SUMMARY
Spontaneous T cell responses against tumors occur frequently and have prognostic value in patients. The mechanism of innate immune sensing of immunogenic tumors leading to adaptive T cell responses remains undefined, although type I interferons (IFNs) are implicated in this process. We found that spontaneous CD8+ T cell priming against tumors was defective in mice lacking stimulator of interferon genes complex (STING), but not other innate signaling pathways, suggesting involvement of a cytosolic DNA sensing pathway. In vitro, IFN-β production and dendritic cell activation were triggered by tumor-cell-derived DNA, via cyclic-GMP-AMP synthase (cGAS), STING, and interferon regulatory factor 3 (IRF3). In the tumor microenvironment in vivo, tumor cell DNA was detected within host antigen-presenting cells, which correlated with STING pathway activation and IFN-β production. Our results demonstrate that a major mechanism for innate immune sensing of cancer occurs via the host STING pathway, with major implications for cancer immunotherapy.
INTRODUCTION
Spontaneous T cell responses against human cancers are believed to contribute to the control of tumor growth, based on the observed prognostic benefit of an immune infiltrate in the tumor microenvironment in patients. In metastatic disease, a preexisting T cell-inflamed tumor microenvironment appears to be associated with clinical responses to therapeutic vaccines and other immunotherapies and is being explored as a predictive biomarker (Gajewski et al., 2010; Hamid et al., 2011; Harlin et al., 2009). Preliminary data exploring clinical responses to anticytotoxic T-lymphocyte-associated protein 4 (CTLA-4) or antiprog-rammed cell death protein 1 (PD-1) mAbs also have suggested that patients with clinical benefit have a preexisting CD8+ T cell infiltrate and associated gene signature (Ji et al., 2012; Spranger et al., 2013; Topalian et al., 2012). In early-stage colon cancer, the presence of effector-memory CD8+ T cells has powerful prognostic importance, having been reported to be more predictive of outcome than tumor-node-metastasis (TNM) stage (Pagès et al., 2009). Similar positive prognostic import has been observed in breast cancer (Mahmoud et al., 2011) and in ovarian cancer (Hwang et al., 2012). However, the mechanism by which the host immune system initiates innate immune sensing of tumors and thereby bridges to induction of an adaptive tumor-specific T cell response is largely unknown. It has been suggested that endogenous adjuvants released from dying cells are capable of initiating innate immune cell activation (Jounai et al., 2012; Kono and Rock, 2008; Marichal et al., 2011; McKee et al., 2013). In chemotherapy and radiotherapy models, treated cancer cells were shown to release ATP or/and high-mobility group protein B1 (HMGB1) and activate dendritic cells (DCs) via the inflammasome or Toll-like receptor 4 (TLR4) pathways, respectively, which in turn contributed to activation of antitumor T cells (Apetoh et al., 2007; Ghiringhelli et al., 2009). These data have indicated that tumor cell-derived factors can facilitate induction of antitumor immunity that contributes to tumor control with conventional cancer therapeutics. However, in the context of a spontaneous natural antitumor T cell response, the factors and mechanisms necessary to induce innate immune sensing might be distinct and have not been defined. This represents a critical knowledge gap, because strategies to trigger this innate immune activation and generate an endogenous T cell response might be necessary to expand the fraction of patients who can derive clinical benefit from current immunotherapies.
Spontaneous tumor antigen-specific T cell priming, when it does occur, appears to be dependent on host type I IFN production and signaling on host cells, via a mechanism that involves promotion of cross-presentation by CD8α+ DCs (Diamond et al., 2011; Fuertes et al., 2011). In the current report, we investigated upstream pathways that might trigger this type I IFN production in response to tumors. In vivo, we found no evidence for a major role for host myeloid differentiation primary response gene 88 (MyD88), Toll/interleukin-1 (IL-1) receptor (TIR) domaincontaining adaptor (TRIF), Toll-like receptor 4 (TLR4), Toll-like receptor 9 (TLR9), P2X purinoreceptor (P2×7R), or mitochondrial antiviral-signaling protein (MAVS) for spontaneous priming of antitumor CD8+ T cells. In contrast, spontaneous CD8+ T cell priming was severely blunted in Tmem173−/− (STING-deficient) and lrf3−/− (IRF3-deficient) mice, and rejection of immunogenic tumors was also ablated. In vitro, the only tumor-derived substance that could induce interferon-β (IFN-β) production was DNA, which was mediated through cGAS, STING, and IRF3. At a single cell level, we observed transfer of tumor-derived DNA into host APCs in vivo, which was associated with TANK-binding kinase 1 (TBK1) and IRF3 phosphorylation, and IFN-β production. Our results demonstrate that a major mechanism for innate immune sensing of cancer is via a cytosolic DNA-STING pathway. These results open up new opportunities for understanding the mechanisms explaining a natural immune response in cancer patients, as well as for developing new therapeutic strategies through facilitating this innate immune response.
RESULTS
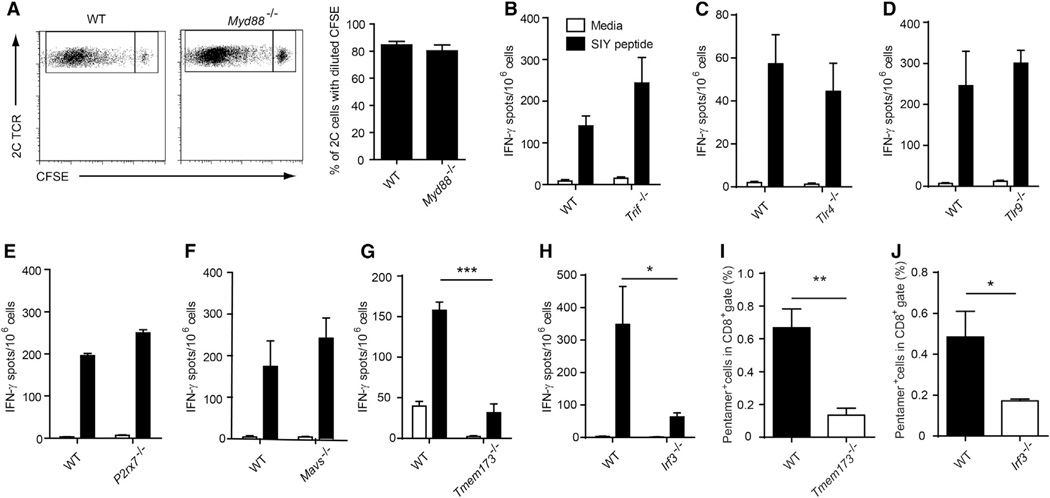
STING and IRF3 Are Required for Spontaneous T Cell Activation against Tumors In Vivo
Recent work has indicated that host type I IFN production serves as a bridge between tumor sensing and a spontaneous adaptive T cell response, via a mechanism involving CD8α+ dendritic cells (Diamond et al., 2011; Fuertes et al., 2011). We therefore pursued a working model in which innate immune sensing pathways might detect tumor-derived factors, induce type I IFN production, and lead to cross-priming of tumor antigen-specific CD8+ T cells (Diamond et al., 2011; Fuertes et al., 2011). To begin to address this question, we utilized gene-targeted mice deficient in specific innate pathways. To determine whether host TLR pathways were required for spontaneous CD8+ T cell priming, we utilized Myd88−/− or Trif−/− mice. Because MyD88 can function in a T cell-intrinsic fashion (Zhou et al., 2009), we performed adoptive transfer of wild-type (WT) CFSE-labeled 2C TCR Tg T cells (that are specific for the model antigen SIY) into WT or Myd88−/− mice and challenged with B16.SIY tumors (Blank et al., 2004). No defect in T cell proliferation or accumulation of divided cells was observed in Myd88−/− hosts (Figure 1A). For interrogation of the other innate immune pathways, we implanted B16. SIY melanoma cells and measured endogenous immune response using IFN-γ ELISPOT and SIY peptide/Kb pentamer flow cytometry. Endogenous CD8+ T cell priming against tumor-derived SIY was intact in Trif−/− mice (Figure 1B), suggesting that the TLR system (Ishii et al., 2006; Stetson and Medzhitov, 2006) is not mandatory. We also examined CD8+ T cell responses in mice lacking TLR4 or TLR9, and no defect was observed using either IFN-γ ELISPOT (Figure 1C and 1D) or SIY peptide/Kb pentamer staining (see Figure S1A and S1B available online). A second candidate mechanism of innate immune sensing is through extracellular ATP, as it has been suggested that dying tumor cells might release ATP, which could be sensed by P2×7R on antigen-presenting cells (APCs) (Ghiringhelli et al., 2009). However, we found no defect in spontaneous priming of CD8+ T cells against tumor-associated antigens in P2×7r−/− mice (Figure 1E). We also examined a role for the defined RNA sensing pathway using Mavs−/− mice, which lack the critical adaptor molecule for RIG−/− and MDA5-dependent innate immune activation. However, no defect of CD8+ T cell priming in Mavs−/− mice was observed (Figure 1F).
Figure 1. STING and IRF3 Are Required for CD8+ T Cell Priming In Vivo.
(A) CFSE-labeled 2C T cell were transferred into WT (n = 6) or Myd88−/− (n = 6) mice and B16.SIY melanoma cells were inoculated 1 day later. After 6 days, mice were sacrificed and splenocytes were stained with anti-CD8 and the clonotypic mAb 1B2 and analyzed by flow cytometry for CFSE dilution.
(B) Trif−/− (n = 5), (C) Tlr4−/− (n = 4), (D) Tlr9−/− (n = 5), (E) P2×7r−/− (n = 5), (F) Mavs−/− (n = 5), (G) Tmem173−/− (STING-deficient) (n = 5), (H) Irf3−/− (IRF3-deficient) (n = 5) mice were inoculated with 106 B16.SIY melanoma cells. After 7 days, splenocytes were analyzed for SlY-specific IFN-γ -producing CD8+ T cell frequencies by ELISPOT assay.
(I and J) SIY peptide-specific pentamer staining was performed in Tmem173−/− (n = 5) and lrf3−/− (n = 5) mice, respectively. WT mice were used as controls. *p < 0.05, ***p < 0.001 (Student’s t test). Data represent mean ± SEM and are representative of two to three independent experiments. See also Figures S1 and S2.
We therefore turned to the other remaining defined pathway for innate immune sensing that can lead to type I IFN production, which is cytosolic DNA sensing via the STING pathway. Recent studies of pathogen sensing have identified a pathway involving an endoplasmic reticulum resident protein called STING leading to IRF3 activation and IFN-β transcription (Ishikawa et al., 2009). STING has been shown to function as an adaptor molecule for DNA recognition pathways and likely occurs indirectly through binding of cyclic dinucleotides generated by the enzyme cGAS (Abe et al., 2013; Burdette et al., 2011; Wu et al., 2013). Strikingly, in both STING-deficient and IRF3-deficient mice, we observed a substantially diminished CD8+ T cell response against tumor-associated antigen in vivo (Figure 1G and 1H), using both IFN-γ ELISPOT and SIY/Kb pentamer analysis (Figure 1I and 1J). These data suggest that STING and IRF3 in host cells are required for spontaneous CD8+ T cell priming response against immunogenic tumors.
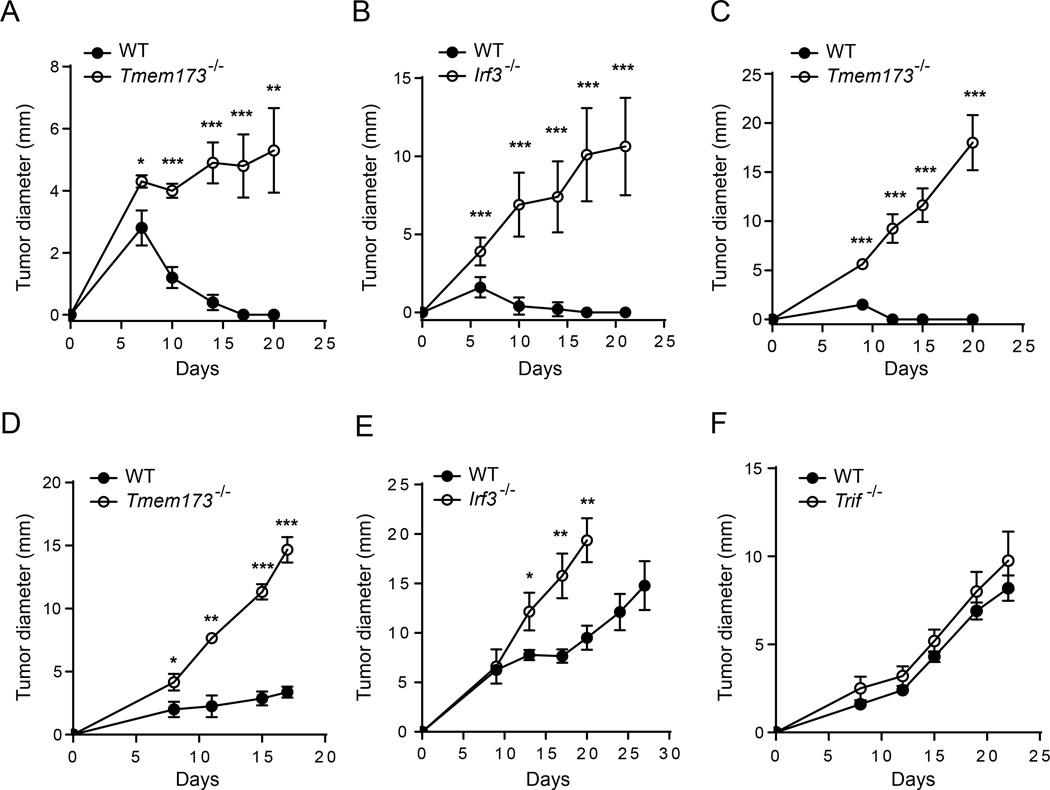
STING-Deficient Mice and IRF3-Deficient Mice Show Defective Tumor Control
To assess the role of STING in tumor growth control, we focused on model systems in which immunogenic tumors were spontaneously rejected. Methylcholanthrene (MCA)-induced sarcomas generated in immune-deficient mice are rejected when implanted in immune-competent mice due to lack of having been subjected to immune editing (Matsushita et al., 2012). This was the case for the MCA-induced sarcoma 1969; however, rejection of this tumor was eliminated in STING-deficient mice and IRF3-deficient mice (Figure 2A and 2B). As another approach, in order to have complete rejection of B16.SIY tumor cells, they were implanted into WT or STING-deficient mice on a 129 genetic background, which is still H-2b but allows tumor rejection likely due to a contribution of minor histocompatibility antigens. In contrast to rejection in WT mice, tumors grew progressively in STING-deficient 129 mice (Figure 2C). We also measured tumor growth of B16.SIY tumors in syngeneic C57BL/6 mice, and while these tumors are not rejected in WT mice, they nonetheless grew faster in STING-deficient and IRF3-deficient hosts, indicating a degree of natural immune control that was similarly STING-dependent (Figure 2D and 2E). In contrast, there was no difference of B16.SIY tumor growth in between WT and Trif−/−mice (Figure 2F).
Figure 2. Tmem173−/−Mice andIrf3−/−Mice Show Defective Tumor Control.
(A and B) 1969 tumor cells were inoculated into WT, Tmem173−/−, or Irf3−/− mice and tumor growth was recorded on the indicated days.
(C) WT or Tmem173−/− mice (129 background) were inoculated with 106 B16.SIY melanoma cells and tumor size was measured over time.
(D–F) B16.SIY tumor cells (106 cells per mouse) were injected into the indicated mice subcutaneously, and tumor growth was measured on the indicated days.
*p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). Data represent mean ± SEM and representative of at least two independent experiments.
To determine whether the failed tumor rejection in STING-deficient mice was relatively unique to the tumor context versus a general property of tissue rejection, we investigated skin graft rejection across minor histocompatibility antigen differences (Molinero et al., 2008). Interestingly, the rate of rejection of STING-deficient male skin into STING-deficient female recipients was identical compared to WT donor-recipient pairs (Figure S2). Thus, not all tissue-based T cell rejection processes are STING-dependent; rather, the tumor context has special properties that render host T cell priming particularly dependent on the STING pathway.
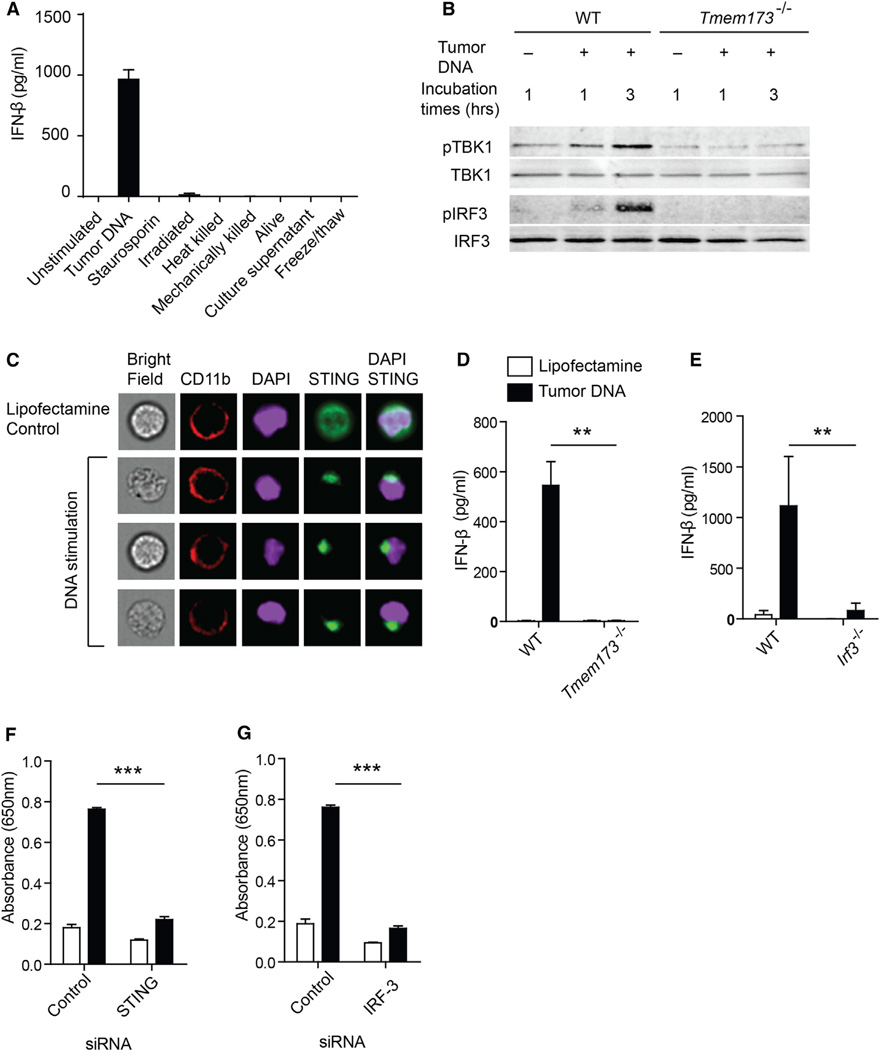
Tumor-Derived DNA Induces IFN-β Production by a cGAS-, STING-, and IRF-3-Dependent Pathway
We turned to an in vitro system to screen fractions of B16 tumor cell extracts and tumor cells killed using a variety of approaches, to determine which preparation might be capable of inducing IFN-β from DCs. Tumor cells killed in multiple ways, including by mechanical disruption, or supernatants from spent B16 cultures failed to induce IFN-β production by bone-marrow-derived DCs (Figure 3A). Based on recent reports characterizing a cytosolic DNA sensing pathway that can detect intracellular viruses, bacteria, and Plasmodium falciparum and drive type I IFN production (Henry et al., 2007; Sharma et al., 2011; Takaoka et al., 2007; Unterholzner et al., 2010), we examined whether tumor-derived DNA might act similarly. Indeed, B16 melanoma-derived total DNA combined with Lipofectamine provoked IFN-β production by DCs (Figure 3A). The inclusion of Lipofectamine was necessary, suggesting that the DNA needed to gain entry to the cytosol, as has been reported previously (Stetson and Medzhitov, 2006). Treatment of the tumor-derived DNA preparation with DNase I abolished this stimulatory effect, supporting DNA as the active component (data not shown). Tumor-derived RNA was minimally stimulatory (data not shown). In immortalized macrophages cells, tumor-derived DNA in combination with Lipofectamine also induced production of IFN-β (Figure S3A). In addition to tumor-derived DNA, normal cell-derived DNA isolated from splenocytes also induced production of IFN-β by mouse BMDCs when combined with Lipofectamine in vitro (Figure S5B), suggesting that there is unlikely to be a unique property of DNA derived from transformed cells that make it more stimulatory. Rather, there must be some characteristic of the tumor cell context that favors DNA transfer to host APCs as tumors become established in vivo.
Figure 3. Tumor-Derived DNA Activates STING Pathway and Induces IFN-β via cGAS-, STING-, and IRF3-Dependent Mechanism.
(A) Cultured B16 melanoma tumor cells were manipulated or used as a source of material as indicated, and incubated with BM-DCs for 18 hr. The amount of secreted IFN-β was measured by ELISA.
(B) BM-DC cells from WT or Tmem173−/− mice were stimulated with 1 µg/ml of tumor-derived DNA for the indicated time points. The amount of pTBK1, total TBK1, plRF3, and total IRF3 was measured by immunoblot.
(C) Asc−/− macrophages that overexpress STING-HA tag were stimulated with tumor-derived DNA for 1 hr and stained for CD11b, HA-tag, and DAPI. Single cell images were acquired using the ImageStream instrument and analyzed with IDEAS software.
(D and E) BM-DCs were generated from WT, Tmem173−/−or lrf3−/− mice and stimulated with tumor DNA in the presence of Lipofectamine. The amount of IFN-β was measured by ELISA.
(F and G) IFN-β reporter cells were transfected with siRNAs specific for STING or IRF3 followed by stimulation with tumor DNA. Reporter activity was measured as described in Experimental Procedures. *p<0.05, **p < 0.01, ***p< 0.001 (Student’s t test). Data represent mean ± SEM and representative of three independent experiments. See also Figure S3.
To assess activation of the STING pathway, we performed immunoblot analysis to assess phosphorylation of TBK1 and IRF3 after tumor-derived DNA stimulation of bone-marrow-derived DCs. We indeed observed increased phosphorylation of TBK1 and IRF3 in DCs from WT mice that was not seen in DCs from STING-deficient mice. The amount of each protein was normalized with GAPDH loading control and the ratio of phosphorylated to total proteins was quantified (pTBKI/TBKI: WT [2.076] versus STING-deficient [0.705], pIRF3/IRF3: WT [0.308] versus STING-deficient [0.009]; p < 0.051, p < 0.0001) (Figure 3B). This amount of phosphorylation of TBK1 and IRF3 was comparable to what we observed with LPS stimulation, although the latter was preserved in STING-deficient DCs (Figure S3B). It has been reported that activated STING accumulates as aggregates in a perinuclear region (Ishikawa et al., 2009). Induced aggregation of STING was also observed in murine macrophage cells in response to tumor DNA (Figure 3C).
Recent data have indicated that the ultimate direct ligand of STING is cyclic dinucleotides, generated in response to cytosolic DNA binding the enzyme cGAS (Sun et al., 2013; Wu et al., 2013). In the present model, siRNA knockdown of cGAS in macrophages also significantly reduced production of IFN-β in response to tumor DNA stimulation (Figure S3C and S3D). IFN-β production was severely blunted with STING-deficient or IRF3-deficient DCs (Figure 3D and 3E), confirming a requirement for this defined pathway for DC activation. As a confirmatory approach, we utilized a reporter cell line expressing the secreted embryonic alkaline phosphatase (SEAP) enzyme driven by the IFN-β-inducible IFN-induced 54 kDa protein (ISG54) promoter. In this system, specific siRNAs for STING or IRF3 resulted in substantial inhibition of IFN-β-inducible ISG54 promoter activity after stimulation with tumor-derived DNA (Figure 3F and 3G). STING- or IRF3- and IRF7-deficient macrophages also did not produce IFN-β after tumor-derived DNA stimulation (Figure S3E). Reintroduction of STING into STING-deficient macrophages restored production of IFN-β after tumor-DNA stimulation (Figure S3F). These data indicate that tumor-derived DNA can induce production of IFN-β when introduced into APCs in vitro, via a mechanism dependent upon cGAS, STING, and IRF3.
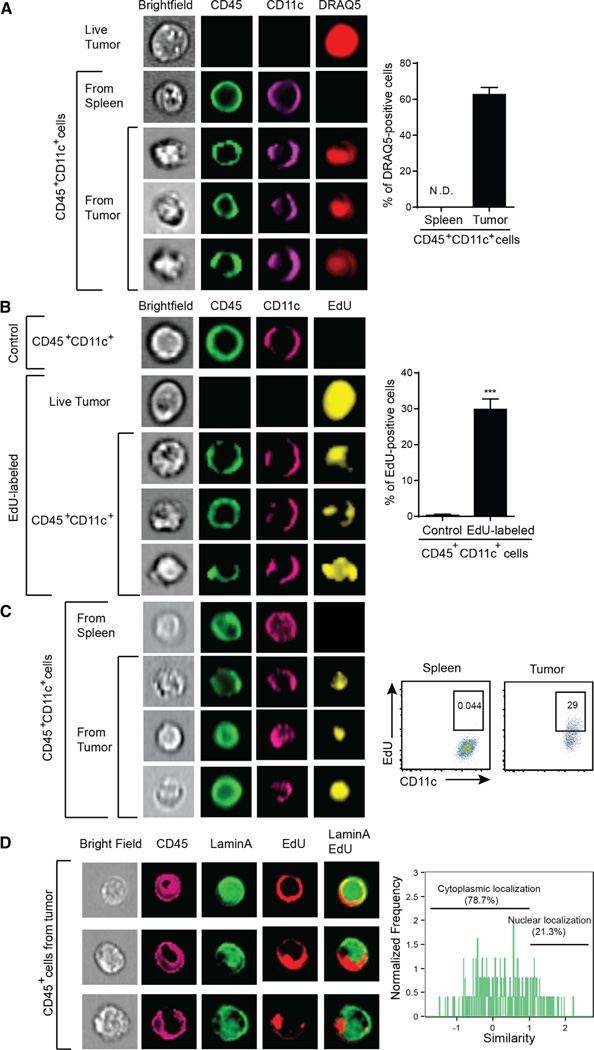
Tumor-Derived DNA Is Transferred to Host APCs In Vivo
Because killed tumor cells in vitro could not recapitulate the IFN-β production triggered upon tumor implantation in vivo, we pursued a direct in vivo approach to determine whether tumor-derived DNA was indeed transferred to host APCs within the tumor microenvironment and whether this resulted in STING pathway activation. B16 tumor cells were stained in vitro with DNA-intercalating dye DRAQ5 prior to implantation in vivo. In order to avoid dilution of the dye as a consequence of cell proliferation, we performed most of these experiments analyzing host inflammatory cells 1 day after tumor injection. The early tumor bump was harvested, disrupted into a single cell suspension, and analyzed by cytometry. In order to ensure that the analysis focused exclusively on host myeloid cells and not fusion heterokaryons or cell aggregates, single cell analysis using the Amnis ImageStream instrument was employed. Host DCs were identified based on staining for CD11c and CD45. Indeed, approximately 60% of CD45+CD11c+ cells showed positive staining with tumor-cell-derived DRAQ5, in a diffuse staining pattern (Figure 4A), whereas controls were negative. Using an in vitro trans-well system, we found that DRAQ5 transfer was not detected in nonlabeled cells separated by a membrane, arguing that detection in DCs was not a consequence of leaking of the DRAQ5 dye from the tumor cells (data not shown).
Figure 4. Tumor-Infiltrating Host APCs Uptake Tumor-Derived DNA In Vivo.
(A) B16.SIY tumor cells were stained with DRAQ5, extensively washed, and then inoculated into mice subcutaneously. The next day, isolated single-cell suspensions were stained and single cell images were acquired using ImageStream. Acquired images were analyzed with IDEAS software.
(B) B16.SIY tumor cells were labeled with EdU, extensively washed, and then inoculated into mice. The next day, tumor bumps were harvested and EdU staining was analyzed by ImageStream. Nonlabeled tumor cells were used as a negative control.
(C) B16 melanoma cells were stained with EdU and injected into mice subcutaneously. At day 5, when tumors were becoming palpable, tumors were isolated including any infiltrating host cells and stained to exclude dead cells, along with anti-CD11 c, anti-CD45, and for EdU. Stained cells were acquired by ImageStream or flow cytometry and analyzed by IDEAS or FlowJo software.
(D) At day 3 after injection of EdU-labeled B16 melanoma cells, tumor bumps were isolated and stained with anti-CD45, anti-Lamin A, and for EdU. Images of stained cells were acquired by Amnis ImageStream, and colocalization of EdU and Lamin A signals was analyzed using IDEAS software. ***p < 0.001 (Student’s t test). Data indicate mean ± SEM and representative of at least three (A, B, and C) or two (D) independent experiments. See also Figure S4.
As a second approach, B16 tumor cells were labeled with the nucleotide analog EdU prior to injection into mice. EdU would be covalently incorporated into DNA and would eliminate concern for potential dissociation of the DRAQ5 dye and independent uptake. Similar to DRAQ5, we observed EdU staining on a large population of tumor-infiltrating CD45+CD11c+ cells (Figure 4B). We also observed tumor-derived EdU-labeled DNA transfer to host APCs using the 1969 tumor cell system, arguing that this phenomenon is not unique to B16 melanoma (Figure S4A). Compared to DRAQ5 staining, the percentage of EdU-positive cells in host APCs was consistently lower than that of DRAQ5-positive cells. This difference could be because only a subset of the tumor DNA incorporates EdU with this strategy, or because the EdU labeling is more specific for DNA.
We were concerned that the evidence for DNA transfer to host APCs might be restricted to the first day of tumor cell implantation, when solid tumors have not yet been established. We therefore examined whether tumor-derived EdU could be identified within APCs at later time points, when the tumor was palpable. Indeed, EdU persisted in tumor-infiltrating CD11c+ cells at day 5, after tumors were clearly palpable (Figure 4C). We also examined the intracellular localization of the tumor-derived EdU label by costaining tumor-infiltrating CD45+ cells for the nuclear marker Lamin A or the lysosomal marker LAMP-1. The majority of the signal did not colocalize with either marker (Figure 4D and S4B). These latter data also suggest that the DNA label being seen in host APCs is not simply present in a phagocytic compartment, but rather appears to be localized in the cytosol, which would provide access to the STING pathway for engagement.
Tumor-Infiltrating Hosts APCs Produce IFN- β via a STING-Dependent Mechanism In Vivo
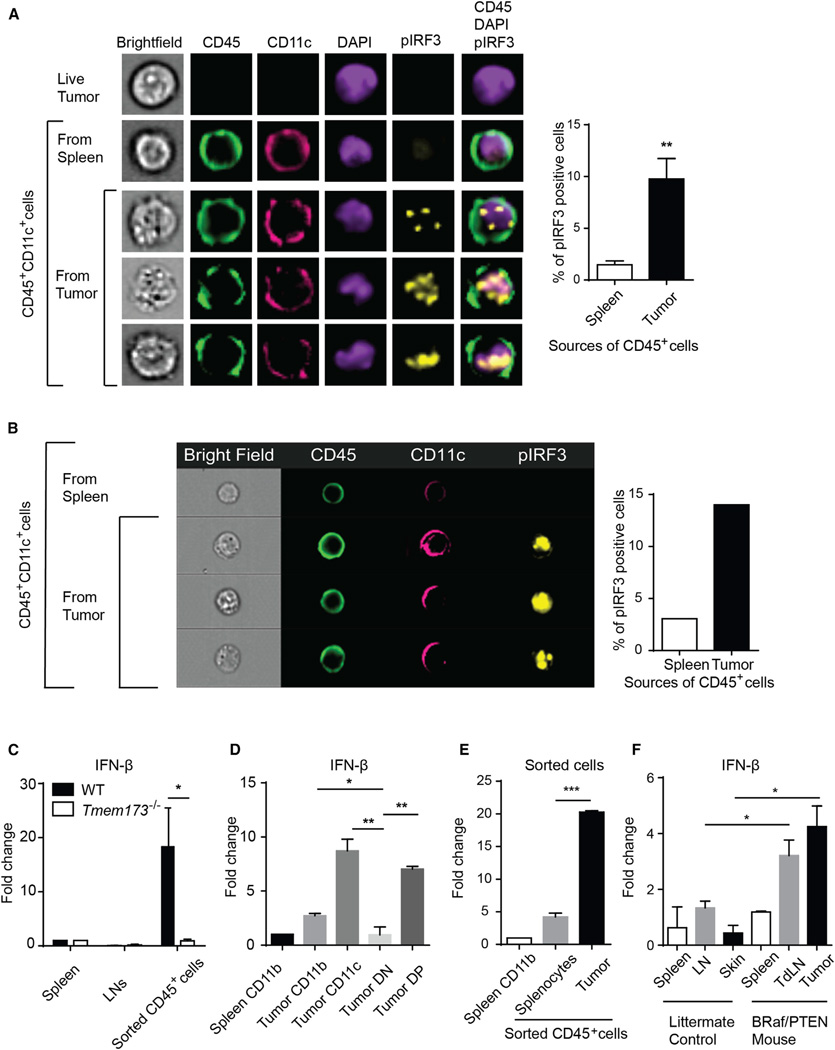
We investigated whether host APCs activated the STING pathway and produced IFN-β within the same time frame as detection of DNA uptake. B16 melanoma cells were implanted subcutaneously, and 1 day later the tumor-infiltrating CD45+ cells were analyzed for phospho-IRF3 induction by ImageStream. As shown in Figure 5A, despite this being a snapshot in time at an early time point, approximately 10% of tumor-infiltrating CD45+ cells showed pIRF3 staining, which appeared to be translocated to nucleus (Figure 5A). To assess whether the pathway remained activated at later time points within the stable tumor microenvironment in a palpable tumor, pIRF3 staining was also examined within CD11c+ cells in 7 day established B16 melanomas and was detected in similar amounts (Figure 5B). In parallel, activation of the upstream kinase TBK1 was similarly assessed by phosphorylation status and confirmed to be induced in a subset of CD11c+ cells (Figure S5A).
Figure 5. Tumor-Infiltrating Host APCs Produce IFN-β via a STING-Dependent Fashion In Vivo.
(A) B16.SIY tumor cells were inoculated into mice subcutaneously. The next day, tumor bumps were harvested and the suspended cells were fixed, per-meabilized, and stained with the indicated antibodies. Acquired images with ImageStream were analyzed using IDEAS software.
(B) B16.SIY melanoma cells were injected into mice. After 1 week, tumors were harvested and host immune cells were isolated and stained with the indicated antibodies. Acquired images were analyzed by Amnis software (IDEAS).
(C and D) B16.SIY tumor cells were inoculated into WT or Tmem173−/−mice. The next day, tumor cells, lymph nodes, and spleens were isolated as above and stained with anti-mouse CD45 antibody (C) and CD11 b and CD11 c (D) antibodies. Stained cell populations were isolated by cell sorting. Expression of IFN-β was measured by q-RT-PCR. CD11b+ or CD11c+ cells from lymph nodes or spleen were used as controls.
(E) Splenocytes were isolated from CD45.1 mice and injected into CD45.2+ C57BL76 mice. After 1 day, associated infiltrating CD45.2+ cells were stained and sorted by flow cytometry for analysis of IFN-β transcript expression. B16 melanoma cells were used as a control with the same procedure.
(F) CD45+ CD19− CD3− live cells were isolated from skin or tumor, LN and spleen of either healthy, nontamoxifen treated littermates or tamoxifen-treated Srafv600EPten−/− tumor-bearing mice using cell sorting. Expression of IFN-β transcript was measured by qPCR. *p = 0.05 (one-sided Mann-Whitney U test, n = 3 each samples) (F). *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). Data indicate mean ± SEM and representative of two (B and G) or three (A and C–F) independent experiments. See also Figures S5 and S6.
To assess whether IFN-β was produced by the early tumor-infiltrating APCs, we isolated tumor-infiltrating CD45+ cells from WT or STING-deficient mice after injection of B16.SIY melanoma by flow cytometric sorting, and then performed quantitative RT-PCR (qRT-PCR). A significant induction of IFN-β transcripts was observed in CD45+ cells from WT mice but not from STING-deficient mice (Figure 5C). Flow cytometric sorting revealed that CD11c+CD11b− or CD11c+CD11b+ cells appeared to be major source of IFN-β production (Figure 5D). These data are consistent with our previous observations demonstrating IFN-β production by DCs in the tumor-draining lymph node with multiple transplantable tumor cell lines (Fuertes et al., 2011).
To further evaluate whether the tumor context is preferential for the ability to induce IFN-β from host APCs, we injected the same number of normal splenocytes subcutaneously, which resulted in minimal induction of IFN-β transcripts compared to B16 tumor cells (Figure 5E). This is despite the fact that purified DNA from splenocytes and B16 cells induced similar amounts of IFN-β production from BM-DCs when combined with Lipofectamine in vitro (Figure S5B). These data suggest that the simple presence of a cell suspension in the subcutaneous space is not sufficient to activate the STING path way. We also observed that the splenocyte injection site evoked markedly diminished DC accumulation compared to tumor cell injection (Figure S5C), suggesting that tumor cells are more capable of initiating innate immune activation via APCs.
We were also concerned with the possibility that STING pathway activation and IFN-β production might only occur with transplantable tumor models. Therefore, a genetic melanoma model was employed, driven by conditional active B-Raf expression and PTEN deletion in the melanocyte lineage using tyrosinase-CreER (Dankort et al., 2009). Once tumors were induced with 4-OH tamoxifen, tumors were harvested and CD45+ host cells were isolated by flow cytometric sorting. Indeed, IFN-β transcripts were detected in CD45+ host cells within the tumor and in the tumor-draining lymph nodes but not in skin or lymph node of tumor-free control littermates (Figure 5F). These tumors also showed presence of a modest T cell infiltrate, consistent with host immune activation (data not shown). Additional cell sorting for CD11c+ cells also revealed IFN-β transcripts (Figure S6A), and plRF3 induction was detected in approximately 8% of DCs by ImageStream (Figure S6B). Combined with our observation that the STING pathway is dispensable for skin allograft rejection, this observation reinforces the notion that the context of a growing tumor in vivo is particularly predisposed toward induction of STING pathway activation and IFN-β production in vivo and separates that property from the technical aspects of percutaneous injection.
STING-Deficient Hosts Fail to Maintain Specific T Cell Expansion Consistent with an Effect on Costimulation
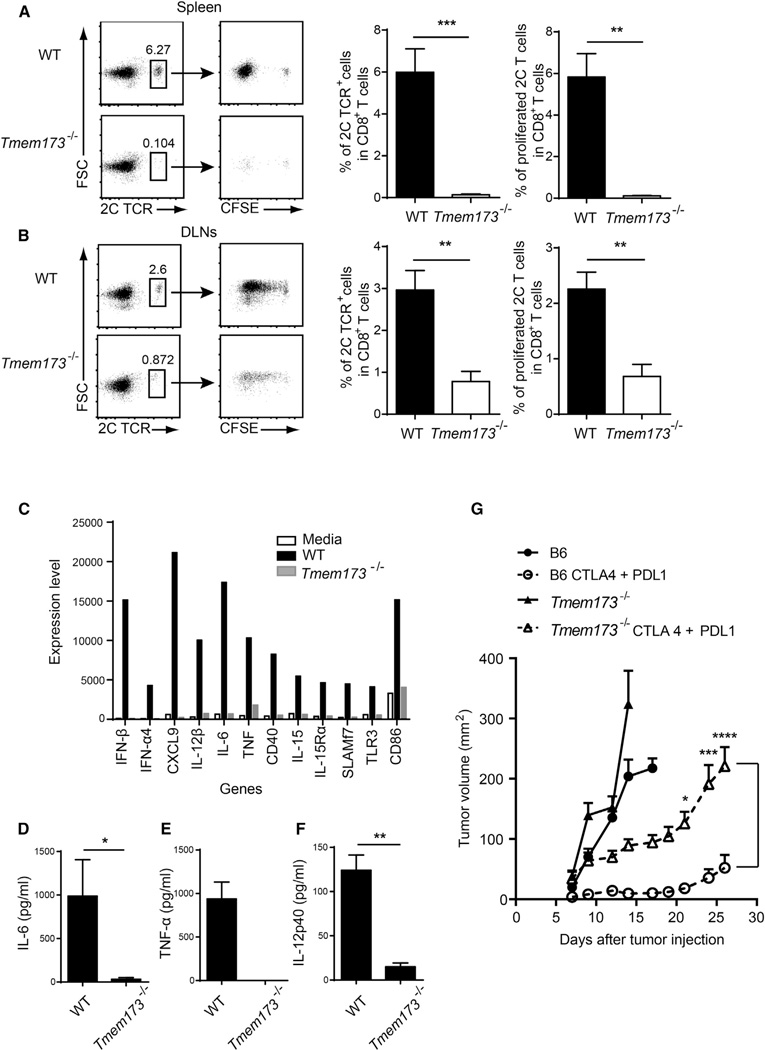
To evaluate in more detail the mechanism by which absence of host STING resulted in impaired antitumor T cell responses in vivo, we adoptively transferred CFSE-labeled 2C TCR Tg T cells into WT and STING-deficient mice and measured T cell proliferation by CFSE dilution after B16.SIY challenge. Interestingly, a similar number of cell divisions were observed in both recipients, but the CFSE-diluted 2C cells failed to accumulate in STING-deficient mice, in the spleen and lymph nodes (Figure 6A and 6B). This is a pattern that has been seen in other models of poor T cell costimulation leading to nonproductive T cell activation (Abe et al., 2013; Hoebe et al., 2003) and suggests that the STING pathway might be required not only for IFN-β production but also for expression of other T cell costimulatory factors. To evaluate a potentially broader DC activation property of DNA, we generated bone-marrow-derived DCs from WT or STING-deficient mice, stimulated them with tumor-derived DNA, and performed gene-expression profiling. Tumor-derived DNA induced expression of a broad spectrum of genes encoding multiple critical cofactors for T cell activation, including cytokines (e.g., IL-12), chemokines (e.g., CXCL9), and costimulatory molecules (e.g., CD86 and CD40; Figure 6C) in WT, but not STING-deficient, DCs. ELISA confirmed STING-dependent induction of IL-6, tumor necrosis factor alpha (TNF-α), and IL-12 (Figure 6D–6F) by tumor DNA. Induction of these factors by DNA was intact in bone-marrow-derived DCs from both Myd88−/− and Trif−/− mice, confirming a TLR-independent mechanism of this DC activation (data not shown). This broader DC activation pattern might explain ability of this DNA sensing pathway to fully prime tumor antigen-specific CD8+ T cells in vivo.
Figure 6. Tmem173−/−Mice Show Defective Accumulation of Antitumor T Cells.
(A and B) CFSE-labeled 2CT cell were transferred into WT or Tmem 173−/− mice and B16.SIY melanoma cells were inoculated into recipient mice after 1 day. On day 6, mice were sacrificed and spleens and tumor-draining lymph nodes were removed. Cells were stained with anti-CD8 and the clonotypic mAb 1B2 and analyzed by flow cytometry for CFSE dilution. Data were analyzed by FlowJo software and quantitated as shown in the panels on the right.
(C) BM-DCs from WT or Tmem173−/− mice were stimulated with tumor DNA for 7 hr and RNA was isolated. Isolated RNA was analyzed by Affymetrix GeneChip analysis.
(D-F) BM-DCs from WT or Tmem173−/− mice were stimulated with tumor DNA and the indicated cytokines were measured by ELISA.
(G) Mice were injected with 1 × 106 B16-SIY cells on day 0. On day 4, combinatorial antibody therapy was initiated. Anti-CTLA-4 mAb (100 ng/mouse) was administered intraperitoneally on days 4,7, and 10, and anti-PD-L1 mAb (100 ng/mouse) was given every other day starting on day 4 and ending on day 16. Tumor growth was measured on the indicated days. Data represent mean ± SEM (n = 10, each group) of combined two independent experiments. Statistical significance was determined using two-way ANOVA test. *p < 0.05, **p< 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t test or two-way ANOVA test). Data indicate mean ± SEM (n = 5) and representative of two (A and B) or three (C-F) independent experiments.
If our model is correct and innate immune sensing via the host STING pathway is necessary for maximal spontaneous T cell priming against tumor antigens, and if that spontaneous immune response is a critical determinant of activity of immunotherapies, then immunotherapies should lose efficacy in STING-deficient hosts. To test this notion, we treated tumor-bearing WT or STING-deficient mice with anti-CTLA-4 and anti-PDL1 mAbs, which we have shown to have strong synergistic therapeutic efficacy (Spranger et al., 2014). In fact, most of the therapeutic effect was lost in STING-deficient mice (Figure 6G). These data suggest that the host STING pathway can play a critical role in the therapeutic efficacy of cancer immunotherapies.
DISCUSSION
Our results indicate that the host STING pathway is particularly critical for innate immune sensing of immunogenic tumors, a process that results in APC activation, IFN-β production, and priming of CD8+ T cells against tumor antigens in vivo. Our data suggest that tumor-derived DNA is the likely ligand for this pathway, which becomes taken up by host APCs much like protein antigens, becomes localized to the cytosol, and is associated with phosphorylation of TBK1 and IRF3 and STING-dependent production of IFN-β in vivo. The failure of immunogenic tumors to be rejected in STING-deficient mice indicates the critical role for this overall process of immune-mediated tumor control in vivo. With most transplantable tumors, this initial STING-dependent priming occurs yet the tumor nonetheless eventually grows progressively. Other work has indicated that immune escape in these cases is mediated by immune inhibitory mechanisms that function at within the tumor microenvironment (Gajewski and Schumacher, 2013; Spranger et al., 2013). Blockade of these immune inhibitory factors, such as CTLA-4 and PD-L1-PD-1 interactions, is efficacious in WT mice, but we found that most of the therapeutic effect is lost in STING-deficient mice. Thus, innate immune sensing via the host STING pathway is critical for optimal tumor control by checkpoint blockade therapies.
An unresolved question in our current work is by what mechanism DNA can gain access to the cytosol of DCs under physiologic conditions in vivo. Presumably this occurs when a subset of tumor cells dies, perhaps in areas of hypoxia or nutrient starvation that are known to occur in solid tumors in vivo. One candidate mechanism of DNA transfer is through binding to the antimicrobial peptide LL37 (Lande et al., 2007). Another possibility is through the CLEC9A receptor that can mediate uptake of material from dying cells by DCs, particularly the CD8α+ subset (Sancho et al., 2009). Chaperoning of tumor-derived DNA by high-mobility group box 1 (HMGB1) is another potential mechanism (Yanai et al., 2009), which based on our results would likely be via a TLR-independent fashion. A fourth possibility is through uptake of autophagosomes, which have been shown to contain DNA (Oka et al., 2012) or through released exosomes. Knockdown of Beclin1 in tumor cells has been shown to blunt subsequent T cell priming in vivo (Li et al., 2008), and purified autophagosomes from tumors have been shown to serve as a potent vaccine platform in preclinical models (Twitty et al., 2011). The uncertain cell biology of the process of DNA transfer to APCs is reminiscent of the still poorly characterized process by which protein antigens are taken up from extracellular sources and gain access to the cytosol for proteasomal degradation and presentation by class IMHC molecules, which also occurs preferentially by selected APCs in vivo. It is attractive to speculate that there might be common features between these two processes.
The question arises as to why free DNA is not highly immune stimulatory in vitro or in vivo. It seems likely that extracellular DNA will be degraded rapidly by DNase I and DNA taken up by phagocytosis would be degraded by DNase II. Knockout mice lacking either of these DNases have been shown to be predisposed toward autoimmunity (Kawane et al., 2006; Napirei et al., 2000), indicating a particular importance for host control of extracellular DNA content in order to regulate endogenous immune responses. Consistent with this model, DNase II conditional knockout mice develop an inflammatory syndrome that leads to death of the animals, which has been reported to be eliminated upon crossing to a STING-deficient background (Ahn et al., 2012).
It is worth contrasting the STING-dependent mechanism we have observed as critical for spontaneous T cell priming against tumors with mechanistic data regarding immunogenic cell death induced by certain chemotherapeutic agents (Kroemer et al., 2013). In those studies, the adaptive immune response was facilitated upon tumor cell exposure to chemotherapy. Dying tumor cells were shown to expose calreticulin (CRT) and release high-mobility group box 1 (HMGB1) and ATP. CRT interacts with CD91 on dendritic cells, serving as an “eat-me” signal. According to this model, HMGB1 serves as a ligand for TLR4 on DCs and activates them via MyD88. ATP released from disrupted tumor cells also has been reported to activate the inflammasome pathway to produce IL-1|3 via P2×7 receptors (Apetoh et al., 2007; Ghiringhelli et al., 2009; Obeid et al., 2007). In our own studies, we have found that multiple transplantable tumor models induce spontaneous IFN-β production from host cells (Fuertes et al., 2013) and the host STING pathway is required. However, spontaneous T cell priming appears to be independent from MyD88 and P2×7R. Thus, the mechanisms of innate immune sensing might be partially distinct based on whether spontaneous or chemotherapy-induced immune responses are being evaluated. Identifying strategies to optimize the triggering of multiple innate immune sensing pathways concurrently might be important to consider from the therapeutic perspective.
A subset of human patients with melanoma shows spontaneous generation of an antitumor T cell response, whereas another major subset shows no such spontaneous immunity (Gajewski, 2006). A subset of patients with ovarian cancer (Hwang et al., 2012), breast cancer (Mahmoud et al., 2011), and colorectal cancer (Galon et al., 2006) displays spontaneous tumor infiltration with T cells. In the colorectal cancer example, the presence of activated CD8+ T cells in the tumor site has been reported to be a more powerful prognostic factor than tumor stage (Pagès et al., 2009), arguing for the critical functional importance of this natural antitumor immune response in patients. It seems plausible that an underlying mechanism explaining the presence or absence of this natural immune response might be whether appropriate innate immune sensing of the presence of tumor cells is occurring. Knowledge of the importance of the STING pathway will provide tools for probing human cancers for candidate defects correlating with the lack of spontaneous T cell infiltration. Facilitating this process when it does not occur de novo through the use of STING agonists could provide a therapeutic strategy to initiate endogenous T cell priming.
EXPERIMENTAL PROCEDURES
Mice and Cells
C57BL/6 mice and 129 mice were obtained from Jackson and Taconic. Myd88−/−, Trif−/−, P2×7r−/−, and Mavs−/−mice were purchased from Jackson ImmunoResearch Laboratories. Tlr4−/−mice were provided by Dr. Cathryn Nagler and Dr. Haochu Huang (University of Chicago), and Tlr9−/− mice were purchased from Oriental Bioservice. Tmem173−/− (STING-deficient) mice were generated by Dr. G. Barber (University of Miami). Irf3−/− (IRF3-deficient) mice were obtained from RIKEN Bio Resource Center. The C57BL6-derived melanoma cell line B16.F10.SIY (henceforth referred to as B16.SIY) was generated as described (Blank et al., 2005). 1969 sarcomas tumor cells were gifted by Dr. Robert Schreiber (Washington University). All cells were cultured in complete DMEM media supplemented 10% heat-inactivated FCS. For measurement of type I IFN reporter activity, B16-Blue IFN reporter cells were purchased from InvivoGen and maintained according to the manufacturer’s instructions. Animals were used according to protocols approved by the IACUC of University of Chicago according to NIH guidelines for animal use.
2C CD8+ T Cell Purification, CFSE Staining, and Adoptive Transfer
2C TCR Tg CD8+ T cells were isolated from spleens and lymph nodes of 2C/RAG2−/− mice by using magnetic beads. T cells were loaded with 2.5 mM CFSE and transferred into WT or designated gene-targeted mice (4 × 106 cells per mouse). After 1 day, recipient mice received 106 B16.SIY cells, and 5 days later splenocytes from recipient mice were analyzed after staining with antimouse CD8α-APC and biotin-labeled anti-2C–TCR (1B2) with SA-PE by flow cytometry to assess CFSE dilution.
IFN-γ ELISPOT and Pentamer Staining
Splenocytes were plated at 106 cells per well and stimulated overnight with SIY peptide. Spots were developed using the BD mouse IFN-γ kit, and the number of spots was measured using an Immunospot Series 3 Analyzer and analyzed using ImmunoSpot software (Cellular Technology). For pentamer staining, cells were labeled with PE-MHC class I pentamer (Proimmune) consisting of murine H-2Kb complexed to SIYRYYGL (SIY) peptide, anti-CD8-APC (53-6.7), anti-CD19-PerCP-Cy5.5 (6D5), and anti-CD4-PerCP-Cy5.5 (RM4–5). Stained cells were analyzed using FACSCanto or LSR II cytometers with FACSDiva software (BD). Data analysis was conducted with FlowJo software (Tree Star).
Preparation of B16 Melanoma Extracts for Potential IFN-β Induction In Vitro
For generation of B16 melanoma-derived extracts, cultured tumor cells were treated with staurosporin (0.5 µM) for 4 hr, irradiated (15,000 rad), incubated for 1 hr at 55°C for heat killing, mechanically killed using 10 passages through a syringe and needle, or treated 3 times by freezing and thawing cycles using liquid nitrogen and water bath at 37°C. For tumor-derived genomic DNA isolation, B16 tumor cells were washed with PBS and DNA was isolated using Blood &Cell Culture DNA Midi Kit (QIAGEN). The concentration and purity of DNA was determined by NanoDrop 1000 (Thermo Scientific). Each cell extract was added into BM-DCs and incubated for 18 hr at 37°C and amount of IFN-β was measured by ELISA.
In Vitro IFN-β Measurement
The IFN-β reporter cell line was cultured in 96-well plates and stimulated with tumor-derived DNA (20 ng/well) with Lipofectamine 2000 (0.75 µl/well) (Invitrogen) for 18 hr. Bone-marrow-derived dendritic cells (BM-DCs) were generated by culturing bone-marrow cells in the presence of rmGM-CSF (20 ng/ml; Bio-Legend) for 9 days followed by stimulation with tumor-derived DNA (20 ng/well) for 7 hr. After incubation, supernatant was collected and IFN-β was measured by ELISA (PBL IFN source) or adding substrate (QUANTI-Blue; InvivoGen) for the reporter cell line.
Statistical Analysis
The Student’s t test, two-way ANOVA and one-sided Mann-Whitney U test were used for statistical analysis. p values < 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Kline for critical reading of the manuscript; C. Nagler and H. Huang for providing Tlr4−/− mice; R. Schreiber for the 1969 tumor cells; Y. Zheng for help with data analysis; and M. Gao for technical assistance. This work was supported by P01 CA97296 and R01 CA181160 from the National Cancer Institute and AI067497 to K.A.F.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE61835.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2014.10.017.
REFERENCES
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Blank C, Brown I, Peterson AC, Spiotto M, Iwai Y, Honjo T, Gajewski TF. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140–1145. doi: 10.1158/0008-5472.can-03-3259. [DOI] [PubMed] [Google Scholar]
- Blank C, Brown I, Kacha AK, Markiewicz MA, Gajewski TF. ICAM-1 contributes to but is not essential for tumor antigen cross-priming and CD8+ T cell-mediated tumor rejection in vivo. J. Immunol. 2005;174:3416–3420. doi: 10.4049/jimmunol.174.6.3416. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8alpha+ dendritic cells. J. Exp. Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34:67–73. doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF. Identifying and overcoming immune resistance mechanisms in the melanoma tumor microenvironment. Clin. Cancer Res. 2006;12:2326s–2330s. doi: 10.1158/1078-0432.CCR-05-2517. [DOI] [PubMed] [Google Scholar]
- Gajewski TF, Schumacher T. Cancer immunotherapy. Curr. Opin. Immunol. 2013;25:259–260. doi: 10.1016/j.coi.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J. 2010;16:399–403. doi: 10.1097/PPO.0b013e3181eacbd8. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagés C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat. Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- Hamid C, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gomez H, Bastholt L, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J. Transl. Med. 2011;9:204. doi: 10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, McKee M, Gajewski TF. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 2003;4:1223–1229. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2012;124:192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Kobiyama K, Takeshita F, Ishii KJ. Recognition of damage-associated molecular patterns related to nucleic acids during inflammation and vaccination. Frontiers in cellular and infection microbiology. 2012;2:168. doi: 10.3389/fcimb.2012.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- Kono H, Rock KL. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–6895. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- Marichal T, Ohata K, Bedoret D, Mesnil C, Sabatel C, Kobiyama K, Lekeux P, Coban C, Akira S, Ishii KJ, et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat. Med. 2011;77:996–1002. doi: 10.1038/nm.2403. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AS, Burchill MA, Munks MW, Jin L, Kappler JW, Friedman RS, Jacobelli J, Marrack P. Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells. Proc. Natl. Acad. Sci. USA HO. 2013:E1122–E1131. doi: 10.1073/pnas.1300392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero LL, Zhou P, Wang Y, Harlin H, Kee B, Abraham C, Alegre ML. Epidermal Langerhans cells promote skin allograft rejection in mice with NF-kappa B-impaired T cells. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2008;8:21–31. doi: 10.1111/j.1600-6143.2007.02038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moray T. Features of systemic lupus erythematosus in Dnasel-deficient mice. Nat. Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini JL, Castedo M, Mignot G, Panaretakis T, Casares N, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 2007;13:54–61. doi: 10.1038/nm1523. [DOI] [PubMed] [Google Scholar]
- Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagés F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 2009;27:5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- Sancho D, Joffre OP, Keller AM, Rogers NC, Martínez D, Hernanz-Falcón P, Rosewell I, Reis e, Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Koblish HK, Horton B, Scherle PA, Newton R, Gajewski TF. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. Journal for immunotherapy of cancer. 2014;2:3. doi: 10.1186/2051-1426-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twitty CG, Jensen SM, Hu HM, Fox BA. Tumor-derived au-tophagosome vaccine: induction of cross-protective immune responses against short-lived proteins through a p62-dependent mechanism. Clin. Cancer Res. 2011;17:6467–6481. doi: 10.1158/1078-0432.CCR-11-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- Zhou S, Kurt-Jones EA, Cerny AM, Chan M, Branson RT, Finberg RW. MyD88 intrinsically regulates CD4 T-cell responses. J. Virol. 2009;83:1625–1634. doi: 10.1128/JVI.01770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.