Summary
Enhanced amino acid catabolism is a common response to inflammation, but the immunologic significance of altered amino acid consumption remains unclear. The finding that tryptophan catabolism helped maintain fetal tolerance during pregnancy provided novel insights into the significance of amino acid metabolism in controlling immunity. Recent advances in identifying molecular pathways that enhance amino acid catabolism and downstream mechanisms that affect immune cells in response to inflammatory cues support the notion that amino acid catabolism regulates innate and adaptive immune cells in pathologic settings. Cells expressing enzymes that degrade amino acids modulate antigen-presenting cell and lymphocyte functions and reveal critical roles for amino acid- and catabolite-sensing pathways in controlling gene expression, functions, and survival of immune cells. Basal amino acid catabolism may contribute to immune homeostasis that prevents autoimmunity, whereas elevated amino acid catalytic activity may reinforce immune suppression to promote tumorigenesis and persistence of some pathogens that cause chronic infections. For these reasons, there is considerable interest in generating novel drugs that inhibit or induce amino acid consumption and target downstream molecular pathways that control immunity. In this review, we summarize recent developments and highlight novel concepts and key outstanding questions in this active research field.
Keywords: IDO, eif2ak4, D-1MT, AHR, mTOR, tolerance
Introduction
Nutrient availability is a major determinant of cell behavior that can override other environmental stimuli when nutrients are in short supply. Free-living microorganisms acquire nutrients (e.g. amino acids, carbohydrates, salts, co-factors, etc.) directly from their microenvironments, and they can sense and respond rapidly to changes in nutrient availability. In multicellular organisms the situation is more complicated involving direct sensing of nutrient levels in tissues and blood, distal influences (e.g. hormonal) that modulate cell metabolism, and centralized control of dietary intake (e.g. in liver) that affects systemic nutrient pools. In each setting, cells must respond rapidly to changes in environmental and intracellular nutrient availability by fine tuning cellular metabolism and to avoid cell death caused by energy deficits and reduced ability to make new macromolecules when cells prepare to divide or differentiate. Cells can synthesize some of the 20 amino acids required to make proteins from other compounds. Higher vertebrates cannot make nine ‘essential’ amino acids (ILKMFTWVH), and dietary intake or protein salvage pathways are the only sources of these amino acids. Tryptophan (Trp) synthesis requires more energy than any other amino acid; this may explain why some microorganisms that can make Trp only do so using a set of inducible genes (e.g. the Trp operon) when Trp is unavailable in local microenvironments and why Trp is the least frequent amino acid found in proteins. Trp is also the substrate for serotonin synthesis in higher vertebrates, providing a rationale for research on neurologic and behavioral effects of altered Trp metabolism. Enhanced Trp catabolism in settings of chronic inflammation correlates with neurologic dysfunction and aberrant behaviors suggesting that altered Trp metabolism may be a common underlying cause of immunologic and neurologic dysfunction in some chronic inflammatory disease syndromes.
Like all cells, immune cells must have continuous access to amino acids to maintain basal metabolism and remain viable. When immune cells are activated by inflammatory and antigenic cues, their demand for amino acids increases rapidly. Limited access to amino acids during immune cell activation may compromise immune responses by inhibiting immune cell division, differentiation, maturation, migration, and acquisition of new effector functions. Although amino acid deprivation per se may attenuate immune responses under some conditions, cells also possess amino acid-sensing pathways that trigger profound changes in cell metabolism when activated by reduced levels of amino acids. Such ‘stress response’ pathways provide sensitive mechanisms to control how immune cells respond under conditions of amino acid depletion. This rationale may explain how amino acid-sensing pathways present in all cell types provide operational specificity as increased demand for amino acids occurs only when immune cells respond to inflammatory and antigenic signals. Altered amino acid metabolism can also influence immunologic responses to inflammatory and antigenic cues by generating new compounds, amino acid catabolites, with immune modulatory properties.
In this review, we focus on pathways that sense and catabolize amino acids and affect immunologic outcomes relevant to clinical syndromes. Initially, we describe key pathways and how these pathways alter cellular functions. Subsequently, we discuss how these pathways modulate immune cell functions in response to inflammatory and antigenic cues and in distinct settings of clinical relevance. Finally, we consider how therapeutic manipulations that target amino acid catabolism may be used to influence immunologic outcomes for clinical benefit. More is known about the control of immunity by Trp catabolism than for any other amino acid and this article reflects this bias. Our primary goal is to examine the effects of altered amino acid metabolism on immune responses using Trp catabolism as a well-documented example of this phenomenon. Control of access to other amino acids may also be relevant to immune regulation, particularly with regard to microorganisms that possess a range of enzymes that higher vertebrates do not possess to synthesize and catabolize amino acids. These aspects of amino acid metabolism are beyond the scope of this article.
Pathways of amino acid catabolism
Although all amino acids can be catabolized into simpler compounds or serve as substrates to make other molecules, not all cell types express enzymes that catalyze such changes, especially in response to inflammation. Higher vertebrates possess three distinct hemoprotein enzymes that catabolize Trp, two isoforms of indoleamine 2,3-dioxygenase (IDO1, IDO2) and tryptophan 2,3-dioxygenase (TDO), and three enzymes that catabolise Arg, inducible nitric oxide synthase (iNOS) and two arginase isoforms (ARG1, ARG2). Genes encoding IDO, iNOS, and ARG are induced by inflammatory cues such as cytokines, a key feature that distinguishes them from enzymes that catabolize other amino acids. Largely for this reason, we focus on the immunologic effects of enhanced Trp and Arg metabolism. Although reduced access to other amino acids may also impact immunologic responses, perhaps via downstream pathways in common with or distinct from Trp and Arg catabolism, our goal is to use the specific examples of Trp and Arg metabolism to illustrate key concepts that relate to the general paradigm that enhanced amino acid catabolism regulates immune responses.
Oxidative Trp catabolism by IDO
IDO catalyzes oxidative catabolism of Trp into N-formylkynurenine, the rate-limiting step of oxidative Trp degradation. Two closely linked, homologous genes encoding two isoforms of IDO enzyme (IDO1, IDO2) are located in syntenic regions of chromosome 8 in humans and mice (1, 2). There is little overlap in IDO1 and IDO2 gene expression patterns and, unlike IDO1 (see below), IDO2 gene expression is not induced by interferons (IFNs) suggesting distinct functional roles (3). Moreover, studies using IDO1 deficient (IDO1−/−) mice suggest that IDO2 has a limited role (if any) in regulating immunity. The IDO1 gene contains 10 exons in humans and mice, and IDO genes are highly conserved in both sequence and genome organization over approximately 600 million years (4). As this period predates the development of adaptive immunity, IDO may have evolved as a component of the primitive mononuclear phagocyte system in invertebrates. IDO is expressed as an intracellular heme-containing enzyme and is not secreted, although Trp consumption mediated by IDO affects both intracellular and microenvironmental Trp stores through Trp transporter systems active in all cells (Fig. 1). IDO degrades L-Trp by catalyzing oxidative cleavage of the indole ring generating N-formylkynurenine (5). Structural studies (X-ray analyses) on IDO enzymes have revealed precise details about the juxta-position of oxygen radicals, hemetetrapyrrole groups, and Trp substrate during catalysis (6). IDO is not expressed under basal conditions by most cell types, and constitutive IDO enzyme activity following gene transduction may be cytotoxic (T. L. McGaha, unpublished data). However, in monocytic lineages and certain stromal cell types, IFNγ strongly induces IDO expression via a signal transducer and activator of transcription-1 (STAT-1) and an IFN-regulatory factor (IRF-1)-dependent mechanism (7, 8). Lipopolysaccharide (LPS) or tumor necrosis factor-α (TNFα) augment IDO induction providing a rationale for the observation that strong IDO induction at sites of inflammation is caused by many infectious pathogens (9–11). Soluble cytotoxic T-lymphocyte antigen 4-immunoglobulin fusion protein (CTLA4Ig) and CpG oligonucleotides that ligate B7 (CD80/86) and Toll-like receptor 9 (TLR9) molecules, respectively, both upregulate IDO in certain dendritic cell (DC) subsets but via IFNα-dependent pathways (12, 13). Thus, IFNγ and IFNα may induce IDO enzyme activity in cell type and context-specific fashion at sites of inflammation. Another complication is that transforming growth factor β (TGFβ) can induce IDO expression directly in some DCs via non-canonical nuclear factor-κB activity (14). Moreover, IDO expression patterns differed profoundly in IFN versus TGFβ-induced cultures, underscoring the point that inflammatory conditions may determine how IDO shapes immune outcomes.
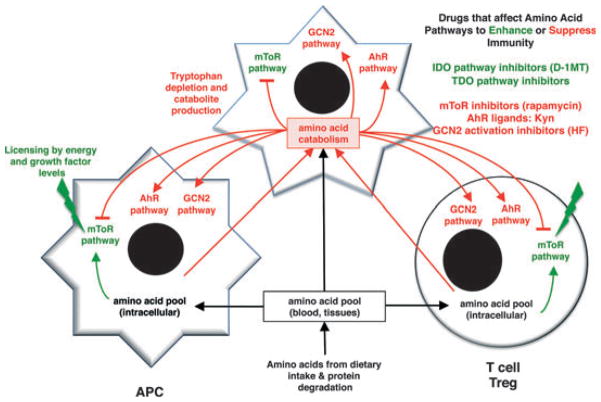
Fig. 1. Amino acid catabolism and altered immune cell functions.
Cells expressing enzymes that catablize amino acids (top) create local conditions where immune cells (bottom) experience reduced access to amino acids triggering effects on mTOR and GCB2 pathways that sense levels of amino acids inside cells. Trp depletion and Kyn production trigger cellular responses via GCN2- and AhR-signaling pathways, respectively, to promote regulatory responses by immune cells as indicated.
Strictly speaking, the IDO pathway comprises all proteins that directly or indirectly contribute to immunosuppressive functions dependent on IDO activity, including proteins that mediate induction of IDO expression (7, 15–17), activation of enzymatic activity by reductases (18–20), post-translational modifications that regulate activity (21), protein degradation (22), and the interpretation and transmission of the signals elicited by low concentrations of Trp and the presence of Trp catabolites [collectively known as kynurenines (Kyns)] including catabolic stress sensors integrated into the general control nonrepressed-2 (GCN2), the aryl hydrocarbon receptor (AhR), and possibly the mammalian target of rapamycin (mTOR) pathways (Fig. 1). This concept of integrated downstream regulatory pathways with IDO at the center has emerged from studies on multiple model systems by many groups; this notion may be critically important for understanding how the IDO pathway is induced, how IDO exerts downstream effects, and the mechanism of action of IDO pathway inhibitors that target IDO directly or target other components of the IDO pathway.
Oxidative Trp catabolism by tryptophan dioxygenase
TDO is a Trp-catabolizing hemoprotein enzyme expressed in the liver (23, 24). Although structurally unrelated to IDO, TDO also catalyzes oxidative cleavage of the indole ring in L-Trp. Unlike IDO genes, the TDO gene does not respond to inflammatory signals but is induced by stress-related glucocorticoids. TDO serves a critical housekeeping function by catabolizing Trp from dietary intake to limit systemic Trp levels in serum. Increased systemic Trp levels in mice lacking intact TDO genes resulted in neurologic dysfunction and anxiety-related behavior suggesting that excess Trp availability compromises neurologic functions in the central nervous system (CNS), possibly by accentuating serotonin production as Trp is the substrate for serotonin synthesis (25). The immunologic significance of TDO was extended by a recent study showing that TDO expressed by tumor cells created a potent barrier to anti-tumor immunity (26, 27).
Arginine catabolism
Arg catabolism is mediated by iNOS and ARG1/2, which are induced by inflammatory cytokines. Arg catabolism has many parallels with Trp catabolism mediated by the IDO pathway and is involved in similar biological processes. Myeloid-derived suppressor cells (MDSCs) often found in tumor microenvironments express high levels of ARG1 activity. In these settings, Arg catabolism induces anergy in tumor-specific effector T cells by a mechanism involving reduced half-life of mRNA encoding the T-cell receptor (TCR) CD3ζ chain (28–30). Arg depletion is sensed in T cells by a GCN2-dependent mechanism, similar to sensing Trp depletion mediated by IDO (31, 32). Suppression of ARG enzyme activity by direct or indirect pharmacologic inhibitors has immune-mediated anti-tumor effects that inhibit regulatory T-cell (Treg) proliferation and promote tumor antigen-specific T-cell tolerance (33–37). Like IDO, ARG1 activity also plays a critical role in immune suppression that maintains allogeneic pregnancies (38).
Expression and function of enzymes that catabolize Trp or Arg promote different immunologic effects in some inflammatory settings suggesting that these enzymes may have context specific, non-overlapping roles. For example, macrophages (MΦs) and endothelial cells express iNOS in response to pro-inflammatory cytokines such as IFNs (IFNα, IFNγ), TNFα, and microbial ligands such as LPS, whereas ARG1 is induced in the same cell types in response to anti-inflammatory mediators that suppress iNOS expression such as TGFβ and interleukin-4 (IL-4). Similarly, nitric oxide released by cells expressing iNOS binds to hemetetrapyrrole groups at the active sites of IDO (and iNOS) in a reversible fashion to form a ferrous iron adduct that blocks oxidative catalysis needed for enzyme functions (5). Thus, co-induction of iNOS and IDO at sites of inflammation may lead to loss of enzyme functions that can be misleading unless enzyme activity is measured. Indeed, post-transcriptional (as well as post-translational) control of enzyme expression and functions may also complicate experimental analysis of links between inflammation, amino acid catabolism, and effects on immune cells that attenuate immune responses.
Effects of amino acid catabolism on immune cells
Cells sense amino acids via at least two distinct pathways involving the serine/threonine kinases mTOR and GCN2 (Fig. 1). Both pathways are associated with control of protein translation and trigger profound changes in cellular metabolism, particularly translational activity, in response to changes in amino acid levels detected at the cell surface or in the cytoplasm. Cells can also sense amino acid catabolites that modulate innate and adaptive immune responses. For example, cells expressing IDO release Kyns into local tissue environments and into general circulation. Some Kyns possess immune regulatory properties that inhibit T-cell responses to mitogenic and antigenic stimuli. The Trp catabolite 3-hydroxyanthranilic acid (3-HAA) modulated T-cell responses in several settings (see below). Moreover, 3-HAA inhibited activation of the master kinase PDPK1 (3-phosphoinositide-dependent protein kinase-1) in activated T cells and suppressed experimental asthma in a mouse model by inducing T-cell apoptosis (39). More recently, Kyn itself was shown to be a natural ligand for the AhR, a member of the basic helix-loop-helix (bHLH) family of transcription factors (see below). In this section, we discuss downstream mechanisms that detect and respond to amino acid catabolism.
The mTOR pathway
mTOR is an evolutionarily conserved member of the phosphatidylinositol kinase-related kinase family (40). mTOR exists in the cell associated with two large, distinct multi-protein complexes, mTOR complex 1 (mTORC1) and mTORC2. mTOR is expressed by all cells and is a central control node downstream of phosphatidylinositol 3-kinase (PI3K)–AKT, WNT–GSK3 (glycogen synthase kinase 3), and adenosine monophosphate-activated protein kinase (AMPK) signaling governing multiple functions of cellular biogenesis including proliferation, adhesion, cytokine production, antigen presentation, and maturation. Not surprisingly, mTOR is critical for immune reactivity. As there have several recent comprehensive reviews on the function of mTOR in immunity (41, 42), our discussion is limited to mTOR regulation by amino acid availability.
mTOR is an important sensor of intracellular nutrient availability and lack of essential amino acids inactivates mTOR resulting in a translational block due to reduced activity of the mTOR target p70 S6 kinase (PS6K) and increased activity of the translational inhibitor 4E-EBP1. Multiple proteins and mechanisms sense the sufficiency and deficiency of particular amino acids, and transmit signals to mTOR: the energy-sensing AMPK pathway, the amino acid-sensing GCN2 pathway (see next section), and mTOR itself. Each pathway recognizes nutritional deficiencies and alters gene expression to allow cells to adapt to extrinsic conditions controlling overlapping as well as distinct sets of genes in response to different nutritional stresses.
It is not known precisely how amino acid availability is sensed by mTOR. However, emerging evidence suggests that mTOR detects amino acid levels via transfer RNA (tRNA) synethase-dependent sensing of intracellular amino acid stores. Essential amino acids are transported from the extracellular microenvironment by membrane transporters that are functionally modulated by internal amino acid availability. Thus, it has been proposed that amino acid transporters may transmit signals regulating mTOR activity. Miotto and colleagues (43–45) demonstrated that the treatment of rat hepatocytes with Leu8-MAP, a non-transportable leucine peptide, inhibited autophagy (an mTOR-regulated function) and induced phosphorylation of S6K. However, other reports suggested that mTOR is not responsive to Leu8-MAP in rat adipocytes (46). A concern with this hypothesis is the relative promiscuity of most amino acid transporters (17, 47, 48). Thus, it is difficult to envision how mTOR sensitivity to depletion of particular amino acids could manifest via transporter-dependent mechanisms. Recent data suggest a more likely scenario in which mTOR senses amino acid levels in cells by interaction with specific cytoplasmic sensors. RAG GTPases, a family of small Ras-related GTPase enzymes, physically interact with mTORC1 and are mechanistically responsible for amino acid-mediated control of mTORC1 activity (49, 50). Recently, Han et al. (51) reported that enzymatic sensing of intracellular leucine by leucine tRNA synthetase (LRS) is critical in regulation of mTORC1 function, suggesting that leucine-charged LRS binds to the C-terminal domain of the RAG GTPase RagD functioning as a GTPase-activating protein culminating in the activation of mTORC1. This function requires amino acid charging as mutation of the amino acid binding domain of LRS-abrogated leucine-induced mTORC1 activity (51). It is important to note that this is not a function of all tRNA synthetases as isoleucine tRNA synthetase did not mediate mTORC1 activation after charging (51). However, as isoleucine can bind to LRS, promiscuity in charging may allow a limited set of tRNA synthetases to generate the full mTOR response to amino acid availability.
Inhibition of mTOR (i.e. by depleting amino acids) stops protein synthesis and promotes cell autophagy under certain conditions. In preliminary studies, cell lines cultured in Trp-free media underwent autophagy and treatment with the reversible IDO pathway inhibitor 1-methyl-[D]-tryptophan (D-1MT) was sufficient to prevent this effect by activating mTOR; similar results were obtained when cells lines were treated with IFNγ to induce IDO1 and deplete Trp (R. Metz and G. C. Prendergast, unpublished data). mTOR activation by D-1MT was specific to Trp withdrawal and did not affect GCN2 activity. These findings are somewhat counterintuitive, as GCN2 is thought to be essential for the IDO-mediated effects of Trp withdrawal to manifest; however, the degree of crosstalk between these pathways has yet to be fully understood. Though D-1MT may activate the mTOR amino acid-sensing pathway directly (in a GCN2-independent fashion), there is reason to think that the GCN2 and mTOR pathways are inter-connected, cooperating with each other to assess nutrient ‘deficiency’ (GCN2) and ‘sufficiency’ (mTOR) to navigate conditions of nutritional stress that would be needed to fine tune the control of cell growth and effector functions appropriately. In the case of D-1MT treatment, mTOR may detect the signal for amino acid ‘sufficiency’ when Trp-starved cells are exposed to D-1MT. Thus, a present working hypothesis is that GCN2 activation acts as a molecular switch to inform the mTOR pathway of ‘insufficient levels’ of a particular amino acid. If so, then GCN2 activation may be required to inhibit the mTOR pathway, and thus both pathways may play a role in establishing an immunosuppressive environment.
The IDO-GCN2 pathway
Studies linking IDO activity with modified immunologic outcomes (see below) prompted a search for molecular pathways that explained these observations. mTOR and GCN2 were potential candidates for biochemical pathways that linked increased IDO activity to suppression of T-cell proliferation in response to mitogenic or antigenic stimulation. The first indication that IDO regulated T-cell responses by activating the GCN2 pathway emerged in 2005 from studies showing that C/EBP homologous protein-10 (CHOP), a GCN2-responsive gene, expression was elevated significantly when T cells were activated in cultures lacking Trp in media (52). Moreover, DCs from lymph nodes (LNs) of tumor-bearing mice and expressing functional IDO enzyme activity inhibited antigen-specific CD8+ T-cell proliferation ex vivo by a GCN2-dependent mechanism, and T cells from mice lacking intact GCN2 genes were completely refractory to the T-cell inhibitory effects of IDO-expressing DCs ex vivo and following cell transfers in vivo (52). These findings suggested that Trp withdrawal due to local IDO activity in cultures or in tumor-draining LNs leading to GCN2 kinase activation in T cells was both necessary and sufficient to explain how IDO blocked T-cell proliferation following antigenic stimulation. Subsequently, Rodriguez et al. (31) used analogous approaches to demonstrate that T cells were also susceptible to Arg withdrawal. Thus, Trp and Arg withdrawal had similar inhibitory effects on T-cell proliferation via a common downstream pathway involving GCN2 activation. The report by Rodriguez et al. (31) also provided a potential rationale for the known links between increased ARG1 activity in MDSCs and regulation of T-cell-mediated anti-tumor immunity by MDSCs, which are commonly associated with tumor progression. It is still unclear if ARG1 activity is essential for the tumor-promoting effects of MDSCs. However, a recent report showed that ARG1 activity induced by IL-13 in MDSCs was essential for inhibition of graft-versus-host disease (GvHD) in a murine bone marrow engraftment model, although requirements for intact GCN2 pathways were not tested in this study (53).
Like mTOR, GCN2 is a serine/threonine kinase that senses reduced cellular access to amino acids. GCN2 is one of four known kinases that phosphorylate the α subunit of the eukaryotic translation initiation factor 2 (eIF2) (54). These kinases respond to a diverse range of environmental stresses including endoplasmic reticulum (ER) stress (e.g. the unfolded protein response) resulting in activation of the double-stranded RNA (dsRNA) activated protein kinase-like ER kinase (PERK), dsRNA-activated protein kinase (PKR), and heme-regulated inhibitor kinase (HRI). As all four stress-activated kinases converge on eIF2α, the downstream pathway is often called the integrated stress response (ISR) (Fig. 2). GCN2-mediated activation of eIF2α suppresses protein synthesis generally but allows translation of select mRNAs involved in autophagy, amino acid uptake, and bio-synthesis and amino acid tRNA re-charging such as Trp-tRNA synthetase (WARS). Phosphorylation of eIF2α delays regeneration of the active form of eIF2 by inhibiting guanosine diphosphate (GDP)/guanosine triphosphate (GTP) exchange, which reduces the efficiency of ribosomal assembly on nascent mRNA (55). This delay, coupled with ribosomal scanning, results in bypassing of initial translational start sites to reduce translational efficiency and global protein synthesis dramatically. However, translation of some mRNA species such as activating transcription factor-4 (ATF4) are selectively increased due to recognition of alternative open-translational reading frames downstream of the initial translational initiation site (55). In particular, ATF4 functions as a master transcription factor regulating cellular responses under conditions of limited amino acid availability. ATF4 forms homo- or heterodimers with several members of the AP-1 and C/EBP families (56), and as the DNA binding site recognized by ATF4 dimers is determined by the binding partner, ATF4 regulates expression of a large number of genes (57).
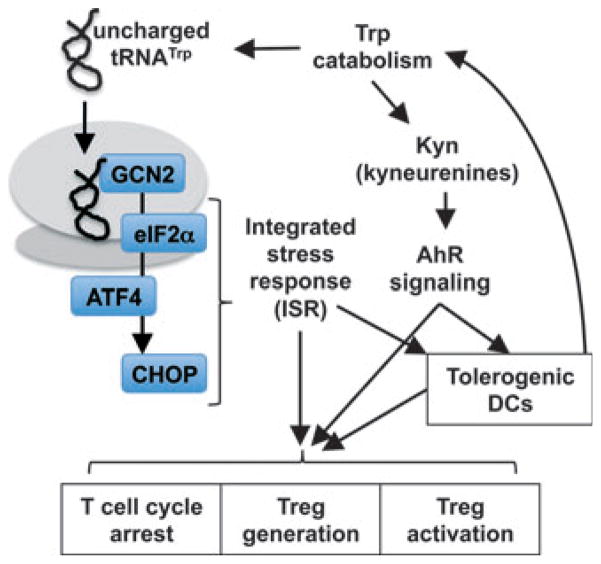
Fig. 2. GCN2 and AhR signaling.
Trp depletion and Kyn production trigger cellular responses via GCN2- and AhR-signaling pathways, respectively, to promote regulatory responses by immune cells. See text for details.
As depicted in Fig. 2, GCN2 kinase activates ATF4-mediated CHOP expression in cells that have restricted access to amino acids, especially when immune cells are activated by exogenous insults such as inflammatory and antigenic stimuli. Unlike mTOR complexes that likely detect amino acids indirectly by GTPases, GCN2 senses reduced amino acids levels directly as uncharged tRNA molecules bind to a regulatory region in GCN2 homologous to HisRS (histidyl-tRNA synthetase) (54, 58). Moreover, unlike mTOR, which primarily senses the nine essential amino acids, GCN2 appears responsive to depletion of any amino acid suggesting a broader role for GCN2 in regulating cellular responses under conditions of limited access to amino acids (17, 48, 59, 60). However, T cells activated ex vivo by mitogens in medium lacking both isoleucine and leucine entered S-phase (defined by thymidine incorporation) prior to complete metabolic arrest due to amino acid starvation; in striking contrast, T cells activated in medium lacking Trp did not enter S-phase (61), suggesting that access to particular amino acids such as Trp may be an essential checkpoint for entry into S-phase, at least in T cells. Hence, the GCN2 pathway may be selectively responsive to withdrawal of particular amino acids in specific cell types, and further studies will be necessary to clarify this point.
Regulation of immune cell behavior by the IDO-GCN2 pathway
Intact GCN2 genes in CD4+ T cells are essential for naive CD4+ T cells to undergo conversion into forkhead box protein 3 (Foxp3)-lineage Tregs through the combined effects of Trp withdrawal and Trp catabolites (62). The GCN2 pathway has also been shown to have a pivotal role in activating pre-formed, functionally quiescent (resting) CD4+ FoxP3-lineage Tregs, which exhibit little, if any, suppressor activity and must be activated by mitogenic signals via the TCR to acquire potent suppressor function (63). Sharma et al. (64) showed that resting (splenic) Tregs acquired potent regulatory phenotypes when cultured with DCs from tumor draining LNs expressing IDO. This regulatory response by Tregs was abrogated completely when Tregs were obtained from mice lacking intact GCN2 genes, indicating a critical requirement for the GCN2 pathway in Tregs for them to acquire regulatory phenotypes in response to IDO-mediated Trp withdrawal. This finding was corroborated in a model of rapid Treg activation in response to high doses of TLR9 ligands (CpGs), which stimulate a small cohort of DCs in mouse spleen to express IDO (65); in this model, Tregs in mice lacking intact GCN2 genes not only failed to acquire regulatory phenotypes but instead underwent functional re-programming to acquire helper/effector functions as indicated by rapid expression of the pro-inflammatory cytokines IFNγ, TNFα, IL-17, and IL-2 following TLR9 ligation (67, A. L. Mellor, unpublished data). Moreover, activation of the IDO-GCN2 pathway in tumor bearing and CpG-treated mice suppressed IL-6 production by plasmacytoid DCs (pDCs) (65, 66). Presumably, locally induced IDO activity triggered GCN2 activation in pDCs that blocked translation of IL-6 mRNA in these settings, although other explanations for IDO-mediated suppression of IL-6 expression have not been excluded. In another study, the ability of DCs to express IDO in response to treatment with CTLA4Ig, which ligates B7 (CD80/CD86) molecules on DCs, was shown to be GCN2-dependent as DCs from GCN2-deficient mice failed to produce IFNα, the obligate IDO inducer in DCs (67). In striking contrast, DCs from GCN2-deficient mice displayed no detectable defect in IFNα production or IDO induction in response to CpG stimulation (TLR9 ligation), suggesting that GCN2 is downstream of B7 ligation but not TLR9 ligation. It is not known whether GCN2 activation in DCs after B7 ligation was mediated by IDO activity. Following UV exposure in HeLa cells, GCN2 kinase was activated by methionyl-tRNA synthetase, suggesting that B7 ligands can also trigger GCN2 activation in DCs via stress pathways independent of upstream IDO induction to deplete Trp (60).
Further supporting a role for stress in the regulation of T-cell immunity, Scheu et al. (68) demonstrated an uncoupling of cytokine mRNA production from protein synthesis in primed T-helper 2 (Th2) cells, an effect that was mediated by the ISR, raising the possibility that stress responses influence T-cell differentiation and effector functions. However, while eIF2α phosphorylation was observed, it is unlikely that GCN2 activation was responsible for the functional block in cytokine protein synthesis. In view of this finding, it seems likely that ‘stress’ from disparate sources modulates lymphocyte behavior in distinctive ways. Moreover, T-cell sensitivity to IDO-induced GCN2 kinase activation is likely lineage and developmental stage dependent as naive CD8+ T cells exhibited more robust GCN2 kinase activity relative to CD4+ T cells in co-cultures with DCs expressing IDO (69). Thus, Trp and perhaps Arg withdrawal induce cell cycle arrest in naive T cells, and a divergent, more complex effect on fully differentiated T cells, reflecting the differing nutritional requirements of naïve and differentiated effector T cells. Although naive T cells must undergo rapid and extensive clonal expansion before differentiating, fully differentiated T cells must manifest effector functions only. However, the effect of GCN2 activation on the behavior of fully differentiated T cells has not been studied beyond its critical role in IDO-dependent Treg activation and will need further clarification.
Although GCN2 activation has been linked to immune regulatory responses in several model systems, evidence suggests that GCN2 activation leading to expression of CHOP may also promote pro-inflammatory responses in some settings. For example, CHOP can form dimers with either the full length or splice variants of the transcription factor CCAAT/enhancer binding protein β (C/EBPβ), and studies on human melanoma cells revealed that CHOP enhanced IL-6 gene promoter activity by sequestering the inhibitory C/EBPβ isoform away from the IL-6 promoter (70). In contrast, CHOP was reported to suppress IL-6 gene transcription in B-cell lines (71), suggesting that cell-type specific factors may influence the direction of GCN2-mediated effects that lead to pro-inflammatory or anti-inflammatory outcomes. In our hands, IL-6 expression in response to LPS, zymosan, or CpG stimulation was enhanced significantly in primary macrophages and DCs expressing IDO (or in macrophages and DCs cultured in the absence of exogenous Trp), but increased IL-6 expression did not occur when macrophages were from mice lacking intact CHOP genes (T. L. McGaha, unpublished data). Although elevated IL-6 mRNA levels were compensated to some extent by reduced mRNA translational efficiency, the overall effect was a significant increase in IL-6 protein synthesis and release. These outcomes imply that induced IDO activity in myeloid cells can amplify pro-inflammatory cytokine responses to microbial stimuli, a rather counterintuitive outcome in view of the overwhelming volume of evidence showing that IDO activity regulates immunity in other settings. One potential explanation is that GCN2 activation is just one of several signals that antigen-presenting cells (APCs) integrate to determine appropriate responses to inflammatory stimuli, and other pathways may cause the GCN2-mediated ISR to trigger distinct immunologic responses appropriate to prevailing conditions. Thus under some conditions (e.g. B7 ligation), the GCN2 pathway may dominantly suppress cytokine gene expression, whereas, under other conditions (e.g. TLR ligation), GCN2 may enhance cytokine production. It should be kept in mind that a positive role for GCN2 likely applies to a subset of cytokines as the global function of GCN2-mediated ISR signaling is to suppress protein translation. In this context, cytokine expression positively regulated by stress-induced transcription factors such as ATF4, CHOP, or C/EBPβ may be increased, whereas others would be reduced due to translational blockade.
Considerable evidence supports the hypothesis that Trp withdrawal, and consequent GCN2 pathway activation have profound effects on T cells and DCs belonging to the adaptive and innate immune systems, respectively. It is likely that Arg withdrawal also stimulates similar downstream responses via the GCN2 pathway in T cells, Tregs, and DCs that promote immune regulation and tolerance. In principle, withdrawal of other amino acids may incite similar immunologic responses, although lack of evidence linking elevated amino acid catabolism with inflammatory signals makes it unlikely that such effects occur in physiologic settings on inflammation relevant to clinical disease syndromes.
Exterznal versus cell autonomous effects of IDO on immune cell behavior
The biochemical effects of IDO on T cells and Tregs are indirect because lymphocytes do not express IDO themselves. Thus, other cells expressing IDO regulate T-cell and Treg behavior by restricting their local access to external pools of Trp (Fig. 1). Cells expressing IDO may create regions of reduced Trp concentration in their immediate local tissue microenvironments (Fig. 3). Consequently, target T cells and Tregs experience reduced access to Trp, which manifests as increased levels of uncharged tRNATrp that triggers GCN2 activation and potentially inhibits mTOR stimulatory functions. It is not clear if cell–cell contact between cells expressing IDO and target T cells (or Tregs) is essential to trigger this effect. Studies performed with T cells/Tregs activated in conditioned media lacking defined amino acids show that cell contact is not critical to trigger T-cell cycle arrest and other behaviors attributable to amino acid withdrawal; however, these studies do not exclude the possibility that cell–cell contact is essential for more subtle ‘threshold’ effects on T-cell/Treg behavior to manifest under physiologic conditions. Even though the initiating events may be subtle in such settings, their consequences may have profound effects on immune cell behavior and mediate pivotal effects on immunologic outcomes. Another idea to emerge from this model is that amino acid transporters and tRNA synthetases that charge tRNA molecules with aminoacyl moieties may be critical for effective control of T-cell behavior in response to amino acid withdrawal. In this regard, it may be significant that human macrophages that mediated T-cell suppression by expressing IDO also co-expressed a high-affinity Trp transporter activity (72), and that Trp-tRNA synthetase (WARS) is the only tRNA synthetase upregulated by IFNγ (73). These findings suggest that increased Trp transporter activity may facilitate Trp removal from local tissues and that some cells types may become more resistant to reduced Trp levels by enhancing WRS expression.
Fig. 3. Potential physiologic effects of IDO-expressing DCs on Trp levels in lymphoid follicles.
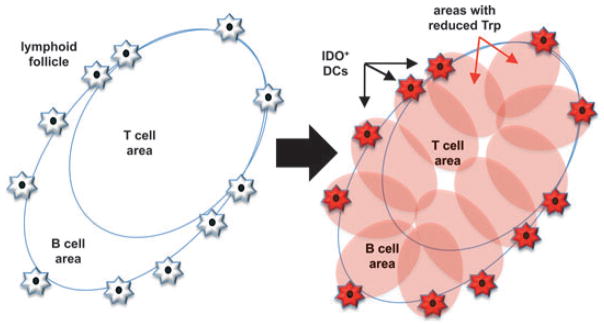
The model posits how small cohorts of splenic MZ DCs expressing IDO may create regions of low Trp reserves in lymphoid follicles effecting T cells, B cells, and macrophages and DCs inside the follicle to generate immune suppression and tolerance.
The model described above implies that the cells most likely to be affected by IDO activity are the cells expressing IDO themselves. A range of stromal and hematopoietic cell types can express functional IDO in response to IFN type I (IFNαβ) or IFN type II (IFNγ), which are both potent IDO inducers, generated at sites of inflammation. Stromal cells such as epithelial and endothelial cells expressing IDO may inhibit T-cell proliferation in LNs where naive T cells first encounter new antigens and may also impede T-cell effector functions at inflammatory lesions where effector T cells accumulate following T-cell differentiation and circulation. However, APCs expressing IDO that present cognate antigens to naive T cells (or Tregs) in LNs are likely to have more profound regulatory effects on T-cell responses than stromal cells. Among APCs, discrete subsets of DCs and macrophages are specialized to express IDO in response to IFNs, while B cells, like T cells, have not been reported to express IDO. It is not clear if IDO expression by APCs alters their ability to function as effective APCs to present antigens and activate T cells. As mentioned above, some evidence suggests that induced IDO expression in DCs compromises their ability to produce pro-inflammatory cytokines such as IL-6 (65, 66), which may adversely affect the maturation status of DCs and their ability to process and present antigens to T cells efficiently. The finding that GCN2 gene ablation blocked the ability of B7 ligands to induce DCs to upregulate IFNα and IDO also suggested that GCN2 activation may be a pivotal factor controlling APC functions (67). Nevertheless, it is still not clear if coordinated antigen presentation and IDO activity by the same APCs is essential for efficient T-cell regulatory effects in physiologic settings of inflammation, as opposed to ex vivo studies with cultured cells.
Like T cells that respond in a bystander fashion to IDO activity in other cells to activate GCN2, IDO-mediated bystander effects may affect APC functions, even if they do not express IDO themselves. In a recent study, we showed that DCs not expressing IDO themselves nevertheless exhibited evidence of ISR activation when IDO was induced in neighboring cells after in vivo challenge with apoptotic cells; this bystander response in APCs upregulated TGFβ expression in spleen, and this response did not manifest when mice lacked intact IDO1 and CHOP genes (74). These data suggest that cell-intrinsic and -extrinsic activation of the ISR by cells expressing IDO may be critical to create local immune-suppressive microenvironments.
A related issue that created some confusion in the literature is the precise identity of physiologic DC and macrophage subsets in humans and mice that can be induced to express functional IDO. For our studies identifying DCs that mediated IDO-dependent T-cell suppression, we used a flow cytometer to selectively sort defined subsets of DCs to high degrees of purity (usually >95%) not achievable using other methods to enrich specific cell types such as monoclonal antibody-mediated affinity selection methods (at least in our experience). By sorting DC subsets from lymphoid tissues in mice such as spleen and inflamed LNs draining sites of tumor growth or skin exposed to tumor-promoters, we identified rare but highly distinctive populations of cells with attributes of DCs and B cells (CD19+ DCs) from these sources (13, 75–78). However, in a recent study to identify splenic cells expressing IDO after mice were exposed to apoptotic thymocytes, we found that CD169+ marginal zone (MZ) macrophages, not splenic DCs, were induced to express functional IDO in this setting (74). Previous studies by Grohmann, Puccetti, and colleagues (22, 79) revealed that IDO expression in most murine DCs was blocked by factors such as the SOCS3 (suppressor of cytokine signaling-3) and DAP12 (DNAX-activating protein-12) that interfere with cytokine signaling and promoted proteosomal degradation of IDO, and ablating these inhibitory pathways led to IDO expression in a wider range of DC subsets. Thus, under normal physiologic conditions, only a few DC and macrophage subsets are specialized to express IDO in response to inflammatory stimuli due to the effects of inhibitory pathways that restrain IDO expression in most DC and macrophage subsets.
DCs expressing functional IDO were identified by their ability to suppress T-cell responses elicited ex vivo in the absence of IDO inhibitor but not when IDO inhibitor was present in cultures. These assays were essential to identify rare cells in lymphoid tissues that possessed the relevant functional properties. In some experiments, we showed that large excesses of T-cell stimulatory DCs failed to stimulate T-cell responses ex vivo when mixed with small cohorts of cells expressing IDO (76, 77). As small cohorts of cells expressing IDO are unlikely to directly suppress the T-cell stimulatory functions of a large excess of DCs that do not express IDO, bystander effects of IDO on other DCs and Tregs may explain why regulatory outcomes were dominant. Small cohorts of cells expressing IDO may be sufficient to create ‘immune privileged’ regions in lymphoid tissues by reducing Trp levels significantly relative to normal (basal) levels. As CD19+ DCs are consistently found dispersed in the perifollicular/MZ regions adjacent to lymphoid follicles, it is possible that when these cells are induced to express IDO they reduce Trp levels in lymphoid follicles below thresholds required (i) for mTOR signaling to stimulate T-cell responses, and (ii) to trigger GCN2 kinase activation in T cells and Tregs located in follicles (Fig. 3). However as discussed above, an alternative model is that direct cell–cell contact may be essential for regulatory effects to manifest due to IDO expression in CD19+ DCs that suppress naive T-cell clonal expansion, effector T-cell differentiation and functions, and activate Tregs in response to inflammatory and antigenic stimuli.
Kyn sensing via the AhR pathway
In inactive form, AhR is located in the cell cytoplasm bound to chaperone molecules. On ligand binding, AhR dissociates from cytoplasmic chaperones, associates with the AhR nuclear translocator, and migrates to the cell nucleus where the dimeric complex modulates gene expression. Several exogenous ligands including dioxins trigger AhR signaling to provoke highly toxic effects in eukaryotic cells. AhR signaling is a pivotal factor that shapes immune cell responses in a number of settings (80), although the large number of potential exogenous AhR ligands and, until recently, the lack of known physiologic ligands made it difficult to evaluate the biologic significance of AhR signaling. The recent discovery that Kyn is a natural physiologic AhR ligand provides a completely novel insight into potential roles for AhR signaling at sites of inflammation where IDO activity is co-induced (see next section). For example, Kyn released by IDO-expressing stromal cells and innate immune cells such as macrophages and DCs may dictate how T cells differentiate in response to antigenic signals received at the same time in such settings. The emerging paradigm that IDO provokes downstream AhR signaling in immune cells via Kyn release is given further complexity by the finding that some AhR ligands also stimulate DCs to express IDO and acquire immune regulatory phenotypes as a consequence (81). Thus, IDO is both downstream and upstream of the AhR pathway, although it is not yet clear if the same AhR ligands induce IDO and mediate responses to IDO; if so, positive feedback loops that induce and amplify IDO activity may manifest in some settings of inflammation (Fig. 1). It is not yet clear if distinct AhR ligands provoke differential effects on innate and adaptive immune cells to stimulate or suppress immunity. This point notwithstanding, Kyn mediates immune regulation by promoting Treg differentiation, possibly by inducing DCs to acquire regulatory phenotypes dependent on IDO (82, 83). AhR signaling downstream of Kyn release may influence innate and adaptive immunity.
Immune regulation by amino acid catabolism in inflammatory settings
In this section, we discuss evidence supporting the paradigm that amino acid catabolism regulates immune responses in inflammatory settings relevant to clinical disease syndromes. As above, we focus on the role of enhanced Trp catabolism in regulating immunity in such settings.
Trp catabolism and regulation of adaptive immunity
Our colleague David Munn was the first to discover that cultured human macrophages inhibited T-cell proliferation by expressing IDO and that IDO-mediated T-cell suppression was mediated by Trp withdrawal as adding excess Trp to media rescued T-cell proliferation (84). The biological significance of this initial finding was not fully appreciated until the subsequent discovery that IDO activity was essential to protect developing fetal tissues from destruction by maternal T cells during pregnancy in mice (85). Since this initial report, numerous studies, in most cases employing IDO inhibitors in murine models of inflammatory syndromes, have shown that IDO mediates immune regulation in diverse settings relevant to a range of clinical syndromes, including infectious, allergic, and autoimmune diseases, cancer, and transplantation.
Although the paradigm that IDO regulates T-cell-mediated immunity is now supported by multiple studies from a number of groups using a variety of models, several key questions regarding the mechanism of IDO-mediated regulation of adaptive immunity remain unresolved. First, the relative contributions of Trp withdrawal and Trp catabolite production in physiologic settings are not fully understood. Second, the critical cell types that are induced to express IDO and mediate Trp catabolism at sites on inflammation have been defined in only a few physiologic settings. Third, the upstream pathways that stimulate IDO induction in specific cell types, and fourth, the downstream pathways that inhibit innate and adaptive immunity have not been fully defined. Resolving these key questions has been complicated by technical difficulties in identifying cells that express IDO (a highly conserved intracellular protein) and cells that exhibit functional IDO enzyme activity in physiologic settings; these cell types are not necessarily equivalent, as posttranscriptional and post-translational factors that promote or inhibit IDO enzyme activity mean that some cells expressing IDO genes and protein may not exhibit IDO enzyme activity. For example, the only methods available to assess IDO enzyme activity are indirect methods to detect Kyn levels in serum or tissues and by measuring IDO enzyme activity mediated by cell-free tissue homogenates (also assessed by detecting Kyn production); both methods are relatively insensitive and are likely to underestimate IDO enzyme activity in small cohorts of physiologic cells that may nevertheless have profound effects on immune cell behavior in response to inflammatory and antigenic stimulation that occurs in local tissue microenvironments. Another complication is that mice lacking intact IDO1 genes exhibited no spontaneous phenotypic abnormalities consistent with the loss of a critical immune regulatory mechanism, including the ability to carry allogeneic pregnancies to term as normal (86). However, the abortifacient effects of the IDO pathway inhibitor 1-methyl tryptophan (1MT) used to demonstrate the role of IDO enzyme activity in maintaining that murine allogeneic pregnancies did not manifest in IDO1-deficient mice, confirming the critical importance of IDO activity during allogeneic pregnancy in wildtype mice (86). Presumably, other immune regulatory mechanisms that are normally redundant (or sub-dominant) in wildtype mice became dominant in IDO1-deficient mice to compensate for developmental loss of the IDO pathway. If so, it is not known what alternative and normally redundant mechanisms compensated for loss of IDO1 gene function in mice. This point notwithstanding, several studies have revealed that IDO1-deficient mice failed to compensate for loss of IDO1 gene function when subjected to specific inflammatory insults that elicited defined responses in wildtype mice. Some examples that illustrate this key point are that IDO1-deficient mice exhibited (i) increased resistance to tumor progression in a murine model of inflammation-dependent skin carcinogenesis (77), (ii) inability to activate Tregs or suppress Th1 responses to immunizing antigens when co-treated with high doses of TLR9 ligands (65, 87), (iii) resistance to development of neurologic depressive-like behavior in response to mycobacterial Bacillus Calmette-Guerin (BCG) infection (88), and (iv) increased susceptibility to lethal T. gondii infection (89).
Trp catabolism and host defense against microbial infection
Trp catabolism by cells expressing IDO is increased at sites of many infections due to release of IFNs during the inflammatory response. The notion that IDO activity in innate immune cells such as macrophages has a role in host defense emerged soon after IDO was discovered based on experiments showing that IDO activity inhibited certain microbial infections. For example, IFNγ-treated human cells expressed IDO and were resistant to Staphylococcus aureus, Toxoplasma gondii, Herpes Simplex virus, and Vaccinia virus infections (90–93). Similarly, control of Chlamydia infection was dependent on IFNγ-mediated Trp depletion (94). Inhibition of IDO or addition of excess Trp abrogated the protective effects of IDO, suggesting that anti-microbial functions of IDO were mediated by Trp withdrawal restricting microbial replication in infected cells. In cultured cells infected with West Nile virus (WNV), IDO was induced in infected and adjacent non-infected cells, but IDO activity did not inhibit WNV yields from infected cells but protected non-infected cells from subsequent infection limiting viral replication in culture (95). The inhibitory effects of IDO on microbial infections in vivo may depend on contributions from Trp withdrawal and Trp catabolite production. For example, the Trp catabolites 3-hydroxy-kynurenine (3-HKyn) and 3-HAA were toxic to Trypanosoma cruzi amastigotes, and 3-HKyn affected the trypomastigote stage and treating acutely T. cruzi-infected mice with 3-HKyn led to effective control of parasite burdens and improved the survival rates of infected mice (96). 3-HKyn and another Trp catabolite, α-picolinic acid, also mediated inhibitory effects on growth of methicillin-resistant Staphylococcus aureus, E. coli, and vancomycin-resistant Enterococcus faecalis (97). Thus, Trp depletion and production of Trp catabolites by infected cells induced to express IDO may work synergistically to limit microbial growth in inflamed tissues.
Trp catabolism and regulation of host immunity to pathogen infections
Diametrically opposed to a role in host defense, IDO activity attenuates host immunity in response to some microbial infections. For example, IDO1-deficient mice were less susceptible to cutaneous Leishmania major infection, indicating a counterregulatory role for IDO; that is, reduced host immunity and parasite clearance when IDO was active (89, 98). Moreover, IDO ablation promoted the generation of Th17 cells in L. major-infected mice, and reduced local inflammation and parasite burdens, as did pharmacologic inhibition of IDO (98). Induced IDO activity may also promote the persistence of lentiviral infections such as human immunodeficiency virus-1 (HIV-1), the cause of AIDS (acquired immune-deficiency syndrome). IDO induced in human DCs following HIV-1 infection attenuated effector T-cell responses, and IDO induction may be induced by IFN type I released by chronically activated pDCs during HIV-1 infection (99–101). Moreover, treating humanized, HIV-infected mice with D-1MT or SIV-infected rhesus macaques with combined anti-retroviral therapy and D-1MT led to significant reduction in viral burdens, suggesting that IDO facilitated viral infection and persistence in these animal models of HIV-1 infection (102, 103). This finding suggested that D-1MT, which is currently being tested as a potential anticancer adjuvant in early-phase clinical trials, may interfere with HIV-1/host interactions to offer potentially novel opportunities to treat HIV-1 infections. These outcomes and data from other similar studies suggest that IDO not only plays key roles in host responses to diverse pathogen infections but also suggest that the dominant nature of these roles is likely to be pathogen-specific.
Although IDO activity may attenuate host immunity to microbial infection to the benefit of pathogens, attenuating host immunity may also limit collateral damage to infected tissues triggered by infections to benefit infected hosts. Treatment with the Trp catabolite 3-HAA suppressed lung pathology mediated by effector Th17 cells in mice with mTB (mycobacterium tuberculosis) infections (104). Moreover in mice lacking M2 macrophages (due to ablation of IL-4-receptor genes) IDO induction in macrophages protected mice from lethal Schistosoma mansoni infection, and inhibiting IDO in infected mice led to enhanced liver pathology and mortality (105). In Candida albicans-infected mice, IDO was induced at primary lesions and in DCs and neutrophils via IFNγ and CTLA-4-dependent mechanisms, and IDO inhibition exacerbated infection and local pathology in this fungal infection model (106). A more recent study using human peripheral blood mononuclear cells suggested that C. albicans modulated Trp metabolism away from the Kyn pathway by inhibiting IDO expression and enhancing the production of 5-hydroxytryptophan metabolites that inhibit IL-17 production and dampen host defense to infection (107). Another example of the tissue protective role of IDO activity in response to infections emerged from a seminal study using mice that develop a chronic inflammatory disorder related to chronic granulomatous disease (CGD); these mice cannot generate superoxide substrate required for IDO enzyme activity, and consequent lack of IDO activity led to failure to suppress generation of pathogenic effector T cells that caused acute inflammatory lung injury in response to pulmonary aspergillosis (108).
Infections that enhance and sustain local Trp catabolism create conditions where protective immunologic responses are dysfunctional, leading to increased risk of clinical disease following primary infections. Sustained elevation of IDO activity in lungs may be a reason why higher susceptibility to secondary respiratory infections such as lethal pneumonia manifests following primary influenza infection. Influenza A virus infection induces substantial increases in IDO enzyme activity in lungs, and pharmacologic inhibition of IDO after virus clearance protected mice from lethal secondary pneumococcal infections, resulting in significantly reduced pneumococcal outgrowth and much lower levels of IL-10 and TNFα in infected lungs (109). In a similar vein, systemic CpG treatment after sublethal Rickettsia infection caused mice to die, and this lethal response to TLR9 ligands was IDO-dependent, as IDO1 gene ablation protected mice from lethality and led to substantial reduction in serum cytokine levels induced by CpG treatment (110). The ability of MuLV (murine leukemia virus) to establish stable, chronic infections in mice was abolished by genetic and pharmacologic IDO ablation, and protective responses in the absence of IDO activity correlated with heightened production of IFN type 1 (111). Similarly, increased protection against persistent encephalomyocarditis virus (EMCV) infection leading to lethal myocarditis was observed in mice after IDO gene ablation or treatment with IDO inhibitors; increased protection also correlated with enhanced IFN type 1 production in this infection model, suggesting that IDO compromises the anti-microbial effects of IFN type I (112). Suppressed IFN type 1 production in response to ECMV infection was mediated, at least in part, by Trp catabolites made by IDO-expressing cells, as administering Kyn to IDO1-deficient mice reversed the protective effect of ablating IDO1 and lowered levels of IFN type 1 induced by ECMV infection (112). Interplay between IDO and IFN type I has also been proposed as a critical factor driving the chronic immune activation syndrome underlying persistent HIV-1 infection that causes fundamental immune dysfunction over extended periods in HIV-1-infected patients (101). A recent report highlights another effect of sustained IDO activity in chronic infections that may be important clinically. IDO induced by IFNγ and TNFα in mice chronically infected with BCG caused neuronal pathology and incited neurologic dysfunction manifesting as depression-like behavior (88, 113, 114). Indeed, increased risk of neurologic dysfunction is a common feature of some infections, particularly when pathogens persist and incite chronically elevated Trp catabolism that sustains increased levels of Trp catabolites in circulation, several of which have toxic neurologic activity. Thus persistent malarial, WNV, and HIV-1 infections incite dementia in a proportion of patients, and sustained exposure of the CNS to neurotoxic Trp catabolites may in part explain this important aspect of microbial pathogenesis. These diverse examples of the immunologic effects of IDO induced in response to microbial infections, and the findings reported in the previous section, demonstrate the complex and potentially confusing roles played by the IDO pathway in (i) promoting innate host immunity, (ii) regulating adaptive immunity that may facilitate pathogen persistence in immunocompetent hosts, (iii) limiting collateral damage of inflamed tissues at sites of infection mediated by immune effector cells, and (iv) driving immunologic and (v) neurologic dysfunction. Further studies are needed to understand how these various effects contribute to microbial persistence, regulation of host immunity, and pathology in response to infections. Another key point is that immunotherapies that target the IDO pathway may have unanticipated potentially detrimental consequences in clinical settings of chronic infectious disease syndromes. Clearly, IDO is a common feature of host–pathogen that may benefit one or both parties. Although targeting the IDO pathway for clinical benefit holds considerable promise for improving treatment of chronic infectious diseases, it is worth bearing in mind that interfering with host–pathogen interactions that evolved over extended periods may have unintended and undesirable consequences in some cases.
Trp catabolism and cancer
Amino acid catabolism is one of several regulatory pathways that attenuate natural and vaccine-induced anti-tumor immunity following immunotherapy. Local immunosuppression is mediated in part by enhanced Trp (or Arg) degradation in response to tumor-induced inflammation that creates potent barriers to anti-tumor immunity. Trp catabolism in such settings is mediated by IDO1 in APCs and/or by IDO1 or TDO in tumor or stromal (parenchymal) cells. Transfection of immunogenic tumor cell lines with recombinant IDO1 or TDO rendered them immunosuppressive and lethally progressive in murine tumor models (27, 115). In humans, expression of IDO1 in numerous types of tumor cells is a significant predictor of poor prognosis. For example, high IDO1 mRNA levels in patients with acute myeloid leukemia correlated negatively with survival (116). Histochemical analysis of colorectal cancer samples indicated that tumors expressing high levels of IDO showed lower levels of infiltrating T cells and enhanced liver metastases that correlated with lower overall patient survival (117). Similarly, biopsies from ovarian carcinomas with abnormally high IDO expression levels were also associated with lower CD8+ T-cell infiltrates (118). In a study on non-small cell lung cancer, enzymatically active IDO was expressed by normal eosinophil granulocytes in the peritumoral stroma, and levels of IDO expression and numbers of local IDO+ cells correlated negatively with overall patient survival (119). Similarly, increased Kyn:Trp ratios in plasma of lung cancer patients (indicative of higher IDO activity somewhere in the body) correlated negatively with tumor progression and cancer stage (120). Enhanced expression of IDO is also seen commonly in prostate cancer but not in benign prostate hyperplasia (121). Poor prognosis correlated with increased levels of IDO expression in serous ovarian carcinoma cells but not in other types of ovarian carcinoma (122, 123). Patients with endometrial cancer or esophageal cancer displaying abnormally high IDO expression also had reduced progression-free and overall survival (124, 125). In contrast to these findings, abnormally high IDO expression levels by tumor endothelial cells in renal cell carcinoma patients correlated positively with survival, suggesting that Trp catabolism may mediate anti-tumor effects in this particular setting, possibly by limiting Trp availability to tumor cells or by generation of toxic Trp metabolites (126). Similarly, hepatocellular carcinoma is another example where elevated that IDO expression was a favorable prognostic indicator (127, 128). As with microbial infections, the complexity of potential roles for cells expressing IDO in tumor microenvironments and the potential for diametric effects on regulation of anti-tumor immunity versus direct inhibitory effects on tumor progression complicate interpretation of the effects of Trp catabolism at sites of tumor growth, and more research is needed to further address these critical issues.
IDO1 is also expressed by some APC subsets at tumor lesions and in inflamed draining LNs affected by local tumor growth. In transplantable murine tumor models, cohorts of IDO+ DCs with plasmacytoid morphology and exhibiting phenotypes attributes of both pDCs and B cells (CD11c+B220+CD19+) were present in tumor-draining LNs (76). These DCs suppressed T-cell responses ex vivo and created antigen-specific T-cell anergy following in vivo DC transfer (52, 76). In humans, IDO+ cells with phenotypic attributes of pDCs were identified in draining LNs of patients with melanoma, breast cancer, and other tumors (76, 129, 130). In patients with malignant melanoma, the presence of IDO-expressing DCs in sentinel LN biopsies correlated with significantly worse clinical prognosis. Similarly, immunohistochemical analysis of sentinel LNs in breast cancer and metastatic pancreatic ductal adenocarcinoma patients showed positive correlations between elevated IDO expression and the number of FoxP3+ Tregs in LNs, suggesting that IDO activity in DCs may promote Treg accumulation and regulatory functions that protect developing tumors from anti-tumor immunity at local tumor lesions (131, 132). Thus, enhanced Trp catabolism mediated by immune cells and/or tumor cells expressing IDO is strongly associated with poor outcome in a number of animal models and clinical settings and may constitute a key target for pharmacological intervention to enhance anti-tumor immunity and promote tumor destruction. As discussed previously, similar processes may also occur in settings of tumor progression where elevated Trp or Arg catabolism by tumor cells expressing TDO and MDSCs expressing ARG1 are found (26).
Trp catabolism and regulation of hyper-immune syndromes
Failure to regulate adaptive immunity leads damage or complete destruction of healthy tissues and specialized cell types in autoimmune and allergic syndromes, and following life-saving transplantation of healthy donor organs, tissues, and cells. The finding that IDO activity during mouse pregnancy protected developing fetal tissues from lethal maternal immunity mediated by T cells and complement prompted considerable interest in examining potential roles for the IDO pathway in suppressing the destruction of healthy tissues in clinically significant hyper-immune syndromes (85, 133). Consistent with the paradigm that elevated Trp catabolism suppresses T-cell-mediated immunity at sites of inflammation, studies performed in a range of animal models of hyper-immune syndromes reveal that IDO is often induced at inflammatory lesions, and genetic or pharmacologic IDO ablation frequently accelerates disease progression and exacerbates disease severity in these models (134). In experimental autoimmune encephalitis (EAE), a murine model of multiple sclerosis, IDO expression increased substantially in the late ‘disease remission’ phase, and estrogen and bone marrow stem cells ameliorated EAE severity by a mechanism dependent on IDO expression in DCs (135–137). Increased IDO activity, detected as abnormally high Kyn:Trp ratios in serum, also correlated positively with stages of rheumatoid arthritis (RA), suggesting that IDO activity increases as disease progresses (138). Increased WARS levels were also detected in T cells extracted from synovial fluid of RA patients; this response may confer increased resistance to IDO-mediated regulation in effector T cells that incite joint pathology (139). Additionally, human umbilical cord mesenchymal stem cells suppressed the inflammatory effects of fibroblast-like synoviocytes and T cells from RA patients ex vivo and attenuated the development of collagen-induced arthritis in mice in an IDO-dependent manner (140). In contrast to these studies, IDO activity promoted differentiation of autoreactive B cells into antibody-secreting cells and promoted arthritis progression in the K/BxN mouse model of RA (141); it is not clear why IDO had diametrically opposing effects on RA disease progression in this model, but the involvement of autoreactive B cells rather than, or as well as, T cells may be significant and may also help explain, at least in part, the role of IDO in promoting allergic airway inflammation, a Th2/B cell-mediated inflammatory disease (142). Glucocorticoids, the most commonly prescribed drug used to treat allergic and autoimmune syndromes, may act, at least in part, by stimulating increased Trp catabolism in DCs by upregulating IDO activity via a ‘reverse signaling’ mechanism involving interactions between the glucocorticoid-induced tumor necrosis factor receptor (GITR) and its ligand (GITRL) on T cells and DCs, respectively (143). Recently, we reported that IDO restrains spontaneous progression of autoimmunity that resembles systemic lupus erythematosus in MRLlpr mice prone to this syndrome (74). Collectively, most studies support the hypothesis that increased Trp catabolism protects tissues from damage caused by dysregulated immunity. However, it is important to emphasize that increased IDO activity, although commonly detected at inflamed sites where pathology manifests in hyper-immune syndromes, rarely (if ever) prevents autoimmune disease progression but often attenuates progression in these syndromes. A potentially useful analogy to bear in mind is that firefighters are always found near burning buildings, irrespective of whether they succeed in bringing fires under control before the building is completely destroyed.
Naturally and artificially induced tolerogenic processes that protect transplanted tissues from immune-mediated rejection are complex and involve many different regulatory mechanisms that may predominate in distinct settings of transplantation and methods to promote transplant tolerance (144). However, Cobbold et al. (145) identified multiple biochemical pathways involving amino acid catabolism and synthesis that were upregulated in a well-documented murine model of ‘infectious tolerance’ that protects skin allografts from T-cell-mediated rejection. In this model, Tregs that mediated allograft protection (tolerance) induced and maintained elevated activities of enzymes that catabolize at least five essential amino acids to create robust T-cell tolerance associated with reduced mTOR signaling. Moreover, inhibition of the mTOR pathway led to increased Foxp3 expression via a mechanism dependent on TCR-signaling activation and TGFβ. Because constitutive Treg-mediated suppression was essential to maintain infectious tolerance to skin allografts in this model, these finding suggest that amino acid sensing and signaling via mTOR fine-tunes the delicate balance between sustained suppression and persistent potential to activate graft-specific effector T cells that allows long-term allograft acceptance. Use of immunosuppressant drugs also revealed critical roles for amino acid metabolism in regulating hyper-immunity. For example, Rodriguez-Hernandez et al. (146) showed that use of the immunosuppressant drug tacrolimus (FK506) to enhance organ transplantation survival correlated with induction of sustained Trp depletion that activated GCN2 kinase activity and inhibited protein synthesis by eIF2a phosphorylation. The authors of this study suggested that GCN2 activation was an undesirable toxicity peripheral to the desired inhibitory effects of tacrolimus on calcineurin, a protein phosphatase; however in light of recent data, GCN2 activation likely promotes allograft survival. Enhanced Trp catabolism mediated by IDO in DCs has also been linked to increased Treg numbers and activity that improved allograft survival, attenuated acute rejection, and suppressed GvHD lethality (145, 147–150). The mechanisms that explain these links have not been fully defined but it is likely that Trp withdrawal to activate the GCN2 pathway and/or release of Trp metabolites with immune modulatory properties are involved (also see next section).
Immunotherapies that target amino acid metabolism to treat immunologic diseases
In this concluding section, we discuss emerging opportunities to target biochemical pathways that regulate amino acid metabolism as potential strategies to treat inflammatory and immunologic disease syndromes. We recently reviewed this general topic (134), but the field is developing rapidly, and we discuss novel, controversial, and outstanding topical issues relating to immunotherapies that target pathways of amino acid metabolism.
Inhibiting amino acid catabolism to enhance immunity
Because amino acid catabolism drives immune regulation that contributes to tumor progression and pathogen persistence blocking amino acid catabolism may be an effective strategy to break established tolerance in patients with cancer and chronic infections. Based on this rationale, the D-1MT is under scrutiny as a potential anti-tumor adjuvant in ongoing clinical trials, and ‘second class’ IDO-specific enzymatic inhibitors are currently under development. Selection of the D-isomer of 1MT for clinical trials excited some controversy because D-1MT does not inhibit IDO enzyme activity in many cell lines and tumor cells, whereas L-1MT is an effective IDO inhibitor in these models (151). For example, D-1MT did not and L-1MT did inhibit Kyn production by cell extracts from primary colorectal tumors expressing IDO1 and an inactive form of IDO2 (152). These findings suggested that L-1MT would be the preferred isomer to rescue T-cell-proliferative responses suppressed by cells expressing IDO. However against expectation, D-1MT was substantially more potent than L-1MT (or D,L-1MT) in rescuing proliferation of T cells cultured with human monocyte-derived DCs or murine physiologic DCs from tumor-draining LNs that expressed IDO (129, 151). Moreover, D-1MT also rescued primary CD4+ and CD8+ T-cell proliferation blocked by bystander IDO+ fibroblast cells (69). Consistent with these findings, D-1MT was more efficacious than L-1MT as an anti-cancer agent in chemo-immunotherapy regimens using cyclophosphamide, paclitaxel, or gemcitabine, when tested in mouse models of transplantable melanoma and breast tumor (4T1), and autochthonous (mmTV-neu) breast tumor formation (151). Additionally, D-1MT (and excess Trp) inhibited de novo Treg generation, prevented IDO-mediated activation of pre-formed Tregs by pDCs, and enhanced functional reprogramming of suppressive (activated) Tregs into effector Th17 cells (64, 66, 153). As a component of anti-tumor vaccination protocols, D-1MT also synergized with vaccines to enhance Th17 generation and consequent anti-tumor effects (67). The beneficial effects of D-1MT treatment in tumor-bearing mice were lost in mice lacking intact IDO1 genes (151). In addition, other well documented IDO pathway inhibitors, including 5-Br-brassinin, menadione, methyl-thiohydantoin-tryptophan, phenylimidazole analogs, and new classes of IDO1 inhibitors (some of which block the enzymatic activity of IDO1), all phenocopied the anti-tumor and T-cell stimulatory effects of D-1MT therapy, validating D-1MT as a drug that effectively targets the IDO pathway to block tumor-induced T-cell suppression (154–161). Taken together, these observations suggest that under experimental co-reductant conditions, D-1MT may not inhibit IDO1 enzymatic activity directly but it is nevertheless an effective drug to block IDO effector signaling activity that suppresses antigen-specific T-cell responses in physiologic settings. In other words, D-1MT treatment phenocopies biological effects resulting from inhibition of IDO1 enzyme activity at lower effective concentrations than predicted from estimates based on direct inhibition of enzymatic activity by D-1MT; as such, D-1MT can be considered to be an effective inhibitor of the IDO pathway. Greater understanding of physiological reductants used by IDO that can affect inhibitor profiles along with elucidation of the mechanism of action of D-1MT by targeted disruption of genes suspected to be part of the IDO pathway will help unmask the components in the IDO pathway targeted by D-1MT.
Some potential targets that may account for the activity of D-1MT as an inhibitor of the IDO pathway are as follows. (i) D-1MT may target a Trp transporter. Monocyte-derived macrophages readily deplete Trp from culture medium to nanomolar levels via IDO1 activity. Trp transport is mediated by two different systems: one consistent with the known system for L-amino acid transporter and a second system with 100-fold higher affinity for L-Trp (Km < 300 nM) (72). One possible mechanism of action of D-1MT could be to block the transport of L-Trp into the cell, thereby mimicking the effects of IDO1 or IDO2 enzyme inhibition. Competition studies showed that L-1MT, DL-1MT, 5-OH-D-Trp, and D-Trp blocked the high-affinity Trp transport system or the L-aminoacid transporter at concentrations in the same range (10–100 mM) at which D-1MT promotes immunostimulatory effects (162). Thus, it is a possibility that D-1MT inhibits the high-affinity Trp transporter to phenocopy the effects of direct IDO enzyme inhibitors. (ii) D-1MT may inhibit unique IDO isoforms expressed by physiologic cells. Distinct IDO1 isoforms may be expressed by physiologic human and murine DCs but not by tumor cells or IDO1-transfected (non-DC) cell types. It is important to note that there remains little knowledge of the physiological reductants used by IDO in cells, which can affect inhibitor profiles and may vary in different tissues (R. Metz and G. C. Prendergast, unpublished observations). Some evidence suggests that IDO1 protein does undergo enzymatic activation in immature DCs after IFNγ stimulation (129). Also, IDO1 gene transcripts are subject to alternative splicing in macrophagess after IFNγ stimulation to encode IDO proteins of different lengths, perhaps also due to post-translational modifications (151). (iii) D-1MT may inhibit IDO-2. D-1MT blocked IDO enzyme activity (EC50 approximately 40 μM) encoded by human IDO2 genes in transfected human HEK cells suggesting that IDO2 is a target of D-1MT (2). DCs co-express IDO1 and IDO2 but the much lower enzyme activity of IDO2 is unlikely to explain how D-1MT blocks Trp degradation just by inhibiting IDO2 activity in DCs. Also, IDO2 may be functionally inactive in DCs, whereas L-1MT blocked IDO1 expression in human DCs (152, 163); however, these studies evaluated IDO1 and IDO2 expression solely based on the analysis of RNA levels not active IDO enzyme. Moreover, D-1MT did not inhibit enzymatic activity of recombinant IDO2 in vitro whereas L-1MT did, and IDO1 enzyme may accept L-1MT but not D-1MT as a substrate based on metabolite analyses (R. Metz and M. Mautino, unpublished data). Here again, understanding of the physiological co-reductants required for IDO2 activity in vivo may affect inhibitor profiles and interpretation of their effects. Finally, as D-1MT treatment phenocopied the effects of selective IDO1 gene ablation in mice in several models (see above), it is unlikely that selective inhibition of IDO2 enzyme activity by D-1MT explains the observed effects of D-1MT treatment as a T-cell stimulant. (iv) D-1MT may undergo racemization. D-1mT may be a chiral prodrug of L-1MT. Administering D-Trp increased L-Trp and serotonin levels in animals, probably via enzymatic conversion of D-Trp into L-Trp (164–166). Racemization of D-1MT did occur in mice and humans but at low levels (<10% and <1%, respectively); these low levels make it unlikely that D-1MT racemization explains the observed inhibitory effects of D-1MT on the IDO pathway. (v) D-1MT may block GCN2 activation. Blocking GCN2 activity or other proteins that activate GCN2 downstream of IDO would phenocopy the effects of IDO1 inhibition. For example, tRNATrp synthetases (WARS) may charge tRNATrp with D-1MT to prevent the activation of GCN2. However, we found no evidence that D-1MT is charged onto tRNATrp by WARS1, nor competed with Trp charging onto tRNATrp (R. Metz and M. Mautino, unpublished). Other WARS isoforms exist that retain Trp-binding specificity, even in the absence of tRNATrp charging activity. Short interfering RNA (siRNA)-mediated knockdown of one of these isoforms in Trp-starved cells abolished the stimulatory effects of D-1MT on the phosphorylation status of S6K (R. Metz and M. Mautino, unpublished), suggesting that a WARS variant of may provide amino acid-sensing activity that could be a functional component of the IDO pathway targeted by D-1MT.
These putative mechanisms are not mutually exclusive, and they are all potential points of interest for future pharmacological intervention. Several studies have focused on developing alternative strategies to attenuate IDO-mediated suppression in tumor microenvironments using genetic methods to knockdown IDO activity (167, 168).
Enhancing amino acid catabolism to suppress hyperimmunity
Reagents that enhance amino acid catabolism or activate downstream pathways triggered as a consequence of increased amino acid catabolism are two potential strategies to suppress immune-mediated pathology in autoimmune, allergic, and transplant settings where clinical goals are to ameliorate the tissue destructive effects of hyper-immunity. Hence, transducing IDO genes, use of pharmacologic reagents that induce IDO (IDO inducers) or that mimic the downstream effects of enhanced Trp catabolism are promising strategies to treat a range of hyper-immune syndromes.
Genetic manipulation of IDO expression
Several studies have shown that IDO gene transduction into cells and tissues to enhance IDO activity is an effective strategy to inhibit T-cell-mediated pathology. For example, transducing IDO genes into rats prior to transplanting lungs across histocompatibility barriers led to significantly extended lung allograft survival with minimal indications of immune-mediated pathology in the absence of any immunosuppressant drugs (169, 170). Increased IDO activity in transplanted lungs inhibited clonal expansion of allograft-specific T cells substantially; moreover, the few cytotoxic CD8+ T cells that still infiltrated lung tissues were profoundly anergized due to selective inhibition of the mitochondrial electron transfer pathway (171). This anergizing effect of enhanced IDO activity on graft-infiltrating T cells in reminiscent of the effect of tumor microenvironments on tumor-infiltrating T cells and may represent a hitherto unknown biochemical attribute of IDO-expressing cells. Similarly, several studies have shown that DCs and other cell types transduced to overexpress IDO-attenuated hyper-immunity and decreased disease severity in a range of hyper-immune syndromes of clinical significance. In principle, these approaches may lead to undesirable complications such as increased risk of tumor formation, chronic infection, or altered cellular functions due to constitutively enhanced IDO activity in transplanted tissues or transduced cell types. It remains to be seen if such complications will manifest and whether they can be minimized to facilitate clinical application of IDO gene transduction methods in clinical settings.
Reagents that induce IDO
Reagents that selectively induce IDO offer alternative strategies to enhance IDO activity that are potentially more flexible than IDO gene transduction, because the timing and potency of induced IDO activity can be manipulated by varying dosing regimens and IDO inducers can be combined easily with treatments such as antigen delivery, vaccines, and transplantation. As IDO induction in DCs promotes and sustains robust and stable regulatory phenotypes in Foxp3-lineage Tregs, IDO inducers that stimulate IDO activity in DCs are also likely to be effective drugs to selectively activate Tregs. Enhancing Treg functions to alleviate or prevent progression and onset of autoimmune syndromes such as type I diabetes and inflammatory bowel disease and GvHD syndromes is a major focus of current research (172). IDO inducers may offer better approaches for clinical application than (i) ex vivo Treg generation and transfer or (ii) the use of relatively unselective reagents such as anti-CD3 monoclonal antibodies to activate Tregs, which risks inciting instead of suppressing immunity by activating effector T cells (173). Several known inducers of IDO activity are unsuited for clinical application due to potential toxicities and immune stimulatory properties that may incite rather suppress hyper-immunity (174). For example, CpGs administered systemically (intravenously) at relatively high doses to mice induce rapid and potent IDO activity via TLR9 ligation in DCs that subsequently active Tregs, but CpGs elicit diametrically opposite pro-inflammatory, immune stimulatory, and anti-tumor effects when administered at lower doses via different routes. Similarly, type I and type II IFNs that stimulate increased IDO activity in many cell types (via STAT-signaling pathways) elicit potentially toxic collateral immune stimulatory responses that likely preclude the use of these reagents to induce IDO and activate Tregs in clinical settings of hyper-immune disease. Nevertheless, several IDO-inducing reagents that may be more acceptable for clinical use have been described, including the biological reagents soluble CTLA4 (CTLA4Ig, Abatacept™) and soluble CD83, sCD83 (174). By ligating B7 molecules CTLA4Ig treatment stimulates IDO induction in selected subsets of APCs in humans and mice; additionally, type I diabetes-prone non-obese diabetic female mice were protected from diabetes onset by CTLA4Ig treatment in the pre-diabetic period (175, 176), suggesting that enhanced IDO activity may contribute to the immunesuppressive properties of CTLA4Ig to supplement the regulatory effects of costimulatory blockade. However, the ability of CTLA4Ig preparations to stimulate IDO activity in selected APC subsets depends critically on poorly defined modalities in the Fc (Ig) domain (177), and modifications introduced into CTLA4Ig isoforms currently available for clinical use may have removed IDO-inducing functionalities. sCD83 is a potent immune suppressant that prevented EAE onset and ameliorated established EAE (178), The mode of action of sCD83 has not been defined, but this reagent may bind to certain APCs and promote immune regulation, as some mature APCs express surface CD83. Recently, Lan et al. (179) reported that sCD83-mediated induction of kidney allograft tolerance was IDO-dependent and that DCs from mice that accepted renal allografts due to sCD83 treatment transferred donor antigen-specific tolerance to naive mice in an IDO-dependent fashion. Some histone deacetylase inhibitors possessing anti-tumor activities also induce IDO and have been used to suppress tissue pathology in murine models of GvHD and arthritis (180, 181). Recently, we discovered that systemic administration of nanoparticles containing DNA complexed with the polycationic polymer polyethylenimine (PEI) stimulated rapid upregulation of IDO enzyme activity in lymphoid and mucosal (lung, colon) tissues of mice (182). Administering DNA or PEI polymer alone did not induce IDO, and IDO induction in DCs following DNA nanoparticle treatment was completely dependent on upstream IFN type I signaling but was not dependent on TLR9 or IFNγ signaling. Moreover, DNA nanoparticle treatment blocked vaccine-induced responses to defined antigens by CD8+ and CD4+ T cells in vivo, and ameliorated immune-mediated pathology in inflamed limb joints in a murine model of arthritis. These findings and several other similar studies in the literature demonstrate emphatically that engineering biologic reagents to induce IDO, especially in DCs, in a viable and efficacious approach to treat clinically relevant models of autoimmune disease syndromes, and to slow or prevent rejection of transplanted tissues.
GCN2 pathway inhibitors
Several immune modulatory reagents mimic the effects of increased amino acid catabolism by triggering downstream responses caused by amino acid withdrawal or catabolite production. Thus, Sundrud et al. (183) reported that halofuginone (HF), a derivative of the bioactive ingredient extracted from a Chinese herbal medicine with immunesuppressive properties, triggered GCN2 pathway activation, which inhibited differentiation of pathologic Th17 effector T cells and suppressed inflammation associated with induction of EAE. More recently, Keller et al. (184) identified glutamyl–prolyl-tRNA synthetase (EPRS) as the molecular target of HF and showed that adding exogenous proline or EPRS reversed the inhibitor effects of HF on the GCN2 pathway. These findings demonstrate that HF acts by mimicking the effects of amino acid withdrawal by blocking tRNA charging to trigger the ISR response in immune cells and indicate that activation of the amino acid withdrawal pathway response is beneficial in restraining Th17-driven immune-pathologies.
Immune-suppressive Trp catabolites
Immune modulatory effects of some Trp catabolites or derivatives that resemble such compounds have also been described in several studies, some of which were mentioned above. Thus, PDKP1 was identified as a molecular target of 3-HAA in activated T cells that drove experimental asthma in mice, and 3-HAA treatment suppressed asthma in this model (39). Treatment with the downstream Trp catabolite 3-HAA also ameliorated the severity of EAE significantly, inhibited effector Th1 and Th17 cell generation, and promoted Treg accumulation in diseased mice (185). Kyn administration also phenocopied the therapeutic effects of increasing IDO activity by suppressing inflammation and accumulation of arthritogenic T cells in joints and draining LNs in rodent models of arthritis (140, 186, 187). On the other hand, administering Trp or Trp catabolites potentiated the therapeutic effects of suboptimal tolerogenic immunotherapy to suppress airway eosinophilia, while ablating IDO activity during immunotherapy but not during inhalation challenge, partially reversed the suppressive effects of immunotherapy on some (but not all) disease parameters in a Th2-driven model of allergic asthma (188, 189). Thus, IDO may promote allergen tolerance but exacerbate effector responses when tolerance was absent and active airway allergic inflammation occurs. However, the effects of increased Trp catabolism may vary in different models as studies on allergic responses to Aspergillus fumigatus revealed that IDO activity in DCs led to Treg expansion and activation to produce IL-10 and TGFβ and inhibited pathologic Th2 effector cells to suppress allergy to the fungus. This findings nicely complement the study by Romani et al. (discussed above) (108) in which failure to provide IDO substrate led to effective ‘biochemical’ ablation of IDO activity to confer increased susceptibility to aspergillosis driven Th17-mediated pathology in lungs of mice with a CGD-like genetic defect.
Future perspectives
The last decade has seen an explosion in our knowledge of the role amino acid sensing and catabolism plays in regulation of virtually every aspect of immunity. These novel insights into the biochemical pathways underpinning immune regulation raise the exciting prospect that these mechanisms can be manipulated for therapeutic benefit. As amino acid consumption affects immunity in varied and often diametrically opposed ways, the current challenge is to elucidate the diverse mechanisms by which amino acid catabolizing pathways influence immune responses. This is no small task, for as highlighted above, amino acid catabolism influences cell behavior in a myriad of ways. However, the identification of major response elements in the system (i.e. mTOR, GCN2, AhR, amino acid catabolites) provides targets for focused research.
Acknowledgments
M. M. and R. M. are employees of NewLink Genetics Corporation that is developing IDO inhibitors for clinical use. A. L. M. and G. C. P. receive compensation from the company as consultants and shareholders as well as research grants to their laboratories.
Footnotes
T.L.M., L.H., and H.L. have no conflicts to declare.
References
- 1.Ball HJ, Yuasa HJ, Austin CJ, Weiser S, Hunt NH. Indoleamine 2,3-dioxygenase-2; a new enzyme in the kynurenine pathway. Int J Biochem Cell Biol. 2009;41:467–471. doi: 10.1016/j.biocel.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67:7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- 3.Croitoru-Lamoury J, et al. Interferon-gamma regulates the proliferation and differentiation of mesenchymal stem cells via activation of indoleamine 2,3 dioxygenase (IDO) PLoS ONE. 2011;6:e14698. doi: 10.1371/journal.pone.0014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T, Yokouchi K, Kawamichi H, Yamamoto Y, Uda K, Yuasa HJ. Comparison of the sequences of Turbo and Sulculus indoleamine dioxygenase-like myoglobin genes. Gene. 2003;308:89–94. doi: 10.1016/s0378-1119(03)00467-0. [DOI] [PubMed] [Google Scholar]
- 5.Thomas SR, et al. Post-translational regulation of human indoleamine 2,3-dioxygenase activity by nitric oxide. J Biol Chem. 2007;282:23778–23787. doi: 10.1074/jbc.M700669200. [DOI] [PubMed] [Google Scholar]
- 6.Sugimoto H, Oda S, Otsuki T, Hino T, Yoshida T, Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc Natl Acad Sci U S A. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-gamma-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271:17247–17252. doi: 10.1074/jbc.271.29.17247. [DOI] [PubMed] [Google Scholar]
- 8.Konan KV, Taylor MW. Importance of the two interferon-stimulated response element (ISRE) sequences in the regulation of the human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271:19140–19145. doi: 10.1074/jbc.271.32.19140. [DOI] [PubMed] [Google Scholar]
- 9.Babcock TA, Carlin JM. Transcriptional activation of indoleamine dioxygenase by interleukin 1 and tumor necrosis factor alpha in interferon-treated epithelial cells. Cytokine. 2000;12:588–594. doi: 10.1006/cyto.1999.0661. [DOI] [PubMed] [Google Scholar]
- 10.Currier AR, Ziegler MH, Riley MM, Babcock TA, Telbis VP, Carlin JM. Tumor necrosis factor-alpha and lipopolysaccharide enhance interferon-induced antichlamydial indoleamine dioxygenase activity independently. J Interferon Cytokine Res. 2000;20:369–376. doi: 10.1089/107999000312306. [DOI] [PubMed] [Google Scholar]
- 11.Robinson CM, Shirey KA, Carlin JM. Synergistic transcriptional activation of indoleamine dioxygenase by IFN-gamma and tumor necrosis factor-alpha. J Interferon Cytokine Res. 2003;23:413–421. doi: 10.1089/107999003322277829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellor AL, Baban B, Chandler PR, Manlapat A, Kahler DJ, Munn DH. Cutting Edge: CpG oligonucleotides induce splenic CD19+ dendritic cells to acquire potent indoleamine 2,3-dioxygenase-dependent T cell regulatory functions via IFN gype 1 signaling. J Immunol. 2005;175:5601–5605. doi: 10.4049/jimmunol.175.9.5601. [DOI] [PubMed] [Google Scholar]
- 13.Baban B, et al. A minor population of splenic dendritic cells expressing CD19 mediates IDO-dependent T cell suppression via type I IFN signaling following B7 ligation. Int Immunol. 2005;17:909–919. doi: 10.1093/intimm/dxh271. [DOI] [PubMed] [Google Scholar]
- 14.Belladonna ML, Orabona C, Grohmann U, Puccetti P. TGF-beta and kynurenines as the key to infectious tolerance. Trends Mol Med. 2009;15:41–49. doi: 10.1016/j.molmed.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Hassanain HH, Chon SY, Gupta SL. Differential regulation of human indoleamine 2,3-dioxygenase gene expression by interferons-gamma and -alpha. Analysis of the regulatory region of the gene and identification of an interferon-gamma-inducible DNA-binding factor. J Biol Chem. 1993;268:5077–5084. [PubMed] [Google Scholar]
- 16.Dai W, Gupta SL. Regulation of indoleamine 2,3-dioxygenase gene expression in human fibroblasts by interferon-gamma. Upstream control region discriminates between interferon-gamma and interferon-alpha. J Biol Chem. 1990;265:19871–19877. [PubMed] [Google Scholar]
- 17.Nakajo T, et al. Glutamine is a key regulator for amino acid-controlled cell growth through the mTOR signaling pathway in rat intestinal epithelial cells. Biochem Biophys Res Commun. 2005;326:174–180. doi: 10.1016/j.bbrc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Pearson JT, Siu S, Meininger DP, Wienkers LC, Rock DA. In vitro modulation of cytochrome P450 reductase supported indoleamine 2,3-dioxygenase activity by allosteric effectors cytochrome b(5) and methylene blue. Biochemistry. 2010;49:2647–2656. doi: 10.1021/bi100022c. [DOI] [PubMed] [Google Scholar]
- 19.Vottero E, et al. Cytochrome b(5) is a major reductant in vivo of human indoleamine 2,3-dioxygenase expressed in yeast. FEBS Lett. 2006;580:2265–2268. doi: 10.1016/j.febslet.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 20.Maghzal GJ, Thomas SR, Hunt NH, Stocker R. Cytochrome b5, not superoxide anion radical, is a major reductant of indoleamine 2,3-dioxygenase in human cells. J Biol Chem. 2008;283:12014–12025. doi: 10.1074/jbc.M710266200. [DOI] [PubMed] [Google Scholar]
- 21.Fujigaki H, Seishima M, Saito K. Posttranslational modification of indoleamine 2,3-dioxygenase. Anal Bioanal Chem. 2012;403:1777–1782. doi: 10.1007/s00216-012-5946-2. [DOI] [PubMed] [Google Scholar]
- 22.Orabona C, et al. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc Natl Acad Sci U S A. 2008;105:20828–20833. doi: 10.1073/pnas.0810278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrocksnadel K, Wirleitner B, Winkler C, Fuchs D. Monitoring tryptophan metabolism in chronic immune activation. Clin Chim Acta. 2006;364:82–90. doi: 10.1016/j.cca.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Forouhar F, et al. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2007;104:473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanai M, et al. Tryptophan 2,3-dioxygenase is a key modulator of physiological neurogenesis and anxiety-related behavior in mice. Mol Brain. 2009;2:8. doi: 10.1186/1756-6606-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 27.Pilotte L, et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zea AH, et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell Immunol. 2004;232:21–31. doi: 10.1016/j.cellimm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Zea AH, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez PC, Zea AH, Culotta KS, Zabaleta J, Ochoa JB, Ochoa AC. Regulation of T cell receptor CD3zeta chain expression by L-arginine. J Biol Chem. 2002;277:21123–21129. doi: 10.1074/jbc.M110675200. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez PC, Quiceno DG, Ochoa AC. L-Arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–1573. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez PC, Ochoa AC. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol Rev. 2008;222:180–191. doi: 10.1111/j.1600-065X.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez PC, Zea AH, Ochoa AC. Mechanisms of tumor evasion from the immune response. Cancer Chemother Biol Response Modif. 2003;21:351–364. doi: 10.1016/s0921-4410(03)21018-8. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez PC, et al. L-arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J Immunol. 2003;171:1232–1239. doi: 10.4049/jimmunol.171.3.1232. [DOI] [PubMed] [Google Scholar]
- 36.Serafini P, Mgebroff S, Noonan K, Borrello I. Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res. 2008;68:5439–5449. doi: 10.1158/0008-5472.CAN-07-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Kropf P, et al. Arginase activity mediates reversible T cell hyporesponsiveness in human pregnancy. Eur J Immunol. 2007;37:935–945. doi: 10.1002/eji.200636542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi T, et al. 3-Hydroxyanthranilic acid inhibits PDK1 activation and suppresses experimental asthma by inducing T cell apoptosis. Proc Natl Acad Sci U S A. 2007;104:18619–18624. doi: 10.1073/pnas.0709261104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 41.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miotto G, Venerando R, Khurana KK, Siliprandi N, Mortimore GE. Control of hepatic proteolysis by leucine and isovaleryl-L-carnitine through a common locus. J Biol Chem. 1992;267:22066–22072. [PubMed] [Google Scholar]
- 44.Miotto G, Venerando R, Marin O, Siliprandi N, Mortimore GE. Inhibition of macroautophagy and proteolysis in the isolated rat hepatocyte by a nontransportable derivative of the multiple antigen peptide Leu8-Lys4-Lys2-Lys-beta Ala. J Biol Chem. 1994;269:25348–25353. [PubMed] [Google Scholar]
- 45.Xu G, Kwon G, Marshall CA, Lin TA, Lawrence JC, Jr, McDaniel ML. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J Biol Chem. 1998;273:28178–28184. doi: 10.1074/jbc.273.43.28178. [DOI] [PubMed] [Google Scholar]
- 46.Lynch CJ, Fox HL, Vary TC, Jefferson LS, Kimball SR. Regulation of amino acid-sensitive TOR signaling by leucine analogues in adipocytes. J Cell Biochem. 2000;77:234–251. doi: 10.1002/(sici)1097-4644(20000501)77:2<234::aid-jcb7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 47.Hyde R, Taylor PM, Hundal HS. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem J. 2003;373:1–18. doi: 10.1042/bj20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 49.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 52.Munn DH, et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Highfill SL, et al. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood. 2010;116:5738–5747. doi: 10.1182/blood-2010-06-287839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 55.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 59.Peng W, et al. Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci Transl Med. 2012;4:118ra111. doi: 10.1126/scitranslmed.3002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon NH, et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc Natl Acad Sci U S A. 2011;108:19635–19640. doi: 10.1073/pnas.1103922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee GK, Park HJ, Macleod M, Chandler P, Munn DH, Mellor AL. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology. 2002;107:452–460. doi: 10.1046/j.1365-2567.2002.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fallarino F, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 63.Thornton AM, Piccirillo CA, Shevach EM. Activation requirements for the induction of CD4 + CD25+ T cell suppressor function. Eur J Immunol. 2004;34:366–376. doi: 10.1002/eji.200324455. [DOI] [PubMed] [Google Scholar]
- 64.Sharma MD, et al. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest. 2007;117:2570–2582. doi: 10.1172/JCI31911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baban B, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol. 2009;183:2475–2483. doi: 10.4049/jimmunol.0900986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma MD, et al. Indoleamine 2,3-dioxygenase controls conversion of Foxp3+ Tregs to TH17-like cells in tumor-draining lymph nodes. Blood. 2009;113:6102–6111. doi: 10.1182/blood-2008-12-195354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manlapat AK, Kahler DJ, Chandler PR, Munn DH, Mellor AL. Cell-autonomous control of interferon type I expression by indoleamine 2,3-dioxygenase in regulatory CD19+ dendritic cells. Eur J Immunol. 2007;37:1064–1071. doi: 10.1002/eji.200636690. [DOI] [PubMed] [Google Scholar]
- 68.Scheu S, Stetson DB, Reinhardt RL, Leber JH, Mohrs M, Locksley RM. Activation of the integrated stress response during T helper cell differentiation. Nat Immunol. 2006;7:644–651. doi: 10.1038/ni1338. [DOI] [PubMed] [Google Scholar]
- 69.Forouzandeh F, Jalili RB, Germain M, Duronio V, Ghahary A. Differential immunosuppressive effect of indoleamine 2,3-dioxygenase (IDO) on primary human CD4+ and CD8+ T cells. Mol Cell Biochem. 2008;309:1–7. doi: 10.1007/s11010-007-9635-y. [DOI] [PubMed] [Google Scholar]
- 70.Hattori T, Ohoka N, Hayashi H, Onozaki K. C/EBP homologous protein (CHOP) up-regulates IL-6 transcription by trapping negative regulating NF-IL6 isoform. FEBS Lett. 2003;541:33–39. doi: 10.1016/s0014-5793(03)00283-7. [DOI] [PubMed] [Google Scholar]
- 71.Gao H, Schwartz RC. C/EBPzeta (CHOP/Gadd153) is a negative regulator of LPS-induced IL-6 expression in B cells. Mol Immunol. 2009;47:390–397. doi: 10.1016/j.molimm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Seymour RL, Ganapathy V, Mellor AL, Munn DH. A high-affinity, tryptophan-selective amino acid transport system in human macrophages. J Leukoc Biol. 2006;80:1320–1327. doi: 10.1189/jlb.1205727. [DOI] [PubMed] [Google Scholar]
- 73.Mellor AL, Munn DH. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation? Immunol Today. 1999;20:469–473. doi: 10.1016/s0167-5699(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 74.Ravishankar B, et al. Tolerance to apoptotic cells is regulated by indoleamine 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2012;109:3909–3914. doi: 10.1073/pnas.1117736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munn DH, et al. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114:280–290. doi: 10.1172/JCI21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mellor AL, et al. Specific subsets of murine dendritic cells acquire potent T cell regulatory functions following CTLA4-mediated induction of indoleamine 2,3 dioxygenase. Int Immunol. 2004;16:1391–1401. doi: 10.1093/intimm/dxh140. [DOI] [PubMed] [Google Scholar]
- 77.Muller AJ, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc Natl Acad Sci U S A. 2008;105:17073–17078. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson BA, Jr, et al. B-lymphoid cells with attributes of dendritic cells regulate T cells via indoleamine 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2010;107:10644–10648. doi: 10.1073/pnas.0914347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orabona C, et al. Cutting edge: silencing suppressor of cytokine signaling 3 expression in dendritic cells turns CD28-Ig from immune adjuvant to suppressant. J Immunol. 2005;174:6582–6586. doi: 10.4049/jimmunol.174.11.6582. [DOI] [PubMed] [Google Scholar]
- 80.Stockinger B, Hirota K, Duarte J, Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Semin Immunol. 2011;23:99–105. doi: 10.1016/j.smim.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 81.Vogel CF, Goth SR, Dong B, Pessah IN, Matsumura F. Aryl hydrocarbon receptor signaling mediates expression of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun. 2008;375:331–335. doi: 10.1016/j.bbrc.2008.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen NT, et al. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munn DH, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 86.Baban B, Chandler P, McCool D, Marshall B, Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. J Reprod Immunol. 2004;61:67–77. doi: 10.1016/j.jri.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Baban B, et al. Physiologic control of IDO competence in splenic dendritic cells. J Immunol. 2011;187:2329–2335. doi: 10.4049/jimmunol.1100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Connor JC, et al. Induction of IDO by bacille Calmette-Guerin is responsible for development of murine depressive-like behavior. J Immunol. 2009;182:3202–3212. doi: 10.4049/jimmunol.0802722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Divanovic S, et al. Opposing biological functions of tryptophan catabolizing enzymes during intracellular infection. J Infect Dis. 2012;205:152–161. doi: 10.1093/infdis/jir621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daubener W, et al. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect Immun. 2001;69:6527–6531. doi: 10.1128/IAI.69.10.6527-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heseler K, Spekker K, Schmidt SK, MacKenzie CR, Daubener W. Antimicrobial and immunoregulatory effects mediated by human lung cells: role of IFN-gamma-induced tryptophan degradation. FEMS Immunol Med Microbiol. 2008;52:273–281. doi: 10.1111/j.1574-695X.2007.00374.x. [DOI] [PubMed] [Google Scholar]
- 92.Adams O, Besken K, Oberdorfer C, MacKenzie CR, Russing D, Daubener W. Inhibition of human herpes simplex virus type 2 by interferon gamma and tumor necrosis factor alpha is mediated by indoleamine 2,3-dioxygenase. Microbes Infect. 2004;6:806–812. doi: 10.1016/j.micinf.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 93.Terajima M, Leporati AM. Role of indoleamine 2,3-dioxygenase in antiviral activity of interferon-gamma against vaccinia virus. Viral Immunol. 2005;18:722–729. doi: 10.1089/vim.2005.18.722. [DOI] [PubMed] [Google Scholar]
- 94.Byrne GI, Lehmann LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect Immun. 1986;53:347–351. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeung AW, Wu W, Freewan M, Stocker R, King NJ, Thomas SR. Flavivirus infection induces indoleamine 2,3-dioxygenase in human monocyte-derived macrophages via tumor necrosis factor and NF-kappaB. J Leukoc Biol. 2012;91:657–666. doi: 10.1189/jlb.1011532. [DOI] [PubMed] [Google Scholar]
- 96.Knubel CP, et al. Indoleamine 2,3-dioxigenase (IDO) is critical for host resistance against Trypanosoma cruzi. FASEB J. 2010;24:2689–2701. doi: 10.1096/fj.09-150920. [DOI] [PubMed] [Google Scholar]
- 97.Narui K, et al. Anti-infectious activity of tryptophan metabolites in the L-tryptophan-L-kynurenine pathway. Biol Pharm Bull. 2009;32:41–44. doi: 10.1248/bpb.32.41. [DOI] [PubMed] [Google Scholar]
- 98.Makala LH, et al. Leishmania major attenuates host immunity by stimulating local indoleamine 2,3-dioxygenase expression. J Infect Dis. 2011;203:715–725. doi: 10.1093/infdis/jiq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boasso A, et al. HIV inhibits CD4+ T-cell proliferation by inducing indoleamine 2,3-dioxygenase in plasmacytoid dendritic cells. Blood. 2007;109:3351–3359. doi: 10.1182/blood-2006-07-034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Malleret B, et al. Primary infection with simian immunodeficiency virus: plasmacytoid dendritic cell homing to lymph nodes, type I interferon, and immune suppression. Blood. 2008;112:4598–4608. doi: 10.1182/blood-2008-06-162651. [DOI] [PubMed] [Google Scholar]
- 101.Boasso A, et al. Overactivation of plasmacytoid dendritic cells inhibits antiviral T-cell responses: a model for HIV immunopathogenesis. Blood. 2011;118:5152–5162. doi: 10.1182/blood-2011-03-344218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Potula R, et al. Inhibition of indoleamine 2,3-dioxygenase (IDO) enhances elimination of virus-infected macrophages in an animal model of HIV-1 encephalitis. Blood. 2005;106:2382–2390. doi: 10.1182/blood-2005-04-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boasso A, et al. Combined effect of antiretroviral therapy and blockade of IDO in SIV-infected rhesus macaques. J Immunol. 2009;182:4313–4320. doi: 10.4049/jimmunol.0803314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31:974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rani R, Jordan MB, Divanovic S, Herbert DR. IFN-γ-driven IDO production from macrophages protects IL-4Rα-deficient mice against lethality during Schistosoma mansoni infection. Am J Pathol. 2012;180:2001–2008. doi: 10.1016/j.ajpath.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bozza S, et al. A crucial role for tryptophan catabolism at the host/Candida albicans interface. J Immunol. 2005;174:2910–2918. doi: 10.4049/jimmunol.174.5.2910. [DOI] [PubMed] [Google Scholar]
- 107.Cheng SC, et al. Candida albicans dampens host defense by downregulating IL-17 production. J Immunol. 2010;185:2450–2457. doi: 10.4049/jimmunol.1000756. [DOI] [PubMed] [Google Scholar]
- 108.Romani L, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–215. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- 109.Van der Sluijs KF, et al. Influenza-induced expression of indoleamine 2,3-dioxygenase enhances interleukin-10 production and bacterial outgrowth during secondary pneumococcal pneumonia. J Infect Dis. 2006;193:214–222. doi: 10.1086/498911. [DOI] [PubMed] [Google Scholar]
- 110.Xin L, et al. Systemic treatment with CpG-B after sublethal rickettsial infection induces mouse death through indoleamine 2,3-dioxygenase (IDO) PLoS ONE. 2012;7:e34062. doi: 10.1371/journal.pone.0034062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoshi M, et al. The absence of IDO upregulates type I IFN production, resulting in suppression of viral replication in the retrovirus-infected mouse. J Immunol. 2010;185:3305–3312. doi: 10.4049/jimmunol.0901150. [DOI] [PubMed] [Google Scholar]
- 112.Hoshi M, et al. L-Tryptophan-kynurenine pathway metabolites regulate type I IFNs of acute viral myocarditis in mice. J Immunol. 2012;188:3980–3987. doi: 10.4049/jimmunol.1100997. [DOI] [PubMed] [Google Scholar]
- 113.Moreau M, et al. Inoculation of Bacillus Calmette-Guerin to mice induces an acute episode of sickness behavior followed by chronic depressive-like behavior. Brain Behav Immun. 2008;22:1087–1095. doi: 10.1016/j.bbi.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’Connor JC, et al. Interferon-gamma and tumor necrosis factor-alpha mediate the upregulation of indoleamine 2,3-dioxygenase and the induction of depressive-like behavior in mice in response to bacillus Calmette-Guerin. J Neurosci. 2009;29:4200–4209. doi: 10.1523/JNEUROSCI.5032-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 116.Chamuleau ME, et al. High INDO (indoleamine 2,3-dioxygenase) mRNA level in blasts of acute myeloid leukemic patients predicts poor clinical outcome. Haematologica. 2008;93:1894–1898. doi: 10.3324/haematol.13113. [DOI] [PubMed] [Google Scholar]
- 117.Brandacher G, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 118.Inaba T, et al. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol Oncol. 2009;115:185–192. doi: 10.1016/j.ygyno.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 119.Astigiano S, et al. Eosinophil granulocytes account for indoleamine 2,3-dioxygenase-mediated immune escape in human non-small cell lung cancer. Neoplasia. 2005;7:390–396. doi: 10.1593/neo.04658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suzuki Y, et al. Increased serum kynurenine/tryptophan ratio correlates with disease progression in lung cancer. Lung Cancer. 2010;67:361–365. doi: 10.1016/j.lungcan.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 121.Feder-Mengus C, et al. High expression of indoleamine 2,3-dioxygenase gene in prostate cancer. Eur J Cancer. 2008;44:2266–2275. doi: 10.1016/j.ejca.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 122.Okamoto A, et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin Cancer Res. 2005;11:6030–6039. doi: 10.1158/1078-0432.CCR-04-2671. [DOI] [PubMed] [Google Scholar]
- 123.Takao M, et al. Increased synthesis of indoleamine-2,3-dioxygenase protein is positively associated with impaired survival in patients with serous-type, but not with other types of, ovarian cancer. Oncol Rep. 2007;17:1333–1339. [PubMed] [Google Scholar]
- 124.Ino K, et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br J Cancer. 2006;95:1555–1561. doi: 10.1038/sj.bjc.6603477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sakurai K, et al. Study of indoleamine 2,3-dioxygenase expression in patients of esophageal squamous cell carcinoma. Gan To Kagaku Ryoho. 2004;31:1780–1782. [PubMed] [Google Scholar]
- 126.Riesenberg R, et al. Expression of indoleamine 2,3-dioxygenase in tumor endothelial cells correlates with long-term survival of patients with renal cell carcinoma. Clin Cancer Res. 2007;13:6993–7002. doi: 10.1158/1078-0432.CCR-07-0942. [DOI] [PubMed] [Google Scholar]
- 127.Ishio T, Goto S, Tahara K, Tone S, Kawano K, Kitano S. Immunoactivative role of indoleamine 2,3-dioxygenase in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2004;19:319–326. doi: 10.1111/j.1440-1746.2003.03259.x. [DOI] [PubMed] [Google Scholar]
- 128.Pan K, et al. Expression and prognosis role of indoleamine 2,3-dioxygenase in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:1247–1253. doi: 10.1007/s00432-008-0395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Munn DH, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–1870. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- 130.Lee JR, et al. Pattern of recruitment of immunoregulatory antigen-presenting cells in malignant melanoma. Lab Invest. 2003;83:1457–1466. doi: 10.1097/01.lab.0000090158.68852.d1. [DOI] [PubMed] [Google Scholar]
- 131.Mansfield AS, Heikkila PS, Vaara AT, vonSmitten KA, Vakkila JM, Leidenius MH. Simultaneous Foxp3 and IDO expression is associated with sentinel lymph node metastases in breast cancer. BMC Cancer. 2009;9:231. doi: 10.1186/1471-2407-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Witkiewicz A, et al. Expression of indoleamine 2,3-dioxygenase in metastatic pancreatic ductal adenocarcinoma recruits regulatory T cells to avoid immune detection. J Am Coll Surg. 2008;206:849–854. doi: 10.1016/j.jamcollsurg.2007.12.014. discussion 854–846. [DOI] [PubMed] [Google Scholar]
- 133.Mellor AL, et al. Prevention of T cell-driven complement activation and inflammation by tryptophan catabolism during pregnancy. Nat Immunol. 2001;2:64–68. doi: 10.1038/83183. [DOI] [PubMed] [Google Scholar]
- 134.Johnson BA, 3rd, Baban B, Mellor AL. Targeting the immunoregulatory indoleamine 2,3 dioxygenase pathway in immunotherapy. Immunotherapy. 2009;1:645–661. doi: 10.2217/IMT.09.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sakurai K, Zou JP, Tschetter JR, Ward JM, Shearer GM. Effect of indoleamine 2,3-dioxygenase on induction of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2002;129:186–196. doi: 10.1016/s0165-5728(02)00176-5. [DOI] [PubMed] [Google Scholar]
- 136.Xiao BG, Liu X, Link H. Antigen-specific T cell functions are suppressed over the estrogen-dendritic cell-indoleamine 2,3-dioxygenase axis. Steroids. 2004;69:653–659. doi: 10.1016/j.steroids.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 137.Matysiak M, Orlowski W, Fortak-Michalska M, Jurewicz A, Selmaj K. Immunoregulatory function of bone marrow mesenchymal stem cells in EAE depends on their differentiation state and secretion of PGE2. J Neuroimmunol. 2011;233:106–111. doi: 10.1016/j.jneuroim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 138.Schroecksnadel K, Winkler C, Duftner C, Wirleitner B, Schirmer M, Fuchs D. Tryptophan degradation increases with stage in patients with rheumatoid arthritis. Clin Rheumatol. 2006;25:334–337. doi: 10.1007/s10067-005-0056-6. [DOI] [PubMed] [Google Scholar]
- 139.Schroecksnadel K, et al. Increased degradation of tryptophan in blood of patients with rheumatoid arthritis. J Rheumatol. 2003;30:1935–1939. [PubMed] [Google Scholar]
- 140.Liu Y, et al. Therapeutic potential of human umbilical cord mesenchymal stem cells in the treatment of rheumatoid arthritis. Arthritis Res Ther. 2010;12:R210. doi: 10.1186/ar3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pigott E, Mandik-Nayak L. Addition of an indoleamine-2,3,-dioxygenase inhibitor to B cell depletion therapy blocks autoreactive B cell activation and recurrence of arthritis. Arthritis Rheum. 2012 doi: 10.1002/art.34406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu H, et al. Indoleamine 2,3-dioxygenase in lung dendritic cells promotes Th2 responses and allergic inflammation. Proc Natl Acad Sci U S A. 2008;105:6690–6695. doi: 10.1073/pnas.0708809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grohmann U, et al. Reverse signaling through GITR ligand enables dexamethasone to activate IDO in allergy. Nat Med. 2007;13:579–586. doi: 10.1038/nm1563. [DOI] [PubMed] [Google Scholar]
- 144.Cobbold SP, Adams E, Nolan KF, Regateiro FS, Waldmann H. Connecting the mechanisms of T-cell regulation: dendritic cells as the missing link. Immunol Rev. 2010;236:203–218. doi: 10.1111/j.1600-065X.2010.00913.x. [DOI] [PubMed] [Google Scholar]
- 145.Cobbold SP, et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rodriguez-Hernandez CJ, et al. The immunosuppressant FK506 uncovers a positive regulatory cross-talk between the Hog1p and Gcn2p pathways. J Biol Chem. 2003;278:33887–33895. doi: 10.1074/jbc.M305220200. [DOI] [PubMed] [Google Scholar]
- 147.Reddy P, et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest. 2008;118:2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sucher R, et al. IDO and regulatory T cell support are critical for cytotoxic T lymphocyte-associated Ag-4 Ig-mediated long-term solid organ allograft survival. J Immunol. 2012;188:37–46. doi: 10.4049/jimmunol.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun X, et al. IDO-competent-DCs induced by IFN-γ attenuate acute rejection in rat liver transplantation. J Clin Immunol. 2012 doi: 10.1007/s10875-012-9681-4. [DOI] [PubMed] [Google Scholar]
- 150.Jasperson LK, Bucher C, Panoskaltsis-Mortari A, Mellor AL, Munn DH, Blazar BR. Inducing the tryptophan catabolic pathway, indoleamine 2,3-dioxygenase (IDO), for suppression of graft-versus-host disease (GVHD) lethality. Blood. 2009;114:5062–5070. doi: 10.1182/blood-2009-06-227587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hou DY, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 152.Lob S, et al. IDO1 and IDO2 are expressed in human tumors: levo-but not dextro-1-methyl tryptophan inhibits tryptophan catabolism. Cancer Immunol Immunother. 2009;58:153–157. doi: 10.1007/s00262-008-0513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen W, Liang X, Peterson AJ, Munn DH, Blazar BR. The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J Immunol. 2008;181:5396–5404. doi: 10.4049/jimmunol.181.8.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Banerjee T, et al. A key in vivo antitumor mechanism of action of natural product-based brassinins is inhibition of indoleamine 2,3-dioxygenase. Oncogene. 2008;27:2851–2857. doi: 10.1038/sj.onc.1210939. [DOI] [PubMed] [Google Scholar]
- 155.Kumar S, et al. Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based inhibitors. J Med Chem. 2008;51:1706–1718. doi: 10.1021/jm7014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 157.Mautino MR, JF, Collier S, Marcinowicz-Flick A, Kesharwani T. PCT/US2008/085167 Patent for IDO inhibitors.
- 158.Mautino MR, KS, et al. PCT/US2009/041609. Patent for IDO inhibitors.
- 159.Mautino MR, KS, Jaipuri F, Waldo J, Kesharwani T, Zhang X. PCT/US2010/054289. Patent for IDO inhibitors.
- 160.Mautino MR, KS, et al. PCT/US2012/033245. Patent for IDO inhibitors.
- 161.Yue EW, et al. Discovery of potent competitive inhibitors of indoleamine 2,3-dioxygenase with in vivo pharmacodynamic activity and efficacy in a mouse melanoma model. J Med Chem. 2009;52:7364–7367. doi: 10.1021/jm900518f. [DOI] [PubMed] [Google Scholar]
- 162.Kudo Y, Boyd CA. Characterisation of L-tryptophan transporters in human placenta: a comparison of brush border and basal membrane vesicles. J Physiol. 2001;531:405–416. doi: 10.1111/j.1469-7793.2001.0405i.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lob S, Konigsrainer A, Schafer R, Rammensee HG, Opelz G, Terness P. Levo-but not dextro-1-methyl tryptophan abrogates the IDO activity of human dendritic cells. Blood. 2008;111:2152–2154. doi: 10.1182/blood-2007-10-116111. [DOI] [PubMed] [Google Scholar]
- 164.Yuwiler A. Conversion of D- and L-tryptophan to brain serotonin and 5-hydroxyindoleacetic acid and to blood serotonin. J Neurochem. 1973;20:1099–1109. doi: 10.1111/j.1471-4159.1973.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 165.Ohara I, Otsuka SI, Yugari Y, Ariyoshi S. Inversion of D-tryptophan to L-tryptophan and excretory patterns in the rat and chick. J Nutr. 1980;110:641–648. doi: 10.1093/jn/110.4.641. [DOI] [PubMed] [Google Scholar]
- 166.Triebwasser KC, Swan PB, Henderson LM, Budny JA. Metabolism of D- and L-tryptophan in dogs. J Nutr. 1976;106:642–652. doi: 10.1093/jn/106.5.642. [DOI] [PubMed] [Google Scholar]
- 167.Huang TT, et al. Skin delivery of short hairpin RNA of indoleamine 2,3 dioxygenase induces antitumor immunity against orthotopic and metastatic liver cancer. Cancer Sci. 2011;102:2214–2220. doi: 10.1111/j.1349-7006.2011.02094.x. [DOI] [PubMed] [Google Scholar]
- 168.Sioud M. Modulation of dendritic cell maturation and function by siRNA-bearing 5′-triphosphate. Methods Mol Biol. 2010;629:395–404. doi: 10.1007/978-1-60761-657-3_26. [DOI] [PubMed] [Google Scholar]
- 169.Swanson KA, Zheng Y, Heidler KM, Mizobuchi T, Wilkes DS. CDllc+ cells modulate pulmonary immune responses by production of indoleamine 2,3-dioxygenase. Am J Respir Cell Mol Biol. 2004;30:311–318. doi: 10.1165/rcmb.2003-0268OC. [DOI] [PubMed] [Google Scholar]
- 170.Liu H, Liu L, Fletcher BS, Visner GA. Novel action of indoleamine 2,3-dioxygenase attenuating acute lung allograft injury. Am J Respir Crit Care Med. 2006;173:566–572. doi: 10.1164/rccm.200509-1413OC. [DOI] [PubMed] [Google Scholar]
- 171.Liu H, Liu L, Liu K, Bizargity P, Hancock WW, Visner GA. Reduced cytotoxic function of effector CD8+ T cells is responsible for indoleamine 2,3-dioxygenase-dependent immune suppression. J Immunol. 2009;183:1022–1031. doi: 10.4049/jimmunol.0900408. [DOI] [PubMed] [Google Scholar]
- 172.Leslie M. Immunology. Regulatory T cells get their chance to shine. Science. 2011;332:1020–1021. doi: 10.1126/science.332.6033.1020. [DOI] [PubMed] [Google Scholar]
- 173.Phillips B, Trucco M, Giannoukakis N. Current state of type 1 diabetes immunotherapy: incremental advances, huge leaps, or more of the same? Clin Dev Immunol. 2011;2011:432016. doi: 10.1155/2011/432016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li L, Huang L, Lemos H, Mautino M, Mellor AL. Altered tryptophan metabolism as a paradigm for good and bad aspects of immune privilege in chronic inflammatory diseases. Front Immun. 2012;3:109. doi: 10.3389/fimmu.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Munn DH, Sharma MD, Mellor AL. Ligation of B7-1/B7-2 by human CD4+ T cells triggers indoleamine 2,3-dioxygenase activity in dendritic cells. J Immunol. 2004;172:4100–4110. doi: 10.4049/jimmunol.172.7.4100. [DOI] [PubMed] [Google Scholar]
- 176.Fallarino F, et al. CTLA-4-Ig activates forkhead transcription factors and protects dendritic cells from oxidative stress in nonobese diabetic mice. J Exp Med. 2004;200:1051–1062. doi: 10.1084/jem.20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mellor AL, et al. Cutting edge: induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J Immunol. 2003;171:1652–1655. doi: 10.4049/jimmunol.171.4.1652. [DOI] [PubMed] [Google Scholar]
- 178.Kotzor N, Lechmann M, Zinser E, Steinkasserer A. The soluble form of CD83 dramatically changes the cytoskeleton of dendritic cells. Immunobiology. 2004;209:129–140. doi: 10.1016/j.imbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 179.Lan Z, et al. Induction of kidney allograft tolerance by soluble CD83 associated with prevalence of tolerogenic dendritic cells and indoleamine 2,3-dioxygenase. Transplantation. 2010;90:1286–1293. doi: 10.1097/TP.0b013e3182007bbf. [DOI] [PubMed] [Google Scholar]
- 180.Misaki K, et al. Histone deacetylase inhibition alters dendritic cells to assume a tolerogenic phenotype and ameliorates arthritis in SKG mice. Arthritis Res Ther. 2011;13:R77. doi: 10.1186/ar3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Choi S, Reddy P. HDAC inhibition and graft versus host disease. Mol Med. 2011;17:404–416. doi: 10.2119/molmed.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang L, et al. Engineering nanoparticles as immunomodulatory reagents that activate regulatory T cells. J Immunol. 2012;188:4913–4920. doi: 10.4049/jimmunol.1103668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sundrud MS, et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Keller TL, et al. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat Chem Biol. 2012;8:311–317. doi: 10.1038/nchembio.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yan Y, et al. IDO upregulates regulatory T cells via tryptophan catabolite and suppresses encephalitogenic T cell responses in experimental autoimmune encephalomyelitis. J Immunol. 2010;185:5953–5961. doi: 10.4049/jimmunol.1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Criado G, Simelyte E, Inglis JJ, Essex D, Williams RO. Indoleamine 2,3 dioxygenase-mediated tryptophan catabolism regulates accumulation of Th1/Th17 cells in the joint in collagen-induced arthritis. Arthritis Rheum. 2009;60:1342–1351. doi: 10.1002/art.24446. [DOI] [PubMed] [Google Scholar]
- 187.Jaen O, Rulle S, Bessis N, Zago A, Boissier MC, Falgarone G. Dendritic cells modulated by innate immunity improve collagen-induced arthritis and induce regulatory T cells in vivo. Immunology. 2009;126:35–44. doi: 10.1111/j.1365-2567.2008.02875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Taher YA, et al. Indoleamine 2,3-dioxygenase-dependent tryptophan metabolites contribute to tolerance induction during allergen immunotherapy in a mouse model. J Allergy Clin Immunol. 2008;121:983–991. doi: 10.1016/j.jaci.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 189.Gordon JR, Li F, Nayyar A, Xiang J, Zhang X. CD8 alpha+, but not CD8 alpha-, dendritic cells tolerize Th2 responses via contact-dependent and -independent mechanisms, and reverse airway hyperresponsiveness, Th2, and eosinophil responses in a mouse model of asthma. J Immunol. 2005;175:1516–1522. doi: 10.4049/jimmunol.175.3.1516. [DOI] [PubMed] [Google Scholar]