Abstract
YscC is the integral outer membrane component of the type III protein secretion machinery of Yersinia enterocolitica and belongs to the family of secretins. This group of proteins forms stable ring-like oligomers in the outer membrane, which are thought to function as transport channels for macromolecules. The YscC oligomer was purified after solubilization from the membrane with a nonionic detergent. Sodium dodecyl sulfate did not dissociate the oligomer, but it caused a change in electrophoretic mobility and an increase in protease susceptibility, indicating partial denaturation of the subunits within the oligomer. The mass of the homo-oligomer, as determined by scanning transmission electron microscopy, was approximately 1 MDa. Analysis of the angular power spectrum from averaged top views of negatively stained YscC oligomers revealed a 13-fold angular order, suggesting that the oligomer consists of 13 subunits. Reconstituted in planar lipid bilayers, the YscC oligomer displayed a constant voltage-independent conductance of approximately 3 nS, thus forming a stable pore. However, in vivo, the expression of YscC did not lead to an increased permeability of the outer membrane. Electron microscopy revealed that the YscC oligomer is composed of three domains, two stacked rings attached to a conical domain. This structure is consistent with the notion that the secretin forms the upper part of the basal body of the needle structure of the type III secreton.
The outer membrane of gram-negative bacteria not only protects the cell against harmful compounds in the extracellular environment, such as antibiotics, detergents, and digestive enzymes, but also forms a barrier to the secretion of proteins and uptake of nutrients. The passage of small hydrophilic molecules across the outer membrane is facilitated by the presence of a large number of proteinaceous channels, formed by general and substrate-specific porins and TonB-dependent receptors (31, 41). Gram-negative bacteria have evolved several specialized pathways for the secretion of proteins. These pathways all require one or more integral outer membrane proteins, which in the cases of the type II and III protein secretion systems are related and belong to the family of secretins (19). Other members of this family are involved in the assembly of type IV pili or in the secretion of filamentous phages.
The best-studied secretins with respect to structure and channel formation are XcpQ of Pseudomonas aeruginosa and PulD of Klebsiella oxytoca, both components of type II secretion pathways, the filamentous phage protein pIV, and the PilQ protein of Neisseria meningitidis that is involved in the assembly of type IV pili. Secretins exist in the outer membrane as stable oligomeric complexes that have been purified and visualized by electron microscopy (EM). Top views of the oligomers show them to be large ring-like structures with an internal cavity that might function as an export channel (2, 4, 7, 35, 42). The fact that the PilQ protein of Neisseria gonorrhoeae is essential for a periplasmically assembled pilus to traverse the outer membrane (55) is consistent with this notion. Further, on some micrographs, the internal cavity of the purified secretin oligomers is occluded (4, 8, 43, 44). Side views of the oligomers reveal cylindrical structures composed of at least two stacked ring-like regions (35, 43).
Purified pIV (37), XcpQ (4), and PulD (42) oligomers have been shown to display pore-forming activity when inserted in phospholipid bilayers. The conductance of the channels was voltage dependent, suggesting that the secretins change their conformation in response to the transmembrane potential. The level of conductance was very irregular, especially for XcpQ and PulD, which also indicates that the channels are gated. In contrast, in vivo expression of secretins seems to have no effect on the permeability of the outer membrane (4, 37), which makes it likely that in the cell the secretin is closed. How the channel is opened to facilitate export is unknown, but probably interactions with substrates or other components of its cognate system induce a conformational change.
The homology between the members of the secretin family is particularly marked in their C-terminal halves. Deletion mutagenesis (20) and limited proteolysis (4, 43) have shown that this domain participates in oligomerization and pore formation, while the N terminus is probably involved in system-specific interactions (12, 49). A small nonconserved domain C terminal to the conserved part of the protein functions as the binding site for a small lipoprotein in a number of secretins (11, 13, 47, 49). These so-called pilot proteins protect the secretin against proteolysis and mediate its stable insertion into the outer membrane (10, 13, 16, 23, 32, 49). In the Pul system of K. oxytoca, the PulS pilot protein remains associated with the secretin after its insertion into the membrane (42).
Secretins have been shown to be major components of type III secretion systems (33, 48, 51). These systems, which include approximately 20 proteins (9, 28), play a central role in the interaction between gram-negative bacteria and their eukaryotic hosts by allowing the bacteria to inject proteins directly into the cytosol of eukaryotic target cells (28). Secretins of two type III systems have been purified and visualized by EM and are ring-shaped oligomeric molecules (10, 32). However, these purified oligomers have not been analyzed further. In this study, we describe the pore formation and EM analysis of the YscC secretin of the type III secretion system of Yersinia enterocolitica.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strains DH5α (21), S17-1b (50), and BL21(DE3) (Novagen) were used for routine gene cloning, conjugational transfer of plasmids to Y. enterocolitica, and expression of genes under T7 promoter control, respectively. Y. enterocolitica KNG22703 is a blaA mutant derivative of strain W22703, in which the gene encoding β-lactamase has been replaced by the luxAB genes (30). KNG22703(pAA203) carries the aphA-3 cassette in the yscC gene creating a nonpolar mutation (32). The pYV-cured Y. enterocolitica strain CE1525 was obtained by growing strain W22703 on calcium-depleted medium at 37°C. Fast-growing colonies were screened, and a strain from which no plasmid could be isolated was selected.
E. coli strains were routinely grown at 37°C in a modified Luria-Bertani broth (LB) (52), and Y. enterocolitica strains were grown at room temperature in LB supplemented with 0.4% glucose. To induce expression of yscC, Y. enterocolitica strains were inoculated at an optical density at 600 nm of 0.1 in brain heart infusion broth supplemented with 0.4% glucose, 20 mM MgCl2, and 20 mM sodium oxalate (BHI-OX). Alternatively, cells were grown in brain heart infusion broth supplemented with 5 mM CaCl2 (BHI-Ca2+). The cultures were grown for 2 h at room temperature, isopropyl-thio-β-d-galactopyranoside (IPTG) (final concentration, 0.1 mM) was added to the cultures, and the cultures were grown for 2 h at 37°C. For Y. enterocolitica, ampicillin (1 mg/ml), kanamycin (50 μg/ml), nalidixic acid (25 μg/ml), and tetracycline (10 μg/ml for cultures grown in LB and BHI-Ca2+ and 20 μg/ml for cultures grown in BHI-OX). For E. coli, ampicillin (100 μg/ml), kanamycin (25 μg/ml), and tetracycline (10 μg/ml for cultures grown in LB and 20 μg/ml for cultures grown in BHI-OX) were used.
Cloning of the yscC and yscW genes.
Recombinant DNA methods were performed essentially as described previously (45). Plasmids were introduced into E. coli by transformation using the CaCl2 procedure (45) and into Y. enterocolitica by conjugation on LB agar plates overnight at room temperature.
The 3,077-bp EcoRI-PstI fragment of plasmid pSM3 (32), carrying the beginning of the virC operon including the full-length yscC gene, was cloned into the vector pUR6500 (18), resulting in pSM3km. To obtain a construct that encodes a six-histidine-tagged YscW (His6-YscW), the part of the yscW gene encoding YscW without its signal sequence and N-terminal cysteine residue was amplified by PCR. PCR was performed using the pYV plasmid of strain W22703 as the template and using primers MCK16 (5′-CATCATTTCTTTTCATATGGTAGATCTCGCCACCCCCCCA-3′) and MCK17 (5′-CCAATATAATAAATCACC-3′). The amplified sequence was cloned into the SmaI site of pBC18R (6) in the direction opposite that of the lac promoter, resulting in pEW1. The EcoRI-SalI fragment of this plasmid was cloned into vector pBluescript (Stratagene) to create pEW2. The BamHI-NdeI fragment of pEW2 was cloned into pET16b (Novagen), thereby placing the yscW gene in pEW3 behind an N-terminal histidine tag and the T7 promoter.
Purification of YscC.
Unless stated otherwise, the entire YscC purification protocol was performed on ice or at 4°C. The pYV-cured Y. enterocolitica strain CE1525 carrying plasmids pSM3km and pRS6 (1) containing the yscC and yscW genes, respectively, was grown in 2.5 liters of BHI-OX, and production of YscC was induced by the addition of IPTG as described above. Cells were washed with 0.9% NaCl solution, resuspended in 50 ml of lysis buffer (1 mM EDTA, 50 mM Tris-HCl [pH 8.5]), and frozen at −20°C. After thawing and sonication, cells were removed by centrifugation for 5 min at 3,000 × g. The cell envelopes were pelleted by centrifugation for 1 h at 20,000 × g, and the YscC oligomer was solubilized with 8.5 ml of extraction buffer containing 3% Elugent (Calbiochem), 250 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl (pH 7.8) in the presence of protease inhibitor (complete protease inhibitor; Boehringer) for 30 min at room temperature. Insoluble debris was removed by centrifugation for 60 min at 150,000 × g. After the addition of sucrose to a final concentration of 15% (wt/wt), the extracted cell envelope proteins were layered on top of a 20 to 40% (wt/wt) sucrose gradient in gradient buffer (0.3% Elugent, 250 mM NaCl, protease inhibitor, 5 mM EDTA, 50 mM Tris-HCl [pH 7.8]) and centrifuged for 28 h at 38,000 rpm in an SW41 rotor (Beckman). Fractions from the gradient containing the YscC oligomer were pooled, dialyzed against chromatography buffer (0.1% Elugent, 100 mM NaCl, 0.1 mM EDTA, 10 mM Tris-HCl [pH 7.8]) and concentrated to 2 ml in a microcentrifuge YM-100 tube (Centricon). One milliliter of concentrated protein solution was loaded on a MonoQ HR 5/5 ion-exchange column, which was eluted with a salt gradient ranging from 100 mM to 1 M NaCl in chromatography buffer. The proteins of the column fractions that contained the YscC oligomer, which eluted at 450 to 500 mM NaCl, were combined and concentrated to 2 ml in a microcentrifuge YM-100 tube and stored at −20°C.
Antibody preparation against YscW.
For overproduction and purification of His6-YscW, a culture of E. coli BL21(DE3) carrying pEW3 that had been grown overnight was diluted 1:20 in LB and grown for 1 h at 37°C. After the addition of IPTG to a final concentration of 1 mM, the bacteria were grown for another 2 h at 37°C. Cells were collected by centrifugation, and the cell pellet was resuspended in a solution consisting of 100 mM NaCl, 1 mM EDTA, and 50 mM Tris-HCl (pH 8.0) and sonicated. The lysate was centrifuged for 25 min at 3,000 × g at 4°C, and the pelleted inclusion bodies were solubilized in 8 M urea-50 mM Tris-HCl (pH 8.0). After centrifugation for 1 h at 100,000 × g at 15°C, solubilized His6-YscW from the supernatant was purified with Ni-nitrilotriacetic acid beads (Qiagen), following the manufacturer's instructions. A rabbit was injected subcutaneously with 0.15 mg of purified protein in Freund's complete adjuvant. Seven booster doses of the same protein preparation in Freund's incomplete adjuvant were given, one every 4 weeks.
SDS-PAGE and immunoblotting.
Proteins were routinely separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (34) with 0.2% SDS in the running gel. For seminative SDS-PAGE, the loading buffer contained no β-mercaptoethanol and only 0.1% SDS instead of 2% SDS, the gels contained no SDS, and electrophoresis was performed at 4°C. The detergent-solubilized YscC oligomer was dissociated by treatment with hot phenol (22). Alternatively, the YscC oligomer was dissociated by treatment with trichloroacetic acid (TCA) and trifluoroacetic acid (TFA). One volume of cell culture was mixed with 1 volume of 10% TCA, and the mixture was incubated for 20 min on ice. Insoluble material was collected by centrifugation at 20,000 × g for 20 min at 4°C, washed twice with 2 volumes of ice-cold acetone, and dried. To increase the dissociation efficiency, 50 μl of TFA was added to the pellet, which was then immediately evaporated by a stream of air. Protein bands were stained either with Coomassie brilliant blue or with silver (38). Alternatively, proteins were transferred onto nitrocellulose membranes by semidry electroblotting. After incubation with primary antibodies and with horseradish peroxidase-coupled anti-rabbit immunoglobulin G (diluted 1:3,000) (Biosource), the immunoblots were developed by enhanced chemiluminescence (Pierce). The primary antibodies used were 1:1,000 dilutions of rabbit antisera directed against a synthetic YscC peptide (32) or YscW.
Stability assay for the purified YscC oligomer.
The stability of the purified YscC oligomer in the presence of detergents was evaluated by incubating the oligomer for 1 h at 40°C in 2% Elugent, octyl-polyoxyethylene (OPOE) (Alexis), sulfobetaine 12 (SB12) (Sigma), SDS (J. T. Baker), Triton X-100 (Serva), or Zwittergent 3-14 (Calbiochem) in chromatography buffer. Similarly, the stability of the oligomer in the presence of 20 mM dithiothreitol (DTT) was evaluated. After the incubations, the samples were placed on ice. The loading buffer for seminative gel electrophoresis was added just before the gel was loaded.
Protease accessibility of the YscC oligomer.
Purified YscC oligomer was diluted 1:10 in chromatography buffer containing 2% SDS, and the solution was either boiled for 10 min or left untreated. The samples were incubated with different concentrations of proteinase K for 10 min at room temperature. Subsequently, SDS-PAGE loading buffer was added, the samples were boiled for 10 min, and proteolytic degradation was immediately evaluated by SDS-PAGE.
Mass determination by STEM.
The protein stock was diluted 1:2 with chromatography buffer without Elugent, and 6-μl aliquots were adsorbed for 60 s to glow-discharged scanning transmission electron microscopy (STEM) films, which are thin carbon films that span a thick fenestrated carbon layer covering 200-mesh/in., gold-plated copper grids. The grids were then blotted, washed on 8 drops of quartz double-distilled water, and freeze-dried at −80°C and 5 × 10−8 torr overnight in the microscope. Tobacco mosaic virus (TMV) particles served as mass standard. These particles were similarly adsorbed (45 s) to separate microscopy grids, washed on 4 droplets of 10 mM ammonium acetate, and air dried.
A STEM HB-5 vacuum generator interfaced to a modular computer system (Tietz Video and Image Processing Systems GmbH, Gauting, Germany) was employed (39). Series of dark-field images (512 by 512 pixels) were recorded from the unstained samples at an accelerating voltage of 80 kV and a nominal magnification of ×200,000. The recording dose was 345 ± 45 electrons/nm2. The digital images were evaluated using the program package IMPSYS (39). Accordingly, the projections were selected in circular boxes, and the total scattering of each region was calculated. The background scattering of the carbon support film was then subtracted, and the mass was calculated. The results were scaled according to the mass measured for TMV and corrected for beam-induced mass loss on the basis of the behavior of proteins in a similar mass range (40; unpublished data). The mass values were then displayed in histograms and described by Gauss curves.
Negative stain EM.
The grids were prepared as described above for mass measurement except that after the washing step, they were negatively stained on 2 droplets of 2% uranyl acetate (pH 4.5), blotted, and air dried. The grids were examined in the STEM, and digital, dark-field images (512 by 512 pixels) were recorded at an accelerating voltage of 100 kV and a nominal magnification of ×500,000. Bright-field images of larger regions of the same grids were subsequently recorded on photographic film using an Hitachi 7000 TEM at an accelerating voltage of 100 kV and a magnification of ×50,000. The negatives were digitalized using a Primescan D7100 scanner at a sampling step size of 0.4 nm/pixel.
Projections were manually selected from the STEM images and examined; averaging procedures were not employed. Top- and side-view projections of the complexes were also manually selected from the TEM images and placed in galleries using the EMAN (36) and SEMPER image-processing packages (46). The galleries were analyzed individually. The side-view projections were angularly and translationally aligned to arbitrarily selected reference particles using the SEMPER programs. Projections with the specified correlation factor (≥0.3 and ≥0.5 for side views of the single and double complexes, respectively) to the reference were subsequently superimposed and averaged. The SPIDER image-processing package (17) was employed to align the top-view projections without the use of a reference. Repeated alignment cycles yielded several class averages. Their rotational power spectra were determined using the SEMPER programs.
Planar lipid bilayer experiments.
Planar lipid bilayer experiments were performed essentially as described previously (54) with 5 mM CaCl2 in the electrode buffer. Briefly, after formation of the lipid bilayer, 50 ng of YscC was added to the aqueous subphase. After a potential of 100 mV was applied over the membrane, insertions of the protein into the bilayer were monitored as stepwise increments of the current. To study the effect of calcium on channel activity, the electrolyte buffer was prepared without CaCl2. Voltage-ramp experiments were performed by applying an increasing potential from 0 to +200 or −200 mV over a time span of 100 s. Zero-current membrane potentials were determined after establishing a 10-fold KCl gradient (1 M in the cis cell and 100 mM in the trans cell) across the bilayer.
Uptake of chromogenic β-lactam antibiotics.
The pYV-cured Y. enterocolitica strain CE1525 carrying plasmid pSM3 (containing the yscC gene) with or without plasmid pRS6 (containing the yscW gene) were grown in BHI-OX or BHI-Ca2+, and expression of yscC was induced by the addition of IPTG as described above. Cells were washed and resuspended to an optical density at 600 nm of 0.5 in phosphate-buffered saline (45). After the addition of CENTA β-lactamase substrate (Merck) or nitrocefin (Calbiochem) to a final concentration of 50 μg/ml, the conversion of these substrates by periplasmic β-lactamase was monitored for 1 min by determining the optical density at 400 or 486 nm. Transport across the outer membrane is the rate-limiting step in the degradation of these antibiotics in this assay (53). As a control for possible leakage of periplasmic β-lactamase, the conversion of the β-lactam antibiotics in the supernatant of the cell suspension was measured.
RESULTS
Purification of the YscC oligomer under nondenaturing conditions.
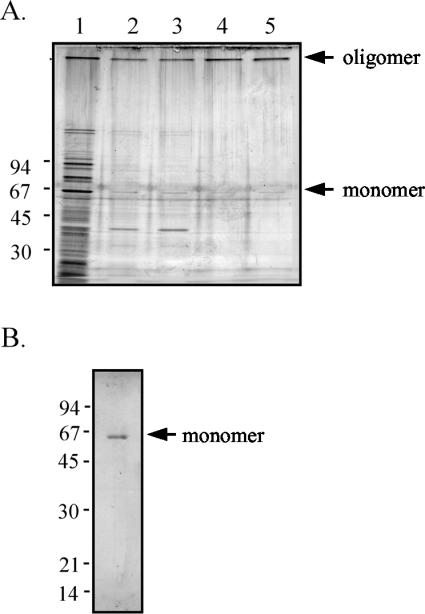
In the protocol for the purification of the YscC oligomer of Koster et al. (32), SDS was used for its extraction from cell envelopes of the Y. enterocolitica wild-type strain KNG22703. Although the oligomer did not dissociate, it is possible that the protein was partially denatured during the extraction. Therefore, a new purification method was developed in which the nonionic detergent Elugent was used to solubilize the cell membranes instead of SDS. A pYV-cured Y. enterocolitica strain (the virulence plasmid pYV carries the genes for all the components of the secretion machinery) was used to produce the YscC oligomer. Two plasmids, pSM3km encoding YscC and pRS6 encoding YscW, the pilot protein required for the production of high levels of YscC and proper localization of the YscC oligomer (5, 32), were introduced into the strain. Membrane proteins from this strain were solubilized in 3% Elugent, and YscC was purified using sucrose gradient centrifugation followed by ion-exchange chromatography (see Materials and Methods). A NaCl concentration of at least 100 mM had to be present in the purification buffers to prevent aggregation of the solubilized secretin. The final preparation contained almost exclusively the oligomeric form of YscC (Fig. 1A). Similar to what was reported previously (32), a small amount of the secretin was purified in its 64-kDa monomeric form (Fig. 1A), as confirmed by immunoblot analysis (data not shown). It is possible that unstable assembly intermediates of the YscC oligomer copurified during the procedure. Upon treatment with hot phenol, the oligomer dissociated into the constituent YscC monomers (Fig. 1B). No other proteins were detected, showing purified YscC to be homo-oligomeric. Immunoblot analysis confirmed that YscW fractionated differently than YscC on the sucrose gradient (data not shown). The yield of the purification procedure was approximately 0.25 mg/liter of culture, which was approximately 30% of the initial amount of YscC present in the preparation.
FIG. 1.
Purification of the YscC oligomer with the nonionic detergent Elugent. (A) Protein fractions of the different steps of the purification procedure were analyzed on a 3 to 15% polyacrylamide gradient gel and stained with silver. Samples were adjusted to contain equal amounts of YscC oligomer. Lane 1, protein fraction from whole cells (lane 1); lane 2, protein fraction from cell envelopes; lane 3, soluble protein fraction obtained after membrane extraction with Elugent; lane 4, protein fraction obtained after sucrose gradient centrifugation; lane 5, purified YscC oligomer after ion-exchange chromatography. (B) The purified YscC oligomer was dissociated with hot phenol, loaded on an 11% polyacrylamide gel, and visualized with Coomassie brilliant blue staining. The positions of the molecular mass markers (in kilodaltons) and of the YscC oligomer and monomer are indicated to the left and right of the gels, respectively.
Modification of the YscC oligomer by temperature and SDS.
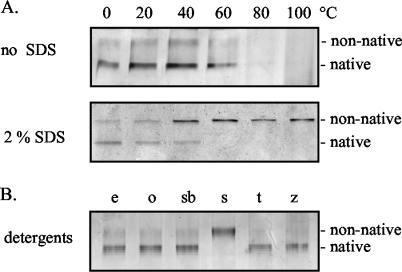
Interestingly, analysis of the purified secretin by seminative SDS-PAGE revealed two high-molecular-weight oligomers (Fig. 2, lane 1), which were both recognized by antibodies directed against YscC (data not shown) and thus represent two forms of the secretin oligomer. When boiled in the presence of 2% SDS, the more- abundant, faster-migrating form was completely converted into the slower-migrating form (Fig. 2, lane 2). This may be the result of SDS partially unfolding the polypeptides within the oligomer without dissociating it but thereby affecting its electrophoretic mobility. A small proportion of the purified YscC oligomer was always in the slower-migrating form after seminative SDS-PAGE (Fig. 2, lane 1) possibly due to the presence of 0.1% SDS in the sample buffer. The fast-migrating YscC oligomer was also the most prominent form in cell envelopes that were directly analyzed by seminative SDS-PAGE (Fig. 2, lane 3), indicating that this form represents the most native configuration of YscC. These results indicate that the YscC oligomer normally has a densely packed structure, which partially unfolds in the presence of SDS. Thus, even though SDS does not dissociate the YscC oligomer, it appears to strongly affect its structure.
FIG. 2.
Effects of heat and SDS on the purified YscC oligomer. The seminative 3 to 9% polyacrylamide gradient gel was loaded with purified YscC oligomer (lane 1), purified YscC oligomer that had been incubated for 10 min at 100°C in the presence of 2% SDS (lane 2), and cell envelopes from the pYV-cured Y. enterocolitica strain CE1525 carrying plasmids pSM3km and pRS6. Proteins were visualized by staining with silver. The positions of the native and partially denatured (nonnative) forms of the YscC oligomer are indicated to the right of the gel.
Stability of the native YscC oligomer.
The stability of the native form of the YscC oligomer was studied in more detail. In a buffer containing 0.1% Elugent, 100 mM NaCl, 0.1 mM EDTA, and 10 mM Tris-HCl (pH 7.8), the native form of the YscC oligomer remained stable during incubation for 10 min at temperatures up to 60°C (Fig. 3A). After incubation at 80 or 100°C, neither form of the oligomer was detected (Fig. 3A), and there was no concomitant increase in the amount of monomeric YscC observed (data not shown). Therefore, heat modification of the oligomer in this buffer most likely caused aggregation. When the YscC oligomer was incubated in the presence of 2% SDS, conversion of the native YscC oligomer into the slow-migrating form was already observed at temperatures above 20°C, and this conversion was detected at all higher temperatures tested (Fig. 3A). Hence, the presence of SDS promotes partial denaturation at lower temperatures and prevents aggregation of the oligomer at higher temperatures.
FIG. 3.
Stability of the native YscC oligomer. (A) Elugent-solubilized YscC oligomer was incubated in the absence or presence of 2.0% SDS for 10 min at the indicated temperatures. (B) Elugent-solubilized YscC oligomer was incubated in the presence of 2% Elugent (e), OPOE (o), SB12 (sb), SDS (s), Triton X-100 (t), or Zwittergent 3-14 (z) for 1 h at 40°C. All samples were loaded on seminative 3 to 9% polyacrylamide gradient gels, subjected to electrophoresis, and stained with silver. The positions of the native and nonnative forms of the YscC oligomer are indicated to the right of the gels.
The stability of the native YscC oligomer was also determined in other detergents that are often used for the extraction of cell envelope proteins. The native form of the oligomer was still present after incubation for 1 h at 40°C in 2% Elugent, OPOE, SB12, Triton X-100, or Zwittergent 3-14 (Fig. 3B).
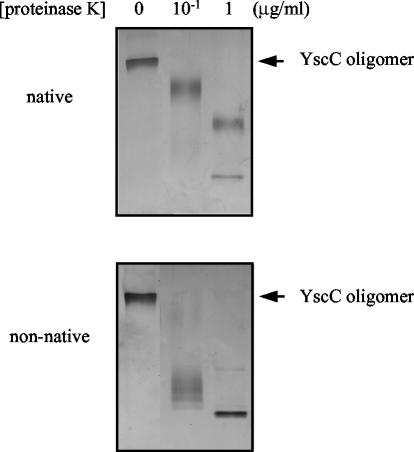
The susceptibility of the two forms to proteolytic degradation was studied by incubating the purified YscC oligomer with 2% SDS either at room temperature or at 100°C to obtain the native or nonnative form, respectively, followed by treatment with proteinase K. The nonnative form showed increased sensitivity to the protease compared to the native form (Fig. 4), which substantiates the notion that SDS induces partial unfolding of the YscC oligomer at high temperatures.
FIG. 4.
Protease susceptibility of the purified YscC oligomer. Purified YscC oligomers were incubated with 2% SDS for 10 min at room temperature for analysis of the native form or at 100°C for analysis of the nonnative form. The oligomers were then incubated with the indicated concentrations of proteinase K for 10 min at room temperature. Samples were loaded on a 3 to 9% polyacrylamide gradient gel. The position of the YscC oligomer to indicated on the right of the gels. Proteins were visualized by staining with silver.
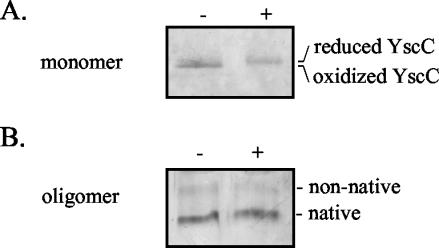
The nonconserved C-terminal domain of YscC contains four cysteine residues that form at least one intramolecular disulfide bond (29). To investigate whether this bond helps to stabilize the native oligomeric form of YscC, the purified oligomer was incubated in the presence of 20 mM DTT for 10 min at 40°C. The minor shift in electrophoretic mobility of the monomers obtained after dissociation of these oligomers with TCA and TFA indicates that at least one disulfide bond was broken by the DTT treatment (Fig. 5A). In its reduced state, the oligomeric secretin was still in its native form after incubation at 40°C (Fig. 5B), which implies that the disulfide bond is not essential to retain the YscC oligomer in this conformation.
FIG. 5.
Role of disulfide bonds in the stability of the SDS-sensitive YscC oligomer. Purified YscC oligomer was incubated in the absence (−) or presence (+) of 20 mM DTT for 10 min at 40°C and was either dissociated into monomers by treatment with TCA and TFA and loaded on a 8% polyacrylamide gel (A) or directly loaded on a seminative 3 to 9% polyacrylamide gradient gel (B). The positions of the reduced and oxidized forms of the YscC monomer and the native and nonnative forms of the YscC oligomer are indicated to the right of the gels. Proteins were visualized by staining with silver.
Mass analysis of the purified YscC oligomers.
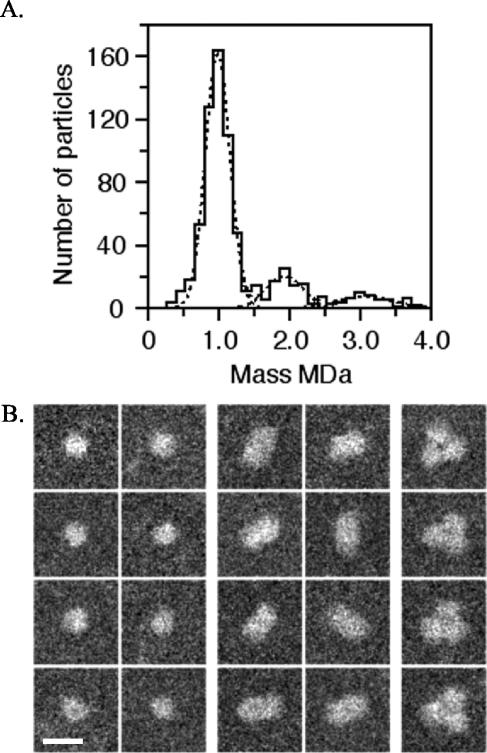
The purified YscC oligomers were adsorbed to thin carbon films and imaged by STEM for mass measurements. Evaluation of the images yielded a histogram displaying a major mass peak and two minor mass peaks (Fig. 6A) and a few higher mass values (data not shown). More than 75% of all the particles had a mass of 985 ± 175 kDa (n = 546; standard error [SE] of ±7 kDa), the overall uncertainty being ±50 kDa considering the SE of the measurement and the 5% calibration uncertainty of the instrument. As shown in the left gallery of Fig. 6B, these particles generally had circular projections ∼14 nm in diameter. The following smaller peak on the histogram indicates a mass of 1,928 ± 254 kDa (n = 97; SE of ±26 kDa) and arises from the association of two oligomers. The corresponding projections were typically both elongated and of uniform mass density, implying that the oligomers were viewed side on (Fig. 6B, center gallery). The third peak indicates a mass of 3,079 ± 374 kDa (n = 53; SE of ±51 kDa) and arises from the association of three oligomers to form aggregates with distinctly triangular projections (Fig. 6B, right gallery).
FIG. 6.
STEM mass analysis of unstained, Elugent-purified YscC oligomers. (A) Histogram of the mass values. The different populations have masses of 985 ± 175 kDa (n = 546), 1,928 ± 254 kDa (n = 97), and 3,079 ± 374 kDa (n = 53), indicating the presence of one, two, and three YscC oligomers, respectively. Correction was made for beam-induced mass loss using the factor 1.029 (see Materials and Methods). (B) Galleries showing particles with masses in the range of the first, second, and third mass peaks (left, middle, and right galleries, respectively). Protein is displayed in white. Bar, 25 nm.
Purely on the basis of the mass of 64.25 kDa predicted from the sequence of the mature YscC monomer, particles in the main population yielding the peak at 985 kDa are composed of 15 ± 1 YscC monomers. However, as YscC is a membrane protein, detergent will also be present, making this number the upper limit.
Negative stain EM.
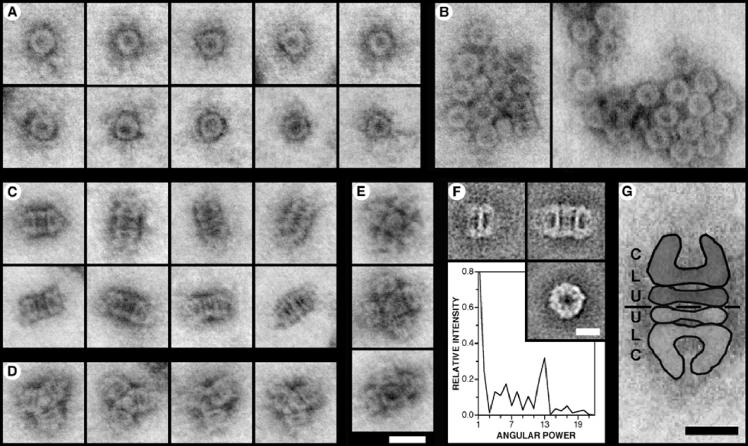
Negatively stained samples of the purified YscC oligomers were first examined by STEM. Most projections were ring-shaped, top views of the particles, which from the mass measurement results and image galleries are single oligomers (Fig. 6). The projections have outer and inner diameters of ∼14 and ∼4.5 nm, respectively (Fig. 7A). There was a considerable tendency to aggregate; large rosette-shaped aggregates were frequently detected (Fig. 7B). Various side views of the oligomer were also observed, and again there was a high tendency for aggregation. The overall shape of these projections was rectangular (Fig. 7C), star-like (Fig. 7D), or square (Fig. 7E). The rectangular structures have a total length of ∼23.5 nm, whereas the star-like and square projections are composed of three or four ∼12-nm-long units, respectively. In accordance with the mass measurements (Fig. 6), these dimensions suggest that the rectangular projections arise from dimers formed by head-to-head or tail-to-tail interactions and that the star-like structures are trimers. Further, they imply that the single YscC oligomer is approximately 12 nm long and 14 nm wide. The square projections may arise from both tetramers and pentamers, as a central density with dimensions consistent with a top view of the oligomer was sometimes observed (Fig. 7E).
FIG. 7.
STEM analysis of Elugent-purified YscC oligomers. (A to E) STEM images of YscC oligomers that have been negatively stained with uranyl acetate. The contrast of the dark-field images has been inverted to show protein as gray. (A) Ring-like top views. (B) Rosette-shaped aggregates showing predominantly top views of the oligomer. (C) Rectangular side views that correspond to dimers of the oligomer. The thick band at the center of the oligomer resolves into two in some projections. The outer ends of the oligomer display various orientations. (D) Star-shaped side views of trimeric aggregates. (E) Essentially square side views formed by the association of four or five oligomers. The micrographs in panels A to E are all shown at the same magnification. Bar, 20 nm. (F) TEM analysis. (Top left) Average side-view projection of a single oligomer (n = 8 of 16); (top right) average side-view projection of two oligomers associated end-to-end (n = 16 of 16); (bottom right) average top view projection (n = 58 of 81). Bar, 10 nm. The graph shows the angular power spectrum of the top-view average, illustrating the presence of a strong 13-fold angular harmonic. (G) Interpretation of the rectangular side views demonstrated using a STEM image. The protein structure is highlighted, and a discontinuous black line indicates the interface between the two oligomers. The different domains within the structure, i.e., the conical domain (C), the lower ring (L), and the upper ring (U), are labeled. Bar, 10 nm.
A few side views of the single oligomers (n = 16) and further rectangular side views from dimers (n = 16) were detected when large areas of the grids were scanned by TEM. The average dimensions of the projections (Fig. 7F, top micrographs) are 10 to 11 and 20 to 23.5 nm long, respectively, and ∼14 nm wide. These dimensions agree well with the STEM results and confirm the proposed assignments. Both the single oligomer and dimer side views reveal internal stain-filled regions up to 6 nm in cross-section. The reference-free alignment and classification of 81 top-view projections selected from the TEM images yielded the average projection shown in Fig. 7F (bottom right micrograph). The average projection was calculated from 58 projections, i.e., 72% of those selected, and has outer and inner diameters of ∼14 and ∼4 nm, respectively. Further, the calculated angular power spectrum displays a strong 13-fold angular harmonic (Fig. 7F, graph).
Both the individual STEM and the average TEM side-view projections reveal that the oligomeric secretin is composed of three domains as indicated in the cartoon (Fig. 7G): (i) a conical domain that shows some variation in form as illustrated in Fig. 7C, (ii) a lower ring), and (iii) a less prominent upper ring that is located at the interface of the dimer.
Channel formation of YscC in lipid bilayers.
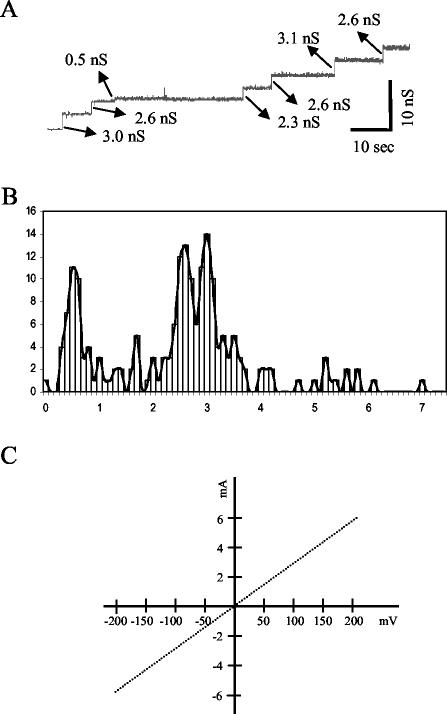
The pore-forming activity of the purified secretin was investigated in planar lipid bilayer experiments. Channel activity with conductance steps of three different size classes were detected at a potential of 100 mV (Fig. 8A and B). Interestingly, only openings and no closings were recorded. Two types of openings, having conductance increments of approximately 2.6 and 3.0 nS, were only slightly different from each other. The third type of opening had a conductance increment of approximately 0.5 nS and was often recorded after a 2.6-nS transition. This result suggests that the YscC oligomer has two conductance states and can undergo a conformational change, leading to the transition from the lower to higher conductance level. To determine whether this conformational change is similar to the one induced by SDS treatment, the purified YscC oligomer was incubated for 10 min in the presence of 2% SDS at 95°C prior to its addition to the aqueous subphase of the planar lipid bilayer system. The SDS-treated oligomer did not show any channel activity, suggesting that either the channel is blocked upon denaturation or the SDS-treated oligomer is unable to insert into the phospholipid bilayer.
FIG. 8.
Pore activity of the YscC oligomer. (A) Channel recording of multiple insertions of the purified YscC oligomer in a planar lipid bilayer at a membrane potential of 100 mV. The sizes of the transitions are indicated. (B) Amplitude histogram of channel openings (n = 204) at 100 mV. (C) Voltage-ramp analysis of multiple YscC channels from 0 to 200 mV and 0 to −200 mV over a total time span of 200 s.
Voltage-ramp recordings of multiple channels showed that the conductance of the pores was constant over the entire voltage range tested, i.e., from 0 to 200 mV, and was identical at positive and negative potentials (Fig. 8C). A zero-current transmembrane potential of 39 ± 3 mV was measured after the application of a 10-fold KCl gradient across the membrane (the trans compartment more diluted), indicating that the channels are anion selective. Application of the Goldman-Hodgkin-Katz equation (26) revealed an eightfold preference for chloride over potassium.
The planar lipid bilayer experiments were routinely conducted in a buffer containing 5 mM CaCl2. In vivo, calcium depletion of the growth medium induces type III secretion in Y. enterocolitica. To test the possibility that calcium directly influences the structure of the YscC secretin, its effect on the conductance of secretin in planar lipid bilayers was determined. No differences in channel behavior were observed when the experiments were performed in the presence or absence of calcium (data not shown).
In vivo pore activity.
The uptake of chromogenic β-lactam antibiotics by intact cells was used to study the pore-forming activity of the YscC oligomer in vivo. In this assay, the diffusion of the antibiotics across the outer membrane is the rate-limiting step in their degradation by periplasmic β-lactamase. Simultaneous expression of yscC and yscW from pSM3 and pRS6, respectively, in the pYV-cured Y. enterocolitica strain CE1525 did not increase the rate of degradation of the antibiotics (data not shown). Also, when these cells were grown in calcium-depleted medium, the rate of degradation of the antibiotics was not affected (data not shown). Apparently, the permeability of the outer membrane did not change upon yscC expression, suggesting that the YscC oligomer was closed. Hence, in vivo, the secretion channel formed by YscC remains closed in the absence of the other components of the type III secretion system.
DISCUSSION
Here, we report structural and functional data for the YscC secretin of the type III protein secretion system of Y. enterocolitica. The use of a nonionic detergent for solubilization allowed the YscC secretin to be purified in its oligomeric form. Analysis of the oligomer by seminative SDS-PAGE always revealed two bands (Fig. 2), both recognized by YscC antibodies. The faster-migrating form was completely converted into the slower-migrating one upon incubation in SDS at temperatures above 20°C. In the absence of SDS, the faster-migrating form was stable at temperatures up to 60°C. The drastic shift in electrophoretic mobility may be explained by the denaturation of a normally tightly folded YscC domain. This finding that unfolding affects the electrophoretic behavior has been observed for other outer membrane proteins (25) and is often used to monitor the refolding of outer membrane proteins in vitro (15). Moreover, the shift in mobility was accompanied by an increase in protease sensitivity, which is also indicative of a change in the folding state of the YscC oligomer. Thus, although the oligomer does not dissociate in SDS, its structure appears to be drastically changed. Importantly, the SDS sensitivity of the YscC oligomer allowed us to monitor the conservation of the folded state during the purification procedure. Since the highly conserved C-terminal halves of secretins are involved in oligomerization, it is probably the N-terminal part of YscC that unfolds in response to heat and SDS. We did not detect a similar SDS-induced change in the electrophoretic mobility of the P. aeruginosa XcpQ and the N. meningitidis PilQ secretins that are involved in type II secretion and type IV pili assembly, respectively (unpublished observations), which could indicate that the N termini of these secretins already denature under the mild conditions of seminative SDS-PAGE.
As documented by Fig. 1B, the purified secretin is homo-oligomeric, being formed from YscC alone. Interestingly, in contrast to the pilot protein PulS, which copurifies with the secretin PulD (42), the pilot protein YscW did not copurify with YscC and is therefore not a stable constituent of the oligomeric structure.
A mass of 600 kDa was reported earlier for the YscC complex (32). However, in that case the size of the oligomer was estimated by SDS-PAGE, where it migrated well above the highest molecular mass marker protein, preventing the accurate determination of its size. The mass of the Elugent-purified, YscC oligomers determined by STEM was significantly higher, 985 kDa (Fig. 6). Since YscC is a membrane protein, this value will include a contribution from the detergent, making the indicated number of monomers (15 ± 1) an overestimate. The amount of detergent retained by a membrane protein depends on both the size of the latter's hydrophobic domain and the detergent used. STEM mass measurements made on approximately 250-kDa photosystem II complexes solubilized in the nonionic detergent n-dodecyl-β-maltoside indicated the association of one 50-kDa detergent micelle (24). Since the detergent mixture used, Elugent, is of the same type, several micelles are probably required to solubilize the much larger YscC oligomer. Accordingly, the latter is most likely composed of 13 or 14, rather than 15, YscC monomers.
As confirmed by mass measurements of unstained samples, the single YscC oligomers adsorbed preferentially with their long axis perpendicular to the air-glow-discharged carbon microscopy film, resulting in circular (unstained samples; Fig. 6) or ring-shaped (negatively stained samples; Fig. 7C and F, bottom right micrograph) projections. Subsequent analysis of the latter recorded from negatively stained samples by TEM yielded an average with strong 13-fold angular order (Fig. 7F, graph). In agreement with the mass measured, this implies the presence of 13 YscC monomers in the oligomer. While the presence of 14 subunits has been reported for the pIV secretin oligomer (37, 44), a dodecameric structure has been proposed for the PilQ (7) and PulD (42) secretins. Thus, despite their homology, the number of subunits in the oligomer seems to differ between members of the family of secretins.
The top views of negatively stained, Elugent-purified YscC oligomers recorded by STEM (Fig. 7A) and the TEM projection average (Fig. 7F, bottom right micrograph) show the secretin to have a stain-accessible central cavity and an outer diameter of ∼14 nm. The latter is considerably smaller than the 20 nm reported earlier for the SDS-purified YscC oligomer (32). This suggests that the presence of SDS resulted in conformational rearrangements at the outer rim of the oligomer, which is in agreement with the observed effect of SDS on the electrophoretic mobility. From the average side-view projection of the single oligomer (Fig. 7F) and the side-view projections of two (Fig. 7C and F) or three (Fig. 7D) associated YscC oligomers, the secretin has a total length of 11 to 12 nm and is composed of two stacked rings and a conical domain (Fig. 7G) that can be in a more or less open state as illustrated in Fig. 7C. Similar conical domains have been found at the ends of other secretin dimers (7, 35, 43).
Secretins of type III systems, such as YscC, are part of a much larger structure, the needle complex, which comprises an external needle and a basal body that traverses both cell membranes and the peptidoglycan layer (14, 33, 48, 51). The latter domain is formed by two upper and two lower stacked rings linked by a rod-like structure, which seems to protrude through the upper rings to form the needle-like extension. The two stacked rings identified in the side views of the purified YscC secretin (Fig. 7C and D; annotated U and L in Fig. 7G) would appear to correspond to the upper rings, which, depending on the bacterial host, have an outer diameter of 15 to 20 nm and a length of approximately 10 nm. This assignment is strengthened by the observation that without their needle-like extensions, the basal bodies of isolated secretion complexes aggregate through their upper rings (33, 51), just as the purified YscC oligomer tends to aggregate. On the basis of two-dimensional averages of Shigella needle complexes (3), the conical domain sometimes visible in the side views of the purified oligomer (Fig. 7C) probably corresponds to the region between the two upper and two lower rings of the basal body.
The purified YscC oligomer inserted into planar lipid bilayers to form stable high-conductance channels. These channels had two conductance levels, 2.6 and 3.0 nS. Conductance increments of about 0.5 nS were often recorded after a 2.6-nS transition, indicating that the 2.6-nS channels can convert into the 3.0-nS form. It remains unknown whether this conversion is related to the partial unfolding of the oligomer, since the inability of SDS-treated oligomers to give channel activity could be the result of a failure to insert into the bilayer. The channel activity of YscC has several unique characteristics compared to those of three other secretins, XcpQ, pIV, and PulD. First, the conductance was very regular, and no closings, only openings, were observed. Other secretins, especially XcpQ (4), display large variations in their conductance level. Second, the current passing through the YscC channel increased linearly as the transmembrane potential was increased. In contrast, the channel activity of the other secretins is nonlinear at higher potentials, which suggests that these pores change in a voltage-dependent manner (37, 42). Third, the direction of the potential field had no influence on the conductance of the YscC channel, while the other secretins show an asymmetrical response (4, 37). The different electrophysiological characteristics of the YscC channel compared to those of the other secretins may be a consequence of YscC being part of the type III protein secretion machinery, which is a stable structure. Once incorporated into the needle complex, the secretin may remain continuously open, gating occurring at a different level within the machinery. In other systems, such as type II secretion, the secreton might be a more dynamic structure, necessitating the secretin to close when not actively involved in secretion of substrates.
In vivo, the expression of YscC alone did not lead to increased permeability of the outer membrane, suggesting that the channel was closed. This result is not consistent with the electrophysiological data discussed above. One explanation might be that the conformation of the YscC oligomer is slightly altered during purification. Alternatively, interactions with components of the cell envelope, such as lipopolysaccharide or peptidoglycan, might affect the structure of the secretin in vivo. However, the channel of the purified YscC oligomer does not seem to have been in a completely open conformation in the lipid bilayer either. The theoretical pore diameter can be derived from the channel activity in planar lipid bilayers, assuming that the channel is an ideal cylinder (26). Since the overall length of the YscC oligomer is ∼12 nm (see above) and the bulk conductance of the electrolyte is 19.6 S/m, the YscC channel with a conductance of 3.0 nS would have a pore size of 1.6 nm. This is much smaller than the ∼4-nm channel diameter indicated by electron microscopy (Fig. 7F, bottom right micrograph). However, it is clear from both the individual and average side views (Fig. 7C and F, top micrographs) that the secretin does not form an ideal cylinder and that the conical domain restricts the channel diameter. Furthermore, protein structures in the cavity of the oligomer might be covered by the negative stain and therefore remain undetected. Studies performed on the purified needle complexes of Salmonella and Shigella suggest that the needle traverses the secretin (3, 33, 51). In order to accommodate the needle, the diameter of the YscC channel should be 6 to 7 nm (27), which is larger than both the electrophysiological and STEM data suggest. Therefore, possibly, the YscC oligomer adopts its final conformation only upon contact with another component of the system, which could be the needle itself. Our attempts to open or widen the pore in vivo and in vitro by calcium depletion were unsuccessful.
Several secretins have been studied so far, and they share many characteristics. However, there are also important differences in the number of subunits forming the oligomer, protein folding, oligomer stability, and electrophysiological behavior of the channel formed. These differences probably reflect the role of the secretin in the various unrelated systems. This makes the study of secretins from different systems essential to obtain insight in the structure and function of this group of important and seemingly versatile transporters.
Acknowledgments
We thank Vesna Olivieri for STEM, Marco Gregorini for help with the TEM image processing, and Andreas Engel for many helpful discussions and critical assessment of the manuscript. We thank R. Diaz-Avalos of the Institute of Molecular Biophysics at Florida State University for kindly providing the TMV particles.
This research was supported in part by grant 700-97-012 from the Council for Chemical Sciences of The Netherlands Organization of Scientific Research (CW-NWO), grant HPRN-CT-2000-00075 from the European Community, grant NF 31-59 415.99 from the Swiss National Foundation, and the Maurice E. Müller Foundation of Switzerland.
REFERENCES
- 1.Allaoui, A., R. Scheen, C. Lambert de Rouvroit, and G. R. Cornelis. 1995. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J. Bacteriol. 177:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 4.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 5.Burghout, P., F. Beckers, E. de Wit, R. van Boxtel, G. R. Cornelis, J. Tommassen, and M. Koster. Role of the pilot protein YscW in the biogenesis of the secretin YscC in Yersinia enterocolitica. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 6.China, B., T. Michiels, and G. R. Cornelis. 1990. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol. Microbiol. 4:1585-1593. [DOI] [PubMed] [Google Scholar]
- 7.Collins, R. F., L. Davidsen, J. P. Derrick, R. C. Ford, and T. Tonjum. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, R. F., R. C. Ford, A. Kitmitto, R. O. Olsen, T. Tonjum, and J. P. Derrick. 2003. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185:2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 10.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47-56. [DOI] [PubMed] [Google Scholar]
- 11.Daefler, S., L. Guilvout, K. R. Hardie, A. P. Pugsley, and M. Russel. 1997. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol. Microbiol. 24:465-475. [DOI] [PubMed] [Google Scholar]
- 12.Daefler, S., M. Russel, and P. Model. 1997. Module swaps between related translocator proteins pIV(f1), pIV(IKe) and PulD: identification of a specificity domain. J. Mol. Biol. 266:978-992. [DOI] [PubMed] [Google Scholar]
- 13.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28:1367-1380. [DOI] [PubMed] [Google Scholar]
- 14.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865-871. [DOI] [PubMed] [Google Scholar]
- 15.De Jonge, M. I., M. P. Bos, H. J. Hamstra, W. Jiskoot, P. van Ulsen, J. Tommassen, L. van Alphen, and P. van der Ley. 2002. Conformational analysis of opacity proteins from Neisseria meningitidis. Eur. J. Biochem. 269:5215-5223. [DOI] [PubMed] [Google Scholar]
- 16.Drake, S. L., S. A. Sandstedt, and M. Koomey. 1997. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23:657-668. [DOI] [PubMed] [Google Scholar]
- 17.Frank, J., B. Shimkin, and H. Dowse. 1981. SPIDER: a modular software system for electron image processing. Ultramicroscopy 6:343-358. [Google Scholar]
- 18.Frenken, L. G., A. de Groot, J. Tommassen, and C. T. Verrips. 1993. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol. Microbiol. 9:591-599. [DOI] [PubMed] [Google Scholar]
- 19.Genin, S., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243:112-118. [DOI] [PubMed] [Google Scholar]
- 20.Guilvout, I., K. R. Hardie, N. Sauvonnet, and A. P. Pugsley. 1999. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 181:7212-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, R. E., and H. Nikaido. 1978. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 24.Hasler, L., D. Ghanotakis, B. Fedtke, A. Spyridaki, M. Miller, S. A. Müller, A. Engel, and G. Tsiotis. 1997. Structural analysis of photosystem II: comparative study of cyanobacterial and higher plant photosystem II complexes. J. Struct. Biol. 119:273-283. [DOI] [PubMed] [Google Scholar]
- 25.Heller, K. B. 1978. Apparent molecular weights of a heat-modifiable protein from the outer membrane of Escherichia coli in gels with different acrylamide concentrations. J. Bacteriol. 134:1181-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hille, B. 1992. Ionic channels of excitable membranes. Sinauer, Sunderland, Mass.
- 27.Hoiczyk, E., and G. Blobel. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. USA 98:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson, M. W., and G. V. Plano. 1999. DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 181:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 31.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 32.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 33.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Linderoth, N. A., M. N. Simon, and M. Russel. 1997. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science 278:1635-1638. [DOI] [PubMed] [Google Scholar]
- 36.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 37.Marciano, D. K., M. Russel, and S. M. Simon. 1999. An aqueous channel for filamentous phage export. Science 284:1516-1519. [DOI] [PubMed] [Google Scholar]
- 38.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 39.Müller, S. A., K. N. Goldie, R. Bürki, R. Häring, and A. Engel. 1992. Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy 46:317-334. [Google Scholar]
- 40.Müller, S. A., and A. Engel. 2001. Structure and mass analysis by scanning transmission electron microscopy. Micron 32:21-31. [DOI] [PubMed] [Google Scholar]
- 41.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opalka, N., R. Beckmann, N. Boisset, M. N. Simon, M. Russel, and S. A. Darst. 2003. Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J. Mol. Biol. 325:461-470. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Saxton, W. O., J. T. Pitt, and M. Horner. 1979. The SEMPER image processing system. Ultramicroscopy 4:343-354. [Google Scholar]
- 47.Schuch, R., and A. T. Maurelli. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183:6991-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, R., V. Priefer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 51.Tamano, K., S. I. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tommassen, J., H. van Tol, and B. Lugtenberg. 1983. The ultimate localization of an outer membrane protein of Escherichia coli K-12 is not determined by the signal sequence. EMBO J. 2:1275-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Gelder, P., N. Saint, R. van Boxtel, J. P. Rosenbusch, and J. Tommassen. 1997. Pore functioning of outer membrane protein PhoE of Escherichia coli: mutagenesis of the constriction loop L3. Protein. Eng. 10:699-706. [DOI] [PubMed] [Google Scholar]
- 54.Van Gelder, P., F. Dumas, J. P. Rosenbusch, and M. Winterhalter. 2000. Oriented channels reveal asymmetric energy barriers for sugar translocation through maltoporin of Escherichia coli. Eur. J. Biochem. 267:79-84. [DOI] [PubMed] [Google Scholar]
- 55.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]