Abstract
Insects provide experimentally tractable and cost-effective model systems to investigate the molecular basis of animal-bacterial interactions. Recent research is revealing the central role of the insect innate immune system, especially anti-microbial peptides and reactive oxygen species, in regulating the abundance and composition of the microbiota in various insects, including Drosophila and the mosquitoes Aedes and Anopheles. Interactions between the immune system and microbiota are, however, bidirectional with evidence that members of the resident microbiota can promote immune function, conferring resistance to pathogens and parasites by both activation of immune effectors and production of toxins. Antagonistic and mutualistic interactions among bacteria have also been implicated as determinants of the microbiota composition, including exclusion of pathogens, but the molecular mechanisms are largely unknown. Some bacteria are crucial for insect nutrition, through provisioning of specific nutrients (e.g. B vitamins, essential amino acids) and modulation of the insect nutritional sensing and signaling pathways (e.g. insulin signaling) that regulate nutrient allocation, especially to lipid and other energy reserves. A key challenge for future research is to identify the molecular interaction between specific bacterial effectors and animal receptors, and to determine how these interactions translate into microbiota-dependent signaling, metabolism and immune function in the host.
Keywords: bacteriocyte, endosymbiont, gut microbiota, nutrition, immunity
Introduction
Insects are the most successful animals, accounting for >90% of known animal species and dominating a variety of terrestrial habitats. Many insect lifestyles are founded on associations with microorganisms. These include the termites, which thrive on a diet of wood or soil through the metabolic capabilities of microorganisms in the hindgut “paunch”1; the leaf-cutting ants, whose apparent herbivory is based on the fungal gardens maintained in their nests2; and the plant sap feeding habit of many hemipterans (aphids, whiteflies etc.), made possible by nutrient exchange with intracellular microorganisms that have been transmitted faithfully from mother to offspring for up to 100–200 million years3. More generally, all insects investigated to date bear resident microorganisms and, although some taxa are not obligately dependent on their microbiota, there is increasing evidence that these microorganisms influence many insect traits.
The ubiquity of microbial associations is not peculiar to insects. Rather, it is the normal condition for animals and other eukaryotes4. The important implication is that the capacity to interact with microorganisms has ancient evolutionary roots, even though the taxonomic composition of the microbiota can vary widely. We should therefore expect that various aspects of the signaling pathways and regulatory circuits controlling animal function have evolved, and function optimally, in the context of continual interactions with microorganisms, and that the fundamental molecular principles dictating interactions with resident microorganisms are likely to be conserved across the animal kingdom. Thus, the study of insect interactions with the resident microbiota is not only of intrinsic interest, but can also shed light on equivalent processes in other animals, including humans. In other words, just as studies on Drosophila and other insects have revealed the fundamental principles of many processes from embryonic development and hormone function to innate immunity and gene expression, so research on the relationship between insects and microorganisms has the potential to identify the basic ground-rules of how animals negotiate with their resident microbiota.
This article provides an overview of recent advances in our understanding of the molecular basis of insect interactions with resident bacteria. Although these associations have been investigated from the perspectives of morphology and whole-insect physiology for many decades, it is only in recent years that the underlying molecular processes have started to be dissected. Much of the molecular research has been conducted on Drosophila melanogaster, but important insights have also been obtained from studies of other insect species, including mosquitoes and aphids (Fig. 1). This review addresses how the microbial populations are regulated, and the molecular basis of their contributions to insect nutrition and defense against natural enemies.
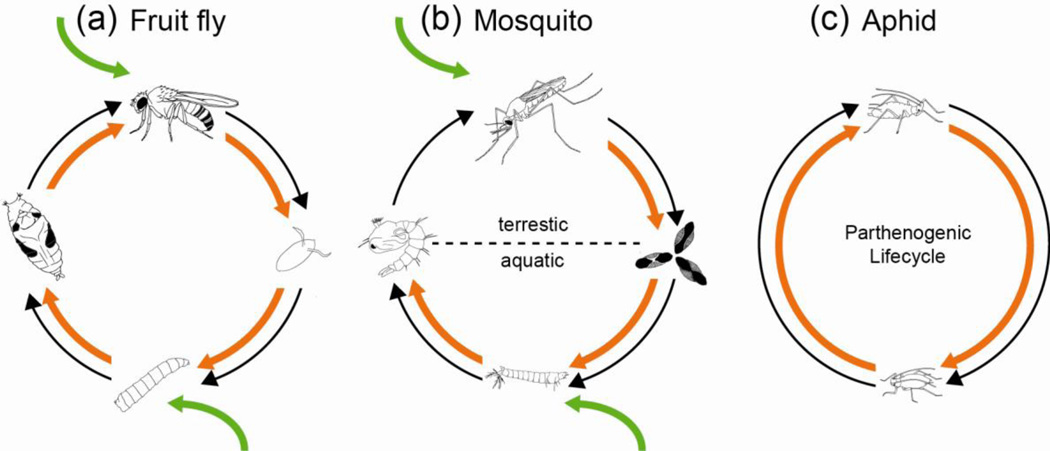
Fig. 1.
Interactions between insects and resident microbiota. (a) Gut microbiota of Drosophila melanogaster, an open system in which both the larval and adult stages gain microorganisms from the environment and previous life stages.(b) Gut microbiota of the mosquito, comparable to Drosophila, except that the microbiota is not retained in the pupal stage. (c) Intracellular bacteria in the aphid, an example of a bacteriocyte symbiosis (see text) in which the microorganisms are transmitted vertically to the female ovaries and inserted into the developing embryo; aphids have no pupal stage and summer generations (depicted) are viviparous. Black arrows, insect life stages; orange arrows, transmission of bacteria across life stages; green arrows, acquisition of microorganisms from the environment. [Figure prepared by S. Villareal and S. Franzenburg]
Regulation of the microbial populations associated with insects
The resident microbiota of insects is regulated, meaning that the abundance, composition and location of the microorganisms fall within certain bounds. Most of the research on the molecular mechanisms that determine the abundance and composition of the insect-associated microorganisms have focused on the role of insect immunity, especially in Drosophila and mosquitoes. The fundamental question posed by this line of research is: How is the immune system structured, such that pathogens are generally eliminated and other microorganisms are spared? In principle, the microbial community may be dominated by taxa that are variously resistant to host immune effectors, lack immune elicitors, or trigger negative regulators of the immune response. The data are fragmentary and often contradictory, but instances of all three modes of interaction with the host immune system have been identified. The key immune effectors that have been studied are anti-microbial peptides (AMPs) and reactive oxygen species, and they are considered below.
Drosophila is an amenable system to study interactions between AMPs and the gut microbiota because the profile of AMPs and the regulation of their production are well-understood. The expression of genes for two complementary sets of AMPs are induced by the IMD (immune-deficiency) and Toll signaling pathways, with activity predominantly against Gram-negative and Gram-positive bacteria, respectively5. Only the IMD pathway is expressed in the midgut of the adult fly, and genetic deletion of this pathway results in a ten-fold increase in numbers of gut bacteria6. The simplest interpretation of these data is that AMPs suppress, but do not eliminate, the populations of symbiotic bacteria. In apparent contradiction with these results, the bacterial populations are also elevated in flies with chronically activated IMD pathway, as obtained by RNAi-knockdown of expression of PGRP-SC2, a negative regulator of IMD that is strongly expressed in the midgut7. The underlying mechanisms are not understood, but one possibility is that the AMPs have differential effects on different members of the microbiota, and the suppression of susceptible taxa could result in the loss of community stability and overgrowth by resistant taxa8. Interestingly, the expression of AMP genes is increased in flies with either mutations or RNAi-expression knockdown of various transcription factors9; 10; 11, and this is associated with changes in the abundance and composition of the gut microbiota for the two transcription factors tested, Caudal and ATF38; 11. These effects have been attributed to the role of transcription factor Caudal in the negative regulation of the IMD immune signaling pathway8 and to a generalized perturbation of gut homeostasis caused by imbalance of transcription factors that regulate gut structure and compartmentalization10. Taken together, these data are consistent with the possibility that the interplay between the suite of AMPs expressed by the host and profile of AMP susceptibility of different community members may play an important role in shaping the composition of the microbiota, as has also been suggested in other animal-microbial systems12.
A second important immune effector is reactive oxygen species (ROS). ROS are produced in the midgut of Drosophila and mosquitoes by dual-oxidases (DUOX), enzymes with both NADPH oxidase and peroxidase domains13; 14; 15. DUOX-derived ROS may play a central role in the control of the gut microbiota in the mosquito Aedes aegypti, as indicated by the significantly elevated gut bacterial populations in individuals with the duox gene silenced by RNAi16. Furthermore, the gut bacteria increase 100–1000-fold over 12 h after the insect takes a blood-meal, and this pattern has been linked to reduced DUOX activity, by a mechanism that involves the activation of protein kinase C by heme in the blood-meal16. Paradoxically, DUOX in the mosquito Anopheles gambiae has been implicated in protecting gut microbiota. The site of the likely protective effect is the peritrophic membrane, which separates the ingested food and microbes from the epithelial cells of the midgut. DUOX functions in conjunction with a peroxidase to reduce the permeability of the peritrophic membrane, probably by catalyzing dityrosine cross links in the mucin proteins; and this has been suggested to reduce the traffic of microbial immune elicitors from the food bolus to the epithelial cells and, in reverse, the transfer of immune effectors produced by the epithelial cells to the microorganisms in the food bolus17. Yet another pattern of response to DUOX-derived ROS is displayed in Drosophila. In this system, the gut DUOX activity is high in flies that have ingested live food yeasts or certain bacterial pathogens, but not symbiotic bacteria18; 19. The activation of the DUOX by bacteria in this system has been linked to bacterial release of uracil, a trait displayed, for example, by the pathogen Erwinia caratova, but not by most of the symbiotic bacteria in the gut19. Further research is required to establish why uracil release is apparently a trait restricted to pathogenic bacteria, and how the uracil interacts with the DUOX activation system.
Various insects possess intracellular bacteria within specialized cells, known as bacteriocytes, whose sole function appears to be to house and maintain the bacteria20. These associations are maintained by obligate vertical transmission, usually by transfer from the bacteriocytes to the eggs developing in the female ovary, and the association is required by both the insect and bacterial partners. The bacteriocyte symbioses have no parallel in mammals, for which all described intracellular bacteria are pathogens. (The only known beneficial intracellular microorganisms in vertebrates are algal cells in the embryos of Ambystoma salamanders21.) Nevertheless, the bacteriocyte symbioses can provide valuable general insights into host mechanisms that regulate the resident microbiota. The bacteria in insect bacteriocytes are very tightly regulated, with uniform densities in replicate insects of a given developmental age, and consistent patterns of variation in abundance with host age, sex, morph etc22; 23; 24; 25. Insect immune effectors have been implicated in the regulation of the bacteria in bacteriocytes of the weevil Sitophilus. The Sitophilus bacteriocytes express a cationic AMP, coleoptericin-A, at high levels. When coleoptericin-A expression is suppressed by RNAi, the bacteria overgrow the bacteriocytes and colonize the insect hemocoel (body cavity), suggesting that this AMP plays a crucial role in controlling the bacterial populations26. However, the negative regulator of the IMD pathway, PGRP-LB, is strongly expressed in both the Sitophilus bacteriocytes and also the symbiosis between the tsetse fly Glossina and its bacteriocyte symbiont Wigglesworthia27; 28, suggesting that IMD-dependent immune effectors are suppressed in the bacteriocytes.
Research on the interactions between the insect immune system and resident microbiota is increasingly being complemented by analyses of among-microbe interactions and their impacts on community composition. These studies are revealing a diversity of interactions. For example, when bacteria of the genera Acetobacter and Lactobacillus are administered in pairs to Drosophila, their abundance can be increased, decreased or unaffected, depending on the identity of the partner bacterium29. Asaia and Acinetobacter promote each other’s growth in the gut of the mosquito Aedes albopictus30, but the interaction between Serratia and other gut bacteria in the locust Schistocerca gregaria is antagonistic31. Many of the interactions may be direct, involving competition or cross-feeding of nutrients, competition for or facilitation of micro-site colonization in the insect gut, or the production of biologically-active molecules, such as antibiotics. How these interactions are integrated to shape the abundance of different members of the microbial communities is an important topic for future research. Nevertheless, interactions between resident bacteria and invading taxa have been addressed in a few systems. For example, ROS produced by a resident gut bacterium Enterobacter in Anopheles mosquitoes function to depress Plasmodium acquisition by the insect32; and possible competition between Wolbachia and RNA viruses for limiting cholesterol in Drosophila may block RNA virus replication33.
Interestingly, not all interactions among the bacteria in an insect host are direct. There is increasing evidence that some interactions between microorganisms are indirect, mediated via the host, particularly the insect immune system34. For instance, one microorganism may stimulate (or suppress) the production of immune effectors with high activity against other microorganisms, so depressing (or promoting) the abundance of the latter. A vivid example is provided by the demonstration that elimination of the bacteriocyte symbiont Wiggleworthia from its tsetse fly Glossina host results in major perturbation of the immune system, including a dramatic reduction of hemocytes, and susceptibility of the insect to opportunistic infection by E. coli. When either hemocytes or Wigglesworthia cell extract were administered to the treated insects, hemocyte titers rebounded, together with recovery of resistance to E. coli35, demonstrating that these effects cannot be assigned to generalized malaise of the insects deprived of Wigglesworthia. These data indicate that a Wigglesworthia product (to be identified) activates host immune function against other microorganisms. This type of interaction can contribute to microbial protection the insect host against pathogens, which is considered next.
Molecular basis of microbial protection of insects against their natural enemies
Resident microorganisms can reduce the susceptibility of their insect hosts to natural enemies by multiple mechanisms. As well as symbiont-mediated stimulation of host immune effectors active against nature enemies (discussed above), symbionts can competitively exclude pathogens and parasites from microsites in the host, and produce toxic secondary compounds that complement the insect immune effectors. Most available information relates to the role of symbiont toxins. For example, Pseudomonas sp. in Pederus rove beetles synthesizes the polyketide “pederin” which protects the beetles from predation36. Another bacterium, Hamiltonella defensa, can protect its aphid host from the parasitic wasp Aphidius ervi but, in this case, the protective effect is perfectly correlated with a bacteriophage bearing a gene for a proteinaceous toxin, e.g. Shiga-like toxin, cytolethal distending toxin, and YD-repeat toxins37; 38. Various other protective functions of resident microorganisms have been identified but the molecular basis of these interactions is not yet fully established. Examples include the protective role of Spiroplasma for both Drosophila hydei against parasitic wasps39, and D. neotestacea against Howardula nematode parasites40, and protection of aphids from entomopathogenic fungi by multiple bacteria41.
Some of the most exciting recent discoveries in the field of bacterial protection of insects against invading microorganisms are emerging from research on insect competence as vectors of medically-important pathogens. This area is founded on the observations that Wolbachia protect Drosophila melanogaster from viruses42,43. When the Wolbachia is experimentally transferred from Drosophila to mosquito species that do not naturally bear this bacterium, transmission of Plasmodium and viruses (e.g. dengue virus, Chikungunya virus, yellow fever virus) is reduced9; 44; 45;46. The depressed vector competence of the Wolbachia-infected mosquitoes may be attributable to heightened immune function, including induction of AMPs, melanization and ROS47; 48. The interactions can, however, be multifaceted. For example, the negative interaction between Wolbachia and dengue virus in Aedes aegypti has also been linked to Wolbachia-mediated induction of a microRNA that suppresses expression of cytosine methyltransferase gene, with consequent global reduction in genome methylation49; 50. These studies raise important questions about how the impacts of Wolbachia on the methylation status of the genome, gene expression patterns and antiviral response of the host mesh together to depress vector competence.
This section has focused principally on microbial traits that confer protection against natural enemies by complementing or activating host immune function. However, insect capacity to resist and tolerate pathogens and parasites is strongly influenced by other physiological factors, particularly nutrition: nutritional health generally promotes resistance to pathogens and parasites51, although reverse effects are known52. These considerations raise the expectation that microbiota-dependent effects on insect nutrition may contribute to the host immunological response to natural enemies. Dissection of the interactive effects of microbiota, immune function and nutrition is an emerging research priority that will be facilitated by a sure foundation of understanding of microbiota effects on insect nutrition, which is reviewed in the next section.
Molecular basis of microbial impacts on insect nutrition
The resident microbiota can influence insect nutrition by two processes, which are not mutually-exclusive. Microorganisms can, first, be a source of nutrients, made available to the insect by lysis of the microbial cells (this applies particularly to microorganisms in the digestive tract) and by the specific release of metabolites from living microbial cells. Second, microorganisms can modulate the host signaling networks that regulate nutrient allocation. Microbial impacts on nutrient acquisition and nutrient allocation are considered in turn, below.
The evidence for microbial provisioning of nutrients is particularly persuasive for bacteriocyte symbioses. It has been appreciated for decades that these associations are largely restricted to insect taxa living on nutrient-poor or unbalanced diets, notably plant sap (deficient in essential amino acids) and vertebrate blood (deficient in B vitamins), and multiple studies conducted over many decades have demonstrated that the nutritional status of these insects is compromised by experimental elimination of the microorganisms20; 53. These interactions involve the two-way transfer of multiple metabolites between the bacteriocyte cytoplasm and intracellular bacteria.
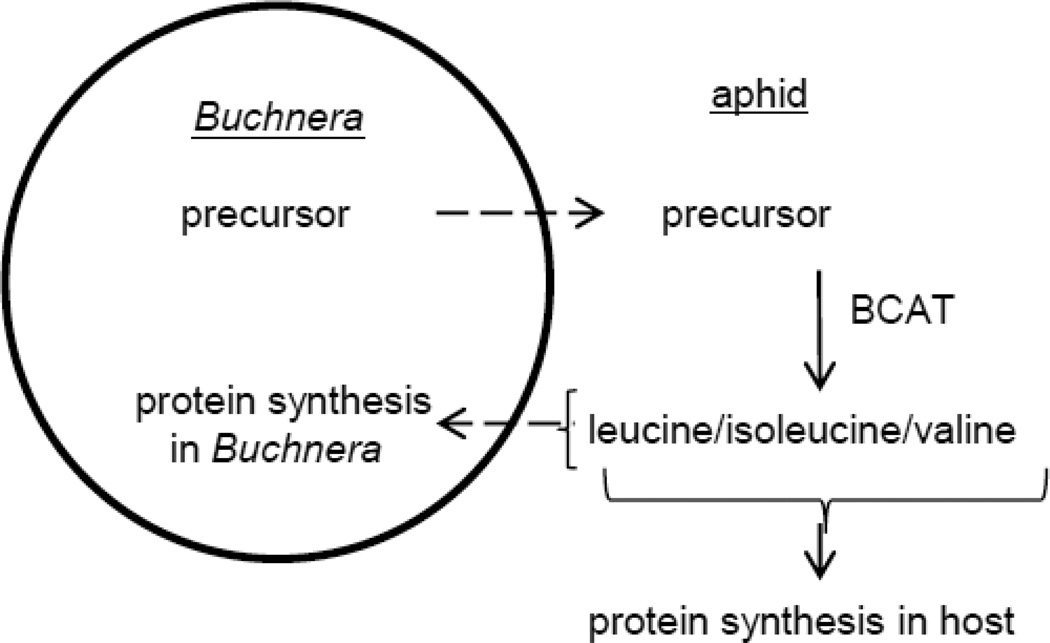
The molecular basis of these interactions has been investigated in the symbiosis between the pea aphid Acyrthosiphon pisum and bacterium Buchnera, facilitated by fully sequenced genomes for both partners. The interface between the Buchnera and the surrounding cell cytoplasm is metabolically very dynamic, with the transport of 58 metabolites between the partners inferred from in silico genome-scale metabolic modeling54. These metabolites include all the nutritional requirements of the Buchnera, the essential amino acids synthesized by the Buchnera, and also metabolic intermediates in essential amino acid synthesis. The last arises from evidence that the synthesis of 5 of the 10 essential amino acids is shared between the Buchnera and the bacteriocyte55, through the evolutionary loss of Buchnera genes coding for reactions that are also coded in the genomes of animals, including the pea aphid host. Fig. 2 provides an exemplar: Buchnera lacks the gene ilvE, coding for branched chain amino acid aminotransferase (BCAT), which mediates the terminal reaction in the synthesis of the branched chain amino acids (BCAs: isoleucine, leucine and valine), and this reaction is mediated by aphid BCAT in the bacteriocyte cytoplasm56–58. Although coupled metabolism through shared metabolic pathways has been demonstrated experimentally only in the aphid-Buchnera symbiosis, it may be widely distributed, at least among bacteriocyte symbioses, because the same genes are known to be missing from phylogenetically different bacteria in some other plant sap feeding insects, including whiteflies and mealybugs59; 60.
Fig. 2.
Metabolite exchange between the bacteriocyte (host cell) and intracellular bacterium Buchnera aphidicola. Insect-mediated terminal reaction in synthesis of branched chain amino acids (BCAs) from immediate precursors: (S)-3-methyl-2-oxopentanoate to isoleucine, 4-methyl-2-oxopentanoate to leucine and 2-oxoisovalerate to valine). The reaction catalyzed by BCAT is reversible (terminal reaction in BCA synthesis /first reaction in BCA degradation) such that, although BCAT functions in most animal tissues in the degradation of BCAs, it is recruited to the bacteriocyte to mediate the final reaction in BCA synthesis.
As well as providing nutrients, resident microorganisms can influence the nutrient allocation patterns of insects. These effects are studied most readily by investigating the effects of eliminating the microbiota on host nutrient content, especially of insects that do not depend on their microbiota; for insects with high dependence, it can be difficult to disentangle the effects of microbial elimination and generalized malaise on the nutrient profiles and underlying signaling circuits. To date, most research has been conducted on Drosophila, which can be raised from the egg under sterile conditions with minor effects on growth and developmental rates61. The resultant axenic insects have elevated levels of lipid and glucose, together with reduced basal metabolic rates, indicative of enhanced energy harvesting29; 62. These effects have been linked to altered insulin signaling63, including reduced expression of key insulin-like peptides (dilp-3 and dilp-5) in neurosecretory cells of the brain64. Although the detail of the mechanisms are unclear, acetic acid produced by Acetobacter gut bacteria has been implicated in promoting insulin signaling and reduced lipid storage in this system64.
Concluding comments
The study of the molecular mechanisms underlying the interactions between insects and their resident microorganisms is a “young” field. As this review illustrates, this research is facilitated by the small size and rapid generation time of many insects, enabling large sample sizes, sophisticated experimental designs and rapid data throughput. For example, the impact of multiple factors on host-microbial interactions can be quantified with sample sizes of insects that would be prohibitive in a mammalian biomedical model. Drosophila is emerging as a model system, especially for the study of interactions with the gut microbiota. Other insects can also make important contributions to understanding the molecular basis of many aspects of animal-microbial interactions, and they will become increasingly tractable to study with the rapidly expanding insect genomic resources65 and improving technologies for insect genetic manipulation66.
Nevertheless, the field faces multiple challenges. The literature on insect-bacterial symbioses is replete with pattern at the whole-insect level, i.e. how elimination or perturbation of the microbiota affects multiple insect traits, ranging from insect morph determination and dispersal behavior to body color, mate choice and food choice67; 68; 69; 70;71. An important priority is to understand the molecular interactions between the microbiota and host that underpin these diverse phenotypes. This understanding can then be used to test and illuminate the core prediction that the fundamental principles of animal-microbial interactions are highly conserved, such that insect-bacterial symbioses can come to offer a valuable model for the mammalian, including human, symbioses.
Highlights.
Insects, like other animals, are colonized by benign and beneficial microorganisms
Insect immune factors affect the composition and abundance of the microbiota
Microbial effectors can both promote and complement host immune function
Microbiota provides nutrients and modulates insect nutrient allocation patterns
Acknowledgements
I thank Dr Susan Villareal and Dr Soeren Franzenburg who prepared Figure 1, and Dr Nicolas Buchon for helpful comments on the manuscript. This work was supported by RO1 GM095372 (NIH) and BIO 1241099 (NSF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014 doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 2.Currie CR. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 3.Douglas AE. Phloem-sap feeding by animals: problems and solutions. J Exp Bot. 2006;57:747–754. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- 4.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 6.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 9.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Rynes J, Donohoe CD, Frommolt P, Brodesser S, Jindra M, Uhlirova M. Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol Cell Biol. 2012;32:3949–3962. doi: 10.1128/MCB.00429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mergaert P, Kondorosi E. Controlling symbiotic microbes with antimicrobial peptides. In: Rajasekaran K, Cary JW, Jaynes JM, Montesinos E, editors. Small Wonders: Peptides for Disease Control. Washington DC, USA: Americal Chemical Society; 2012. pp. 215–223. [Google Scholar]
- 13.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 14.Bae YS, Choi MK, Lee WJ. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 2010;31:278–287. doi: 10.1016/j.it.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, Barillas-Mury C. Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J Biol Chem. 2008;283:3217–3223. doi: 10.1074/jbc.M705873200. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira JH, Goncalves RL, Lara FA, Dias FA, Gandara AC, Menna-Barreto RF, Edwards MC, Laurindo FR, Silva-Neto MA, Sorgine MH, Oliveira PL. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011;7:e1001320. doi: 10.1371/journal.ppat.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327:1644–1648. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ. Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev Cell. 2009;16:386–397. doi: 10.1016/j.devcel.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. 2013;153:797–811. doi: 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 21.Kerney R, Kim E, Hangarter RP, Heiss AA, Bishop CD, Hall BK. Intracellular invasion of green algae in a salamander host. Proc Natl Acad Sci U S A. 2011;108:6497–6502. doi: 10.1073/pnas.1018259108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoll S, Feldhaar H, Fraunholz MJ, Gross R. Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 2010;10:308. doi: 10.1186/1471-2180-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1:E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikori K, Morioka K, Kubo T, Morioka M. Age- and morph-dependent activation of the lysosomal system and Buchnera degradation in aphid endosymbiosis. J Insect Physiol. 2009;55:351–357. doi: 10.1016/j.jinsphys.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Weiss BL, Aksoy S. Tsetse fly microbiota: form and function. Front Cell Infect Microbiol. 2013;3:69. doi: 10.3389/fcimb.2013.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci U S A. 2009;106:12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol. 2006;72:6766–6772. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newell PD, Douglas AE. Interspecies Interactions Determine the Impact of the Gut Microbiota on Nutrient Allocation in Drosophila melanogaster. Appl Environ Microbiol. 2014;80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minard G, Tran FH, Raharimalala FN, Hellard E, Ravelonandro P, Mavingui P, Valiente Moro C. Prevalence, genomic and metabolic profiles of Acinetobacter and Asaia associated with field-caught Aedes albopictus from Madagascar. FEMS Microbiol Ecol. 2013;83:63–73. doi: 10.1111/j.1574-6941.2012.01455.x. [DOI] [PubMed] [Google Scholar]
- 31.Dillon R, Charnely K. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Research in Microbiology. 2002;153:503–509. doi: 10.1016/s0923-2508(02)01361-x. [DOI] [PubMed] [Google Scholar]
- 32.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caragata EP, Rances E, Hedges LM, Gofton AW, Johnson KN, O'Neill SL, McGraw EA. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 2013;9:e1003459. doi: 10.1371/journal.ppat.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292:1099–1012. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- 35.Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol. 2012;188:3395–3403. doi: 10.4049/jimmunol.1103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci U S A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol. 2008;74:6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci U S A. 2005;102:12795–12800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One. 2010;5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 41.Lukasik P, van Asch M, Guo H, Ferrari J, Godfray HC. Unrelated facultative endosymbionts protecte aphids against a fungal. Ecol Lett. 2013;16:214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 42.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 45.van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, Higgs S, O'Neill SL. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis. 2012;6:e1892. doi: 10.1371/journal.pntd.0001892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, Xu Y, Dimopoulos G, Xi Z. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 47.Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2012;109:E23–E31. doi: 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rances E, Ye YH, Woolfit M, McGraw EA, O'Neill SL. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 2012;8:e1002548. doi: 10.1371/journal.ppat.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang G, Hussain M, O'Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A. 2013;110:10276–10281. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M, Frentiu FD, Moreira LA, O'Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A. 2011;108:9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katona P, Katona-Apte J. The interaction between nutrition and infection. Clin Infect Dis. 2008;46:1582–1588. doi: 10.1086/587658. [DOI] [PubMed] [Google Scholar]
- 52.Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF, Jr, Wang J, Ramalingam TR, Bhandoola A, Wynn TA, Belkaid Y. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchner P. Endosymbioses of Animals with Plant Microorganisms. Chichester, UK: John Wiley & Sons; 1965. [Google Scholar]
- 54.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. The central role of the host cell in symbiotic nitrogen metabolism. Proc Biol Sci. 2012;279:2965–2973. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson AC, Ashton PD, Calevro F, Charles H, Colella S, Febvay G, Jander G, Kushlan PF, Macdonald SJ, Schwartz JF, Thomas GH, Douglas AE. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19(Suppl 2):249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 56.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79:6117–6123. doi: 10.1128/AEM.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, van Wijk KJ. Large-scale label-free quantitative proteomics of the pea aphid- Buchnera symbiosis. Mol Cell Proteomics. 2011;10:M110 007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sloan DB, Moran NA. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. 2012;8:986–989. doi: 10.1098/rsbl.2012.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ridley EV, Wong AC, Westmiller S, Douglas AE. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One. 2012;7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 65.The i5k Consortium. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 2013;104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter H, Randau L, Plagens A. Exploiting CRISPR/Cas: interference mechanisms and applications. Int J Mol Sci. 2013;14:14518–14531. doi: 10.3390/ijms140714518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodacre SL, Martin OY, Bonte D, Hutchings L, Woolley C, Ibrahim K, George Thomas C, Hewitt GM. Microbial modification of host longdistance dispersal capacity. BMC Biol. 2009;7:32. doi: 10.1186/1741-7007-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leonardo TE, Mondor EB. Symbiont modifies host life-history traits that affect gene flow. Proc Biol Sci. 2006;273:1079–1084. doi: 10.1098/rspb.2005.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon JC, Fukatsu T. Symbiotic bacterium modifies aphid body color. Science. 2010;330:1102–1104. doi: 10.1126/science.1195463. [DOI] [PubMed] [Google Scholar]
- 73.Tsuchida T, Koga R, Matsumoto S, Fukatsu T. Interspecific symbiont transfection confers a novel ecological trait to the recipient insect. Biol Lett. 2011;7:245–248. doi: 10.1098/rsbl.2010.0699. [DOI] [PMC free article] [PubMed] [Google Scholar]