Abstract
The horizontal transfer and acquisition of virulence genes via mobile genetic elements have been a major driving force in the evolution of Salmonella pathogenicity. Serovars of Salmonella enterica carry variable assortments of phage-encoded virulence genes, suggesting that temperate phages play a pivotal role in this process. Epidemic isolates of S. enterica serovar Typhimurium are consistently lysogenic for two lambdoid phages, Gifsy-1 and Gifsy-2, carrying known virulence genes. Other serovars of S. enterica, including serovars Dublin, Gallinarum, Enteritidis, and Hadar, carry distinct prophages with similarity to the Gifsy phages. In this study, we analyzed Gifsy-related loci from S. enterica serovar Abortusovis, a pathogen associated exclusively with ovine infection. A cryptic prophage, closely related to serovar Typhimurium phage Gifsy-2, was identified. This element, named Gifsy-2AO, was shown to contribute to serovar Abortusovis systemic infection in lambs. Sequence analysis of the prophage b region showed a large deletion which covers genes encoding phage tail fiber proteins and putative virulence factors, including type III secreted effector protein SseI (GtgB, SrfH). This deletion was identified in most of the serovar Abortusovis isolates tested and might be dependent on the replicative transposition of an adjacent insertion sequence, IS1414, previously identified in pathogenic Escherichia coli strains. IS1414 encodes heat-stable toxin EAST1 (astA) and showed multiple genomic copies in isolates of serovar Abortusovis. To our knowledge, this is the first evidence of intergeneric transfer of virulence genes via insertion sequence elements in Salmonella. The acquisition of IS1414 (EAST1) and its frequent transposition within the chromosome might improve the fitness of serovar Abortusovis within its narrow ecological niche.
Salmonella enterica serovar Abortusovis is a pathogen that occurs only in ovines, and it ranks as the most frequently isolated serovar in cases of ovine salmonellosis in Italy and other European countries (33). Serovar Abortusovis causes a systemic infection that can be lethal in newborn lambs and that can result in abortion in pregnant ewes. However, infection of adult animals occurs without clinical symptoms (33). Like other host-restricted serovars (i.e., S. enterica serovar Typhi), serovar Abortusovis infection induces a low level of mucosal inflammation (31). During the systemic phase of infection, serovar Abortusovis bacteria reach high numbers in the placenta and tissues of the aborted fetus, and they are shed by vaginal discharges up to 12 days following abortion, allowing dissemination in the environment and infection of new hosts. In this respect, the pathophysiology of serovar Abortusovis appears very different from that of S. enterica serovars Typhimurium and Enteritidis, which are common causes of gastroenteritis in a broad range of animal species.
Recent studies suggest that the ability of S. enterica serovars to gain diverse virulence traits and to adapt to a variety of animal hosts may be due partly to the variable distribution of effector protein genes carried by temperate bacteriophages (8, 26). Upon lysogenic conversion, virulence functions provided by prophages may ameliorate the fitness of pathogenic salmonellae within the host tissues or increase transmissibility and survival in the host population.
Functional phages containing genes that encode virulence effectors have been isolated, to date, only from strains of serovar Typhimurium. P2-like SopEΦ was isolated from a few serovar Typhimurium strains, including SL1344 and epidemic strain DT204 (8, 10). This phage carries the gene that encodes SopE, an effector protein translocated by the Salmonella pathogenicity island 1 type III secretion system (TTSS). In other sopE-positive Salmonella strains belonging to S. enterica serovars Gallinarum, Hadar, Dublin, and Enteritidis, the sopE gene is carried by a cryptic lambdoid prophage similar to the Gifsy-1 and Gifsy-2 phages (20). Lambdoid phage Gifsy-3 has been isolated uniquely from serovar Typhimurium strain ATCC 14028s (8). This phage also carries the gene that encodes a TTSS-translocated effector, SspH1. In addition, sspH1 has been detected in Salmonella bongori and in a few serotypes of S. enterica subspecies houtenae and indica (30). Lambdoid phages Gifsy-1 and Gifsy-2, in contrast, are present in all serovar Typhimurium epidemic isolates tested so far (8).
Given the variable distribution of phage-carried virulence genes, the comparative analysis of these genes in serovars of S. enterica with distinct pathogenic traits and host adaptations may help to define the relative roles of these virulence effectors. In this study, we examined whether epidemic strains of serovar Abortusovis carry lambdoid prophages or prophage-like elements carrying virulence genes. We found that serovar Abortusovis carries a Gifsy-2-related cryptic prophage, named Gifsy-2AO, inserted in the same chromosomal position as serovar Typhimurium Gifsy-2 prophage. The deletion of Gifsy-2AO had no effect on intestinal invasion but attenuated virulence in the systemic phase of lamb infection. Sequence analysis of Gifsy-2AO showed a deletion of ca. 6 kbp within the prophage b region, corresponding to tail fiber genes stfT and tfaT and putative virulence genes sseI (gtgB), gtgC, and gtgD carried by Gifsy-2. This deletion is consistent for all European strains tested, and the deleted region is adjacent to an insertion sequence (IS) element that is 99% identical to IS1414 described for enterotoxigenic Escherichia coli (ETEC) strain 27D (17).
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains studied are listed in Table 1. SS44 and 15/5 are reference strains of serovar Abortusovis. SS44 is an isolate from sheep originating from Sardinia, Italy, that has been used extensively in virulence studies (27, 31, 34). 15/5 is a French isolate that has been used to analyze the immune response of sheep to serovar Abortusovis (9, 14, 22, 23). Additional S. enterica strains analyzed by PCR and Southern blot hybridization were from our personal collection of clinical isolates. Twenty-two serovar Agona strains were isolated from pigs in Brazil (provided by M. Cardoso, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil). Sixty-six epidemic strains were human isolates from Zimbabwe. This collection includes 28 strains of serovar Typhimurium; 9 strains of serovar Enteritidis; 4 strains of serovar Typhi; 2 strains each of serovars Decatur, Bovismorbificans, Infantis, Isangi, Tennessee, and Otomarscens; and 1 strain each of serovars Vejle, Weltevreden, Gaminara, Heidelberg, Rissen, Scheissheim, Senftenberg, Shwarzengrung, Bonn, Blegdam, II 16:−:1,7, II 16:g,t:z42, and II 9,12:m,t:−. An epidemic strain of ETEC isolated in Pakistan (strain SSM3422) was used as a positive control in astA gene detection assays. Finally, we made use of a collection of 67 serovar Abortusovis epidemic strains, of which 35 originated from Italy, 2 from France, 6 from Albania, 1 from Russia, 1 from the United Kingdom, and 22 from Iran.
TABLE 1.
Strains used in this study
| Serovar and strain | Relevant genotype | Reference |
|---|---|---|
| S. enterica serovar Abortusovis | ||
| SS44 | Wild type | 1 |
| 15/5 | Wild type | 14 |
| SSM2992 | SS44 ΔGifsy-2AO::Km | This study |
| SSM2993 | SS44 ΔgtgE::Km | This study |
| SSM2994 | SS44 ΔsodC1::Km | This study |
| SSM3227 | SS44 ΔgtgE ΔsodC1::Km | This study |
| MA6776 | SS44 araBA-5463 araD-2939::MudJ lac::Tn10Tc | This study |
| MA6777 | SS44 araBA-5463 araD-2939::MudJ lac | This study |
| SSM3239 | SSM2992 araBA-5463 araD-2939::MudJ lac | This study |
| SSM3448 | SSM2993 araBA-5463 araD-2939::MudJ lac | This study |
| SSM3449 | SSM2994 araBA-5463 araD-2939::MudJ lac | This study |
| SSM3474 | SSM3227 araBA-5463 araD-2939::MudJ lac::Tn10Tc | This study |
| SSM916 | SS44 invH201::TnphoA | 31 |
| S. enterica serovar Typhimurium | ||
| ATCC 14028s | Wild type | 6 |
| SL1344 | Wild type | 13 |
Genetic procedures.
Genetic manipulations and PCR amplifications were performed according to standard methods. Primers used in this study are given in Table 2. Southern hybridizations were performed by using EcoRI- or HindIII-digested chromosomal DNA electrophoresed through 0.8% agarose and transferred onto a nylon membrane. Primer pairs REC1 and REC2, 03-01 and 03-02, 03-03 and 03-04, 02-37 and 02-38, SOD1 and SOD2, 02-11 and 02-12, and 02-20 and 02-21 were used to amplify the recE, orfH, stfT, gogB, sodC1, gtgE, and astA genes, respectively. PCR products were labeled with fluorescein (Amersham) and used as probes to hybridize digested genomic DNA from serovar Abortusovis strains SS44 and 15/5. Serovar Typhimurium strain ATCC 14028s was used as a control. Analysis of Gifsy-1 and Gifsy-2 integration into the respective attB sites was performed as described in Results. In brief, primers pp12 and pp16 were used to amplify a 460-bp fragment corresponding to the Gifsy-1 attB site and adjacent chromosomal sequences (phage-free strains). Primer pair pp12 and pp38 were used to amplify a 645-bp fragment corresponding to the junction between the chromosome and the “left” end of the inserted Gifsy-1 prophage (lysogenic strains). The same strategy was applied to analyze the Gifsy-2 attB site with primer pair pp30 and pp27 (303 bp) and primer pair pp27 and pp35 (632 bp). The amplification of Gifsy-2AO b region sequences from serovar Abortusovis strains SS44 and 15/5 was performed by arbitrary PCR (15). PCR products were then cloned by using TA-cloning technology (Invitrogen). Sequencing was performed with an ABI 3100 automated DNA sequencer (Applied Biosystems). We made use of the technique described by Datsenko and Wanner to obtain deletion mutants SSM2993 (ΔgtgE), SSM2994 (ΔsodC1), and SSM2992 (ΔGifsy-2AO) (2). PCR primers of 60 nucleotides (nt) were synthetized with 40 nt at the 5′ end corresponding to the endpoints of the deletion required. The deletion of gtgE and sodC1 coincided with nt 52 to 622 and nt 61 to 520 of the respective open reading frames. To obtain a gtgE sodC1 double mutant (SSM3227), we made use of the Flp resolvase method to delete the kanamycin cassette inserted into the gtgE mutant strain SSM2992. The sodC1 deletion was then moved in by P22 transduction. To obtain strain SSM2992, oligonucleotides were synthetized to achieve a deletion encompassing the whole Gifsy-2AO element (bp 1098220 to 1143961 as defined in the S. enterica serovar Typhimurium LT2 genome sequence at the National Center for Biotechnology Information). The 20 nt at the primers' 3′ ends anneal to priming sites P1 and P2 of pKD4 (2).
TABLE 2.
Primers used in this study
| Primer | Sequence 5′—3′ |
|---|---|
| 02-11 | AGGAGGAGTGTAAAGGT |
| 02-12 | GTAGAACTGGTTTATGAC |
| 02-20 | GCGAAGTTCTGGCTCAATGT |
| 02-21 | AGCGACTCGATGGCATTCGT |
| 02-37 | GCTCATCATGTTACCTCTAT |
| 02-38 | AGGTTGGTATTTCCCATGCA |
| 03-01 | ATGACGGAAGCGCATAAGCA |
| 03-02 | AAGGTCAGCCAGGATGGAGA |
| 03-03 | ATCCGAAACCGCCGCGAAGA |
| 03-04 | TGCTGTTGGCGTACCAGTCA |
| pp12 | GCCCTCCCGCCTGACCTTG |
| pp16 | CCGCGCTCACGCTCAAGATCC |
| pp38 | GCACCCACAGACCGTCCC |
| pp30 | CGAGCGCATTTCGTGCCCC |
| pp27 | GGCGAAGAGGCGCATCAGG |
| pp35 | CGAGTCCTTCAACATTACGC |
| REC1 | TGTACTCTGGTGCAGTGA |
| REC2 | TCTACGTCCAGAGTATCG |
| SOD1 | TATTGTCGCTGGTAGCTG |
| SOD2 | CAGGTTTATCGGAGTAAT |
Lamb infections.
One- to two-month-old Sarda lambs with no cultural or serological evidence of Salmonella infection were used. Competitive infections were performed as previously described (32). Lambs were infected orally with a total of 1 × 108 to 5 × 108 CFU or intravenously (i.v.) with 2 × 107 to 5 × 107 CFU. Inocula were obtained by growing strains statically at 37°C for 18 h. For oral infection, bacterial suspensions (2.5 ml) were mixed 1:1 with antacid [5% (wt/vol) Mg(SiO3)3, 5% (wt/vol) NaHCO3, 5% (wt/vol) MgCO3] and administered orally to animals immediately before the morning feeding. At 3 to 5 days postinfection, the animals were killed by pentobarbitone overdose. Samples of approximately 1 g were taken in triplicate from all tissues analyzed. The systemic samples (from the liver, spleen, and mesenteric lymph nodes [MLN]) were taken first to avoid contamination with the intestinal contents. Tissues were homogenized, and dilutions were plated, in triplicate, on Luria-Bertani (LB) agar plates supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 80 μg/ml), l-arabinose (1 mM), and kanamycin (50 mg/ml). The competitiveness index (CI) was calculated as (percentage of strain A recovered/percentage of strain B recovered)/(percentage of strain A inoculated/percentage of strain B inoculated).
Ovine ileal loop invasion assay.
The ovine ileal loop invasion assay was performed as previously described (35). Four- to five-month-old ewes were anesthetized with pentobarbitone (0.44 mg/kg of body weight) for the duration of the experiment. The abdominal wall of each animal was opened by a mid-line incision, the distal ileum was exteriorized, and the lumen was flushed with phosphate-buffered saline (PBS). Loops 9 cm in length with 1-cm spaces were constructed by using braided surgical silk. Loop inocula were prepared as follows. Log-phase cultures were harvested by centrifugation and resuspended in LB broth. Approximately 109 CFU (in 5 ml of LB broth) was injected into each loop. Sterile LB broth was used as a negative control. At 1 h postinoculation, loops were again exteriorized, and 5 ml of PBS containing 300 μg of gentamicin per ml was injected. The loops were returned to the abdominal cavity, and the wound was repaired. After two further hours, the animal was killed with an overdose of pentobarbitone, the ileum was exteriorized, and the individual loops were cut out. The tissue was gently washed with saline, and six circular 6-mm-radius biopsy specimens were removed from the central area of the loop. Each biopsy specimen was placed in 3 ml of PBS and homogenized, and counts of viable organisms were performed on LB agar plates.
Nucleotide sequence accession numbers.
The sequences reported here were submitted to the GenBank database and assigned accession numbers AY502962 (strain SS44) and AY502963 (strain 15/5).
RESULTS
Prophage sequences in strains of serovar Abortusovis.
Gifsy-1 and Gifsy-2 prophages are widely distributed within serovar Typhimurium strains, and Gifsy-2-related loci have been described for other serovars of S. enterica, including serovars Dublin, Enteritidis, Pullorum, and Choleraesuis (3, 28). Here, we analyzed the distribution of sequences associated with lambdoid prophages in two serovar Abortusovis reference strains, SS44 and 15/5. Specific probes were derived from the recE locus, carried by Gifsy-1 and Gifsy-2 phages of serovar Typhimurium, and from Gifsy-2-carried orfH and stfT genes that are conserved between the Gifsy and Fels-1 (lambdoid) phages. Southern hybridization analysis demonstrated that these loci are absent in SS44 and 15/5 strains (data not shown). Overall, these data suggested a lack of lambdoid prophages related to those of serovar Typhimurium or a loss of prophage regions in otherwise lysogenic strains. Thus, we examined the presence of virulence genes (or genes putatively involved in virulence) carried by Salmonella lambdoid prophages, including gogB (Gifsy-1), sodC1 and gtgE (Gifsy-2), and sopE (SopEΦ and cryptic lambdoid prophages). Strains SS44 and 15/5 were found to be positive only for gtgE and sodC1 (data not shown). These genes are the major Gifsy-2 phage-carried contributors to virulence in serovar Typhimurium (11). Since the SS44 and 15/5 strains carry sodC1 and gtgE genes, we analyzed whether a prophage equivalent to Gifsy-2 occurred in serovar Abortusovis epidemic strains. In lysogenic strains of serovar Typhimurium, Gifsy-1 and Gifsy-2 prophages are integrated, respectively, within the lepA coding region (centisome 57) and between genes pncB and pepN (centisome 24). Analysis of the attL and attR sequences and of the relative attachment sites on the Salmonella chromosome (attB) allowed us to design a set of oligonucleotides to specifically detect the insertion of prophages at these sites (see Materials and Methods). A first pair of primers amplifies a fragment including the attB site and the regions adjacent to both sides, unless a large sequence (i.e., a prophage) is inserted. A second pair amplifies the sequence corresponding to the junction between the chromosome and a boundary of the inserted prophage (lysogenic state). This method was applied to a collection of 65 epidemic strains of serovar Abortusovis originating from Italy (35), France (2), Albania (6), and Iran (21). None of the Abortusovis strains examined carried Gifsy-1 or any other element at the lepA site. On the other hand, these strains carried a large region between pepN and pncB (i.e., at the location of serovar Typhimurium Gifsy-2 phage). Following the deletion of this region, serovar Abortusovis strain SS44 was no longer positive for genes sodC1 and gtgE (strain SSM2992). Taken together, these data indicate that serovar Abortusovis carries an element, named Gifsy-2AO, related to serovar Typhimurium prophage Gifsy-2. Efforts to induce viable phage particles by exposing wild-type strains to mitomycin C and H2O2 did not yield any plaques on strain SSM2992 (data not shown), suggesting that Gifsy-2AO is a cryptic prophage.
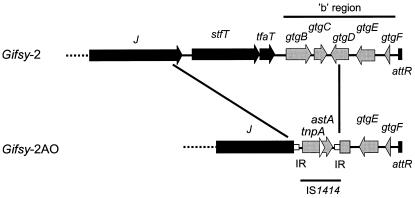
Sequence analysis of Gifsy-2AO b region.
In serovar Typhimurium, SodC1 and GtgE are the major virulence determinants carried by Gifsy-2 (11). However, the b region of Gifsy-2 encodes other putative virulence proteins: SseI, GtgC, GtgD, GtgE, and GtgF (Fig. 1A). SseI, a type III-secreted protein, is of particular interest since it is induced in macrophages and is regulated by the Salmonella pathogenicity island 2-encoded SsrB activator protein (36). Having shown that serovar Abortusovis Gifsy-2AO shares the integration site of serovar Typhimurium Gifsy-2 phage and that it carries the gtgE and sodC1 virulence genes, we examined the sequence of the Gifsy-2AO b region to further compare the repertoires of virulence-associated loci in the two prophages. We obtained a set of fragments corresponding to the prophage b region and the adjacent chromosomal DNA (pepN) from strains SS44 and 15/5 by means of arbitrary PCR. Alignment of these sequences with those of serovar Typhimurium strain LT2 (16) showed 99% identity from the pepN gene up to the 5′ end of gene gtgD (Fig. 1B). Interestingly, the gtgD sequence is interrupted by an insertion sequence with 99% identity to that of IS1414 of ETEC strain 27D (17). As previously described, IS1414 carries two overlapping genes: tnpA, encoding a transposase, and, in a +1 reading frame, astA, encoding heat-stable enterotoxin EAST1 (17). No flanking direct repeats were identified at the borders of the IS element. Upstream from IS1414, the Gifsy-2AO b region showed a large deletion (5,868 bp) encompassing the 3′ end (72 bp) of gtgD, gtgC, sseI, and the tail fiber protein genes tfaT and stfT (Fig. 1B). This adjacent deletion probably originated from a replicative IS1414 transposition involving cointegrate formation and deletion of the contiguous chromosomal segment. This deletion was consistently found in all European serovar Abortusovis isolates. However, strains originating from Iran (Asia) harbored sseI within Gifsy-2AO, and only two Iranian isolates were IS1414 positive. Cotransduction frequency analysis of the gtgE, sseI, and sodC1 genes showed that in Iranian serovar Abortusovis strains SSM2026 (IS1414 negative) and SSM2046 (IS1414 positive), the order of and the distance between the genes described above are virtually identical to those of serovar Typhimurium strain ATCC 14028s (data not shown). These data suggest that the acquisition of IS1414 and the subsequent deletion of the sseI locus occurred in serovar Abortusovis strains that eventually become more widely distributed in European countries.
FIG. 1.
Gene organization within the “right” end regions of Gifsy-2 (serovar Typhimurium) and Gifsy-2AO (serovar Abortusovis) prophages. Black arrows show genes with a role in production of phage particles, named according to their phage lambda orthologs. Gray arrows show genes within the b region. Sequence data are from reference 16 and this study. IR, IS1414 inverted repeated sequences.
Gifsy-2AO plays a pathogenic role during serovar Abortusovis systemic infection in lambs.
Serovar Typhimurium strains cured of the Gifsy-2 phage are significantly attenuated after oral or intraperitoneal inoculation of mice (7). This important attenuation by both routes of infection points to a role for Gifsy-2 during the systemic phase of the infective process. To evaluate the contribution of Gifsy-2AO to serovar Abortusovis virulence in ovines, we determined the virulence of strain SSM3239 (ΔGifsy-2AO) in infection competition assays. One- to two-month-old lambs were infected with an equal mixture of strain SSM3239 and wild-type strain MA6776. Three to 4 days later, bacteria were recovered from livers, spleens, MLN, and intestinal walls. In i.v. infected lambs, mutant strain SSM3239 was consistently outcompeted by the wild-type strain in the liver, spleen, and MLN (Table 3). However, the two strains showed equivalent levels of colonization of the intestinal wall. Similar results were obtained with orally infected lambs (data not shown). Taken together, these data suggest that the attenuation of strain SSM3239 is due to the Gifsy-2AO contribution to the systemic phase of infection.
TABLE 3.
Competition assays with strains SSM3239 (ΔGifsy-2AO) and MA6776 (wild type) in lambsa
| Lamb tissue | No. of lambs | Median CI | Pb |
|---|---|---|---|
| Liver | 7 | 0.17 | <0.0001 |
| Spleen | 9 | 0.06 | <0.0001 |
| MLN | 5 | 0.16 | <0.0001 |
| Intestine | 5 | 1.97 | NS |
Assays were performed by i.v. inoculating 1- to 2-month-old lambs. The CI was calculated according to the formula given in Materials and Methods.
Student's t test was used to compare output and inoculum. NS, not significant.
Previous studies have shown that SodC1 and GtgE are major contributors to the virulence induced by Gifsy-2 in serovar Typhimurium (11). Hence, sodC1 and gtgE are likely to contribute to Gifsy-2AO-induced virulence in serovar Abortusovis-infected lambs. To test this hypothesis, we constructed gtgE and sodC1 mutants and tested their virulence phenotypes in competition assays against the wild-type serovar Abortusovis strain SS44 (MA6777). We found that both genes contribute to virulence in lambs (Table 4). Furthermore, a sodC1 gtgE double mutant (SSM3474) competed evenly with the ΔGifsy-2AO mutant strain (SSM3239), suggesting that the two genes are the major virulence determinants carried by serovar Abortusovis Gifsy-2AO (Table 4).
TABLE 4.
Competition assays with sodC1 and gtgE deletion strainsa
| Strain A | Strain B | Tissue | No. of lambs | Median CI | Pb |
|---|---|---|---|---|---|
| SSM3448 ΔgtgE | MA6777 WT | Liver | 3 | 0.59 | 0.008 |
| Spleen | 3 | 0.42 | 0.006 | ||
| MLN | 3 | 0.46 | 0.005 | ||
| SSM3449 ΔsodC1 | MA6777 WT | Liver | 4 | 0.43 | 0.001 |
| Spleen | 4 | 0.33 | <0.001 | ||
| MLN | 4 | 0.31 | <0.001 | ||
| SSM3239 ΔGifsy- | SSM3474 ΔgtgE | Liver | 4 | 1.13 | NS |
| 2AO | ΔsodC1 | Spleen | 4 | 0.96 | NS |
| MLN | 4 | 1.27 | NS |
Assays were performed by i.v. inoculating 1- to 2-month-old lambs. The CI was calculated according to the formula given in Materials and Methods.
Student's t test was used to compare output and inoculum. NS, not significant.
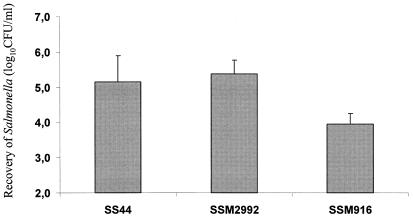
To further evaluate whether Gifsy-2AO had any effect on intestinal invasion, we made use of ovine ileal loops to quantify the invasion of intestinal mucosa. Three hours after loop inoculation, intracellular bacteria were enumerated by a gentamicin protection assay. The degree of mucosal invasion of Gifsy-2AO-cured strain SSM2992 was compared to that of wild-type strain SS44 and an invH isogenic derivative (SSM916). Strains SSM2992 and SS44 were recovered in comparable numbers, demonstrating that Gifsy-2AO does not affect intestinal invasion. In contrast, the invH mutant was recovered in significantly lower numbers (Fig. 2).
FIG. 2.
Relative levels of invasiveness of S. enterica serovar Abortusovis strains SS44 (wild type), SSM2992 (ΔGifsy-2AO), and SSM916 (ΔinvH) in ovine ileal loops; each bar represents the mean ± the standard error of the mean of results from six loops tested. Three samples from each loop were analyzed. For the results for SS44 versus those for SSM916, the P value was <000.1.
Multiple copies of astA are associated with epidemic strains of serovar Abortusovis.
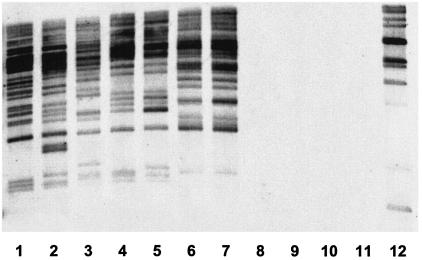
The astA gene has been shown to be distributed among several categories of pathogenic E. coli (18). Furthermore, multiple genomic copies of the gene have been observed in ETEC strains (17). These data have prompted us to determine whether European isolates of serovar Abortusovis carry other copies of this element in addition to the one carried by Gifsy-2AO. Southern blot analysis revealed numerous restriction fragments hybridizing with the IS1414-astA probe (Fig. 3). The number of genomic copies and the variety of IS1414-astA profiles, even between serovar Abortusovis strains that are epidemically related, suggest that IS1414 can transpose actively. In a recent study, an epidemic strain of S. enterica serovar Agona isolated in Brazil was also found by PCR to be astA positive (21). In this study, we used PCR and Southern blot analysis to identify the astA gene in 22 serovar Agona epidemic strains isolated from pigs in Brazil. None of the strains examined was found to be astA positive (data not shown). We also extended the analysis of the distribution of this element to diverse S. enterica serovars. A collection of 63 epidemic strains belonging to 20 serovars of S. enterica subsp. enterica and 3 epidemic strains belonging to 3 serovars of S. enterica subsp. salamae, including those most frequently responsible for salmonellosis worldwide (see Materials and Methods for a list), was analyzed by PCR for the presence of the astA or IS1414 element. Our results showed that none of these strains was astA positive.
FIG. 3.
Southern blot hybridization of serovar Abortusovis epidemic strains with the IS1414 (astA) probe. Multiple genomic copies of IS1414 (astA) were detected in the chromosomes of serovar Abortusovis epidemic isolates from Albania (strains SSM0074 [lane 1] and SSM0075 [lane 2]), Sardinia (strains SSM0078 [lane 3], SSM0088 [lane 4], and SSM0096 [lane 5]), Iran (strains SSM2045 [lane 6], SSM2046 [lane 7], SSM2026 [lane 8], and SSM2027 [lane 9]), and Russia (strain SSM0041 [lane 10]). Serovar Typhimurium strain ATCC 14028s (lane 11) and ETEC strain SSM3422 (lane 12) were used as negative and positive controls, respectively.
DISCUSSION
Phage-mediated transfer favors the reassortment of effector proteins in epidemic strains of Salmonella spp. Under laboratory conditions, the lysogeny of phages carrying virulence genes occurs efficiently, even in strains from different S. enterica serovars (8, 25, 37), suggesting that required host factors are conserved (i.e., Gifsy-1 and Gifsy-2 receptor protein OmpC) (12). In addition, the exchange of gene cassettes (morons) between unrelated phages might have further increased the efficiency of gene transfer among the broad variety of S. enterica serovars (20). However, phage-carried genes for virulence effectors show a variable distribution among strains of Salmonella. Such divergence may be related to the evolution of bacterial fitness within diverse biological niches. In this context, phage-mediated acquisition of a specific virulence effector by a large group of S. enterica serovars would imply a beneficial effect on a common pathogenic step (i.e., survival against macrophage cytotoxicity). In agreement with this hypothesis, the acquisition of Gifsy-2-encoded SodC1, a superoxide dismutase induced in macrophages and required during systemic infection in mice, is associated with highly virulent serovars causing extraintestinal infections (4, 5, 28, 32).
In all S. enterica serovars analyzed so far, sodC1-positive serovars also harbor Gifsy-2-carried sseI and gtgE genes, and with the exception of serovar Typhi (sodC1 and gtgE negative; sseI positive) and European isolates of serovar Abortusovis (sodC1 and gtgE positive; sseI negative), the three genes always occur together (S. Uzzau and G. Falchi, unpublished observations). The lack of sodC1 and gtgE virulence genes in serovar Typhi is not surprising, since in this pathogen, which occurs only in humans, a number of functions involved in host interaction have been inactivated, probably as part of its adaptation toward a very narrow ecological niche (24). Overall, these data strongly suggest that Gifsy-2-related prophages are widely distributed in highly virulent S. enterica serovars and that they share an ancestor phage. Evolutionary selection of such lysogens might have been driven by their enhanced potential to cause systemic diseases.
The data presented here confirm and extend these results. We identified a cryptic prophage, Gifsy-2AO, consistently associated with serovar Abortusovis epidemic strains. Sequence analysis showed that this element is located at the same attB site as serovar Typhimurium Gifsy-2 (i.e., between pepN and pncB) and that it carries both the sodC1 and gtgE virulence genes. The deletion of Gifsy-2AO reduces extraintestinal infection 10- to 100-fold in oral and intraperitoneal mixed infections in lambs. In contrast, Gifsy-2AO showed no significant contribution to intestinal invasion in lambs as measured by an ileal loop assay (3 h postinfection) and a competition assay (3 to 4 days postinfection). Lysogenic conversion by Gifsy-2-related prophages, therefore, appears to be associated with the enhancement of systemic virulence in serovars of S. enterica. Our data also showed that gtgE and sodC1 contribute to the Gifsy-2AO-encoded virulence of serovar Abortusovis in ovines. These results are consistent with findings from previous studies demonstrating that SodC1 and GtgE are the major contributors to virulence induced by Gifsy-2 during serovar Typhimurium infection in mice (11).
Following the horizontal acquisition of IS1414 (possibly from a pathogenic E. coli strain), a number of serovar Abortusovis genomic loci, including Gifsy-2AO, have been targeted by this insertion element. Strikingly, all European isolates of serovar Abortusovis showed an IS1414-dependent deletion of the sseI gene. Given the fact that these strains originate from areas where serovar Abortusovis is endemic (i.e., Sardinia, Italy, and Albania), the lack of TTSS-translocated effector SseI does not appear to have reduced the fitness of this serovar as an ovine pathogen. Studies from different laboratories failed to identify the role of SseI in virulence in animals (8, 19, 36). While we cannot rule out a pathogenic role for SseI, our data suggest that this TTSS-translocated effector may have a small impact on serovar Abortusovis infection and persistence in ovines. In addition to the SseI gene, we were unable to identify in strains of serovar Abortusovis a number of other phage-carried genes previously described for other pathogenic serovars (see Results for a list). Adaptation to a unique animal species (ovines) may have required the maintenance of a limited number of effectors and strategies compared to those of broad-host-range serovars like serovar Typhimurium.
IS1414 has been previously identified in an ETEC strain (17). This IS carries two overlapping genes encoding a transposase (tnpA) and EAST1 (astA), a heat-stable toxin. To our knowledge, the presence of IS1414 in serovar Abortusovis represents a unique example of intergeneric horizontal transfer of a virulence gene (astA) via a transposable element. The analysis of the Gifsy-2AO b region clearly showed that the insertion of IS1414 may give rise to adjacent deletions, possibly via a replicative transposition mechanism. The acquisition of IS1414 appears to be relatively recent, since a number of epidemic strains isolated in Iran carry sseI within the Gifsy-2AO element and are negative for IS1414. The potential relevance of this element in serovar Abortusovis pathogenicity is unclear. IS1414 encodes EAST1, a virulence marker of pathogenic E. coli whose association with pathogenicity is a matter of controversy (18). Savarino et al. have clearly demonstrated that EAST1 is structurally related to E. coli Shiga toxin and that it shares the Shiga toxin function of binding membrane-associated guanylate cyclase, causing an intracellular increase of cyclic GMP (29). Yet there is no evidence of EAST1-dependent fluid secretion in animal studies (17) or of an association of EAST1 with epidemic strains of E. coli with high diarrheagenic potential (18). The clarification of EAST1's contribution to host interaction will require further studies. However, the very frequent transposition of IS1414 may provide, by itself, plasticity to the chromosome and the selection of novel virulence forms in serovar Abortusovis.
Acknowledgments
We are grateful to Valentino Petruzzi for assistance in ligated ileal loop experiments.
This work was financially supported by grants from the Cofin-2000 National Research Program “Mechanisms of pathogenicity of intracellular bacteria” and the Cofin-2002 National Research Program “Virulence determinants of intracellular bacteria” from MURST (Ministero dell'Università e della Ricerca Scientifica e Tecnologica, Italy), and by grant Ricerca Corrente, IZSSAO11/99, from Ministero della Salute, Italy.
REFERENCES
- 1.Colombo, M. M., G. Leori, S. Rubino, A. Barbato, and P. Cappuccinelli. 1992. Phenotypic features and molecular characterization of plasmids in Salmonella abortusovis. J. Gen. Microbiol. 138:725-731. [Google Scholar]
- 2.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 5.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 8.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 9.Guilloteau, L., D. Buzoni-Gatel, F. Bernard, I. Lantier, and F. Lantier. 1993. Salmonella abortusovis infection in susceptible BALB/cby mice: importance of Lyt-2+ and L3T4+ T cells in acquired immunity and granuloma formation. Microb. Pathog. 14:45-55. [DOI] [PubMed] [Google Scholar]
- 10.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, T. D., and J. M. Slauch. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 14.Lantier, F., P. Pardon, and J. Marly. 1983. Immunogenicity of a low-virulence vaccinal strain against Salmonella abortus-ovis infection in mice. Infect. Immun. 40:601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 16.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 17.McVeigh, A., A. Fasano, D. A. Scott, S. Jelacic, S. L. Moseley, D. C. Robertson, and S. J. Savarino. 2000. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 68:5710-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menard, L. P., and J. D. Dubreuil. 2002. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit. Rev. Microbiol. 28:43-60. [DOI] [PubMed] [Google Scholar]
- 19.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 21.Paiva de Sousa, C., and J. D. Dubreuil. 2001. Distribution and expression of the astA gene (EAST1 toxin) in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 291:15-20. [DOI] [PubMed] [Google Scholar]
- 22.Pardon, P., J. Marly, F. Lantier, and R. Sanchis. 1990. Vaccinal properties of Salmonella abortusovis mutants for streptomycin: screening with an ovine model. Ann. Rech. Vet. 21:57-67. [PubMed] [Google Scholar]
- 23.Pardon, P., R. Sanchis, J. Marly, F. Lantier, L. Guilloteau, D. Buzoni-Gatel, I. P. Oswald, M. Pepin, B. Kaeffer, and P. Berthon. 1990. Experimental ovine salmonellosis (Salmonella abortusovis): pathogenesis and vaccination. Res. Microbiol. 141:945-953. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebahiha, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrel. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 25.Pelludat, C., S. Mirold, and W.-D. Hardt. 2003. The SopEΦ phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 185:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knuppel, W. Rabsch, W. D. Hardt, and H. Tschape. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 27.Rubino, S., G. Leori, P. Rizzu, G. Erre, M. M. Colombo, S. Uzzau, G. Masala, and P. Cappuccinelli. 1993. TnphoA Salmonella abortusovis mutants unable to adhere to epithelial cells and with reduced virulence in mice. Infect. Immun. 61:1786-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansone, A., P. R. Watson, T. Wallis, P. R. Langford, and J. S. Kroll. 2002. The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis. Microbiology 148:719-726. [DOI] [PubMed] [Google Scholar]
- 29.Savarino, S. J., A. Fasano, J. Watson, B. M. Martin, M. M. Levine, S. Guandalini, and P. Guerry. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc. Natl. Acad. Sci. USA 90:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzzau, S., G. S. Leori, V. Petruzzi, P. R. Watson, G. Schianchi, D. Bacciu, V. Mazzarello, T. S. Wallis, and S. Rubino. 2001. Salmonella enterica serovar-host specificity does not correlate with the magnitude of intestinal invasion in sheep. Infect. Immun. 69:3092-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uzzau, S., L. Bossi, and N. Figueroa-Bossi. 2002. Differential accumulation of Salmonella[Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol. Microbiol. 46:147-156. [DOI] [PubMed] [Google Scholar]
- 33.Uzzau, S., D. J. Brown, T. Wallis, S. Rubino, G. Leori, S. Bernard, J. Casadesus, D. J. Platt, and J. E. Olsen. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzzau, S., P. A. Gulig, B. Paglietti, G. Leori, B. A. D. Stocker, and S. Rubino. 2000. Role of the Salmonella abortusovis virulence plasmid in the infection of BALB/c mice. FEMS Microbiol. Lett. 188:15-18. [DOI] [PubMed] [Google Scholar]
- 35.Watson, P. R., S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 1995. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect. Immun. 63:2743-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, S., R. L. Santos, R. M. Tsolis, S. Mirold, W. D. Hardt, L. G. Adams, and A. J. Baumler. 2002. Phage mediated horizontal transfer of the sopE1 gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for calves. FEMS Microbiol. Lett. 217:243-247. [DOI] [PubMed] [Google Scholar]