Significance
Gene therapy has made significant strides in the treatment and even cure of single-gene defects. However, the maturation of this field will require more sophisticated vehicles capable of cell-selective delivery of large genetic payloads whose regulated expression will restore or enhance cellular functionality. High-capacity herpes simplex virus vectors have the potential to meet these challenges but have been limited by the need to preserve one particularly cytotoxic viral product, infected cell polypeptide 0, to maintain transgene transcriptional activity. Our study describes a vector design that solves this conundrum, thereby promising the near-term availability of viral vectors that can efficiently deliver large or multiple regulated transgenes to a diversity of cells without attendant cytotoxicity.
Keywords: HSV vector, gene therapy, ICP0, insulator, dystrophin
Abstract
The design of highly defective herpes simplex virus (HSV) vectors for transgene expression in nonneuronal cells in the absence of toxic viral-gene activity has been elusive. Here, we report that elements of the latency locus protect a nonviral promoter against silencing in primary human cells in the absence of any viral-gene expression. We identified a CTCF motif cluster 5′ to the latency promoter and a known long-term regulatory region as important elements for vigorous transgene expression from a vector that is functionally deleted for all five immediate-early genes and the 15-kb internal repeat region. We inserted a 16.5-kb expression cassette for full-length mouse dystrophin and report robust and durable expression in dystrophin-deficient muscle cells in vitro. Given the broad cell tropism of HSV, our design provides a nontoxic vector that can accommodate large transgene constructs for transduction of a wide variety of cells without vector integration, thereby filling an important void in the current arsenal of gene-therapy vectors.
One of the limitations of promising viral-gene transfer vectors, including lentivirus, adeno-associated virus (AAV), and adenovirus vectors, is their small genome-packaging capacity (≤8 kb) (1), which precludes the incorporation of large or multiple therapeutic transgene cassettes (2–4). The large, double-stranded DNA genome of herpes simplex virus (HSV; ∼152 kb) contains a substantial number of genes that are not required for infection, and redundant genes located in the repeats flanking the unique long (UL) and short (US) segments of the viral genome represent additional regions that can be deleted without significant loss of infectivity. Although HSV is a neurotropic virus, inasmuch as it establishes natural latency in neurons, it is capable of infecting a wide range of cells with high efficiency, suggesting its potential for broad use as a gene-therapy vector. The goal of this research was to create a vector capable of long-term persistence and extended transgene expression in nondividing, nonneuronal cells without toxicity for the transduced cell or potential rescue of replicating virus.
HSV-1 initiates a cascade of viral-gene expression that begins with the expression of five immediate early (IE or α) genes. Two of these genes, infected cell polypeptide (ICP) 4 and ICP27, are essential for entry into the lytic-virus replication cycle, and HSV gene-delivery vectors are therefore typically deleted for one or both of these genes to mimic the latent state in sensory neurons without the possibility of reactivation. In addition, the expression of toxic nonessential IE genes, ICP0 and ICP22, has been down-regulated by promoter manipulation or blocked by deletion (5–11). The complete elimination of vector toxicity for nonneuronal cells achieved by silencing all four of these IE genes results in quiescent intranuclear episomal genomes that are diluted over time in dividing cells but persist in nondividing cells (12–14). However, transgene expression is also rapidly extinguished. Quiescence is due to global epigenetic silencing of the viral genome and can be reversed by ectopic expression of ICP0 (15, 16). To allow foreign gene expression without restoring ICP0, the transgene must be selectively protected against epigenetic silencing.
Viruses that are deleted for the ICP4 and ICP27 genes can be grown on complementing cells engineered to produce the cognate essential gene products upon virus infection (7). However, robust amplification of viruses that are in addition deleted for the ICP0 gene requires further complementation. Stable maintenance of cell lines that inducibly express all three IE genes has been a challenge due to leaky expression of the highly toxic ICP0 protein (10).
In the present study, we used HSV–bacterial artificial chromosome (BAC) recombineering to reconstruct a previous replication-defective vector that expressed a low level of ICP0, was capable of robust transient transgene expression, and displayed minimal disturbance of mouse embryonic stem cell developmental gene expression (17). In agreement with studies by others (10, 13, 18), we found that complete elimination of ICP0 expression was required to limit vector toxicity for human fibroblasts and that both viral-gene expression and transgene expression were essentially abolished in this situation. To facilitate consistent vector growth, we constructed a cell line that stably and efficiently complements the multiple IE gene defects in this type of vector. We show that a heterologous promoter–reporter gene expression cassette remains transcriptionally active in human dermal fibroblasts (HDFs) when located in the latency-associated transcript (LAT) locus but is silenced on deletion of a neighboring element rich in CTCF-binding motifs, designated CTRL [CCCTC binding protein (CTCF) repeat long (CTRL)] (19), and the enhancer-like latency active promoter 2 (LAP2 or LATP2) region. Reporter-gene expression was also observed when the transgene cassette with LAT elements was moved to other positions in a silent-vector genome, but omission of the LAT elements abolished expression. We present evidence that these results are not unique to HDFs but also apply to other nonneuronal primary human cells. Lastly, we show that residual cytotoxicity observed with these vectors is eliminated by deletion of the BAC region, and we illustrate the potential of our findings by documenting vector-mediated expression of full-length dystrophin through at least 7 d. Together, these results hold the promise of significantly expanding the utility of HSV gene-transfer vectors to include a wide range of nonneuronal cells.
Results
Vector Engineering and Virus Growth.
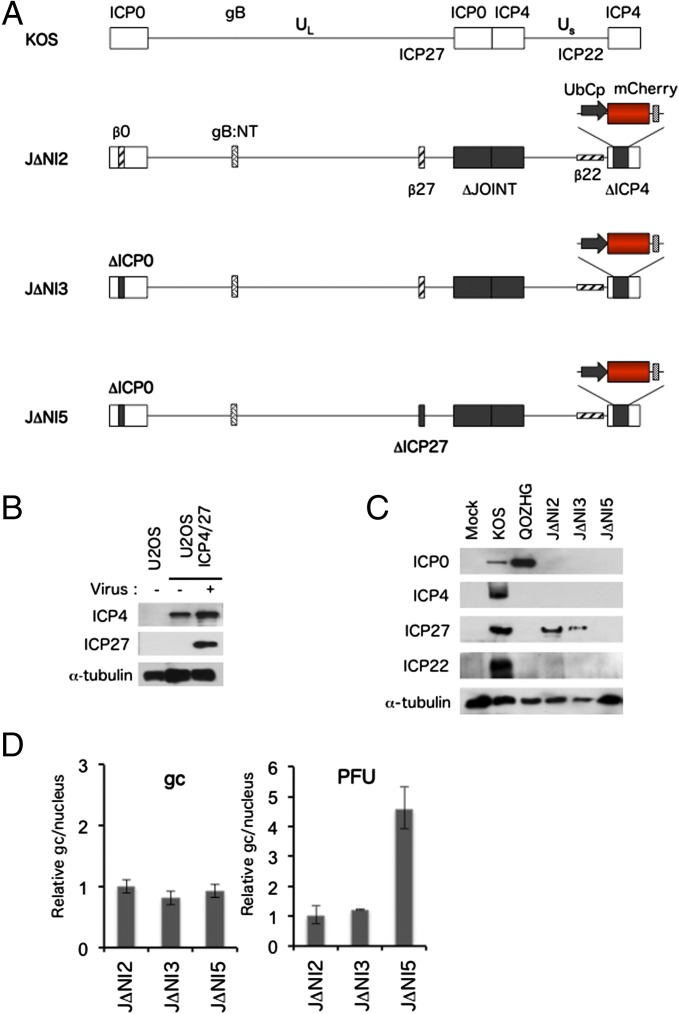
We used Red-mediated recombineering in bacteria (20) to generate an HSV–BAC construct, J∆NI5, that was deleted for the ICP0, ICP4, and ICP27 IE genes, the promoter and translation initiation codon of the ICP47 IE gene as part of a deletion that included the entire internal repeat (joint) region between the two unique segments (UL, US) of the viral genome, and the consensus VP16-binding (TAATGARAT) motifs in the ICP22 IE gene regulatory region to change the expression kinetics of this gene to that of an early (E or β) gene (Fig. 1A). To convert this BAC construct to infectious virus, we created a novel U2OS-based cell line, U2OS-ICP4/27, that permanently expresses ICP4 and produces ICP27 in response to infection with an ICP4/ICP27-deleted virus (Fig. 1B); U2OS cells naturally complement ICP0 (21), eliminating the need to express this toxic protein. We performed a series of experiments to confirm the anticipated characteristics of J∆NI5 in comparison with two control viruses, J∆NI2 and J∆NI3, that had been produced as less debilitated predecessors of J∆NI5. J∆NI2 contained the intact coding regions of ICP0 and ICP27 under the control of copies of the early HSV thymidine kinase (TK) promoter (β0/β27) whereas, in J∆NI3, the ICP0 gene was deleted (Fig. 1A). In all three J∆NI viruses, an mCherry reporter-gene expression cassette was present at the position of the deleted ICP4 locus in the US terminal repeat, and the glycoprotein B gene was replaced with a hyperactive allele, gB:N/T (22), to enhance virus entry into cells. The genotypes of these virus constructs and all other HSV vectors generated in this study are summarized in Table 1. The biological titers in plaque-forming units (pfu) per milliliter and physical titers in viral genome copies (gc)/mL of the virus stocks used in this work are listed in Table 2.
Fig. 1.
Vector genome structures and complementing cells for virus production. (A) Schematic representations of the genomes of WT HSV-1 KOS, as present in KOS-37 BAC (57), and derivatives J∆NI2, J∆NI3, and J∆NI5. UL, unique long segment; US, unique short segment. The US region in KOS-37 BAC and its derivatives is inverted compared with the standard representation of the HSV genome. Open boxes, terminal and internal inverted repeats. Black boxes, deletions (∆); the ICP47 promoter and translation initiation codon are removed as part of the joint deletion. Cross-hatched horizontal box, the ICP22 IE gene converted to early-expression kinetics by promoter TAATGARAT deletion (β22). All J∆NI recombinants described in this study contain the hyper-activating N/T mutations in the gB gene (gB:NT) (22) and a ubiquitin C promoter (UbCp)-mCherry cassette with the SV40 polyA region in the ICP4 locus. The BAC elements, including a chloramphenicol-resistance gene and β-galactosidase expression cassette, are located between loxP sites in the UL37-UL38 intergenic region (57). (B) Western blot analysis of U2OS-ICP4/27 cells. Uninfected cells and cells infected with J∆NI5 virus at an MOI of 1 were harvested at 24 hpi, and extracts were prepared for gel electrophoresis; extracts from uninfected U2OS cells were used as control. Blots were probed with antibodies for ICP4, ICP27, or α-tubulin as a loading control. (C) Immunoblot analysis of IE gene products in HDFs. Cells were infected with KOS, QOZHG, or J∆NI viruses at 1 pfu per cell, and extracts were prepared at 24 hpi. Blots were probed with antibodies for the indicated IE gene products or α-tubulin as a loading control. (D) Relative nuclear viral DNA levels after infection with equal gc or pfu. HDFs were infected with the indicated J∆NI vectors at 5,000 gc per cell (Left) or 1 pfu per cell (Right). At 2 hpi, nuclear DNA was isolated, and relative viral gc numbers were determined by qPCR for the gD gene normalized to the cellular 18S rRNA genes.
Table 1.
Selected gene modifications in vector constructs
| Vector | ICP0 | ICP4* | ICP22 | ICP27 | ICP47 | gB† | LAT‡ | UL3/4§ | UL45/46§ | UL50/51§ |
| KOS | + | + | + | + | + | + | + | — | — | — |
| KNTc | + | + | + | + | + | N/T | + | UbC-mCherry | — | — |
| QOZHG | + | ∆ | β | CMV-GFP | β | + | + | — | — | — |
| J∆NI2 | β | ∆ | β | β | ∆p | N/T | 3′∆ | — | — | — |
| J∆NI3 | ∆ | ∆ | β | β | ∆p | N/T | 3′∆ | — | — | — |
| J∆NI5 | ∆ | ∆ | β | ∆ | ∆p | N/T | 3′∆ | — | — | — |
| J∆NI6GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | 3′∆ | CAG-GFP | — | — |
| J∆NI7GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP | — | — | — |
| J∆NI7GFP∆C1 | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP, ∆C1 | — | — | — |
| J∆NI7GFP∆C2 | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP, ∆C2 | — | — | — |
| J∆NI7GFP∆LP2 | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP, ∆LP2 | — | — | — |
| J∆NI7GFP∆C12 | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP, ∆C12 | — | — | — |
| J∆NI7GFP∆C12LP2 | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP, ∆C12LP2 | — | — | — |
| J∆NI9GW | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | GW¶ | — |
| J∆NI9GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | CAG-GFP | — |
| J∆NI9LAT-GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | LAT-CAG-GFP | — |
| J∆NI10GW | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | — | GW¶ |
| J∆NI10GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | — | CAG-GFP |
| J∆NI10LAT-GFP | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | — | LAT-CAG-GFP |
| J∆NI10LAT-GFP∆C12LP2 | ∆ | ∆ | β | ∆ | ∆p | N/T | ∆ | — | — | LAT∆C12LP2-CAG-GFP |
| J∆NI5∆B#1# | ∆ | ∆ | β | ∆ | ∆p | N/T | 3′∆ | — | — | — |
| J∆NI5∆B#2# | ∆ | ∆ | β | ∆ | ∆p | N/T | 3′∆ | — | — | — |
| J∆NI7GFP∆B# | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-GFP | — | — | — |
| J∆NI7-GWL1 | ∆ | ∆ | β | ∆ | ∆p | N/T | GW¶ | — | — | — |
| J∆NI7mDMD∆B# | ∆ | ∆ | β | ∆ | ∆p | N/T | CAG-mDMD | — | — | — |
+, intact WT gene; β, promoter modifications to change expression kinetics from immediate-early (IE) to early (E); ∆, complete gene deletion; ∆p, promoter/start codon deletion. All J∆NI constructs are deleted for the internal repeat (joint) region.
All J∆NI viruses have the same UbC promoter-mCherry expression cassette in the deleted ICP4 locus.
N/T, gB:D285N/A549T allele referred to herein as gB:N/T.
3′∆, deletion in the LAT primary transcript region resulting from deletion of the ICP0 promoter and coding region (GenBank JQ673480, positions 1,522–5,500) or promoter alone (J∆NI2, GenBank positions 1,522–2,234). Replacement insertions within the remaining portion of the LAT 2-kb intron are specified.
Insertions are specified; —, no insertion.
GW, gateway recombination cassette.
BAC-deleted viruses.
Table 2.
Virus titers
| Virus | gc/mL (× 1011) | pfu/mL (× 108) | gc/pfu |
| KOS | 5.33 | 130.00 | 41.02 |
| KNTc | 8.15 | 29.60 | 275.23 |
| QOZHG | 1.55 | 2.33 | 663.54 |
| J∆NI2 | 3.28 | 4.00 | 819.98 |
| J∆NI3 | 3.46 | 3.36 | 1,028.43 |
| J∆NI5 | 11.74 | 5.00 | 2,347.65 |
| J∆NI6GFP | 7.30 | 2.66 | 2,744.20 |
| J∆NI7GFP | 3.99 | 1.86 | 2,144.88 |
| J∆NI7GFP∆C1 | 1.66 | 0.50 | 3,310.18 |
| J∆NI7GFP∆C2 | 8.42 | 3.50 | 2,405.25 |
| J∆NI7GFP∆LP2 | 4.75 | 1.70 | 2,796.75 |
| J∆NI7GFP∆C12 | 5.97 | 1.86 | 3,211.54 |
| J∆NI7GFP∆C12LP2 | 8.03 | 3.00 | 2,676.55 |
| J∆NI9GFP | 6.70 | 2.00 | 3,352.08 |
| J∆NI9LAT-GFP | 5.58 | 2.66 | 2,097.43 |
| J∆NI10GFP | 10.50 | 1.66 | 6,328.27 |
| J∆NI10LAT-GFP | 2.30 | 0.93 | 2,460.11 |
| J∆NI10LAT-GFP∆C12LP2 | 2.69 | 1.23 | 2,186.17 |
| J∆NI5∆B#1 | 6.50 | 8.66 | 750.98 |
| J∆NI5∆B#2 | 10.18 | 14.60 | 697.53 |
| J∆NI7GFP∆B | 26.55 | 14.30 | 1,856.67 |
| J∆NI7mDMD∆B | 7.01 | 3.66 | 1,916.53 |
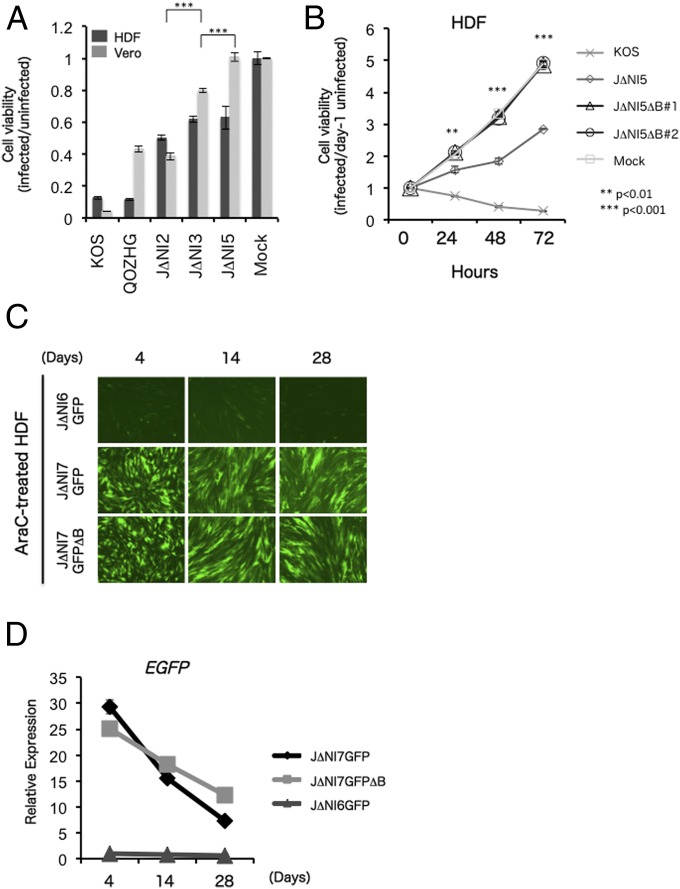
BAC transfection and virus-growth curves confirmed the distinct complementation requirements of the three viruses (Fig. S1). Also consistent with the genetic differences between these three viruses, immunoblotting for IE gene products after infection of noncomplementing HDFs detected ICP27 in both J∆NI2- and J∆NI3-infected cells, a low level of ICP0 only in J∆NI2-infected cells, and no products in J∆NI5-infected cells (Fig. 1C). At 2 h postinfection (hpi), viral DNA levels in the nuclei of HDFs infected with equal gc of the 3 J∆NI viruses were similar while infections with equal pfu resulted in dissimilar nuclear gc levels (Fig. 1D), validating the use of gc titers to standardize virus input for comparisons of gene expression between different viruses in the remainder of this study.
Viral- and Reporter-Gene Expression in Noncomplementing Cells.
Expression analysis by quantitative reverse transcription-PCR (qRT-PCR) of several IE, E, and late (L or γ) genes in infected HDFs confirmed that the ICP0 and ICP27 gene deletions in J∆NI5 reduced viral-gene expression dramatically from the already low levels detected in HDFs infected with J∆NI2 or J∆NI3 at the same gc doses (Fig. S2). Expression of the mCherry reporter gene from its ectopic promoter was also dramatically reduced in J∆NI5-infected HDFs (Fig. S3A), ∼100-fold by the ICP0 gene deletion (Fig. S3B, compare J∆NI2- and J∆NI3-infected cells) and another approximately eightfold by the additional deletion of the ICP27 gene (J∆NI3- vs. J∆NI5-infected cells). These observations confirmed that the residual ICP0 and ICP27 expression from J∆NI2 is sufficient to prevent global transcriptional silencing of the viral genome whereas deletion of these two genes essentially eliminated all native and exogenous promoter activity. Consistent with published studies (13), mCherry expression could be induced in J∆NI5-infected HDFs by superinfection with a replication-defective ICP0+ virus [QOZHG; ICP4−/ICP27−/β-ICP22/β-ICP47 (23)] (Fig. S3C). Thus, ICP27 was not required to derepress the silent J∆NI5 genome, and the small amount of ICP0 expressed in J∆NI2-infected HDFs was sufficient to support limited transcriptional activity throughout the viral genome.
The LAT Locus Supports High-Level Transgene Expression from the Silent J∆NI5 Genome.
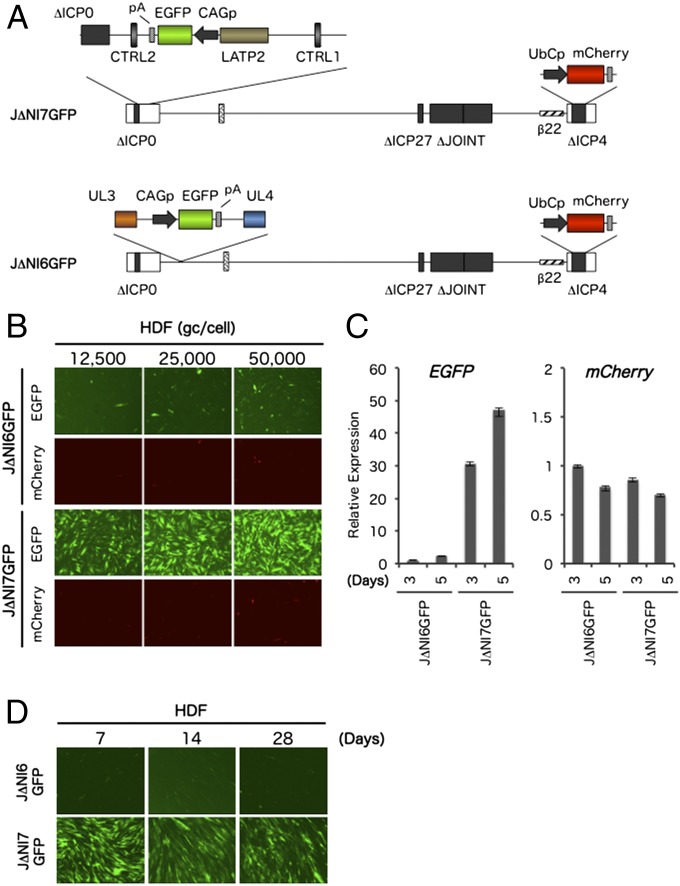
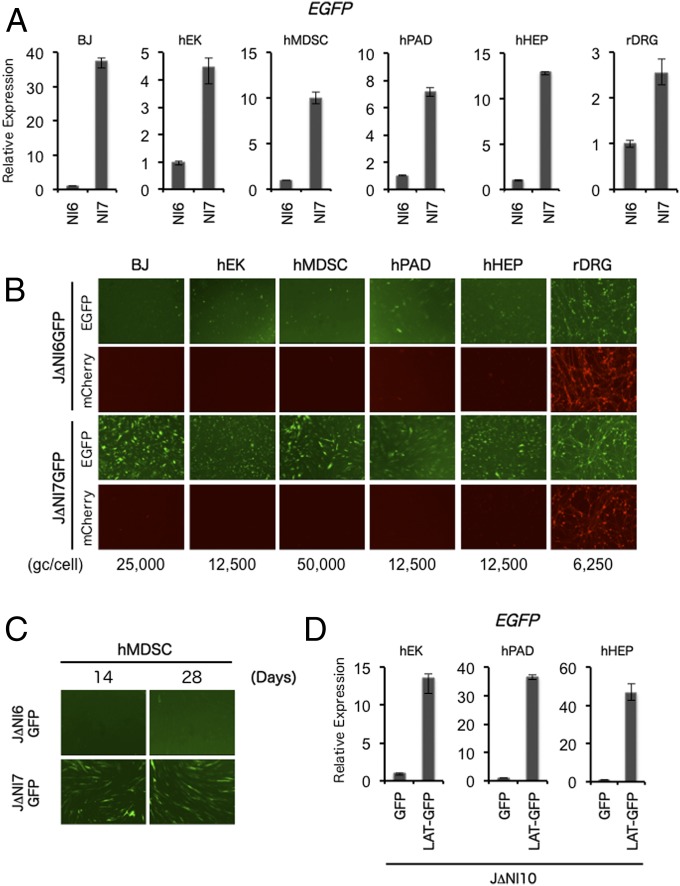
Upon infection of neuronal cells, HSV enters a latent state in which the viral genome is transcriptionally silent except for the LAT locus. We explored whether the LAT locus would similarly allow a cellular promoter to remain active in nonneuronal cells in the context of an otherwise silent viral genome. To this end, we introduced an expression cassette consisting of the CAG enhancer/promoter and EGFP gene (CAGp-GFP) into the LAT 2-kb intron region of J∆NI5, creating a vector construct referred to as J∆NI7GFP (Fig. 2A). As a control, we introduced the same CAGp-GFP cassette into the UL3-UL4 intergenic region of J∆NI5, producing vector J∆NI6GFP (Fig. 2A); the UL3-UL4 intergenic region has been used by others for nondisruptive insertion of transgene cassettes (24, 25) and is close to, but outside, the LAT locus. Infectious viruses were produced on U2OS-ICP4/27 cells, and the gc:PFU ratios of the new virus stocks were similar to that of J∆NI5 (Table 2). Both J∆NI6GFP and J∆NI7GFP produced abundant green and red fluorescence during amplification in U2OS-ICP4/27 cells, confirming the integrity of their transgene-expression cassettes. To examine the abilities of these viruses to express the EGFP transgene in noncomplementing cells, HDFs were infected with increasing gc per cell, and EGFP fluorescence was recorded at 3 dpi (Fig. 2B). J∆NI7GFP-infected cells showed abundant, viral dose-dependent EGFP expression whereas J∆NI6GFP infection produced limited expression, even at the highest dose. Although we used a higher virus input here than in earlier J∆NI5 infections, mCherry expression remained minimal. These results were confirmed by qRT-PCR measurements of EGFP and mCherry mRNA levels at 3 and 5 dpi (Fig. 2C) and were consistent with the suggestion that the J∆NI6GFP and J∆NI7GFP virus genomes, like the J∆NI5 genome, were silent in infected HDFs whereas the LAT locus maintained transcriptional activity from a non-HSV promoter. Because EGFP mRNA levels in J∆NI7GFP-infected cells were at least as high at 5 dpi as at 3 dpi (Fig. 2C), we asked whether expression could be detected at later times when the cells are fully contact-inhibited. The results showed that expression could persist for at least 4 wk in some of the cells (Fig. 2D). Together, these observations indicated that the LAT locus is a privileged site for durable transgene expression in nonneuronal cells in the background of an HSV genome that lacks all IE gene expression.
Fig. 2.
J∆NI6GFP and J∆NI7GFP genome structures and reporter-gene expression. (A) J∆NI7GFP contains a CAG promoter-EGFP expression cassette with the rabbit β-globin polyA (pA) region in the 2-kb LAT intron region between the LATP2 long-term expression/enhancer region and a downstream series of CTCF-binding motifs (CTRL2) in the intron. LATP2 extends from the LAT transcription initiation site to within the 2-kb intron. J∆NI6GFP contains the same CAG promoter-EGFP expression cassette between the UL3 and UL4 genes. (B–D) EGFP and mCherry expression in infected HDFs. (B) Cells were infected with J∆NI6GFP or J∆NI7GFP virus at different gc per cell, and fluorescence was visualized at 3 dpi. (C) HDFs were infected with J∆NI6GFP or J∆NI7GFP vector at 12,500 gc per cell and harvested 3 or 5 d later for mRNA extraction and qRT-PCR analysis for the two reporter genes. Expression normalized to 18S rRNA is shown relative to J∆NI6GFP-infected cells on day 3. (D) HDFs were infected with J∆NI6GFP or J∆NI7GFP virus at 25,000 gc per cell, and EGFP fluorescence was photographed at 7, 14, and 28 dpi.
CTRLs and LATP2 Support Transgene Expression from the LAT Locus.
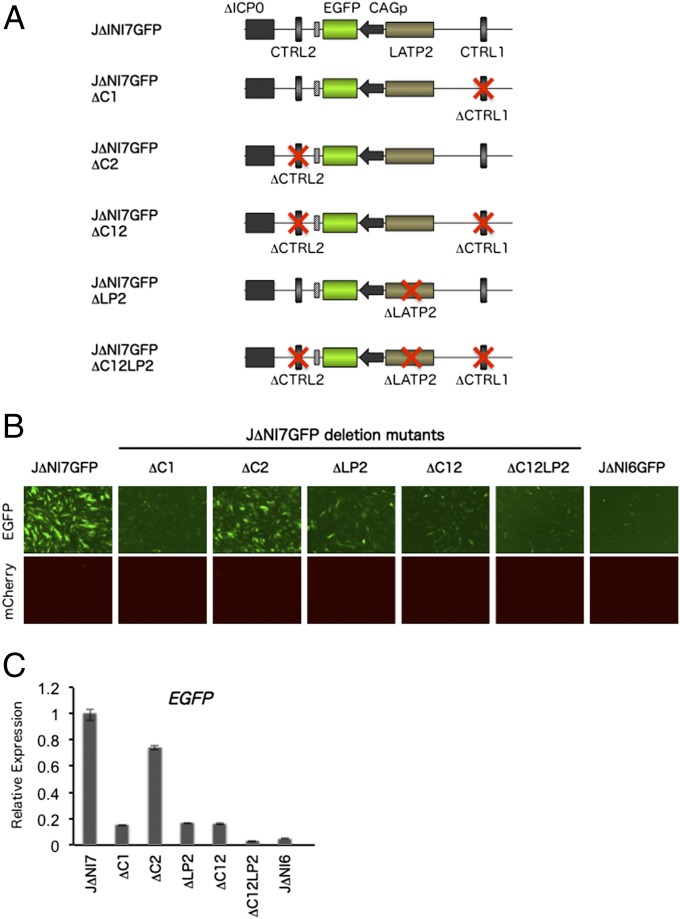
It has been demonstrated that LAT expression during latency is controlled by the latency-associated promoters (LAPs) 1 and 2 (26, 27). LAP1 is located upstream of the transcription initiation site of the ∼8.3-kb unstable primary LAT transcript that is processed to a stable 2-kb LAT intron whereas LAP2 is located downstream of LAP1 in the first exon and extending into the 2-kb intron region. A LAP2-containing region extending somewhat further into the intron has been referred to as LATP2 (8). In addition to acting as a promoter, it has been shown that the LAP2/LATP2 region can act as a position-independent long-term expression/enhancer element for transgene expression in neurons (8, 28). However, it has also been reported that the LAT locus is protected from global silencing of the viral genome during latency by regions rich in CTCF-binding sites, termed CTRLs, one located upstream of LAP1 (CTRL1) and the other in the 2-kb intron just downstream of LATP2 (CTRL2) (29). We therefore explored the potential roles of the LATP2 and CTRL elements in enabling the observed EGFP expression in J∆NI7GFP-infected HDFs. We deleted the three elements separately and in combination from the J∆NI7GFP genome (Fig. 3A) and examined reporter-gene expression in infected HDFs (Fig. 3 B and C). Deletion of either CTRL1 (ΔC1) or LATP2 (ΔLP2) caused a marked decrease in green fluorescence and EGFP mRNA levels whereas the deletion of CTRL2 (ΔC2) had only a minor effect. Deletion of both CTRLs (ΔC12) had the same effect as the deletion of CTRL1 alone whereas deletion of all three elements (ΔC12LP2) reduced expression further to the level observed in J∆NI6GFP-infected cells. As expected, mCherry fluorescence was visually undetectable in any of the infected cells (Fig. 3B), suggesting that the different deletions did not cause derepression of other sites in the viral genome. These results indicated that both CTRL1 and the LATP2 region play significant roles in protecting the linked transgene from transcriptional silencing in the context of a viral genome that is functionally devoid of all IE genes.
Fig. 3.
Effect of LAT locus elements on EGFP expression from J∆NI7GFP. (A) Genome representations of J∆NI7GFP and derivatives deleted for CTRL1 (ΔC1), CTRL2 (ΔC2), or LATP2 (ΔLP2) individually or in combinations. (B) Reporter-gene expression in infected HDFs. Cells were infected with the indicated viruses at 12,500 gc per cell, and fluorescence was recorded at 3 dpi. (C) Relative EGFP mRNA levels in HDFs infected with J∆NI7GFP, derivatives deleted for LAT elements, or J∆NI6GFP; viruses are identified by abbreviated names. Cells were infected at 12,500 gc per cell and processed at 3 dpi for qRT-PCR analysis. Expression levels were normalized to 18S rRNA and are presented relative to the level in J∆NI7GFP-infected cells.
LAT Locus Protection of Transgene Expression Is Position-Independent.
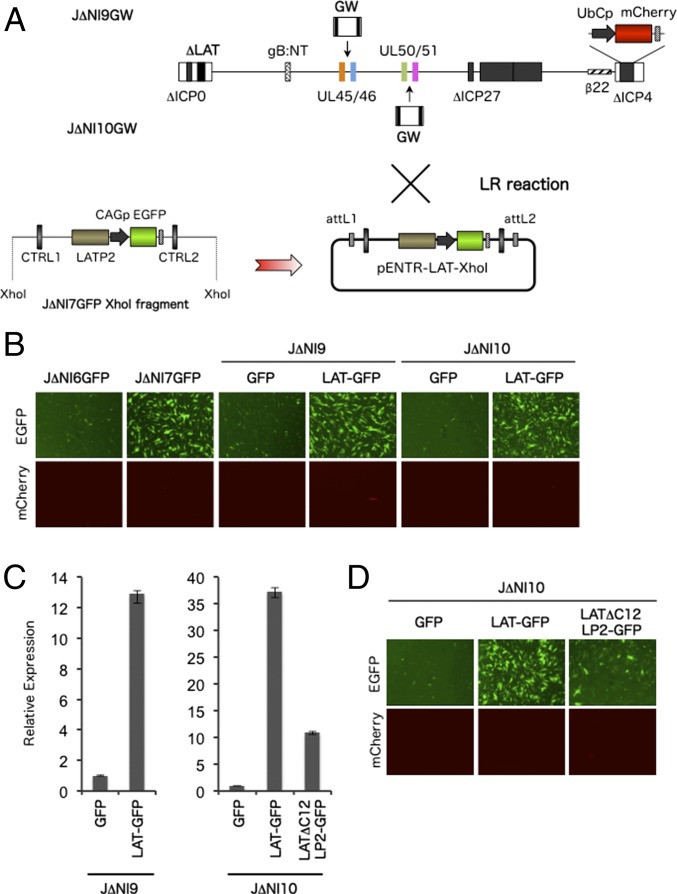
To determine whether sequences associated with the LAT locus are sufficient to protect an embedded transgene expression cassette against silencing in the absence of IE gene products, we inserted a restriction fragment corresponding to the LAT:CAGp-GFP region of J∆NI7GFP, including the two CTRLs, LAP1 and LATP2, at one of two ectopic positions in a J∆NI5 derivative that was deleted for the same LAT region to avoid recombination between the native and new sites. We first engineered a Gateway (GW) recombination cassette into the intergenic region between UL45 and UL46 (J∆NI9GW) or between UL50 and UL51 (J∆NI10GW) of the LAT-deleted J∆NI5 genome (J∆NI5∆L) (SI Materials and Methods) and then introduced the LAT:CAGp-GFP fragment or just CAGp-GFP without LAT sequences by recombination with the GW cassette (Fig. 4A). After virus production on U2OS-ICP4/27 cells (Table 2), the vectors were tested for reporter-gene expression in infected HDFs. At 3 dpi, the vectors that lacked LAT sequences surrounding the reporter cassette (J∆NI9GFP and J∆NI10GFP) showed low levels of EGFP fluorescence (Fig. 4B) and mRNA (Fig. 4C) in infected cells, similar to the J∆NI6GFP control vector, suggesting that the two intergenic insertion sites were transcriptionally repressed, like the intergenic region between UL3 and UL4 (Fig. 2). However, when the reporter cassette was flanked at both sides by LAT sequences (vectors J∆NI9LAT-GFP and J∆NI10LAT-GFP), EGFP expression increased to the level observed in J∆NI7GFP-infected cells (Fig. 4B), indicating that the antisilencing activity of the LAT-derived regions is functional in a position-independent manner. To confirm the dependence of this activity on LATP2 and either or both CTRLs, we introduced a LATP2- and CTRL-deleted version of the LAT:CAGp-GFP fragment by GW recombination into the J∆NI10GW genome; the deletions were the same as those in the LAT region of the earlier JΔNI7GFPΔC12LP2 vector (Fig. 3A). We found that the deletions reduced EGFP expression in infected HDFs, although not fully down to the level observed with the LAT-free J∆NI10GFP vector (Fig. 4 C and D); it is unclear why the ΔC12LP2 deletions seemed to have a smaller effect here (∼3.5-fold) than in JΔNI7GFP (∼50-fold) (Fig. 3C). However, our results clearly demonstrate that a portion of the LAT locus that includes the two CTRLs, LAP1 and LATP2, can protect an embedded transgene expression cassette in a position-independent manner against global silencing of the viral genome in the absence of IE gene expression and indicate that at least CTRL1 and LATP2 play a role in this activity. We also determined whether the effect of the transferred LAT sequences was limited to the embedded transgene. We observed moderate increases in the levels of ICP22, ICP6, and glycoprotein C (gC) mRNA produced by J∆NI10LAT-GFP compared with J∆NI10GFP (Fig. S4), but these increases were substantially smaller than the increase in GFP expression (Fig. 4B). Of interest, expression of the three distal genes was also somewhat higher from J∆NI10LAT-GFP than from J∆NI5 (Fig. S4), suggesting that the reach of the LAT elements may be influenced by their context.
Fig. 4.
Anti-silencing activity of LAT sequences positioned elsewhere in the viral genome. (A) Construction of J∆NI9 and J∆NI10 vectors. A LAT region spanning CTRL1, LATP2, and CTRL2 was removed from the J∆NI5 genome (∆LAT), and a GW recombination cassette was introduced between UL45 and UL46 to generate J∆NI9GW or between UL50 and UL51 to produce J∆NI10GW (Upper). XhoI restriction sites were used to isolate a CAGp-GFP–containing LAT fragment from J∆NI7GFP (Lower Left). The XhoI fragment was cloned into pENTR1A (Lower Right) and transferred into J∆NI9GW or J∆NI10GW by attL/attR recombination with the respective GW cassettes (LR reaction) to produce J∆NI9LAT-GFP and J∆NI10LAT-GFP, respectively. As controls, we also recombined the CAGp-GFP cassette without LAT sequences via a pENTR1A intermediate into the GW locus of J∆NI9GW or J∆NI10GW, producing J∆NI9GFP and J∆NI10GFP. (B) Reporter-gene expression in HDFs infected with J∆NI9 or J∆NI10 viruses. HDFs were infected with the indicated viruses at 12,500 gc per cell. EGFP and mCherry fluorescence were recorded at 3 dpi. (C) EGFP mRNA levels in infected HDFs determined by qRT-PCR as in earlier figures. Levels are shown relative to J∆NI9GFP- or J∆NI10GFP-infected cells. J∆NI10ΔC12LP2-GFP was constructed by transfer of the XhoI LAT fragment from J∆NI7ΔC12LP2-GFP into the GW site of J∆NI10GW, similar to the construction of J∆NI10LAT-GFP above. (D) Effect of the deletion of both CTRLs and LATP2 from J∆NI10LAT-GFP on transgene expression. HDFs were infected at 12,500 gc per cell, and EGFP and mCherry fluorescence were recorded at 3 dpi.
LAT Locus Elements Protect Transgene Expression in Other Cell Types.
To evaluate the applicability of our observations beyond HDFs, we tested EGFP and mCherry expression from selected vectors in other cell types. By qRT-PCR, we observed higher EGFP mRNA levels in JΔNI7GFP-infected than in JΔNI6GFP-infected human cells at 3 dpi (Fig. 5A). The greatest difference was observed in BJ human foreskin fibroblasts (∼35-fold), similar to the difference in HDFs (∼30-fold) (Fig. 2C). Human neonatal keratinocytes (hEKs) showed the smallest difference (∼4.5-fold), with intermediate values seen in human muscle-derived stem cells (hMDSCs) (∼10-fold), human preadipocytes (hPADs) (approximately sevenfold), and human hepatocytes (hHEPs) (∼13-fold). We also compared the two vectors in fetal rat dorsal root ganglion (rDRG) neurons and found only a ∼2.5-fold difference. Fig. 5B shows representative fluorescence images at 3 dpi from an independent experiment performed under the same infection conditions. The results were largely consistent with the qRT-PCR data. Of interest, whereas none of the human cells showed significant mCherry fluorescence, abundant mCherry expression was observed in both the JΔNI6GFP- and JΔNI7GFP-infected rat DRG cultures. It is not yet known whether this observation is unique to neuronal cells and may be a function of the promoter driving mCherry-gene expression, the location of the expression cassette in the viral genome, and/or the rat origin of the cells. We were able to maintain hMDSCs for an extended period, allowing the monitoring of EGFP expression over time in JΔNI7GFP-infected cells. As shown in Fig. 5C, EGFP remained detectable for at least 4 wk after infection, similar to our observation with JΔNI7GFP-infected HDFs (Fig. 2D). Lastly, we determined for some of the cells whether CAGp-GFP activity in the UL50/51 intergenic region was enhanced by flanking LAT sequences as in HDFs. The results in Fig. 5D show a substantial enhancement in all three cell types that may, for unknown reasons, exceed the difference between JΔNI6GFP and JΔNI7GFP in the same cells. Overall, these results indicated that the position-independent antisilencing activity of genetic elements in the LAT locus is not limited to HDFs but is operative in a variety of nonneuronal human cell types.
Fig. 5.
Reporter-gene expression from J∆NI vectors in other noncomplementing cells. (A) The cells listed at the top were infected with J∆NI6GFP or J∆NI7GFP at the gc per cell indicated below the panels of B. EGFP and mCherry fluorescence were recorded at 3 dpi. (B) EGFP gene expression in cells infected as in A was measured at 3 dpi by qRT-PCR analysis. Results normalized to 18S rRNA are shown relative to J∆NI6GFP-infected cells. (C) hMDSCs were infected with J∆NI6GFP or J∆NI7GFP virus at 50,000 gc per cell, and EGFP fluorescence was photographed at 14 and 28 dpi. (D) qRT-PCR determination of EGFP mRNA levels in hEK, hPAD, and hHEP cells 3 d after infection with J∆NI10GFP or J∆NI10LAT-GFP at 12,500 gc per cell. Normalized expression is shown relative to J∆NI10GFP-infected cells.
Elimination of Residual Vector Cytotoxicity by BAC Removal.
Although J∆NI5 and its derivatives displayed no overt toxicity for noncomplementing cells, we evaluated this perception by quantitative cell viability (MTT) assay. HDFs and Vero cells were infected with KOS, QOZHG, J∆NI2, J∆NI3, or J∆NI5 virus at 25,000 gc per cell, and assays were performed at 5 dpi. Surprisingly, whereas toxicity for Vero cells was significantly reduced between J∆NI2 and J∆NI3 (P < 0.001) and was no longer detectable in J∆NI5-infected cells, the number of viable cells in J∆NI5-infected HDF cultures remained well below the number in mock-infected cultures and was not dramatically greater than that in J∆NI2- or J∆NI3-infected cultures (Fig. 6A); QOZHG, which overexpresses ICP0, was highly toxic for HDFs, similar to replication-competent virus (KOS). A significant difference between J∆NI5 and a fully IE gene-inactivated vector such as d109 that was previously shown to be devoid of any toxicity for human fibroblasts (13) is the presence of the loxP-flanked 11-kb BAC region in the J∆NI5 genome. To eliminate this region from our vectors, we used retroviral transduction to produce a J∆NI5-complementing cell line expressing Cre recombinase, U2OS-ICP4/27-Cre cells (Fig. S5A and SI Materials and Methods). After infection of these cells with J∆NI5 virus, plaques were screened by staining for LacZ activity expressed from the BAC cassette, two negative viral clones were purified (J∆NI5∆B#1 and J∆NI5∆B#2), and accurate excision of the BAC region was confirmed by PCR and sequencing through the remaining loxP site (SI Materials and Methods). Cell-viability assays showed unimpaired growth of J∆NI5∆B-infected HDFs compared with mock-infected HDFs whereas the BAC-positive J∆NI5 virus again inhibited HDF growth (Fig. 6B). We next determined whether the BAC region affects short- and long-term transgene expression. We generated a BAC-deleted version of J∆NI7GFP (J∆NI7GFP∆B) (Fig. S5B) and recorded EGFP expression over time in infected HDFs compared with infections with BAC-positive J∆NI6GFP and J∆NI7GFP (Fig. 6C); cytosine arabinoside (AraC) was included in the postinfection maintenance media in this experiment to block cell division to prevent dilution of vector genomes. No dramatic differences were observed in EGFP fluorescence levels between J∆NI7GFP∆B- and J∆NI7GFP-infected cells through 28 d postinfection. In the same experiment, qRT-PCR measurements showed a moderately slower decline in EGFP gene expression in J∆NI7GFP∆B- than in J∆NI7GFP-infected HDFs (Fig. 6D). Together, these results indicated that BAC deletion is advantageous for sustained transgene expression in nondividing cells whereas the opposite effect may be observed in dividing cells in vitro due to a higher rate of cell division and thus faster dilution of viral genomes.
Fig. 6.
Residual J∆NI5 cytotoxicity and effects of BAC excision. (A) In vitro cytotoxicity assay. HDFs and Vero cells were infected at 25,000 gc per cell, and cell viability in triplicate wells was measured at 5 dpi by MTT assay. Plotted values represent the mean ratios of virus-infected to mock-infected cells. Differences between J∆NI2- and J∆NI3-infected Vero cells and between J∆NI3- and J∆NI5-infected Vero cells (brackets) were statistically highly significant (***P < 0.001). (B) BAC deletion eliminates J∆NI5 vector toxicity for HDFs. HDFs (2 × 103) were infected with KOS, J∆NI5, or two different J∆NI5∆B isolates at 25,000 gc per cell, and cell viability in triplicate wells was measured by MTT assay at the indicated hpi. Plotted values represent the mean ratios of virus-infected to day-1 mock-infected cells. Statistically significant differences between J∆NI5-infected and both J∆NI5∆B#1- and J∆NI5∆B#2-infected cells are indicated by asterisks (**P < 0.01; ***P < 0.001). (C) Effect of BAC excision on J∆NI7GFP reporter-gene expression. HDFs were infected with J∆NI6GFP, J∆NI7GFP, or BAC-deleted J∆NI7GFP (J∆NI7GFP∆B) at 25,000 gc per cell, and EGFP fluorescence was visualized at 4, 14, and 28 dpi. To prevent vector dilution by cell division, 1 μM AraC (Sigma) was included in the postinfection maintenance media. (D) EGFP mRNA levels in cells infected with J∆NI6GFP, J∆NI7GFP, or J∆NI7GFP∆B viruses. HDFs were infected at 25,000 gc per cell and harvested on the indicated days postinfection for mRNA extraction and qRT-PCR quantification of EGFP gene expression. Data were normalized to viral gc numbers determined in the same cultures and is presented relative to the normalized value of J∆NI6GFP-infected cells on day 4.
Efficient Transduction of Dystrophin with the J∆NI5 System.
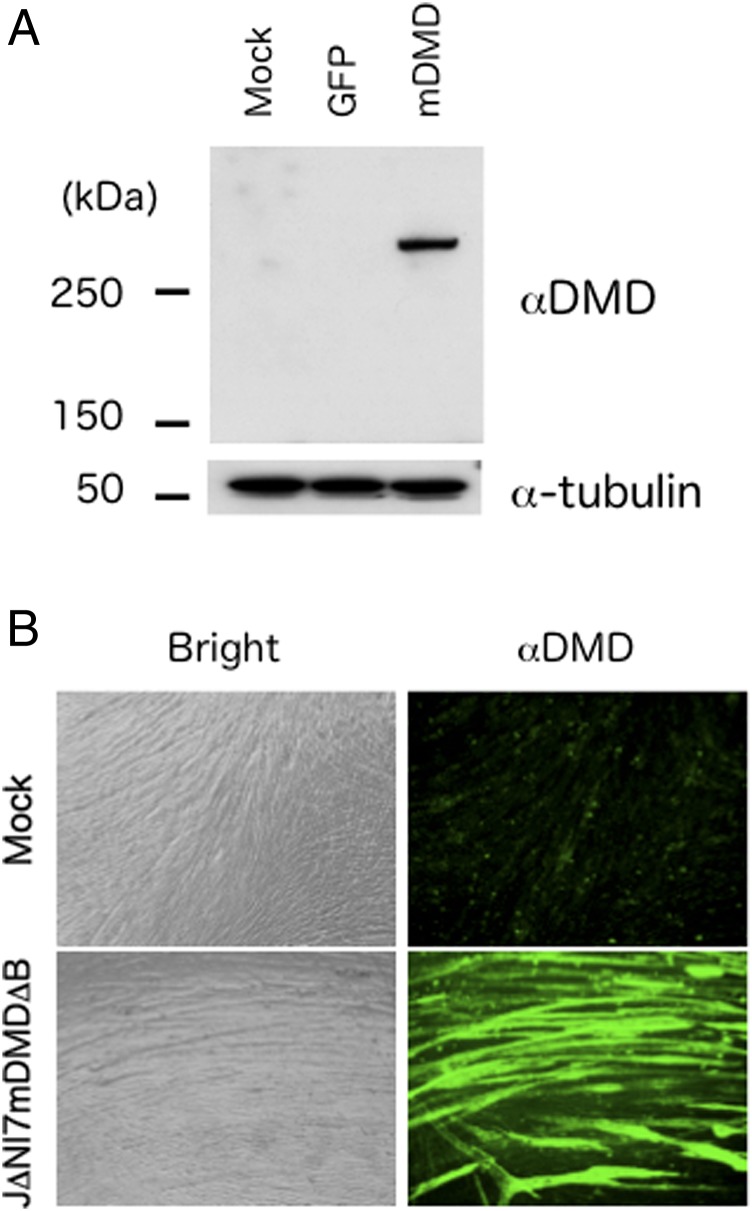
To illustrate the capacity of the J∆NI5 vector system, we chose to insert the complete 14-kb cDNA encoding the murine version of dystrophin, the defective protein in Duchenne muscular dystrophy (DMD) (30). Currently no helper-independent viral-vector system exists that is capable of vigorous production of full-length murine or human dystrophin. We modified the J∆NI5 genome by insertion of a GW cassette into the LAT region between LATP2 and CTRL2, creating J∆NI7-GWL1, and then recombined the cassette with the 16.5-kb insert of a pENTR construct containing the complete mouse dystrophin cDNA between the CAG promoter and the rabbit β-globin polyadenylation region (Fig. S6). Including the mCherry expression cassette, the resulting vector construct, J∆NI7-mDMD, carried ∼20 kb of transgene sequences in addition to the BAC region. A small virus stock was generated by transfection of U2OS-ICP4/27 cells, the BAC region was deleted as described above by passage through U2OS-ICP4/27-Cre cells, and a clonal isolate referred to as J∆NI7-mDMD∆B was amplified to high titer (Table 2). Dystrophin-deficient mdx mouse-derived muscle progenitor cells infected with J∆NI7-mDMD∆B showed the presence of full-length dystrophin at 3 dpi (Fig. 7A) and wide-spread expression in differentiated cultures at 7 dpi (Fig. 7B), supporting the utility of the J∆NI5 vector backbone for efficient transduction of nonneuronal cells with large genes that cannot be accommodated by current vector systems.
Fig. 7.
Efficient J∆NI-mediated transduction by a large transgene. (A) Western blot analysis of dystrophin expression in mock-, J∆NI7GFP∆B-, and J∆NI7mDMD∆B-infected mdx mouse-derived muscle progenitor cells (25,000 gc per cell) at 3 dpi. (Upper) Anti-dystrophin antibody (αDMD). (Lower) Anti–α-tubulin antibody (α-tubulin) as loading control. (B) Dystrophin immunofluorescence (αDMD) and bright-field (Bright) images of mock- and J∆NI7mDMD∆B-infected mdx muscle progenitor cells (50,000 gc per cell) at 7 dpi.
Discussion
HSV offers a number of important features as a gene-therapy vector, including its ability to infect a wide range of cells and establish the viral genome as a stable extrachromosomal element, efficient low-dose transduction that helps reduce inflammation and the induction of antiviral immunity, and a very large payload capacity that easily exceeds that of most vectors in current use. However, whereas delivery of large payloads represents an important unmet need in the gene-therapy field, it has not been possible to create HSV vectors that provide robust and persistent transgene expression in the absence of cytotoxic viral IE gene expression.
Our study reports the engineering of HSV vectors that are both noncytotoxic and capable of persistent transgene expression. We created an HSV backbone that does not produce any IE proteins in noncomplementing cells and explored the possibility that the latency locus could be exploited to protect an embedded cellular promoter against global silencing of the viral genome in nonneuronal cells, similar to the apparent protection of LAT-promoter activity in latently infected sensory neurons.
A critical limitation of most existing replication-defective HSV vectors is the presence of one or more expressed IE genes that are activated on infection in a manner independent of other viral-gene expression. ICP0 expression has proved especially important for transgene activity because the elimination of ICP0 activity led to complete genome silencing (13, 16). Although ICP0 has the attractive feature of preventing heterochromatin remodeling and consequent transgene silencing (15), it can also induce cell-cycle arrest in dividing cells and apoptosis in postmitotic cells (18, 31). To overcome this problem, HSV vectors have been engineered to dramatically reduce but not eliminate ICP0 expression in the hope that low levels of the protein would maintain transgene activity without loss of cell proliferation or viability (8, 9, 17, 32). However, these strategies do not universally eliminate vector toxicity for nonneuronal cells, which varies with the target cell and multiplicity (7, 11). Our study confirms that even low levels of ICP0 can reduce cell viability despite the upside of maintaining some transgene expression. This reduced viability may be a result of the action of ICP0 alone or possibly in combination with leaky expression of other viral functions. Thus, we and others have searched for a strategy to express transgenes when all IE gene expression is shut down.
In one approach to achieve this goal, ICP0 has been substituted with similar IE genes from other herpesviruses, including the pp71 gene of human cytomegalovirus (HCMV) (33, 34), a betaherpesvirus, with the notion that the products of these genes may provide the antisilencing functions of ICP0 without toxicity (35). However, it is well-known that pp71 degrades a chromatin-remodeling factor, hDaxx, followed by disruption of the hDaxx–ATRX complex (36, 37) and members of the retinoblastoma tumor-suppressor family of proteins (38). The disruption of the hDaxx–ATRX complex can cause genomic instability (39) whereas the loss of retinoblastoma family proteins dysregulates normal cell-cycle progression (40, 41). Indeed, pp71 overexpression seems to induce malignant alteration of the cell. Everett et al. have used an inducible cell-line system to demonstrate that the ICP0 equivalents of the alphaherpesviruses, bovine herpes virus type 1 (BHV-1), equine herpesvirus type 1 (EHV-1), and pseudorabies virus (PRV), can effectively derepress silent HSV-1 genomes (42). However, they also reported that all three of these proteins adversely affect cell growth, although less dramatically than ICP0. Thus, prior results argue that the strategies of reducing ICP0 expression or substituting ICP0 with other herpesvirus transactivators will not solve the problem of genome silencing in the absence of ICP0 without deleterious effects on the cells.
Because the latency locus remains active in neuronal cells at a time when all viral lytic genes are silenced, including the ICP0 gene, we and others reasoned that this locus might be a preferred site for transgene expression from quiescent HSV genomes in nonneuronal cells. Lilley et al. (32) used a highly debilitated vector to demonstrate reporter-gene expression in Vero cells at 23 dpi from a LATP2-linked expression cassette in the UL41 locus, but residual ICP0 expression could not be ruled out. Our results show that a foreign promoter inserted into the LAT locus can drive robust short-term reporter-gene expression in the complete absence of IE gene products in a variety of primary human nonneuronal cells and that this expression can persist in postmitotic cells. These attributes of our platform create a gene-transfer vector with broad utility beyond the nervous system. We proceeded to examine which elements within the LAT locus provided for this remarkable transgene expression.
Recent findings have shown that, during latency, the cellular CTCF protein is enriched on the two HSV CTRLs and that acetylated histone H3, an epigenetic beacon of active transcription, accumulates in the LAT region between these elements (19, 43–45). These findings suggested that CTRLs may act as boundary elements to shield the LAT locus against the spreading of repressive epigenetic modifications (29), and it was shown in short-term plasmid transfection assays that a fragment encompassing CTRL2 combines the functions of a typical insulator (19). In the context of a complete viral genome, we found that CTRL1, along with LATP2, was essential for preventing the rapid silencing of an ectopic promoter in the LAT locus in nonneuronal cells in the absence of all viral IE gene products (Fig. 3). Furthermore, we showed that the LAT locus provides protection against the silencing of an embedded promoter when moved away from its native position in the viral genome (Fig. 4) and supports long-term expression in postmitotic human fibroblasts (Figs. 2D and 6C) and striatal muscle cells (Fig. 5C). CTRL1 consists of 46 copies of two separate, but overlapping, CTCF-binding motifs whereas CTRL2 contains 9 copies of only one of these motifs (19), perhaps explaining the greater effect of CTRL1 than CTRL2 deletion on transgene expression in the LAT locus. Recent evidence indicates that the function of insulators is dependent on additional sequences aside from CTCF-binding motifs, and thus a more complete analysis of these viral elements will be required to determine whether they have specific limitations (46). LAP2 (LATP2) has been reported to contain chromatin-binding elements that bend DNA, and such activity could enhance the function of nearby foreign promoters (47). Although some chromatin regulatory elements located within the LAT promoter regions have shown tissue specificity, it has been unclear whether LATP2 can be broadly useful to counteract transgene silencing in nonneuronal cells in the absence of ICP0 (48, 49). Our data indicate that, in combination with CTRL1 and to a lesser extent alone, it can.
We used a ubiquitin C promoter-mCherry cassette in the ICP4 locus to monitor the effect of sequential deletion of IE genes on transgene expression. As expected, expression in HDFs declined to essentially undetectable with the progressive removal of all IE gene functions, but we could readily detect mCherry fluorescence during growth of the different vectors in complementing cells, as well as on infection of unmodified U2OS cells. These findings were consistent with the designs of the sequential J∆NI backbones, and mCherry expression from J∆NI5 derivatives was not demonstrably altered or elevated in other noncomplementing cells, with one notable exception, DRG neurons. Although we do not yet know the cause of this difference, a recent report by Harkness et al. noted greater persistence of transcriptional activity within repeat regions of the IE gene-depleted d109 vector than elsewhere along the genome in trigeminal ganglion neurons, but not in MRC5 human embryonic lung cells (50). The mCherry cassette in our vectors replaces the ICP4 gene in the remaining repeat bordering US, a region flanked on each side by a cluster of CTCF-binding motifs referred to by Bloom and coworkers as CTaS (or B4) and CTRS3′ (or B6), respectively (19, 29). Because the ICP4 gene is severely repressed in latently infected neurons relative to the LAT region, it seems unlikely that these elements alone are responsible for the enhanced mCherry expression we observed in rat DRGs, and thus other factors, including the nature of the transgene promoter, may play an important role.
The application of highly defective HSV vectors for gene therapy will require methods for efficient vector engineering and subsequent production minimizing defective particles. We found that our highly defective vectors can be readily produced as infectious particles by transfection of the corresponding BAC constructs into complementing U2OS-ICP4/27 cells and can be amplified to high titers on these cells (Table 2). Importantly, because there is no sequence overlap between the complementing genes in U2OS-ICP4/27 cells and the genome of J∆NI5 and its derivatives, the risk of generating replication-competent revertants is minimal. We used U2OS cells for vector complementation because these cells naturally complement ICP0 function (21), thus eliminating the need to express this toxic protein. In the past, Vero cells engineered to complement ICP0 in addition to ICP4 and ICP27 have proven difficult to maintain (10) whereas our U2OS-ICP4/27 line is stable. Moreover, we found that U2OS cells can tolerate constitutive expression of ICP4 whereas the kinetics of ICP4 induction in Vero-based cells in response to defective virus infection can be slow (51). We have also added Cre recombinase to our complementing line to eliminate the BAC region during vector production. Removal of the 11-kb BAC insert had the advantage of recreating space for recombinant sequences and eliminating residual J∆NI5 toxicity for HDFs. Although bacterial sequences have been shown to contribute to transgene silencing in different vector systems (52), we observed only a modest increase in long-term reporter-gene expression as a result of BAC excision. It is also noteworthy that none of our BAC vector constructs challenged the size limit for efficient viral-genome packaging into virions, suggesting that the consistent growth acceleration we have noticed after BAC excision is due to other factors, perhaps including the elimination of possible recognition of the bacterial sequences by pattern-recognition receptors such as TLR9. Together, our U2OS-ICP4/27-Cre and U2OS-ICP4/27 cells provide an effective and reliable means to produce safe stocks of highly defective HSV vectors from engineered BAC constructs, important attributes for application to patient therapies.
In summary, we describe for the first time, to our knowledge, a nontoxic HSV-vector platform that can accommodate large transgenes, exemplified by the 14-kb mouse dystrophin cDNA, with continued expression in nonneuronal cells. Our results indicate that proximity to certain elements from the LAT locus protects transgene expression against viral-genome silencing. This new generation of HSV vector represents a potent tool for delivery of large payloads to both neuronal and nonneuronal cells, suggesting that it will fill an important niche in the gene-therapy vector arsenal.
Materials and Methods
Cells.
Human osteosarcoma U2OS cells (ATCC) were grown in DMEM with 10% (vol/vol) FBS and penicillin-streptomycin (P/S). HDFs (PCS-201-010; ATCC) and BJ human foreskin fibroblasts (CRL-2522; ATCC) were grown in DMEM with 10% (vol/vol) Embryonic Stem Cell Qualified FBS (Invitrogen) and P/S. Vero, Vero-7b (7), and 293T cells were cultured in DMEM containing 5% (vol/vol) FBS and P/S. hHEPs were isolated and cultured as described (53, 54). hPADs (PT-5020; Lonza) were maintained with PBM-2 Basal Medium (Lonza). hMDSCs (kindly provided by J. Huard, University of Pittsburgh, Pittsburgh, PA) were cultured as described (55). hEKs (Invitrogen) were cultured in EpiLife Medium supplemented with Human Keratinocyte Growth Supplement (both from Invitrogen). DRGs were microdissected from day-15 rat embryos, dissociated with 3 mg/mL type-I collagenase (Sigma) in Leibovitz’s L-15 media for 30 min at 37 °C with constant shaking, and plated on poly-d-lysine (Sigma)–coated coverslips at ∼105 cells per well in 24-well plates in 500 μL of defined Neurobasal medium with B27 supplement, Glutamax-I, Albumax-II, and P/S (Gibco/Invitrogen), supplemented with 100 ng/mL 7.0S NGF (Sigma). At 1–3 d post-plating, cultures were treated with 10 µM uridine and 10 µM fluorodeoxyuridine (Sigma) in the above media for 1–2 d to limit the expansion of dividing cells, such as fibroblasts and glia. Cells were then washed with PBS and incubated with NGF-supplemented Neurobasal medium as above. Virus infections were performed at 10–15 d after plating. Differentiation-prone muscle progenitor cells from dystrophin-deficient mdx mice (kindly provided by J. Huard and B. Wang, University of Pittsburgh, Pittsburgh, PA) were isolated and cultured as described (56).
U2OS-ICP4 cells (Fig. S1A) were generated by infection of U2OS cells with purified ICP4 lentivirus as described in SI Materials and Methods. Puromycin (2 μg/mL)-resistant clones were isolated and screened by infection with ICP4-deficient virus (QOZHG) at a multiplicity of infection (MOI) of 0.5 before immunofluorescent staining for ICP4. U2OS-ICP4/27 cells were similarly generated by infection of U2OS-ICP4 cells with purified ICP27 lentivirus, selection for resistance to puromycin (2 μg/mL) and blasticidin (10 μg/mL), and screening of QOZHG-infected clones for ICP27 immunofluorescence. We tested the stability of the J∆NI5-complementing properties of the U2OS-ICP4/27 clone used in this study by plaque assay at different cell passages and found no significant decline in plaquing efficiency through at least 20 passages (Table S1).
Additional materials and methods are provided in SI Materials and Methods and Tables S2 and S3.
Supplementary Material
Acknowledgments
We are grateful to Hiroyuki Nakai for the pBluescriptUB-Flag-mArt plasmid; Klaus Osterrieder for plasmids pEPkan-S2 and pBAD-I-sceI; J. Thomson (Morgridge Institute) for pEP4-EO2SCK2M-EN2L (Addgene plasmid 20924); Greg Smith for Escherichia coli strain GS1783; David Leib for KOS37-BAC DNA; Tsuyoshi Akagi for pCX4hyg; Nobutaka Kiyokawa, Hajime Okita and Akihiro Umezawa for retroviral packaging plasmids; Patricia Spear for pPEP100; John Blaho for anti-ICP22; T. J. Ley (Washington University) for pTurbo-Cre; Thomas Caskey for pCCL-DMD; Johnny Huard and Bing Wang for hMDSCs and mdx muscle progenitor cells; and Mingdi Zhang for rat DRG culturing. This work was supported by NIH Grants NS064988 and DK044935, CHDI Foundation Grant A3777, and Commonwealth of Pennsylvania Grant SAP 4100061184 (to J.C.G.). P.M. was supported by an Atlante fellowship from the University of Ferrara, Italy, under the mentorship of Michele Simonato. G.V. was supported by a grant from the Italian Ministry for University and Research (PRIN 2010-11 2010N8PBAA) (to Michele Simonato).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423556112/-/DCSupplemental.
References
- 1.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 2.Allocca M, et al. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J Clin Invest. 2008;118(5):1955–1964. doi: 10.1172/JCI34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: Renewed optimism from genetic approaches. Nat Rev Genet. 2013;14(6):373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 4.Monahan PE, et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: A strategy for broad clinical application. Mol Ther. 2010;18(11):1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krisky DM, et al. Rapid method for construction of recombinant HSV gene transfer vectors. Gene Ther. 1997;4(10):1120–1125. doi: 10.1038/sj.gt.3300497. [DOI] [PubMed] [Google Scholar]
- 6.Krisky DM, et al. Development of herpes simplex virus replication-defective multigene vectors for combination gene therapy applications. Gene Ther. 1998;5(11):1517–1530. doi: 10.1038/sj.gt.3300755. [DOI] [PubMed] [Google Scholar]
- 7.Krisky DM, et al. Deletion of multiple immediate-early genes from herpes simplex virus reduces cytotoxicity and permits long-term gene expression in neurons. Gene Ther. 1998;5(12):1593–1603. doi: 10.1038/sj.gt.3300766. [DOI] [PubMed] [Google Scholar]
- 8.Palmer JA, et al. Development and optimization of herpes simplex virus vectors for multiple long-term gene delivery to the peripheral nervous system. J Virol. 2000;74(12):5604–5618. doi: 10.1128/jvi.74.12.5604-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preston CM, Mabbs R, Nicholl MJ. Construction and characterization of herpes simplex virus type 1 mutants with conditional defects in immediate early gene expression. Virology. 1997;229(1):228–239. doi: 10.1006/viro.1996.8424. [DOI] [PubMed] [Google Scholar]
- 10.Samaniego LA, Wu N, DeLuca NA. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71(6):4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu N, Watkins SC, Schaffer PA, DeLuca NA. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70(9):6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson SA, DeLuca NA. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc Natl Acad Sci USA. 2003;100(13):7871–7876. doi: 10.1073/pnas.1230643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72(4):3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terry-Allison T, Smith CA, DeLuca NA. Relaxed repression of herpes simplex virus type 1 genomes in Murine trigeminal neurons. J Virol. 2007;81(22):12394–12405. doi: 10.1128/JVI.01068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferenczy MW, DeLuca NA. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J Virol. 2011;85(7):3424–3435. doi: 10.1128/JVI.02263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferenczy MW, Ranayhossaini DJ, Deluca NA. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J Virol. 2011;85(10):4993–5002. doi: 10.1128/JVI.02265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craft AM, et al. Herpes simplex virus-mediated expression of Pax3 and MyoD in embryoid bodies results in lineage-Related alterations in gene expression profiles. Stem Cells. 2008;26(12):3119–3129. doi: 10.1634/stemcells.2008-0417. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs WE, 2nd, DeLuca NA. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J Virol. 1999;73(10):8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amelio AL, McAnany PK, Bloom DC. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol. 2006;80(5):2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40(2):191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 21.Yao F, Schaffer PA. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69(10):6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida H, et al. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol. 2010;84(23):12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Xp, et al. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J Virol. 2000;74(21):10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baines JD, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65(2):938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menotti L, Casadio R, Bertucci C, Lopez M, Campadelli-Fiume G. Substitution in the murine nectin1 receptor of a single conserved amino acid at a position distal from the herpes simplex virus gD binding site confers high-affinity binding to gD. J Virol. 2002;76(11):5463–5471. doi: 10.1128/JVI.76.11.5463-5471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goins WF, et al. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol. 1994;68(4):2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwaagstra JC, Ghiasi H, Nesburn AB, Wechsler SL. Identification of a major regulatory sequence in the latency associated transcript (LAT) promoter of herpes simplex virus type 1 (HSV-1) Virology. 1991;182(1):287–297. doi: 10.1016/0042-6822(91)90672-x. [DOI] [PubMed] [Google Scholar]
- 28.Berthomme H, Lokensgard J, Yang L, Margolis T, Feldman LT. Evidence for a bidirectional element located downstream from the herpes simplex virus type 1 latency-associated promoter that increases its activity during latency. J Virol. 2000;74(8):3613–3622. doi: 10.1128/jvi.74.8.3613-3622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloom DC, Giordani NV, Kwiatkowski DL. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta. 2010;1799(3-4):246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichavant C, et al. Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol Ther. 2011;19(5):830–840. doi: 10.1038/mt.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanfilippo CM, Blaho JA. ICP0 gene expression is a herpes simplex virus type 1 apoptotic trigger. J Virol. 2006;80(14):6810–6821. doi: 10.1128/JVI.00334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilley CE, et al. Multiple immediate-early gene-deficient herpes simplex virus vectors allowing efficient gene delivery to neurons in culture and widespread gene delivery to the central nervous system in vivo. J Virol. 2001;75(9):4343–4356. doi: 10.1128/JVI.75.9.4343-4356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Homer EG, Rinaldi A, Nicholl MJ, Preston CM. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J Virol. 1999;73(10):8512–8518. doi: 10.1128/jvi.73.10.8512-8518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston CM, Nicholl MJ. Human cytomegalovirus tegument protein pp71 directs long-term gene expression from quiescent herpes simplex virus genomes. J Virol. 2005;79(1):525–535. doi: 10.1128/JVI.79.1.525-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preston CM, Rinaldi A, Nicholl MJ. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J Gen Virol. 1998;79(Pt 1):117–124. doi: 10.1099/0022-1317-79-1-117. [DOI] [PubMed] [Google Scholar]
- 36.Lukashchuk V, McFarlane S, Everett RD, Preston CM. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J Virol. 2008;82(24):12543–12554. doi: 10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80(8):3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol. 2003;23(6):1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De La Fuente R, Baumann C, Viveiros MM. Role of ATRX in chromatin structure and function: Implications for chromosome instability and human disease. Reproduction. 2011;142(2):221–234. doi: 10.1530/REP-10-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14(23):3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14(23):3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett RD, Boutell C, McNair C, Grant L, Orr A. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J Virol. 2010;84(7):3476–3487. doi: 10.1128/JVI.02544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ertel MK, Cammarata AL, Hron RJ, Neumann DM. CTCF occupation of the herpes simplex virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner. J Virol. 2012;86(23):12741–12759. doi: 10.1128/JVI.01655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol. 2004;78(3):1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang QY, et al. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci USA. 2005;102(44):16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Recillas-Targa F, Valadez-Graham V, Farrell CM. Prospects and implications of using chromatin insulators in gene therapy and transgenesis. BioEssays. 2004;26(7):796–807. doi: 10.1002/bies.20059. [DOI] [PubMed] [Google Scholar]
- 47.French SW, Schmidt MC, Glorioso JC. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol Cell Biol. 1996;16(10):5393–5399. doi: 10.1128/mcb.16.10.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emery DW, Chen H, Li Q, Stamatoyannopoulos G. Development of a condensed locus control region cassette and testing in retrovirus vectors for A gamma-globin. Blood Cells Mol Dis. 1998;24(3):322–339. doi: 10.1006/bcmd.1998.0200. [DOI] [PubMed] [Google Scholar]
- 49.Sadelain M, Wang CH, Antoniou M, Grosveld F, Mulligan RC. Generation of a high-titer retroviral vector capable of expressing high levels of the human beta-globin gene. Proc Natl Acad Sci USA. 1995;92(15):6728–6732. doi: 10.1073/pnas.92.15.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harkness JM, Kader M, DeLuca NA. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J Virol. 2014;88(12):6847–6861. doi: 10.1128/JVI.00516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant KG, Krisky DM, Ataai MM, Glorioso JC., 3rd Engineering cell lines for production of replication defective HSV-1 gene therapy vectors. Biotechnol Bioeng. 2009;102(4):1087–1097. doi: 10.1002/bit.22123. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80(7):3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueki S, et al. Hepatic B7 homolog 1 expression is essential for controlling cold ischemia/reperfusion injury after mouse liver transplantation. Hepatology. 2011;54(1):216–228. doi: 10.1002/hep.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida O, et al. CD39 expression by hepatic myeloid dendritic cells attenuates inflammation in liver transplant ischemia-reperfusion injury in mice. Hepatology. 2013;58(6):2163–2175. doi: 10.1002/hep.26593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao X, et al. BMP2 is superior to BMP4 for promoting human muscle-derived stem cell-mediated bone regeneration in a critical-sized calvarial defect model. Cell Transplant. 2013;22(12):2393–2408. doi: 10.3727/096368912X658854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gharaibeh B, et al. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc. 2008;3(9):1501–1509. doi: 10.1038/nprot.2008.142. [DOI] [PubMed] [Google Scholar]
- 57.Gierasch WW, et al. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J Virol Methods. 2006;135(2):197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.