Summary
The causes of childhood cancer have been systematically studied for several decades, but apart from high-dose radiation and prior chemotherapy there are few or no strong external risk factors. On the other hand, inherent risk factors including birth weight, parental age, and congenital anomalies are consistently associated with most types of pediatric cancer. Rare, highly-penetrant syndromes have long been known to cause a small proportion of cancers but recently the contribution of common genetic variation to etiology has come into focus through genome wide association studies. These have highlighted genes not previously implicated in childhood cancers and, surprisingly, have suggested that common variation explains a larger proportion of childhood cancers than adult. Rare variation and non-Mendelian inheritance, such as through maternal genetic effects or de novo germline mutations, may also contribute to childhood cancer risk but have not been widely examined to date.
Keywords: Epidemiology, etiology, genome-wide association studies, case-control studies, pediatric cancer
Introduction
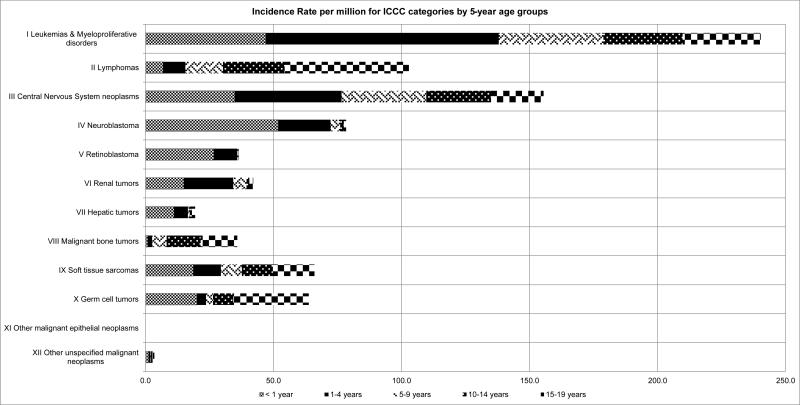
The causes of childhood cancer have been systematically studied for several decades. The incidence of all cancers occurring in children under 20 years of age is about 175 cases per million in the United States (1), with the incidence of many individual types [typically grouped by the International Classification of Childhood Cancer (ICCC) schema(2)], in the low dozens (Figure 1). Rarity is thus a central fact which dictates the quality and quantity of evidence for causal associations between putative risk factors and childhood cancers. Most etiologic investigations of childhood cancer have thus of necessity used the case-control study design (3), in which the characteristics of patients with a disease are compared to those of a carefully selected group of disease-free controls. When they require participant involvement, as in the many studies of childhood cancer which have collected exposure information through parental interview, case-control studies are susceptible to both recall and selection biases.
Fig. 1.
Incidence rate per million for International Classification of Childhood Cancer (ICCC) categories by 5-year age groups
Given the milieu for childhood cancer epidemiology, evidence regarding causal associations has accumulated slowly. However, for the most common types of cancer, particularly acute lymphoblastic leukemia (ALL), the body of literature is now sufficiently large to allow data synthesis through meta-analyses and data pooling. This review will only briefly discuss such analyses of external, or environmental, risk factors, as they have mainly demonstrated weak or null associations. In contrast, intrinsic characteristics or conditions of childhood cancer patients have shown stronger, more consistent associations and in the last half-decade the application of genome wide single nucleotide variant (SNV) arrays to several childhood cancers has generated surprising insights into their biology. Hence, the majority of this review for the general pediatrician will cover these topics in a broad overview to reflect their increased importance in the current understanding of childhood cancer etiology.
Demographic risk factors
Childhood cancer incidence has long been noted to vary by age, sex, and race/ethnicity. Overall incidence is highest in infancy at about 240 cases per million per year. This rate drops to a nadir of 128 cases per million at 5-9 years of age before rising to 220 cases per million at 15-19 years of age (1). Grouped incidence however obscures interesting patterns among individual cancers (Figure 1). For instance, all the embryonal tumors (neuroblastoma, Wilm's tumor, retinoblastoma, etc.) share a downward sloping incidence which starts high at birth and dissipates after about 5 years of age. ALL is notable for the incidence peak which occurs between 2 to 5 years of age, while bone sarcoma incidence peaks sharply around the time of the pubertal growth spurt in early-tomid adolescence.
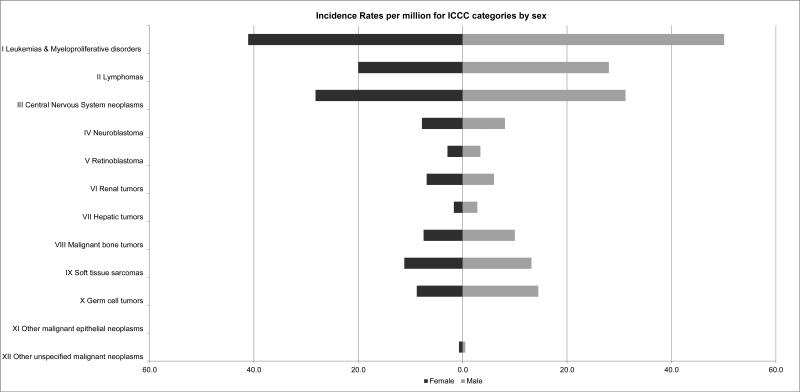
For most childhood cancers there is a slight male preponderance (Figure 2). The male-to-female ratio ranges from 1.04 to 1.64 in neuroblastoma and germ cell tumors, respectively, in cases 0-19 years of age but varies considerably by age group and more specific diagnosis. Wilm's tumor is notable for being the one major childhood cancer which is more common in females.
Fig. 2.
Incidence rates per million for International Classification of Childhood Cancer (ICCC) categories by sex.
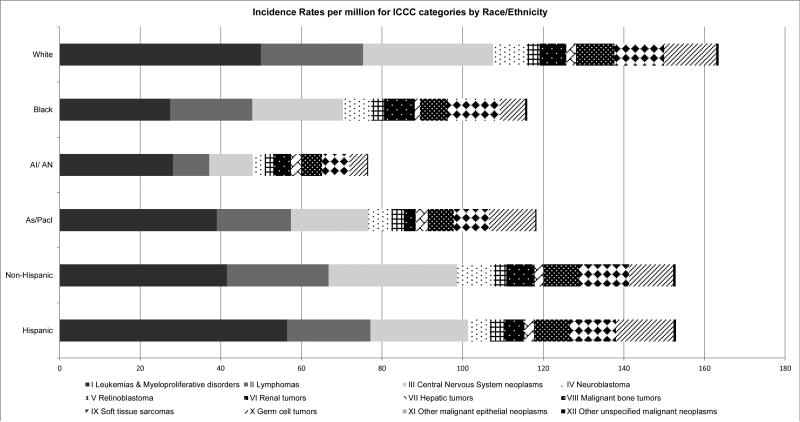
Childhood cancer risk also differs by race/ethnicity (Figure 3). Relative to white children in the United States the incidence of most types of cancer is lower in black, Asian, and Hispanic children. In some cases, such as the near complete lack of Ewing sarcoma among black and Asian children, the disparity is rather dramatic. In a few notable instances cancer incidence is higher in other groups compared to white children. That acute leukemia incidence is about 10% higher in Hispanic children compared to white children is particularly notable. The extent to which racial/ethnic differences are attributable to genetic versus environmental differences has yet to be determined but will surely come into focus as the genetic architecture of childhood cancer continues to be elucidated.
Fig. 3.
Incidence rates per million for International Classification of Childhood Cancer (ICCC) categories by race/ethnicity. AI/AN, American Indian/Alaska Native; As/Pacl, Asian/Pacific Islander.
Environmental risk factors
High dose ionizing radiation and prior chemotherapy are accepted causes of childhood cancers, each raising risk several fold (4-7). No other environmental risk factors, by which we mean any exposure which originates outside the body, have emerged as definitively causal for childhood cancer.
Measurement of environmental exposures poses a challenge in elucidating their effects on childhood cancer risk. Prospective studies would require hundreds of thousands, if not millions, of children to identify enough cases to generate statistically meaningful results. Thus most childhood cancer studies must rely on the case-control design, which is particularly problematic for evaluating certain types of exposures. Demographic and intrinsic factors are unambiguous, relatively easy to obtain via questionnaire, and, in such cases as parental age, race/ethnicity, and birth defects, generally not subject to recall error. In contrast, environmental factors such as parental diet, maternal medication, caffeine, and alcohol use, and pesticide and air pollution exposure, among others, are very difficult to measure accurately in a retrospective design. Although use of registries, birth and medical records and other data sources reduces some sources of bias, accurate exposure assessment remains a major barrier to determining the causal impact of environmental factors on childhood cancer risk.
For many childhood cancers findings are inconsistent or studies too few to conduct meta-analysis; moreover synthesis is hampered by the need to separately examine exposures during the preconceptional, pregnancy, and postnatal periods and the progressively finer classifications of tumors. ALL, being the most common childhood cancer, has however been the subject of several meta-analyses of putative environmental risk factors (Table 1)(8-15).
Table 1.
Results of selected meta-analyses of environmental risk factors and childhood ALL.
| Exposure | Time period | N studies | Findings | Ref |
|---|---|---|---|---|
| Maternal alcohol use | Pregnancy | 10 | No association of any alcohol use during pregnancy with ALL [mOR* = 1.10 (0.93-1.29) | (11) |
| Maternal coffee use | Pregnancy | 5 | Small association of any coffee consumption during pregnancy with ALL [mOR = 1.16 (1.00-1.34)] | (8) |
| Daycare attendance | Postnatal | 14 | Small reduced risk of ALL associated with daycare attendance [mOR = 0.76 (0.67-0.87)] | (13) |
| Electromagnetic field exposure | Postnatal | 9 | No association of electromagnetic field exposure >=0.2 μT with ALL [mOR = 1.25 (0.97-1.60)] | (15) |
| Occupational pesticide exposure | Pregnancy | 5 | Strong association of maternal occupational exposure to pesticides during pregnancy and ALL [mOR = 2.64 (1.40-5.00)] | (14) |
| Maternal prenatal vitamins | Pregnancy | 3 | Small reduced risk of ALL associated with maternal prenatal vitamin consumption [mOR = 0.61 (0.50-0.74)] | (9) |
| Paternal smoking | Preconception | 10 | Small association of any paternal preconceptional smoking with ALL [mOR = 1.15 (1.06-1.24)] | (12) |
| Maternal smoking | Pregnancy | 20 | No association of any maternal smoking during pregnancy with ALL [mOR = 1.03 (0.95-1.12)] | (10) |
mOR = meta-analytic odds ratio
Exposure to infections has been one of the most commonly examined environmental exposures in relation to ALL risk, and there are two main hypotheses regarding the nature of this relationship. Kinlen proposes that previously isolated, and therefore immunologically naïve, populations are susceptible when exposed to specific infectious agents due to population mixing (16). A recent meta-analysis estimated an increased risk of ALL in rural settings of population mixing (17). Greaves’ hypothesized that an immature and unchallenged immune system, resulting from delayed exposure to common infections, produces an unregulated immune response and leads to ALL in the presence of susceptible cells (18, 19). While direct measurement of exposure to infections and the resulting immune response is generally not feasible, several proxies have been employed, including birth order(20-22), daycare attendance(23, 24), breastfeeding(25), infectious illness histories(26), and vaccinations(27). Meta-analyses have shown protective effects for both breastfeeding (28) and daycare attendance (13), although because these are indirect exposure measures, it is unclear whether infection exposure or some other factor is driving these associations.
Several recent meta-analyses have identified increased risk for both residential (29, 30) and maternal occupational (14) exposure to pesticides. Studies of residential pesticide exposure have generally relied on self-report, which is subject to recall bias that may inflate risk estimates. Some recent studies have used residential proximity to pesticide applications (31-33), a method which is less prone to bias but still prone to measurement error which may attenuate risk estimates (34). Occupational studies typically rely on either self-report or record data. While an association between pesticide exposure and ALL is supported by the available meta-analytic data, it is difficult to estimate the true magnitude of effect, if there is indeed a causal relationship, given the varying exposure assessment methods and the inherent biases and measurement errors therein.
Associations for other exposures examined using meta-analyses, including maternal alcohol(11), coffee(35), and vitamin use(9), and both paternal (12) and maternal (10) smoking have yielded mostly null (10, 11) or slightly elevated (12, 35) results. Maternal prenatal vitamin use was associated with a decreased risk of ALL in offspring, although the meta-analysis was based on only three studies (9). . While a causal role for these risk factors is possible, observational epidemiology is not conclusive in these circumstances. High-quality studies with a focus on accurate exposure assessment are necessary to evaluate environmental risk factors for childhood cancer.
Intrinsic risk factors
Several intrinsic characteristics of children or their parents have been consistently associated with childhood cancers. Risk of ALL(36), central nervous system tumors(37), neuroblastoma(38), and Wilm's tumor(39), among others, rises as a linear function of birth weight, to varying degrees, and recent analyses that have used alternate measures of birth size (e.g. size for gestational age, percent of optimal birth weight) have found similar results (40). Risk of acute myeloid leukemia is elevated with both low and high birth weight(41), while risk of hepatoblastoma is inversely related to birth weight and strikingly elevated among the smallest infants (42). The reasons behind the association of higher birth weight with childhood cancers have not been explored in detail, but may include prenatal growth hormone exposure (43), the underlying genetics of birth weight (44), and the greater number of cells at risk for carcinogenic transformation. The strong inverse association of hepatoblastoma with birth weight has been thought to be related to neonatal treatment, but no culprit exposure has been identified to date(45).
Advanced parental age has also been associated with most childhood cancers. A large pooled analysis of population-based record-linkage studies found significant positive linear trends in leukemia, lymphoma, brain tumor, neuroblastoma, Wilm's tumor, bone tumors, and soft tissue sarcomas with 6-15% increased risk per five years of maternal age(46). Paternal age was not associated with these cancers after adjustment for maternal age, however since the two are highly correlated it is not clear that maternal age was solely responsible. As with birth weight, the reasons behind these findings are unclear, but may include genetic or epigenetic mutations associated with advanced parental age (47).
Structural birth defects have consistently been found to increase the risk of childhood cancers, as a group, about threefold (48-50) although due to the rarity of both individual birth defects and individual childhood cancers more specific associations have not been reported to date. Undoubtedly some of this association is explained by underlying genetic causes, but as most birth defects appear sporadic(51) genetics are not likely the sole explanation for cooccurrence.
Genetic risk factors
Inherited syndromes, caused by high-penetrance germline DNA mutations(52, 53), chromosomal aneuploidy(54), or epigenetic disorders(55), are known to cause a minority of childhood cancers. Although the proportion attributable to syndromes has rarely been precisely quantified for common childhood cancers the estimate is typically 5-10%. For especially rare cancers, such as pediatric adrenocortical carcinoma, the proportion can be much higher (56). Specific syndromes predisposing to particular childhood cancers are covered elsewhere in this issue.
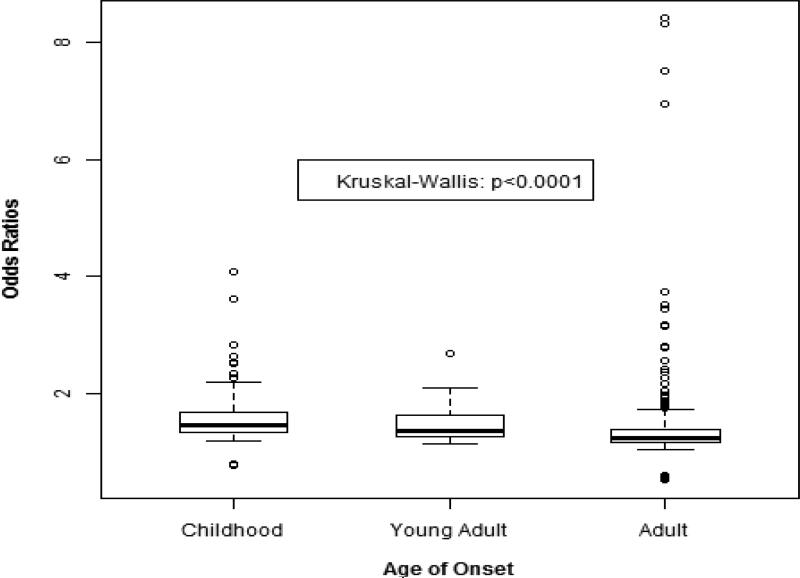
Genome wide association studies (GWAS) compare the frequency of hundreds of thousands of common SNV's in those with a disease to those without(57). Due to the large number of comparisons made in GWAS a SNV-disease association must reach a high degree of statistical significance (generally p < 5 × 10−8) to be convincing. This requires large sample sizes not readily achievable for rare diseases. Yet, despite the a priori presumption that the GWAS design could not be successfully applied to childhood cancers investigations of ALL(58-64), neuroblastoma(65-74), Wilm's tumor(75), osteosarcoma(76), and Ewing's sarcoma (77), each have identified multiple variants associated with each disease (Table 2). The unexpected success of GWAS to studies of these rare cancers appears to be due to the larger magnitude of SNV-disease association among young onset cancers compared to those with adult onset, which was recently formally quantified (Figure 4) (78). An implication of this finding, besides reaffirming the applicability of GWAS to other childhood cancers not yet studied thusly, is that common genetic variation explains a greater proportion of the population attributable risk for childhood than adult cancers.
Table 2.
SNV's identified by GWAS of childhood cancers.
| Cancer | Gene | SNV | Population | Subtype | OR | 95% CI | P-value | Ref |
|---|---|---|---|---|---|---|---|---|
| ALL | ARID5B | rs10821936 | European | Total | 1.91 | 1.6-2.2 | 1.4 × 10−15 | (39) |
| ALL | ARID5B | rs10740055 | European | Total | 1.53 | 1.4-1.6 | 5.35 × 10−14 | (38) |
| ALL | ARID5B | rs10821936 | African-American | Total | 1.52 | 1.1-2.0 | 0.004 | (40) |
| ALL | ARID5B | rs10821936 | Hispanic | Total | 1.95 | 1.6-2.4 | 3.78 × 10−11 | (40) |
| ALL | ARID5B | rs10821936 | European | B-hyperdiploid | 2.17 | 1.5-3.1 | 1.62 × 10−5 | (39) |
| ALL | CDK2NA | rs3731217 | European | Total | 0.71 | 0.6-0.8 | 3.01 × 10−11 | (41) |
| ALL | CDK2NA | rs17756311 | European | Total | 1.43 | 1.2-1.7 | 3.25 × 10−5 | (40) |
| ALL | CEPBE | rs2239633 | European | Total | 1.34 | 1.2-1.5 | 2.88 × 10−7 | (38) |
| ALL | CEBPE | rs4982731 | Hispanic | Total | 1.58 | 1.3-1.9 | 2.32 × 10−6 | (40) |
| ALL | COMMD3/BMI1 | rs4266962 | European | Total | 1.41 | 1.2-1.7 | 4.35 × 10−8 | (40) |
| ALL | GATA3 | rs3824662 | European | Total | 1.31 | 1.2-1.4 | 8.62 × 10−12 | (44) |
| ALL | GATA3 | rs3824662 | European | Philadelphia-like | 3.85 | 2.7-5.47 | 2.17 × 10−14 | (43) |
| ALL | IKZF1 | rs11978267 | European | Total | 1.69 | 1.4-1.9 | 8.8 × 10−11 | (39) |
| ALL | IKZF1 | rs4132601 | European | Total | 1.69 | 1.6-1.8 | 1.2 × 10−19 | (38) |
| ALL | PIP4K2A | rs10828317 | European | Total | 1.23 | 1.2-1.3 | 2.3 × 10−9 | (44) |
| Ewing sarcoma | TARDBP | rs9430161 | European | Total | 2.20 | 1.8-2.7 | 1.4 × 10−20 | (57) |
| Ewing sarcoma | EGR2 | rs224278 | European | Total | 1.66 | 1.4-1.9 | 4 × 10−17 | (57) |
| Neuroblastoma | FLJ22536 | rs6939340 | European | Total | 1.40 | 1.3-1.6 | 5.82 × 10−8 | (52) |
| Neuroblastoma | BARD1 | rs6435862 | European | Total | 1.64 | 1.4-1.9 | 3.19 × 10−9 | (46) |
| Neuroblastoma | BARD1 | rs6435862 | African-American | Total | 1.44 | 1.2-1.7 | 1.8 × 10−5 | (46) |
| Neuroblastoma | HACE1 | rs4336470 | European | Total | 1.26 | 1.2-1.4 | 2.7 × 10−11 | (49) |
| Neuroblastoma | LIN28B | rs17065417 | European | Total | 1.38 | 1.2-1.5 | 1.2 × 10−8 | (49) |
| Neuroblastoma | LMO1 | rs110419 | European | Total | 1.34 | 1.3-1.4 | 5.2 × 10−16 | (54) |
| Osteosarcoma | GRM4 | rs1906953 | European | Total | 1.57 | 1.4-1.8 | 8.0 × 10−9 | (56) |
| Osteosarcoma | Intergenic at 2p25.2 | rs7591996 | European | Total | 1.39 | 1.2-1.5 | 1.0 × 10−8 | (56) |
| Wilm's tumor | Intergenic at 2p24 | rs3755132 | European | Total | 1.48 | 1.3-1.7 | 1.03 × 10−14 | (55) |
| Wilm's tumor | Intergenic at 11q14 | rs790356 | European | Total | 1.43 | 1.3-1.6 | 4.25 × 10−15 | (55)I |
Fig. 4.
Boxplot of SNV odds ratios from GWAS of cancer by age group. Dark horizontal lines represent the median and the box represents the 25th and 75th percentiles. (From Raynor LA, Pankratz N, Spector LG, et al. An analysis of measures of effect size by age of onset in cancer genomewide association studies. Genes Chromosomes Cancer 2013;52(9): 857; with permission.)
The GWAS and replication studies of ALL and neuroblastoma include diverse populations and subtype-specific analyses, giving a more mature picture of the genetic architecture of each disease than is available for those with a single GWAS to date. Two recent GWAS of ALL conducted with African-American and Hispanic cases and controls replicated many of the SNV's first identified in studies of subjects with European ancestry; SNV's in ARID5B, IKZF1, and PIP4K2A were associated with ALL in both ethnicities, and CEBPE as well in Hispanics (60). OR's per allele were similar in each group, in line with the generally high trans-ethnic replicability of GWAS results (79), however frequencies varied in directions that suggest several SNVs may explain a substantial proportion of lower incidence of ALL in African-Americans and the higher one in Hispanics compared to Europeans. ARID5B rs10821936 was present in 33% of Europeans, 24% of African-Americans, and 47% of Hispanics; the equivalent numbers for IKZF1 rs11978267 were 28%, 19%, and 26%. CEBP rs4982731 was present in 28% of Europeans and 39% of Hispanics, as well as 38% of African-Americans in which this SNV did not replicate. Similarly, SNVs in BARD1 replicated in a GWAS of neuroblastoma among African-Americans, while no others did, possibly due to small sample size (71).
Several SNVs in both diseases show far stronger OR's with specific subtypes, demonstrating that lumping disparate cases can dilute associations. In ALL, ARID5B SNVs have been more strongly associated with hyperdiploid disease(59, 80, 81) and GATA3 SNV's with leukemias displaying a Philadelphia-chromosome-like expression pattern (63); the latter is an especially dramatic instance, with subtype-specific ORs per allele of about 3.5 versus 1.3 in total ALL. BARD1 and LMO1 SNVs are associated with aggressive disease in neuroblastoma (66, 67, 73), and SNVs in or near DUSP12, DDX4, IL31RA, and HSD17B12 with low-risk disease (73). It seems reasonable to speculate that similarly specific associations with subtypes of other childhood cancers will emerge as the GWAS literature expands.
Although the progress in identifying common variants associated with several childhood cancers in the past several years has been remarkable, a large portion of heritability remains unexplained. For instance, estimates indicate that about 25% of genetic variation in ALL risk was due to common variants identified by GWAS available in 2012 (82). The remainder of genetic risk may be attributable to several other plausible mechanisms which will more often be evaluated as next-generation sequencing and the inclusion of parental samples are adopted by the field. Rare variation is generally defined as that with a population allele frequency of <0.01. A recent exome-sequencing study of infant leukemia, among the first of its kind, identified compound heterozygosity for rare pathogenic variants in the MLL3 gene as risk factors (83). Sequencing of parents and children and comparing exomes or genomes can identify de novo mutations which will not be apparent by other technologies(47). Lastly, since many childhood cancers are presumed to initiate in utero(84), maternal genetic effects may be relevant(85), but have only been examined in a candidate gene rather than genome wide context to date (86, 87).
Conclusion
The rarity of childhood cancer slows the search for their causes, but the accumulation of case-control studies and advancement of genomic technology have improved our knowledge in recent years. Few environmental risk factors for childhood cancer have been identified that exceed the capacity of observational epidemiology to distinguish causal associations from those due to bias. Inherent risk factors such as birth weight, parental age, and birth defects- as well as common genetic variation- are on the other hand consistently associated with childhood cancers.
Key Points.
- Apart from high-dose radiation and prior chemotherapy there are few or no strong external risk factors with relative risks >2.
- Inherent risk factors including birth weight, parental age, and congenital anomalies are consistently associated with most types of pediatric cancer.
- Common genetic variation has been associated with several childhood cancers in genome wide association studies, often with subtype-specificity, and explains a larger proportion of childhood than adult cancers.
Contributor Information
Logan G. Spector, Division of Epidemiology/Clinical Research Department of Pediatrics University of Minnesota 420 Delaware Street, SE, MMC 715 Minneapolis, MN 55455.
Nathan Pankratz, Department of Lab Medicine and Pathology University of Minnesota.
Erin L. Marcotte, Division of Epidemiology/Clinical Research Department of Pediatrics University of Minnesota.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekreuse SF, et al. SEER Cancer Statistics Review, 1975-2011. National Cancer Institute; Bethesda, MD: 2014. [Google Scholar]
- 2.Steliarova-Foucher E, Stiller C, Lacour B, Kaatsch P. International Classification of Childhood Cancer, third edition. Cancer. 2005;103(7):1457–67. doi: 10.1002/cncr.20910. [DOI] [PubMed] [Google Scholar]
- 3.Wacholder S. Design issues in case-control studies. Stat Methods Med Res. 1995;4(4):293–309. doi: 10.1177/096228029500400403. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins MM, Wilson LM, Stovall MA, Marsden HB, Potok MH, Kingston JE, et al. Epipodophyllotoxins, alkylating agents, and radiation and risk of secondary leukaemia after childhood cancer. BMJ. 1992;304(6832):951–8. doi: 10.1136/bmj.304.6832.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109(1):13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 6.Tucker MA, Meadows AT, Boice JD, Jr., Stovall M, Oberlin O, Stone BJ, et al. Leukemia after therapy with alkylating agents for childhood cancer. J Natl Cancer Inst. 1987;78(3):459–64. [PubMed] [Google Scholar]
- 7.Ron E, Modan B, Boice JD, Jr., Alfandary E, Stovall M, Chetrit A, et al. Tumors of the brain and nervous system after radiotherapy in childhood. N Engl J Med. 1988;319(16):1033–9. doi: 10.1056/NEJM198810203191601. [DOI] [PubMed] [Google Scholar]
- 8.Cheng J, Su H, Zhu R, Wang X, Peng M, Song J, et al. Maternal coffee consumption during pregnancy and risk of childhood acute leukemia: a metaanalysis. Am J Obstet Gynecol. 2014;210(2):151.e1–151.e10. doi: 10.1016/j.ajog.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Goh YI, Bollano E, Einarson TR, Koren G. Prenatal multivitamin supplementation and rates of pediatric cancers: a meta-analysis. Clin Pharmacol Ther. 2007;81(5):685–91. doi: 10.1038/sj.clpt.6100100. [DOI] [PubMed] [Google Scholar]
- 10.Klimentopoulou A, Antonopoulos CN, Papadopoulou C, Kanavidis P, Tourvas AD, Polychronopoulou S, et al. Maternal smoking during pregnancy and risk for childhood leukemia: a nationwide case-control study in Greece and meta-analysis. Pediatr Blood Cancer. 2012;58(3):344–51. doi: 10.1002/pbc.23347. [DOI] [PubMed] [Google Scholar]
- 11.Latino-Martel P, Chan DS, Druesne-Pecollo N, Barrandon E, Hercberg S, Norat T. Maternal alcohol consumption during pregnancy and risk of childhood leukemia: systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1238–60. doi: 10.1158/1055-9965.EPI-09-1110. [DOI] [PubMed] [Google Scholar]
- 12.Milne E, Greenop KR, Scott RJ, Bailey HD, Attia J, Dalla-Pozza L, et al. Parental prenatal smoking and risk of childhood acute lymphoblastic leukemia. Am J Epidemiol. 2012;175(1):43–53. doi: 10.1093/aje/kwr275. [DOI] [PubMed] [Google Scholar]
- 13.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int J Epidemiol. 2010;39(3):718–32. doi: 10.1093/ije/dyp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wigle DT, Turner MC, Krewski D. A systematic review and meta-analysis of childhood leukemia and parental occupational pesticide exposure. Environ Health Perspect. 2009;117(10):1505–13. doi: 10.1289/ehp.0900582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L, Liu X, Wang C, Yan K, Lin X, Li S, et al. Magnetic fields exposure and childhood leukemia risk: A meta-analysis based on 11,699 cases and 13,194 controls. Leuk Res. 2014;38(3):269–274. doi: 10.1016/j.leukres.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Kinlen L. Evidence for an infective cause of childhood leukaemia: comparison of a Scottish new town with nuclear reprocessing sites in Britain. Lancet. 1988;2(8624):1323–7. doi: 10.1016/s0140-6736(88)90867-7. [DOI] [PubMed] [Google Scholar]
- 17.Kinlen LJ. An examination, with a meta-analysis, of studies of childhood leukaemia in relation to population mixing. Br J Cancer. 2012;107(7):1163–8. doi: 10.1038/bjc.2012.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greaves MF. Aetiology of acute leukaemia. Lancet. 1997;349(9048):344–9. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 19.Greaves MF. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia. 1988;2(2):120–5. [PubMed] [Google Scholar]
- 20.Roman E, Simpson J, Ansell P, Lightfoot T, Mitchell C, Eden TO. Perinatal and reproductive factors: a report on haematological malignancies from the UKCCS. Eur J Cancer. 2005;41(5):749–59. doi: 10.1016/j.ejca.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Jourdan-Da Silva N, Perel Y, Mechinaud F, Plouvier E, Gandemer V, Lutz P, et al. Infectious diseases in the first year of life, perinatal characteristics and childhood acute leukaemia. Br J Cancer. 2004;90(1):139–45. doi: 10.1038/sj.bjc.6601384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dockerty JD, Draper G, Vincent T, Rowan SD, Bunch KJ. Case-control study of parental age, parity and socioeconomic level in relation to childhood cancers. Int J Epidemiol. 2001;30(6):1428–37. doi: 10.1093/ije/30.6.1428. [DOI] [PubMed] [Google Scholar]
- 23.Gilham C, Peto J, Simpson J, Roman E, Eden TO, Greaves MF, et al. Day care in infancy and risk of childhood acute lymphoblastic leukaemia: findings from UK case-control study. BMJ. 2005;330(7503):1294. doi: 10.1136/bmj.38428.521042.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neglia JP, Linet MS, Shu XO, Severson RK, Potter JD, Mertens AC, et al. Patterns of infection and day care utilization and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2000;82(1):234–40. doi: 10.1054/bjoc.1999.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Infante-Rivard C, Fortier I, Olson E. Markers of infection, breast-feeding and childhood acute lymphoblastic leukaemia. Br J Cancer. 2000;83(11):1559–64. doi: 10.1054/bjoc.2000.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Steensel-Moll HA, Valkenburg HA, van Zanen GE. Childhood leukemia and infectious diseases in the first year of life: a register-based case-control study. Am J Epidemiol. 1986;124(4):590–4. doi: 10.1093/oxfordjournals.aje.a114431. [DOI] [PubMed] [Google Scholar]
- 27.Dockerty JD, Skegg DC, Elwood JM, Herbison GP, Becroft DM, Lewis ME. Infections, vaccinations, and the risk of childhood leukaemia. Br J Cancer. 1999;80(9):1483–9. doi: 10.1038/sj.bjc.6690548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwan ML, Buffler PA, Abrams B, Kiley VA. Breastfeeding and the risk of childhood leukemia: a meta-analysis. Public Health Rep. 2004;119(6):521–35. doi: 10.1016/j.phr.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Maele-Fabry G, Lantin AC, Hoet P, Lison D. Residential exposure to pesticides and childhood leukaemia: a systematic review and meta-analysis. Environ Int. 2011;37(1):280–91. doi: 10.1016/j.envint.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Turner MC, Wigle DT, Krewski D. Residential pesticides and childhood leukemia: a systematic review and meta-analysis. Environ Health Perspect. 2010;118(1):33–41. doi: 10.1289/ehp.0900966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rull RP, Gunier R, Von Behren J, Hertz A, Crouse V, Buffler PA, et al. Residential proximity to agricultural pesticide applications and childhood acute lymphoblastic leukemia. Environ Res. 2009;109(7):891–9. doi: 10.1016/j.envres.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds P, Von Behren J, Gunier RB, Goldberg DE, Harnly M, Hertz A. Agricultural pesticide use and childhood cancer in California. Epidemiology. 2005;16(1):93–100. doi: 10.1097/01.ede.0000147119.32704.5c. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds P, Von Behren J, Gunier RB, Goldberg DE, Hertz A, Harnly ME. Childhood cancer and agricultural pesticide use: an ecologic study in California. Environ Health Perspect. 2002;110(3):319–24. doi: 10.1289/ehp.02110319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rull RP, Ritz B. Historical pesticide exposure in California using pesticide use reports and land-use surveys: an assessment of misclassification error and bias. Environ Health Perspect. 2003;111(13):1582–9. doi: 10.1289/ehp.6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng J, Su H, Zhu R, Wang X, Peng M, Song J, et al. Maternal coffee consumption during pregnancy and risk of childhood acute leukemia: a metaanalysis. Am J Obstet Gynecol. 2014;210(2):151, e1–151, e10. doi: 10.1016/j.ajog.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Hjalgrim LL, Westergaard T, Rostgaard K, Schmiegelow K, Melbye M, Hjalgrim H, et al. Birth weight as a risk factor for childhood leukemia: a meta-analysis of 18 epidemiologic studies. Am J Epidemiol. 2003;158(8):724–35. doi: 10.1093/aje/kwg210. [DOI] [PubMed] [Google Scholar]
- 37.Harder T, Plagemann A, Harder A. Birth weight and subsequent risk of childhood primary brain tumors: a meta-analysis. Am J Epidemiol. 2008;168(4):366–73. doi: 10.1093/aje/kwn144. [DOI] [PubMed] [Google Scholar]
- 38.Harder T, Plagemann A, Harder A. Birth weight and risk of neuroblastoma: a meta-analysis. Int J Epidemiol. 2010;39(3):746–56. doi: 10.1093/ije/dyq040. [DOI] [PubMed] [Google Scholar]
- 39.Chu A, Heck JE, Ribeiro KB, Brennan P, Boffetta P, Buffler P, et al. Wilms' tumour: a systematic review of risk factors and meta-analysis. Paediatr Perinat Epidemiol. 2010;24(5):449–69. doi: 10.1111/j.1365-3016.2010.01133.x. [DOI] [PubMed] [Google Scholar]
- 40.Milne E, Greenop KR, Metayer C, Schuz J, Petridou E, Pombo-de-Oliveira MS, et al. Fetal growth and childhood acute lymphoblastic leukemia: findings from the childhood leukemia international consortium. Int J Cancer. 2013;133(12):2968–79. doi: 10.1002/ijc.28314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caughey RW, Michels KB. Birth weight and childhood leukemia: a meta-analysis and review of the current evidence. Int J Cancer. 2009;124(11):2658–70. doi: 10.1002/ijc.24225. [DOI] [PubMed] [Google Scholar]
- 42.Spector LG, Puumala SE, Carozza SE, Chow EJ, Fox EE, Horel S, et al. Cancer risk among children with very low birth weights. Pediatrics. 2009;124(1):96–104. doi: 10.1542/peds.2008-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross JA, Perentesis JP, Robison LL, Davies SM. Big babies and infant leukemia: a role for insulin-like growth factor-1? Cancer Causes Control. 1996;7(5):553–9. doi: 10.1007/BF00051889. [DOI] [PubMed] [Google Scholar]
- 44.Lunde A, Melve KK, Gjessing HK, Skjaerven R, Irgens LM. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol. 2007;165(7):734–41. doi: 10.1093/aje/kwk107. [DOI] [PubMed] [Google Scholar]
- 45.Turcotte LM, Georgieff MK, Ross JA, Feusner JH, Tomlinson GE, Malogolowkin MH, et al. Neonatal medical exposures and characteristics of low birth weight hepatoblastoma cases: A report from the Children's Oncology Group. Pediatr Blood Cancer. 2014 doi: 10.1002/pbc.25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson KJ, Carozza SE, Chow EJ, Fox EE, Horel S, McLaughlin CC, et al. Parental age and risk of childhood cancer: a pooled analysis. Epidemiology. 2009;20(4):475–83. doi: 10.1097/EDE.0b013e3181a5a332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565–75. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 48.Botto LD, Flood T, Little J, Fluchel MN, Krikov S, Feldkamp ML, et al. Cancer risk in children and adolescents with birth defects: a population-based cohort study. PLoS One. 2013;8(7):e69077. doi: 10.1371/journal.pone.0069077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carozza SE, Langlois PH, Miller EA, Canfield M. Are children with birth defects at higher risk of childhood cancers? Am J Epidemiol. 2012;175(12):1217–24. doi: 10.1093/aje/kwr470. [DOI] [PubMed] [Google Scholar]
- 50.Fisher PG, Reynolds P, Von Behren J, Carmichael SL, Rasmussen SA, Shaw GM. Cancer in children with nonchromosomal birth defects. J Pediatr. 2012;160(6):978–83. doi: 10.1016/j.jpeds.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hobbs CA, Chowdhury S, Cleves MA, Erickson S, MacLeod SL, Shaw GM, et al. Genetic epidemiology and nonsyndromic structural birth defects: from candidate genes to epigenetics. JAMA Pediatr. 2014;168(4):371–7. doi: 10.1001/jamapediatrics.2013.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2(4):475–84. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seif AE. Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis. Cancer Genet. 2011;204(5):227–44. doi: 10.1016/j.cancergen.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Ross JA, Spector LG, Robison LL, Olshan AF. Epidemiology of leukemia in children with Down syndrome. Pediatr Blood Cancer. 2005;44(1):8–12. doi: 10.1002/pbc.20165. [DOI] [PubMed] [Google Scholar]
- 55.DeBaun MR, Tucker MA. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr. 1998;132:398–400. doi: 10.1016/s0022-3476(98)70008-3. 3 Pt 1. [DOI] [PubMed] [Google Scholar]
- 56.Choong SS, Latiff ZA, Mohamed M, Lim LL, Chen KS, Vengidasan L, et al. Childhood adrenocortical carcinoma as a sentinel cancer for detecting families with germline TP53 mutations. Clin Genet. 2012;82(6):564–8. doi: 10.1111/j.1399-0004.2012.01841.x. [DOI] [PubMed] [Google Scholar]
- 57.Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356(11):1094–7. doi: 10.1056/NEJMp068126. [DOI] [PubMed] [Google Scholar]
- 58.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–10. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–5. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105(10):733–42. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–4. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellinghaus E, Stanulla M, Richter G, Ellinghaus D, te Kronnie G, Cario G, et al. Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia. 2012;26(5):902–9. doi: 10.1038/leu.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45(12):1494–8. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298–307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 65.Bosse KR, Diskin SJ, Cole KA, Wood AC, Schnepp RW, Norris G, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72(8):2068–78. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capasso M, Diskin SJ, Totaro F, Longo L, De Mariano M, Russo R, et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34(3):605–11. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diskin SJ, Capasso M, Diamond M, Oldridge DA, Conkrite K, Bosse KR, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106(4):dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44(10):1126–30. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987–91. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Latorre V, Diskin SJ, Diamond MA, Zhang H, Hakonarson H, Maris JM, et al. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol Biomarkers Prev. 2012;21(4):658–63. doi: 10.1158/1055-9965.EPI-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen le B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7(3):e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469(7329):216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turnbull C, Perdeaux ER, Pernet D, Naranjo A, Renwick A, Seal S, et al. A genome-wide association study identifies susceptibility loci for Wilms tumor. Nat Genet. 2012;44(6):681–4. doi: 10.1038/ng.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savage SA, Mirabello L, Wang Z, Gastier-Foster JM, Gorlick R, Khanna C, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45(7):799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44(3):323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 78.Raynor LA, Pankratz N, Spector LG. An analysis of measures of effect size by age of onset in cancer genomewide association studies. Genes Chromosomes Cancer. 2013;52(9):855–9. doi: 10.1002/gcc.22081. [DOI] [PubMed] [Google Scholar]
- 79.Marigorta UM, Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013;9(6):e1003566. doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chokkalingam AP, Hsu LI, Metayer C, Hansen HM, Month SR, Barcellos LF, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Causes Control. 2013;24(10):1789–95. doi: 10.1007/s10552-013-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Linabery AM, Blommer CN, Spector LG, Davies SM, Robison LL, Ross JA. ARID5B and IKZF1 variants, selected demographic factors, and childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Leuk Res. 2013;37(8):936–42. doi: 10.1016/j.leukres.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Enciso-Mora V, Hosking FJ, Sheridan E, Kinsey SE, Lightfoot T, Roman E, et al. Common genetic variation contributes significantly to the risk of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26(10):2212–5. doi: 10.1038/leu.2012.89. [DOI] [PubMed] [Google Scholar]
- 83.Valentine MC, Linabery AM, Chasnoff S, Hughes AE, Mallaney C, Sanchez N, et al. Excess congenital non-synonymous variation in leukemia-associated genes in MLL- infant leukemia: a Children's Oncology Group report. Leukemia. 2014;28(6):1235–41. doi: 10.1038/leu.2013.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spector LG, Hooten AJ, Ross JA. Ontogeny of gene expression: a changing environment for malignancy. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1021–3. doi: 10.1158/1055-9965.EPI-08-0275. [DOI] [PubMed] [Google Scholar]
- 85.Santure AW, Spencer HG. Influence of mom and dad: quantitative genetic models for maternal effects and genomic imprinting. Genetics. 2006;173(4):2297–316. doi: 10.1534/genetics.105.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nousome D, Lupo PJ, Okcu MF, Scheurer ME. Maternal and offspring xenobiotic metabolism haplotypes and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2013;37(5):531–5. doi: 10.1016/j.leukres.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lupo PJ, Nousome D, Kamdar KY, Okcu MF, Scheurer ME. A case-parent triad assessment of folate metabolic genes and the risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2012;23(11):1797–803. doi: 10.1007/s10552-012-0058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]