Abstract
The oxygen-limiting (hypoxic) microenvironment of tumors induces metabolic reprogramming and cell survival, but the underlying mechanisms involving mitochondria remain poorly understood. We previously demonstrated that hypoxia-inducible factor 1 mediates the hyperfusion of mitochondria by inducing Bcl-2/adenovirus E1B 19-kDa interacting protein 3 and posttranslational truncation of the mitochondrial ATP transporter outer membrane voltage-dependent anion channel 1 in hypoxic cells. In addition, we showed that truncation is associated with increased resistance to drug-induced apoptosis and is indicative of increased patient chemoresistance. We now show that silencing of the tumor suppressor TP53 decreases truncation and increases drug-induced apoptosis. We also show that TP53 regulates truncation through induction of the mitochondrial protein Mieap. While we found that truncation was independent of mitophagy, we observed local microfusion between mitochondria and endolysosomes in hypoxic cells in culture and in patients' tumor tissues. Since we found that the endolysosomal asparagine endopeptidase was responsible for truncation, we propose that it is a readout of mitochondrial-endolysosomal microfusion in hypoxia. These novel findings provide the framework for a better understanding of hypoxic cell metabolism and cell survival through mitochondrial-endolysosomal microfusion regulated by hypoxia-inducible factor 1 and TP53.
INTRODUCTION
Hypoxia is a natural occurring stress that results in compensatory changes in metabolism and cell survival during embryonic development and tumor growth. Hypoxia stabilizes and activates the transcription factor hypoxia-inducible factor (HIF) through inhibition of oxygen-dependent hydroxylases that earmark the alpha subunit of HIF for proteasomal degradation (1). HIF induces or represses the expression of genes implicated in a myriad of functions, including those regulating metabolism and resistance to drug-induced cell death. Genes coding for the enzymes of the glycolytic pathway, including hexokinase, are highly induced by HIF-1, and this is in part responsible for the switch in metabolism from mitochondrial respiration to glycolysis in cancer cells. Considerable studies have pointed to the Warburg effect, also termed aerobic glycolysis, as the major adaptive response of cancer cells, but mitochondrial metabolism and mitochondrial dynamics are also starting to be recognized as important adaptive strategies of cancer cells (2). Mitochondria are critical organelles that regulate both metabolism and cell death. They are dynamic organelles that continuously undergo fission and fusion during cell growth (3, 4). Under stress conditions, such as nutrient depletion or hypoxia, mitochondria either fragment or are degraded by HIF-dependent mitophagy (mitochondrial removal by autophagy) (5) or hyperfuse together to form elongated or rounded structures that optimize ATP production and promote cell survival (6–11).
We reported previously that certain cell lines exposed to hypoxia contained enlarged mitochondria (6). We found that the mechanism was HIF-1 and Bcl-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3/BNIP3L) dependent but that it was independent of mitophagy. In addition, the hypoxic cells were more resistant to stimulated cell death than normoxic cells (12). Furthermore, we reported that the mitochondrial outer membrane protein voltage-dependent anion channel 1 (VDAC1) was posttranslationally cleaved at the C terminus in these cells in a HIF-1-dependent manner and in human lung adenocarcinoma tissue (12). VDAC mediates the transport of ions and small metabolites, such as ADP/ATP, from and into mitochondria (13). Three mammalian isoforms of VDAC exist in eukaryotic cells. VDACs bind hexokinase, the first enzyme of the glycolytic pathway, and in so doing provide ATP for conversion of glucose to glucose-6-phosphate. VDACs also play a key role in apoptosis through Ca2+ regulation of VDAC1 expression and binding of antiapoptotic proteins of the Bcl-2 family (14, 15).
The TP53 transcription factor plays an important role in the response to and regulation of metabolic stress in cancer (16, 17). It is known that a TP53-inducible protein, Mieap (also referred to as Spata18) (18), controls mitochondrial quality through interaction with the HIF-1-inducible protein BNIP3 (19). In addition, Mieap has been proposed to induce the accumulation of lysosomal proteins within mitochondria by way of repairing damaged mitochondria (20).
In the present study, we investigated further the mechanism behind the hypoxic regulation of the truncation of VDAC1. We propose that enlarged hypoxic mitochondria make fusional contact with late endolysosomes through TP53-induced Mieap in promoting cell survival. Furthermore, we report that VDAC1 is cleaved at loop 14 by the endolysosomal protease asparagine endopeptidase (also termed legumain). Intimate contact between mitochondria and vacuoles has been described only in Saccharomyces cerevisiae yeast (21, 22) and in erythroid cells (23). This cross talk between organelles was found to regulate lipid transport, cellular metabolism, and iron transport. We now show that a spatial and functional interorganellar connection also exists in eukaryotes, both in cells in culture and in lung adenocarcinomas of patients. We propose, finally, that VDAC posttranslational modification marks mitochondria for protection from mitophagy and reflects a survival strategy of hypoxic cancer cells in vitro and in vivo in patients.
MATERIALS AND METHODS
Cells and hypoxic conditions.
Cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) supplemented with 5 or 10% inactivated fetal bovine serum, as appropriate. M. van de Wetering provided the LS174 cells. p53+/+ and p53−/− mouse embryonic fibroblasts (MEFs) were provided by P. Roux. An Invivo2 200 anaerobic workstation (Ruskinn Technology Biotrace International Plc) set at 1% O2, 94% N2, and 5% CO2 was used for hypoxic incubation. HeLa cells were incubated for 48 h in hypoxia, while the other cells, unless otherwise indicated, were incubated for 72 h.
Pharmacological inhibitors and chemicals.
Bafilomycin A1 was purchased from Calbiochem. Brefeldin A, chloroquine diphosphate, LY-294002, 3-methyladenine, rapamycin, valinomycin, pepstatin A, E64d, rotenone antimycin, oligomycin, and trifluorocarbonylcyanide phenylhydrazone (FCCP) were from Sigma.
Transfection of plasmids and RNA interference.
The plasmid carrying yellow fluorescent protein (YFP)-tagged Parkin (YFP-Parkin) was purchased from Addgene (24). The 21-nucleotide RNAs were synthesized by Eurogentec or OriGene. HeLa or LS174 cells were transfected with 40 or 100 nM small interfering RNA (siRNA) twice at an interval of 24 h prior to normoxia or hypoxia, as described previously (6).
siRNA sequences were as follows: siRNA against VDAC1 (forward), 5′-GAUACACUCAGACUCUAAA-3′; siRNA against VDAC1 3′ untranslated region (forward), 5′-CUCCAGGUUAAAGUUGAUUCA-3′; siRNA against VDAC2 (forward), 5′-GAAGAUUUGGCCUUAAUAU-3′; siRNA against TP53#1 (forward), 5′-GACUCCAGUGGUAAUCUAC-3′; siRNA against TP53#3 (forward), 5′-UAUGGCGGGAGGUAGACUG-3′; siRNA against autophagy-related gene 5 (ATG5)/ATG6/ATG7/beclin, the sequences were described previously (25); siRNA against ATG4B (forward), 5′-CUGAAGAUGACUUCAAUGA-3′; siRNA against Lamp2 (forward), 5′-GAAAAUGCCACUUGCCUUU-3′; siRNA against Mieap#1 (forward), 5′-GUAGCAGUGACUUAAGGCUAAG-3′; siRNA against Mieap#2 (forward), 5′-GGUGCAGGGACAACUCUUUGGG-3′; siRNA against Mieap#3 (forward), 5′-GAGAUAUGUUGCAUUGCCUUUGC-5′; siRNA against Mieap#4 (forward), 5′-GCACUAUCUGCCUAGGUAACUGC-5′; siRNA against Lamp1A (forward), 5′-AGAAAUGCAACACGUUA-5′; siRNA against Lamp2A (forward), 5′-GCUGUGCGGUCUUAUGCAU-5′; siRNA against BNIP3 and B3L, the sequences were described previously (25); and siRNA against mitofusin 1 (Mfn1), the sequence was previously described (12). Three siRNAs against asparagine endopeptidase and a scrambled siRNA were purchased from Origene; the sequences were not disclosed.
Caspase activation and cell death induced by STS.
Quantification of caspase 3/7 activity was done using a luciferin/luciferase-based assay (Caspase-Glo 3/7 kit; Promega) according to the manufacturer's instructions. Tests under each condition were performed eight times, and the entire experiment was performed three times. Significant differences (P < 0.005) were based on the Student t test. Staurosporine (STS; 1 μM) was added 4 h prior to the assay for caspase 3/7 activity. Cell death was also determined by trypan blue exclusion and confirmed with an Adam cell counter.
Transmission electron microscopy.
Cells were fixed in situ with 1.6% glutaraldehyde in 0.1 M phosphate buffer at room temperature for at least 1 h and then conserved at 4°C. Samples were rinsed in the same buffer and then postfixed with 1% osmium tetroxide and 1% potassium ferrocyanide in 0.1 M cacodylate buffer for 1 h at room temperature to enhance the staining of cytoplasmic membranes. Cells were rinsed with distilled water, dehydrated in alcohol, and embedded in epoxy resin. Embedded samples were then processed for ultrathin sectioning and observed with a JEM1400 transmission electron microscope (JEOL, Tokyo, Japan) equipped with a Morada charge-coupled-device camera (Olympus SIS, Rungis, France).
For immunogold staining, cells were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M phosphate buffer (PB; pH 7.4) for 2 h and were processed for ultracryomicrotomy according to a slightly modified Tokuyasu method. In brief, a cell suspension was spun down in 10% gelatin. After immersion in 2.3 M sucrose (in pH 7.4 0.1 M PB) overnight at 4°C, the samples were rapidly frozen in liquid nitrogen. Ultrathin (70-nm-thick) cryosections were prepared with an ultracryomicrotome (Leica EMFCS, Austria) and mounted on Formvar-coated nickel grids (Electron Microscopy Sciences, Fort Washington, PA). Immunostaining was performed with an automated immunogold labeling system (Leica EM IGL) as follows: the grids were incubated successively in phosphate-buffered saline (PBS) containing 50 mM NH4Cl (2 times for 5 min each time), PBS containing 1% bovine serum albumin (BSA; 2 times for 5 min each time), PBS containing the relevant primary antibody (for VDAC1, antibody ab15895; for Lamp1, antibody ab25630; for Lamp2, antibody ab25631 [all antibodies were from Abcam]) in 1% BSA for 1 h, PBS containing 0.1% BSA (3 times for 5 min each time), PBS containing 1% BSA and 15-nm colloidal gold-conjugated protein AG (Cell Microscopy Core, University Medical Center, Utrecht, The Netherlands), PBS containing 0.1% BSA for 5 min, and PBS twice for 5 min each time. Lastly, the samples were fixed for 10 min with 1% glutaraldehyde, rinsed in distilled water, and contrasted with a mixture of methylcellulose-sucrose and 0.3% uranyl acetate on ice. After drying in air, the sections were examined under a JEOL 1400 transmission electron microscope. For Lamp1 and Lamp2 staining with mouse primary antibodies, an additional step was added before the incubation with the colloidal gold protein AG: the grids were incubated with a secondary rabbit antibody raised against mouse IgG (antibody Z0259; Dako).
Respirometry and extracellular acidification.
The cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were obtained using a Seahorse XF96 extracellular flux analyzer from Seahorse Bioscience (North Billerica, MA). The final concentrations of the agents are given in the appropriate figure legends. The experiments were performed according to the manufacturer's instructions. Protein standardization was performed after each experiment, and no noticeable differences in protein concentration and cell phenotype were detected.
Immunoblotting.
For immunoblotting, cells were lysed in 1.5× SDS buffer and the protein concentration was determined using a bicinchoninic acid assay. Forty micrograms of protein from whole-cell extracts was resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore). The membranes were blocked in 5% nonfat milk in TN buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) and incubated in the presence of the primary antibody and then secondary antibodies in 5% nonfat milk in TN buffer. The rabbit polyclonal antibody to the central regions of VDAC1 was purchased from Abcam (antibody ab15895). Rabbit polyclonal anti-HIF-1α antibody (antiserum 2087) was produced and characterized in our laboratory (26). Anti-phospho-TP53 (Ser15) (Cell Signaling Technology, Boston, MA), anti-transactivation TP73 (anti-TA TP73) (catalog number IMG-246; Imgenex), anti-Mieap (Novus Biologicals, Cambridge, United Kingdom), anti-p62 (Sigma-Aldrich, St. Louis, MO), and anti-TP63 and anti-beclin 1 (Novus Biological, Cambridge, United Kingdom) were obtained from the indicated manufacturers. Mouse anti-β-tubulin and β-actin were from Sigma, and anti-asparaginyl endopeptidase (anti-AEP; EC 3.4.22.34; Sigma Prestige) was from Sigma. Anti-BNIP3 and anti-BNIP3L were described previously (25). After washing in TN buffer containing 1% Triton X-100 and then in TN buffer, immunoreactive bands were visualized with an ECL system (Amersham Biosciences). The enhanced chemiluminescence (ECL) signal for β-tubulin or β-actin was used as a loading control.
Pulldown with BNIP3 and mass spectrometry.
HepG2 cells with stable expression of the tetracycline repressor (pcDNA6/TR; Life Technologies) were transiently transfected with the pcDNA 4/TO plasmid expressing the HaloTag-BNIP3 fusion protein using the Lipofectamine reagent. At 24 h after the transfection, the cells were then incubated in the presence of 1 μg/ml tetracycline to induce HaloTag-BNIP3 expression and incubated without or with etoposide 50 μM in hypoxia (1% O2) for 16 h.
Protein pulldown was performed according to the HaloTag mammalian pulldown system protocol (Promega). Tobacco etch virus protease cleavage (30 units of Protev proteases in 50 μl of Protev cleavage buffer for 1 h at 25°C) was used to isolate the entire complex, including the bait protein BNIP3 fused to the HaloTag. After elution, proteins were boiled for 5 min with 3-[3-(1,1-bisalkyloxyethyl)pyridin-1-yl]propane-1-sulfonate (PPS; final concentration, 0.8%; silent surfactant; Protein Discovery), reduced for 30 min at 50°C with dithiothreitol (5 mM), and then alkylated for 30 min in the dark using iodoacetamide (15 mM). Samples were then digested overnight at 37°C with trypsin (Trypsin Gold, mass spectrometry [MS] grade; Promega). Prior to MS, samples were acidified with 1 μl of 12 N HCl, and PPS detergent was hydrolyzed after a 45-min incubation at 37°C, followed by centrifugation (10 min, 16,000 × g) at 4°C. Peptides were analyzed using a nano-liquid chromatography (LC)-electrospray ionization-tandem MS maXis Impact ultra-high-resolution time of flight system (Bruker, Bremen, Germany) coupled with a nano-LC UltiMate 3000 system (Thermo). The digests were separated by reverse-phase liquid chromatography using a reverse-phase Thermo column (75 μm by 250 mm; Acclaim PepMap 100 C18) in an Ultimate 3000 liquid chromatography system. Mobile phase A was 95% water, 5% acetonitrile, and 0.1% formic acid. Mobile phase B was 20% water, 79.9% acetonitrile, and 0.1% formic acid. The digest (8 μl) was injected, and the organic content of the mobile phase was increased linearly from 5% mobile phase to 40% mobile phase B in 85 min and from 40% mobile phase B to 100% mobile phase B in 10 min. The column effluent was connected to a Captive Spray apparatus (Bruker). In the survey scan, MS spectra were acquired for 0.5 s in the mass-to-charge (m/z) ratio range of from 50 and 2,200. The 15 most intense peptide ions with charges of +2 or +3 were sequenced. The collision-induced dissociation (CID) energy was automatically set according to the m/z ratio and charge state of the precursor ion. The maXis and Thermo systems were piloted by Compass HyStar (version 3.2) software (Bruker).
Peak lists were created using the DataAnalysis (version 4.0) program (Bruker) and saved as an MGF file for use with the ProteinScape (version 3.1) program (Bruker) and Mascot (version 2.4) as the search engine (Matrix Science). Enzyme specificity was set to trypsin, and the maximum number of missed cleavages per peptide was set to 1. Carbamidomethylation, oxidation of methionine, and Gln–pyro-Glu were allowed as variable modifications. The mass tolerance for the monoisotopic peptide window was 7 ppm, and the MS/MS tolerance window was set to 0.05 Da. The peak lists were searched against the NCBInr mammalian database (entries 02122011 and 2075986) with an automatic decoy database search. Scaffold (version 4.3) program (Proteome Software) was used to validate the MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 95% probability by use of the PeptideProphet algorithm (27) with Scaffold delta-identified peptides. Protein identifications were accepted if they could be established at greater than 5.0% probability to achieve a false discovery rate (FDR) of less than 1.0% and contained at least 2 identified peptides. Protein probabilities were assigned with the ProteinProphet algorithm (28).
Mass spectrometry of VDAC1 and VDAC1-ΔC.
Imperial blue-stained bands excised from SDS-polyacrylamide gels, corresponding to full-length VDAC1 from normoxic and hypoxic cell mitochondria and a truncated form of VDAC1 (VDAC1-ΔC) from hypoxic HeLa cell mitochondria, were digested in gel with trypsin, as recently described (29). For LC-MS analysis, peptides were separated on a reverse-phase LC column (75 μm by 20 cm) packed in-house with C18 resin (Magic C18 AQ; particle size, 3 μm; Michrom BioResources, Auburn, CA) using a linear gradient from 95% solvent A (98% water, 2% acetonitrile, 0.15% formic acid) and 5% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 30% solvent B over 40 min at a flow rate of 0.2 μl/min. Each survey scan acquired in the Orbitrap analyzer at 60,000 full width at half maximum was followed by 20 MS/MS scans of the most intense precursor ions in the linear ion trap with dynamic exclusion enabled for 20 s. Charge state screening was employed to select for ions with at least two charges and rejecting ions with an undetermined charge state. The normalized collision energy was set to 32%, and one microscan was acquired for each spectrum. The ion accumulation time was set to 300 ms (MS) and 50 ms (MS/MS).
For peptide identification, raw files were converted to MGF format using the MassMatrix conversion tool (version 3.9; www.massmatrix.org) and searched against a decoy human Swiss-Prot database consisting of forward and reverse protein sequences (download date, 16 May 2012) containing VDAC and known contaminants, resulting in a total of 41,251 protein sequences using the Mascot (version 2.4) search engine (Matrix Science). The search parameters were set as follows; semitryptic specificity was required, up to two missed cleavages were allowed, carbamidomethyl of cysteine residues was set as fixed, oxidation of methionine residues was set as a variable modification, the precursor mass tolerance was 10 ppm, and the fragment mass tolerance for CID tandem mass spectra was 0.6 Da. After importing the data to Scaffold software (version 4.2.1; Proteome Software), the FDR was set to <1% for peptide identifications by the Scaffold local FDR algorithm.
For peptide quantification, raw files were loaded into the Skyline software tool (version 2.4) to generate extracted ion chromatograms and determine peak abundances for all identified peptide ions generated by the VDAC protein. All integrated peaks/transitions were manually inspected and validated, if required.
SDS-PAGE of recombinant AEP incubated with recombinant VDAC1 or hypoxic HeLa cell mitochondria.
Recombinant VDAC1 expressed in Escherichia coli was reconstituted into either N,N-dimethyldodecylamine N-oxide (LDAO) micelles or large unilamellar vesicles (LUVs) following published protocols (30, 31) and incubated with low-pH-autoactivated (0.1 M citrate buffer, pH 5.5) recombinant AEP (rAEP) (32) for 180 min at room temperature. Samples of inactive or low-pH-autoactivated rAEP and recombinant VDAC1, together with samples of active AEP incubated with VDAC1 LUVs or LDAO micelles, were analyzed by SDS-PAGE. Gels were stained with Coomassie blue. Mitochondria from HeLa cells were isolated using a mitochondrion isolation kit (MitoSciences). Mitochondria were resuspended in 0.1 M citrate buffer (pH 5.5), and rAEP was added. AEP-expressing plasmid pcDNA3.1 was used for overexpression of AEP.
Patients and tissue sample preparation.
Ten patients who underwent surgery for lung adenocarcinoma between May 2007 and May 2010 at the Pasteur Hospital (Department of Thoracic Surgery, CHU of Nice, Nice, France) were selected. The patients received the necessary information concerning the study, and consent was obtained. The study obtained the approval of the Ethics Committee of the CHU of Nice. The main clinical and histopathological data are summarized in Table S2 in the supplemental material. Morphological classification of the tumors was assigned according to the WHO criteria (33). The tumors were staged according to the international tumor-node-metastasis system (34). Tumors with at least 80% tumor cells were selected. Protein and microRNA were extracted from the same tissue sample using an AllPrep DNA/RNA/protein kit according to the protocol described by the manufacturer (Qiagen).
Statistics.
All values are the means ± standard deviations (SDs). Significant differences are based on the Student t test, and P values are indicated in the figures. All statistical tests were two-sided, and P values of <0.05 indicated statistical significance.
RESULTS
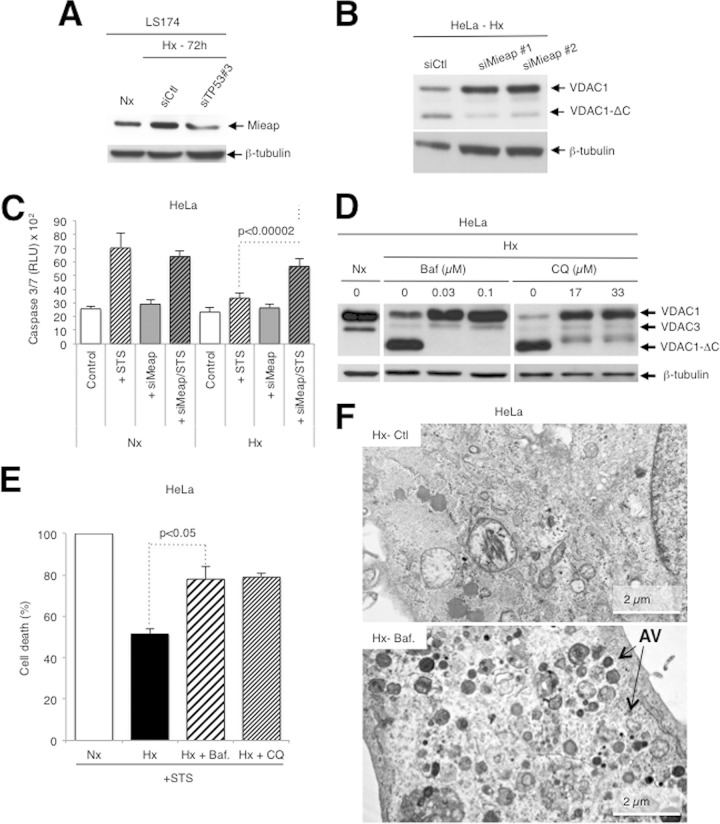
Cleavage of VDAC1 is regulated by TP53.
We previously demonstrated that in hypoxia (1% O2 for 48 to 72 h) certain cell lines (the CCL39, LS174, A549, 786-O, HeLa, and MEF cell lines) showed enlarged functional mitochondria and enhanced resistance to staurosporine (STS)-induced apoptosis, while others (the PC3, SkMel, MDA-MB, and HT29 cell lines) showed a normal tubular mitochondrial network and were sensitive to apoptosis (6). More recently, we showed that hypoxic cells with enlarged mitochondria contained a truncated form of VDAC1 (VDAC1-ΔC), the production of which was HIF-1 dependent (12). We now confirm the HIF-1 dependence of VDAC processing with the pharmacological inhibitor dimethyl-oxalylglycine (DMOG), which stabilizes HIF-1α in normoxia through inhibition of HIF dioxygenases (Fig. 1A). In contrast to the findings in hypoxia, DMOG treatment led to more rapid VDAC1 processing (24 versus 48 h). However, unlike the findings in hypoxia, high concentrations of DMOG (1 mM) induced cell death. So, subsequent experiments were done under the more physiological condition of hypoxia. VDAC1-ΔC was not detected or only minimally detected in normoxia in any of the cell lines tested (Fig. 1B). We now note that the cells with enlarged mitochondria were TP53 wild type, while the other cell lines were either TP53 null or mutant (Fig. 1B). Some of these cell lines also showed increased expression in hypoxia. We showed that in LS174 cells transfected with TP53-specific siRNA, the level of VDAC1-ΔC diminished (Fig. 1C). The amount of full-length VDAC1 was substantially increased, which further confirmed that VDAC1-ΔC is a product of full-length VDAC1, as we previously demonstrated (12). In addition, we examined mouse embryonic fibroblasts either wild type or null for TP53 and noted that the former showed VDAC1-ΔC in hypoxia, while the latter did not (Fig. 1D). Since we found previously that the formation of VDAC1-ΔC is associated with increased resistance to STS-induced apoptosis (12), we examined if silencing of TP53 modified the level of resistance. Indeed, in hypoxia, LS174 cells transfected with TP53-specific siRNA showed more STS-induced cell death than control transfected cells challenged with STS (Fig. 1E). Knowing that HeLa cells are wild type for TP53 but that the level of TP53 is low due to degradation by the human papillomavirus E6 protein (35), as confirmed here (see Fig. S1A in the supplemental material), we questioned further the involvement of TP53. In mammals, the TP53 family comprises two additional proteins, TP63 and TP73, that activate TP53 target genes (36). So, we hypothesized that in HeLa cells these proteins might substitute for TP53 and thereby promote VDAC cleavage and protection from induced apoptosis. However, TP63 was present at only very low levels (see Fig. S1B in the supplemental material) and TP63 siRNA (DeltaNp63- and Tap63-specific siRNAs) did not modify the level of VDAC1-ΔC (see Fig. S1C in the supplemental material) in hypoxic HeLa cells. In contrast, siRNA against TP73 diminished the level of VDAC1-ΔC (see Fig. S1D in the supplemental material) and enhanced sensitivity to STS-induced cell death (Fig. 1F) in HeLa cells. So, TP73 was able to substitute for TP53 in HeLa cells. We also noted that the LS174 cells were mutated in TP73 (37), which further confirmed the involvement of TP53 in truncation, as described above. Silencing of TP53 or of HIF-1α in hypoxic LS174 cells diminished survival, and silencing of the two together further diminished survival in the presence of STS (Fig. 1G).
FIG 1.
Truncation of VDAC1 requires wild-type TP53. (A) HeLa cells were treated for 24 h with dimethyl-oxalylglycine, and cell lysates were analyzed by immunoblotting for HIF-1α, VDAC, or β-actin. (B) Wild-type (wt) or TP53 null or mutated cells were analyzed by immunoblotting for VDAC, HSP90, or β-actin. (C) (Top) LS174 cells were transfected with control siRNA (siCtl) or TP53-specific siRNA (siTP53; 40 nM) in normoxia (Nx) or hypoxia (Hx), and cell lysates were analyzed by immunoblotting for HIF-1α, TP53, phospho-TP53 (P-TP53), VDAC, or β-tubulin. (Bottom) Quantification of TP53 is shown. (D) wt or TP53 null MEFs in hypoxia were analyzed by immunoblotting. (E) LS174 cells were transfected with either control siRNA or TP53-specific siRNA in normoxia or hypoxia and challenged or not challenged with STS (1 mM) for 4 h or not challenged. Apoptosis was evaluated with caspase 3/7 (three independent experiments were performed in quadruplicate, and values are means ± SDs). RLU, relative light units. (F) HeLa cells were transfected with either control siRNA or TP73 siRNA in normoxia or hypoxia and analyzed by immunoblotting. (G) LS174 cells were transfected with control siRNA (siCont) or siRNA against TP53, HIF-1α (siHIF-1α), or both TP53 and HIF-1α and incubated in hypoxia. Cells were then challenged or not challenged with STS (1 mM) for 4 h or not challenged. Apoptosis was evaluated with an Adam system (independent experiments were performed in triplicate, and values are means ± SDs).
TP53 regulation of VDAC1 cleavage occurs through mitochondrial Mieap and is dependent on the endolysosomal pH.
It is known that a TP53-inducible protein, Mieap (18), controls mitochondrial quality (19). Using TP53-specific siRNA, we confirmed, as published previously (19), that Mieap is TP53 inducible (Fig. 2A). Silencing of Mieap expression (see Fig. S1E in the supplemental material) diminished the truncation of VDAC1 (Fig. 2B) and increased STS-induced cell death in hypoxia (Fig. 2C). HeLa cells express only TP73, while LS174 cells express only TP53, yet a similar level of expression of Mieap was noted (Fig. 2A; see also Fig. S1E in the supplemental material). This suggests exchangeable gene activation by TP53/TP73. These results demonstrate that TP53 regulates VDAC1 truncation via Mieap. In fact, Mieap has been proposed to induce accumulation of lysosomal proteins within mitochondria by way of repairing damaged mitochondria (20). However, we were unable to detect the lysosomal membrane protein Lamp1 or Lamp2 in mitochondria by immunoelectron microscopy (see Fig. S2A to C in the supplemental material), and siRNA against Lamp1 and/or Lamp2 did not modify the truncation of VDAC1 (see Fig. S2D in the supplemental material). Instead, we questioned whether lysosomes could contribute to mitochondrial quality control or metabolism by acting from the outside of the mitochondria rather than from the inside. HeLa cells were first incubated in hypoxia in the presence of agents that increase the lysosomal pH, including bafilomycin A1, an inhibitor of lysosomal V-ATPase, as well as chloroquine and NH4Cl, which are weak bases. All these compounds efficiently inhibited the cleavage of VDAC1 to VDAC1-ΔC, confirming an important lysosomal function in VDAC1 cleavage (Fig. 2D; see also Fig. S2E in the supplemental material). Treatment with bafilomycin A1 or chloroquine was also associated with an increase in drug-induced cell death (Fig. 2E).
FIG 2.
Truncation of VDAC1 requires TP53-induced Mieap and an increase in endosomal-lysosomal pH-inhibited VDAC1-ΔC formation. (A) LS174 cells were transfected with either control siRNA (siCtl) or TP53-specific siRNA (siTP53#3; one of three TP53-specific siRNAs; 40 nM), incubated in hypoxia (Hx), and lysed for analysis by immunoblotting. (B) HeLa cells were transfected with either control siRNA or two different Mieap-specific siRNAs (siMieap#1 and siMieap#2; 40 nM), incubated in hypoxia, and lysed for analysis by immunoblotting. (C) HeLa cells were transfected with control siRNA or Mieap-specific siRNA (siMieap; 40 nM), incubated in normoxia (Nx) or hypoxia, and then challenged or not challenged with STS (1 mM) for 4 h. Apoptosis from caspase 3/7 activity was evaluated. Data are from at least three independent experiments performed in quadruplicate, and values are means ± SDs. (D) HeLa cells were incubated in normoxia or hypoxia in the absence or presence of bafilomycin A1 (Baf) or chloroquine (CQ) at the indicated concentrations. The formation of VDAC1-ΔC was detected by immunoblotting. (E) HeLa cells were incubated in normoxia or hypoxia without or with bafilomycin A1 (0.03 mM) or chloroquine (17 mM) and then incubated without or with STS (1 mM) for 24 h. Cell death was evaluated by trypan blue exclusion. (F) Electron micrographs of hypoxic HeLa cells incubated without (control) or with bafilomycin A1 (0.03 μM). AV, autophagic vacuole.
However, as bafilomycin A1 and chloroquine are also known to inhibit the maturation and degradation of autophagic vacuoles, as confirmed here (Fig. 2F; see also Fig. S2F in the supplemental material), and as hypoxia also induces autophagy through HIF-1α stabilization, as seen from the enhanced expression of p62/SQSTM1, the autophagic adaptor protein (see Fig. S3A in the supplemental material), and BNIP3/BNIP3L(NIX) expression (5, 25), we investigated the role of autophagy in VDAC1 processing. However, known inhibitors (LY-294002 and 3-methyladenine) or activators (rapamycin) of autophagy (38) did not modify the processing of VDAC1 (see Fig. S3B in the supplemental material). The inhibitors pepstatin A and E64d (38) did not and did inhibit truncation, respectively (see Fig. S3C in the supplemental material). In addition, siRNA against autophagy-related gene (ATG) proteins, key components of the autophagic machinery, did not modify VDAC processing but did lower the expression of p62 (see Fig. S3D in the supplemental material). Although we have already demonstrated in HeLa cells that hypoxia did not induce mitophagy, we examined further the possible implication of mitophagy, using the model of trifluorocarbonylcyanide phenylhydrazone (FCCP)-depolarized mitochondria. Exogenous overexpression of YFP-Parkin in HeLa cells (see Fig. S3E in the supplemental material) without or with FCCP, used as previously described (24), did not induce the truncation of VDAC1 in normoxia (see Fig. S3F in the supplemental material). Taken together, these results suggest that lysosomes are implicated in VDAC processing but that the mechanism does not involve autophagy and/or mitophagy.
Truncation of VDAC1 is dependent on BNIP3 and mitochondrial fusion but not on the mitochondrial membrane potential.
As Mieap has been reported to interact with BNIP3/BNIP3L at the mitochondrial outer membrane (19), we investigated the role of BNIP3 and BNIP3L in VDAC1 truncation. Using siRNA against BNIP3 and BNIP3L (25), we found that these proteins are necessary for processing of VDAC1 into VDAC1-ΔC (Fig. 3A). In addition, BNIP3L-specific siRNA diminished oxidative phosphorylation (OXPHOS) (Fig. 3B) to a level similar to that obtained with VDAC1-specific siRNA (Fig. 3C), whereas BNIP3-specific siRNA significantly reduced OXPHOS. As we previously showed that enlarged mitochondria resulted from hyperfusion through BNIP3 and BNIP3L (6), we first tested valinomycin, a K+ ionophore that uncouples OXPHOS, as confirmed here (see Fig. S4 in the supplemental material), and prevents the fusion of mitochondria (39). Valinomycin inhibited truncation (Fig. 3D), while neither oligomycin (see Fig. S5A in the supplemental material) nor FCCP (see Fig. S5B and C in the supplemental material) affected truncation in hypoxic cells. However, valinomycin-treated cells partially maintained glycolysis in both normoxia and hypoxia, thereby allowing cells to survive. These results suggest that mitochondrial fusion is a prerequisite to VDAC1 truncation. In addition, siRNA against mitofusin 1 (Mfn1), a protein that we previously showed is essential for hypoxic mitochondrial hyperfusion (6), partially silenced Mfn1 and diminished the level of VDAC1-ΔC (Fig. 3E).
FIG 3.
BNIP3 and mitochondrial fusion are implicated in VDAC1 truncation. (A) HeLa cells were transfected with a control siRNA (siCtl), BNIP3-specific siRNA (siB3; 40 nM), BNIP3L-specific siRNA (siB3L; 40 nM), or siRNAs against both BNIP3 and BNIP3L (siB3/siB3L; 40 nM each), incubated in hypoxia (Hx), and lysed for analysis by immunoblotting for the indicated proteins. (B) HeLa cells were transfected with a control siRNA or BNIP3- or BNIP3L-specific siRNA, and the oxygen consumption rate (OCR) was evaluated with a Seahorse apparatus. Glucose (Glu.; 10 mM), oligomycin (Oligo.; 1 μM), FCCP (1 μM), and rotenone (Rot.; 1 μM) plus antimycin (AA; 1 μM) were injected at the indicated times. (C) HeLa cells were transfected with control siRNA or two different VDAC1-sprecific siRNAs (siVDAC#1 and siVDAC#2), and the oxygen consumption rate was evaluated with a Seahorse apparatus. Glucose (10 mM), oligomycin (1 μM), FCCP (1 μM), and rotenone (1 μM) plus antimycin (1 μM) were injected at the indicated times. (D) Valinomycin was added to HeLa cells and then placed in hypoxia, and lysates were analyzed by immunoblotting. (E) HeLa cells were transfected with control siRNA or Mfn1-specific siRNA (siMfn1#1) and incubated in hypoxia, and lysates were analyzed by immunoblotting. (F) HepG2 cells expressing a tetracycline-inducible HaloTag-BNIP3 fusion protein were incubated in hypoxia (16 h) and then incubated without or with etoposide (Eto; 50 mM). Cell lysates were pulled down using the HaloTag mammalian pulldown system protocol, and proteins were identified by mass spectrometry. The major proteins pulled down are presented for each condition. Results are representative of those from three experiments. Details concerning the individual proteins are given in Table 1. (G) HeLa cells were incubated in hypoxia without or with brefeldin A, and cell lysates were analyzed by immunoblotting.
To further examine the role of BNIP3, we used the HaloTag mammalian pulldown system protocol with BNIP3 as the bait in HepG2 cells. Indeed, these liver-derived cells, which are enriched in mitochondria, presented high levels of VDAC1 and VDAC1-ΔC, which were even upregulated in hypoxia (see Fig. S1A in the supplemental material). We identified by mass spectrometry the binding of several proteins, including clathrin, a structural protein involved in vesicle trafficking (40), in hypoxic HepG2 cells. When apoptosis was induced with etoposide in hypoxic cells, which we previously found induced the same level of cell death as STS (12), BNIP3 also pulled down the lysosomal protein Lamp1 (Fig. 3F; Table 1), which further implicates lysosome involvement in resistance to apoptosis in hypoxia. However, Mieap was not pulled down, suggesting that BNIP3 and Mieap did not bind directly, which contrasts with the findings of a previous report (19). Since pulldown experiments showed that clathrin bound BNIP3 in hypoxic cells and that clathrin is involved in vesicle trafficking, the effect of brefeldin A, an inhibitor of transport of secretory proteins from the endoplasmic reticulum to the Golgi apparatus, was examined. Brefeldin A inhibited VDAC1-ΔC formation (Fig. 3G), which further suggests that secretory vesicles are involved in truncation.
TABLE 1.
Details of major proteins pulled down with a HaloTag-BNIP3 fusion protein expressed in hypoxic HepG2 cells challenged or not challenged with etoposidea
| Accession no. | Protein (source species) | Function | Molecular size (kDa) |
|---|---|---|---|
| gi 17064083 | p25 (Homo sapiens) | A component of the cytoplasmic dynein motor machinery involved in minus-end-directed transport | 25.0 |
| gi 4505773 | Prohibitin (Homo sapiens) | Plays a role in human cellular senescence and tumor suppression | 29.8 |
| gi 30353925 | Clathrin protein (Homo sapiens) | Major protein component of the cytoplasmic face of intracellular organelles, called coated vesicles and coated pits; these specialized organelles are involved in the intracellular trafficking of receptors and endocytosis of a variety of macromolecules | 187.8 |
| gi 28386260 | Pgmc2 protein (Mus musculus) | 23.0 | |
| gi 1020320 | CDM protein (Homo sapiens) | 14.1 | |
| gi 39644554 | Lamp1 protein (Homo sapiens) | Presents carbohydrate ligands to selectins; also implicated in tumor cell metastasis | 33.4 |
| gi 4757900 | Calreticulin precursor (Homo sapiens) | A multifunctional protein that acts as a major Ca2+-binding (storage) protein in the lumen of the endoplasmic reticulum | 48.1 |
| gi 15529966 | Transmembrane protein 205 (Homo sapiens) | Plays a role in resistance to the chemotherapeutic agent cisplatin | 21.2 |
HepG2 cells were in hypoxia for 16 h, and etoposide was used at 50 mM. Results are representative of those from three experiments.
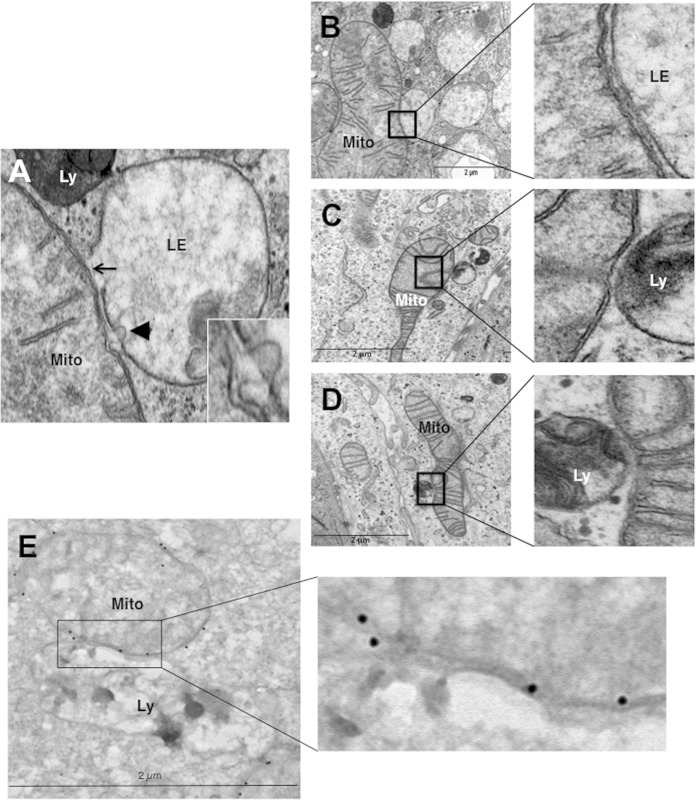
Mitochondria make contact with endolysosomal compartments in hypoxia.
We observed on electron micrographs of hypoxic LS174 cells that hyperfused mitochondria make contact with late endosomes and lysosomes (Fig. 4A). The membranes of mitochondria and endolysosomes showed fusional contact, where some late endosomes puckered up to mitochondria (arrow) or pinched into mitochondria (arrowhead) (Fig. 4A, inset). Although it was difficult to quantify, we frequently noted an interaction between these organelles (Fig. 4B to D) and confirmed the identity of mitochondria by performing immunogold labeling of VDAC (Fig. 4E). In addition, we did not observe enlarged mitochondria in the proximity of autolysosomes. So, these data further support a process other than mitophagy in the transformation of mitochondria in hypoxia.
FIG 4.
Hypoxic LS147 cells show close interactions between mitochondria and late endosomes and lysosomes. (A) A representative image from an electron micrograph of an LS174 cell showing an interaction between a mitochondrion (Mito) and a late endosome (LE) showing membrane rupture (arrow) and a late endosome with a lysosome (Ly) showing membrane rupture (arrowhead; see the inset). (Inset) Enlarged view of a late endosome showing blebbing at the mitochondrion-late endosome contact site. (B to D) (Left) Representative electron micrographs of LS174 cells showing the interaction of a mitochondrion with late endosomes and lysosomes. (Right) Enlarged views of the interaction sites. (E) (Left) Representative electron micrograph of LS174 cells showing immunogold-stained VDAC, allowing detection of mitochondria making contact with a lysosome. (Right) Enlarged view of the contact site.
VDAC1 is C-terminally truncated at asparagine 214 by endolysosomal asparagine endopeptidase.
Mass spectrometric analyses showed that the site of cleavage in VDAC1 was at asparagine 214 (major cleavage site) and glycine 213 (minor cleavage site) (Fig. 5A and B; see also Fig. S6A in the supplemental material). These residues are located in loop 14, which is accessible to the cytosol. The asparagine residue is not conserved in VDAC2 or VDAC3, but the glycine is (Fig. 5B), and these residues are highly conserved in VDAC1 proteins ranging from Xenopus to humans (see Fig. S6B in the supplemental material). Since we suspected the involvement of late endosomes and lysosomes in truncation, we screened siRNAs against some endolysosomal enzymes and found that transfection of asparaginyl endopeptidase (AEP)-specific siRNA lowered the level of expression of AEP and the amount of VDAC1-ΔC (Fig. 5C). We did not detect any difference in the expression levels of the AEP proteins in normoxia compared to hypoxia by immunoblotting. Drug-induced apoptosis was also higher in hypoxic cells transfected with AEP-specific siRNA (Fig. 5D). Incubation of low-pH-autoactivated recombinant AEP (32) with recombinant VDAC1 in large unilamellar vesicles led to minimal degradation; however, in micelles, VDAC1 was degraded, but degradation was substantial (Fig. 5E). Addition of low-pH-autoactivated recombinant AEP to mitochondria that were isolated from normoxic cells and that thus contained only full-length VDAC showed the production of a signal at the level of VDAC1-ΔC (Fig. 5F). Incubation of hypoxic mitochondria with recombinant AEP only minimally increased the amount of the truncated form of VDAC1, VDAC1-ΔC (data not shown). This may suggest that there is some posttranslational modification to VDAC1 that is required for processing but that not all of the full-length form is modified and thus remains insensitive to AEP, though it may be sensitive to another lysosomal enzyme. Note that incubation with trypsin did not modify the ECL profile of samples of normoxic or hypoxic mitochondria (data not shown). However, overexpression of AEP in HeLa cells did not result in either the substantial truncation of VDAC1 in normoxia or an increase in truncation in hypoxia (see Fig. S6C in the supplemental material). Since AEP is normally localized in endolysosomes and active only at low pH, this may suggest that hypoxia-induced local fusion is a prerequisite to truncation and that the degree of local fusion is limiting. In addition, the time frame required to obtain VDAC1 processing in hypoxia is relatively long, 48 h in HeLa cells and 72 h in LS174 cells, which is much longer than the time required to stabilize HIF-1α. We speculate from these results that AEP is responsible for the major cleavage of VDAC1 during microfusional contact of intact mitochondria with endolysosomes in hypoxia, but since we detected two adjacent sites of cleavage, another enzyme or partner protein yet to be identified is also required for efficient truncation.
FIG 5.
Mitochondria-endolysosome contact in hypoxia leads to truncation of VDAC1 at asparagine 214 by the endolysosomal asparagine endopeptidase. (A) Ratios of the abundance of human VDAC1 (hVDAC1) peptides from the normoxic sample to the abundance from the hypoxic sample. Residue numbers are indicated together with the peptide amino acid sequence. The y axis displays the decimal logarithm of the relative abundance of a given peptide in normoxia versus hypoxia. (B) Structure of VDAC1 showing the major cleavage site of VDAC1 C terminal to asparagine 214 (site C) and a minor cleavage site at glycine 213 (site C′). The consensus sequence between human VDAC1, VDAC2, and VDAC3 at the cleavage site is shown by boldface circles. (C) HeLa cells transfected with two different AEP-specific siRNAs (siAEP#1 and siAEP#2) incubated in hypoxia (Hx) and analyzed by immunoblotting (Ctl, control; siCtl, scrambled control siRNA). (D) HeLa cells were transfected with either a control siRNA or asparagine endopeptidase-specific siRNA (siAEP; 100 nM), incubated in normoxia (Nx) or hypoxia, and challenged with STS (1 mM) for 4 h. Apoptosis was evaluated from the level of caspase 3/7. Data from two independent experiments performed in quadruplicate are given as means ± SDs. (E) rAEP was activated by low pH and incubated with recombinant VDAC1 (rVDAC1) reconstituted into either large unilamellar vesicles (LUVs) or N,N-dimethyldodecylamine N-oxide (LDAO) micelles for 180 min at room temperature. The samples were analyzed by SDS-PAGE. Mk, molecular mass marker; rAEP inact., inactive rAEP; rAEP act., pH-activated rAEP. (F) Autoactivated rAEP was added to mitochondria isolated from normoxic HeLa cells, and the mixture was incubated for 30 min at 37°C. The samples were analyzed by immunoblotting for VDAC.
Truncated VDAC1 is present in tumor tissue from patients with wild-type TP53 lung adenocarcinomas, and enlarged mitochondria made contact with endolysosomes.
Using a small cohort of 10 patients with lung adenocarcinomas for whom we had the medical history, we found a correlation between the expression of phosphorylated-TP53 and cleavage of VDAC1 (Fig. 6A). However, this needs to be confirmed with a larger cohort. This would also allow correlation between the levels of the activity of TP53, the amount of VDAC1 processing, and the activation of HIF-induced targets. Nonetheless, the findings obtained with this small cohort allow us to propose the basic principle of cross talk between mitochondria and lysosomes in higher eukaryotes.
FIG 6.
Truncated VDAC is present in lysates of lung adenocarcinoma tissue from patients, and tumor tissue sections show a close interaction between mitochondria and lysosomes. (A) Representative immunoblots of protein extracts of control tissue (lanes C) and tumor tissue (lanes T) from patients' lungs. (B) (Top) A representative electron micrograph of a cell from a patient's lung adenocarcinoma showing the interaction between a mitochondrion (Mito) and lysosomes (Ly); (bottom) enlarged view of the contact site. (C) A representative electron micrograph of tumor tissue showing the interaction of a mitochondrion with lysosomes. (D) (Top) A representative electron micrograph of tumor tissue showing the interaction of a mitochondrion with lysosomes; (bottom) enlarged view of the interaction site.
As in our previous study with a cohort of 46 patients with lung adenocarcinoma (12), VDAC1 truncation tended to be associated with a higher tumor stage and more metastatic relapse in an independent population of lung adenocarcinomas (Tables 2 and 3). Enlarged mitochondria in close contact with lysosomes were detected by electron microscopy of tumor sections (Fig. 6B to D). This was observed in all the patients showing truncated VDAC1. Even though it was not a rare event, it was difficult to quantify. We did not confirm that the regions in the tissue were hypoxic. This would require doing investigations using light and electron microscopy. Contact between mitochondria and lysosomes was not found in patients not showing truncated VDAC1. While these findings are for a small cohort, they do allow us to propose the basic principle of cross talk between mitochondria and lysosomes in higher eukaryotes.
TABLE 2.
Characteristics of patients and their tumors and patient outcomes
| Characteristic | Value |
|---|---|
| No. (%) of patients | 10 (100) |
| No. (%) of patients by gender | |
| Female | 2 (20) |
| Male | 8 (80) |
| Median (range) age (yr) at time of surgery | 62.7 (38–74) |
| No. (%) of patients with tumor size of: | |
| T1 | 5 (50) |
| T2 | 2 (20) |
| T3 | 3 (30) |
| No. (%) of patients with node stage of: | |
| N0 | 5 (50) |
| N1 | 2 (20) |
| N2 | 3 (30) |
| No. (%) of patients with the following stage at diagnosis: | |
| IA | 4 (40) |
| IIB | 1 (10) |
| IIIA | 4 (40) |
| IV | 1 (10) |
| No. (%) of patients with the following histology: | |
| Adenocarcinoma | 10 (100) |
| Relapse (locoregional or metastatic) | 3 (30) |
| No. (%) of patients who died | 5 (50) |
| Median (range) length of follow-up (mo) | 57.5 (38.4–72.0) |
TABLE 3.
Phospho-TP53 and VDAC1-ΔC status of 10 patients with lung adenocarcinomas, determined by immunoblotting, and their tumor grade, adjuvant treatment, relapse, and metastases
| Patient no. | Phospho-TP53 | VDAC1-ΔC | Stage | Adjuvant treatmenta | Relapse | Metastasis | Alive |
|---|---|---|---|---|---|---|---|
| 2 | − | − | IIB | No | − | No | |
| 3 | − | − | IA | Chemo. | − | No | |
| 6 | − | − | IA | No | − | Yes | |
| 1 | + | + | IA | No | − | Yes | |
| 4 | + | + | IV | Cisp. + Nav. | + | Brain | No |
| 5 | + | + | IIIA | Chemo. | + | Lung | No |
| 7 | + | + | IA | No | − | Yes | |
| 8 | + | + | IA | No | − | Yes | |
| 9 | + | + | IIIA | + | + | Brain | No |
| 10 | + | + | IIIA | Cisp. + Nav. | − | Yes |
Chemo, chemotherapy; Cisp, cisplatin; Nav, navelbine.
DISCUSSION
The tumor suppressor TP53 has been reported to function not only as a guardian of the genome but also as a guardian of mitochondrial integrity and function (2). It regulates both mitophagy and mitochondrial metabolism through the induction of genes that regulate, respectively, autophagy and components of the electron transport chain, glycolysis, and the pentose phosphate pathway (41). In addition, TP53 has been reported to activate necrosis by opening the mitochondrial permeability transition pore (42). Although our results point to the involvement of TP53-induced Mieap and BNIP3-mediated regulation of the hypoxic processing of VDAC1, we do not conclude, as suggested previously (19, 20), that lysosome-like organelles or lysosomal proteins accumulate in mitochondria. However, the latter studies examined exogenous overexpression of adenovirus-infected Mieap in normoxia, which contrasts with the conditions of the present study, which examined endogenous proteins in hypoxic cells. We also did not find evidence for the production of mitochondrion-derived vesicles, which are involved in a pathway for shuttling cargo from mitochondria to lysosomes that is an alternative to mitophagy (43, 44). However, this pathway was followed after short-term oxidative stress rather than long-term hypoxia, as in our study. Instead, our results suggest that after long-term hypoxia, late endosomes and lysosomes dock onto mitochondria and bring the outer mitochondrial membrane proteins into contact with their contents. This is in agreement with the findings of the study by Gomes et al., who found that mitochondrial elongation is not dependent on autophagosome formation (8). Our data suggest that mitophagy is not the basis for the mechanism behind VDAC1 processing. This finding confirmed data from our previous study, in which we showed that in hypoxia the area occupied by mitochondria, determined by electron microscopy, was equivalent to or even larger than that in normoxia and that the mass of mitochondria determined by fluorescence-activated cell sorter analysis remained constant (6). So, we propose a novel mechanism in which enlarged mitochondria escape mitophagy and promote metabolic efficiency. Why some mitochondria undergo mitophagy in hypoxia (5) while others persist is still not clear, but it has been suggested that it may be simply sterical; that is, elongated mitochondria may not fit into autophagosomes (8). Our results provide an additional or complementary explanation. We suggest that after long-term stresses, posttranslational modification of VDAC1 marks mitochondria for protection from autophagy and metabolic efficiency. Under such stress conditions, cells maximize ATP production to promote cell survival (6), as do the stress-induced hyperfused mitochondria of MEFs (10), which also rendered them more resistant to cell death. The cleavage of VDAC1 at residues 213 and 214 would result in a protein without β strands 15 to 19 and probably a change in conformation, as suggested by our previous results obtained by immunofluorescence of hypoxic cells with an anti-VDAC antibody (12). This C-terminal segment of the protein contains the binding site for β-NADH (30), which promotes the closed state of VDAC1. If the residual 14 strands resided in the membrane and formed a pore, VDAC1-ΔC would remain constitutively open, allowing optimal efficiency in hypoxia to promote glycolysis through its binding to hexokinases I and II, which we previously showed still bind to VDAC1-ΔC (12). VDAC1-ΔC reconstituted into a planar lipid bilayer also interacted with Bcl-xL and reduced its channel conductance (12). At this stage, we do not know if the C-terminal fragment of VDAC1 is further degraded or if it plays a functional role in the mitochondrial membrane.
We showed that VDAC1 is cleaved by AEP (EC 3.4.22.34), a cysteine protease mainly located in endolysosomes. AEP plays important roles in regulation of the immune system (45) and in cancer (46, 47) and has recently been shown to cleave the protein tau in Alzheimer's disease (48, 49). In addition, its expression and activity are regulated by TP53 (50), and it cleaves peptide bonds carboxy terminal to asparagine (51). The asparagine residue of VDAC1 cleaved by AEP is exposed on the cytoplasmic side of the mitochondria, so this residue could come into contact with the contents of lysosomes. We suggest that BNIP3 acts as a docking site for lysosomes. We also observed that drug-induced apoptosis in hypoxia involves the interaction of BNIP3 with clathrin and Lamp1.
Finally, in the present study we have provided experimental evidence supporting the conclusion that cleavage of VDAC1 reflects a survival response of hypoxic cells that is the readout of an interaction between hyperfused mitochondria with endolysosomes. We show that this phenomenon exists not only in vitro but also in vivo in patients with lung adenocarcinomas. This cross talk between organelles is mediated by TP53-induced Mieap and binding to hypoxia-induced BNIP3. Further understanding of the function of cancer cell mitochondria should stimulate investigation into pharmacological approaches to modulate mitochondrial function to design better cancer treatments.
Supplementary Material
ACKNOWLEDGMENTS
We thank C Pierreux (de Duve Institute, UC Louvain, Louvain, Belgium) for his help with HaloTag protein purification and G. Chinnadurai (Saint Louis University Health Sciences Center, St. Louis, MO) for providing the HA-BNIP3 cDNA-containing plasmid.
This research was supported by grants from the Fondation ARC, Fondation de France, ANR, INCA, la Ligue Nationale Contre le Cancer (Équipe Labelisée LNCC), METOXIA and MOMP (FP7-EU programs), MRT, and Canceropôle PACA. The Institute for Research on Cancer and Aging of Nice is funded by the Centre A. Lacassagne, CNRS, and INSERM.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript. We disclose no potential conflicts of interest.
The authors' contributions are as follows: conception and design, M. C. Brahimi-Horn and N. M. Mazure; development of methodology, M. C. Brahimi-Horn, C. Michiels, S. Hiller, and N. M. Mazure; acquisition of data (acquired and managed patients, provided facilities, etc.), S. Lacas-Gervais, R. Adaixo, K. Ilc, M. Rouleau, A. Notte, M. Dieu, T. Voeltzel, V. Maguer-Satta, M. Ilie, P. Hofman, and A. Schmidt; analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis), M. C. Brahimi-Horn, S. Lacas-Gervais, A. Schmidt, S. Hiller, and N. M. Mazure; writing, review, and/or revision of the manuscript, M. C. Brahimi-Horn, J. Pouysségur, and N. M. Mazure; technical or material support, J. Pelletier and B. Manoury; and study supervision, N. M. Mazure.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01402-14.
REFERENCES
- 1.Brahimi-Horn MC, Bellot G, Pouyssegur J. 2011. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev 21:67–72. doi: 10.1016/j.gde.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. 2014. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland ML, Chourasia AH, Macleod KF. 2013. Mitochondrial dysfunction in cancer. Front Oncol 3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman JR, Nunnari J. 2014. Mitochondrial form and function. Nature 505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. 2008. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Chiche J, Rouleau M, Gounon P, Brahimi-Horn MC, Pouyssegur J, Mazure NM. 2010. Hypoxic enlarged mitochondria protect cancer cells from apoptotic stimuli. J Cell Physiol 222:648–657. doi: 10.1002/jcp.21984. [DOI] [PubMed] [Google Scholar]
- 7.Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. 2012. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem 287:42379–42388. doi: 10.1074/jbc.M112.413682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes LC, Di Benedetto G, Scorrano L. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A 108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, Martinou JC. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wappler EA, Institoris A, Dutta S, Katakam PV, Busija DW. 2013. Mitochondrial dynamics associated with oxygen-glucose deprivation in rat primary neuronal cultures. PLoS One 8:e63206. doi: 10.1371/journal.pone.0063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahimi-Horn MC, Ben-Hail D, Ilie M, Gounon P, Rouleau M, Hofman V, Doyen J, Mari B, Shoshan-Barmatz V, Hofman P, Pouyssegur J, Mazure NM. 2012. Expression of a truncated active form of VDAC1 in lung cancer associates with hypoxic cell survival and correlates with progression to chemotherapy resistance. Cancer Res 72:2140–2150. doi: 10.1158/0008-5472.CAN-11-3940. [DOI] [PubMed] [Google Scholar]
- 13.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. 2010. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med 31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 14.De Stefani D, Bononi A, Romagnoli A, Messina A, De Pinto V, Pinton P, Rizzuto R. 2012. VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death Differ 19:267–273. doi: 10.1038/cdd.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisthal S, Keinan N, Ben-Hail D, Arif T, Shoshan-Barmatz V. 2014. Ca-mediated regulation of VDAC1 expression levels is associated with cell death induction. Biochim Biophys Acta 1843:2270–2281. doi: 10.1016/j.bbamcr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. 2013. Metabolic regulation by p53 family members. Cell Metab 18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb E. 2011. p53 guards the metabolic pathway less travelled. Nat Cell Biol 13:195–197. doi: 10.1038/ncb2177. [DOI] [PubMed] [Google Scholar]
- 18.Bornstein C, Brosh R, Molchadsky A, Madar S, Kogan-Sakin I, Goldstein I, Chakravarti D, Flores ER, Goldfinger N, Sarig R, Rotter V. 2011. SPATA18, a spermatogenesis-associated gene, is a novel transcriptional target of p53 and p63. Mol Cell Biol 31:1679–1689. doi: 10.1128/MCB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura Y, Kitamura N, Shinogi D, Yoshida M, Goda O, Murai R, Kamino H, Arakawa H. 2012. BNIP3 and NIX mediate Mieap-induced accumulation of lysosomal proteins within mitochondria. PLoS One 7:e30767. doi: 10.1371/journal.pone.0030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto Y, Kitamura N, Nakamura Y, Futamura M, Miyamoto T, Yoshida M, Ono M, Ichinose S, Arakawa H. 2011. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS One 6:e16054. doi: 10.1371/journal.pone.0016054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell 30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell 30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. 2007. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood 110:125–132. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 24.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. 2009. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. 1999. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem 274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 27.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 29.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W. 2007. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell 28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. 2008. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varnier A, Kermarrec F, Blesneac I, Moreau C, Liguori L, Lenormand JL, Picollet-D'hahan N. 2010. A simple method for the reconstitution of membrane proteins into giant unilamellar vesicles. J Membr Biol 233:85–92. doi: 10.1007/s00232-010-9227-8. [DOI] [PubMed] [Google Scholar]
- 32.Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C. 2003. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J Biol Chem 278:38980–38990. doi: 10.1074/jbc.M305930200. [DOI] [PubMed] [Google Scholar]
- 33.Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC(ed). 2004. Pathology and genetics of tumours of the lung, pleura, thymus and heart. WHO histological classification of tumors of the lung. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 34.Mountain CF. 1997. Revisions in the international system for staging lung cancer. Chest 111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 35.Hietanen S, Lain S, Krausz E, Blattner C, Lane DP. 2000. Activation of p53 in cervical carcinoma cells by small molecules. Proc Natl Acad Sci U S A 97:8501–8506. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candi E, Agostini M, Melino G, Bernassola F. 2014. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum Mutat 35:702–714. doi: 10.1002/humu.22523. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa H, Nagashima M, Khan MA, McMenamin MG, Hagiwara K, Harris CC. 1999. Mutational analysis of p73 and p53 in human cancer cell lines. Oncogene 18:3415–3421. doi: 10.1038/sj.onc.1202677. [DOI] [PubMed] [Google Scholar]
- 38.Barth S, Glick D, Macleod KF. 2010. Autophagy: assays and artifacts. J Pathol 221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombes A, Rojo M. 2005. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep 6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, Cao Z, Li Y, Ren H, Zhang C, Li L, Chen S, Xi J, Yu L. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol 14:924–934. doi: 10.1038/ncb2557. [DOI] [PubMed] [Google Scholar]
- 41.Bensaad K, Vousden KH. 2007. p53: new roles in metabolism. Trends Cell Biol 17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. 2012. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. 2014. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J 33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. 2012. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol 22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 45.Maschalidi S, Hassler S, Blanc F, Sepulveda FE, Tohme M, Chignard M, van Endert P, Si-Tahar M, Descamps D, Manoury B. 2012. Asparagine endopeptidase controls anti-influenza virus immune responses through TLR7 activation. PLoS Pathog 8:e1002841. doi: 10.1371/journal.ppat.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo P, Zhu Z, Sun Z, Wang Z, Zheng X, Xu H. 2013. Expression of legumain correlates with prognosis and metastasis in gastric carcinoma. PLoS One 8:e73090. doi: 10.1371/journal.pone.0073090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Chen S, Zhang M, Li N, Chen Y, Su W, Liu Y, Lu D, Li S, Yang Y, Li Z, Stupack D, Qu P, Hu H, Xiang R. 2012. Legumain: a biomarker for diagnosis and prognosis of human ovarian cancer. J Cell Biochem 113:2679–2686. doi: 10.1002/jcb.24143. [DOI] [PubMed] [Google Scholar]
- 48.Basurto-Islas G, Grundke-Iqbal I, Tung YC, Liu F, Iqbal K. 2013. Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease. J Biol Chem 288:17495–17507. doi: 10.1074/jbc.M112.446070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Song M, Liu X, Kang SS, Kwon IS, Duong DM, Seyfried NT, Hu WT, Liu Z, Wang JZ, Cheng L, Sun YE, Yu SP, Levey AI, Ye K. 2014. Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer's disease. Nat Med 20:1254–1262. doi: 10.1038/nm.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamane T, Murao S, Kato-Ose I, Kashima L, Yuguchi M, Kozuka M, Takeuchi K, Ogita H, Ohkubo I, Ariga H. 2013. Transcriptional regulation of the legumain gene by p53 in HCT116 cells. Biochem Biophys Res Commun 438:613–618. doi: 10.1016/j.bbrc.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Halfon S, Patel S, Vega F, Zurawski S, Zurawski G. 1998. Autocatalytic activation of human legumain at aspartic acid residues. FEBS Lett 438:114–118. doi: 10.1016/S0014-5793(98)01281-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.