Significance
How organisms monitor external environmental signals and integrate these measurements into their development is a fundamental question. Here, we investigate this issue in the context of vernalization, the perception and memory of long-term cold temperature exposure in plants. We use a combination of mathematical modeling coupled with experiments on plants exposed to an interrupted cold temperature regime. We find that cold temperature exposure is likely to be registered in an all-or-nothing (digital) manner at the relevant gene FLOWERING LOCUS C. Such a mechanism allows for integration of noisy, interrupted temperature signals and for subsequent switching into a stable, digital epigenetic memory state. This analysis is a key step toward uncovering the biochemical elements behind long-term cold temperature monitoring.
Keywords: vernalization, digital, analog, temperature, FLC
Abstract
A fundamental property of many organisms is an ability to sense, evaluate, and respond to environmental signals. In some situations, generation of an appropriate response requires long-term information storage. A classic example is vernalization, where plants quantitatively sense long-term cold and epigenetically store this cold-exposure information to regulate flowering time. In Arabidopsis thaliana, stable epigenetic memory of cold is digital: following long-term cold exposure, cells respond autonomously in an all-or-nothing fashion, with the fraction of cells that stably silence the floral repressor FLOWERING LOCUS C (FLC) increasing with the cold exposure duration. However, during cold exposure itself it is unknown whether vernalizing cold is registered at FLC in individual cells in an all-or-nothing (digital) manner or is continuously varying (analog). Using mathematical modeling, we found that analog registration of cold temperature is problematic due to impaired analog-to-digital conversion into stable memory. This disadvantage is particularly acute when responding to short cold periods, but is absent when cold temperatures are registered digitally at FLC. We tested this prediction experimentally, exposing plants to short periods of cold interrupted with even shorter warm breaks. For FLC expression, we found that the system responds similarly to both interrupted and uninterrupted cold, arguing for a digital mechanism integrating long-term temperature exposure.
In many circumstances, organisms react to external environmental signals through altered gene expression programs (1–3). In some cases, these responses are maintained even without the originating signals. This outcome can be achieved through information storage in epigenetic memory (4–7). Such memory, whether implemented by cis- or trans-acting positive feedback loops, is typically digital, encoding an all-or-nothing response at a cellular level (6, 8). Such a digital mechanism permits reliable information storage even in the presence of large fluctuations in, e.g., protein concentration levels. However, although the eventual memory is digital, mechanisms for monitoring noisy external signals could be analog or digital, before their information content is stored in long-term digital memory. Here, we study constraints that might favor or disfavor digital vs. analog signal registration.
The best-characterized system in which an environmental signal is converted into quantitative, digital epigenetic memory is vernalization in Arabidopsis thaliana (9, 10). In vernalization, memory of exposure to long-term cold is epigenetically stored in a cell-autonomous process that involves histone modifications (specifically trimethylation at lysine 27 of histone H3, H3K27me3) and Polycomb complexes, at the floral repressor gene FLOWERING LOCUS C (FLC). An important phase of the process involves cold-dependent accumulation of H3K27me3 in a localized “nucleation” region (no more than two or three nucleosomes) within the transcribed region of FLC (10–12). H3K27me3 accumulation at the nucleation region (NR) therefore acts as a register of cold exposure duration. The occurrence of such an H3K27me3 peak then triggers spreading of H3K27me3, which increases across the entire FLC locus at the end of cold exposure, resulting in epigenetic maintenance of the silencing. According to FLC-GUS assays (12), this subsequent memory is both quantitative and digital, with the fraction of cells stably silenced via H3K27me3 determining the overall level of FLC expression, which quantitatively regulates flowering time (10, 12, 13). Importantly, the cold-dependent H3K27me3 NR accumulation is also quantitative, increasing with longer cold exposure. However, current measurements of FLC histone modification levels are population based and do not reveal the status of individual cells. Consequently, it is unclear whether H3K27me3 NR accumulation during cold temperature exposure is either an all-or-nothing response in each cell (digital) or a gradual increase in all cells (analog). Here, we exploit vernalization to probe how organisms register external signals, with the goal of distinguishing between analog and digital mechanisms for cold-induced H3K27me3 nucleation at FLC.
Using simulations and mathematical reasoning, we show that analog cold temperature registration is deficient due to inadequate analog-to-digital conversion for stable information storage. This difficulty is not present with digital cold temperature registration and is particularly problematic when responding to short cold periods. We investigate this effect experimentally by exposing plants to multiple short cold periods interspersed with even shorter warm interruptions. The observed response to interrupted vs. uninterrupted cold treatment is similar, providing strong evidence that cold is registered at FLC digitally rather than in an analog fashion, before digital epigenetic information storage. Nevertheless, we also elaborate how the observed integration of the temperature signal is still imperfect. This finding suggests a multistep cold-sensing mechanism that may help to generate appropriate responses to a noisy environment.
Results
H3K27me3 Nucleation at FLC Can Potentially Be Digital or Analog.
The quantitative nature of vernalization involves the epigenetic silencing of FLC in an increasing fraction of cells. Postcold FLC silencing and H3K27me3 accumulation over the whole FLC gene occur in a digital (all-or-nothing) cell-autonomous fashion, with increasing cold exposure increasing the fraction of cells with high levels of H3K27me3 over FLC. In our modeling we assume both alleles of FLC behave identically, both during and after cold exposure, coordinated by their physical clustering during cold exposure (14). Previous modeling of FLC involved a “tug of war” between opposing, mutually exclusive “M” (silencing) and “A” (activating) histone marks (12), with M representing H3K27me3. Furthermore, within the NR, the A mark has been shown to be consistent with H3K36me3 (15), although outside this region its identity is still unclear. Each modification is assumed to be self-reinforcing, recruiting factors that tend to add the same, but remove the opposing, mark (16), consistent with recent data (15). This feedback ensures robust bistability with the system tending to be locked in either an M- or an A-rich state. We implemented these dynamics in a stochastic Monte Carlo simulation, which keeps track of the number of histones modified in the M or A states or not modified (“U”). Effects of DNA replication are incorporated, as well as nucleosome swap out and noisy addition/removal of marks (details in SI Text).
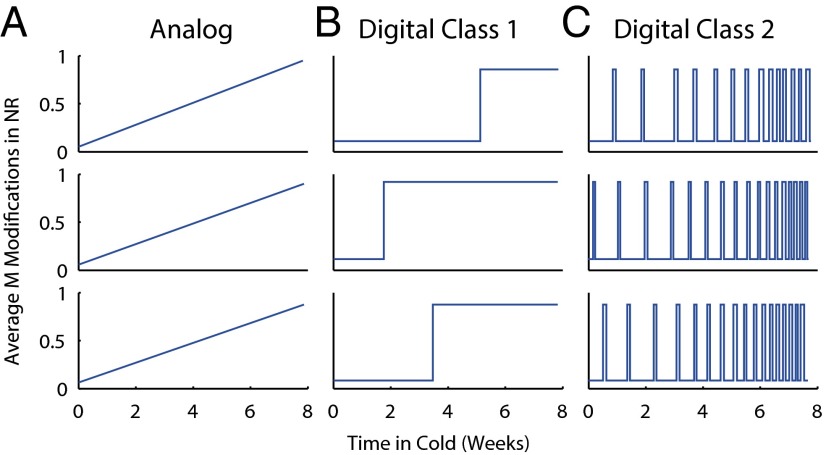
During cold exposure itself, H3K27me3 accumulates at a localized NR in FLC, registering the duration of cold exposure. Consequently, the H3K27me3 NR peak provides a cold-dependent “push” toward the silenced state, as the peak causes the addition of H3K27me3 across the locus. Self-reinforcement can cause the entire locus to become covered with H3K27me3, enforcing a switch to an epigenetically silenced state. This switch occurs at the end of cold exposure with unknown factors preventing H3K27me3 spreading during the cold. Note that, for FLC, the switching element (NR) is physically contained within the downstream bistable object. However, this physical arrangement is not important for our conclusions. Conceptually, there are two ways in which a cold-induced H3K27me3 peak can arise (Fig. 1), analog or digital.
Fig. 1.
(A–C) Schematic plots of NR peak profile shown for three representative cells in the population for (A) analog, (B) digital class 1, and (C) digital class 2 models as a function of cold exposure duration.
Analog.
Here, H3K27me3 nucleation accumulates essentially equally in all cells, with amounts increasing over time during cold exposure (Fig. 1A). Due to the fast and noisy dynamics of histones and their modifications, a straightforward way to induce an analog, slowly rising H3K27me3 NR peak is to slowly change the probabilities per unit time of histone modification addition/removal as a function of cold exposure duration.
Digital.
In a digital system, the H3K27me3 NR peak is an all-or-nothing phenomenon. However, there are two ways in which this digital peak can be realized. In class 1, more cells stochastically but irreversibly acquire this peak as the duration of cold exposure increases (Fig. 1B). This could occur through the probability per unit time of H3K27me3 addition at the NR irreversibly undergoing a sudden, large increase at a random time during the cold. Hence, the H3K27me3 modifications would be present for an overwhelming fraction of the time in nucleating cells (digital: on) but only for a small fraction of time in nonnucleating cells (digital: off). Due to the stochastic nature of the switch, the fraction of cells exhibiting a digital NR peak will increase with increasing cold duration. However, due to the long time periods (months) over which vernalizing cold is registered, the probability per second of irreversible switching must be very low (on the order of 10−7 s−1) to prevent premature saturation. In class 2, the digital peak itself appears and disappears stochastically within a cell with a certain frequency. This is due to the probabilities per unit time of histone modification addition/removal changing stochastically back and forth from values inducing digital high or low levels of H3K27me3 nucleation, with the fraction of time the digital peak is present in a cell increasing as the duration of cold exposure increases (Fig. 1C).
The H3K27me3 peak is believed to be responsible for switching the epigenetic state of the locus from active to silenced with high H3K27me3 across the whole locus at the end of cold exposure (12). In an analog model, cells switch with a probability that positively correlates with the H3K27me3 NR peak height. In a digital class 1 model, only cells that have stochastically acquired a digital H3K27me3 NR peak during cold can switch (with high probability). In a digital class 2 model, cells switch with a probability that positively correlates with the fraction of time the digital NR peak is present. In this study, we restrict ourselves to studying each model separately, although in principle the different mechanisms could be combined.
Model of H3K27me3 Dynamics Reveals Difficulties with an Analog Mechanism.
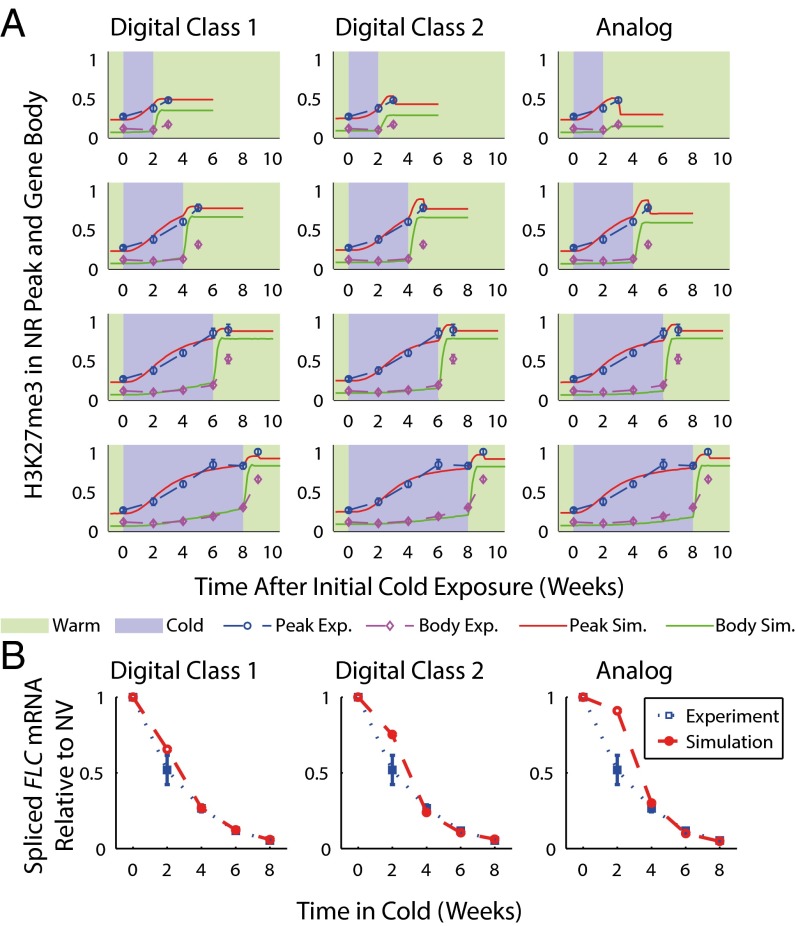
To study the issue of digital vs. analog nucleation, we adapted our original model of epigenetic memory in vernalization (12) to function with either analog or digital temperature-registering nucleation (SI Text). The models were parameterized using new and existing cell population-level ChIP (12, 15) and FLC expression data (Fig. 2), with our previous parameterization as a starting point (SI Text and Table S1). Using a χ2 test statistic, all of the models formally provided a satisfactory fit to the data, due to the relatively large SDs, although with the digital models providing an overall superior fit (Fig. 2). However, considering the SE of the mean (SEM), the analog model performed poorly for the expression level at 2 wk cold (plus a 7-d period of warm), compared with both digital models, and poorly in the ChIP at 4 wk cold (plus a 7-d period of warm), compared with the digital class 1 model. Using the SEM, we found the probabilities that the true 2-wk mean would be as far, or farther from, the experimental relative expression data mean as in the analog or digital models, using a one-tailed t test and approximating simulated data as having zero error. These probabilities were 0.003 (analog), 0.1 (digital, class 1), and 0.02 (digital, class 2). We also found similar probabilities for the ChIP at 4 wk cold (plus a 7-d period of warm), giving 0.04 (analog), 0.37 (digital, class 1), and 0.02 (digital, class 2). As the analog and digital class 1 models have equal numbers of parameters (with the digital class 2 model having one more) (Table S1), the analog model is disfavored, especially compared with the digital class 1 model.
Fig. 2.
(A) Experimentally measured H3K27me3 levels in NR (peak), and elsewhere across FLC (body), during and 7 d after cold for uninterrupted cold periods of 2 wk, 4 wk, 6 wk, and 8 wk. Also shown are simulated H3K27me3 levels for digital and analog models. (B) Experimentally measured levels of spliced FLC mRNA (expression) 7 d after cold relative to nonvernalized (NV), i.e., with no exposure to cold, values for uninterrupted cold periods of 2 wk, 4 wk, 6 wk, and 8 wk. Also shown are simulated relative mRNA levels for digital and analog models. Experimental error bars are SEM, with n = 5 for body H3K27me3 levels; n = 7, 7, 9, 5, and 8 for peak levels immediately after 0 wk, 2 wk, 4 wk, 6 wk, and 8 wk cold; n = 7, 8, 5, and 7 for peak levels following 2 wk, 4 wk, 6 wk, and 8 wk cold and a 7-d period of warm; and with n = 8, 13, 10, and 8 for relative expression 7 d after 2 wk, 4 wk, 6 wk, and 8 wk cold, respectively.
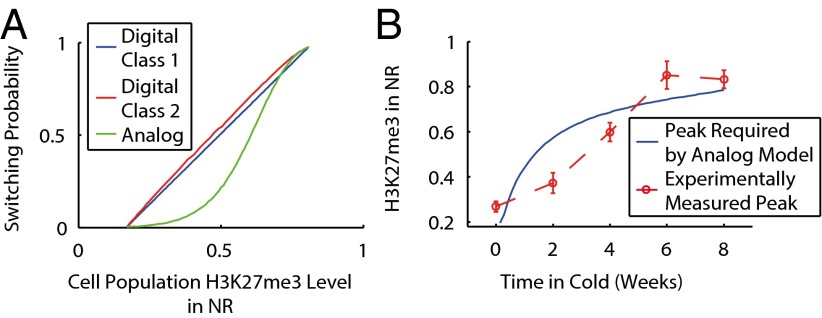
From model simulations (SI Text), we then extracted the relationships between the probability for each cell to switch to a silenced state at the end of cold exposure and the cell population level of H3K27me3 in the NR. For the digital class 1 model, this relationship was almost linear, with each cell that digitally nucleates subsequently having a high probability (∼0.98) to switch into a silenced state at the end of the cold period (Fig. 3A). For the digital class 2 model, the relationship was also linear. Interestingly, for the analog version of the model, we found a sigmoidal form that could not be significantly altered by reparameterization. As a result, low levels of nucleating H3K27me3 from short periods of cold exposure could only inefficiently induce a switch (Fig. 3A). This observation implies that vernalization with analog but not digital temperature-registering nucleation is insensitive to short cold periods, in agreement with the above poor fit of the analog model to our 2-wk cold exposure expression data. Note that this sigmoidal form of the switching probability as a function of cell population nucleating H3K27me3 levels is distinct from the rise of nucleating H3K27me3 as a function of cold duration, which is also, but separately, sigmoidal (Fig. 2A).
Fig. 3.
(A) Probability that the cell switches to the silenced state following cold as a function of population-averaged H3K27me3 NR peak level for digital and analog models. Plots start at the basal level of H3K27me3 present in all simulated nonvernalized cells in the nucleation region and end at maximum H3K27me3 nucleation level. (B) Experimental H3K27me3 NR peak profile as function of cold exposure duration (red dashed line and data points, as in Fig. 2). Solid blue line is simulated H3K27me3 NR peak profile, making the analog model as effective as the digital class 1 model.
Conceptually, the reason for the insensitivity of the analog model to short periods of cold exposure is as follows. A short period of cold gives rise only to a relatively small analog H3K27me3 NR peak in each cell. The self-reinforcing nature of the histone modifications then means that such a small NR peak is ineffective in switching the state of the locus to one covered by silencing H3K27me3 modifications; otherwise switching would occur frequently due to noise. Hence, for short periods of cold, analog-to-digital conversion of information about cold temperature exposure is ineffective. Digital nucleation, with an all-or-nothing H3K27me3 NR peak inside each cell, is not affected by this issue. Intuitively, to make the analog model as effective in switching after a short period of cold as the digital models, the NR peak would have to rise rapidly for short cold periods before slowing. To test this reasoning, we constructed the nucleation profile that would be required to make the analog model perform as well as the digital class 1 model in fitting to the expression data (SI Text). We found that the necessary profile is as described above, i.e., the opposite of that observed experimentally, where the (cell population averaged) NR peak rises slowly with time for short cold exposures before accelerating (Fig. 3B). For example, performing the same statistical analysis as above, we found a probability <0.01 that the true 2-wk ChIP mean would be as far, or farther from, the experimental ChIP data mean as in the refitted analog model. Accordingly, this analysis again does not support an analog nucleation hypothesis.
In addition to the problem of weak switching from a small NR peak in the analog model, there is a second difficulty. Experiments show that the cell population NR peak level rises slowly with cold exposure duration (Figs. 2A and 3B), before accelerating. Both analog and digital models therefore have a reduced switching probability for short cold exposure periods. However, the analog model will perform worse for short cold periods as weak nucleation is compounded by weak switching.
Plants Respond Similarly to Both Interrupted and Uninterrupted Cold, Favoring a Digital Mechanism.
Our data have so far favored digital rather than analog nucleation. To further test this hypothesis we subjected Arabidopsis plants to multiple short cold periods broken up by even shorter warm interruptions. The results of these experiments were compared with uninterrupted cold treatments with the same overall duration of cold. This protocol was designed to reveal any weakness in responding to short interrupted periods of cold, as expected with an analog mechanism. Through the use of such warm interruptions, the differences found previously between the analog and digital mechanisms for short periods of cold exposure should be amplified, compared with equivalent uninterrupted cold treatments. These experiments were also able to probe how well the FLC system could cope with variable long-term temperature profiles more similar to those found in natural conditions.
We exposed Arabidopsis plants to cold treatments including one, two, and three interruptions: For one interruption, two periods of 7 d, 14 d, or 21 d cold were compared against 14 d, 28 d, and 42 d of uninterrupted cold; for two interruptions, three periods of 11 d, 14 d, or 18 d of cold were compared against 33 d, 42 d, and 54 d of uninterrupted cold; for three interruptions, four periods of 7 d cold were compared with 28 d of uninterrupted cold. The warm interruptions were of 4 d duration, as this period was sufficient to allow for a switch from nucleation to silencing with high levels of H3K27me3 across the FLC locus (12). Plants subjected to interrupted cold treatment experienced additional days of warm compared with plants given continuous cold, due to the warm interruptions. Accordingly, plants subjected to continuous cold were, at the end of cold treatment, grown in the warm for a period such that the total warm exposure duration was the same for the corresponding uninterrupted vs. interrupted cold experiments. FLC spliced mRNA levels relative to those without cold exposure were determined by quantitative RT-PCR. In interrupted treatments, these measurements were taken 7 d after the end of cold exposure. In uninterrupted treatments, measurements were taken following cold exposure after 7 d plus the summed duration of the warm interruptions in the corresponding interrupted treatment.
Strikingly, the vernalization response was similar in both interrupted and uninterrupted cold treatments (Fig. 4). On average, we found interrupted cold generated spliced mRNA levels only 1.5 ± 0.2 (SEM) times that of the equivalent uninterrupted cold. Compared to our simulations, this fold change was closer to that of the digital class 1 [1.9 ± 0.02 (SEM)] and class 2 [2.5 ± 0.03 (SEM)] models, rather than to that of the analog model [4.0 ± 0.07 (SEM)] (Fig. 4 A–C). For reasons that are unclear, however, even the digital models underestimated the degree of FLC silencing relative to that in nonvernalized conditions in all cases.
Fig. 4.

(A) Postcold spliced mRNA relative to nonvernalized (NV) levels from analog (an) and digital models (both classes, d1 and d2), compared with experiments (ex). Uninterrupted cold is compared with (Left) one, (Center) two, and (Right) three interruptions; e.g., 2 × 7 d refers to two periods of 7 d cold duration, with a warm interruption in between, compared with 14 d of uninterrupted cold. Error bars are SEM, with n = 3 (Left) and n = 2 (Center and Right) for experiments and n = 10 for simulations. (B) Leaf number as a measurement of flowering time, for two and three interruption treatments. Box and whisker plots show median, 25th, and 75th percentiles and range of data excluding any outliers, which are represented as individual points; n = 12 for each set of measurements.
To confirm our results, we also measured leaf number as a quantitative measure of flowering time, with a higher leaf number indicating delayed flowering. Interrupted cold treatments of three periods of 14 d and four periods of 7 d were compared with 42 d and 28 d of uninterrupted cold, respectively. These results showed only a 1.3 ± 0.1 (SEM) fold increase in leaf number when comparing interrupted cold to the equivalent period of uninterrupted cold (Fig. 4D). Overall, these results demonstrate that plants respond similarly to both interrupted and uninterrupted cold. This finding supports the hypothesis that quantitative, digital memory of FLC expression is written via quantitative, digital H3K27me3 nucleation.
An Improved Integration over the Temperature Signal Is Possible but Is Not Implemented in Arabidopsis.
The above results demonstrate that Arabidopsis is able to integrate over an interrupted cold temperature signal. However, as illustrated by measurements of spliced mRNA, the response to interrupted cold is still slightly reduced compared with uninterrupted cold exposure. mRNA levels differed by 1.5-fold between interrupted and uninterrupted cold treatments (P < 0.05 paired-sample one-tailed t test on the natural logarithm of the ratios). Our experiments on leaf number also showed a similar trend, although it is difficult to be quantitative due to the uncertain effect of warm interruptions during cold exposure on leaf number. This finding is intriguing because, theoretically, it is straightforward in the digital nucleation case to achieve an improved integration over interrupted cold, with an identical response to uninterrupted cold, as we now describe.
Using a digital class 1 model, we assume that a fraction f of cells acquire a digital nucleation peak by a time t after the beginning of cold exposure, with cold registration beginning immediately once cold exposure starts. Assuming that all cells with a digital NR peak switch to the silenced state at the end of a cold period, then the fractional silencing after a single cold period will be f(t). We require that two cold periods interrupted by a short warm period should lead to an identical vernalization response to an equal total duration of uninterrupted cold. Defining the fraction of unsilenced cells g(t) = 1 − f(t), this reasoning leads to g(t1 + t2) = g(t1) g(t2). Assuming continuity, the unique solution is g(t) = exp(−αt) for constant α and hence f(t) = 1 − exp(−αt). To satisfy the boundary condition f(t) → 1 as t → ∞, we require α > 0. This corresponds to a one-step, stochastic, irreversible Poisson switching process in individual cells.
From the above, we assume our experimental H3K27me3 data reflect the fraction of cells with a digital NR peak. Generalizing, we now allow for some cells to be silenced before cold exposure, as well as for a small NR peak in cells that have not responded to cold (SI Text). We then fit to the cell population H3K27me3 nucleation and expression levels (Fig. S1), with relatively poor agreement. For example, a one-tailed t test as before on the 2-wk ChIP data yielded a probability <0.01 that the true 2-wk ChIP mean would be as far, or farther from, the experimental ChIP data mean as in the perfect buffering model. This result accords with our experiments showing a weakened response to interrupted cold, with an interrupted to uninterrupted spliced mRNA ratio of 1.5.
Digital Memory Imposes Generic Constraints on the Registration of Information.
The above analysis has exposed a deficiency in analog temperature-registering nucleation in our current model, with poor fits to the data. However, there could be specific features of this model that cause this behavior, features that might not be present in other potential models for epigenetic switching/memory. In particular, the above model assumes cis-acting digital epigenetic memory with self-sustaining histone modifications. However, other forms of memory, for example trans-acting via a protein with strong positive transcriptional feedback, are also possible in principle. To elucidate the generality of our results, we therefore introduce a simple conceptual model. The model represents a generic bistable system with controllable cold-induced switching from one state to the other. Here, we use the conceptual model to compare the analog model specifically with the digital class 1 model, which is sufficient to expose the conceptual difficulties of an analog mechanism.
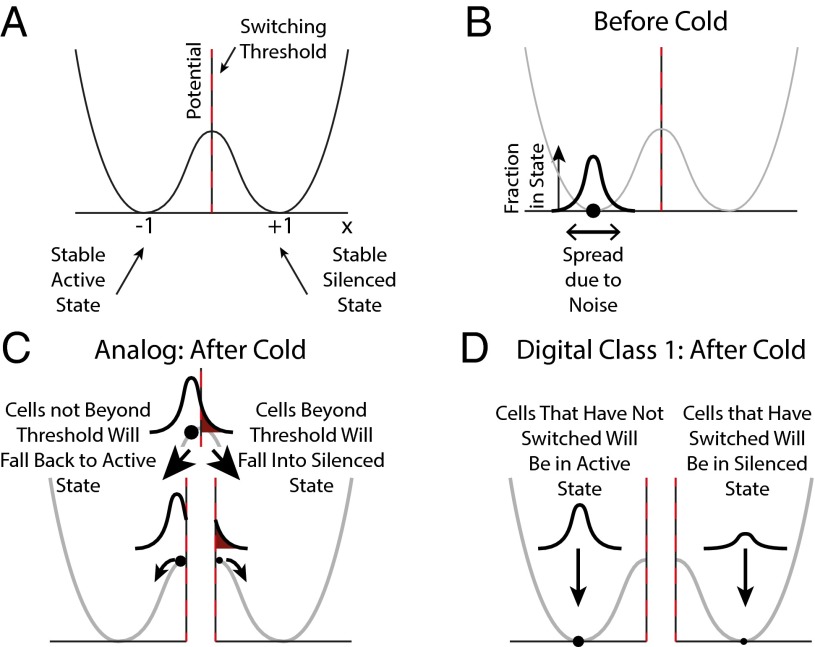
The model uses a variable x that represents the system expression state. A stable, active (silenced) transcriptional state corresponds to values around −1 (+1), respectively. To stabilize these steady states, we assume the existence of a symmetric double-well potential (Fig. 5A). The dynamics of the system can be illustrated pictorially: When placed in a certain state (value of x), the system will (for x ≠ 0) “roll” downward until it reaches one of the minima at x = ±1. To switch from one state to another, it is necessary to move the system state x past the local maximum at x = 0, after which the system will relax back to the other state. At the start of cold exposure, x takes a value around −1 in most cells, consistent with an active state (Fig. 5B). During cold exposure, cold temperature registration occurs, such that at the end of cold exposure, the population level of x has increased. If at this time x > 0 in a cell, then a switch occurs to a stable silenced state around x = 1 (Fig. 5 C and D).
Fig. 5.
Conceptual model for switching in a symmetric bistable system. (A) Potential landscape showing two stable expression states (x = ±1), with switching threshold (x = 0). (B) Configuration before cold exposure, with distribution of states around the most stable value. (C) In the analog model, at end of the cold period, cells corresponding to the part of the distribution beyond threshold (dark red) “roll” down to the silenced state. (D) In the digital class 1 model, cells that have digitally nucleated (not nucleated) will be distributed around x = 1 (x = −1), respectively.
The model imposes a potential landscape with two stable states. However, the mechanism that generates this landscape is unspecified. Hence, our conclusions should be applicable regardless of how the states are stabilized (e.g., through cis-acting self-reinforcing histone marks or trans-acting transcriptional feedback). For a discussion linking the conceptual model with our specific cis-acting histone-based model, see SI Text.
So far, the conceptual model is deterministic whereas biological systems are noisy. Thus, we assume that there is a distribution of system states described by x around its deterministic value. At the end of the cold the part of the distribution at x > 0 will switch to the silenced state. Hence, the overall switching probability is given by the fraction of the distribution at the end of cold exposure that lies at x > 0. In the analog case, at the end of cold exposure, a cold-induced increase in x will have occurred in all cells with the fraction of the distribution lying at x > 0 positively correlated with cold exposure duration. Hence, the fraction of cells that switch depends on the duration of cold exposure (Fig. 5C). In the digital class 1 case, near-perfect switching occurs stochastically but irreversibly in individual cells, as the value of x shifts from near −1 to near +1 (Fig. 5D). Hence, it is the number of cells that digitally register cold that determines how many enter the silenced state after cold exposure. Thus, both the analog and digital models can produce quantitative increases in silencing with increasing cold exposure duration.
However, as in the detailed model discussed previously, the analog implementation suffers from an issue not present in the digital version. Due to the potential landscape and distribution of system states, the analog model switching probability behaves differently for different cold exposure durations. At short (and long) durations, the switching probability increases more slowly with increasing cold duration, whereas for intermediate durations, it increases more rapidly. This means that few cells will switch for short cold periods in the analog system, and there is a region of saturation after long cold periods. In the former case this problem arises because, for short cold periods, few cells acquire a large enough change in x to carry them over the maximum at x = 0 and hence cannot switch. Although the conceptual model is simple, this feature is general for any system exhibiting bistability and that must be controllably switched from one state to another. The bistability requirement necessitates a potential landscape as in Fig. 5A, ensuring two stable states. As a result, small analog changes (here, short cold periods) will be unable to induce appropriately efficient switching. Hence, any analog model will fail to respond properly to small changes. The digital class 1 model clearly does not suffer from this deficiency.
Overall, we conclude that analog cold registration is problematic when confronted with short periods of cold. This difficulty arises from an inability to convert small analog changes into stable digital memory, regardless of whether this memory is held in cis or in trans. The cold-induced switching element that sets up the appropriate fraction of postcold digitally silenced cells should therefore also be digital. Hence, the conclusions reached for our detailed model are generic and should moreover apply to any system that needs to digitally store quantitative information.
Discussion
The mechanism of epigenetic memory storage is poorly understood and intensely debated (17, 18). Even less is known about how epigenetic memory states are switched in response to external signals. Here, we find evidence that long-term cold temperature registration at FLC during vernalization is digital. Such a mechanism can effectively integrate over interruptions to cold exposure due to robust switching of FLC into a stable, digital epigenetic memory state.
The possibility of digital temperature monitoring has not been generally considered, but provides insight into the identification of the relevant molecular components. However, H3K27me3 nucleation need not be the direct temperature-perceiving element. Rather, digital nucleation of H3K27me3 could be directed by upstream temperature sensor(s). For example, temperature could be sensed by discrete change(s) to a protein or RNA structure (19, 20). Such change(s) could be cold-temperature sensitive by occurring at much higher probabilities at low as opposed to high temperatures. One consequence of such a temperature-dependent change could be the digital, cell-autonomous induction of the plant homeodomain protein VIN3, whose expression is known to increase quantitatively with cold exposure duration (21). In a digital class 1 model, this switch could then induce a stable, digital H3K27me3 NR peak, which subsequently mediates switching of the FLC locus to high levels of H3K27me3 coverage at the end of cold exposure. In principle an analog component to vernalizing cold temperature response is also possible. For example, a protein concentration level could continuously increase with cold exposure duration. Such an altered protein concentration could then, in a digital class 2 model, alter the fraction of time during which a digital NR peak is present at FLC. Nevertheless, as our results demonstrate, regardless of prior analog circuitry, the NR switching element for FLC that sets up the appropriate fraction of postcold silenced cells is likely to be digital. We emphasize, however, that ambient temperature sensing, without a need for subsequent digital storage, will not be subject to the same constraints.
Overall, we find that the digital class 1 model fits our data better than the class 2 model (Fig. 4 A–C), with both classes being superior to the analog model. Moreover, there are additional theoretical reasons to favor the class 1 over the class 2 digital model. With digital class 1 H3K27me3 nucleation, it is more straightforward to obtain the appropriate switching than in either the class 2 model or, especially, the analog model. If, in a digital class 1 model, a very strong H3K27me3 signal is imposed in the NR, near-perfect switching will result due to the strength of the change, with the self-reinforcing H3K27me3 signal spreading over the entire locus. In this case, there is substantial leeway in the height of the H3K27me3 NR peak with which this can be achieved. By contrast, in the analog mechanism, the height of the H3K27me3 NR peak is necessarily critical and fluctuations in its level will compromise the quantitative vernalization response. Moreover, the digital class 1 model cannot suffer from the problem of weak switching from a small H3K27me3 NR peak. For the digital class 2 model, however, the results are sensitive to the residence time of the nucleating on state, not just to the fraction of time it is present. The residence time must be of a similar duration to the time it takes at the end of the cold to complete a switch to a fully silenced state with H3K27me3 covering the locus. If the residence time is too long, the system will switch too effectively from a NR peak present for only a small fraction of the time. This is because, although the NR peak is rarely present, when the peak does appear, it resides for an extended period, allowing efficient switching. In this case, the response of the system will saturate too soon in the cold. However, if the residence time is too short, switching will be ineffective from NR peaks present for a small fraction of the time. In either case, the outcome is a nonlinear response similar to that in the analog model (Fig. 3A). These properties make the digital class 1 model need less fine-tuning and make it more robust to parameter variations. In addition, the digital class 1 model has one fewer parameters than the class 2 model, although fitting better (Fig. 4 A–C). Such a digital model may also perform better than analog models when confronted with circadian variation in temperature.
Our experiments in Arabidopsis have shown that plants can integrate over interruptions in long-term cold exposure. As plants in the field are exposed to a highly fluctuating temperature environment (22), this ability is probably an important component driving adaptation. Nevertheless, the response to interrupted cold is not the same as the response to uninterrupted cold. Our data are consistent with the evolution of a strategy ensuring plants do not respond too quickly to short periods of cold, thus requiring a full winter and reducing the risk of premature flowering after a short, cold autumn. This strategy also ensures reliable flowering after a winter interrupted by frequent warm interludes.
Interestingly, within our favored digital class 1 model, a perfect interruption-buffering system is a Poisson nucleation process with a constant probability per unit time in the cold of digitally and irreversibly inducing nucleation. However, the lessened response we have found for short cold periods indicates that such a system is not implemented in Arabidopsis. Within a digital class 1 model, the probability per unit time of digitally nucleating early on during cold exposure is reduced, before later recovering. This could be achieved by one or more stochastic, digital switches before digital nucleation at FLC can occur, i.e., a multistep cold-sensing mechanism. In this case, the probability of digital class 1 nucleation at FLC would be suppressed for short cold periods. It will be interesting to see whether this or other similar possibilities are borne out by future experiments.
Finally, we emphasize that our conclusions are not restricted to vernalization or even to cases with Polycomb-based epigenetic memory, but will apply to any biological situation where quantitative information is digitally stored. Analog switching elements will have limitations with low-level signals due to inherent limitations in subsequent analog-to-digital conversion. Digital switching elements, which do not suffer from this deficiency, may therefore be more widespread.
Materials and Methods
Plant Material and Growth Conditions.
The Columbia line FRI-Sf2 was described previously (23). Sterilized seeds were sown on MS media (no glucose), stratified at 4 °C for 2 d, and pregrown for 7 d in warm conditions (20 °C, 16 h light). Seedlings were transferred to 4 °C under short-day conditions (8 h light) for appropriate cold treatments. T0 seedlings were harvested immediately after cold treatment, whereas T7 seedlings were harvested after a further 7 d or 10 d of growth on MS media (no glucose) following transfer back to the warm condition (16 h light). Nonvernalized control seedlings continued to grow in the warm condition after pregrowth and were harvested for expression analysis at between 3 d and 15 d. For flowering-time recording, plants were pricked out to grow on Arabidopsis compost until bolting. The total numbers of leaves (rosette and cauline) were recorded to indicate flowering time.
Real-Time Quantitative PCR and ChIP Analysis.
cDNA was synthesized using SuperScript III (Invitrogen) with a mixture of oligo d(T) and analyzed by quantitative PCR on a LightCycler 480 II instrument (Roche), using the Universal Probe Library assay (UPL) (Roche). FLC transcripts (5′-gctacttgaacttgtggatagcaa-3′ and 5′-ggagagggcagtctcaaggt-3′, UPL probe 65) were normalized against the Arabidopsis UBC gene (At5g25760, 5′-tcctcttaactgcgactcagg-3′ and 5′-gcgaggcgtgtatacatttg-3′, UPL probe 9). ChIP assays were performed as previously described (12). Tables S2 and S3 contain details of primers used for ChIP and samples taken, respectively. SHOOT MERISTEMLESS (STM) was used as the internal control for the ChIP experiments. H3K27me3 data are the ratio (H3K27me3 FLC/H3 FLC)/(H3K27me3 STM/H3 STM).
Supplementary Material
Acknowledgments
We thank Robert Ietswaart, Scott Berry, and members of the C. Dean group for discussions. This work was supported by The Royal Society (A.A.), by Grant BB/J004588/1 from the Biotechnology and Biological Sciences Research Council (to C.D. and M.H.), and by the European Research Council Grant ENVGENE (to C.D. and M.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503100112/-/DCSupplemental.
References
- 1.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9(8):583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 2.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12(12):833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 3.Franklin KA, Toledo-Ortiz G, Pyott DE, Halliday KJ. Interaction of light and temperature signalling. J Exp Bot. 2014;65(11):2859–2871. doi: 10.1093/jxb/eru059. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. 2010;22(3):284–290. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11(4):285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15(5):340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 7.Steffen PA, Fonseca JP, Ringrose L. Epigenetics meets mathematics: Towards a quantitative understanding of chromatin biology. BioEssays. 2012;34(10):901–913. doi: 10.1002/bies.201200076. [DOI] [PubMed] [Google Scholar]
- 8.Casadesús J, Low DA. Programmed heterogeneity: Epigenetic mechanisms in bacteria. J Biol Chem. 2013;288(20):13929–13935. doi: 10.1074/jbc.R113.472274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61(6):1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 10.Song J, Angel A, Howard M, Dean C. Vernalization - a cold-induced epigenetic switch. J Cell Sci. 2012;125(Pt 16):3723–3731. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 11.Finnegan EJ, Dennis ES. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol. 2007;17(22):1978–1983. doi: 10.1016/j.cub.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 13.Satake A, Iwasa Y. A stochastic model of chromatin modification: Cell population coding of winter memory in plants. J Theor Biol. 2012;302(0):6–17. doi: 10.1016/j.jtbi.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Rosa S, et al. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013;27(17):1845–1850. doi: 10.1101/gad.221713.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Howard M, Dean C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol. 2014;24(15):1793–1797. doi: 10.1016/j.cub.2014.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129(4):813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 17.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conaway JW. Introduction to theme “Chromatin, epigenetics, and transcription”. Annu Rev Biochem. 2012;81:61–64. doi: 10.1146/annurev-biochem-090711-093103. [DOI] [PubMed] [Google Scholar]
- 19.Storz G. An RNA thermometer. Genes Dev. 1999;13(6):633–636. doi: 10.1101/gad.13.6.633. [DOI] [PubMed] [Google Scholar]
- 20.Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev. 2006;30(1):3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 21.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427(6970):159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 22.Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc Natl Acad Sci USA. 2010;107(25):11632–11637. doi: 10.1073/pnas.0914293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I, et al. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6(6):903–909. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.