SUMMARY
Adipose tissue is an essential regulator of metabolic homeostasis. In contrast with white adipose tissue, which stores excess energy in the form of triglycerides, brown adipose tissue is thermogenic, dissipating energy as heat via the unique expression of the mitochondrial uncoupling protein UCP1. A subset of UCP1+ adipocytes develop within white adipose tissue in response to physiological stimuli, however, the developmental origin of these “brite” or “beige” adipocytes is unclear. Here, we report the identification of a BMP7-ROCK signaling axis regulating beige adipocyte formation via control of the G-actin-regulated transcriptional coactivator myocardin related transcription factor A, MRTFA. White adipose tissue from MRTFA–/– mice contains more multilocular adipocytes and expresses enhanced levels of brown-selective proteins, including UCP1. MRTFA–/– mice also show improved metabolic profiles and protection from diet-induced obesity and insulin resistance. Our study hence unravels a central pathway driving the development of physiologically functional beige adipocytes.
Keywords: Bone Morphogenetic Protein 7, RhoA kinase, actin, Myocardin-related transcription factor A (MRTFA), Serum response factor (SRF), Beige adipocytes, Uncoupling protein 1 (UCP1)
INTRODUCTION
Adipose tissue is an essential regulator of metabolic homeostasis and is composed of mesenchymal stem cells, vascular cells, preadipocytes, and mature fat cells. At least two types of adipocytes exist with distinct phenotypes and function. White adipocytes are morphologically distinct from brown adipocytes and store lipids in unilocular droplets. In comparison, brown adipocytes contain multilocular lipid droplets and numerous mitochondria. Whereas white adipocytes store excess energy in the form of triglycerides, brown adipocytes are thermogenic, oxidative cells that, when activated by appropriate stimuli, dissipate energy as heat (Cannon and Nedergaard, 2004). Energy release is achieved due to the activity of UCP1, a brown adipose tissue (BAT)-specific mitochondrial uncoupling protein (Nedergaard et al., 2005). Because of its unique function, enhancing BAT mass and/or activity is an attractive therapeutic approach for individuals suffering from obesity and/or type 2 diabetes.
A distinct population of UCP1+ adipocytes develops within the white adipose tissue (WAT) of mice in response to cold exposure (Cousin et al., 1996; Cousin et al., 1992; Guerra et al., 1998; Loncar, 1991a, b), β3 adrenergic receptor (AR) agonist treatment (Collins et al., 1997; Ghorbani and Himms-Hagen, 1997; Granneman et al., 2005; Guerra et al., 1998; Nagase et al., 1996), PPARγ agonist administration (Petrovic et al., 2010; Vernochet et al., 2009; Wilson-Fritch et al., 2004; Xue et al., 2005), or exposure to FGF21, irisin or cardiac natriuretic peptides (Bordicchia et al., 2012; Bostrom et al., 2012; Fisher et al., 2012). The developmental origins of these cells designated “brite” or “beige” adipocytes, are not well understood (Enerback, 2009; Ishibashi and Seale, 2010; Petrovic et al., 2010; Young et al., 1984). Current hypotheses propose that they arise from the conversion of existing white adipocytes and/or the differentiation of WAT resident progenitor cells (Himms-Hagen et al., 2000; Jimenez et al., 2003; Lee et al., 2012; Long et al., 2014; Rosenwald et al., 2013; Wang et al., 2013; Wu et al., 2012; Zingaretti et al., 2009). Lineage tracing studies revealed that interscapular BAT arises from the dermomyotome (Atit et al., 2006; Seale et al., 2008). Brown and beige adipocytes in vitro possess a distinct gene expression signature (Wu et al., 2012) suggesting divergent processes regulate prenatal development of classical BAT and postnatal formation of brite/beige adipocytes within WAT.
Discovering the origins of adipocyte progenitors is of intense interest. A population of white adipocyte progenitors resident in the adult WAT stroma were characterized by specific cell surface markers (Rodeheffer et al., 2008). PPARγ lineage tracing studies indicated that WAT progenitors reside in the mural cell compartment of adipose vasculature (Tang et al., 2008) and at least a population of beige cells have a smooth muscle-like origin (Long et al., 2014). These observations suggest that white and beige adipocytes arise from progenitors closely associated with the vasculature. Physiological signals that regulate the fate of these progenitors, and their tissues of origin, have yet to be determined.
Members of the TGFβ superfamily are intimately involved in the development and maintenance of the vasculature (Jakobsson and van Meeteren, 2013; Patel-Hett and D'Amore, 2011). TGFβ promotes smooth muscle differentiation and coordinates the expression of SMC genes (Hautmann et al., 1997; Li et al., 2012; Sinha et al., 2004; Wang et al., 2010). TGFβ also inhibits adipocyte differentiation via its co-effector, Smad3, which complexes with C/EBPβ and represses activation of adipogenic target genes (Choy and Derynck, 2003). Interestingly, Smad3 knockout mice develop brown-like adipocytes in WAT depots and are protected from obesity, illustrating the role of the TGFβ/Smad3 pathway in the negative regulation of browning of WAT (Yadav et al., 2011).
In contrast with TGFβ, Bone Morphogenetic Proteins (BMPs), promote adipocyte formation (Schulz and Tseng, 2009). Exposure of multipotent MSCs to BMP2 or BMP4 gives rise to a population of preadipocyte-like cells, which differentiate to mature adipocytes (Ahrens et al., 1993; Bowers and Lane, 2007; Tang et al., 2004; Wang et al., 2009; Wang et al., 1993). BMP7 initiates the commitment of MSCs to the brown adipocyte lineage (Tseng et al., 2008) by promoting the expression of the brown adipocyte factors PRDM16, PGC-1α and UCP1 and mitochondrial biogenesis (Tseng et al., 2008). Importantly, the absence of BMP7 in mice attenuates the formation of BAT (Tseng et al., 2008); BMP7 is also able to induce the conversion of progenitors isolated from WAT, BAT and muscle to brown-like adipocytes (Schulz et al., 2011). BMP4 and BMP8b have also been implicated in the browning of WAT and enhancing energy expenditure and insulin sensitivity (Qian et al., 2013;Whittle et al., 2012).
Additional downstream effectors of TGFβ and BMPs include members of the Rho-GTPase family, which mediate the dynamic control of monomeric and filamentous actin (Moustakas and Heldin, 2008). Monomeric G-actin can regulate the nucleus-cytoplasm shuttling of SRF (serum response factor) coregulators, MRTFs (myocardin related transcription factors) and, thereby, influence the expression of SRF target genes including smooth muscle actin (SMA) (Miralles et al., 2003; Olson and Nordheim, 2010). Several studies have reported on the involvement of Rho-GTPase in regulating the fate of MSCs (McBeath et al., 2004; Sordella et al., 2003), but there are no studies of the potential role of MRTFs. Here, we identified a novel BMP7-controlled signaling and transcriptional circuit involving MRTFA, which enhances the development of beige adipocytes in WAT, resulting in protection from diet-induced obesity and insulin resistance.
RESULTS
BMP7 and TGFβ1 Mediate Distinct Effects on Lineage Commitment of MSCs
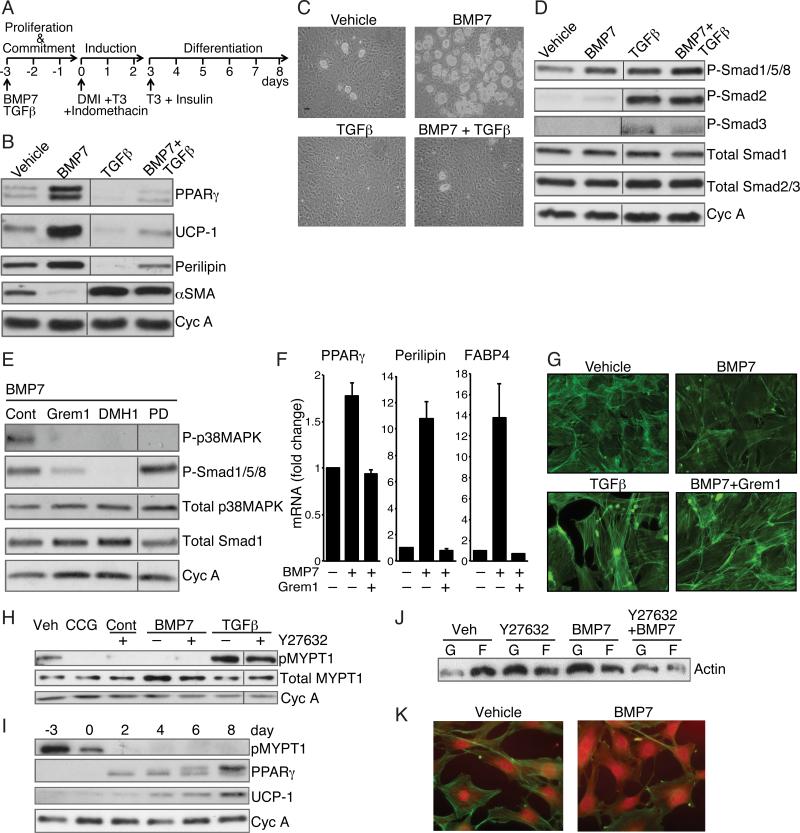
To investigate the effects of BMP7 and TGFβ1 on lineage commitment, subconfluent multipotent C3H/10T1/2 MSCs were exposed to each effector for 3 days until reaching confluence and then exposed to a brown adipogenic cocktail (illustrated in Figure 1A). As anticipated, BMP7-treated cells developed into brown-like adipocytes, noted by elevated expression of UCP1, PPARγ and Perilipin (Figure 1B). BMP7 also down regulated SMA in proliferating MSCs (Figure 1B). MSCs exposed to TGFβ1, however, failed to undergo adipogenesis but instead produced elevated levels of SMA, suggesting the cells committed to a smooth muscle-like lineage. Exposure to both BMP7 and TGFβ1 resulted in a mixed population of differentiated cells, as indicated by large numbers of fibroblastic cells, and to a lesser extent, Oil Red O positive adipocytes (data not shown). Heterogeneity of the population was confirmed by the abundant expression of SMA and a low level of adipocyte proteins (Figure 1B). To confirm that the effects of BMP7 and TGFβ were not unique to C3H/10T1/2 MSCs, these factors also regulated the differentiation of bone marrow-derived MSCs (Figure 1C).
Figure 1. BMP7 and TGFβ Mediate Distinct Effects on Lineage Commitment of MSCs through alteration of ROCK activity.
(A) Differentiation protocol used for mesenchymal stem cells.
(B and C) Subconfluent C3H10T1/2 cells (B) or mouse bone marrow derived MSCs (Life Technologies, Gibco) (C) were exposed to BMP7 (6.3 nM) and/or TGFβ1 (1 nM) for 3 days, then induced to differentiate without BMP7 or TGFβ as in (A) Differentiation of cells was assessed by Western blot (B) or phase contrast microscopy (C).
(D and E) Western blot analysis of C3H10T1/2 cell proteins after a 10 min exposure to vehicle, BMP7, TGFβ or BMP7 and TGFβ (D) or BMP7 alone following a 2 h pretreatment with either 125 nM Gremlin1 or 200 nM Dorsmorphin homologue (DMH1) or 50 μM PD169316 (E).
(F) Bone marrow MSCs were induced to differentiate in response to BMP7 as in A in presence or absence of Gremlin1 and RNA was subjected to qPCR analysis.
(G) Subconfluent C3H10T1/2 MSCs were exposed to TGFβ1 (1 nM) alone or BMP7 (6.3 nM) with or without Gremlin1 for 48 h, then stained with Alexa Fluor 488 phalloidin and visualized at 40x magnification.
(H) Subconfluent C3H10T1/2 MSCs were exposed to BMP7 or TGFβ with or without Y27632 (10 μM) or CCG1423 (10 μM) for 2 h at which stage protein lysates were analyzed for phospho-MYPT1.
(I) Protein extracts from C3H10T1/2 MSCs at the indicated times of differentiation (as illustrated in 1A) were analyzed by Western blot.
(J and K) Subconfluent, proliferating C3H10T1/2 MSCs were treated with Y27632 with or without BMP7 for 48 h and levels of F-actin versus G-actin were analyzed by Western blot (J) and immunofluorescence (K). Non-adjacent lanes on western blots (B, D, E, H) are indicated by a vertical line. See also Figure S1.
We next analyzed the signaling pathways activated in response to these effectors. As expected, TGFβ1 in the presence or absence of BMP7 activated the phosphorylation of SMAD2 and SMAD3. BMP7, however, stimulated the phosphorylation of SMADs 1/5/8, with negligible phosphorylation of SMADs 2 or 3 (Figure 1D). Interestingly, TGFβ1 also activated SMADs 1/5/8 indicating that BMP7-induced commitment is not mediated exclusively by activation of the SMADs, but by additional downstream effectors of the BMP receptor. In fact, BMP7 activated p38MAPK signaling (Figure S1A), which was required for BMP7-induced brown adipogenesis since treatment of the MSCs with a p38MAPK inhibitor (PD169316) suppressed UCP1 expression (Figure S1B). Inhibition of the BMP type I receptors with dorsomorphin homologue (DMH1) or inhibition of BMP7 directly with Gremlin1, (Hsu et al., 1998) attenuated the activation of SMADs 1/5/8 and p38MAPK confirming that stimulation of both pathways occurs through BMP7 and its receptor (Figure 1E). Notably, DMH1 diminished C3H/10T1/2 MSCs differentiation in a concentration-dependent manner (Figure S1C) and Gremlin1 attenuated the differentiation of bone marrow-derived MSCs in response to BMP7 (Figure 1F).
BMP7 represses ROCK activity and alters actin dynamics in MSCs
Cells exposed to BMP7 or TGFβ1, exhibited marked changes in morphology. BMP7-treated cells exhibited a broadened shape and a less prominent actin network when compared to control cells (Figure 1G). This phenomenon is controlled by BMP activity, as it was reversed by co-incubation with Gremlin1. TGFβ1, in contrast, promoted an elongated morphology and formation of actin microfilaments. We hypothesized that these changes in actin dynamics were mediated by Rho-associated protein kinase (ROCK), a major effector of the RhoA pathway (Schofield and Bernard, 2013). Following exposure of MSCs to BMP7, ROCK activity was repressed below basal levels, as assessed by a reduction in the phosphorylation of one of its targets, MYPT1 (Figure 1H). In fact, BMP7 inhibited ROCK as effectively as the ROCK-specific inhibitor, Y27632. As expected, TGFβ1 strongly induced MYPT1 phosphorylation, which was only partially inhibited by a dose of Y27632 that was sufficient to block endogenous TGFβ1 activity. Importantly, ROCK activity declined rapidly during BMP7-induced brown adipogenesis reaching undetectable levels in mature adipocytes (Figure 1I).
Coincident with modulation of the RhoA pathway are changes in the relative amounts of G-actin and F-actin. Consistent with its inhibition of ROCK, biochemical and cell staining assays revealed that BMP7 increased the G/F-actin ratio (Figure 1J, K). The ROCK inhibitor Y27632 similarly enhanced G-actin levels relative to F-actin (Figure 1J).
Inhibition of the ROCK/SRF pathway enhances brown adipogenesis in MSCs
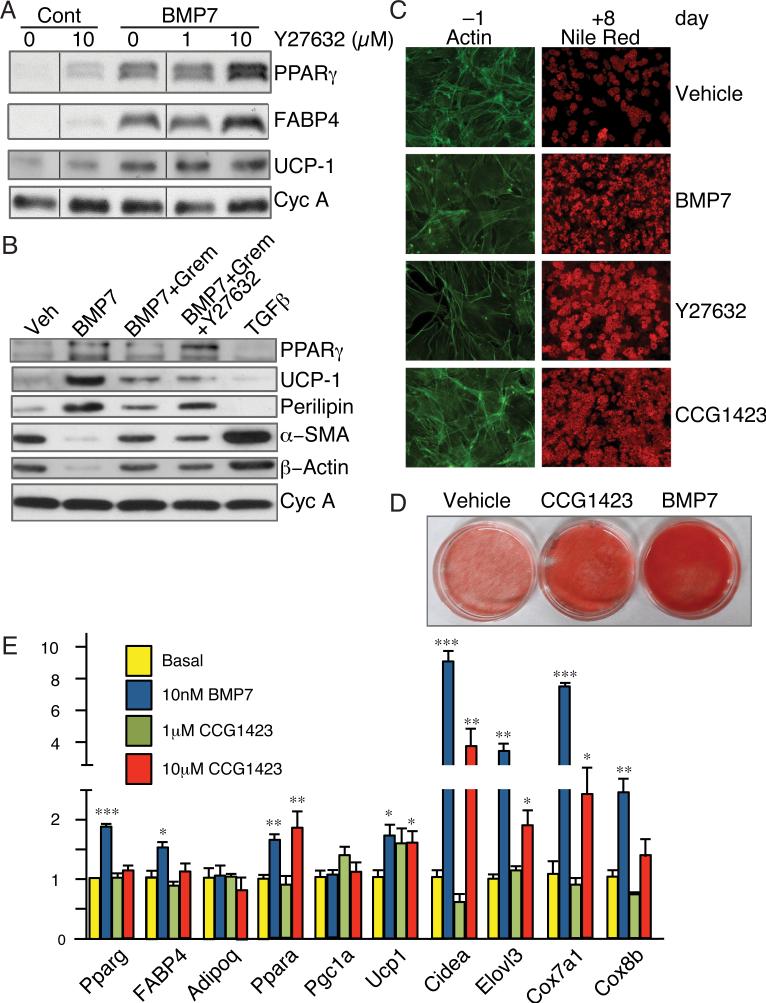
The pro-adipogenic properties of BMP7 closely correlated with inhibition of ROCK activity, which led us to question whether Y27632 could mimic the effects of BMP7. Although, Y27632 alone had a modest effect on adipocyte gene expression (Figure 2A) and on the number of cells converting to Nile Red-positive adipocytes (Figure 2C), it did potentiate the action of BMP7 as revealed by enhanced expression of PPARγ and FABP4 (Figure 2A). ROCK inhibition did not alter BMP7-induced UCP1 expression. Interestingly, ROCK inhibition partially overcame the inhibitory effect of Gremlin on adipocyte gene expression (Figure 2B, compare lanes 3 and 4) and differentiation (Figure S2) while having limited effects on UCP1 expression.
Figure 2. Inhibition of ROCK or SRF activity promotes commitment to the adipogenic lineage.
(A) C3H/10T1/2 cells were treated with the indicated concentrations of Y27632 in the presence or absence of BMP7 and induced to differentiate for 8 days and extracted proteins analyzed by Western blot.
(B) C3H/10T1/2 cells were treated with Gremlin1 and/or Y27632 2 h prior to the addition of BMP7 or TGFβ, as indicated. Following adipocyte differentiation (day 8), total cellular proteins were analyzed.
(C) C3H10T1/2 cells were exposed to either BMP7, Y27632 or CCG1423 (10 μM) and actin filaments were analyzed after 2 days as in Figure 1G (left panels) or allowed to mature to adipocytes and stained with Nile Red (right panels).
(D) Subconfluent C3H10T1/2 cells were treated with either CCG1423 or BMP7 for 3 d prior to induction of differentiation for 8 d, at which stage cultures were visualized following Nile Red staining.
(E) C3H/10T1/2 cells were treated with 1 μM or 10 μM CCG1423, or BMP7 for 3 d and induced to differentiate. Total RNA was subjected to q-PCR analysis. Non-adjacent lanes on Western blots in panel A are indicated by a vertical line. See also Figure S2.
Cytoplasmic G-actin regulates SRF activity by associating with or dissociating from its coregulators, MRTFs (Posern and Treisman, 2006). When effectors activate ROCK and drive G-actin into filamentous actin (F-actin), MRTFs translocate to the nucleus and promote transcription of SRF-target genes (Olson and Nordheim, 2010). Since BMP7 inhibits ROCK, we questioned whether its pro-adipogenic activity involves suppression of MRTF/SRF activity. In fact, exposure of MSCs to CCG1423 (an inhibitor of SRF activity (Evelyn et al., 2007)) prior to stimulation of adipogenesis enhanced their conversion to adipocytes (Figure 2C and 2D) while inducing expression of brown adipocyte genes (Figure 2E). Importantly, unlike Y27632, CCG1423 did not alter the actin cytoskeleton, (Figure 2C, phalloidin stained cells) indicating that it blocks SRF downstream of F-actin polymerization.
Genetic Manipulation of MRTFA-SRF Transcriptional Activity Regulates Commitment of MSCs to an Adipogenic Lineage
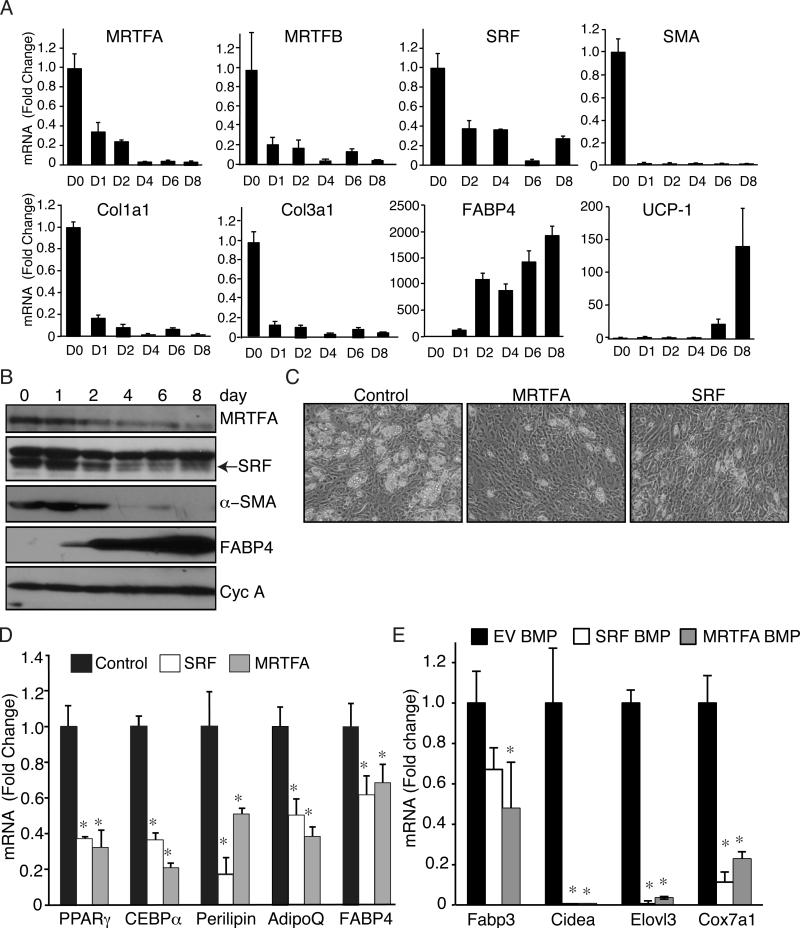
Our observation that CCG1423 could mimic BMP7 suggests that MRTF/SRF antagonizes the commitment of MSCs to an adipogenic lineage. Other studies have identified SRF as an inhibitor of adipogenesis in 3T3-L1 preadipocytes (Mikkelsen et al., 2010), but little is known about its role in regulating the fate of MSCs. Using C3H/10T1/2 MSCs, adipogenesis was accompanied by a decrease in MRTFA, MRTFB, SRF and select SRF target genes, SMA, collagen 1α1 and collagen 3α1 (Figure 3A and 3B). The lower band in Figure 3B corresponds to SRF (Figure S3A). Cell lines were generated to ectopically express SRF or MRTFA (Luchsinger et al., 2011; Wang et al., 2012a), which resulted in reduced conversion of the MSCs to adipocytes (Figure 3C) and inhibited expression of select white and brown adipogenic genes (Figure 3D and 3E). In addition, treatment with CCG1423 partially rescued the inhibitory effect of MRTFA or SRF on adipocyte formation (Figure S3B). Together, these results demonstrate that the ROCK-SRF pathway is an important BMP7 target and its repression facilitates adipogenesis.
Figure 3. MRTFA and SRF overexpression prevent differentiation of MSCs to adipocytes.
(A and B) qPCR and Western blot analysis of indicated mRNAs (A) and proteins (B) in C3H/10T1/2 MSCs undergoing brown adipogenesis.
(C) C3H/10T1/2 cell lines ectopically expressing SRF or MRTFA were induced to differentiate then visualized by phase contrast microscopy.
(D and E) The cell lines (C) were differentiated in absence (D) or presence (E) of BMP7 and isolated RNA subjected to qPCR analysis. See also Figure S3.
Increased production of beige adipocytes in WAT of MRTFA-deficient mice
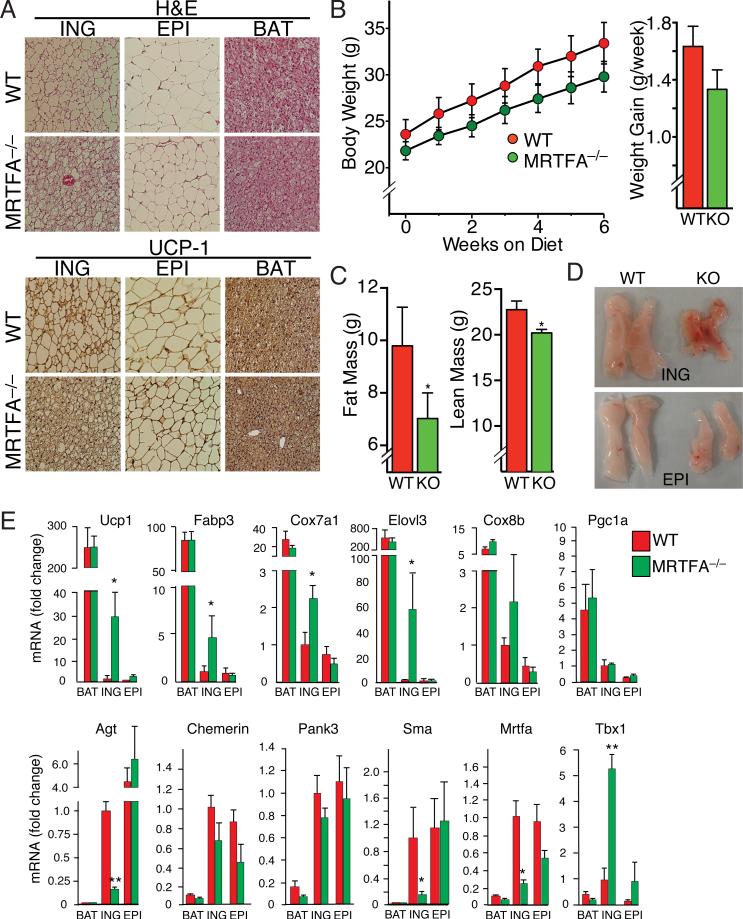
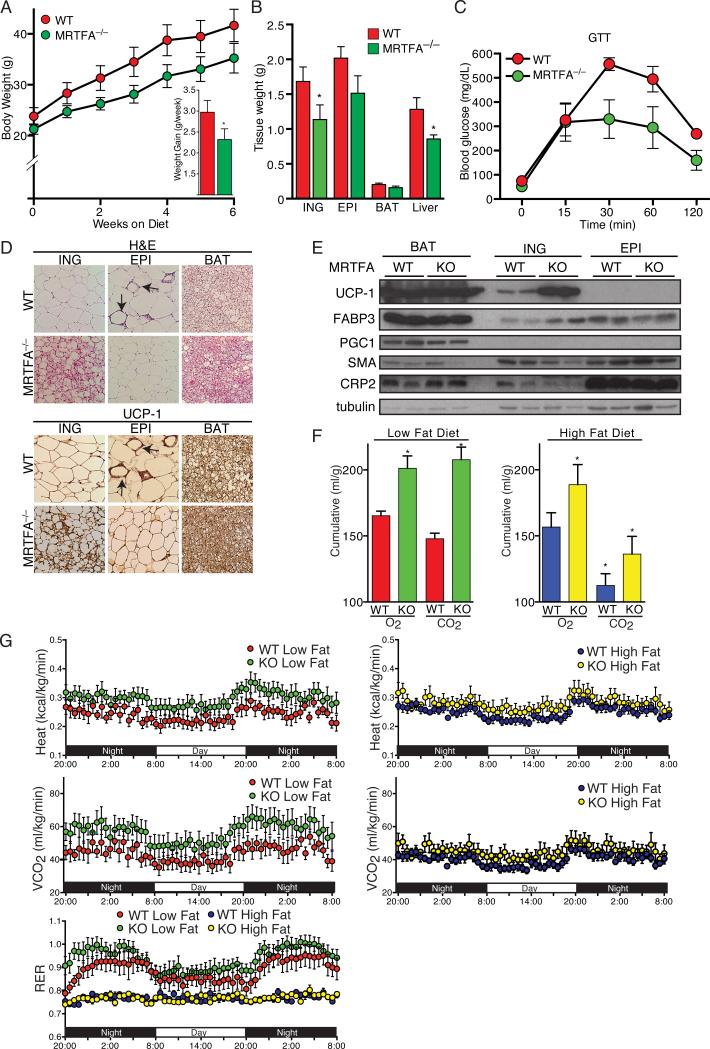
To investigate the potential role of MRTFA/SRF in adipose development in vivo, we analyzed different adipose tissues in MRTFA-deficient mice (Li et al., 2006) and observed dramatic increases in multilocular adipocytes (Figure S4B) within the subcutaneous (inguinal, iWAT) fat depot of these mice compared to their WT littermates (Figure 4A). The epididymal (eWAT) depot of the KO mouse had few multilocular cells, but the size of the adipocytes in this depot was smaller than in the corresponding depot of WT littermates. Immunohistochemical staining of multiple tissue sections demonstrated that the multilocular adipocytes in the iWAT express UCP1 in mice maintained at room temperature, suggesting that MRTFA deficiency alters an early developmental pathway (Figure 4A). We did not detect any morphological or UCP1 expression changes in the BAT of KO mice. With maturation, iWAT from WT mice underwent hypertrophy leading to an increase in the size of individual unilocular adipocytes (Figure S4A). In contrast to WT mice, the iWAT of MRTFA–/– mice consisted of more multilocular cells and the extent of hypertrophy with age was less pronounced (Figure S4A). Body weight also correlated with the differences in depot morphology. MRTFA–/– mice gained less weight during 6 weeks of low fat diet and contained less overall fat mass relative to lean mass when compared to WT littermates (Figure 4B-D and Figure S4C). MRTFA-deficient mice also had lower fasting levels of glucose than WT mice but there were no differences in glucose measured during a GTT (Figure S4D and S4E). MRTFA–/– mice also produced lower circulating levels of leptin and higher levels of adiponectin and insulin (Figure S4F).
Figure 4. MRTFA deficiency in mice enhances recruitment of beige adipocytes to the white adipose depots.
(A) H&E and UCP1 staining of representative sections (n=7) of ING (Inguinal) WAT, EPI (Epididymal) WAT and BAT from 11-13 wk-old WT and MRTFA–/– (KO) mice on rodent diet with 10% kcal% fat.
(B) Body weight and average weight gain per wk of WT and MRTFA–/– mice on rodent diet with 10% kcal% fat (n=5/group, *p<0.05).
(C) Body composition determined by NMR (described in Experimental Procedures) analysis of mice from (B) (Data presented mean ± SEM, n=5/group, *p<0.05).
(D) Gross morphology of representative ING and EPI fat pads of mice from (B).
(E) Relative mRNA levels of BAT and WAT enriched genes as well as other genes in WAT and BAT depots of 15 wk old WT and MRTFA–/– mice on chow diet was analyzed by qPCR (Data presented mean ± SEM, n=4/group, *p<0.05, **p<0.01). See also Figure S4.
RNA analysis revealed that iWAT from MRTFA–/– mice expresses 30 fold more UCP1 mRNA relative to WT littermates (Figure 4E). No significant difference in UCP1 mRNA levels in BAT of WT and KO mice were observed, though the relative amount of UCP1 mRNA in BAT was significantly higher than in iWAT of WT littermates. Expression of other brown adipocyte mRNAs (Fabp3, Cox7a1, Elovl3, and Cox8b) were similarly higher in the iWAT of MRTFA–/– mice compared to WT littermates. Our previous studies identified genes that are abundantly expressed in WAT relative to BAT (Vernochet et al., 2009). Importantly, one of these genes, angiotensinogen (Agt) is expressed at much lower levels in MRTFA–/– iWAT relative to WT iWAT (Figure 4E), consistent with the enhanced development of brown-like adipocytes within iWAT of KO mice. Expression of SMA was also reduced in MRTFA–/– iWAT compared to the WT depot. The transcription factor Tbx1 has recently been assigned as a marker of beige adipocytes (Wu et al., 2012). Importantly, Tbx1 expression was significantly higher in MRTFA–/– iWAT compared to WT iWAT (Figure 4E) suggesting that the appearance of UCP1 positive, multilocular adipocytes in MRTFA–/– mice is likely due to recruitment of beige progenitors.
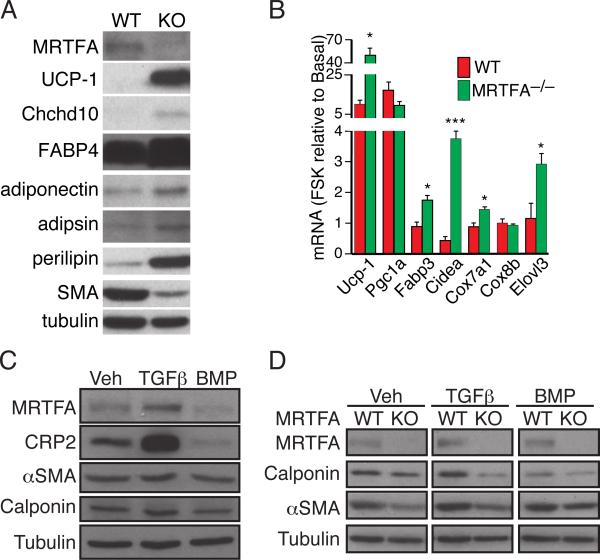
Stromal vascular cells (SVF) from MRTFA–/– iWAT undergo beige adipogenesis more extensively than WT SVF cells
Because the mice used in this study were a total body knockout, we examined whether MRTFA represses beige adipocyte formation directly. SVF cells of WT and KO iWAT differentiated into adipocytes in vitro to approximately the same extent as judged by comparable expression of adiponectin, adipsin, and FABP4 (Figure 5A). MRTFA–/– SVFs, however, expressed higher amounts of select BAT proteins including UCP1 and Chchd10/NDG2 (Figure 5A). Treatment of the adipocytes with forskolin (FSK) for 4 h dramatically induced expression of UCP1 mRNA in both WT and KO cells when normalized to basal untreated levels (Figure 5B). The FSK-dependent induction of UCP1, Fabp3, Cidea, Cox7a1 and Elovl3 were significantly higher in KO versus WT adipocytes (Figure 5B). The induction of PGC-1α mRNA, however, was similar in both WT and KO cells. A recent study suggested that some beige adipocytes have a smooth muscle-like origin; therefore, we analyzed expression of SM genes during early adipogenesis in SVF cells (Long et al., 2014). Treatment of cells with BMP7 for 48 h resulted in a significant decrease in MRTFA, CRP2, SMA and calponin expression (Figure 5C). In contrast, TGFβ1 enhanced SM protein expression (Figure 5C and 5D). In addition, the KO SVF cells expressed significantly lower amounts of the SM proteins under all conditions compared to WT cells (Figure 5D). These data suggest that BMP7 directs SVF progenitors to a beige adipocyte lineage rather than a SM lineage.
Figure 5. Cells from the stromal vascular fraction (SVF) of MRTFA–/– ING WAT express less smooth muscle-like features and undergo brown adipogenesis more extensively than WT SVF cells.
(A) Western blot analysis on BAT and WAT enriched proteins of adipocytes arising from hormonal induction of SVCs isolated from WT and MRTFA–/– inguinal depots.
(B) WT and MRTFA–/– SVC adipocytes were treated with or without forskolin (FSK) for 4 h prior to isolation of total RNA for analysis. Values are presented as fold change in response to treatment with FSK in WT and MRTFA–/– adipocytes.
(C and D) Subconfluent SVCs from ING and EPI WAT of WT and/or MRTFA–/– mice were exposed to TGFβ1 or BMP7 for 2 d before reaching confluence and subjected to Western blot analysis.
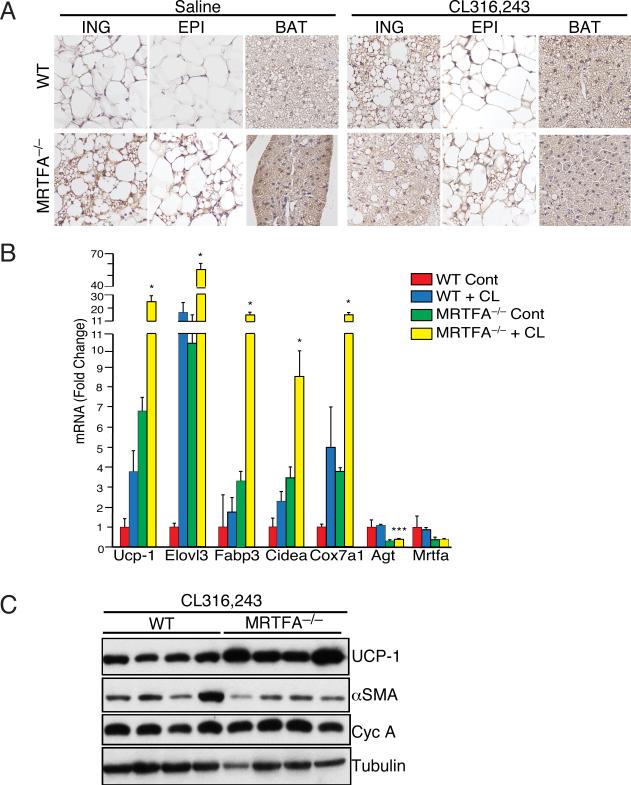
Treatment of mice with a β-adrenoceptor agonist such as CL316,243 results in “browning” of iWAT characterized by enhanced UCP1 expression and appearance of beige/brite adipocytes (Cousin et al., 1992) (Barbatelli et al., 2010) (Wu et al., 2012). We questioned, therefore, whether MRTFA–/– mice respond differently to β-adrenergic stimulation than WT counterparts. Consistent with the earlier observations (Cousin et al., 1992), we show that WT mice induce browning of WAT depots based on more intense UCP1 staining in response to CL316,243 (Figure 6A). Importantly, MRTFA–/– mice exhibited an enhanced response to CL316,243 than their WT littermates by producing larger regions of UCP1 positive multilocular adipocytes in iWAT (Figure 6A) and expressed higher amounts of select brown genes UCP1, Elovl3, Fabp3, Cidea, and Cox7a1 (Figure 6B) and UCP1 protein (Figure 6C).
Figure 6. MRTFA–/– mice are highly responsive to β-adrenergic signaling, which enhances beige adipocyte formation in ING WAT.
(A) UCP1 immunohistochemistry of representative sections of BAT, ING and EPI depots of WT and MRTFA–/– mice following daily intraperitoneal injections of 1mg/kg CL316,243 or saline for 7 d.
(B and C) mRNA and protein levels of BAT and WAT enriched genes as well as other genes in ING WAT of WT and MRTFA–/– mice treated with or without CL316,243 were determined by qPCR (B) and Western blot (C), respectively. Data presented mean ± SEM, n=4 *p<0.05, **p<0.01.
MRTFA-deficient mice are protected from diet-induced obesity and insulin resistance
Mice were fed a high fat diet (HFD, fat 60 kcal%, 20 kcal% carbohydrate, protein 20 kcal%) for 6 weeks starting at 6 weeks of age. MRTFA–/– mice gained less weight on the HFD compared to the WT littermates (Figure 7A). Body composition analysis revealed that KO mice gained less fat mass without any major change in lean mass or food intake (Figure S5B). The iWAT and eWAT of KO mice weighed less than corresponding depots in control littermates while there was no significant change in weight of BAT (Figure 7B). The weight of the liver in KO mice fed a HFD for 6 weeks was much lower than in WT littermates (Figure 7B). This difference in weight was likely due to the extensive accumulation of hepatic lipid in WT mice (Figure S5C). MRTFA–/– mice had significantly lower levels of fasting glucose and exhibited enhanced glucose tolerance compared to WT littermates (Figure S5A and 7C). Additionally, KO mice had lower circulating levels of leptin (Figure S5D) consistent with them having less fat as well as higher circulating levels of adiponectin (Figure S5D). The high fat diet also resulted in hypertrophy of individual adipocytes in the iWAT and eWAT of WT mice (Figure 7D). The corresponding depots in the KO mice showed no sign of excess lipid deposition and the iWAT still produced abundant amounts of UCP1 as well as other brown genes (Figure 7D and 7E). Importantly, KO mice were protected from diet-induced inflammation as observed by a reduction in crown-like structures in the eWAT when compared to WT littermates (Figure 7D). Taken together, these results indicate that MRTFA deficiency reduces HFD-induced obesity, insulin resistance, eWAT inflammation and hepatic steatosis.
Figure 7. MRTFA-deficient mice are protected from diet-induced obesity and its associated insulin resistance and exhibit altered whole body energy expenditure.
(A-B) WT and MRTFA–/– mice, starting from 4-5 wk old, were fed a rodent HFD (60% kcal% fat) for 6 wk. Body weight and weight gain were measured in weekly intervals (A), while weights of ING WAT, EPI WAT, BAT and liver were measured at end of study (B).
(C) After 5 wk on the HFD, mice were then IP injected with glucose at 2 mg/kg of body weight after fasting overnight and whole blood glucose was measured at 15, 30, 60 and 120 min.
(D) H&E and UCP1 staining of sections of ING, EPI and BAT depots.
(E) Western blot analysis of BAT and WAT enriched genes as well as other genes in WAT and BAT depots of WT and MRTFA–/– mice were conducted as in Figure 4. Arrows point to crown-like structures. Data presented are mean ± SEM, n=6-7/group *p<0.05, **p<0.01.
(F-G) WT and MRTFA–/– mice on LFD or HFD for 6 wk were housed individually in metabolic chambers for 3 days and 2 nights. Cumulative O2 consumption and CO2 production were measured by CLAMs (F) Traces of heat production, CO2 production and RER during 12 h dark and light cycles (G) RER (respiratory exchange ratio) were calculated by volume of carbon dioxide produced (exhaled)/volume of oxygen consumed (inhaled). Data presented are mean ± SEM, n=5-7/group, *p<0.05. See also Figures S5 and S6.
MRTFA Regulates Whole Body Energy Expenditure
Indirect calorimetry analysis of mice following 6 weeks on high or low fat diets demonstrated that KO mice produced more heat, consumed more oxygen and carbon dioxide (Figure 7F and 7G) without any change in food intake or physical activity compared to WT animals (Figure S6). Measurement of RER (respiratory exchange ratio) following the 6 week on LFD revealed that KO mice expended significantly larger amounts of carbohydrate than WT mice during the dark period with both sets of mice switching equally to fat expenditure during the day light (Figure 7G). As expected, KO and WT mice metabolized predominantly fat throughout the entire night-day-night period on HFD (Figure 7G).
Discussion
There is an urgent need for effective and safe therapeutics to address the complications arising from excessive adipose tissue expansion. Most anti-obesity drugs presently available have serious side effects including depression and gastrointestinal maladies (Kang and Park, 2012). Similarly, several of the drugs that treat insulin resistance, type 2 diabetes and cardiovascular disease, either have life threatening side effects or are minimally effective (Ahmadian et al., 2013; Stein et al., 2013). With the discovery that humans possess metabolically active brown adipose tissue (van der Lans et al., 2013), it has been suggested that increasing BAT mass and/or activity might be an effective anti-obesity therapy (Farmer, 2009). We report here on the identification of a novel-signaling and transcriptional regulatory pathway that, when inhibited in mice, leads to “beigeing” of iWAT and protection against diet-induced obesity and insulin resistance.
Various aspects of cell morphology control adipocyte differentiation (Rodriguez Fernandez and Ben-Ze'ev, 1989; Spiegelman and Farmer, 1982; Spiegelman and Ginty, 1983). It has recently been shown that cell shape regulates commitment of human MSCs to an adipocyte or osteoblast fate via modulation of RhoA signaling (McBeath et al., 2004). Specifically, dominant-negative RhoA committed hMSCs to the adipogenic lineage, while constitutively active RhoA favored the osteogenic lineage. Our work demonstrates a role for RhoA signaling in controlling the fate of MSCs to an adipogenic versus smooth muscle-like lineage. Furthermore, we demonstrate that BMP7, in contrast to TGFβ suppresses ROCK and MRTFA/SRF activity, facilitating commitment to the adipocyte lineage. The mechanisms by which TGFβ family members control ROCK are still poorly understood. In other developmental systems, BMPs activate ROCK and BMP2 for example stimulates osteogenesis in hMSCs via ROCK-activated changes in cytoskeletal tension (Wang et al., 2012c). In fact, our report is one of very few documenting a BMP-associated suppression of the ROCK-actin-MRTFA signaling pathway. One potential mechanism for the suppression is a BMP-mediated inhibition of basal TGFβ1 activity, as recently reported for control of cardiac fibrosis (Wang et al., 2012b).
Modulation of MRTFA in MSCs highlights its role in regulating BMP7-induced adipocyte formation, but this doesn't necessarily exclude the participation of other actin-responsive transcription factors, including MRTFB. The inhibitory action of MRTFA appears to be mediated by its coactivation of SRF since ectopic expression of SRF also attenuates the adipogenic action of BMP7. Studies by Rosen and coworkers showed that SRF inhibited the differentiation of 3T3-L1 preadipocytes (Mikkelsen et al., 2010) and recently others demonstrated that MKL1 (MRTFA) blocks 3T3-L1 adipogenesis at a step upstream of PPARγ (Nobusue et al., 2014). The precise mechanism by which SRF controls adipogenic commitment might not involve a single target gene/protein but expression of an entire program of genes coding for extracellular matrix and cytoskeleton. Such a process would facilitate extensive cell spreading, which is known to attenuate adipocyte formation (Spiegelman and Ginty, 1983).
The appearance of UCP1+ adipocytes in iWAT of KO mice suggests a role for suppression of MRTF/SRF activity in the development of beige adipocytes. Other studies have demonstrated that disruption of TGFβ signaling in mice similarly enhances beige adipocyte formation in WAT (Yadav et al., 2011). The MRTFA-associated browning of iWAT likely arises through recruitment of beige progenitors, as opposed to transdifferentiation of mature white adipocytes (Barbatelli et al., 2010; Wu et al., 2012; Wu et al., 2013). In fact, iWAT of KO mice express high levels of Tbx1 (Figure 4E), supporting a mechanism of de novo generation of beige adipocytes.
Formation of beige adipocytes could arise from crosstalk between adipose and other organs since MRTFA is absent in all tissues of the knockout mouse (Li et al., 2006). Interestingly, recent studies have shown a role for BMP signaling in regulating adipose formation through central as well as peripheral actions (Townsend et al., 2012; Whittle et al., 2012). The central actions of the BMPs, however, involve pathways other than the ROCK-MRTFA axis (Townsend et al., 2012; Whittle et al., 2012). Consistent with the action of MRTFA being an autonomous process, cells derived from the SVF of KO iWAT differentiate to beige adipocytes more robustly than WT cells (Figure 5). This could be due to expansion of a pool of beige progenitors in the vascular compartment of MRTFA–/– iWAT from various sources including pericytes and/or endothelial cells. In fact, a small set of capillary endothelial cells express the adipogenic commitment factor, ZFP423 suggesting a contribution of specialized endothelial cells to the adipose lineage (Gupta et al., 2012). Furthermore, lineage tracing studies identified vascular endothelial cells as progenitors of both white and brown adipocytes (Tran et al., 2012). Such endothelial progenitors would need to undergo endothelial-mesenchymal transition (End-MT) prior to further adipogenic differentiation. In fact, BMP/TGFβ signaling has been linked to End-MT (Maddaluno et al., 2013; Zeisberg et al., 2007), raising the possibility that MRTFA could be functioning at such an early stage of adipogenic lineage commitment.
Until recently, it was difficult to establish a role for beige adipocytes in controlling energy expenditure (EE) from the dominant role played by BAT. In fact, Cannon and Nedergaard have argued that brite/beige adipocytes have a minimal contribution to EE since the level of UCP1 expression in these cells is too low (Shabalina et al., 2013). Spiegelman and coworkers have demonstrated, that absence of beige adipocyte development in mice leads to DIO and insulin resistance without any detectable change in BAT development or activity; highlighting the potential importance of beige adipose in energy balance (Cohen et al., 2014). Our findings support the concept that beige adipocytes perform a physiological role since lack of MRTFA facilitated beige cell development without any significant effect on BAT activity based on minimal change in BAT mass, morphology or UCP1 expression (Figure 4).
Our studies have uncovered a novel regulatory pathway that attenuates the anti-adipogenic activity of MRTFA/SRF leading preferentially to beige adipocyte formation in iWAT. We suggest that this pathway directs the commitment of adipose stroma progenitors to beige adipocyte over vascular lineages. Small molecules that suppress MRTFA/SRF activity might lead to the development of effective therapeutics to combat obesity-associated comorbidities.
Experimental Procedures
Animals
MRTFA+/– and WT mice (mixed C57BL/6J 129 genetic background) were a gift from Dr. Eric Olson, UT Southwestern Medical Center (Li et al., 2006) and housed at 23°C in a 12 h light/dark cycle with free access to normal chow. All experiments used matched littermates. Experimental procedures were approved by the Boston University Institutional Animal Care and Use Committee.
High Fat Feeding and β-adrenergic stimulation
For diet studies, 4-6 week-old male WT and MRTFA–/– mice were fed a diet with 10% kcal% fat (low fat diet, Research Diets Inc, D12450B) or diet with 60% kcal% fat (high fat diet, Research Diets Inc, D12492) for 6 weeks. For β-adrenergic stimulation, 14 week old male WT and MRTFA–/– mice were IP injected daily for 7 d with 1 mg/kg CL316,243 (Sigma-Aldrich) or vehicle.
Histology
Fat pads were fixed in 10% formalin, paraffin-embedded, and sectioned (7 μm) prior to hematoxylin and eosin (H&E) staining or immunohistochemistry for UCP1 (Abcam; 1:100). Signal was detected using the Vector ABC Elite kit.
Indirect Calorimetry and Body composition (BUMC Metabolic Phenotyping Core)
Animals were individually housed in metabolic chambers maintained at 20-24°C on a 12-h light/12 h dark cycle. Heat, VO2, VCO2 and RER were measured continuously using CLAMS consisting of open circuit calorimeter & motion detectors. Body composition was measured by non-invasive quantitative MRI (EchoMRI700).
Glucose Tolerance Tests
Mice were fasted for 14 h then blood glucose was measured (Bayer Contour) by tail bleeding at 0, 15, 30, 60, and 120 minutes after an injection of glucose (2 mg/kg body weight).
Serum Leptin and Adiponectin
Mice were euthanized, and serum, obtained from heart blood using serum separation tubes (BD), was analyzed using mouse leptin (Millipore EMD), and total and HMW adiponectin mouse adiponectin (ALPCO) ELISA kits.
Adipose Tissue Fractionation and Culture of Stromal Vascular Cells
Inguinal fat pads were dissected from male mice, digested in collagenase, filtered through a 100 μm mesh and centrifuged at 500 xg to isolate stromal vascular cells for differentiation as outlined for C3H/10T1/2 cells stated below.
Cell Culture
Pre-confluent C3H/10T1/2 cells (ATCC) were treated with 6.3 nM rhBMP7 or 1 nM rhTGFβ1 (R&D Systems) for 3 d. The confluent cells were induced to differentiate in 10% FBS, 5 μM dexamethasone, 0.5 mM isobutylmethylxanthine, 860 nM insulin, 1 nM 3,3,5-triiodo-L-thyronine (T3), and 125 μM indomethacin. Two d after induction, cells were maintained in 10% FBS, insulin, and T3 for 6d. Cells were treated with recombinant Gremlin1 (R&D), and the small molecule inhibitors Y27632, PD169316, SB341542 (Sigma), DMH1 (Tocris), or CCG1423 (Cayman) as described in Figure legends. Mouse bone marrow derived MSCs were obtained from Life Technologies.
Plasmids and Viruses
MRTFA and SRF cDNAs (Chang et al., 2003; Luchsinger et al., 2011) were sub-cloned into the pMSCV retroviral vector and packaged using EcoPack cells (Clontech). Cells were incubated with viral supernatants supplemented with 10 μg/mL polybrene and selected with hygromycin.
Gene Expression Analysis
Total RNA was isolated from cells or tissues using TRIzol reagent (Life Technologies). Gene expression was performed using an Applied Biosystems cDNA kit and Maxima SYBR Green qPCR Master Mix (Fermentas) as described (Pino et al. 2012). Primer sequences are provided in Supplemental Table 1.
Western blot analysis
Total cellular protein was extracted and subjected to western blot analysis as described (Vernochet et al., 2009) using antibodies listed in Supplemental Table 2.
Oil Red O and Nile Red Staining
Oil Red O staining was as described (Vernochet et al., 2010). Cover slips were stained with Nile Red for 20 min at room temperature.
Phalloidin/DNAse I staining
Cells on gelatin-coated cover slips were fixed in formaldehyde, permeabilized and stained in Alexa Fluor 488 Phalloidin and/or Deoxyribonuclease I Alexa Fluor® 594 (Molecular Probes).
G-actin/Factin Assay
Relative G-actin/F-actin ratio was assessed using the G-actin/F-actin assay kit (Cytoskeleton Inc.) according to the manufacturer's directions with minor modifications.
Statistical Analysis
Results are presented as mean ± SEM. Statistical differences were determined by a Student two tailed t test with equal variance or paired. Significance was considered as p ≤ 0.05.
Supplementary Material
Acknowledgements
We are grateful to Eric Olson, UT Southwestern Medical Center, TX for the MRTFA+/– mice. We thank Tom Balon of the Mouse Phenotyping Core for his advice and assistance. This work was supported by NIH grants DK051586, DK098830 (SRF), and HL078869, and BNORC pilot grant P30 DK046200 (MDL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
S.R.F., M.D.L., M.E.M. and C.L. conceived of the project. S.R.F, M.D.L., M.E.M, C.L. and H.B. designed the experiments. M.E.M. performed the experiments in Figure 1-2. C.L. performed the experiments in Figure 4-7. H.B. performed the experiments in Figure 3. B.D.S. provided scientific and technical advice. S.R.F., M.E.M. and M.D.L wrote the manuscript. All authors edited the manuscript.
References
- Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. PPARγ signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M, Ankenbauer T, Schroder D, Hollnagel A, Mayer H, Gross G. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol. 1993;12:871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. β-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, Giacobino JP, De Matteis R, Cinti S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab. 2010;298:E1244–1253. doi: 10.1152/ajpendo.00600.2009. [DOI] [PubMed] [Google Scholar]
- Bordicchia M, Liu D, Amri EZ, Ailhaud G, Dessi-Fulgheri P, Zhang C, Takahashi N, Sarzani R, Collins S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. 2012;122:1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers RR, Lane MD. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle. 2007;6:385–389. doi: 10.4161/cc.6.4.3804. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chang YF, Wei J, Liu X, Chen YH, Layne MD, Yet SF. Identification of a CArG-independent region of the cysteine-rich protein 2 promoter that directs expression in the developing vasculature. Am J Physiol Heart Circ Physiol. 2003;285:H1675–1683. doi: 10.1152/ajpheart.00165.2003. [DOI] [PubMed] [Google Scholar]
- Choy L, Derynck R. Transforming growth factor-β inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J Biol Chem. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Daniel KW, Petro AE, Surwit RS. Strain-specific response to β 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology. 1997;138:405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- Cousin B, Bascands-Viguerie N, Kassis N, Nibbelink M, Ambid L, Casteilla L, Penicaud L. Cellular changes during cold acclimatation in adipose tissues. J Cell Physiol. 1996;167:285–289. doi: 10.1002/(SICI)1097-4652(199605)167:2<285::AID-JCP12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- Enerback S. The origins of brown adipose tissue. N Engl J Med. 2009;360:2021–2023. doi: 10.1056/NEJMcibr0809610. [DOI] [PubMed] [Google Scholar]
- Evelyn CR, Wade SM, Wang Q, Wu M, Iniguez-Lluhi JA, Merajver SD, Neubig RR. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6:2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Obesity: Be cool, lose weight. Nature. 2009;458:839–840. doi: 10.1038/458839a. [DOI] [PubMed] [Google Scholar]
- Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, et al. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani M, Himms-Hagen J. Appearance of brown adipocytes in white adipose tissue during CL 316,243-induced reversal of obesity and diabetes in Zucker fa/fa rats. Int J Obes Relat Metab Disord. 1997;21:465–475. doi: 10.1038/sj.ijo.0800432. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Li P, Zhu Z, Lu Y. Metabolic and cellular plasticity in white adipose tissue I: effects of β3-adrenergic receptor activation. Am J Physiol Endocrinol Metab. 2005;289:E608–616. doi: 10.1152/ajpendo.00009.2005. [DOI] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J Clin Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautmann MB, Madsen CS, Owens GK. A transforming growth factor β (TGFβ) control element drives TGFβ-induced stimulation of smooth muscle α-actin gene expression in concert with two CArG elements. J Biol Chem. 1997;272:10948–10956. doi: 10.1074/jbc.272.16.10948. [DOI] [PubMed] [Google Scholar]
- Himms-Hagen J, Melnyk A, Zingaretti MC, Ceresi E, Barbatelli G, Cinti S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am J Physiol Cell Physiol. 2000;279:C670–681. doi: 10.1152/ajpcell.2000.279.3.C670. [DOI] [PubMed] [Google Scholar]
- Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson L, van Meeteren LA. Transforming growth factor β family members in regulation of vascular function: in the light of vascular conditional knockouts. Exp Cell Res. 2013;319:1264–1270. doi: 10.1016/j.yexcr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Jimenez M, Barbatelli G, Allevi R, Cinti S, Seydoux J, Giacobino JP, Muzzin P, Preitner F. β 3-adrenoceptor knockout in C57BL/6J mice depresses the occurrence of brown adipocytes in white fat. Eur J Biochem. 2003;270:699–705. doi: 10.1046/j.1432-1033.2003.03422.x. [DOI] [PubMed] [Google Scholar]
- Kang JG, Park CY. Anti-Obesity Drugs: A Review about Their Effects and Safety. Diabetes Metab J. 2012;36:13–25. doi: 10.4093/dmj.2012.36.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bowens N, Cheng L, Zhu X, Chen M, Hannenhalli S, Cappola TP, Parmacek MS. Myocardin-like protein 2 regulates TGFβ signaling in embryonic stem cells and the developing vasculature. Development. 2012;139:3531–3542. doi: 10.1242/dev.082222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncar D. Convertible adipose tissue in mice. Cell Tissue Res. 1991a;266:149–161. doi: 10.1007/BF00678721. [DOI] [PubMed] [Google Scholar]
- Loncar D. Development of thermogenic adipose tissue. Int J Dev Biol. 1991b;35:321–333. [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger LL, Patenaude CA, Smith BD, Layne MD. Myocardin- related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J Biol Chem. 2011;286:44116–44125. doi: 10.1074/jbc.M111.276931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Dynamic control of TGF-β signaling and its links to the cytoskeleton. FEBS Lett. 2008;582:2051–2065. doi: 10.1016/j.febslet.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Nagase I, Yoshida T, Kumamoto K, Umekawa T, Sakane N, Nikami H, Kawada T, Saito M. Expression of uncoupling protein in skeletal muscle and white fat of obese mice treated with thermogenic β 3-adrenergic agonist. J Clin Invest. 1996;97:2898–2904. doi: 10.1172/JCI118748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Ricquier D, Kozak LP. Uncoupling proteins: current status and therapeutic prospects. EMBO Rep. 2005;6:917–921. doi: 10.1038/sj.embor.7400532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H, Kano K. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat Commun. 2014;5:3368. doi: 10.1038/ncomms4368. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-Hett S, D'Amore PA. Signal transduction in vasculogenesis and developmental angiogenesis. Int J Dev Biol. 2011;55:353–363. doi: 10.1387/ijdb.103213sp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino E, Wang H, McDonald ME, Qiang L, Farmer SR. Roles for peroxisome proliferator-activated receptor γ (PPARγ) and PPARγ coactivators 1α and 1β in regulating response of white and brown adipocytes to hypoxia. J Biol Chem. 2012;287:18351–18358. doi: 10.1074/jbc.M112.350918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Qian SW, Tang Y, Li X, Liu Y, Zhang YY, Huang HY, Xue RD, Yu HY, Guo L, Gao HD, et al. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc Natl Acad Sci U S A. 2013;110:E798–807. doi: 10.1073/pnas.1215236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rodriguez Fernandez JL, Ben-Ze'ev A. Regulation of fibronectin, integrin and cytoskeleton expression in differentiating adipocytes: inhibition by extracellular matrix and polylysine. Differentiation. 1989;42:65–74. doi: 10.1111/j.1432-0436.1989.tb00608.x. [DOI] [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- Schofield AV, Bernard O. Rho-associated coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem Mol Biol. 2013;48:301–316. doi: 10.3109/10409238.2013.786671. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Tseng YH. Emerging role of bone morphogenetic proteins in adipogenesis and energy metabolism. Cytokine Growth Factor Rev. 2009;20:523–531. doi: 10.1016/j.cytogfr.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina IG, Petrovic N, de Jong JM, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013;5:1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Sinha S, Hoofnagle MH, Kingston PA, McCanna ME, Owens GK. Transforming growth factor-β1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am J Physiol Cell Physiol. 2004;287:C1560–1568. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- Sordella R, Jiang W, Chen GC, Curto M, Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell. 2003;113:147–158. doi: 10.1016/s0092-8674(03)00271-x. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Farmer SR. Decreases in tubulin and actin gene expression prior to morphological differentiation of 3T3 adipocytes. Cell. 1982;29:53–60. doi: 10.1016/0092-8674(82)90089-7. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Ginty CA. Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes. Cell. 1983;35:657–666. doi: 10.1016/0092-8674(83)90098-3. [DOI] [PubMed] [Google Scholar]
- Stein SA, Lamos EM, Davis SN. A review of the efficacy and safety of oral antidiabetic drugs. Expert Opin Drug Saf. 2013;12:153–175. doi: 10.1517/14740338.2013.752813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101:9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend KL, Suzuki R, Huang TL, Jing E, Schulz TJ, Lee K, Taniguchi CM, Espinoza DO, McDougall LE, Zhang H, et al. Bone morphogenetic protein 7 (BMP7) reverses obesity and regulates appetite through a central mTOR pathway. FASEB J. 2012;26:2187–2196. doi: 10.1096/fj.11-199067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KV, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, Smorlesi A, Perugini J, De Matteis R, Sbarbati A, et al. The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab. 2012;15:222–229. doi: 10.1016/j.cmet.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lans AA, Hoeks J, Brans B, Vijgen GH, Visser MG, Vosselman MJ, Hansen J, Jorgensen JA, Wu J, Mottaghy FM, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J Clin Invest. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C, Davis KE, Scherer PE, Farmer SR. Mechanisms regulating repression of haptoglobin production by peroxisome proliferator-activated receptor-γ ligands in adipocytes. Endocrinology. 2010;151:586–594. doi: 10.1210/en.2009-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C, Peres SB, Davis KE, McDonald ME, Qiang L, Wang H, Scherer PE, Farmer SR. C/EBPα and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor γagonists. Mol Cell Biol. 2009;29:4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yin S, Cen L, Liu Q, Liu W, Cao Y, Cui L. Differentiation of adipose-derived stem cells into contractile smooth muscle cells induced by transforming growth factor-β1 and bone morphogenetic protein-4. Tissue Eng Part A. 2010;16:1201–1213. doi: 10.1089/ten.TEA.2009.0303. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Yang KD, Ko JY, Huang CC, Huang HY, Wang FS. The effects of shockwave on bone healing and systemic concentrations of nitric oxide (NO), TGF-β1, VEGF and BMP-2 in long bone non-unions. Nitric Oxide. 2009;20:298–303. doi: 10.1016/j.niox.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Wang D, Prakash J, Nguyen P, Davis-Dusenbery BN, Hill NS, Layne MD, Hata A, Lagna G. Bone morphogenetic protein signaling in vascular disease: anti-inflammatory action through myocardin-related transcription factor A. J Biol Chem. 2012a;287:28067–28077. doi: 10.1074/jbc.M112.379487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang EA, Israel DI, Kelly S, Luxenberg DP. Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors. 1993;9:57–71. doi: 10.3109/08977199308991582. [DOI] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun A, Li L, Zhao G, Jia J, Wang K, Ge J, Zou Y. Up-regulation of BMP-2 antagonizes TGF-β1/ROCK-enhanced cardiac fibrotic signalling through activation of Smurf1/Smad6 complex. J Cell Mol Med. 2012b;16:2301–2310. doi: 10.1111/j.1582-4934.2012.01538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YK, Yu X, Cohen DM, Wozniak MA, Yang MT, Gao L, Eyckmans J, Chen CS. Bone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tension. Stem Cells Dev. 2012c;21:1176–1186. doi: 10.1089/scd.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle AJ, Carobbio S, Martins L, Slawik M, Hondares E, Vazquez MJ, Morgan D, Csikasz RI, Gallego R, Rodriguez-Cuenca S, et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell. 2012;149:871–885. doi: 10.1016/j.cell.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Li Y, Choi JH, Giang A-H, Khandekar MJ, Nuutila P, Schaart G, Huang K, et al. Beige Adipocytes are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Mol Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav H, Quijano C, Kamaraju AK, Gavrilova O, Malek R, Chen W, Zerfas P, Zhigang D, Wright EC, Stuelten C, et al. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, Nedergaard J, Cinti S. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.