome data and ran Allegro v1.2c (26) with a pseudo-consanguine pedigree (first-degree cousins), a full penetrant recessive model, and a disease allele frequency of 0.0001. To search for the gene underlying the hearing deficit in patients II:3, II:5 and II:6, chromosomal candidate regions identified in genome-wide linkage analysis with 10 K SNP arrays were used for filtering. The resulting list of candidate genes was then prioritized by taking into account positive rejected substitution scores obtained with GERP++ (33). In addition to the above large-scale sequencing databases consulted by the Varbank pipeline, we searched the Exome Aggregation Consortium (ExAC) database (Cambridge, MA, USA; http://exac.broadinstitute.org, as of 12/2014), which aggregates numerous databases including the current versions of the ESP and the TGP, for the AHI1 variants discussed herein.
Segregation analysis for AHI1 variants and qPCR
Confirmation and segregation analysis for the AHI1 variants c.3196C>T (p.Arg1066*) and c.3263_3264delGG (p.Trp1088Leufs*16) in exon 25 was carried out by Sanger sequencing of 534-bp PCR products generated with primers 5′-TGCTTCTCCTGCTGTGTTCC-3′ (1F) and 5′-CAGAATGGAGCAGCTTCTAATG-3′ (1R), on samples of all family members with indicated genotypes (Figs 1A and 2A). In addition, alternative primers were used for confirmation and segregation analysis of c.3196C>T (2F: 5′-CCATTTCTCAATCCCACAGTC-3′, 2R: 5′-CCATTGGTAAACATATGCAAAGG-3′; PCR product: 488 bp). To exclude allelic dropout in PCR amplification using the above primers, we conducted qPCR with primers flanking the binding sites of the PCR/sequencing primers (B1qF2 5′-GCTTCACAAGTAATTTACTT-3′ with B1qR2 5′-GCAGCTGAAACCAAAGCAGTAC-3′ yielding a 199 bp product around the primer binding sites of 1F and 2F, and B2qF2 5′-GTATAACTGATAGTCCAAG-3′ with B2qR2 5′-TAAGAAAGCTGGTAGCTAG-3′ yielding a 227 bp product around the primer binding sites 1R and 2R, respectively). qPCR primers were used at a final concentration of 0.7 pm/µl in a 14.25 µl reaction mix containing 6.25 µl 2× PowerSybrGreen Master Mix (Life Technologies, Carlsbad, CA, USA) on the StepOnePlus™ System (Life Technologies).
In silico assessment of the p.Ser761Leu mutation
Evolutionary conservation of the p.Ser671 residue was determined by alignment of the respective peptide stretches from five species using Clustal Omega (34). The structure of AHI1 and the effect of p.Ser671Leu were predicted using an algorithm we recently developed, the WD40 structure predictor, WDSP (35). MD simulations were performed using Gromacs version 5.0 at 310 K (36). RSFF1 force field was used (37). Each protein was solvated with ∼7000 TIP4P/Ew61 water molecules. The ionic Arg, Lys, Asp and Glu side chains were neutralized with counterion (Cl− or Na+). The velocity rescaling thermostat with τT = 0.5 ps was used to maintain the NPT ensemble. Electrostatics was treated using the particle-mesh Ewald (PME) method with areal-space cutoff of 0.9 nm. Van der Waals interactions were cut off at 0.9 nm with the long-range dispersion correction for energy and pressure. All bond lengths involving hydrogen atoms were constrained by the LINCS, allowing a time step of 2 fs. Fifty and 90 ns MD simulation was run for the wild-type and for the p.Ser761Leu mutant, respectively.
Western blotting
Protein extracts were obtained by lysing subconfluent primary fibroblast cultures (individual II:4, family 1) with crude lysis buffer (1% SDS, 10 mm EDTA). Samples corresponding to 15 µg protein were run together with a SeeBluePlus2 molecular weight ladder on a 4–12% NuPAGE gradient gel in MOPS-SDS Buffer at 130 V for 2.5 h. The gel was blotted onto a nitrocellulose membrane (Fa Amersham, Hybond-ECL) at 30 V for 2.5 h and subsequently blocked with 6% milk powder for 3 h. Incubation with primary antibodies PA5-30901 and PA5-31846 was 12 h at 4°C in a 1:1000 dilution. The secondary goat anti-rabbit IgG-HRP antibody was used in a 1:10 000 dilution for 1 h at room temperature, followed by 3 × 5 min washes in TBS-T buffer. Detection and exposure were carried out by ECL-sensitive chemiluminescent substrate and film.
Morpholino knockdown in zebrafish
All zebrafish studies were conducted with the Oregon AB wild-type strain crossed against the Tübingen strain to obtain embryos for injection. Animals were raised in a 10 h dark and 14 h light cycle and maintained as described (38). Animals were staged according to the standard series (39) or by hours or days postfertilization (hpf or dpf, respectively). All experiments were conducted in accordance with Institutional Animal Care and Use guidelines. Whole-mount images of live embryos were obtained on a Leica dissecting microscope with a Zeiss Axiocam HRC. Antisense Morpholinos (Gene Tools) directed against the splice acceptor of the 13th exon [SPL8 (11)] or the splice donor of the 23rd exon (e23i23, 5′-AGCAAATTAACGCTTACCGTCTGAT 3′) were microinjected into one-cell stage embryos at concentrations between 2 and 5 µg/µl. Five embryos per experimental condition (injected versus uninjected control) from each time point were pooled for total RNA extraction with TRIzol (Invitrogen). cDNA was synthesized by reverse transcription using the Superscript III kit (Invitrogen) and used in PCR reactions with primers flanking the SPL8 (11) or e23i23 (forward: 5′-tcaagccggacagccaatcaga-3′; reverse: 5′-gaggcttcagcatccgtgtccag-3′) target sites. Individual bands were separated by electrophoresis on 1.5% agarose gels, isolated and purified (Qiagen) for sequencing (GeneWiz).
Supplementary Material
Funding
This work was supported by funding from the Marie-Louise Geissler-Stiftung, the Gertrud Kusen-Stiftung, the Imhoff-Stiftung and Köln Fortune (University Hospital of Cologne) to H.J.B.; the NIH (DC010447 and HD22486) to M.W.; and NSFC 21133002 and KQTD201103 to Y.D.W.
Supplementary Material
Acknowledgements
We are indebted to the families who participated in this study. We thank Jeremy Wegner for technical assistance in sequencing the zebrafish ahi1 gene transcripts and Karin Boß for critical reading and very helpful discussions.
Conflict of Interest statement. H.J.B. is an employee of Bioscientia which is part of a publicly traded diagnostic company. The work described in this study is unrelated to this employment.
References
- 1.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., et al. (2012) A systematic survey of loss-of-function variants in human protein-coding genes. Science, 335, 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valente E.M., Brancati F., Silhavy J.L., Castori M., Marsh S.E., Barrano G., Bertini E., Boltshauser E., Zaki M.S., Abdel-Aleem A., et al. (2006) AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann. Neurol., 59, 527–534. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods, 7, 575–576. [DOI] [PubMed] [Google Scholar]
- 4.de Castro E., Sigrist C.J., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res., 34, W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck B.B., Phillips J.B., Bartram M.P., Wegner J., Thoenes M., Pannes A., Sampson J., Heller R., Gobel H., Koerber F., et al. (2014) Mutation of POC1B in a severe syndromic retinal ciliopathy. Hum. Mutat., 35, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X.H., Zhang H., Wu Y.D. (2010) Is Asp-His-Ser/Thr-Trp tetrad hydrogen-bond network important to WD40-repeat proteins: a statistical and theoretical study. Proteins, 78, 1186–1194. [DOI] [PubMed] [Google Scholar]
- 7.Wu X.H., Chen R.C., Gao Y., Wu Y.D. (2010) The effect of Asp-His-Ser/Thr-Trp tetrad on the thermostability of WD40-repeat proteins. Biochemistry, 49, 10237–10245. [DOI] [PubMed] [Google Scholar]
- 8.Dasen J.S., Martinez Barbera J.P., Herman T.S., Connell S.O., Olson L., Ju B., Tollkuhn J., Baek S.H., Rose D.W., Rosenfeld M.G. (2001) Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev., 15, 3193–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. (2004) Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet., 36, 1008–1013. [DOI] [PubMed] [Google Scholar]
- 10.Ringrose A., Zhou Y., Pang E., Zhou L., Lin A.E., Sheng G., Li X.J., Weng A., Su M.W., Pittelkow M.R., et al. (2006) Evidence for an oncogenic role of AHI-1 in Sezary syndrome, a leukemic variant of human cutaneous T-cell lymphomas. Leukemia, 20, 1593–1601. [DOI] [PubMed] [Google Scholar]
- 11.Simms R.J., Hynes A.M., Eley L., Inglis D., Chaudhry B., Dawe H.R., Sayer J.A. (2012) Modelling a ciliopathy: Ahi1 knockdown in model systems reveals an essential role in brain, retinal, and renal development. Cell. Mol. Life Sci., 69, 993–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet., 133, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., et al. (2013) Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet., 93, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroes H.Y., Van Zanten B.G., De Ru S.A., Boon M., Mancini G.M., Van der Knaap M.S., Poll-The B.T., Lindhout D. (2010) Is hearing loss a feature of Joubert syndrome, a ciliopathy? Int. J. Pediatr. Otorhinolaryngol., 74, 1034–1038. [DOI] [PubMed] [Google Scholar]
- 15.Esmailzadeh S., Jiang X. (2011) AHI-1: a novel signaling protein and potential therapeutic target in human leukemia and brain disorders. Oncotarget, 2, 918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell C.J., Dinwiddie D.L., Miller N.A., Hateley S.L., Ganusova E.E., Mudge J., Langley R.J., Zhang L., Lee C.C., Schilkey F.D., et al. (2011) Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med., 3, 65ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreasen C., Nielsen J.B., Refsgaard L., Holst A.G., Christensen A.H., Andreasen L., Sajadieh A., Haunso S., Svendsen J.H., Olesen M.S. (2013) New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur. J. Hum. Genet., 21, 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cassa C.A., Tong M.Y., Jordan D.M. (2013) Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum. Mutat., 34, 1216–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jabbari J., Jabbari R., Nielsen M.W., Holst A.G., Nielsen J.B., Haunso S., Tfelt-Hansen J., Svendsen J.H., Olesen M.S. (2013) New exome data question the pathogenicity of genetic variants previously associated with catecholaminergic polymorphic ventricular tachycardia. Circ. Cardiovasc. Genet., 6, 481–489. [DOI] [PubMed] [Google Scholar]
- 20.Risgaard B., Jabbari R., Refsgaard L., Holst A.G., Haunso S., Sadjadieh A., Winkel B.G., Olesen M.S., Tfelt-Hansen J. (2013) High prevalence of genetic variants previously associated with Brugada syndrome in new exome data. Clin. Genet., 84, 489–495. [DOI] [PubMed] [Google Scholar]
- 21.Eisenberger T., Di Donato N., Baig S.M., Neuhaus C., Beyer A., Decker E., Murbe D., Decker C., Bergmann C., Bolz H.J. (2014) Targeted and genomewide NGS data disqualify mutations in MYO1A, the ‘DFNA48 gene’, as a cause of deafness. Hum. Mutat., 35, 565–570. [DOI] [PubMed] [Google Scholar]
- 22.Alsalem A.B., Halees A.S., Anazi S., Alshamekh S., Alkuraya F.S. (2013) Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet., 9, e1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser J. (2014) The hunt for missing genes. Science, 344, 687–689. [DOI] [PubMed] [Google Scholar]
- 24.Lim E.T., Raychaudhuri S., Sanders S.J., Stevens C., Sabo A., MacArthur D.G., Neale B.M., Kirby A., Ruderfer D.M., Fromer M., et al. (2013) Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron, 77, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., Fitzpatrick D.R., Nurnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J.L., Chitayat D., et al. (2007) Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet., 80, 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudbjartsson D.F., Jonasson K., Frigge M.L., Kong A. (2000) Allegro, a new computer program for multipoint linkage analysis. Nat. Genet., 25, 12–13. [DOI] [PubMed] [Google Scholar]
- 27.Rüschendorf F., Nürnberg P. (2005) ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics, 21, 2123–2125. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo G., Burge C.B. (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol., 11, 377–394. [DOI] [PubMed] [Google Scholar]
- 31.Via M., Gignoux C., Burchard E.G. (2010) The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med., 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W., O'Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., et al. (2013) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature, 493, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol., 6, e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers F., Higgins D.G. (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol., 1079, 105–116. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Jiang F., Zhuo Z., Wu X.H., Wu Y.D. (2013) A method for WD40 repeat detection and secondary structure prediction. PLoS ONE, 8, e65705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 29, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang F., Zhou C.Y., Wu Y.D. (2014) Residue-specific force field based on the protein coil library. RSFF1: modification of OPLS-AA/L. J. Phys. Chem. B, 118, 6983–6998. [DOI] [PubMed] [Google Scholar]
- 38.Westerfield M. (2007) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- 39.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn., 203, 253–310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.