Abstract
Survival and replication in the intracellular environment are critical components of the ability of Salmonella enterica serovar Typhimurium to establish systemic infection in the murine host. Intracellular survival is mediated by a number of genetic loci, including Salmonella pathogenicity island 2 (SPI2). SPI2 is a 40-kb locus encoding a type III secretion system that secretes effector molecules, which permits bacterial survival and replication in the intracellular environment of host cells. A two-component regulatory system, ssrAB, is also encoded in SPI2 and controls expression of the secretion system and effectors. While the environmental signals to which SPI2 responds in vivo are not known, activation of expression is dependent on OmpR and can be stimulated in vitro by chelation of cations or by a shift from rich to acidic minimal medium. In this work, we demonstrated that SPI2 activation is associated with OmpR in the phosphorylated form (OmpR-P). Mutations in envZ and ackA-pta, which disrupted two distinct sources of OmpR phosphorylation, indicated that SPI2 activation by chelators or a shift from rich to acidic minimal medium is largely dependent on functional EnvZ. In contrast, the PhoPQ pathway is not required for SPI2 activation in the presence of OmpR-P. As in the case of in vitro stimulation, SPI2 expression in macrophages correlates with the presence of OmpR-P. Additionally, EnvZ, but not acetyl phosphate, is required for maximal expression of SPI2 in the intracellular environment, suggesting that the in vitro SPI2 activation pathway is the same as that used in vivo.
Salmonella pathogenicity island 2 (SPI2) is a 40-kb locus required for systemic salmonellosis in mice (18, 36, 38). SPI2 encodes a two-component regulatory system (SsrAB), structural components of a type III secretion system, secreted effector proteins which manipulate the intracellular host environment to allow bacterial persistence and replication, and chaperones which mediate secretion of the effectors. Disruption of the SPI2 regulatory or secretory apparatus results in a dramatic decline in the ability of salmonellae to survive in the intracellular environment of host phagocytes (6, 36), which is a prerequisite for systemic infection (13, 27). Accordingly, SPI2 deficiency leads to diminished virulence during systemic disease in a variety of hosts, including mice, chickens, and cows (4, 18, 21, 36, 38, 47), but not during noninvasive gastrointestinal colonization of cows (42).
Whereas the role of SPI2 in pathogenesis is clear, the molecular mechanisms by which it enhances intracellular survival largely remain elusive. A number of effector proteins, encoded both within and outside SPI2, are secreted through the SPI2 secretion apparatus into the host cell during intracellular infection (46), but their precise functions remain undefined. SPI2 has been implicated in disruption of host processes, including modification of phagosomal trafficking, evasion of host-derived oxidative stress, and induction of late-stage host cell cytotoxicity (recently reviewed by Waterman and Holden [46]), but descriptions of molecular interactions of effectors with host proteins and/or molecular activities are limited.
Our understanding of the environmental signals and regulatory pathways that lead to SPI2 expression is likewise rudimentary. SPI2 is known to be expressed by intracellular Salmonella but not by extracellular bacteria (6, 12, 16, 45), and in vitro conditions that promote expression have also been described. Shifting bacteria from Luria-Bertani (LB) medium to M9 pH 5 acidic minimal medium is known to result in activation of several SPI2 promoters (25) and an increase in transcription of some SPI2 genes (9). Low osmolarity and an acidic pH have therefore been proposed to be the signals that activate SPI2 expression under these conditions. Several groups have also reported upregulation of SPI2 in response to cation chelation when a variety of promoter-reporter fusions (51), Western blots (8), and transcript measurement (23) were used. Magnesium has been proposed to regulate SPI2 (8), although other workers have reported that SPI2 expression is independent of the magnesium concentration (16, 25, 32). Studies of pH as a signal suggest that acidic pH activates SPI2 (25) and alkaline pH represses SPI2 (32). Bafilomycin, which inhibits acidification of the phagosome, also prevents intracellular SPI2 expression, but interpretation of these results is confounded by possible pleiotropic effects on other molecules, such as the divalent cation transporter Nramp1 (19). In addition, SPI2 expression has also been reported to be independent of an acidic pH in vitro (2), indicating that further examination of pH as a signal is necessary. Other proposed signals include phosphate limitation (8), a decrease in osmolarity (25), iron limitation (51), and calcium limitation (16).
Upstream signals which activate SPI2 expression are known to be dependent on a functional OmpR protein in both intracellular and in vitro medium shift conditions (25). OmpR is the response regulator in a two-component regulatory system in which the EnvZ sensor kinase and other molecules are used to detect and respond to the extracellular environment. OmpR is phosphorylated in response to extracellular osmolarity, and the OmpC and OmpF porins are reciprocally regulated in response to the proportion of OmpR in the phosphorylated state (34). The cytoplasmic domain of EnvZ has been well characterized as a phosphodonor to OmpR in vitro, and the location of the envZ gene in the same operon with ompR suggests that this phosphorelay interacts in vivo. However, some reports indicate that EnvZ is not required for OmpR phosphorylation in response to high osmolarity (15, 26, 37), suggesting that OmpR can be phosphorylated by sources other than EnvZ.
One known alternative phosphodonor for a number of response regulators, including OmpR, is the small metabolite acetyl phosphate. Acetyl phosphate serves as an intermediate molecule in the intracellular balance between acetate and acetyl coenzyme A, which are substrates for the reactions catalyzed by acetate kinase (ackA) and phosphotransacetylase (pta), respectively. Acetyl phosphate can directly phosphorylate OmpR in vitro (22, 30), and its ability to control expression of the OmpC and OmpF porins suggests that this phosphotransfer occurs in vivo (30). EnvZ was previously reported to be required for intracellular expression of SPI2 (12), but the role of acetyl phosphate in intracellular induction of SPI2 and the contributions of EnvZ and acetyl phosphate to in vitro expression have not yet been examined.
In addition to being required for expression of SPI2, OmpR has been shown to bind directly to the ssrA and ssrB promoters (12, 25), demonstrating that there is a direct role for OmpR in SPI2 regulation. OmpR and OmpR-P exhibit different binding affinities for the ssrA promoter, as measured by fluorescence anisotropy (12), suggesting that one form may be more active in activating transcription than the other. Mutation of envZ eliminates SPI2 promoter activity (12), suggesting that OmpR-P is the form which activates SPI2 expression, but high osmolarity (which results in OmpR phosphorylation) has also been reported to repress SPI2 expression (25). The phosphorylation state of OmpR during SPI2 activation therefore remains unclear.
Many tools have been developed during two decades of characterization of the EnvZ-OmpR phosphorelay, which is among the most extensively characterized bacterial two-component systems. We employed a number of existing and newly developed genetic tools in order to elucidate upstream events in the SPI2 signaling cascade during in vitro and in vivo activating conditions. We observed that SPI2 expression requires EnvZ but is independent of acetyl phosphate and PhoPQ under all activating conditions.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. LB, M9 pH 7, and M9 pH 5 media were prepared as previously described (9, 25).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) and/or source |
|---|---|---|
| Strains | ||
| SL1344 | xyl hisG rpsL | 40 |
| CK5 | SL1344 ΔhisG::Kan | This study |
| CK500 | SL1344 ΔhisG::FRT | This study |
| CK500/pCP20 | CK500/pCP20 | This study |
| SL1344 ΔenvZ::Kan | SL1344 ΔenvZ::Kan | Corrella Detweiler |
| CK101 | SL1344 ΔenvZ::Kan (P22 transduced) | This study |
| CK121 | SL1344 ΔenvZ | This study |
| CK115 | SL1344 ΔackA-pta::Kan | This study |
| CK116 | CK121 ΔackA-pta::Kan | This study |
| CK117 | SL1344 ΔhisG::pCK60 | This study |
| CK118 | SL1344 ΔhisG::pCK61 | This study |
| CK103 | SL1344 ΔhisG::pCK53 | This study |
| CK104 | SL1344 ΔhisG::pCK54 | This study |
| CK119 | SL1344 ΔackA-pta | This study |
| CK120 | SL1344 ΔenvZ ΔackA-pta | This study |
| CK107 | CK121 ΔhisG::pCK53 | This study |
| SL1344 phoP::Tet | SL1344 phoP::Tet | Bruce Stocker |
| CK122 | CK107 phoP::Tet/pTaz1-1 | This study |
| CK123 | CK107 phoP::Tet/pTazT247R | This study |
| CK136 | SL1344 ΔssrAB | This study |
| SL1344 fur/pFMI10 | SL1344 fur-1 zbf::Tn10/pFMI10 | Ferric Fang; this study |
| CJD359/pFMI10 | SL1344 ompR1009::Tn10/pFMI10 | Anthea Lee; this study |
| CK119/pFMI10 | CK119/pFMI10 | This study |
| CK120/pFMI10 | CK120/pFMI10 | This study |
| CK121/pFMI10 | CK121/pFMI10 | This study |
| CK136/pFMI10 | CK136/pFMI10 | This study |
| CK124 | CK121 ΔhisG::pCK60 | This study |
| CK125 | CK121 ΔhisG::pCK61 | This study |
| CK107 | CK121 ΔhisG::pCK53 | This study |
| CK108 | CK121 ΔhisG::pCK54 | This study |
| CK124/pTaz1-1 | CK124/pTaz1-1 | This study |
| CK124/pTazT247R | CK124/pTazT247R | This study |
| CK125/pTaz1-1 | CK125/pTaz1-1 | This study |
| CK125/pTazT247R | CK125/pTazT247R | This study |
| CK107/pTaz1-1 | CK107/pTaz1-1 | This study |
| CK107/pTazT247R | CK107/pTazT247R | This study |
| CK108/pTaz1-1 | CK108/pTaz1-1 | This study |
| CK108/pTazT247R | CK108/pTazT247R | This study |
| Plasmids | ||
| pFMI10 | pFPV25 with ssaG promoter | 45 |
| pCP20 | FLP recombinase expression vector | 7 |
| pCE37 | Promoter lacZ integration vector | 11 |
| pCK52 | pCE37 rrn multiple cloning site | This study |
| pCK60 | pCK52 with ompC promoter | This study |
| pCK61 | pCK52 with ompF promoter | This study |
| pCK53 | pCK52 with ssrA promoter | This study |
| pCK54 | pCK52 with ssaG promoter | This study |
| pTaz1-1 | ColE1 Taz expression vector | 10, 20 |
| pTazT247R | ColE1 Taz (T247R) mutant expression vector | 10, 20 |
Microarray analysis.
Publicly available data were downloaded from the Stanford Microarray Database (17, 39) with a 60% good data filter and a channel 1 signal intensity greater than 150. The data were analyzed with Significance Analysis for Microarrays (SAM) (43), version 1.21, as a Microsoft Excel plug-in. Significant genes were retrieved from SAM by using SAMster (33). Lexical analysis was performed on the significant gene list by using LACK as previously described (23).
Correction of the pFMI10 (mig-10) sequence.
In order to determine the promoter region contained in pFMI10 (45) for cloning into the lacZ reporter vector pCK52, we performed sequencing on the insert region of pFMI10. The sequence obtained corresponds to coordinates 1488910 to 1489707 of the sequenced LT2 chromosome (31). These coordinates represent the beginning-middle of sseG to the middle-end of ssaG. The insert in pFMI10 was previously reported to be the ssaH promoter (9, 24, 25, 45), but given the location of the sequenced region, it is more likely an ssaG promoter fusion. The construct is referred to as such below.
Green fluorescent protein (GFP) assay of SPI2 induction.
Increased expression of the ssaG-gfp promoter fusion was induced by a shift from LB medium to M9 pH 5 medium as previously described (25). Briefly, overnight LB medium cultures were diluted 1:50 into LB medium with appropriate antibiotics. The bacteria were grown with agitation for 2.5 h and washed twice with phosphate-buffered saline before resuspension in M9 pH 5 medium for 4 h. Chelator inductions were conducted in an identical fashion except that resuspension was in LB medium or M9 medium with a chelator. 2,2′-Dipyridyl (DP) (Sigma D7505), EDTA, trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (CDTA) (Sigma D1383), diethylenetriaminepentaacetic acid (DTPA) (Sigma D6518), triethylenetetraminhexaacetic acid (TTHA) (Sigma T7633), N-(2-hydroxyethyl)ethylenediaminetriacetic acid (HEDTA) (Sigma H8126), and nitriloacetic acid (NTA) (Sigma N7642) were used at the concentrations indicated below.
For tissue culture infections, 2.5 × 105 RAW264.7 cells were seeded into 24-well tissue culture dishes and infected at a multiplicity of infection of 10:1. Extracellular bacteria were removed by washing after 1 h of infection, and the medium was replaced every hour to limit replication of extracellular bacteria as previously described (24). The host cells were lysed at 4 h postinfection, and the collected bacteria were analyzed for levels of GFP expression.
GFP was measured as previously described (25). SPI2 was induced as described above, and GFP was quantitated with a FACS-calibur flow cytometer (Becton Dickenson) and CellQuest software. Averages of the median fluorescence intensities of the replicates are reported below.
Cation rank score calculation.
SL1344/pFMI10 was treated as described above with CDTA, EDTA, DTPA, TTHA, HEDTA, and NTA at concentrations of 1.25, 2.5, 5, and 10 mM. For each concentration, the chelators were ordered with respect to the ability to induce the ssaG-gfp fusion, as measured by flow cytometry. Pairwise comparisons of the rank of each chelator were compared to a similarly ranked list of the chelators ordered by the association constants (Ka) for a given cation. For every comparison in which the induction-based rankings matched the expected order based on Ka, a value of 1 was added to the overall score (zero for incongruous order). The 15 total possible pairwise comparisons were tallied to give the overall rank score for each cation at each concentration of chelator.
Construction of mutant strains.
Gene knockouts were constructed by using the approach and plasmids of Datsenko and Wanner (7). Deletion primers were generated for the entire genome of LT2 by using a Perl script (available from us) and are freely available at http://falkow.stanford.edu. The primer sequences have also been made available by Robert Edwards in a Red Swap primer database, which is accessible at http://www.salmonella.org. Deletions were generated in LT2/pKD46, followed by P22-mediated transduction to SL1344. Detailed protocols for these procedures are available at http://falkow.stanford.edu.
Construction of chromosomally integrated lacZ fusions.
In order to construct chromosomally integrated promoter fusions which did not disrupt the primary locus, we designed an integration vector, pCK52, based on the integration technique of Ellermeier et al. (11). Inverse PCR was used to amplify pCE37 (11) with restriction sites added between the Flp recombination target (FRT) site and the ribosome binding site of lacZ. The rrn terminator of pKD13 (7) was PCR amplified and cloned into the introduced KpnI site between the FRT site and lacZ to obtain pCK52.
Promoter regions of ssrA, ssaG, ompC, and ompF were PCR amplified and cloned into the BamHI-NheI sites of pCK52. CK5 was constructed by the gene knockout procedure of Datsenko and Wanner by using a pKD13 template. The Kanr marker was removed by transformation with pCP20 (7) and antibiotic resistance screening to obtain CK500/pCP20. The promoter-lacZ constructs were integrated into the hisG::FRT site of CK500 by using FLP recombinase-mediated integration as described by Ellermeier et al. (11).
Taz expression and chemiluminescent LacZ activity measurement.
All experiments in which the Taz construct was used were conducted in M9 pH 7 medium. Bacteria were grown for 4 h in the presence of 1 mM aspartate as indicated below. LacZ activity was determined by using the Gal-Screen chemiluminescent substrate as previously described (24). At the time of harvest, the optical density at 630 nm was measured by using a Bio-Tek EL 311SX microplate reader, and this was followed by permeabilization with 5 μl of chloroform and 5 μl of 0.1% sodium dodecyl sulfate with vortexing. Fifty microliters of the permeabilized solution was added to 50 μl of freshly prepared Gal-Screen reagent (Applied Biosystems) in white 96-well plates and incubated for 60 min at room temperature. Luminescence was measured by using a Tropix TR717 microplate luminometer, was quantitated by using the WinGlow software, and was normalized for bacterial density. All experiments were performed in triplicate, and means and standard errors are reported below.
RESULTS
SPI2 expression increases in response to cation chelation.
We observed during studies of iron regulation in Salmonella that SPI2 mRNA abundance increases upon treatment with DP, a cation chelator which preferentially chelates iron (23). We previously reported that 8 of the top 256 genes induced by DP were SPI2-related genes, and a lexical analysis with LACK determined that the genes were overrepresented with high significance (23). We reanalyzed these data using an updated version of SAM (43) and found three additional significantly upregulated genes (ssaQ, ssaP, and ssaD) in addition to the eight previously observed upregulated genes (sseA, ssrB, ssrA, sscA, srfB, ssaQ, ssaB, and ssaC). Moreover, the significance of overrepresentation of SPI2 genes in our data set was an order of magnitude greater when the updated software was used (P = 0.00038 versus the previously reported P = 0.004).
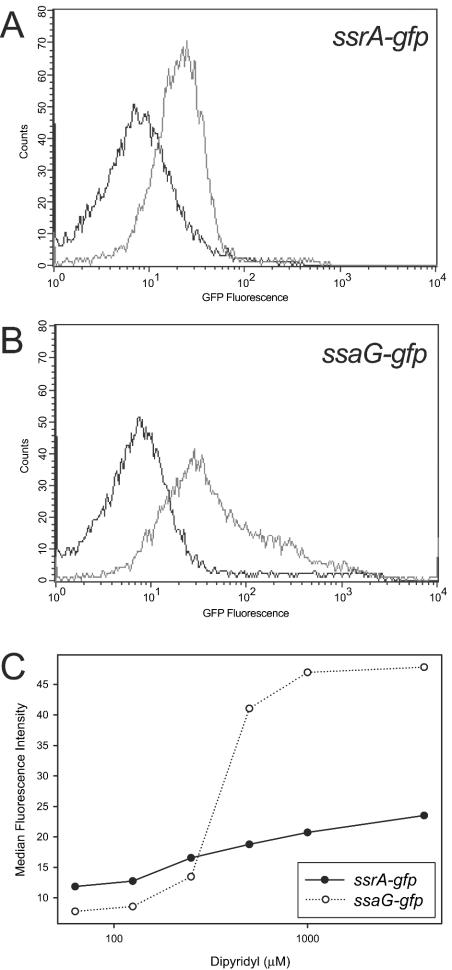
We confirmed the observed upregulation using previously characterized GFP reporter fusions to the promoter regions of ssrA, a SPI2 regulatory gene (6), and ssaG, a SPI2 secretion system structural gene (45) previously reported to be ssaH (see Materials and Methods). We observed that treatment with DP increased GFP expression from these promoter fusions and that the response increased with chelation (Fig. 1). The fusions were induced by chelators in both rich medium (LB medium) (Fig. 1) and minimal medium (M9 medium) (data not shown), suggesting that cation chelation alone is sufficient for SPI2 expression in vitro. These results are in accord with reports that the cation chelators DP, EDTA, and EGTA induce SPI2 expression (8, 23, 51).
FIG. 1.
DP induces SPI2 promoter-gfp fusions. GFP fusions to the ssrA (A) and ssaG (B) promoters showed increased activity upon treatment with 500 μM DP in LB medium (light line) compared to the activity in an untreated LB medium control (dark line). (C) Activities of the ssrA and ssaG promoters increase with increasing DP concentration.
Nature of the regulatory cation.
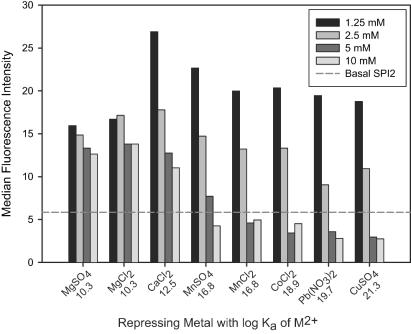
The specific cation(s) which regulates SPI2 has not been identified. We initially employed the add-back experimental design (51), in which specific metal salts are added to chelator-treated medium. The rationale behind this design is that specific cations negate the effects of chelator treatment (in our case SPI2 induction) but that nonspecific cations do not. We used a panel of metal salts to observe their abilities to repress ssaG-gfp expression during treatment of the bacteria with the chelator CDTA. We observed that increasing concentrations of all metals tested repressed CDTA-induced ssaG expression (Fig. 2) but that different metals repressed to different degrees [Mg(II) was the weakest, and Pb(II) and Cu(II) were the strongest]. However, the degree of ssaG repression which we observed correlated strongly with the association constants of different repressing ions for the chelator; ions with stronger affinities for CDTA repressed ssaG more strongly than ions with lower affinities. This suggests that the degree of ssaG repression is more likely to reflect the stability of the cation-chelator complex than specific regulation by a particular cation.
FIG. 2.
Repression of SPI2 expression correlates with chelator-metal ion complex stability. Salmonella containing an ssaG-gfp fusion was grown for 4 h in M9 glucose minimal medium (SPI2 repressing conditions), and this was followed by 4 h of treatment with 10 mM CDTA in the presence of different concentrations of metal salts. Log Ka values are indicated beneath the metals on the x axis. We found that the anion did not appear to have an effect on ssaG expression [chloride versus sulfate for Mg(II) and Mn(II)].
Because of this limitation in the add-back experimental design, we employed a different approach to identify cations which regulate SPI2. The family of complexane-type chelators contains a number of chemically similar members, including the commonly used molecules EDTA and EGTA, with slightly different affinity profiles for different cations (5). In general, the difference in affinity between members of a given pair of complexanes for a cation is smaller than the difference in affinity of a single complexane for two different ions. Because different complexanes generally have similar association constants for a given cation, nonspecific binding of metals to chelators should occur to similar degrees in comparisons of any given pair of complexanes, minimizing the affinity measurement problem of the add-back design. In order to identify the cations that regulate SPI2, we measured ssaG-gfp expression in response to a panel of complexanes at concentrations of 1.25, 2.5, 5, and 10 mM. We observed that EDTA, EGTA, and particularly CDTA showed strong induction of SPI2, while HEDTA, TTHA, DTPA, and NTA showed little or no induction (data not shown). To identify the cations with the most dramatic SPI2 regulatory effects, we constructed a pairwise comparison matrix to compare observed induction of ssaG-gfp by different concentrations of chelators with previously measured Ka values of the chelators for different cations (5). Based on the tabulated scores from the pairwise comparisons (see Materials and Methods), we predicted that Ca(II), Mg(II), Co(II), Mn(II), and Ni(II) are the best candidates for SPI2 regulatory cations. (Table 2). Cd(II), Cu(II), Fe(III), Zn(II), Ag(I), Hg(II), and Fe(II) are less likely to be involved in SPI2 regulation but had moderately high scores in our comparison, and Pb(II) is highly unlikely to be the cation to which SPI2 responds. Because we expect that Pb(II) is unlikely to play a role in SPI2 regulation due to its relative absence in biological systems, these results support the biological relevance of our approach and also our assertion that the add-back design is not appropriate for identification of specific regulatory cations.
TABLE 2.
Scores calculated from a pairwise specificity matrixa
| Cation | Score at a concn of:b
|
|
|---|---|---|
| 1.25 mM | 2.5, 5, or 10 mM | |
| Ca(II) | 14 | 15 |
| Mg(II) | 15 | 14 |
| Co(II) | 14 | 13 |
| Mn(II) | 14 | 13 |
| Ni(II) | 14 | 13 |
| Cd(II) | 13 | 12 |
| Cu(II) | 13 | 12 |
| Fe(III) | 13 | 12 |
| Zn(II) | 13 | 12 |
| Ag(I) | 12 | 11 |
| Hg(II) | 12 | 11 |
| Fe(II) | 11 | 10 |
| Pb(II) | 2 | 3 |
See Materials and Methods.
Higher scores indicate better correlation of previously measured association constants with the observed ssaG-gfp regulatory effects. The results for chelator concentrations of 2.5, 5, and 10 mM were identical and thus are reported together.
SPI2 expression correlates with OmpR in the phosphorylated state.
Previous work indicated that SPI2 was upregulated when bacteria were shifted from LB medium to M9 pH 5 medium and that this upregulation was dependent on OmpR (25). Recently, Feng et al. reported that unphosphorylated OmpR and phosphorylated OmpR exhibit different binding affinities for the ssrA promoter region (12), but the phosphorylation state of OmpR during SPI2 activation was not determined. We employed the sensor kinase Taz in order to examine the role of OmpR phosphorylation in SPI2 regulation. Taz is a hybrid molecule constructed from the periplasmic domain of the aspartate-responsive sensor kinase Tar and the cytoplasmic signaling domain of the sensor kinase EnvZ of E. coli. Taz regulates its kinase activity in response to extracellular aspartate levels, which in turn modulate phosphorylation levels of OmpR (20, 44).
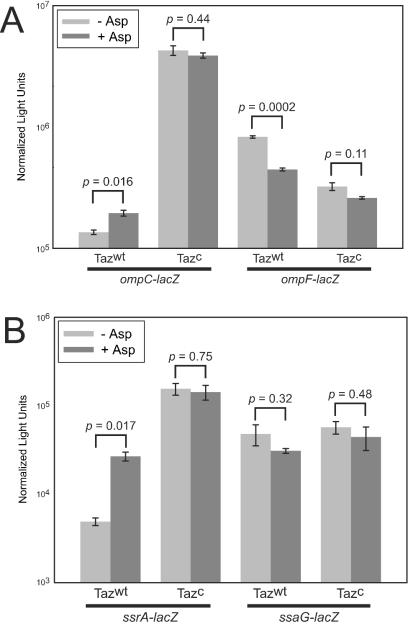
In order to confirm that the system behaves in Salmonella as it does in E. coli, we tested regulation of the ompC and ompF porin gene promoters of Salmonella by Taz of E. coli. We observed that Taz activation reciprocally regulates expression of ompC and ompF as it does in E. coli (10, 20, 44): addition of aspartate upregulates expression from the ompC promoter and downregulates expression from the ompF promoter (Fig. 3A). Moreover, the Taz(T247R) superkinase, which constitutively phosphorylates OmpR (10, 20), exhibits further elevated levels of ompC and repressed levels of ompF promoter activity but is unresponsive to aspartate, indicating that the system behaves in Salmonella as it does in E. coli.
FIG. 3.
Regulation of porin and SPI2 genes in Salmonella by the hybrid sensor kinase Taz. (A) Taz regulation of porin genes behaves in Salmonella as it does in E. coli. Treatment of wild-type Taz (Tazwt) with 1 mM aspartate increased ompC promoter activity, and there was a concomitant decrease in ompF promoter activity. The reciprocal responses of the promoters were amplified upon regulation by the constitutively active superkinase T247R mutant of Taz (Tazc). (B) Taz regulation of SPI2. The ssrA promoter was upregulated in the presence of aspartate or the superkinase, but these conditions were not sufficient for activation of expression from the ssaG promoter. The error bars indicate standard errors (n = 3), and the P values are the result of unpaired t tests assuming unequal variances.
We next used the Taz system to examine regulation of the SPI2 ssrA and ssaG promoters in response to different phosphorylation levels of OmpR. We constructed fusions of the ssrA and ssaG promoters to lacZ and integrated them at the hisG locus of CK500, a derivative of SL1344. Addition of aspartate increased expression of ssrA (Fig. 3B), and the Taz(T247R) superkinase further increased expression of ssrA, indicating that ssrA was upregulated in the presence of OmpR-P. In contrast, the ssaG promoter was not upregulated in response to aspartate or the superkinase. This result indicates that OmpR-P is not sufficient for expression of ssaG under our assay conditions (M9 pH 7 medium). Additionally, because phosphorylated OmpR increased expression of ssrA, we inferred that ssrA expression alone is not sufficient for activation of the ssaG promoter. Together with the observation that the SsrAB two-component system is required for activation of the ssaG promoter (Fig. 4A) (25, 45), these results suggest that SsrA may respond to pH at the posttranscriptional level in order to activate ssaG gene expression.
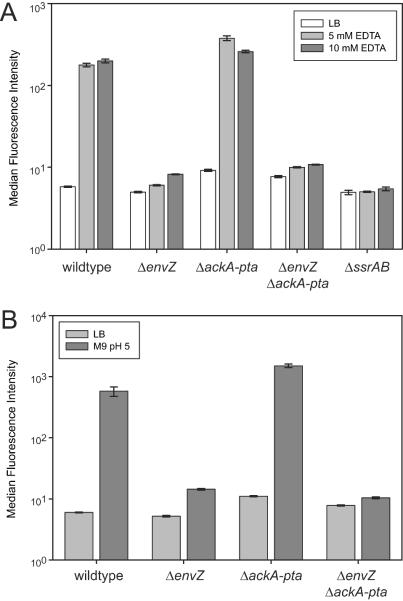
FIG. 4.
(A) EnvZ is required for SPI2 induction by EDTA. An envZ mutant exhibited basal levels of ssaG expression upon treatment with EDTA, in contrast to the wild type and an ackA-pta mutant, in which ssaG expression was increased upon treatment with EDTA. (B) EnvZ is required for medium shift control of ssaG. Shifting wild-type Salmonella from a rich medium (LB medium) to an acidic minimal medium (M9 pH 5 medium) resulted in ssaG-gfp expression in wild-type and ackA-pta backgrounds but not in an envZ mutant.
Expression of SPI2 in vitro requires envZ but not ackA-pta.
SPI2 expression requires functional OmpR (12, 16, 25). OmpR is known to receive signals from at least three sources: the sensor kinase EnvZ (49, 50), the sensor kinase ArcB (under anaerobic conditions) (28), and the metabolite acetyl phosphate (22, 29). We were interested in determining if EnvZ and/or acetyl phosphate could serve as a phosphodonor to OmpR during chelator activation of SPI2 expression. Deletion of the ackA and pta genes eliminates the pathways for generation of acetyl phosphate from acetate and acetyl coenzyme A, respectively, and thereby prevents synthesis of acetyl phosphate (30). We used mutants with deletions in envZ and the ackA-pta operon in order to eliminate EnvZ and acetyl phosphate phosphodonation to OmpR, respectively, and examined regulation of the SPI2 GFP fusions in these mutant backgrounds.
We observed that induction of an ssaG-gfp fusion by EDTA was greatly reduced in the envZ mutant (Fig. 4A), indicating that EnvZ is necessary for induction of SPI2 expression by chelators. This deficiency was not due to the mutant having increased susceptibility to EDTA because expression of a constitutive rpsM-gfp fusion in the envZ background was not affected by EDTA treatment (data not shown). In contrast to the envZ mutant, the ackA-pta mutant was not defective in ssaG-gfp expression, indicating that acetyl phosphate is not required for SPI2 expression in the presence of EnvZ. A double mutant having mutations in envZ and ackA-pta had induction properties similar to those of the envZ mutant, further demonstrating the importance of envZ in SPI2 activation. As in medium shift (25) and macrophage induction (45) experiments, expression of ssaG in response to cation chelation is also dependent on a functional ssrAB (Fig. 4A). The ssrA-gfp fusion displayed a similar pattern of activity: induction by chelators was abolished in the envZ and envZ ackA-pta mutants but was not compromised in the ackA-pta mutant (data not shown). These results indicate that EnvZ, but not acetyl phosphate, is required for SPI2 induction by chelator treatment.
SPI2 promoter fusions to GFP are upregulated (25), and the abundance of SPI2 mRNA transcripts increases (9), in response to a shift from LB medium to M9 pH 5 medium. We examined the role of EnvZ and acetyl phosphate in SPI2 expression under these inducing conditions. As in the case of EDTA induction, we observed that EnvZ is required for induction of ssaG-gfp by medium shift (Fig. 4B). We again observed that the ssaG-gfp reporter exhibited no defect in activation in the ackA-pta background, demonstrating that the absence of acetyl phosphate does not have a significant effect on SPI2 induction by medium shift. The envZ ackA-pta double mutant behaved like the envZ single mutant. The ssrA-gfp fusion exhibited an expression pattern similar to that of the ssaG-gfp fusion (data not shown). These results confirm that EnvZ is required for activation of SPI2 by medium shift and that acetyl phosphate is not required.
PhoP is not required for activation of SPI2 in the presence of OmpR-P.
There have been conflicting reports regarding the role of PhoP in regulation of SPI2 (8, 25, 32). We observed that treatment with 12.5 mM EDTA induced ssaG-gfp expression in wild-type and fur mutant strains but not in ompR and phoP mutant strains (data not shown). However, phoP mutants were highly sensitive to treatment with EDTA compared to wild-type, fur, and ompR mutants (data not shown) and could therefore have been unresponsive due to toxicity rather than to genuine regulatory effects. Because the Taz system can be used under conditions that do not cause any toxicity to a phoP mutant (growth in M9 minimal medium at pH 7), we utilized this system with the chromosomal ssrA-lacZ fusions to examine the role of phoP in SPI2 regulation.
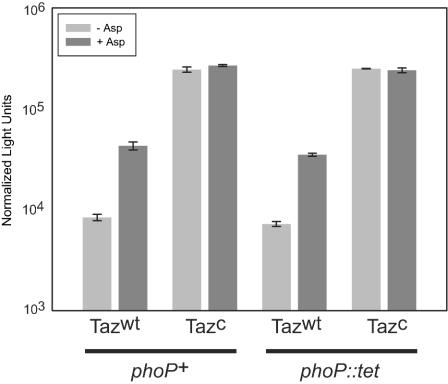
As previously observed, the ssrA-lacZ fusion exhibited increased expression in response to addition of aspartate and in the Taz(T247R) superkinase strain (Fig. 3B and 5). An isogenic strain with a mutation in phoP was constructed and observed for Taz-dependent regulation of the ssrA promoter. We observed almost identical behaviors with the wild-type and phoP mutant backgrounds with regard to aspartate and superkinase responsiveness (Fig. 5) (the Pearson correlation value was 0.994 for the four expression values for each strain), indicating that PhoP is not required for OmpR-dependent induction of ssrA expression.
FIG. 5.
PhoP is not required for SPI2 induction in the presence of OmpR-P. Taz regulation of a ssrA-lacZ fusion was identical in wild-type and phoP backgrounds. The error bars indicate standard errors (n = 3). Tazwt, wild-type Taz; Tazc, superkinase T247R mutant of Taz.
Intracellular SPI2 expression correlates with phosphorylated OmpR and requires envZ.
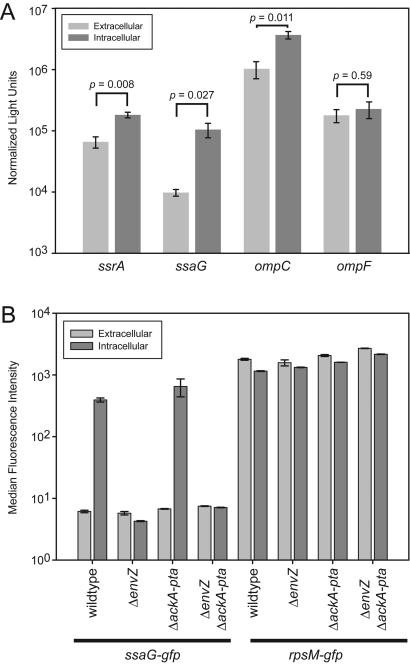
SPI2 expression is essential for replication of Salmonella within mammalian macrophages (6, 36). We therefore wished to determine the phosphorylation state of OmpR during persistence of Salmonella in the intracellular environment. Feng et al. suggested that OmpR is phosphorylated intracellularly based on the observation that EnvZ mutants do not express SPI2 in the intracellular environment (12). Based on our evidence that SPI2 is activated primarily in response to phosphorylated OmpR in vitro and numerous reports that SPI2 is expressed intracellularly, we expected that OmpR-P would be present during intracellular life. We observed that in addition to the ssrA and ssaG promoters, the ompC promoter was active in the phagosomal environment (Fig. 6A). In contrast, the ompF promoter did not show a significant change in activity. These data are consistent with increased levels of OmpR in the phosphorylated form during life in the intracellular environment.
FIG. 6.
Induction of SPI2 in the intracellular environment. (A) RAW264.7 macrophage-like cells were infected for 4 h, and this was followed by lysis and a LacZ assay. The SPI2 and ompC promoters exhibited significantly increased activity, while ompF did not, which correlated with the presence of OmpR-P. (B) EnvZ but not acetyl phosphate is required for full expression of ssaG-gfp in the intracellular environment. The error bars indicate standard errors (n = 3), and the P values are the result of unpaired t tests assuming unequal variances.
We were interested in determining the contributions of EnvZ and acetyl phosphate to the regulation of SPI2 expression in the intracellular environment. As observed for the in vitro conditions, intracellular bacteria exhibited greater ssaG-gfp expression than extracellular bacteria exhibited (Fig. 6B). An envZ mutant displayed basal levels of GFP in the intracellular environment, indicating that ssaG expression in the intracellular environment is dependent on EnvZ. The ackA-pta mutant exhibited no defect in expression of ssaG-gfp in the intracellular environment, which is in accord with our in vitro observations. The envZ ackA-pta double mutant behaved like an envZ mutant, confirming that envZ is essential for SPI2 expression. The decrease in intracellular expression in the envZ and envZ ackA-pta mutants was not due to a decrease in viability, as an rpsM-gfp fusion expressed high constitutive levels of GFP in all mutants and environments (Fig. 6B). In addition, the ackA-pta mutant survived at least as well as the wild type in RAW264.7 cells during the short times used in our assay (data not shown). These results are also recapitulated by the expression pattern displayed by the ssrA-gfp fusion (data not shown). We concluded that EnvZ, but not acetyl phosphate, is required for full expression of SPI2 during intracellular life.
DISCUSSION
The SPI2 secretion system is known to be a critical component of Salmonella's ability to survive and replicate within host cells. Mutations in SPI2 components lead to dramatic decreases in fitness in tissue culture (6, 36) and in host models of systemic disease (4, 18, 21, 36, 38, 47). While the importance of SPI2 for bacterial survival within the murine host is undeniably clear, the mechanisms by which it leads to enhanced virulence are only beginning to be understood (46). Similarly, the precise signals and pathways through which SPI2 is activated upon entry into the intracellular environment are poorly characterized. Because transcriptional responses are often linked to the functions of the genes within the regulon, a better understanding of SPI2 upstream and downstream signaling has the potential to provide insights into the mechanisms by which SPI2 manipulates the host environment, as well as the chemistry of the phagosomal environment.
We previously reported that SPI2 is significantly overrepresented in a set of genes that are upregulated in response to treatment with the cation chelator DP, as detected by microarray analysis (23). We confirmed that SPI2 expression is activated by DP treatment using GFP reporter fusions to a regulatory gene promoter (ssrA) and a structural gene promoter (ssaG) (Fig. 1). Additionally, we observed that these responses are not specific to DP but are also induced by other cation chelators, such as EDTA, EGTA, and CDTA (Table 2 and data not shown). These results are in accord with the reports of Deiwick et al. (8) and Zaharik et al. (51), who described SPI2 responses to 6 mM EGTA and 250 μM DP treatment, respectively.
A number of investigators have examined the nature of the signal which regulates SPI2, but the results have been conflicting. Deiwick et al. reported dependence of SPI2 expression on Mg(II) (8), but more recent results contradict the earlier observations (16, 25, 32). Zaharik et al. reported that Fe(II) was responsible for SPI2 regulation based on its ability to repress SPI2 induction by DP (51). We conducted a similar experiment using the chelator CDTA to induce SPI2 and various metal salts to repress expression. Our data demonstrate that the degree of repression correlates well with the association constant for the cation-chelator complex, which is strong evidence that this experimental design measures complex stability rather than specific regulation by the cation (Fig. 2). This is especially supported by the strong repression of SPI2 expression by Pb(II), which we feel is unlikely to be a specific regulator of SPI2 expression due to the relative absence of Pb(II) in biological systems and our observation obtained with the panel of complexane chelators that Pb(II) is highly unlikely to regulate SPI2 expression (Table 2). The ions tested by Zaharik et al. (51) also follow this trend; Fe(II) repressed SPI2 expression more strongly than Mg(II) or Mn(II) repressed SPI2 expression, which is consistent with our expectations from the Kas of these cations with DP [log Ka(Fe) = 9.55, log Ka(Mg) = 0.5, and log Ka(Mn) = 2.62l (5)]. While it is still possible that iron plays a role in regulating SPI2 expression, our data indicate that there is not enough evidence to draw this conclusion and, moreover, that caution must be exercised in interpreting data obtained with the add-back experimental design.
The add-back experimental design and interpretation are fairly commonly employed. For example, EGTA is often used as a calcium-specific chelator (8), although it has a higher association constant with Fe(III) (log Ka = 20.5) than with Ca(II) (log Ka = 11.00) (5). The source of misinterpretation is presumably an inappropriate extension of the observation that EGTA has a higher affinity for Ca(II) than other common chelators, such as EDTA (log Ka = 10.96), DTPA (log Ka = 10.74), and HEDTA (log Ka = 8.14), have and therefore chelates more Ca(II) at a given chelator concentration. At this time it is not possible to eliminate the bioavailability of individual cation species, so better technical and/or creative methods are needed to precisely identify the cation(s) to which SPI2 gene expression responds. We feel that our complexane-based prediction that Ca(II), Mg(II), Co(II), Mn(II), and/or Ni(II) regulates SPI2 (Table 2), while still only approximate, is the most accurate estimate to date. We note that Garmendia et al. reported upregulation of SPI2 by an absence of Ca(II) (16), which is consistent with our observations.
In addition to cation chelation, a shift from LB medium to M9 pH 5 medium can also activate SPI2 promoter activity. Lee et al. previously demonstrated that OmpR is required for the increase in SPI2 expression and that OmpR binds to the promoter region of ssrA, the regulatory sensor kinase of SPI2 (25). Feng et al. extended these findings by identifying additional OmpR binding sites and demonstrating that OmpR-P binds the ssrA promoter region with higher affinity than OmpR (12). High osmolarity was reported to repress SPI2 expression, suggesting that OmpR-P represses SPI2 expression (25), but Feng et al. recently suggested that OmpR-P activates SPI2 expression based on the observation that an EnvZ mutant does not express SPI2 in macrophages (12). In order to examine the role of OmpR phosphorylation in SPI2 regulation, we employed the well-characterized Taz hybrid sensor kinase, which phosphorylates OmpR in response to an increasing extracellular aspartate concentration (44), and the Taz (T247R) mutant, which possesses constitutive kinase activity (10, 20). In addition to properly regulating the ompC and ompF porins in Salmonella (Fig. 3A), activating Taz kinase activity with aspartate upregulated expression of a chromosomally integrated ssrA-lacZ fusion, suggesting that OmpR-P is the form which is required for SPI2 expression (Fig. 3B). Moreover, the Taz (T247R) superkinase strain further increased ssrA-lacZ expression, which is also consistent with the hypothesis that OmpR-P is the form which activates SPI2 expression. These results provide positive evidence that is complementary to and in accord with speculation that OmpR-P activates SPI2 expression (12). We did not observe any evidence of SPI2 repression in our Taz experiments, but additional work is needed to resolve this possible regulatory function of OmpR-P.
While OmpR-P did activate ssrA expression, an ssaG-lacZ fusion was not coordinately upregulated with ssrA, indicating that OmpR phosphorylation and expression of SsrA alone are insufficient for upregulation of downstream SPI2 genes (Fig. 3B). This observation indicates that an additional signal(s) which is not present in the Taz growth conditions (M9 pH 7 minimal medium) is necessary for the SsrAB phosphorelay to activate expression of downstream SPI2 genes. Our observation that ssaG-gfp is upregulated in M9 pH 5 medium but not in M9 pH 7 medium suggests that pH may activate SsrAB at the posttranscriptional level. However, we note that pH cannot be the only signal for SsrAB activation because acidified LB medium is not sufficient to induce SPI2 expression (25; data not shown). One interpretation of these observations is that SsrA regulation of other SPI2 genes requires both cation starvation and an acidic environment. Other reports also support the hypothesis that there is a role for acid regulation of SPI2; bafilomycin treatment of macrophages, which disrupts acidification of the phagosome, is reported to abrogate expression of SPI2 (6, 16), and secretion of a number of effectors is reported to occur upon medium acidification in vitro (2, 35). However, other data indicate that acid does not play a role in regulation of SPI2; expression of the sseA and sseB SPI2 effectors is not upregulated by acid treatment (2), and vacuole acidification is not required for intracellular replication in certain cell lines (41). Moreover, SPI2 expression can be induced in vitro by cation chelation in the absence of acidification (Fig. 4A). Further work is clearly needed to determine whether environmental acidification is a relevant activator of SPI2 expression in vivo (41). An alternative possibility is that the signal is not acidification per se, but rather a pleiotropic effect of acidification, such as modulation of a host molecule such as Nramp1 (19) or alteration of cation solubility or bioavailability. The host molecule Nramp1 transports cations out of the phagosome in a pH-dependent manner (19) and has been reported to affect SPI2 expression levels (51), suggesting that cation starvation mediated by Nramp1 plays a role in SPI2 regulation during vacuolar life. However, it is important to note that SPI2 is expressed at high levels and is active in Nramp1-defective cells, such as the RAW264.7 macrophage-like cell line (25, 45), and in Nramp1-defective mice, such as BALB/c mice, indicating that Nramp1 cannot be the major factor which regulates SPI2 expression in vivo. Finally, we propose the possibility that cation starvation coupled with acidification might lead to generalized activation of multiple two-component systems. The observations that cations and acidic pH activate EnvZ-OmpR (Fig. 4), PhoQ-PhoP (1), and possibly the SsrA-SsrB phosphorelays are in accord with this proposal. Careful measurement of the intracellular chemistry of the phagosomal environment and detailed genetic studies are essential to distinguish between these possibilities.
OmpR can receive phosphorylation signals from at least three sources: the EnvZ sensor kinase (49, 50), the ArcB sensor kinase (28), and the small molecule acetyl phosphate (22, 29). We observed that EnvZ is required for full expression of SPI2 by all known activating signals, including chelators (Fig. 4A), medium shift (Fig. 4B), and the intracellular environment (Fig. 6); the latter observation is also in accord with results reported by Feng et al. (12). In contrast, a mutant with a mutation in the ackA-pta loci, which results in acetyl phosphate deficiency (30), has no defect in SPI2 expression during intracellular life or under in vitro conditions. These results indicate that while acetyl phosphate may play a role in porin regulation, it does not regulate SPI2 expression in the host environment, in contrast to EnvZ. Moreover, our results indicate that the signaling pathways which induce SPI2 expression under the three known activating conditions are identical.
In a number of reports the authors have reached conflicting conclusions regarding the role of the PhoPQ two-component regulatory system in regulation of SPI2. Deiwick et al. concluded from an analysis of secreted SPI2 proteins that PhoPQ modulates SPI2 activation (8). Lee et al. observed that PhoPQ seemed to play a role in expression of SPI2 under magnesium limiting conditions but that PhoPQ was not required for SPI2 expression in RAW264.7 macrophage-like cells (25). Using a luciferase reporter, Miao et al. observed that SPI2 expression was not affected by a constitutively phosphorylated PhoP in vitro or by a PhoP mutant in RAW264.7 cells (32). On the phenotypic level, PhoPQ and SPI2 appear to contribute to systemic disease independently in mouse disease models (3), suggesting that they act independent of one another. Our results obtained by using the Taz system demonstrate that in the presence of OmpR-P, the PhoPQ regulatory system is not required for SPI2 expression (Fig. 5). Although no instances have been reported, it is formally possible that PhoPQ could play a role in SPI2 regulation by modulating events upstream of OmpR. However, most of the data reported to date indicate that PhoPQ is unlikely to play a direct role in the regulation of SPI2.
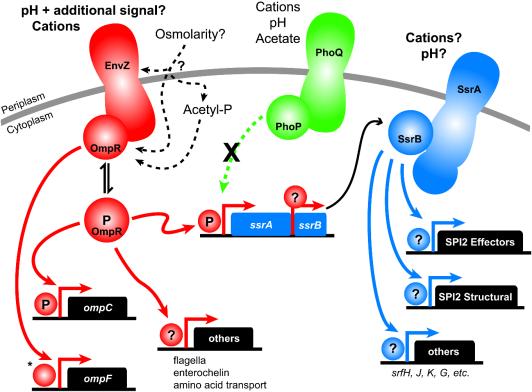
Figure 7 shows a model of our current understanding of the SPI2 regulatory cascade. The first level of SPI2 regulation involves the EnvZ-OmpR phosphorelay, which is typically described as an osmolarity-responsive two-component regulatory system. While the cytoplasmic domain of EnvZ can phosphorylate OmpR in vitro (14), a number of reports have demonstrated that porin regulation in vivo occurs independent of the presence of EnvZ (15, 26, 37), and thus the role of EnvZ in porin regulation by osmolarity, as well as the substrate(s) for EnvZ, remain unclear. The dependence of SPI2 activation on EnvZ in response to the in vitro and in vivo conditions described here suggests that pH and cations should be examined as possible EnvZ substrates. We noted that pH signaling to EnvZ requires at least one additional signal based on the observation that pH and a shift from LB medium to M9 medium do not induce SPI2 alone but do induce SPI2 in combination. Acetyl phosphate does not regulate SPI2 expression under our conditions, but its role in regulating porins (30) suggests that two separate signaling pathways (acetyl phosphate and EnvZ) may feed into OmpR as an intermediate and result in regulation of distinct groups of genes (porins and SPI2, respectively). The majority of the data available also indicate that the PhoPQ phosphorelay is unlikely to be directly involved in SPI2 activation.
FIG. 7.
Model of SPI2 signaling. Different colors indicate different two-component systems. The primary signals are detected by the EnvZ sensor kinase and are independent of acetyl phosphate signaling. PhoPQ is unlikely to directly regulate SPI2. Question marks indicate unknown phosphorylation states of response regulators. Much of the work that was used to synthesize this model is not cited; for this we apologize to the authors of relevant work. The asterisk indicates unphosphorylated OmpR at the ompF promoter in order to reflect the predominance of OmpF expression in conditions under which OmpR is predominantly unphosphorylated; however, the regulation at the porin promoters is considerably more complex than that depicted.
The SsrAB phosphorelay is the second level of regulation in the model; our data suggest that SsrA expression is distinct from phosphorelay activity that leads to transcriptional activation of downstream genes, such as ssaG, indicating that SsrA requires an environmental signal to activate SsrB. Because SsrA-based activation of SsrB presumably occurs in the in vitro conditions which we examined (based on upregulation of ssaG expression), we propose that cations and/or pH is likely to be involved in this step. Finally, there has been only one study of direct downstream activation of SPI2 genes by SsrB, which constitutes the third regulatory tier (48). This model outlines some of the important areas for future research on the SPI2 regulatory cascade.
Acknowledgments
We thank Takeshi Yoshida (Masayori Inouye's lab), Corrie Detweiler, and Bruce Stocker for strains; Thomas Wehrman for advice on chemiluminescent LacZ activity measurement; Anthea Lee for discussions; and Elizabeth Joyce, Trevor Lawley, Denise Monack, Igor Brodsky, and Kaman Chan for critical reading of the manuscript.
C.K. thanks the Howard Hughes Medical Institute and the Stanford Graduate Fellowships Program for support. This work was funded by NIH grant AI 26195.
REFERENCES
- 1.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., K. E. Unsworth, and D. W. Holden. 2001. In vivo genetic analysis indicates that PhoP-PhoQ and the Salmonella pathogenicity island 2 type III secretion system contribute independently to Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 69:7254-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bispham, J., B. N. Tripathi, P. R. Watson, and T. S. Wallis. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun. 69:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, K. L., K. Ueno, and T. Imamura. 1982. CRC handbook of organic analytical reagents. CRC Press, Boca Raton, Fla.
- 6.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 9.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, R., T. Yoshida, and M. Inouye. 2000. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine kinase/phosphatase, in Escherichia coli. J. Biol. Chem. 275:38645-38653. [DOI] [PubMed] [Google Scholar]
- 11.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 12.Feng, X., R. Oropeza, and L. J. Kenney. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48:1131-1143. [DOI] [PubMed] [Google Scholar]
- 13.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forst, S., J. Delgado, and M. Inouye. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc. Natl. Acad. Sci. USA 86:6052-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forst, S., J. Delgado, G. Ramakrishnan, and M. Inouye. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385-2396. [DOI] [PubMed] [Google Scholar]
- 17.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Matese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 19.Jabado, N., A. Jankowski, S. Dougaparsad, V. Picard, S. Grinstein, and P. Gros. 2000. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192:1237-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, T., and M. Inouye. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J. Mol. Biol. 232:484-492. [DOI] [PubMed] [Google Scholar]
- 21.Jones, M. A., P. Wigley, K. L. Page, S. D. Hulme, and P. A. Barrow. 2001. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infect. Immun. 69:5471-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenney, L. J., M. D. Bauer, and T. J. Silhavy. 1995. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:8866-8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, C. C., and S. Falkow. 2003. Significance analysis of lexical bias in microarray data. BMC Bioinformatics 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, C. C., D. Monack, and S. Falkow. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonardo, M. R., and S. Forst. 1996. Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signals in Escherichia coli. Mol. Microbiol. 22:405-413. [DOI] [PubMed] [Google Scholar]
- 27.Leung, K. Y., and B. B. Finlay. 1991. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 88:11470-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsubara, M., S. I. Kitaoka, S. I. Takeda, and T. Mizuno. 2000. Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells 5:555-569. [DOI] [PubMed] [Google Scholar]
- 29.Matsubara, M., and T. Mizuno. 1999. EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci. Biotechnol. Biochem. 63:408-414. [DOI] [PubMed] [Google Scholar]
- 30.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 31.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 32.Miao, E. A., J. A. Freeman, and S. I. Miller. 2002. Transcription of the SsrAB regulon is repressed by alkaline pH and is independent of PhoPQ and magnesium concentration. J. Bacteriol. 184:1493-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller, A., J. O'Rourke, P. Chu, C. C. Kim, P. Sutton, A. Lee, and S. Falkow. 2003. Protective immunity against Helicobacter is characterized by a unique transcriptional signature. Proc. Natl. Acad. Sci. USA 100:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 35.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prohinar, P., S. A. Forst, D. Reed, I. Mandic-Mulec, and J. Weiss. 2002. OmpR-dependent and OmpR-independent responses of Escherichia coli to sublethal attack by the neutrophil bactericidal/permeability increasing protein. Mol. Microbiol. 43:1493-1504. [DOI] [PubMed] [Google Scholar]
- 38.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, B. P., M. Reina-Guerra, S. K. Hoiseth, B. A. Stocker, F. Habasha, E. Johnson, and F. Merritt. 1984. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am. J. Vet. Res. 45:59-66. [PubMed] [Google Scholar]
- 41.Steele-Mortimer, O., M. St-Louis, M. Olivier, and B. B. Finlay. 2000. Vacuole acidification is not required for survival of Salmonella enterica serovar Typhimurium within cultured macrophages and epithelial cells. Infect. Immun. 68:5401-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Utsumi, R., R. E. Brissette, A. Rampersaud, S. A. Forst, K. Oosawa, and M. Inouye. 1989. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245:1246-1249. [DOI] [PubMed] [Google Scholar]
- 45.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 46.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 47.Wigley, P., M. A. Jones, and P. A. Barrow. 2002. Salmonella enterica serovar Pullorum requires the Salmonella pathogenicity island 2 type III secretion system for virulence and carriage in the chicken. Avian Pathol. 31:501-506. [DOI] [PubMed] [Google Scholar]
- 48.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, T., S. Cai, and M. Inouye. 2002. Interaction of EnvZ, a sensory histidine kinase, with phosphorylated OmpR, the cognate response regulator. Mol. Microbiol. 46:1283-1294. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, T., L. Qin, and M. Inouye. 2002. Formation of the stoichiometric complex of EnvZ, a histidine kinase, with its response regulator, OmpR. Mol. Microbiol. 46:1273-1282. [DOI] [PubMed] [Google Scholar]
- 51.Zaharik, M. L., B. A. Vallance, J. L. Puente, P. Gros, and B. B. Finlay. 2002. Host-pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc. Natl. Acad. Sci. USA 99:15705-15710. [DOI] [PMC free article] [PubMed] [Google Scholar]