Abstract
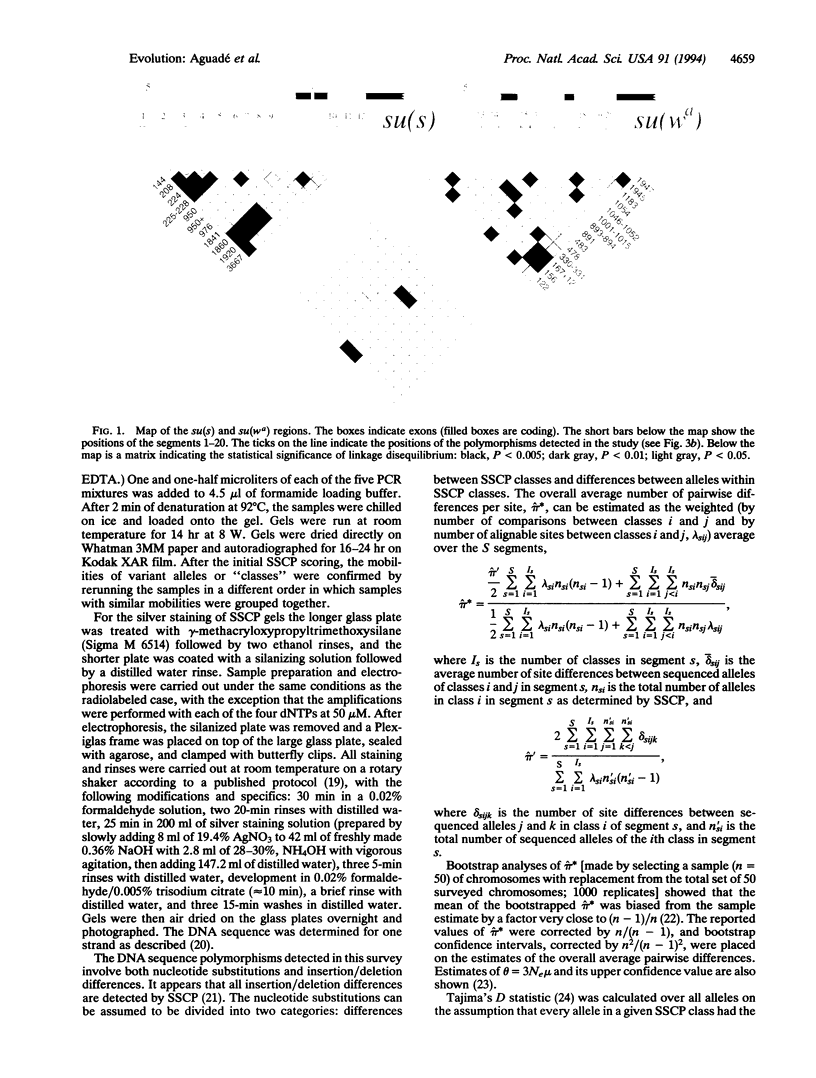
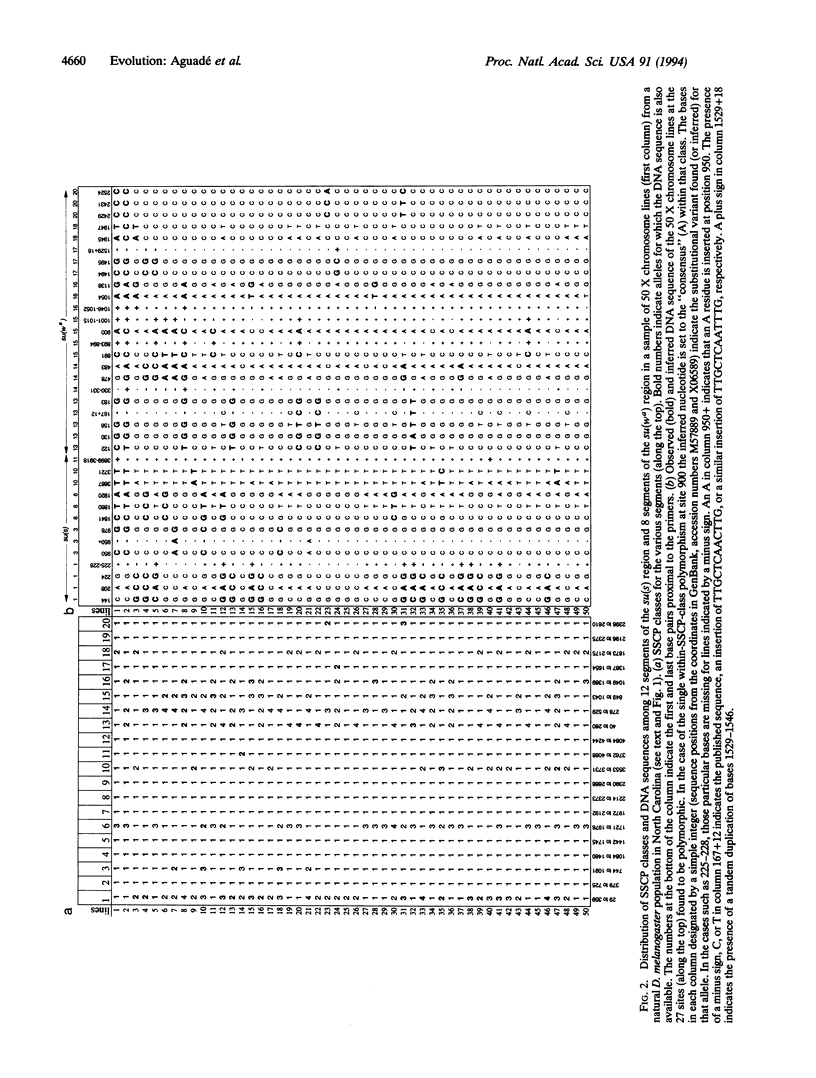
Single-strand conformation polymorphism (SSCP) analysis followed by DNA sequencing of stratified sub-samples was used to survey DNA polymorphism in the su(s) and su(wa) regions in a natural population of Drosophila melanogaster. su(s) and su(wa) are located near the telomere of the X chromosome, where the rate of crossing over per kilobase of DNA monotonically decreases toward the tip. SSCP was assessed in 12 noncoding segments amplified from the su(s) region (3213 bp) and in 8 noncoding segments amplified from the su(wa) region (1955 bp). Sets of segments were multiplexed in a single electrophoretic lane to increase the number of base pairs assayed per lane. Eight segments were monomorphic, and the other 12 segments exhibited two to four SSCP classes. Only four within-SSCP-class DNA sequence differences (a single nucleotide substitution) were observed among 24,360 bp compared within classes. The between-SSCP-class DNA sequence comparisons revealed 27 substitutions and 9 insertion/deletion polymorphisms. The average numbers of substitutional differences per site were 0.0010 and 0.0021 for su(s) and su(wa), respectively. These values are intermediate between those reported for the more distal y-ASC region (0.0004) and the more proximal Pgd locus (0.0024). This observation is consistent with the prediction of the hitchhiking-effect model-i.e., a monotonic increase in polymorphism as a function of crossing over per kilobase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguade M., Miyashita N., Langley C. H. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics. 1989 Jul;122(3):607–615. doi: 10.1093/genetics/122.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé M., Miyashita N., Langley C. H. Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics. 1992 Nov;132(3):755–770. doi: 10.1093/genetics/132.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1993 Oct 7;365(6446):548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992 Apr 9;356(6369):519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F. Molecular population genetics of the distal portion of the X chromosome in Drosophila: evidence for genetic hitchhiking of the yellow-achaete region. Genetics. 1991 Dec;129(4):1147–1158. doi: 10.1093/genetics/129.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A. J., Ajioka J. W., Kreitman M. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics. 1991 Dec;129(4):1111–1117. doi: 10.1093/genetics/129.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993 Aug;134(4):1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. PCR-SSCP: a method for detection of mutations. Genet Anal Tech Appl. 1992 Jun;9(3):73–79. doi: 10.1016/1050-3862(92)90001-l. [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H. The "hitchhiking effect" revisited. Genetics. 1989 Dec;123(4):887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., MacDonald J., Miyashita N., Aguadé M. Lack of correlation between interspecific divergence and intraspecific polymorphism at the suppressor of forked region in Drosophila melanogaster and Drosophila simulans. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1800–1803. doi: 10.1073/pnas.90.5.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984 Aug;107(4):611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Campos J. M., Comerón J. M., Miyashita N., Aguadé M. Intraspecific and interspecific variation at the y-ac-sc region of Drosophila simulans and Drosophila melanogaster. Genetics. 1992 Apr;130(4):805–816. doi: 10.1093/genetics/130.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita N. T., Aguadé M., Langley C. H. Linkage disequilibrium in the white locus region of Drosophila melanogaster. Genet Res. 1993 Oct;62(2):101–109. doi: 10.1017/s0016672300031694. [DOI] [PubMed] [Google Scholar]
- Miyashita N. T. Molecular and phenotypic variation of the Zw locus region in Drosophila melanogaster. Genetics. 1990 Jun;125(2):407–419. doi: 10.1093/genetics/125.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974 Feb;23(1):23–35. [PubMed] [Google Scholar]
- Stephan W., Langley C. H. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila ananassae populations. I. Contrasts between the vermilion and forked loci. Genetics. 1989 Jan;121(1):89–99. doi: 10.1093/genetics/121.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Mitchell S. J. Reduced levels of DNA polymorphism and fixed between-population differences in the centromeric region of Drosophila ananassae. Genetics. 1992 Dec;132(4):1039–1045. doi: 10.1093/genetics/132.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila ananassae populations. II. The Om(1D) locus. Mol Biol Evol. 1989 Nov;6(6):624–635. doi: 10.1093/oxfordjournals.molbev.a040580. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989 Nov;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe T. H., Stephan W. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol Biol Evol. 1993 Jul;10(4):842–854. doi: 10.1093/oxfordjournals.molbev.a040046. [DOI] [PubMed] [Google Scholar]