Abstract
The discovery of cytosolic RNA granule (RG) component proteins associated with human cataract has initiated investigations on post-transcriptional mechanisms of gene expression control in the lens. Application of established mouse lens epithelial cell lines (LECs) can provide rapid insights on RG function in lens cells, especially because mouse mutants in several RG components are not available. However, although these LECs represent potential reagents for such analyses, they are uncharacterized for lens gene expression or RG formation. Therefore, a detailed molecular and cellular characterization of three permanent mouse LECs 17EM15, 21EM15 and αTN4 is performed in this study. Comparative analysis between microarray gene expression datasets on LEC 21EM15 and iSyTE lens tissue demonstrates that 30% of top 200 iSyTE identified lens-enriched genes are expressed in these cells. Majority of these candidates are independently validated to either have lens expression, function or linkage to cataract. Moreover, analysis of microarray data with genes described in Cat-Map, an online database of cataract associated genes and loci, demonstrates that 131 genes linked to cataract loci are expressed in 21EM15 cells. Furthermore, gene expression in LECs is compared to isolated lens epithelium or fiber cells by qRT-PCR and by comparative analyses with publically available epithelium or fiber-specific microarray and RNA-seq (sequencing) datasets. Expression of select candidate genes was validated by regular and real-time quantitative RT-PCR. Expression of lens epithelium-enriched genes Foxe3, Pax6, Anxa4 and Mcm4 is up-regulated in LEC lines, compared to isolated lens fiber cells. Moreover, similar to isolated lens epithelium, all three LECs exhibit down-regulation of fiber cell-expressed genes Crybb1, Mip and Prox1 when compared to fiber cells. These data indicate that the LEC lines exhibit greater similarity to lens epithelium than to fiber cells. Compared to non-lens cell line NIH3T3, LECs exhibit significantly enriched expression of transcription factors with important function in the lens, namely Pax6, Foxe3 and Prox1. In addition to these genes, all three LECs also express key lens- and cataract-associated genes, namely Dkk3, Epha2, Hsf4, Jag1, Mab21l1, Meis1, Pknox1, Pou2f1, Sfrp1, Sparc, Tdrd7 and Trpm3. Additionally, 21EM15 microarrays indicate expression of Chmp4b, Cryab and Tcfap2a among others important genes. Immunostaining with makers for Processing bodies (P-bodies) and Stress granules (SGs) demonstrates that these classes of RGs are robustly expressed in all three LECs. Moreover, under conditions of stress, 17EM15 and αTN4 exhibit significantly higher numbers of P-bodies and SGs compared to NIH3T3 cells. In sum, these data indicate that mouse LECs 21EM15, 17EM15 and αTN4 express key lens or cataract genes, are similar to lens epithelium than fiber cells, and exhibit high levels of P-bodies and SGs, indicating their suitability for investigating gene expression control and RG function in lens-derived cells.
Keywords: Lens, Cataract genes, 21EM15, 17EM15, αTN4, Stress granules, Processing body, iSyTE
It has recently been demonstrated that alterations of cytoplasmic RNA granule (RG) components can cause human eye disorders, including pediatric cataract and glaucoma, as well as other degenerative diseases (Lachke et al., 2011; Ramaswami et al., 2013; Wolozin, 2014). RGs are dynamic ribonucleoprotein (RNP) complexes present in the cytoplasm of eukaryotic cells that are implicated in the regulation of various aspects of mRNA control, including mRNA stability, degradation, intracellular localization and translation into protein (Adjibade and Mazroui, 2014; Anderson and Kedersha, 2009; Buchan, 2014; Lachke and Maas, 2011; Moore, 2005). Eukaryotic cells exhibit at least two classes of RGs, namely, Processing bodies (P-bodies) and stress granules (SGs) (Eulalio et al., 2007; Kedersha et al., 2013; Parker and Sheth, 2007; Sheth and Parker, 2003) whereas metazoan cells may also harbor additional RGs like transport RNP particles or germ cell-specific granules (GCGs) (de Mateo and Sassone-Corsi, 2014; Fritzsche et al., 2013; Gao and Arkov, 2013; Kiebler and Bassell, 2006; Lachke et al., 2011). Therefore, study of RGs in a cell or tissue-specific environment represents a critical first step in understanding their specialized function in post-transcriptional regulation of gene expression.
P-bodies are cytoplasmic ribonucleoprotein complexes that harbor mRNAs that are recruited away from active translation and need to be temporarily stored or degraded (Anderson and Kedersha, 2009). P-bodies associate with the molecular machinery involved in translation repression as well as mRNA decay, such as non-sense mediated decay (NMD), micro (mi) RNA-mediated silencing or decay, and Xrn1-mediated 5′ to 3′ degradation (Anderson and Kedersha, 2009; Eulalio et al., 2007). P-bodies are endogenously found in cells, but can also be stimulated to increase in numbers as a response to stress (Parker and Sheth, 2007). SGs on the other hand, assemble in conditions of stress, and are cytoplasmic aggregates of stalled translation preinitiation complexes (Buchan and Parker, 2009; Kedersha et al., 2013). SGs contain 48S preinitiation complexes as the core components, and also include small ribosomal subunits as well as the early translation initiation factors eIF2, eIF3, eIF4E, and eIF4G (Kedersha et al., 2013; Kedersha and Anderson, 2002). SGs can interact with P-bodies resulting in exchange of mRNAs that are then directed to translational re-initiation or degradation (Kedersha et al., 2005).
Recently it was demonstrated that a novel RG component Tudor domain containing 7 protein (Tdrd7) exhibits a highly enriched expression pattern in vertebrate lens development and its deficiency in human, mouse and chicken causes cataracts (Lachke et al., 2011). Interestingly, Tdrd7 is found to differentially associate with other granule proteins like Stau1 and Dcp1a, a P-body marker, in lens fiber cells. This finding has led to a new interest in investigating post-transcriptional control mechanisms in the lens. However, due to their overall importance to cellular regulation, it is expected that deleting core RG components such as Dcp1a and Ddx6 would cause embryonic lethality in mice. Furthermore, conditional null mouse mutant alleles for these genes or many other RG components are not currently available. Mouse lens epithelial cell lines 17EM15, 21EM15, and αTN4, offer an alternative reagent for these investigations, but their suitability for studying RG-mediated post-transcriptional control is currently undetermined. Moreover, although a number of studies have utilized LEC lines to gain insights into lens biology and disease, few reports have described their detailed characterization or their potential to support lens gene expression (Carper et al., 2001; Dave et al., 2012; Donner et al., 2007; Haque et al., 1999; Krausz et al., 1996; Lachke et al., 2011; Liu et al., 2008; Nakajima et al., 2006; Reddan et al., 1989; Rich et al., 1999; Rowan et al., 2008; Russell et al., 1990; Spector et al., 2002; Tanaka et al., 2004; Yamada et al., 1990). Therefore, we aimed to investigate these mouse LECs for their expression of lens marker genes and their capacity to support formation of distinct RGs.
In this study, we present the characterization of three LECs 17EM15, 21EM15 and αTN4 in comparison to the mouse fibroblast cell line NIH3T3 as well as isolated lens epithelium and fiber cells. Through comparative gene expression analysis as well as regular and quantitative RT-PCR, we find that these LECs exhibit robust expression of several key lens genes. These analyses also indicate that the three LECs are similar to isolated lens epithelium expression compared to lens fiber cells. In addition, these data demonstrate that the LECs express many other genes with lens function as recognized by the online databases iSyTE and Cat-Map, and provide a systematic catalog of their expression levels. Finally, in light of our recent identification of RG components associated with cataract, we present evidence that these LECs support formation of robust levels of P-bodies and SGs, and therefore are suitable for studies on RG-mediated post-transcriptional control of gene expression.
METHODS
Mouse Husbandry
Mice were bred and maintained at the University of Delaware Animal Facility adhering to the ARVO Statement for the use of animals in ophthalmic and vision research. Wild type ICR outbred mice were obtained from Taconic (Hudson, New York) and used for immunostaining analysis. Mice were housed in a 14 hour light to 10 hour dark cycle. Embryos were staged by designating the day that the vaginal plug was observed in the dam as E0.5.
Cell Culture
The mouse LECs 17EM15 and 21EM15 were a generous gift of Dr. John Reddan (Oakland University, Michigan) who originally developed these lines (Reddan et al., 1989). The mouse LEC αTN4, with confirmed original source from Dr. Paul Russell’s laboratory (Yamada et al., 1990), was obtained from Dr. Richard Maas (Brigham and Women’s Hospital and Harvard Medical School, Massachusetts). The mouse fibroblast cell line NIH3T3, with confirmed original source, was obtained from Dr. Gary Laverty (University of Delaware, Delaware). All four cell lines were cultured in 100 mm cell culture treated plates (Thermo Scientific, Waltham, MA; 130182), 10 mL of: DMEM with 4.5 g/L glucose, L-glutamine, and sodium pyruvate included (Corning Cellgro, Manassas, VA; 10-013-CV), 10% Fetal Bovine Serum (Fisher Scientific, Pittsburg, PA; 03-600-511), and 1% penicillin-streptomycin (GE Healthcare Life Sciences, Logan, UT; SV30010). The cells were grown at 37°C, and water saturated atmosphere with 5% CO2. These cells grow well in these conditions and usually are 80% confluent after three days in culture (after 10% seeding). Cells were passaged three times, and grown to 60% or 80% confluence for immunofluorescence or RNA isolation, respectively.
Cell Line Authentication
Genomic DNA was extracted from cell lines using the Gentra Puregene DNA kit (Qiagen, Venlo, Netherlands). Primers were chosen for authentication based on murine and human short tandem repeats (STRs) within their respective genomes as recommended (Almeida et al., 2014). The two human primers D4S2408 and D8S1106 are abbreviated to HD4S and HD8S respectively within this publication. PCR amplification was performed using the Qiagen PCR kit (20 μl ddH2O, 2.5μl 10x Coral Red Buffer, 0.5 μl 10μM dNTP, 0.5 μl of each 10 μM primers, 0.5 μl template cDNA, 0.5 μl taq polymerase). PCR products were amplified using the following program: 95°C for 3 minutes, 94°C for 45 seconds, 59°C for 2 minutes, 72°C for 1 minute, cycled 30 times, final extension at 72°C for 10 minutes. PCR products were size separated on a 1% agarose gel and imaged with UVP GelDoc-It 310 Imager (Upland, California).
Whole-Genome Datasets Analyses
Microarray datasets for LEC 21EM15 cells infected with lentiviral preparations containing GFP and considered as control were generated in a previous study (Lachke et al., 2011) were obtained from GEO (GSM634229, GSM634230 and GSM634231). Analysis of these datasets was performed under ‘R’ Statistical environment [http://www.r-project.org/index.html]. Raw expression datasets (Illumina MouseWG-6 v2.0 chip) were imported and background corrected using lumi (Du et al., 2008) package available within Bioconductor [http://www.bioconductor.org/], followed by normalization using Rank invariant method included in lumi (Du et al., 2008). Three independent biological replicates were considered in these analysis and probes with detection p-value ≤ 0.05 in at least two replicates were considered significantly present and were used to reduce probe-level experiment to gene-level by selecting probes with highest median expression for a gene. Selected candidates of significantly expressed genes from normalized LEC 21EM15 microarray datasets were validated by RT-PCR. Further, expression of select genes in LEC 21EM15 microarrays was compared to isolated lens epithelium and lens fiber cells microarrays and RNA-seq datasets reported in previous studies (Hoang et al., 2014; Nakahara et al., 2007). Briefly, due to platform differences (microarrays on different chips, microarrays vs RNA-seq), we analyzed the epithelium vs fiber cells in each dataset to first identify a signature set of genes that represents each cell type and then compared it with gene expression in 21EM15 cells. These datasets were obtained from NCBI’s GEO (GSE7533) and SRA (SRP040480).
iSyTE-based Lens Gene Expression Analysis
We recently developed a bioinformatics tool called as iSyTE – integrated Systems Tool for Eye gene discovery (Lachke et al., 2012b), which has contributed to the characterization of several important candidates associated with lens development and cataract. Significantly expressed genes from (please see whole-genome dataset analysis for details on significance) LEC 21EM15 microarray datasets were searched in iSyTE data using in-house scripts to identify candidates within top 200 iSyTE enrichment minranks.
Immunostaining Analysis
For immunostaining analysis on cultured cells, cells were grown in 16 well Thermo Scientific Nunc™ Lab-Tek™ glass chamber slides with 200 μl of media (reported above), and seeded at 8.0 × 103 and incubated for approximately 24 hr to reach 60% confluence. Cells were stressed with 0.5 mM aresenite for 45 minutes at 37°C and 5% CO2, washed in 1X PBS, fixed in 4% paraformaldehyde at room temperature for 15 minutes and permeabilized with cold methanol for 10 minutes. Cells were then blocked in 5% chicken serum (Abcam, Cambridge, Massachusetts) in 1X PBS containing 0.3% TritonX100 for 1 hr at room temperature and incubated with primary antibody overnight at 4°C.
For immunostaining analysis on lens tissue, mouse embryonic head tissue embedded in OCT was cryosectioned into 12 μm thick coronal sections after fixation in 4% PFA on ice for 30 min and treatment with 30% sucrose for 4 hr. Similar to immunostaining of cultured cells, chicken serum (Abcam) (5% solution in 1X PBS and 0.3% TritonX100) was used for blocking and antibody dilution. Sections were incubated with primary antibody overnight at 4°C. The appropriate chicken anti-primary antibody conjugated with either Alexa 488 or Alexa 594 fluorophores (Invitrogen, Grand Island, New York) was used at a concentration of 1:500 for 2 hr at room temperature.
To establish the identity of a subset of lens RGs as P-bodies, experiments were performed with the translation elongation inhibitor cycloheximide (CHX) (Sigma-Aldrich, Saint Louis, Missouri), treatment of which is known to disassemble P-bodies (Kedersha and Anderson, 2007). Briefly, mice pregnant with E12.5 embryos were subjected to intra-peritonial injection with 150 μg/gm body weight of CHX solution in DMSO (Sigma-Aldrich) or DMSO only (control), and sacrificed after 4 hr 30 min. Embryonic head tissues were dissected and processed for immunofluorescence analysis of lens sections as described above. Commercial antibodies were obtained from the following sources and used at the indicated dilutions: eIF3η antibody (sc16377, 1:200; Santa Cruz Biotechnology, Dallas, Texas), Dcp1a antibody (H00055802-M06, 1:600; Novus Biochemicals, Littleton, Colorado) and Ddx6 antibody (ab54611, 1:800; Abcam). Images were obtained by confocal microscopy using a Zeiss model 780 instrument (Zeiss, Jena, Germany) and optical sections in the range of 0.6 – 1.0 μm. Three biological replicates were stained for the P-bodies for each time-point in these experiments. Digitized images were processed with Adobe Photoshop.
RNA Granule Quantification
Cells were immunostained with P-body (Ddx6) and SG (eIF3η) specific antibody markers and these RGs were counted under normal and stress (arsenite treated) conditions. At least three microscopic fields on each slide at 40x magnification were using for quantification. Independent RG counts from two researchers were averaged. The difference in RG counts under non-stress and stress conditions was calculated for individual cell lines and between cell lines. P-values were estimated using Student’s t-test.
RT-PCR Analysis
Total RNA was extracted from isolated lens epithelium or fiber cells from P60 stage mouse lens as described, as well as from lens epithelial cell lines 17EM15, 21EM15 and αTN4, and the fibroblast cell line NIH3T3 using the RNeasy Mini Kit (Qiagen). For isolation of epithelium or fiber cells, mice were euthanized, eye balls were removed and immersed in 1X PBS (Fisher Scientific, Hampton, New Hampshire) at room temperature. Lenses were dissected out using a light microscope. Finally the capsule was peeled to separate the lens fibers from the lens epithelium, which is firmly adherent to the lens capsule creating capsular bags. Capsular bags from 5 lenses were pooled per biological replicate to obtain enough tissue for RNA extraction. Similarly lens fibers from 5 lenses were pooled to produce each biological replicate. An iScript cDNA Synthesis Kit (Bio-Rad, Hercules, California) was used for cDNA synthesis followed by both regular PCR as well as real time quantitative PCR as described previously (Lachke et al., 2011). For regular (semi-quantitative) RT-PCR, cDNA was PCR amplified using the Qiagen PCR kit and the following program: 94°C for 2 minutes, 94°C for 15 seconds, 57°C for 30 seconds, 72°C for 30 seconds, cycled 45 times, final extension at 72°C for 7 minutes. PCR products were size separated on a 1% agarose gel and imaged with UVP GelDoc-It 310 Imager (Upland, California). Primer sequences are provided in Table 1. qPCR was performed using a LowRox SYBR green kit and analysis on a 7500 Fast PCR system (Applied Biosystems, Foster City, California). Statistical analysis was performed using a two-level nested analysis of variance to determine the significance of the fold change.
Table 1.
Primers used for RT-PCR analysis.
| Gene Name | Forward Primer | Reverse Primer | Expected Product Size |
|---|---|---|---|
| Actb | AGCCATGTACGTAGCCATCC | CTCTCAGCTGTGGTGGTGAA | 228 |
| Akt2 | TGGCTGGAAAAGGCGGTATT | GCTCGTTCCCGCTCCTTATT | 189 |
| Aldh1a1 | TGCAGGGGCAGCCATCTCCT | TTCCCCCAGCGTCCTCCACC | 447 |
| Anxa2 | CTTCAAGGGAGGCTCTCAGC | TTTGACTGACCCGTAGGCAC | 139 |
| Bach2 | TGCTGGCCGCATGCAGTGAA | ATGGTCCTCTTGGGGGCGCT | 308 |
| Bcl2l13 | TTCGGTGTGATGTACCTGGA | ACGGTAGGGACCTGTGTGAG | 461 |
| Bfsp1 | GTCCTGCAGCAGATCGTACA | TGCTGCCTTCAATCTCAATG | 388 |
| Ccng1 | AATGGCCTCAGAATGACTGC | AGTCGCTTTCACAGCCAAAT | 201 |
| Cdh1 | CGGAGAGGAGAGTCGAAGTG | CATGCTCAGCGTCTTCTCTG | 201 |
| Clu | TGGCCCTCTGGGAGGAGTGC | TGTGGAAGCTCGGAGGCCCA | 405 |
| Cp | ATTTTCAACGGGCTGATGAC | ACTTTCTCGGGTTCAGAGCA | 313 |
| Cpeb1 | GGGCGTTGGGTCCCGAATGG | CCTGAAGCAAGGCCCGGACG | 452 |
| Cryba1 | GGAAACTCTTCCAACCACCA | CCACTGGCGTCCAATAAAGT | 388 |
| Cryba4 | GAAGGCTTCCAGGGCCGACG | GCCCACTTCAGTGCCGTCCC | 363 |
| Crybb1 | CTTTGAGCAATCTGCCTTCC | GTGCCACCAGAGACGGTTAT | 267 |
| Dhx32 | AAGCCTGGCGACTGACGTGC | TGACCACCAGGTCGCCCACA | 356 |
| Dkk3 | TGTGTACACTGCTGGCGGCG | GGTGCAGTGACCCCAGGCAC | 563 |
| Dnajb1 | CCCATATAGCCACCTGCACT | CTGCCTATCGCTCGAAAAAC | 196 |
| Elp2 (Statip1) | TTGTGTCTGGAGCAGACGAG | CAAAGGGCTGAGAGGCTATG | 201 |
| Epha2 | GCCAGTTTAGCCACCACAAT | CACTTCCCACATGACAATGC | 428 |
| Foxe3 | GAAGCCGCCCTACTCATACA | AGGAAGCTACCGTTGTCGAA | 273 |
| Gprc5b | AGTTCAAACGGTGGAAGGTG | CCCAGGCTGCTAGATCTTTG | 418 |
| Gprc5c | TCAGCCCAGTCCTGCTTAGT | GAAGGACCAAGCATCCTCAG | 201 |
| Hmox1 | CACGCATATACCCGCTACCT | TGTGCTTGACCTCAGGTGTC | 199 |
| Hs2st1 | GAGAAGCCCTGTCTGTGTCC | ATTCGCTCCAAAAAGCGATA | 202 |
| Hsf4 | CTTTGTTCGCCAACTCAACA | GTAGGGGTCCTGGAGGAGAG | 508 |
| Inppl1 | GTGGAGAGGAGACAGGCAAC | GGAGGTCTCTGTGGAAGCTG | 200 |
| Jag1 | TAGCTGCCTGCCGAACCCCT | GCTGGAGGCTGGAGGACCGA | 590 |
| Mab21l1 | TGACACGAGCGAAGTGAAAC | TGGGTTGGTCAGGATCTCTC | 601 |
| Man1c1 | CTCGGCCTTCTACCTGACTG | AACTGTCCCCAAGTCCTCCT | 368 |
| Mcm4 | TGCTGCAAAAGGAGTCTGAA | TGGGCTTAGGCTCTCTGTGT | 202 |
| Meis1 | TCTGCACTCGCATCAGTACC | GTTGTCCAAGCCATCACCTT | 628 |
| Meis2 | GCGGTCTTCGCCAAGCAGGT | TGCCTGCGGAGTGCGTTGAG | 325 |
| Mip | GGAAACCTAGCGCTCAACAC | CCATTGGAGTCACTGGGTCT | 401 |
| Myo6 | AGGAGCTGGCAAAACAGAAA | ATTTCCAAGGTGCAGGACAC | 589 |
| Pax6 | AGTTCTTCGCAACCTGGCTA | ACTTGGACGGGAACTGACAC | 846 |
| Pdpn (Gp38) | TCTACTGGCAAGGCACCTCT | CGTTTCATCCCCTGCATTAT | 198 |
| Pitx3 | GAGGAATCGCTACCCTGACA | ATAGCTGAAAGAGGCGTGCT | 592 |
| Pknox1 | AGGGGTGTCCTGCCGAAGCA | GCACCAGCCCGCTGATGTGT | 499 |
| Pou2f1 | GCCTCGGCCTCCACCTCAGA | GGAGGCTGCAGCGGCAGAAA | 507 |
| Prox1 | CTGGGCCAATTATCACCAGT | GCCATCTTCAAAAGCTCGTC | 205 |
| Ptprk | GTGCCATAGGCATTGTTGTG | ATGGATTTCTGCTTGGATGC | 202 |
| Pvrl3 | TTGCCCTTTCCTTTGTCAAC | TGCATGTCTGATGGTGGAAT | 191 |
| Rab36 | GGCTCAGACTCTCCAAGGTG | GCTGGGCCTCTTCTCTAGGT | 545 |
| Sfrp1 | GGACCGGCCCATCTACCCGT | GTGGCAGGGACAGTCGGCAC | 412 |
| Slc3a2 | TCTTCACTCTGCCAGGGACT | GCCACAAAGGGGAACTGTAA | 526 |
| Smyd2 | GTGGAAGTCCGAAAGCTCAG | TGAGGGAGTACACGGGGTAG | 310 |
| Sparc | AGAATTTGAGGACGGTGCAG | AAGTGGCAGGAAGAGTCGAA | 216 |
| Sulf1 | TGTGCCTTTCTTCATTCGTG | TTTGGCCTTCTTGTTTGTCC | 199 |
| Tdrd7 | CTAAGGGCTGTCCTGCAGTC | TGAGAGTTGCCTTTGGCTTT | 340 |
| Trim8 | GACGTGGAGATACGGAGGAA | CGATCTTAGGGGGAGAAAGG | 436 |
| Trpm3 | GGGTCGCCAGGCAAGCCATT | CCGGCAGCACACATGCTGGA | 461 |
| Zfp365 | AAGTCCGGGCAGCCTTTGCG | GCATCCCGGCTCGCTCATCC | 486 |
| Zwint | TGGCGGACGCGGAGAAAAACG | TCCGGCGCCTCTTGACCTCT | 549 |
RESULTS
Authentication of Mouse LECs
For this study, we selected three established mouse LEC lines that have been used in lens research (Carper et al., 2001; Donner et al., 2007; Haque et al., 1999; Krausz et al., 1996; Lachke et al., 2011; Nakajima et al., 2006; Reddan et al., 1989; Rich et al., 1999; Rowan et al., 2008; Russell et al., 1990; Spector et al., 2002; Tanaka et al., 2004; Yamada et al., 1990). Recognizing the difficulties in data interpretation that result due to misidentification of cell lines, the eye community requires that before a cell line is proposed for use in an ocular study, its identity needs to be firmly established by molecular characterization (Beebe, 2013). Therefore, we obtained the mouse LECs 17EM15 and 21EM15 directly from Dr. John Reddan, who originally developed these lines (Reddan et al., 1989). In addition, we obtained αTN4 and fibroblast cell line NIH3T3 with confirmed original sources from experts in the field Drs. Richard Maas and Gary Laverty, respectively. To further determine the authenticity of the cell lines as recommended by the eye research community, we used mouse and human-specific primer sets designed to detect STRs within the respective genomes as previously described (Almeida et al., 2014). As expected, all four cell lines exhibit amplification of mouse STR region-specific PCR products, while no PCR product with the human genome-specific STR primer sets is detected (Figure 1). We note that for the αTN4 line, although amplification of the STR 5-5 marker produced a weak product (Figure 1), it was nevertheless consistently detected in several biological replicates with independent DNA preparations. Although these cell lines have not been karyotyped in detail, RT-PCR gene expression and in particular microarrays on 21EM15 indicate chromosome-wide expression of all mouse genes. This data confirms that all the LECs as well as the NIH3T3 cells used in this study were derived from mouse.
Figure 1. Authentication of lens epithelial cell lines using mouse and human specific STRs.
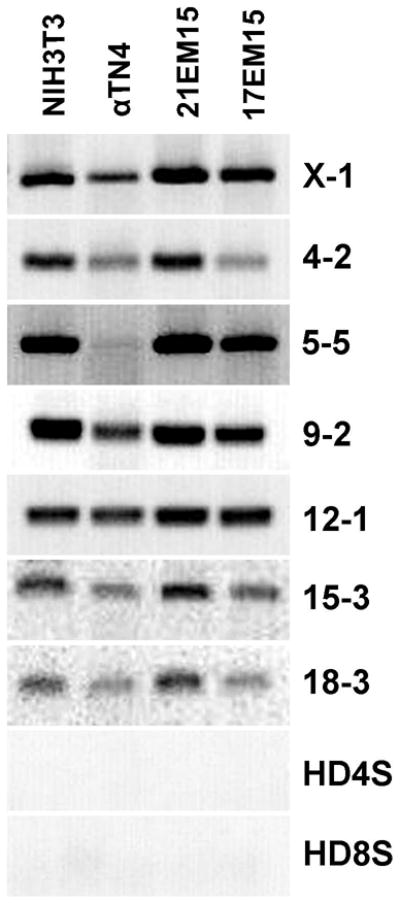
To validate their species identity, PCR on genomic DNA from individual cell lines was used to test the presence or absence of mouse or human-specific short tandem repeats (STRs) as previously recommended (Almeida et al., 2014). Mouse-specific PCR products are indicated by their chromosomal location (X-1 through 18-3). As expected, none of the cell lines amplify human STRs (HD4S – human D4S2408, HD8S – human D8S1106), but exhibit mouse-specific STR amplification, confirming their mouse origin.
Expression Profiling of Mouse LEC 21EM15
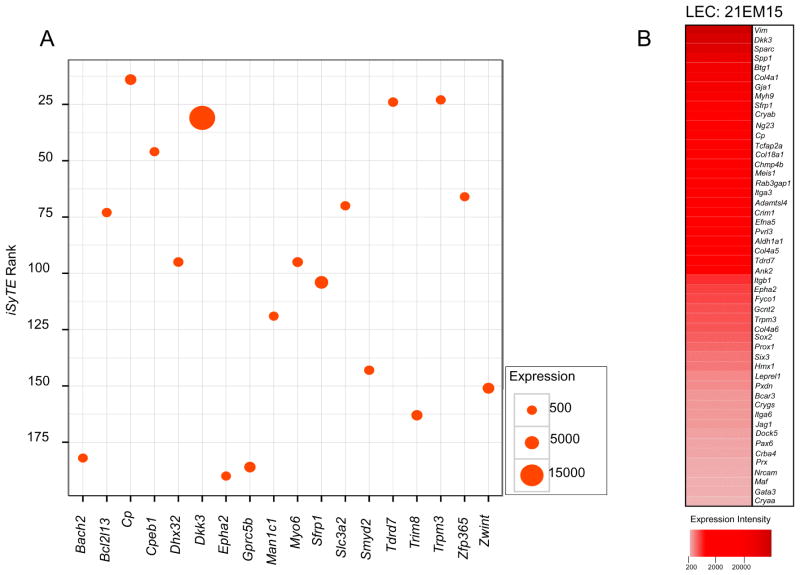
We first sought to gain insight into the gene expression profile of a mouse LEC, 21EM15, for which microarray expression datasets were already available in GEO. We first compared 21EM15 expression data with lens gene expression data in iSyTE, an effective tool we recently developed for lens gene discovery (Lachke et al., 2012b), and identified that ~80% of the top 200 minRank iSyTE genes were significantly expressed in this cell line. This gene list was further interrogated through Cat-map, OMIM and literature-based analysis to identify association of candidate genes with cataract, or function and/or expression in the lens. Through this analysis, we identified 67 genes (~30%) that have established lens function and/or are linked to cataracts, and were significantly expressed in 21EM15 cells (see supplementary Table S1 for list of candidate genes). A scatter plot of select candidate genes from this list serves to indicate their relative expression in 21EM15 cells (Figure 2A).
Figure 2. iSyTE-identified lens-enriched genes and cataract-associated genes in Cat-Map are expressed in the LEC 21EM15.
(A) Scatter plot of select genes within the top 200 minrank in iSyTE indicates expression of several key lens genes in LEC 21EM15 microarrays. The y-axis indicates iSyTE enrichment-based rank. Size of filled circles represents normalized expression levels of genes. (B) Heat map of select genes that exhibit significant expression in LEC 21EM15 microarrays and are either identified as iSyTE enriched genes, have a known function in the lens or found in Cat-Map. Visualization of the candidates as a heat map allows comparative analyses between their relative expression levels in these cells.
Next we analyzed 21EM15 expression data with genes described in Cat-Map, an online database of cataract associated genes and loci (Shiels et al., 2010). At an expression cut-off of 185 fluorescence intensity units, 131 candidate Cat-Map genes are detected as significantly expressed in 21EM15 cells (see supplementary Table S2 for list of candidate genes).
Furthermore, a list of 50 important genes, which are significantly expressed in 21EM15 cells and identified based on iSyTE or Cat-Map filters, is provided in Table 2. Within this list, it is evident that 38% (n=19) candidates represent genes whose functional compromise is directly causative of human cataract. Mouse mutants for 40% (n=20) of candidates where the gene is rendered non-functional exhibit lens defects and cataract, while 7 other candidate genes exhibit enriched or dynamic expression pattern in the lens (Table 2). A heat-map based on Table 2 allows visualization of the candidate genes for comparative analysis between their relative expression levels in 21EM15 cells (Figure 2B). These data demonstrates highly significant expression of several genes important to lens biology, including but not limited to Dkk3, Cryab, Chmp4b, Col4a1, Col5a5, Crim1, Efna5, Epha2, Fyco1, Gcnt2, Pvrl3, Sparc, Spp1, Sfrp1, Tcfap2a, Tdrd7, Trpm3 and Vim. Thus, 21EM15 microarray analysis indicates that these cells exhibit high levels of expression for several important candidate genes that are associated with lens biology and cataract, or are known to be expressed in the lens.
Table 2.
Microarrays-based expression of key lens and cataract genes in LEC 21EM15.
| Gene | Expression | Cat-Map | Association with Cataract and/or Function, Expression in the Lens | Citation |
|---|---|---|---|---|
| Adamtsl4 | 1154 | Yes | Autosomal recessive isolated ectopia lentis and cataract | (Ahram et al., 2009) |
| Aldh1a1 | 824 | - | Essential for lens transparency in human, rat and mouse | (Choudhary et al., 2005) |
| Ank2 | 642 | - | Mouse mutants exhibit lens fiber cell defects and cataract | (Moré et al., 2001) |
| Bcar3 | 217 | - | Mouse mutants exhibit lens defects | (Kamaid and Giráldez, 2008; Near et al., 2009) |
| Btg1 | 7675 | - | Expressed in the lens | (Kamaid and Giráldez, 2008) |
| Chmp4b | 1450 | Yes | Autosomal dominant progressive pediatric posterior subcapsular cataract | (Shiels et al., 2007) |
| Col18a1 | 1608 | Yes | Lens abnormalities; Col18a1:Hspg2 double mutant mice exhibit lens defects | (Menzel et al., 2004; Rossi et al., 2003) |
| Col4a1 | 6207 | Yes | Mouse mutants exhibit vacuolar cataract | (Van Agtmael et al., 2005) |
| Col4a5 | 775 | Yes | Dominant X-linked congenital cataract | (Antignac et al., 1992) |
| Col4a6 | 310 | - | Expressed in the early mouse lens development | (Bai et al., 2009) |
| Cp | 2120 | - | Expressed and secreted by lens epithelial cells in culture | (Harned et al., 2006) |
| Crim1 | 1047 | - | Mouse mutants exhibit smaller lens | (Pennisi et al., 2007) |
| Cryaa | 185 | Yes | Autosomal dominant congenital cataract | (Litt et al., 1998) |
| Cryab | 3594 | Yes | Autosomal dominant congenital cataract | (Berry et al., 2001) |
| Cryba4 | 201 | Yes | Autosomal dominant cataract and microphthalmia | (Billingsley et al., 2006) |
| Crygs | 216 | Yes | Autosomal dominant progressive cortical cataract | (Sun et al., 2005) |
| Dkk3 | 18542 | - | Expressed in early mouse eye and lens development | (Ang et al., 2004) |
| Dock5 | 204 | - | Mouse mutants exhibit lens rupture and cataract | (Omi et al., 2008) |
| Efna5 | 1014 | Yes | Mouse mutants exhibit cataract | (Cooper et al., 2008) |
| Epha2 | 345 | Yes | Autosomal dominant posterior polar cataract | (Shiels et al., 2008) |
| Fyco1 | 334 | Yes | Autosomal recessive congenital cataracts | (Chen et al., 2011) |
| Gata3 | 189 | Yes | Mouse mutants exhibit lens defects | (Maeda et al., 2009) |
| Gcnt2 | 318 | Yes | Autosomal recessive congenital cataracts, adult i phenotype | (Yu et al., 2001) |
| Gja1 | 5930 | Yes | Cataract and other ocular defects | (Paznekas et al., 2003) |
| Hmx1 | 257 | Yes | Cataract and other defects; and mouse mutants exhibit microphthalmia | (Munroe et al., 2009; Schorderet et al., 2008) |
| Itga3 | 1260 | - | Mouse mutants double null for Itga3 and Itga6 exhibit lens defects | (De Arcangelis et al., 1999) |
| Itga6 | 213 | - | Mouse mutants exhibit lens fiber cell differentiation defects | (Basu et al., 2014) |
| Itgb1 | 379 | Yes | Mouse mutants exhibit loss of lens epithelial cells | (Simirskii et al., 2007) |
| Jag1 | 213 | - | Mouse mutants exhibit lens fiber cell differentiation defects | (Le et al., 2012) |
| Leprel1 | 234 | - | Nonsyndromic myopia and cataract | (Mordechai et al., 2011) |
| Maf | 190 | Yes | Autosomal dominant juvenile onset cataract | (Jamieson et al., 2002) |
| Meis1 | 1347 | Yes | Regulates Pax6 in lens; and mouse mutants exhibit eye defects | (Hisa et al., 2004; Zhang et al., 2002) |
| Myh9 | 5094 | Yes | Cataract and other defects; mouse mutants exhibit lens defects | (Kunishima et al., 2001; Suzuki et al., 2013) |
| Ng23 | 2985 | - | Enriched expression in mouse lens development | (Lachke et al., 2012b) |
| Nrcam | 192 | - | Mouse mutants exhibit lens defects | (Moré et al., 2001) |
| Pax6 | 202 | Yes | Congenital cataract, aniridia and corneal dystrophy; and induces ectopic lens | (Altmann et al., 1997; Glaser et al., 1994) |
| Prox1 | 279 | Yes | Mouse mutants exhibit lens fiber cell elongation defects | (Wigle et al., 1999) |
| Prx | 194 | Yes | Required for lens fiber cell membrane organization | (Maddala et al., 2011) |
| Pvrl3 | 907 | Yes | Misregulation of Pvrl3 is associated with congenital cataract | (Weinstein et al., 1982) |
| Pxdn | 229 | Yes | Recessive congenital cataract | (Khan et al., 2011) |
| Rab3gap1 | 1347 | Yes | Cataract and other phenotypes | (Handley et al., 2013) |
| Sfrp1 | 3708 | - | Expressed in lens development | (Chen et al., 2004) |
| Six3 | 260 | Yes | Functions in mouse lens development; and induces ectopic lens | (Liu et al., 2006) |
| Sox2 | 293 | Yes | Functions in mouse lens development; and mutations cause anophthalmia | (Donner et al., 2007; Fantes et al., 2003) |
| Sparc | 14679 | Yes | Mouse mutants exhibit severe cataract and lens defects | (Gilmour et al., 1998) |
| Spp1 | 10706 | - | ECM component in postoperative capsular opacification, expressed by LECs on injury | (Saika et al., 2003) |
| Tcfap2a | 1675 | - | Mouse null mutants exhibit lens defects | (Pontoriero et al., 2008) |
| Tdrd7 | 696 | Yes | Deficiency causes cataract in human, mouse, chicken | (Lachke et al., 2011; Tanaka et al., 2011) |
| Trpm3 | 312 | - | Congenital cataract and glaucoma | (Bennett et al., 2014) |
| Vim | 24701 | Yes | Heterozygous G596A mutation causes pulverulent cataract | (Müller et al., 2009) |
Comparative analysis of LECs to isolated lens epithelium and fiber cells
Although the above comparative analysis of 21EM15 microarrays to iSyTE and CatMap databases provides important insights, the former is based on enriched-expression of genes in the whole embryonic lens, while the latter informs on genes associated with cataract, regardless of their expression in the lens epithelium or fiber cells. Therefore, we next sought to address how these LEC lines compare to postnatal mouse lens epithelium and fiber cells.
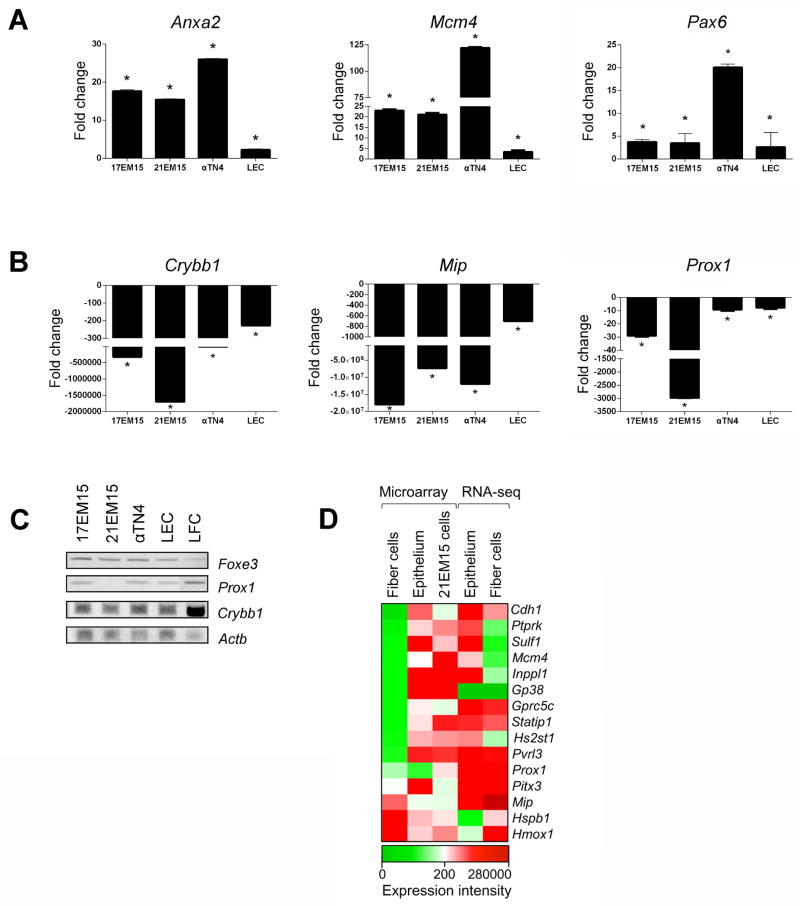
We isolated postnatal mouse lens epithelium and lens fiber cells and compared qRT-PCR and semi-RT-PCR expression of key epithelium-enriched or fiber cell enriched genes with LEC lines. The expression of lens epithelium-enriched genes, Foxe3 and Pax6 as well as others (Anxa4, Mcm4) identified from lens epithelium/fiber cell microarrays and RNA-seq data (see below) follows similar expression trends in lens epithelial cell lines, in contrast to lens fiber cells (Fig. 3A–D). Moreover, we find that following the same trend as in isolated lens epithelium, expression of fiber cell-expressed genes Crybb1, Mip, Prox1 is down-regulated in all three LECs, compared to isolated fiber cells (Fig. 3B, C). These data indicate that these lens epithelial cell lines exhibit greater similarity to lens epithelium than to fiber cells.
Figure 3. Lens epithelial cell lines exhibit gene expression pattern similar to isolated lens epithelium compared to lens fiber cells.
(A) Real time quantitative RT-PCR confirms significant up-regulation of lens epithelium-expressed genes Anxa2, Mcm4, and Pax6 in the LECs and isolated lens epithelium when compared to isolated lens fiber cells. (B) In contrast, real time quantitative RT-PCR exhibits significant down-regulation of lens fiber cell-expressed genes Crybb1, Mip, and Prox1 in the LECs and isolated lens epithelium when compared to isolated lens fiber cells. (C) Regular semi-quantitative RT-PCR demonstrates relative expression levels of the epithelium marker Foxe3, the fiber cell early differentiation marker Prox1 and the fiber cell differentiation marker Crybb1 in isolated lens epithelium and fiber cells as well as in 17EM15, 21EM15 and αTN4 cells. (D) Heat-map of comparative gene expression analysis between isolated lens epithelium and fiber cell microarrays and RNA-seq data and the LEC 21EM15 microarrays for select epithelium- or fiber cell-enriched genes. Asterisk denotes p-values < 0.05.
We also performed comparative analysis between the lens epithelial cell line 21EM15 microarrays and previously generated P14 stage mouse postnatal lens epithelium and fiber cells microarray datasets (Nakahara et al., 2007). Furthermore, we also compared 21EM15 cell line gene expression with RNA-seq data on isolated epithelium and fiber cells from newborn mouse lenses (Hoang et al., 2014). This analysis is represented by a heat-map for a signature set of genes that are lens-epithelium or lens fiber cell enriched (Figure 3D). Although limited because of different platforms (different microarray chips and sequencing), these analyses support the conclusion that the mouse epithelial cell line 21EM15 exhibits a gene expression pattern that is more similar to isolated lens epithelium compared to lens fiber cells.
RT-PCR Validates Lens-expressed Genes in LEC lines
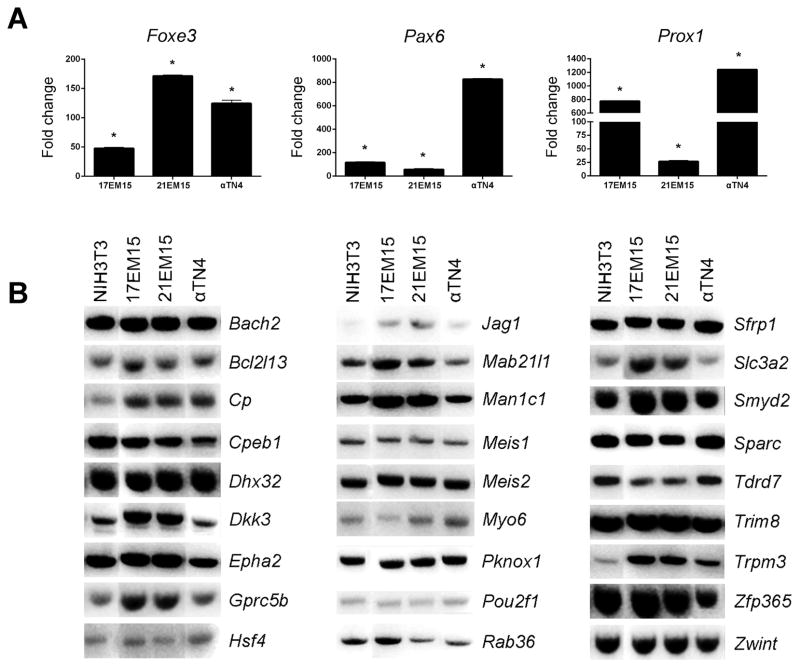
From the above comparative microarray dataset analysis as well as from the lens literature, 30 genes were selected for further validation in the LECs 17EM15, 21EM15 and αTN4. Among these candidates are included some important genes that have significant expression in 21EM15 microarrays but are not identified within the iSyTE or Cat-Map databases (see supplementary Table S3 for list of candidate genes). Also included are candidates with known function in the lens based on literature, as well those that were identified by iSyTE or described in Cat-Map as associated with lens defects (Table S3). In real time qRT-PCR analysis, Pax6, Foxe3 and Prox1 were found to have higher expression in all three LEC lines in comparison to NIH3T3 cells (Figure 4A). Moreover, expression of 27 genes including key genes Dkk3, Epha2, Hsf4, Jag1, Mab21l1, Meis1, Meis2, Pknox1, Pou2f1, Sfrp1, Sparc, Tdrd7 and Trpm3, among others, was analyzed by RT-PCR, which confirmed that all three LECs express these important lens genes (Figure 4B). The full list of genes expressed in 21EM15 cells in provided in supplementary Table S4.
Figure 4. RT-PCR validation of key lens genes in lens epithelial cell lines.
(A) Real time quantitative RT-PCR demonstrates significant up-regulation of Pax6, Foxe3 and Prox1 in these LECs. (B) Regular semi-quantitative RT-PCR validates expression of 27 key lens genes in LEC lines 17EM15, 21EM15 and αTN4. Candidates were selected based on their expression in 21EM15 microarray data, their identification among top ranked genes in iSyTE, or other compelling evidence from literature. Asterisk denotes p-values < 0.05.
P-bodies are Expressed in Embryonic and Postnatal Lens
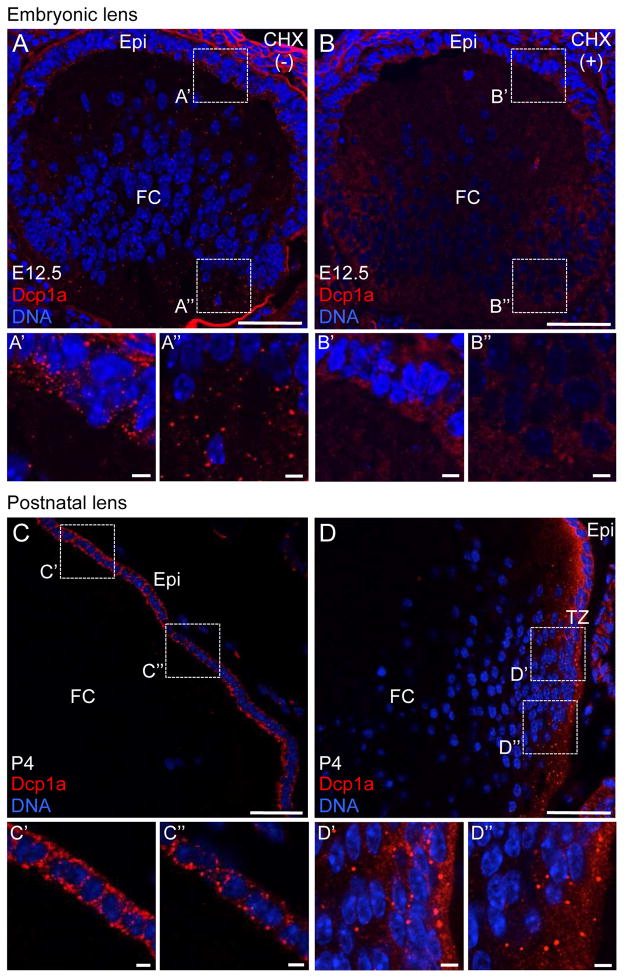
To gain further insight into the relevance of RGs in lens biology, we next sought to investigate the expression of cytoplasmic P-bodies in embryonic and early postnatal stages of lens development. We immunostained E12.5 mouse lens tissue with Dcp1a that serves as a marker for P-bodies. At E12.5, we find that high levels of P-bodies can be detected in both the lens anterior epithelium and fiber cells (Figure 5A, A′, A″). We next sought to validate the identity of these RGs as bona fide P-bodies by challenging them with cycloheximide (CHX) in utero. CHX inhibits protein synthesis by blocking the translocation step during translation of mRNAs and therefore titrates them away from P-bodies resulting in their disassembly. CHX treatment results in dissolution of P-bodies in anterior epithelium and fiber cells at E12.5 (Figure 5B, B′, B″), confirming the identity of these cytoplasmic RGs as P-bodies. We next tested the presence of P-bodies in early postnatal lens at day 4 (P4). Contrary to E12.5, P-bodies were completely absent in fiber cells at P4 (Figure 5C, D), but were detected in the anterior epithelium (Figure 5C′, C″) and the transition zone (Figure 5D′ and D″). These findings were confirmed by a second P-body marker Ddx6 (data not shown) and demonstrate that P-bodies exhibit a dynamic expression pattern in lens development.
Figure 5. Embryonic and early postnatal mouse lens harbors cytoplasmic Processing bodies.
(A) Immunostaining by Dcp1a, a Processing body (P-body) marker, demonstrates that mouse lens at stage embryonic day 12.5 (E12.5) harbors P-bodies in both lens anterior epithelium (Epi) (A′) as well as in fiber cells (FC) (A″). (B) In presence of translational elongation inhibitor cycloheximide (CHX), these cytoplasmic structures can be disassembled in both Epi (B′) and FC (B″), indicating them are bona fide P-bodies. (C) Mouse lens at postnatal day 4 (P4) exhibits P-bodies in the Epi (C′) and (C″) but not in FC. (D) P4 mouse lens also exhibits P-bodies in the transition zone (TZ) (D′) and (D″). Similar results were obtained using Ddx6 as a P-body marker (data not shown). Dotted line box indicates area selected for representation at high magnification. Scale bar in A–D is 50 μm and in A′, A″, B′, B″, C′, C″, D′, D″ is 5 μm.
LECs Support Formation of P-bodies and SGs
To determine the applicability of these LECs to study RGs, we investigated their potential for supporting P-bodies and SGs. We compared 17EM15, 21EM15, αTN4 and NIH3T3 in their ability to form P-bodies and SGs under normal and conditions of oxidative stress induced by arsenite. While eIF3η, an early initiation factor, was used to determine the presence of SGs (Kedersha and Anderson, 2007), Ddx6 was used to determine the presence of P-bodies (Kedersha and Anderson, 2007). In unstressed conditions, P-bodies are detected in the LECs 17EM15, 21EM15 and αTN4 as well as the non-lens cell line NIH3T3 (Figure 6), and their numbers are found to increase under conditions of stress, as has been described for other cell lines (Anderson and Kedersha, 2009). As expected, formation of SGs was detected only under conditions of stress in all four cell lines (Figure 6).
Figure 6. Lens epithelial cell lines support formation of Processing bodies and Stress granules.
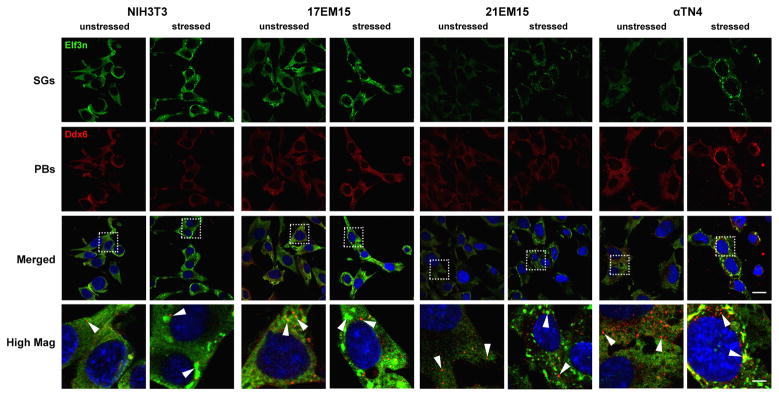
The potential of LECs 17EM15, 21EM15, αTN4 and the non-lens cell line NIH3T3 to support formation of Processing bodies (P-bodies, red) and Stress granules (SGs, green) under normal and unstressed conditions was investigated by immunostaining with P-body and SG markers. Exposure to sodium arsenite (0.5 mM) for 45 min was used as a stress condition and Ddx6 and Elf3η were used as P-body or SG markers, respectively. The LECs 17EM15, 21EM15, αTN4, as well as NIH3T3 cells, in unstressed conditions exhibit P-bodies that appear to increase in number under conditions of stress. Formation of SGs was detected only under conditions of stress in all cell lines. These data indicate that all three LECs support formation of P-bodies and SGs. Dotted line box indicates area selected for representation at high magnification; arrowheads indicate P-bodies in unstressed conditions, and P-bodies and SGs in stressed conditions. Scale bar indicates 22.5 μm in merged images, while that at high magnification indicates 5 μm.
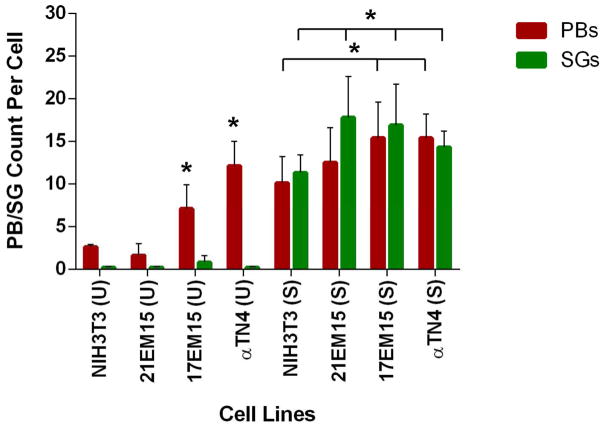
To gain further insights into potential variation between the ability of these various cell lines to harbor these RGs, the numbers of P-bodies and SGs per cell were estimated for each cell line by counting these cytoplasmic RGs under normal and stress conditions. In both unstressed or stress conditions, αTN4 and 17EM15 cells exhibit significantly higher numbers of P-bodies compared to NIH3T3 cells (Figure 7). Interestingly, in conditions of stress, 17EM15, 21EM15 and αTN4 exhibit significantly higher numbers of SGs compared to NIH3T3 cells. These data demonstrate that all three LECs have the potential to form P-bodies and SGs. Furthermore, they suggest that specific LECs, αTN4 and 17EM15, have the potential to form higher levels of P-bodies compared to NIH3T3, while all three LECs form higher number of SGs compared to NIH3T3.
Figure 7. Lens epithelial cell lines exhibit elevated numbers of Processing bodies or Stress granules in comparison to NIH3T3 cells.
Processing bodies (P-bodies) and Stress granules (SGs) were counted under normal (un-stress, U) and stress (S) conditions for all four cell lines. Average numbers of P-bodies and SGs per cell under both unstressed and stressed conditions are shown in the graph. Horizontal bar between cell lines denotes p-values < 0.05 for PB or SG counts per cell. In un-stress or stress conditions, αTN4 and 17EM15 cells exhibit significantly higher numbers of P-bodies in comparison to the NIH3T3 cell line. In conditions of stress, all three LECs exhibit significantly higher numbers of SGs compared to NIH3T3 cells. Asterisk denotes p-values < 0.05.
DISCUSSION
In recent years cytoplasmic RGs have become established as an integral part of post-transcriptional regulation within eukaryotic cells, carrying out various aspects of gene expression control by regulating mRNA localization, stability, decay, or translation into protein (Anderson and Kedersha, 2009). Over the last decade, several distinct types of RGs have been identified and characterized, with SGs and P-bodies being found in all eukaryotic cells tested (Anderson and Kedersha, 2009). The significance of these cytoplasmic RNP complexes in development and disease is beginning to get unraveled and recently it was demonstrated that deficiency of an RG component Tdrd7 results in formation of cataracts in diverse vertebrates, including human (Lachke et al., 2011). This has led to a growing interest in investigating RG-based post-transcriptional control mechanisms in the lens.
Here we have investigated the utility of three mouse LECs as a potential resource for gaining insights into function of RGs in lens cells by testing their potential to express key lens genes as well as to form distinct RGs in culture. In past, several naturally or virally transformed LEC lines have been established from different animal species including human, bovine, rabbit and mouse (Andley et al., 1994; Ibaraki et al., 1998; Reddan et al., 1989; Weinstein et al., 1982; Yamada et al., 1990). However, these have been only partially characterized with regards to their potential to support expression of lens genes and none have been characterized for expression of RGs. The human cell line HLE-B3 is described to express β-crystallin (Andley et al., 1994), while a second human cell line SRA01/04 is reported to express CRYAA and CRYBB2 based on RT-PCR amplification (Ibaraki et al., 1998). The bovine cell line expresses α- and γ-crystallins (Weinstein et al., 1982) and the rabbit cell line N/N1003A expresses Pax6 (Krausz et al., 1996). Western blot and RT-PCR analyses have demonstrated that mouse LEC αTN4 expresses αA- and αB-crystallins as well as Pax6 transcripts (Krausz et al., 1996; Yamada et al., 1990) and while expression studies on 17EM15 and 21EM15 cells are limited (Haque et al., 1999), besides αA- and αB-crystallins they have been described to support Notch1 mediated signaling (Rowan et al., 2008) and expression of Pax6, Sox2, Prox1, Jag1, Epha2, Hspb1 and Crygs (Lachke et al., 2011).
Through comparative gene expression analysis and RT-PCR, we find that LECs 17EM15, 21EM15 and αTN4 express a number of genes important to lens development and homeostasis. We investigated if these cells also express new promising candidates in lens biology by comparative analysis with our recently developed bioinformatics tool iSyTE, which utilizes lens-enriched gene expression for prioritization of candidate genes with lens function (Lachke et al., 2012b). Application of iSyTE has thus far contributed to the characterization of several genes associated with lens development and cataract that include the Tudor family RNA binding protein - Tdrd7 (Lachke et al., 2011), cell adhesion protein- Pvrl3 (Lachke et al., 2012a), selenoprotein- Sep15 (Kasaikina et al., 2011), chromatin modulators- CBP, p300 (Wolf et al., 2013), and the transcription factor- Zeb2 (Manthey et al., 2014). Comparison of 21EM15 microarray datasets with the top 200 iSyTE-enriched candidate genes revealed that ~80% of these genes were significantly expressed in 21EM15 and ~30% have known lens function or expression.
Furthermore, comparative analysis of 21EM15 microarrays with Cat-Map identified 131 genes in this database that are significantly expressed in these cells. These include several key transcription factor genes (e.g. Pax6, Six3, Sox2, Meis1, Foxe3, Maf, Prox1 and Hmx1) and receptor or ligand genes (Efna5, Epha2 and Trpm3) as well as important genes linked to human cataract (Adamtsl4, Chmp4b, Col4a5, Cryaa, Cryab, Cryba4, Crygs, Fyco1, Gcnt2, Gja1, Leprel1, Myh9, Pvrl3, Pxdn, Rab3gap1, Tdrd7 and Vim). However, many other important cataract linked genes, especially those encoding crystallins, e.g. Cryba1, Crybb1, Crybb2, Crybb3, Crygc, and Crygd, as well as other important proteins, e.g. Bfsp1, Bfsp2, Gja3, Gja8, Lim2 and Mip, are not expressed significantly in 21EM15 cells. Furthermore, expression of Crystallin genes, e.g. Cryaa, Cryab, Cryba4, and Crygs, in these cells, although significant in microarray analysis, is not similar to their relative high expression in lens fiber cells.
Moreover, while these data indicate that all three LECs possess a gene expression profile more characteristic of isolated lens epithelium than do NIH3T3 cells (and lens fiber cells), the LECs also harbor specific differences. Interestingly, 17EM15 and 21EM15 follow similar expression trends for many genes tested, but not all (e.g. Bcl2l13, Jag1, Hsf4, Myo6, Rab36, Slc3a2), perhaps reflective of their being derived by the same laboratory. The experimentally transformed cell line αTN4 differs from both these LECs and isolated lens epithelium in that it exhibits significantly higher expression of the epithelium markers Pax6, Anxa1 and Mcm4. Compared to 17EM15 and αTN4, 21EM15 cells have significantly lower expression of Prox1 and Crybb1. Thus, although all three LECs exhibit lens epithelial cell characteristic, they do differ in the expression levels of specific genes and also in formation of specific RGs, which could be a reflection of the underlying genetic or epigenetic changes that may have resulted due to culturing or selection, or their independent derivation from different mouse lens epithelia. Therefore in the absence of whole genome expression profiling data on all of the LECs, it is difficult to conclude if any one of the LECs is similar to isolated lens epithelium.
It should be noted that some genes are missed by the microarrays on 21EM15, but are present in the RT-PCR analysis, which is reflective of the suboptimal hybridization of transcripts to the microarray probesets specific for these genes on the Illumina microarray platform, leading to their being called “absent” in the analysis. From our experience analyzing both Affymetrix 430 2.0 and Illumina WG-6 microarrays, we have noted several such cases wherein the probeset for a specific gene is suboptimal. For example, the Foxe3-specific probeset on the Affy chip correctly reports on the experimentally validated high expression of this gene in the lens. In contrast, the Foxe3 probeset on the Illumina WG-6 microarray platform is not as effective for detecting Foxe3, and calls it “absent” – even in early lens samples, wherein the gene is highly expressed. Thus, microarrays on various different platforms or RNA-Seq analysis will be more informative in future analyses of these cell lines.
In the absence of detailed karyotyping on these cell lines, we sought to gain insight from the RT-PCR and microarrays data on the chromosomal constitution of these cell lines. We first noted that RT-PCR analysis of the genes in represents expression from majority of mouse chromosomes (1 through 11, 16, 17, 19) and thus suggests their presence in all three LECs. Furthermore, analysis of microarray data from 21EM15 confirmed that this cell line expresses genes from all the mouse chromosomes.
Thus, although these cells express key lens genes from iSyTE and Cat-map, and occasionally exhibit spontaneous aggregates of lentoid bodies in culture, they have limitations in their application to study of fiber cells. Although presently there is no evidence of differentiation of these cells into a fiber cell fate, it will be interesting to investigate the effect of conditions used in studies that describe directed differentiation of embryonic stem cell and induced pluripotent stem cells into lens fate (Qiu et al., 2012; Yang et al., 2010). Nevertheless, these LECs offer advantages over primary cell culture that pose challenges such as potential variability between samples or limited lifespan, and in past have been useful in identifying targets that were later validated in the lens tissue (Lachke et al., 2011).
These LECs will be beneficial to gain insights into molecular regulation or function of genes as well as in various high-throughput approaches and screens as described below. Because these cells are easily transfected by plasmid or lentiviral constructs they are amenable to shRNA-mediated gene knockdown approaches (Donner et al., 2007; Lachke et al., 2011; Rowan et al., 2008). Unlike αTN4, the 17EM15 and 21EM15 cell lines are not experimentally transformed, and may potentially harbor spontaneous transformation events as has been described for other cells line (Boukamp et al., 1988; Hughes, 1996; Todaro and Green, 1963). However, all three cell lines grow well in culture conditions and display consistent growth kinetics across passages. Therefore these LECs can be used to investigate the molecular function of Cat-Map genes or new iSyTE-predicted candidate genes with potential function in the lens. Furthermore, similar to previous approaches (Ohn et al., 2008), these LECs can be used in high-throughput screens for identifying candidates that mediate RG assembly or function. For the key lens genes that are expressed, these LECs can be used in approaches to test protein-protein interactions (Mak and Moffat, 2012; Völkel et al., 2010). Recently, protocols have been developed and applied for the successful isolation and biochemical characterization of RGs like chromatoid bodies (Meikar and Kotaja, 2014). Similarly, these cell lines can be a useful resource to isolate protein and RNA components from P-bodies and SGs for further characterization. Given the recent connection of SGs to apoptosis (Thedieck et al., 2013), these approaches have the potential to unravel surprising new regulatory connections in lens biology. Recent findings have indicated a function for miRNAs in lens development. Since P-bodies are linked to miRNA-mediated control, these cell lines may offer a reagent for investigating their potential role in miRNA-mediated regulatory control of lens genes. Finally, detection of high levels of P-bodies and SGs in these LECs make them a useful resource for testing the molecular strategies that lens epithelial cells potentially use to neutralize the effect of stress factors such as ultraviolet radiation.
Thus, our data demonstrate that LECs 17EM15, 21EM15 and αTN4 retain expression of several important lens marker genes and are similar to isolated lens epithelium than to lens fiber cells. These LECs also exhibit elevated levels of P-bodies or SGs when compared to NIH3T3 cells in normal and oxidative stress conditions. In sum, these findings indicate their suitability for investigating the molecular function of these cytosolic ribonucleoprotein complexes in lens cells.
Supplementary Material
Expression of top iSyTE candidate genes in LEC 21EM15 microarrays.
Expression of CatMap genes in LEC 21EM15 microarrays.
Expression of RT-PCR validated genes in LEC 21EM15 microarrays.
Expression of all genes in LEC 21EM15 microarrays.
A detailed molecular and cellular characterization of three permanent mouse lens epithelial cell lines (LEC) 17EM15, 21EM15 and αTN4 was performed
Comparative analysis between microarray gene expression datasets on LEC 21EM15 and iSyTE lens tissue identified several lens-enriched and Cat-Map genes that are expressed in these cells
Compared to non-lens cell line NIH3T3, all three LECs exhibit significantly enriched expression of Pax6, Foxe3 and Prox1
Mouse LECs 21EM15, 17EM15 and αTN4 express key lens cataract genes and exhibit high levels of Processing bodies and Stress granules, indicating their suitability for investigating gene expression control and RNA granule function in lens-derived cells
Acknowledgments
Grant support: Supported by National Institutes of Health (NIH) Grant R01 EY021505 to DCB and SAL. SAL is a Pew Scholar in Biomedical Sciences
Authors thank Dr. Melinda Duncan for helpful discussions and Drs. John Reddan, Richard Maas and Gary Laverty for cell lines. Supported by National Institutes of Health (NIH) Grant R01 EY021505 to DCB and SAL. SAL is a Pew Scholar in Biomedical Sciences.
References
- Adjibade P, Mazroui R. Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Ahram D, Sato TS, Kohilan A, Tayeh M, Chen S, Leal S, Al-Salem M, El-Shanti H. A homozygous mutation in ADAMTSL4 causes autosomal-recessive isolated ectopia lentis. Am J Hum Genet. 2009;84:274–278. doi: 10.1016/j.ajhg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JL, Hill CR, Cole KD. Mouse cell line authentication. Cytotechnology. 2014;66:133–147. doi: 10.1007/s10616-013-9545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Dev Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Andley UP, Rhim JS, Chylack LT, Fleming TP. Propagation and immortalization of human lens epithelial cells in culture. Invest Ophthalmol Vis Sci. 1994;35:3094–3102. [PubMed] [Google Scholar]
- Ang SJ, Stump RJW, Lovicu FJ, McAvoy JW. Spatial and temporal expression of Wnt and Dickkopf genes during murine lens development. Gene Expr Patterns GEP. 2004;4:289–295. doi: 10.1016/j.modgep.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Antignac C, Zhou J, Sanak M, Cochat P, Roussel B, Deschênes G, Gros F, Knebelmann B, Hors-Cayla MC, Tryggvason K. Alport syndrome and diffuse leiomyomatosis: deletions in the 5′ end of the COL4A5 collagen gene. Kidney Int. 1992;42:1178–1183. doi: 10.1038/ki.1992.402. [DOI] [PubMed] [Google Scholar]
- Bai X, Dilworth DJ, Weng YC, Gould DB. Developmental distribution of collagen IV isoforms and relevance to ocular diseases. Matrix Biol J Int Soc Matrix Biol. 2009;28:194–201. doi: 10.1016/j.matbio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Rajakaruna S, De Arcangelis A, Zhang L, Georges-Labouesse E, Menko AS. α6 integrin transactivates insulin-like growth factor receptor-1 (IGF-1R) to regulate caspase-3-mediated lens epithelial cell differentiation initiation. J Biol Chem. 2014;289:3842–3855. doi: 10.1074/jbc.M113.515254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DC. The use of cell lines to “model” ocular tissues: cautionary tales. Invest Ophthalmol Vis Sci. 2013;54 doi: 10.1167/iovs.13-12873. [DOI] [PubMed] [Google Scholar]
- Bennett TM, Mackay DS, Siegfried CJ, Shiels A. Mutation of the Melastatin-Related Cation Channel, TRPM3, Underlies Inherited Cataract and Glaucoma. PloS One. 2014;9:e104000. doi: 10.1371/journal.pone.0104000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry V, Francis P, Reddy MA, Collyer D, Vithana E, MacKay I, Dawson G, Carey AH, Moore A, Bhattacharya SS, Quinlan RA. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–1145. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw J, Héon E. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–709. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014:11. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carper D, John M, Chen Z, Subramanian S, Wang R, Ma W, Spector A. Gene expression analysis of an H(2)O(2)-resistant lens epithelial cell line. Free Radic Biol Med. 2001;31:90–97. doi: 10.1016/s0891-5849(01)00561-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Ma Z, Jiao X, Fariss R, Kantorow WL, Kantorow M, Pras E, Frydman M, Pras E, Riazuddin S, Riazuddin SA, Hejtmancik JF. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011;88:827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stump RJW, Lovicu FJ, McAvoy JW. Expression of Frizzleds and secreted frizzled-related proteins (Sfrps) during mammalian lens development. Int J Dev Biol. 2004;48:867–877. doi: 10.1387/ijdb.041882yc. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Xiao T, Vergara LA, Srivastava S, Nees D, Piatigorsky J, Ansari NH. Role of aldehyde dehydrogenase isozymes in the defense of rat lens and human lens epithelial cells against oxidative stress. Invest Ophthalmol Vis Sci. 2005;46:259–267. doi: 10.1167/iovs.04-0120. [DOI] [PubMed] [Google Scholar]
- Cooper MA, Son AI, Komlos D, Sun Y, Kleiman NJ, Zhou R. Loss of ephrin-A5 function disrupts lens fiber cell packing and leads to cataract. Proc Natl Acad Sci U S A. 2008;105:16620–16625. doi: 10.1073/pnas.0808987105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Craig JE, Sharma S. The status of intercellular junctions in established lens epithelial cell lines. Mol Vis. 2012;18:2937–2946. [PMC free article] [PubMed] [Google Scholar]
- De Arcangelis A, Mark M, Kreidberg J, Sorokin L, Georges-Labouesse E. Synergistic activities of alpha3 and alpha6 integrins are required during apical ectodermal ridge formation and organogenesis in the mouse. Dev Camb Engl. 1999;126:3957–3968. doi: 10.1242/dev.126.17.3957. [DOI] [PubMed] [Google Scholar]
- De Mateo S, Sassone-Corsi P. Regulation of spermatogenesis by small non-coding RNAs: role of the germ granule. Semin Cell Dev Biol. 2014;29:84–92. doi: 10.1016/j.semcdb.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinforma Oxf Engl. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JRO, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, Arn E, Bakosova A, Jungwirth K, Hörmann A, Palfi Z, Sandholzer J, Schwarz M, Macchi P, Colinge J, Superti-Furga G, Kiebler MA. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Gao M, Arkov AL. Next generation organelles: structure and role of germ granules in the germline. Mol Reprod Dev. 2013;80:610–623. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DT, Lyon GJ, Carlton MB, Sanes JR, Cunningham JM, Anderson JR, Hogan BL, Evans MJ, Colledge WH. Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40 develop normally but show severe age-onset cataract formation and disruption of the lens. EMBO J. 1998;17:1860–1870. doi: 10.1093/emboj/17.7.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Handley MT, Morris-Rosendahl DJ, Brown S, Macdonald F, Hardy C, Bem D, Carpanini SM, Borck G, Martorell L, Izzi C, Faravelli F, Accorsi P, Pinelli L, Basel-Vanagaite L, Peretz G, Abdel-Salam GMH, Zaki MS, Jansen A, Mowat D, Glass I, Stewart H, Mancini G, Lederer D, Roscioli T, Giuliano F, Plomp AS, Rolfs A, Graham JM, Seemanova E, Poo P, García-Cazorla A, Edery P, Jackson IJ, Maher ER, Aligianis IA. Mutation spectrum in RAB3GAP1, RAB3GAP2, and RAB18 and genotype-phenotype correlations in warburg micro syndrome and Martsolf syndrome. Hum Mutat. 2013;34:686–696. doi: 10.1002/humu.22296. [DOI] [PubMed] [Google Scholar]
- Haque MS, Arora JK, Dikdan G, Lysz TW, Zelenka PS. The rabbit lens epithelial cell line N/N1003A requires 12-lipoxygenase activity for DNA synthesis in response to EGF. Mol Vis. 1999;5:8. [PubMed] [Google Scholar]
- Harned J, Fleisher LN, McGahan MC. Lens epithelial cells synthesize and secrete ceruloplasmin: effects of ceruloplasmin and transferrin on iron efflux and intracellular iron dynamics. Exp Eye Res. 2006;83:721–727. doi: 10.1016/j.exer.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, Jenkins NA, Copeland NG. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TV, Kumar PKR, Sutharzan S, Tsonis PA, Liang C, Robinson ML. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing [WWW Document] [accessed 12.1.14];2014 http://www.molvis.org/molvis/v20/1491/ [PMC free article] [PubMed]
- Hughes SE. Functional characterization of the spontaneously transformed human umbilical vein endothelial cell line ECV304: use in an in vitro model of angiogenesis. Exp Cell Res. 1996;225:171–185. doi: 10.1006/excr.1996.0168. [DOI] [PubMed] [Google Scholar]
- Ibaraki N, Chen SC, Lin LR, Okamoto H, Pipas JM, Reddy VN. Human lens epithelial cell line. Exp Eye Res. 1998;67:577–585. doi: 10.1006/exer.1998.0551. [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GCM. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- Kamaid A, Giráldez F. Btg1 and Btg2 gene expression during early chick development. Dev Dyn Off Publ Am Assoc Anat. 2008;237:2158–2169. doi: 10.1002/dvdy.21616. [DOI] [PubMed] [Google Scholar]
- Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Natarajan SK, Malinouski M, Schweizer U, Tsuji PA, Carlson BA, Maas RL, Lou MF, Goldstein LE, Hatfield DL, Gladyshev VN. Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J Biol Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. doi:10.1042/ [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K, Rudkin A, Parry DA, Burdon KP, McKibbin M, Logan CV, Abdelhamed ZIA, Muecke JS, Fernandez-Fuentes N, Laurie KJ, Shires M, Fogarty R, Carr IM, Poulter JA, Morgan JE, Mohamed MD, Jafri H, Raashid Y, Meng N, Piseth H, Toomes C, Casson RJ, Taylor GR, Hammerton M, Sheridan E, Johnson CA, Inglehearn CF, Craig JE, Ali M. Homozygous mutations in PXDN cause congenital cataract, corneal opacity, and developmental glaucoma. Am J Hum Genet. 2011;89:464–473. doi: 10.1016/j.ajhg.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Krausz E, Augusteyn RC, Quinlan RA, Reddan JR, Russell P, Sax CM, Graw J. Expression of Crystallins, Pax6, Filensin, CP49, MIP, and MP20 in lens-derived cell lines. Invest Ophthalmol Vis Sci. 1996;37:2120–2128. [PubMed] [Google Scholar]
- Kunishima S, Matsushita T, Kojima T, Amemiya N, Choi YM, Hosaka N, Inoue M, Jung Y, Mamiya S, Matsumoto K, Miyajima Y, Zhang G, Ruan C, Saito K, Song KS, Yoon HJ, Kamiya T, Saito H. Identification of six novel MYH9 mutations and genotype-phenotype relationships in autosomal dominant macrothrombocytopenia with leukocyte inclusions. J Hum Genet. 2001;46:722–729. doi: 10.1007/s100380170007. [DOI] [PubMed] [Google Scholar]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai ACH, Nair KS, Cosma MI, Smith RS, Hodges E, Alfadhli SM, Al-Hajeri A, Shamseldin HE, Behbehani A, Hannon GJ, Bulyk ML, Drack AV, Anderson PJ, John SWM, Maas RL. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Higgins AW, Inagaki M, Saadi I, Xi Q, Long M, Quade BJ, Talkowski ME, Gusella JF, Fujimoto A, Robinson ML, Yang Y, Duong QT, Shapira I, Motro B, Miyoshi J, Takai Y, Morton CC, Maas RL. The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum Genet. 2012a;131:235–250. doi: 10.1007/s00439-011-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Ho JWK, Kryukov GV, O’Connell DJ, Aboukhalil A, Bulyk ML, Park PJ, Maas RL. iSyTE: integrated Systems Tool for Eye gene discovery. Invest Ophthalmol Vis Sci. 2012b;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Maas RL. RNA Granules and Cataract. Expert Rev Ophthalmol. 2011;6:497–500. doi: 10.1586/eop.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Conley KW, Mead TJ, Rowan S, Yutzey KE, Brown NL. Requirements for Jag1-Rbpj mediated Notch signaling during early mouse lens development. Dev Dyn Off Publ Am Assoc Anat. 2012;241:493–504. doi: 10.1002/dvdy.23739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- Liu WB, Li Y, Zhang L, Chen HG, Sun S, Liu JP, Liu Y, Li DWC. Differential expression of the catalytic subunits for PP-1 and PP-2A and the regulatory subunits for PP-2A in mouse eye. Mol Vis. 2008;14:762–773. [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddala R, Skiba NP, Lalane R, Sherman DL, Brophy PJ, Rao PV. Periaxin is required for hexagonal geometry and membrane organization of mature lens fibers. Dev Biol. 2011;357:179–190. doi: 10.1016/j.ydbio.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Moriguchi T, Hamada M, Kusakabe M, Fujioka Y, Nakano T, Yoh K, Lim KC, Engel JD, Takahashi S. Transcription factor GATA-3 is essential for lens development. Dev Dyn Off Publ Am Assoc Anat. 2009;238:2280–2291. doi: 10.1002/dvdy.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AB, Moffat J. A versatile lentiviral expression system to identify mammalian protein-protein interactions. Methods San Diego Calif. 2012;57:409–416. doi: 10.1016/j.ymeth.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Manthey AL, Lachke SA, FitzGerald PG, Mason RW, Scheiblin DA, McDonald JH, Duncan MK. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech Dev. 2014;131:86–110. doi: 10.1016/j.mod.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikar O, Kotaja N. Isolation of chromatoid bodies from mouse testis as a rich source of short RNAs. Methods Mol Biol Clifton NJ. 2014;1173:11–25. doi: 10.1007/978-1-4939-0931-5_2. [DOI] [PubMed] [Google Scholar]
- Menzel O, Bekkeheien RCJ, Reymond A, Fukai N, Boye E, Kosztolanyi G, Aftimos S, Deutsch S, Scott HS, Olsen BR, Antonarakis SE, Guipponi M. Knobloch syndrome: novel mutations in COL18A1, evidence for genetic heterogeneity, and a functionally impaired polymorphism in endostatin. Hum Mutat. 2004;23:77–84. doi: 10.1002/humu.10284. [DOI] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Mordechai S, Gradstein L, Pasanen A, Ofir R, El Amour K, Levy J, Belfair N, Lifshitz T, Joshua S, Narkis G, Elbedour K, Myllyharju J, Birk OS. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. Am J Hum Genet. 2011;89:438–445. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moré MI, Kirsch FP, Rathjen FG. Targeted ablation of NrCAM or ankyrin-B results in disorganized lens fibers leading to cataract formation. J Cell Biol. 2001;154:187–196. doi: 10.1083/jcb.200104038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Bhattacharya SS, Moore T, Prescott Q, Wedig T, Herrmann H, Magin TM. Dominant cataract formation in association with a vimentin assembly disrupting mutation. Hum Mol Genet. 2009;18:1052–1057. doi: 10.1093/hmg/ddn440. [DOI] [PubMed] [Google Scholar]
- Munroe RJ, Prabhu V, Acland GM, Johnson KR, Harris BS, O’Brien TP, Welsh IC, Noden DM, Schimenti JC. Mouse H6 Homeobox 1 (Hmx1) mutations cause cranial abnormalities and reduced body mass. BMC Dev Biol. 2009;9:27. doi: 10.1186/1471-213X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara M, Nagasaka A, Koike M, Uchida K, Kawane K, Uchiyama Y, Nagata S. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–3064. doi: 10.1111/j.1742-4658.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Belusko PB, Walkup RD, Azuma M, Shearer TR. Involvement of Egr-1 in lens epithelial cell death induced by selenite. Exp Eye Res. 2006;82:874–878. doi: 10.1016/j.exer.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Near RI, Smith RS, Toselli PA, Freddo TF, Bloom AB, Vanden Borre P, Seldin DC, Lerner A. Loss of AND-34/BCAR3 expression in mice results in rupture of the adult lens. Mol Vis. 2009;15:685–699. [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi N, Kiyokawa E, Matsuda M, Kinoshita K, Yamada S, Yamada K, Matsushima Y, Wang Y, Kawai J, Suzuki M, Hayashizaki Y, Hiai H. Mutation of Dock5, a member of the guanine exchange factor Dock180 superfamily, in the rupture of lens cataract mouse. Exp Eye Res. 2008;86:828–834. doi: 10.1016/j.exer.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi DJ, Wilkinson L, Kolle G, Sohaskey ML, Gillinder K, Piper MJ, McAvoy JW, Lovicu FJ, Little MH. Crim1KST264/KST264 mice display a disruption of the Crim1 gene resulting in perinatal lethality with defects in multiple organ systems. Dev Dyn Off Publ Am Assoc Anat. 2007;236:502–511. doi: 10.1002/dvdy.21015. [DOI] [PubMed] [Google Scholar]
- Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dyn Off Publ Am Assoc Anat. 2008;237:602–617. doi: 10.1002/dvdy.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Yang J, Liu T, Jiang Y, Le Q, Lu Y. Efficient generation of lens progenitor cells from cataract patient-specific induced pluripotent stem cells. PloS One. 2012;7:e32612. doi: 10.1371/journal.pone.0032612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddan JR, Kuck JF, Dziedzic DC, Kuck KD, Reddan PR, Wasielewski P. Establishment of lens epithelial cell lines from Emory and cataract resistant mice and their response to hydrogen peroxide. Lens Eye Toxic Res. 1989;6:687–701. [PubMed] [Google Scholar]
- Rich A, Farrugia G, Rae JL. Effects of melatonin on ionic currents in cultured ocular tissues. Am J Physiol. 1999;276:C923–929. doi: 10.1152/ajpcell.1999.276.4.C923. [DOI] [PubMed] [Google Scholar]
- Rossi M, Morita H, Sormunen R, Airenne S, Kreivi M, Wang L, Fukai N, Olsen BR, Tryggvason K, Soininen R. Heparan sulfate chains of perlecan are indispensable in the lens capsule but not in the kidney. EMBO J. 2003;22:236–245. doi: 10.1093/emboj/cdg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Conley KW, Le TT, Donner AL, Maas RL, Brown NL. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P, Yamada T, Merola LO. Induction of the enzyme aldose reductase in a lens epithelial cell line from a transgenic mouse. Arch Biochem Biophys. 1990;276:259–264. doi: 10.1016/0003-9861(90)90036-x. [DOI] [PubMed] [Google Scholar]
- Saika S, Miyamoto T, Ishida I, Ohnishi Y, Ooshima A. Osteopontin: a component of matrix in capsular opacification and subcapsular cataract. Invest Ophthalmol Vis Sci. 2003;44:1622–1628. doi: 10.1167/iovs.02-0873. [DOI] [PubMed] [Google Scholar]
- Schorderet DF, Nichini O, Boisset G, Polok B, Tiab L, Mayeur H, Raji B, de la Houssaye G, Abitbol MM, Munier FL. Mutation in the human homeobox gene NKX5-3 causes an oculo-auricular syndrome. Am J Hum Genet. 2008;82:1178–1184. doi: 10.1016/j.ajhg.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Knopf HLS, Maraini G, Li A, Jiao X, Hejtmancik JF. The EPHA2 gene is associated with cataracts linked to chromosome 1p. Mol Vis. 2008;14:2042–2055. [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Knopf HLS, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simirskii VN, Wang Y, Duncan MK. Conditional deletion of beta1-integrin from the developing lens leads to loss of the lens epithelial phenotype. Dev Biol. 2007;306:658–668. doi: 10.1016/j.ydbio.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector A, Li D, Ma W, Sun F, Pavlidis P. Differential amplification of gene expression in lens cell lines conditioned to survive peroxide stress. Invest Ophthalmol Vis Sci. 2002;43:3251–3264. [PubMed] [Google Scholar]
- Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–710. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Kunishima S, Ikejiri M, Maruyama S, Sone M, Takagi A, Ikawa M, Okabe M, Kojima T, Saito H, Naoe T, Matsushita T. Establishment of mouse model of MYH9 disorders: heterozygous R702C mutation provokes macrothrombocytopenia with leukocyte inclusion bodies, renal glomerulosclerosis and hearing disability. PloS One. 2013;8:e71187. doi: 10.1371/journal.pone.0071187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Hosokawa M, Vagin VV, Reuter M, Hayashi E, Mochizuki AL, Kitamura K, Yamanaka H, Kondoh G, Okawa K, Kuramochi-Miyagawa S, Nakano T, Sachidanandam R, Hannon GJ, Pillai RS, Nakatsuji N, Chuma S. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc Natl Acad Sci U S A. 2011;108:10579–10584. doi: 10.1073/pnas.1015447108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Saika S, Ohnishi Y, Ooshima A, McAvoy JW, Liu CY, Azhar M, Doetschman T, Kao WWY. Fibroblast growth factor 2: roles of regulation of lens cell proliferation and epithelial-mesenchymal transition in response to injury. Mol Vis. 2004;10:462–467. [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, Nitschke R, Kuehn EW, Jonker JW, Groen AK, Reth M, Hall MN, Baumeister R. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Agtmael T, Schlötzer-Schrehardt U, McKie L, Brownstein DG, Lee AW, Cross SH, Sado Y, Mullins JJ, Pöschl E, Jackson IJ. Dominant mutations of Col4a1 result in basement membrane defects which lead to anterior segment dysgenesis and glomerulopathy. Hum Mol Genet. 2005;14:3161–3168. doi: 10.1093/hmg/ddi348. [DOI] [PubMed] [Google Scholar]
- Völkel P, Le Faou P, Angrand PO. Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry. Biochem Soc Trans. 2010;38:883–887. doi: 10.1042/BST0380883. [DOI] [PubMed] [Google Scholar]
- Weinstein BI, Schwartz J, Lonial H, Dominguez MO, Gordon GG, Hochstadt J, Southren DB, Dunn MW, Southren AL. Normal and conditionally transformed bovine lens epithelial cell lines containing alpha-and gamma-crystallin. Exp Eye Res. 1982;34:71–81. doi: 10.1016/0014-4835(82)90010-0. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wolf L, Harrison W, Huang J, Xie Q, Xiao N, Sun J, Kong L, Lachke SA, Kuracha MR, Govindarajan V, Brindle PK, Ashery-Padan R, Beebe DC, Overbeek PA, Cvekl A. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17:47–52. [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Nakamura T, Westphal H, Russell P. Synthesis of alpha-crystallin by a cell line derived from the lens of a transgenic animal. Curr Eye Res. 1990;9:31–37. doi: 10.3109/02713689009000052. [DOI] [PubMed] [Google Scholar]
- Yang C, Yang Y, Brennan L, Bouhassira EE, Kantorow M, Cvekl A. Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB J Off Publ Fed Am Soc Exp Biol. 2010;24:3274–3283. doi: 10.1096/fj.10-157255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LC, Twu YC, Chang CY, Lin M. Molecular basis of the adult i phenotype and the gene responsible for the expression of the human blood group I antigen. Blood. 2001;98:3840–3845. doi: 10.1182/blood.v98.13.3840. [DOI] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of top iSyTE candidate genes in LEC 21EM15 microarrays.
Expression of CatMap genes in LEC 21EM15 microarrays.
Expression of RT-PCR validated genes in LEC 21EM15 microarrays.
Expression of all genes in LEC 21EM15 microarrays.