Abstract
Recurrent inactivating mutations in components of SWI/SNF chromatin-remodeling complexes have been identified across cancer types, supporting their roles as tumor suppressors in modulating oncogenic signaling pathways. We report here that SMARCE1 loss induces EGFR expression and confers resistance to MET and ALK inhibitors in non-small cell lung cancers (NSCLCs). We found that SMARCE1 binds to regulatory regions of the EGFR locus and suppresses EGFR transcription in part through regulating expression of Polycomb Repressive Complex component CBX2. Addition of the EGFR inhibitor gefitinib restores the sensitivity of SMARCE1-knockdown cells to MET and ALK inhibitors in NSCLCs. Our findings link SMARCE1 to EGFR oncogenic signaling and suggest targeted treatment options for SMARCE1-deficient tumors.
Keywords: EGFR signaling, drug resistance, RNAi screening, SMARCE1, SWI/SNF
Introduction
Mutations in several components of SWI/SNF complexes have been linked to malignant transformation and progression. For example, ARID1A is found inactivated by mutations at variable frequency (5%–50%) across a broad range of cancer types including subtypes of ovarian cancer, neoplasms of the liver, colon, stomach, pancreas, lung and breast1,2,3,4,5,6,7,8,9. In addition to ARID1A, inactivating mutations in SNF5, PBRM1 and SMARCA4 encoding other SWI/SNF subunits also occur at a high frequencies in several types of cancer10,11. SNF5 is mutated in 98% of rhabdoid tumors12,13,14,15 and PBRM1 is mutated in 41% of renal cell carcinomas16. Expression of SMARCA4 is absent in 15%–50% of primary non-small cell lung cancer (NSCLC) samples17,18 and inactivating biallelic SMARCA4 mutations are found in almost all cases of small cell carcinoma of the ovary, hypercalcaemic type19,20,21. In addition, loss-of-function mutations in SMARCE1 are associated with inherited multiple spinal meningiomas22 and are also found in breast cancer23,24. These examples highlight the tumor suppressor role of the SWI/SNF complexes in human cancer.
The molecular mechanisms by which different SWI/SNF components drive malignant transformation are currently largely unknown. SMARCA4 has been recently shown to regulate expression of MYC-associated factor X gene, MAX, which is inactivated in small cell lung cancer25. However, direct links between other SWI/SNF components and oncogenic signaling remains obscure. This is in part because these multi-component complexes are known to vary in their compositions and associations with other cellular components and transcription factors, which in turn control distinct sets of genes and pathways in different cellular and developmental contexts10,11,26. Thus, the specific mechanisms of different SWI/SNF components are likely context-dependent.
Functional genetic screens provide a powerful tool to uncover novel components of signaling pathways and can help to identify mechanisms of drug resistance in preclinical models of cancer27,28. Using this approach, we previously identified MED12, a component of the transcriptional MEDIATOR complex, as a determinant of responses to inhibitors targeting ALK and EGFR receptor tyrosine kinases in NSCLCs by regulating TGFβ receptor signaling29. As observed for ALK and EGFR inhibitors, the rapid development of drug resistance will also present a major clinical challenge for MET inhibitor-based therapies, which are being tested in clinical trials30. Herein we employ a functional genetic approach to identify genes that modulate drug responses to MET inhibition in NSCLCs. We identify the SWI/SNF component SMARCE1 (also known as BAF57) as a candidate drug resistance gene for MET and ALK inhibitors in NSCLCs and establish its role in regulating EGFR signaling.
Results
Large-scale RNAi screens identify SMARCE1 as a critical determinant of drug responses to MET and ALK kinase inhibitors in NSCLCs
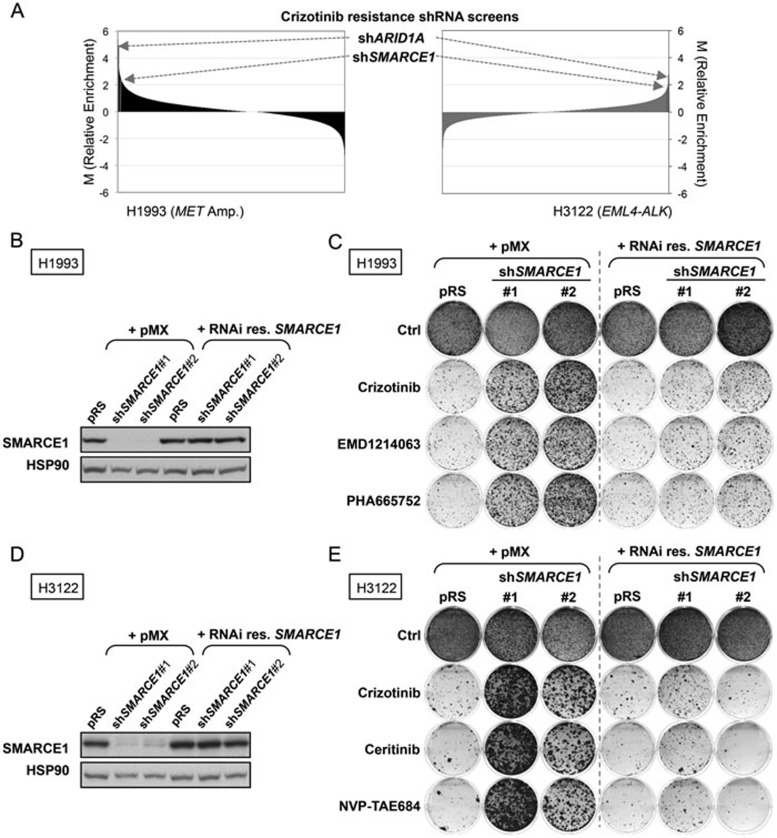
To identify novel genes whose suppression confers resistance to MET inhibition in NSCLC, we performed a large-scale RNAi screen in the H1993 NSCLC cell line, which is driven by MET amplification and is sensitive to the receptor tyrosine kinase (RTK) inhibitor crizotinib (Figure 1A, left). Since crizotinib also effectively targets ALK31, we compared the top candidates from the H1993 screen to those previously identified in our crizotinib resistance screen in the EML4-ALK translocated H3122 NSCLC cell line29 (Figure 1A, right). This analysis identified two shRNAs targeting SWI/SNF chromatin remodeling genes ARID1A and SMARCE1 as the only two common top hits enriched in these different screens (Figure 1A). These results suggest that these two genes are potential modulators of the response to MET and ALK inhibitors.
Figure 1.
Large-scale RNAi screens identify SMARCE1 as a critical determinant of drug responses to MET and ALK kinase inhibitors in NSCLCs. (A) Crizotinib resistance pooled screens performed in MET-amplified H1993 (left panel) and EML4-ALK positive H3122 (as previously described29; right panel) NSCLC cells identify ARID1A and SMARCE1 as the common candidates whose suppression conferred cirzotinib resistance. For the H1993 screen, the NKI human shRNA library was introduced into H1993 cells by retroviral infection, which were then left untreated (control) or treated with 400 nM crizotinib for 14 or 32 days, respectively. After selection, shRNA inserts from both populations were recovered, labeled and hybridized to DNA oligonucleotide arrays for quantification. Data from three independent experiments were normalized, averaged and log2 transformed as previously described50. Relative enrichment values of each shRNA (crizotinib treated vs untreated) are plotted for both screens. The arrow indicates the shARID1A and shSMARCE1 vectors (in Red), which are the only two common hits among the top candidates that were enriched. (B–E) Validation of SMARCE1 in modulating drug responses to MET and ALK inhibitors. Ectopic expression of a RNAi-resistant SMARCE1 cDNA restores the SMARCE1 expression in SMARCE1-knockdown H1993 cells (MET-amplified). H1993 cells expressing pRS control or independent shSMARCE1 vectors were retrovirally infected with viruses containing pMX or pMX-SMARCE1 (RNAi-resistant). The total SMARCE1 protein levels were documented by western blotting. HSP90 was used as a loading control (B). Ectopic expression of a RNAi-resistant SMARCE1 cDNA resensitizes SMARCE1-knockdown cells to MET inhibition in MET-amplified NSCLC cells. The above-described cells (B) were grown in the absence or presence of 300 nM Crizotinib, 150 nM EMD1214063, or 150 nM PHA665752. Cells were then fixed, stained and photographed after 12 days (untreated) or 28 days (treated). Data shown are representative of at least three independent experiments (see Supplementary information, Figure S2 for the quantification and statistical analysis of the independent experiments) (C). Ectopic expression of a RNAi-resistant SMARCE1 cDNA restores the SMARCE1 expression in SMARCE1-knockdown H3122 cells (EML4-ALK positive). H3122 cells expressing pRS control or independent shSMARCE1 vectors were retrovirally infected with viruses containing pMX or pMX-SMARCE1 (RNAi-resistant). The total SMARCE1 protein levels were documented by western blotting. HSP90 was used as a loading control (D). Ectopic expression of RNAi-resistant SMARCE1 cDNA resensitizes SMARCE1-knockdown cells to ALK inhibitors in NSCLC cells harboring EML4-ALK translocation. The above-described cells (D) were grown in the absence or presence of 300 nM Crizotinib, 20 nM Ceritinib or 5 nM NVP-TAE684. Cells were then fixed, stained and photographed after 10 days (untreated) or 21 days (treated). Data shown are representative of at least three independent experiments (see Supplementary information, Figure S2 for the quantification and statistical analysis of the independent experiments) (E).
To rule out the possibility of “off-target” effects of these vectors in causing drug resistance, two independent shRNA vectors for both ARID1A and SMARCE1 were tested in validation assays in multiple cell systems. We found that the expression of these non-overlapping shRNAs efficiently suppressed expression of ARID1A or SMARCE1 and conferred resistance to crizotinib in MET-amplified H1993 cells in long-term colony formation assays (Supplementary information, Figure S1A and S1B). Similarly, these knockdown vectors also conferred resistance to ALK inhibition by crizotinib or NVP-TAE684 in EML4-ALK positive H3122 cells (Supplementary information, Figure S1C and S1D). However, suppression of SMARCE1, but not of ARID1A, conferred crizotinib resistance in an additional NSCLC cell line, EBC1 (MET-amplified; Supplementary information, Figure S1E and S1F). These results validate our initial screens, but also indicate that the effect of ARID1A is potentially context-dependent and suggest a major role for SMARCE1 in modulating drug responses to MET and ALK inhibitors in NSCLCs.
To further validate the role of SMARCE1 in modulating drug responses, we performed rescue experiments using an RNAi-resistant SMARCE1 cDNA, and examined additional inhibitors targeting MET (EMD1214063 and PHA665752) and ALK (Ceritinib). EMD1214063 is currently being tested in clinic and ceritinib has been recently approved by the US Food and Drug Administration to treat ALK-positive metastatic NSCLC patients who have shown disease progression or are intolerant to crizotinib. Reconstitution of SMARCE1 expression in SMARCE1-knockdown cells significantly restored the sensitivity of these cells to all MET and ALK inhibitors in both H1993 and H3122 systems (Figure 1B-1E and Supplementary information, Figure S2). Furthermore, we found that SMARCE1 expression levels correlate with drug responses by using a panel of lentiviral shSMARCE1 vectors with different degrees of knockdown efficiency (Supplementary information, Figure S3). Taken together, our data demonstrate that SMARCE1 is a genuine on-target hit and establish its critical role in regulating responses to MET and ALK inhibition.
SMARCE1 suppression results in activation of AKT and ERK
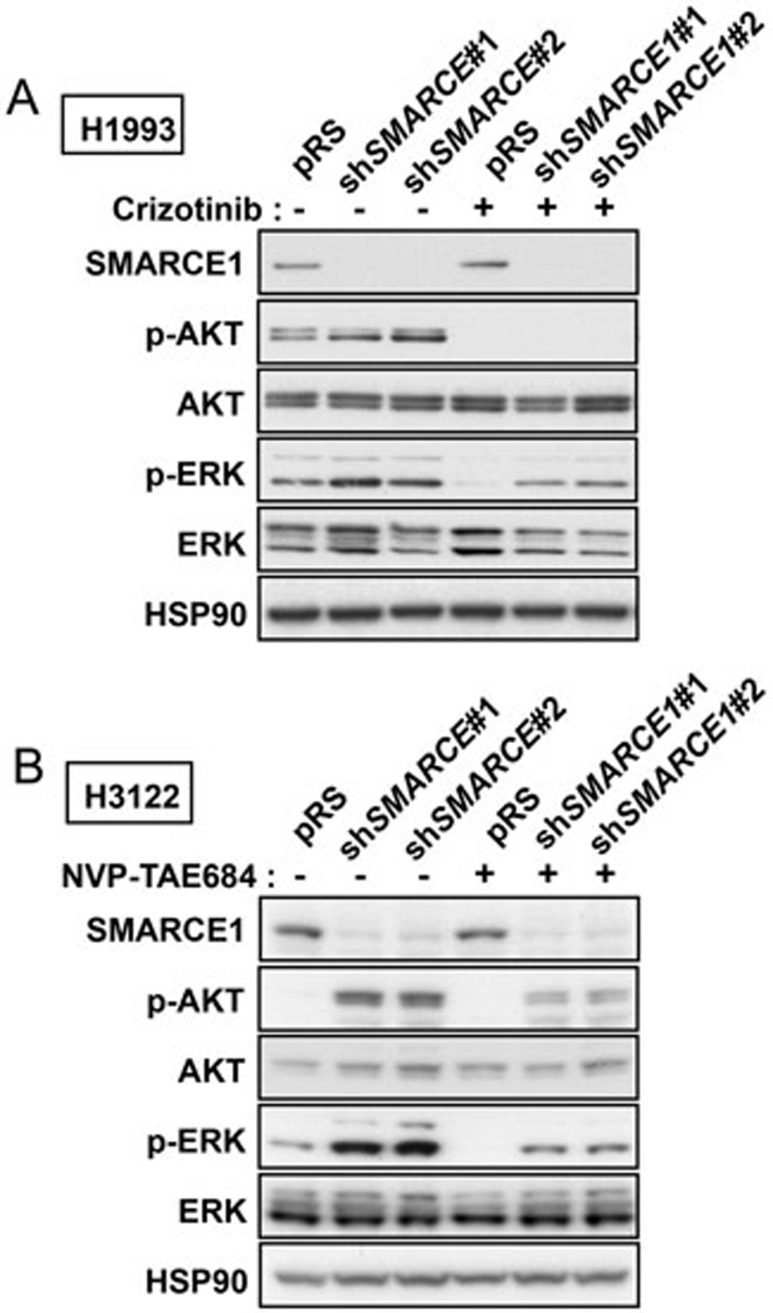
To dissect the underlying mechanisms by which SMARCE1 controls drug resistance, we first analyzed the MAPK/ERK and PI3K/AKT signaling cascades, which represent critical pathways downstream of MET and ALK signaling. We observed that H1993 cells in which SMARCE1 was suppressed maintained significantly higher levels of phosphorylated ERK (p-ERK) in the presence of crizotinib compared to control cells (Figure 2A). Similarly, SMARCE1 suppression also caused H3122 cells to maintain increased levels of both p-ERK and phosphorylated AKT (p-AKT) in the presence of ALK inhibitor (Figure 2B). These results indicate that knockdown of SMARCE1 affects the MAPK/ERK and to a lesser extent the PI3K/AKT signaling routes. Conceivably, activation of these two signaling pathways may contribute to the drug resistance phenotype induced by SMARCE1 knockdown. Consistent with this notion, expression of active alleles of these signaling components demonstrated that MAPK/ERK activation was sufficient to confer resistance to MET and ALK inhibitors in both H3122 and H1993 cells. Activation of PI3K/AKT signaling was also sufficient to bypass the arrest induced by crizotinib although only in H1993 cells (Supplementary information, Figure S4). These data suggest that SMARCE1 loss confers resistance to MET and ALK inhibitors by enhancing the activation of both MAPK/ERK and PI3K/AKT pathways.
Figure 2.
SMARCE1 suppression leads to activation of AKT and ERK. (A) Knockdown of SMARCE1 results in ERK activation in H1993 cells. H1993 cells expressing pRS control or shSMARCE1 vectors were untreated or treated with 500 nM crizotinib for 3 h. Cell lysates were harvested and immunoblotted for SMARCE1, p-AKT (S473), AKT, p-ERK, and ERK. HSP90 was used as a loading control. (B) Knockdown of SMARCE1 leads to activation of AKT and ERK in H3122 cells. H3122 cells expressing pRS control or shSMARCE1 vectors were untreated or treated with 50 nM NVP-TAE684 for 3 h. Cell lysates were harvested and immunoblotted for SMARCE1, p-AKT, AKT, p-ERK, and ERK. HSP90 was used as a loading control.
SMARCE1 suppression leads to induction of EGFR
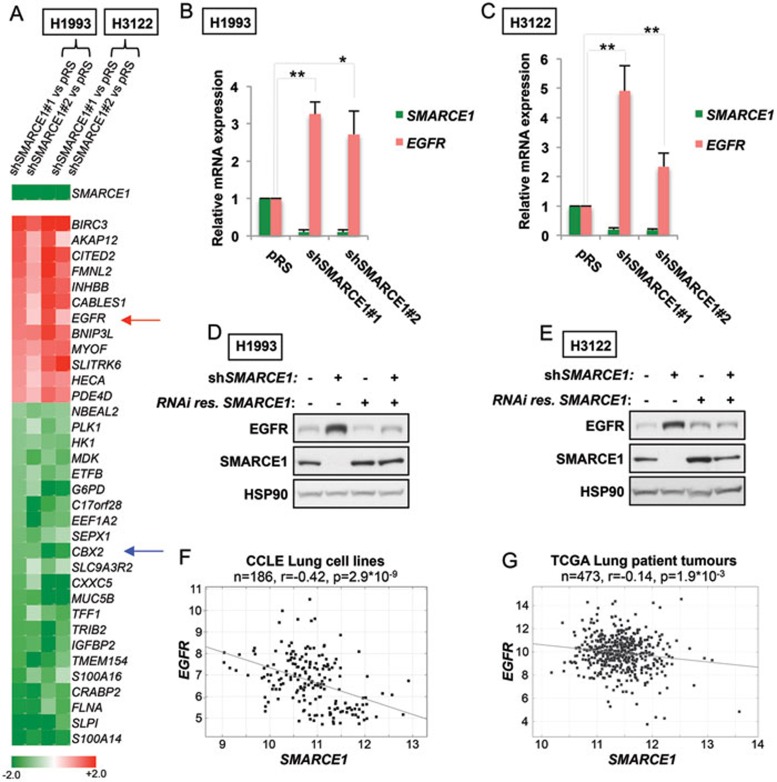
We next sought to determine the mechanism by which SMARCE1 regulates MAPK/ERK and PI3K/AKT pathways. As a component of the SWI/SNF chromatin-remodeling complexes, loss of SMARCE1 may directly impact the transcription machinery. Therefore, a subset of the genes/pathways that are dysregulated upon knockdown of SMARCE1 may contribute to activation of MAPK/ERK and PI3K/AKT pathways in causing drug resistance. To identify target genes of SMARCE1, we performed transcriptome sequencing analysis (RNA-Seq) in H3122 and H1993 control cells and multiple SMARCE1-knockdown derivatives thereof (to avoid potential off-target effects). A heat map representation of the top target genes with altered expression upon SMARCE1 knockdown in both H3122 and H1993 cell lines is shown in Figure 3A. Interestingly, we found EGFR among the genes upregulated upon suppression of SMARCE1.
Figure 3.
Suppression of SMARCE1 leads to induction of EGFR. (A) Expression heat map of the top genes regulated by SMARCE1 (shSMARCE1 vs pRS control; shSMARCE#2 vs pRS control). Transcriptome expression analysis was performed in H1993 and H3122 cells and multiple SMARCE1-knockdown derivatives using next-generation sequencing (RNA-Seq). EGFR is indicated by red arrow and CBX2 is indicated by blue arrow (Unit: log2). (B, C) Suppression of SMARCE1 leads to induction of EGFR expression. SMARCE1 and EGFR mRNA expression was measured by qRT-PCR in H1993 (B) and H3122 (C) cells stably expressing pRS control or shRNA vectors targeting SMARCE1. Error bars denote SD; * and ** denote P values < 0.05 and < 0.01 of three independent biological replicates, respectively. (D, E) Reconstitution of SMARCE1 suppressed EGFR induction driven by SMARCE1 knockdown. H1993 (D) and H3122 (E) cells expressing pRS control or independent shSMARCE1 vectors were retrovirally infected with viruses containing pMX or pMX-SMARCE1 (RNAi-resistant). Cell lysates were immunoblotted for EGFR, SMARCE1, and HSP90. (F, G) Inverse correlation between SMARCE1 and EGFR expression in a panel of 186 lung cancer cell lines (F) and 473 human lung adenocarcinomas (G). Relative gene expression levels of SMARCE1 and EGFR were acquired from the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA). R stands for Pearson product-moment correlation coefficient.
In parallel to the transcriptome analysis, we performed unbiased shRNA re-sensitization screens to identify kinases whose suppression restore sensitivity to crizotinib in SMARCE1-knockdown cells, using a lentiviral shRNA library representing the full complement of 518 human kinases32. We only identified EGFR as the common kinase whose suppression restored sensitivity to crizotinib in two independent screens (Supplementary information, Figure S5).
Our two independent and complementary approaches identified EGFR as a potential SMARCE1 target gene and modulator of drug resistance. EGFR is a well-established oncogenic driver RTK in NSCLCs acting upstream of both ERK and AKT pathways33,34. The upregulation of EGFR in SMARCE1-knockdown cells is consistent with our observation that suppression of SMARCE1 enhances the activation of both MAPK/ERK and PI3K/AKT pathways (Figure 2). Moreover, EGFR activation has been causally linked to resistance to MET and ALK inhibitors in different experimental systems and lung patients35,36,37. Collectively, these observations led us to hypothesize that the induction of EGFR is critical for the drug resistance phenotype driven by SMARCE1 loss.
To test this hypothesis, we first examined the EGFR mRNA expression in H1993 and H3122 control cells and multiple SMARCE1-knockdown derivatives by qRT-PCR. In line with our RNA-Seq results, suppression of SMARCE1 induced EGFR expression in both cell systems (Figure 3A-3C). Consistently, we also observed that EGFR protein levels were elevated in SMARCE1-knockdown cells and that restoration of SMARCE1 expression in SMARCE1-knockdown cells efficiently suppressed the EGFR induction resulting from SMARCE1-knockdown in both cell systems (Figure 3D and 3E). These results strongly support a role for SMARCE1 in suppressing EGFR.
As SMARCE1 is known to function through SWI/SNF complexes, we were next interested in determining whether downregulation of other SWI/SNF components can phenocopy SMARCE1 suppression. We found that knockdown of SMARCE1, ARID1A or SMARCA4 (also known as BRG1, an ATPase SWI/SNF subunit) similarly induces EGFR expression and confers resistance to MET inhibition in H1993 cells (Supplementary information, Figure S6A and S6B). In H3122 cells, knockdown of ARID1A or SMARCA4 also induces EGFR expression but to a lesser extent compared to SMARCE1 suppression, which is consistent with their respective drug responses: SMARCE1 knockdown confers a stronger resistance response compared to ARID1A suppression, and SMARCA4 knockdown fails to confer resistance (Supplementary information, Figure S6C and S6D). These differences could likely reflect the context-dependency of SMARCA4 and ARID1A, similar to what we observed in EBC1 cells (Supplementary information, Figure S1). Taken together, these results support a model that SMARCE1 regulates EGFR expression through SWI/SNF chromatin remodeler complexes, and highlight that SMARCE1 has the dominant role in regulating EGFR expression and drug responses.
Next, it was important to determine whether the regulation of EGFR by SMARCE1 is a general mechanism conserved in heterogeneous lung cancer cell lines and patient tumors. Using mRNA gene expression data (Microarray) from the Cancer Cell Line Encyclopedia (CCLE)38, we found an inverse correlation between SMARCE1 and EGFR expression in a panel of 186 lung cell lines (r = −0.42, P = 2.9 × 10−9; Figure 3F). Furthermore, we also observed a mild but significant inverse correlation between SMARCE1 and EGFR mRNA expression in a tumor data set comprising 473 human lung adenocarcinomas, using RNA-Seq data from The Cancer Genome Atlas (TCGA; r = −0.14, P = 1.9 × 10−3; Figure 3G). Collectively, our data establish a novel role of SMARCE1 in suppressing EGFR expression in NSCLCs.
SMARCE1 suppresses EGFR expression in part through regulating CBX2
Since SWI/SNF complexes are mostly known for their role in activating gene expression10,11,26, it is conceivable that SMARCE1 suppresses EGFR transcription indirectly by regulating the expression of transcriptional repressors. To address this possibility, we surveyed our transcriptome analysis data for potential known transcriptional repressors of EGFR among the top targets genes that were downregulated upon SMARCE1 knockdown (Figure 3A). We found that Chromobox Homolog 2 (CBX2), which encodes a component of the Polycomb Repressive Complex 1 (PRC1), was dependent on SMARCE1 for expression. Previous studies have identified EGFR as a Polycomb target gene in both mouse and human cells39,40, and CBX2 was shown to have the strongest binding to Egfr promoter in mouse ES cells among the different CBX family proteins41. Together, these observations suggest that SMARCE1 may be suppressing EGFR transcription through regulating the expression of CBX2.
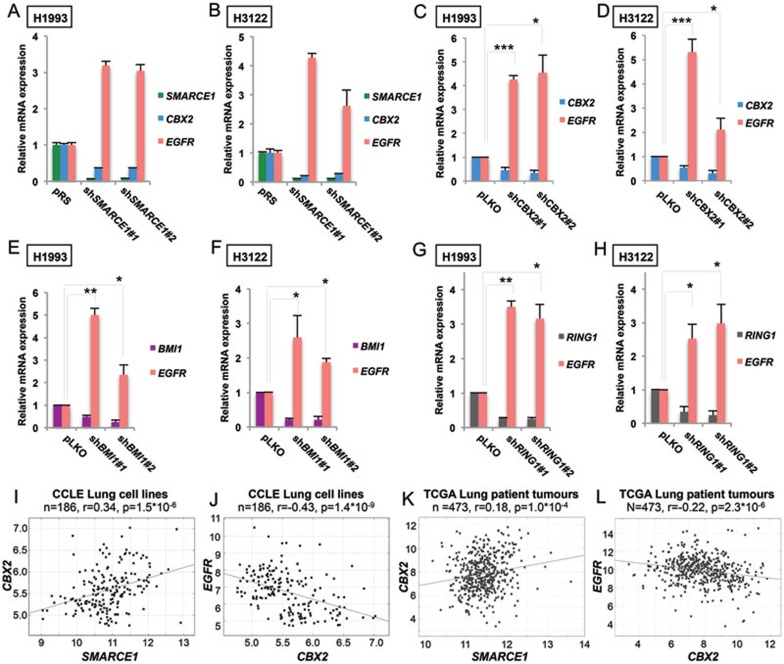
To test this possibility, we first confirmed that CBX2 mRNA was suppressed upon SMARCE1 knockdown and was associated with induction of EGFR mRNA expression in H1993 and H3122 cells using qRT-PCR (Figure 4A and 4B). Next, we found that knockdown of CBX2 with two independent shRNAs resulted in increased levels of EGFR mRNA in both H1993 and H3122 cells (Figure 4C and 4D), indicating that suppression of CBX2 is sufficient to induce EGFR expression. Similarly, downregulation of BMI1 or RING1A, other key PRC1 components, also resulted in increased EGFR expression in both H1993 and H3122 cells (Figure 4E-4H).
Figure 4.
SMARCE1 knockdown suppresses CBX2 expression and PRC1 is required for suppression of EGFR. (A, B) Suppression of SMARCE1 leads to downregulation of CBX2 and upregulation of EGFR expression. SMARCE1, CBX2 and EGFR mRNA expression was analyzed by qRT-PCR in H1993 (A) and H3122 (B) cells stably expressing pRS control or shRNA vectors targeting shSMARCE1. Error bars denote SD. (C, D) CBX2 knockdown causes induction of EGFR expression. CBX2 and EGFR mRNA expression was analyzed by qRT-PCR in H1993 (C) and H3122 (D) cells stably expressing pLKO control or independent shRNAs targeting CBX2. Error bars denote SD; * and *** denote P values < 0.05 and < 0.001 of three independent biological replicates, respectively. (E-H) Suppression of additional PRC1 components, BMI1 and RING, leads to a transcriptional induction of EGFR in NSCLC cells. BMI1, RING1 and EGFR mRNA levels of H1993 (E and G) and H3122 (F and H) cells expressing pLKO control or independent shRNAs targeting BMI1 (E and F) or RING1 (G and H) was measured by qRT-PCR. Error bars denote SD; * and ** denote P values < 0.05 and < 0.01 of three independent biological replicates, respectively. (I, J) SMARCE1 expression is positively correlated with CBX2 expression (I) and CBX2 expression is negatively correlated with EGFR expression (J) in the panel of 186 CCLE lung cell lines. Relative gene expression levels of SMARCE1, CBX2 and EGFR were acquired from CCLE. R stands for Pearson product-moment correlation coefficient. (K, L) SMARCE1 expression is positively correlated with CBX2 expression (K) and CBX2 expression is negatively correlated with EGFR expression (L) in 473 TCGA human lung adenocarcinomas. Relative gene expression levels of SMARCE1, CBX2 and EGFR were acquired from TCGA. R stands for Pearson product-moment correlation coefficient.
Moreover, we performed chromatin immunoprecipitation (ChIP) experiments in H3122 control or SMARCE1-knockdown cells using an antibody against H2AK119ub, which is an established histone mark catalyzed by PRC142, and subjected the ChIP materials along with whole-cell extracts (WCE) to next-generation sequencing. We identified H2AK119ub signals within 110 kb region downstream of EGFR transcription start site within the first intron, and these signals are reduced in SMARCE1-knockdown cells (Supplementary information, Figure S8). The region encompassing the first intron of EGFR gene has been shown to be critical to regulate EGFR transcription and includes possible binding sites for repressor proteins43,44. Taken together, these data support the role of the PRC1 repressor complex in suppressing EGFR.
Consistent with these observations, we found that SMARCE1 expression is positively correlated with CBX2 expression (r = 0.34; P = 1.5 × 10−6) and that CBX2 expression is inversely correlated with EGFR expression (r = −0.43; P = 1.4 × 10−9), in the CCLE lung cell line panel where we observed the inverse correlation between SMARCE1 and EGFR expression (Figures 4I, 4J and 3F). Moreover, a positive correlation between SMARCE1 and CBX2 expression (r = 0.18; P = 1.0 × 10−4), as well as an inverse correlation between CBX2 and EGFR expression (r = −0.22; P = 2.3 × 10−6) were also found in the same set of 473 TCGA lung adenocarcinomas (Figure 4K and 4L), in which we observed the inverse correlation between SMARCE1 and EGFR mRNA expression (Figure 3G). Collectively, these results support a model in which SMARCE1 suppresses EGFR expression in part through regulating CBX2 expression in NSCLCs.
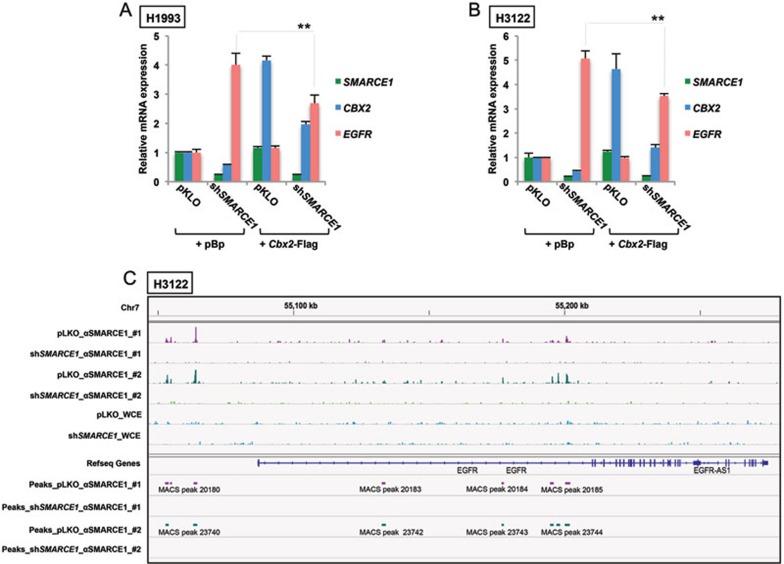
In line with this model, we found that restoration of CBX2 by forced expression of a Cbx2-Flag cDNA partially suppresses EGFR induction driven by SMARCE1 knockdown in both H1993 and H3122 cells (Figure 5A, 5B and Supplementary information, Figure S8). These results further establish the contribution of CBX2 to the regulation of EGFR by SMARCE1, and suggest that SMARCE1 may also regulate EGFR expression through additional mechanisms that are independent of CBX2.
Figure 5.
SMARCE1 suppresses EGFR expression in part through regulating CBX2 and is recruited to regulatory elements of EGFR locus. (A, B) Restoration of CBX2 expression partially suppresses EGFR induction in SMARCE1-knockdown cells. H1993 (A) or H3122 (B) cells stably expressing pLKO control or lentiviral shSMARCE1#3 were retrovirally infected with viruses containing pBabe or Cbx2-Flag cDNA. SMARCE1, CBX2 (total) and EGFR mRNA expression was analyzed by qRT-PCR. Error bars denote SD; ** denote P values < 0.01 of triplicates of representative experiments (See Supplementary information, Figure S7 for additional experiments). (C) SMARCE1 is recruited to regulatory elements of EGFR locus. Chromatin immunoprecipitation (ChIP) experiments were performed in H3122 cells expressing pLKO control or shSMARCE1#3 using an antibody against SMARCE1. ChIP materials along with WCE (whole-cell extract) were sequenced using next-generation sequencing. Enriched regions in the genome were identified by comparing the ChIP samples to mixed input (WCE) using the MACS peak caller. Snapshot of the binding events of SMARCE1 adjacent to or within EGFR gene from two independent biological replicates are shown.
SMARCE1 is recruited to regulatory elements of EGFR locus
Given that some SWI/SNF complexes are also capable of repressing gene expression45,46, it is also possible that SMARCE1 suppresses EGFR expression through direct regulation. To test this possibility, we performed ChIP experiments in H3122 control or SMARCE1-knockdown cells using an antibody against SMARCE1 and subjected the ChIP materials along with WCE to next-generation sequencing. We found strong SMARCE1 binding peaks spanning the 20 kb region upstream of EGFR gene, as well as the 110 kb regulatory region downstream of EGFR transcription start site within the first intron where H2AK119ub signals were also detected (Figure 5C and Supplementary information, Figure S7). Similar results were also obtained using a second antibody against SMARCE1 (data not shown). Importantly, these SMARCE1 binding signals were abolished in SMARCE1-knockdown cells, indicating that they are genuine SMARCE1 binding peaks. These observations suggest that SMARCE1 can also potentially regulate EGFR transcription through these direct interactions with the regulatory regions of EGFR locus.
EGFR inhibition resensitizes SMARCE1-deficient cells to MET and ALK kinase inhibitors in NSCLCs
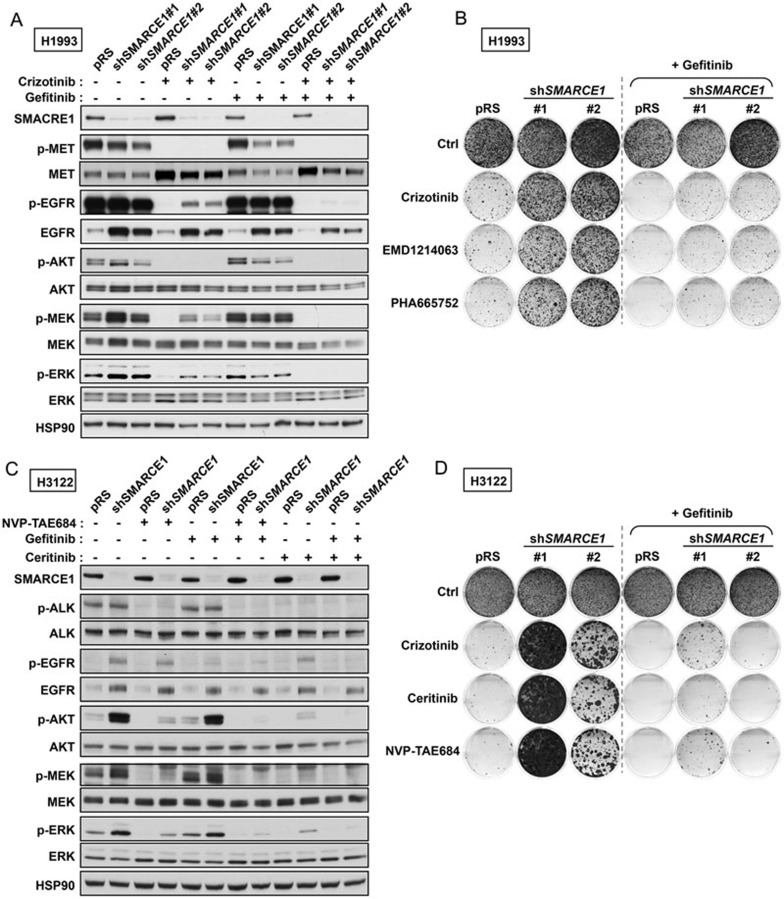
Since EGFR activation has been causally linked to resistance to MET and ALK inhibitors35,36,37, we then asked whether EGFR upregulation is required for the observed drug resistance to MET and ALK inhibitors driven by SMARCE1 knockdown using a selective EGFR inhibitor gefitinib. Consistent with the elevated total protein levels of EGFR induced by SMARCE1 knockdown, H1993 and H3122 cells expressing shSMARCE1 vectors maintained elevated p-EGFR levels in the presence of different inhibitors targeting MET (crizotinib and EMD1214063) or ALK (NVP-TAE684 and ceritinib), coinciding with the elevated downstream p-ERK and pAKT levels in the same cells (Figure 6A, 6C and Supplementary information, Figure S9). We found that addition of the EGFR inhibitor gefitinib to treatments targeting MET or ALK abolished the elevated levels of p-EGFR, p-AKT and p-ERK in SMARCE1-knockdown cells (Figure 6A, 6C and Supplementary information, Figure S9). These results suggest that the elevated MAPK/ERK and PI3K/AKT signaling in SMARCE1-knockdown cells upon inhibition of MET or ALK results from EGFR activation. Consequently, the addition of gefitinib restored the sensitivity of SMARCE1-knockdown cells to MET and ALK inhibitors in H1993 and H3122 cells in long-term growth assays (Figure 6B, 6D and Supplementary information, Figure S10). Similar results were also obtained in EBC1 cells (Supplementary information, Figure S11), confirming the critical role of EGFR activation in the drug resistance driven by SMARCE1 knockdown.
Figure 6.
EGFR inhibition resensitizes SMARCE1-deficient cells to MET and ALK kinase inhibitors in NSCLCs. (A) EGFR inhibitor suppressed the ERK activation driven by SMARCE1 knockdown in MET-amplified NSCLC cells. H1993 cells expressing pRS or shSMARCE1 vectors were cultured in the absence or presence of 500 nM crizotinib, 500 nM gefitinib or their combination for 3 h. Cell lysates were harvested for immunoblot analysis and probed for the indicated proteins. (B) Combination of EGFR and MET inhibitors synergistically inhibits growth of SMARCE1-knockdown NSCLC cells driven by MET amplification. H1993 cells expressing pRS control or shSMARCE1 vectors were cultured in the absence and the presence of 300 nM crizotinib, 150 nM EMD1214063, 150 nM PHA665752, 500 nM gefitinib, or their combinations as indicated. The cells were fixed, stained and photographed after 14 (untreated and gefitinib alone) or 34 days (MET inhibitors alone and gefitinib plus MET inhibitors). Data shown are representative of at least three independent experiments (See Supplementary information, Figure S10 for the quantification and statistical analysis of the independent experiments). (C) EGFR inhibitor suppressed the ERK and AKT activation driven by SMARCE1 knockdown in NSCLC cells harboring EML4-ALK translocation. H3122 cells expressing pRS or shSMARCE1 vectors were cultured in the absence or presence of 100 nM NVP-TAE684, 400 nM Ceritinib, 1 μM gefitinib or their combination for 3 h. Cell lysates were harvested for immunoblot analysis and probed for the indicated proteins. (D) Combination of EGFR and MET inhibitors synergistically inhibits growth of SMARCE1-knockdown NSCLC cells harboring EML4-ALK translocation. H3122 cells expressing pRS or shSMARCE1 vectors were cultured in the absence or presence of 300 nM Crizotinib, 20 nM Ceritinib, 5 nM NVP-TAE684, 250 nM gefitinib or their combination as indicated. The cells were fixed, stained and photographed after 10 (untreated and gefitinib alone) or 28 days (ALK inhibitors alone and gefitinib plus ALK inhibitors). Data shown are representative of at least three independent experiments (See Supplementary information, Figure S10 for the quantification and statistical analysis of the independent experiments).
These results are consistent with our re-sensitization screens identifying EGFR whose suppression restores sensitivity to crizotinib in SMARCE1-knockdown cells (Supplementary information, Figure S5). Collectively, our findings establish that SMARCE1 suppresses EGFR expression, whose activation is required for the drug resistance phenotype induced by SMARCE1 knockdown and suggest a therapeutic strategy to treat lung cancers with SMARCE1 loss.
Discussion
Our study identifies SMARCE1 as a critical modulator of the drug response to MET and ALK inhibitors in NSCLC cells and provides a direct link between this SWI/SNF component and EGFR oncogenic signaling. Mechanistically, SMARCE1 suppresses EGFR expression, in part through regulating the levels of Polycomb Repressive Complex component CBX2. This transcriptional network between SMARCE1, CBX2 and EGFR is supported by gene expression data of the CCLE lung cell line panel and a large number of primary lung tumors. In addition, our ChIP-seq data suggests that SMARCE1 may regulate EGFR expression through direct binding to regulatory elements of EGFR locus.
Suppression of ARID1A, another SWI/SNF component identified in our screens, also confers resistance to MET and ALK inhibitors in H1993 and H3122, but not in EBC1 cells. Similarly, downregulation of SMARCA4 confers drug resistance in H1993 but not in H3122 cells. This may reflect a context-dependent role of SWI/SNF complexes, which are known to vary in their compositions and associations with other cellular components10,11,26. Consistent with this, we did not observe an inverse correlation between ARID1A or SMARCA4 and EGFR expression in the same lung tumor data sets (data not shown), where SMARCE1 expression is negatively correlated with EGFR expression. Given the context-dependency of SWI/SNF complexes, our data do not rule out the possibility that loss of SMARCE1 activates other oncogenic pathways independent of EGFR in other cell types.
Crosstalk between different RTK signalings is important in mediating resistance to single agent-based therapies targeting EGFR, ALK and MET. For example, MET amplification accounts for ∼ 20% of EGFR mutant NSCLC tumors that acquired resistance to EGFR inhibitors47, and EGFR activation results in resistance to MET inhibition in MET-driven gastric cancer cells35,36. In addition, EGFR activation is associated with acquired resistance to crizotinib in EML4-ALK positive NSCLC patients37. These clinical data support our finding that EGFR activation is critically required for drug resistance to MET and ALK inhibitors driven by SMARCE1 loss in NSCLCs. Such treatment combinations targeting EGFR and MET or ALK are currently being explored in clinical trials (NCT01441128, NCT01121575, NCT01801111). While drug resistance patient tumor samples are needed to further validate the role of SMARCE1 in modulating drug responses, the regulation between SMARCE1 and EGFR signaling is likely hard-wired as suggested by our correlation analysis using the large number of lung patient tumors (Figures 3G, 4K and 4I). Since SMARCE1 has not been documented to be mutated in lung cancers, our study suggests that its expression may be a potential predictive marker for drug responses to MET and ALK inhibitors and that EGFR inhibitors may be effective to treat SMARCE1-deficient tumors.
Our study establishes a novel role for SMARCE1 in modulating drug responses through regulating EGFR expression in lung cancers. Given the frequent involvement of SWI/SNF complexes and EGFR signaling in various human cancers, our findings may have extended implications to other tumor types in which these signaling pathways have a key role.
Materials and Methods
Compounds and antibodies
Crizotinib (S1068), NVP-TAE648 (S1108), gefitinib (S1025), EMD1214063 (S7067), PHA665752 (S1070) and Ceritinib (S7083) were purchased from Selleck Chemicals (Houston, TX, USA). Primary antibodies used are as follows: EGFR (1005), HSP90 (H-114), pERK1/2 (T202/Y204; E4) and ERK1/2 (C16) were from Santa Cruz Biotechnology; pEGFR (Y1068) was from AbCam; p-MET(3126), MET(3127), p-ALK(3341), ALK(3333), p-AKT (S473) (4060), AKT(2920), p-MEK(#9154), MEK (4694) and H2AK119ub (8240) antibodies were from Cell Signaling; ARID1A (H00008289-M01) was from Abnova; SMARCE1 (A300-810A) was from Bethyl Laboratories; Anti-Flag M2 was from Sigma.
Plasmids
All retroviral shRNA vectors were generated by ligating synthetic oligonucleotides (Invitrogen) against the target genes into the pRetroSuper retroviral vector as described48. The following RNAi target sequences were used for retroviral shRNA vectors for this study. shGFP, 5′-GCTGACCCTGAAGTTCATC-3′ shARID1A#1, 5′-GGGGTGAGCTGCAACAAAG-3′ shARID1A#2, 5′-AGGAGAAGCTGATCAGTAA-3′ shSMARCE1#1, 5′-GGAAGAAAGTCGACAGAGA-3′ shSMARCE1#2, 5′-GGAGAACCGTACATGAGCA-3′ shSMARCA4#1, 5′-GGGTACCCTCAGGACAACA-3′ shSMARCA4#2, 5′-GATTTGCGAACCAAAGCGA-3′. shSMARCA4 vectors are gifts from Dr Katrien Berns (NKI, Amsterdam).
Individual lentiviral shRNA vectors targeting CBX2 were collected from the TRC library. shCBX2#1, TRCN0000020325; shCBX2#2, TRCN0000020327; shSMARCE1#3, TRCN0000015780; shBMI1#1, TRCN0000020155; shBMI1#2, TRCN0000020157; shRING1#1, TRCN0000021990; shRING1#2, TRCN0000021993.
The human SMARCE1 expression construct and the RNAi resistant forms were generated by PCR amplifying SMARCE1 from H3122 cDNA using the following primers: forward, 5′-GTACGAATTCCACCATGTCAAAAAGACCATCTTATGC-3′ reverse, 3′-GAATAAGTGTTGCCTTGTTTTGTGCTCGAGACTG-5′. The fragment was cloned into the retroviral expression vector pMX-IRES-blasticidin using the EcoRI and XhoI restriction sites in the multiple cloning site and sequence verified. The SMARCE1 that is resistant against shSMARCE1#1 was generated by site-directed mutagenesis using the following primer pair: forward, 5′-GCATGGAGAAAGGAGAGCCATATATGAGCATTCAGCCTG-3′ reverse, 3′-CAGGCTGAATGCTCATATATGGCTCTCCTTTCTCCATGC-5′.
The SMARCE1 that is resistant against shSMARCE1#2 was generated by site-directed mutagenesis using the following primer pair: forward, 5′-GAAGCTGCTTTAGAGGAGGAGAGCCGACAGAGACAATCTC-3′ reverse, 3′-GAGATTGTCTCTGTCGGCTCTCCTCCTCTAAAGCAGCTTC-5′. Both SMARCE1-ND clones were sequence verified. The pBABE-Cbx2-Flag (#1952) was obtained from Addgene.
Cell culture and viral transduction
H1993, H3122 and EBC1 cells were cultured in RPMI with 8% fetal bovine serum (FBS), penicillin and streptomycin at 5% CO2. HEK 293T and Phoenix were cultured in DMEM with 8% FBS, penicillin and streptomycin at 5% CO2. Subclones of each cell line expressing the murine ecotropic receptor were generated and used for all experiments shown. Retroviral and lentiviral infections were performed as described29.
Phoenix cells were used as producers of retroviral supernatants as described at http://www.stanford.edu/group/nolan/retroviral_systems/phx.html.
HEK 293T cells were used as producers of lentiviral supernatants as described at http://www.broadinstitute.org/rnai/public/resources/protocols.
The calcium phosphate method was used for the transfection of Phoenix and 293T cells. Selection of infected cells was performed for 7-10 days in 2 μg/ml puromycin or in 10 μg/ml blasticidin.
For the CBX2 restoration assays, H1993 and H3122 cells were first infected with lentivirus harboring pLKO or shSMARCE1#3. After puromycin selection, cells were then infected with 3 rounds of diluted retrovirus containing pBABE or Cbx2-Flag cDNA in order to mimic endogenous CBX2 expression levels. Transduced cells were plated and collected for qRT-PCR and immunoblot analysis 3-6 days after the last retroviral infection. Experiments with the reverse order of infections were also performed for H3122 cells.
Pooled shRNA screens
The human NKI shRNA library and the pooled shRNA screens were performed as described49,50. Additional details can be found at http://www.screeninc.nki.nl. shRNA “Dropout” Screen using a custom TRC Kinome Library was performed as described32,51.
Colony formation assay
Single cell suspensions of the lung cancer cell lines were seeded into 6-well plates (2-4 × 104 cells/well) and cultured both in the absence and presence of the indicted inhibitors. At the endpoints of colony formation assays, cells were fixed with formaldehyde, stained with crystal violet (0.1% w/v) and photographed. Crystal violet was then extracted with 10% acetic acid and measured at OD 590 nM. All relevant assays were performed independently at least three times.
Protein lysate preparation and immunoblot
Cells were seeded in medium containing 8% FBS. After 24 h, the medium was replaced with fresh medium (8% FBS) containing the inhibitors as indicated in the text. After the drug stimulation, cells were washed with cold PBS, lysed with protein sample buffer and processed with Novex NuPAGE Gel Electrophoresis Systems (Invitrogen).
Quantitative RT-PCR (qRT-PCR)
Cells were first seeded and then harvested for RNA isolation using Trizol (Invitrogen) the next day. qRT-PCR assays were carried out to measure mRNA levels of genes as described52. Relative mRNA levels of each gene shown were normalized to the expression of the house-keeping gene GAPDH. The sequences of the primers for assays using SYBR Green master mix (Roche) are as follows:
GAPDH_forward, 5′-AAGGTGAAGGTCGGAGTCAA-3′
GAPDH_reverse, 3′-AATGAAGGGGTCATTGATGG-5′
ARID1A_forward, 5′-CCAACAAAGGAGCCACCAC-3′
ARID1A_reverse, 3′-TCTTGCCCATCTGATCCATT-5′
SMARCE1_forward, 5′-CGGCTTATCTGGTGGCTTT-3′
SMARCE1_reverse, 3′-AACAACTACAGGCTGGGAGG-5′
EGFR_forward, 5′-TCCTCTGGAGGCTGAGAAAA-3′
EGFR_reverse, 3′-GGGCTCTGGAGGAAAAGAAA-5′
CBX2_Pan-forward, 5′-GCGGCTGGTCCTCCAAACA-3′
CBX2_Pan-reverse, 3′-CTTGCCTCTCTTCCGGTTCT-5′
BMI1_forward, 5′-CTTTCATTGTCTTTTCCGCC-3′
BMI1_reverse, 3′-TCGTTGTTCGATGCATTTCT-5′
RING1_forward, 5′-TTGAGCTCCAGCATTGAGG-3′
RING1_reverse, 3′-CCACTCATCGTTGTGGTCTG-5′
SMARCA4_forward, 5′-GTGAAGAGAAGCGAGACGCC-3′
SMARCA4_reverse, 3′-AGTGGACATCTTCACGGGAG-5′.
RNA-Seq gene expression analysis
Transcriptome sequencing analysis of cell lines were performed using RNA-Seq as described29. To rule out “off-target” effects, we considered genes that are significantly deregulated in the same direction by two independent shSMARCE1 vectors. The top genes regulated by SMARCE1 contain genes that were up- or downregulated by > 2-fold upon SMARCE1 knockdown.
Gene expression statistical analysis
The mRNA gene expression (microarray) data for the lung cell line panel was obtained from the CCLE38 (CCLE_Expression_Entrez_2012-10-18.res). The mRNA gene expression (RNA-seq) data for the 473 lung adenocarcinomas samples was obtained from TCGA. The TCGA Level 3 data (normalized and gene-level summarized) was downloaded in December 2013. TCGA mRNA gene expression levels were log2 transformed. P-values for the correlation coefficients are derived from a Student's t-distribution under the null hypothesis of no correlation.
Chromatin immunoprecipitation-sequencing
ChIP-Seq experiments were performed as described53. For each reaction, 2 ml cell lysate was transferred to a 15 ml tubes and sonicated using Bioruptor (HIGH-power mode, 40 cycles of 30 s ON and 30 s OFF).
Acknowledgments
We thank Ron Kerkhoven and other members of NKI Genomics Core Facility for support. We also thank Iris de Rink, Arno Velds and Roel Kluin for their kind help on data analysis. We are grateful to Cinzia Pochet for support. This work was supported by grants from Canadian Institutes of Health Research (CIHR) (MOP-130540 to SH), Canada Research Chair (CRC), the European Research Council (ERC), the Dutch Cancer Society (KWF), the EU COLTHERES project and grants by the Netherlands Organization for Scientific Research (NWO) to the Cancer Genomics Center Netherlands (CGC.NL) and the Cancer System Biology Center (CSBC). AIP is a recipient of the CIHR funded McGill Chemical Biology Postdoctoral Fellowship Award. SH is supported by CRC in Functional Genomics.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Validation of individual shRNAs targeting ARID1A and SMARCE1 in modulating drug responses to MET and ALK kinase inhibitors in multiple NSCLC cell systems.
Validation of SMARCE1 in modulating drug responses to MET and ALK inhibitors.
Differential SMARCE1 expression correlates with drug response in H3122 cells.
MAPK/ERK activation is sufficient to confer resistance to MET and ALK inhibitors in NSCLCs, whereas AKT activation results in resistance to only MET inhibition.
Crizotinib re-sensitization Kinome screen in SMARCE1 knockdown cells identifies EGFR as the top candidate required for drug resistance driving by SMARCE1 knockdown.
Multiple SWI/SNF components regulate EGFR expression and drug responses to MET and ALK inhibitors in NSCLC cells.
Histone mark processed by PCR1 is detected in the regulatory elements of EGFR locus in a SMARCE1 dependent manner.
Restoration of CBX2 level partially suppresses EGFR induction in SMARCE1 knockdown cells.
EGFR inhibitor suppressed the ERK activation driven by SMARCE1 knockdown in MET-amplified NSCLC cells.
EGFR inhibition resensitizes SMARCE1 deficient cells to MET and ALK kinase inhibitors in NSCLCs.
EGFR inhibition resensitizes SMARCE1 deficient EBC1 NSCLC cells to MET inhibitor.
References
- Wiegand KC, Lee AF, Al-Agha OM, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–333. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih IM, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Lawrence MS, Voet D, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer Nature 2012487330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M. SWI/SNF Chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem. 2012;287:30897–30905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Sevenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N, Lellouch-Tubiana A, Schofield D, et al. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- Duns G, Hofstra RM, Sietzema JG, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33:1059–1062. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- Medina PP, Sanchez-Cespedes M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics. 2008;3:64–68. doi: 10.4161/epi.3.2.6153. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Nieto S, Canada A, Pros E, et al. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat. 2011;32:E1999–E2017. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- Witkowski L, Carrot-Zhang J, Albrecht S, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- Ramos P, Karnezis AN, Craig DW, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–429. doi: 10.1038/ng.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupryjanczyk J, Dansonka-Mieszkowska A, Moes-Sosnowska J, et al. Ovarian small cell carcinoma of hypercalcemic type - evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Pol J Pathol. 2013;64:238–246. doi: 10.5114/pjp.2013.39331. [DOI] [PubMed] [Google Scholar]
- Smith MJ, O'Sullivan J, Bhaskar SS, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- Villaronga MA, Lopez-Mateo I, Markert L, Espinosa E, Fresno Vara JA, Belandia B. Identification and characterization of novel potentially oncogenic mutations in the human BAF57 gene in a breast cancer patient. Breast Cancer Res Treat. 2011;128:891–898. doi: 10.1007/s10549-011-1492-4. [DOI] [PubMed] [Google Scholar]
- Kiskinis E, Garcia-Pedrero JM, Villaronga MA, Parker MG, Belandia B. Identification of BAF57 mutations in human breast cancer cell lines. Breast Cancer Res Treat. 2006;98:191–198. doi: 10.1007/s10549-005-9149-9. [DOI] [PubMed] [Google Scholar]
- Romero OA, Torres-Diz M, Pros E, et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2014;4:292–303. doi: 10.1158/2159-8290.CD-13-0799. [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Martin SA, Ashworth A. RNA interference screening demystified. J Clin Pathol. 2009;62:195–200. doi: 10.1136/jcp.2008.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Bernards R. Understanding resistance to targeted cancer drugs through loss of function genetic screens. Drug Resist Updat. 2012;15:268–275. doi: 10.1016/j.drup.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Huang S, Holzel M, Knijnenburg T, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- Ou SH. Crizotinib: a novel and first-in-class multitargeted tyrosine kinase inhibitor for the treatment of anaplastic lymphoma kinase rearranged non-small cell lung cancer and beyond. Drug Des Devel Ther. 2011;5:471–485. doi: 10.2147/DDDT.S19045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J, Yarden Y, Mayes E, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- Qi J, McTigue MA, Rogers A, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71:1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachleitner-Hofmann T, Sun MY, Chen CT, et al. HER kinase activation confers resistance to MET tyrosine kinase inhibition in MET oncogene-addicted gastric cancer cells. Mol Cancer Ther. 2008;7:3499–3508. doi: 10.1158/1535-7163.MCT-08-0374. [DOI] [PubMed] [Google Scholar]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Chrysogelos SA. Chromatin structure of the EGFR gene suggests a role for intron 1 sequences in its regulation in breast cancer cells. Nucleic Acids Res. 1993;21:5736–5741. doi: 10.1093/nar/21.24.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Meyer-Staeckling S, Schmidt H, Agelopoulos K, Buerger H. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–7260. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- Nagl NG, Zweitzig DR, Thimmapaya B, Beck GR, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- Huang S, Laoukili J, Epping MT, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of individual shRNAs targeting ARID1A and SMARCE1 in modulating drug responses to MET and ALK kinase inhibitors in multiple NSCLC cell systems.
Validation of SMARCE1 in modulating drug responses to MET and ALK inhibitors.
Differential SMARCE1 expression correlates with drug response in H3122 cells.
MAPK/ERK activation is sufficient to confer resistance to MET and ALK inhibitors in NSCLCs, whereas AKT activation results in resistance to only MET inhibition.
Crizotinib re-sensitization Kinome screen in SMARCE1 knockdown cells identifies EGFR as the top candidate required for drug resistance driving by SMARCE1 knockdown.
Multiple SWI/SNF components regulate EGFR expression and drug responses to MET and ALK inhibitors in NSCLC cells.
Histone mark processed by PCR1 is detected in the regulatory elements of EGFR locus in a SMARCE1 dependent manner.
Restoration of CBX2 level partially suppresses EGFR induction in SMARCE1 knockdown cells.
EGFR inhibitor suppressed the ERK activation driven by SMARCE1 knockdown in MET-amplified NSCLC cells.
EGFR inhibition resensitizes SMARCE1 deficient cells to MET and ALK kinase inhibitors in NSCLCs.
EGFR inhibition resensitizes SMARCE1 deficient EBC1 NSCLC cells to MET inhibitor.