Abstract
Glioblastoma multiforme (GBM) is a lethal, therapy-resistant brain cancer consisting of numerous tumor cell subpopulations, including stem-like glioma-initiating cells (GICs), which contribute to tumor recurrence following initial response to therapy. Here, we identified miR-182 as a regulator of apoptosis, growth, and differentiation programs whose expression level is correlated with GBM patient survival. Repression of Bcl2-like12 (Bcl2L12), c-Met, and hypoxia-inducible factor 2α (HIF2A) is of central importance to miR-182 anti-tumor activity, as it results in enhanced therapy susceptibility, decreased GIC sphere size, expansion, and stemness in vitro. To evaluate the tumor-suppressive function of miR-182 in vivo, we synthesized miR-182-based spherical nucleic acids (182-SNAs); i.e., gold nanoparticles covalently functionalized with mature miR-182 duplexes. Intravenously administered 182-SNAs penetrated the blood–brain/blood–tumor barriers (BBB/BTB) in orthotopic GBM xenografts and selectively disseminated throughout extravascular glioma parenchyma, causing reduced tumor burden and increased animal survival. Our results indicate that harnessing the anti-tumor activities of miR-182 via safe and robust delivery of 182-SNAs represents a novel strategy for therapeutic intervention in GBM.
Keywords: miR-182, Bcl2L12, c-Met, HIF2A, glioblastoma, nanotechnology, spherical nucleic acids
Patients diagnosed with the most common and aggressive manifestation of malignant glioma, glioblastoma multiforme (GBM), have a median survival of well under 2 years (Dunn et al. 2012). The current standard of treatment entails surgical resection followed by radiation therapy combined with administration of the DNA-alkylating agent temozolomide (TMZ), which increases median survival by only 2.4 mo (Stupp et al. 2009). Continued advances in surgical practices, the advent of new radiotherapy techniques (Crowley et al. 2006), and the testing of targeted therapies have failed to improve this outcome.
Recently, we identified the atypical Bcl-2 family protein Bcl2-like12 (Bcl2L12) as a potent oncogene and therapeutic RNAi target in GBM that blocked therapy-induced apoptosis and promoted gliomagenesis in vivo by inhibiting effector caspase and p53 activation (Stegh et al. 2007, 2008a,b, 2010; Stegh and DePinho 2011; Jensen et al. 2013). While Bcl2L12 is highly expressed in almost all primary GBM specimens, levels of overexpression vary by GBM subtype. Specifically, in silico oncogenomic analyses revealed robust genomic amplification (nonfocal 19q event) as well as mRNA overexpression of Bcl2L12 in mesenchymal, classical, and neural GBM subtypes and lower expression levels in proneural tumors. Importantly, genomic amplification of the Bcl2L12 gene locus does not account for high Bcl2L12 expression in many GBM tumors (Stegh et al. 2010), indicating additional mechanisms of regulation besides copy number alteration. Thus, the understanding of molecular mechanisms that regulate Bcl2L12 transcript levels will provide important insights for therapeutic approaches aimed at reducing Bcl2L12 expression in established glioma.
MicroRNAs (miRNAs) are small noncoding RNAs (with a length of ∼22 nucleotides) that silence gene expression via mRNA degradation, deadenylation, or translational repression (Kim 2005). Multiple genomic studies have identified miRNAs as critical regulators of GBM pathogenesis and therapy response (Huse and Holland 2009). In addition, miRNA signatures have been described as prognostic and diagnostic factors and as markers for GBM subtype classification (Setty et al. 2012). While the precise role and specific gene targets for most miRNAs remain elusive, several have been determined that target signaling pathways important in regulating glioma growth and survival, including receptor tyrosine kinase (RTK)–PI3K–PTEN, retinoblastoma, Bcl-2, and p53 signaling (Iorio and Croce 2009, 2012).
Despite the expanding knowledge of miRNA function in cancer, inadequate delivery and poor dissemination throughout tumor parenchyma remain intractable problems that have impeded functional studies of miRNAs in vivo and have prevented the implementation of miRNA based therapy into clinical practice (Nana-Sinkam and Croce 2013). To neutralize oncogene expression in glioma orthotopic xenografts, our group developed RNAi-based spherical nucleic acids (SNAs) as a novel siRNA-based nanotechnological platform for biotherapeutic gene silencing. SNAs cross the blood–brain barrier (BBB) and blood–tumor barrier (BTB) upon systemic intravenous administration, trigger robust intratumoral protein knockdown, increase intratumoral apoptosis, reduce tumor cell proliferation, and impair glioma progression with an up to fivefold reduction in tumor burden in the absence of significant toxicity and immunogenicity (Jensen et al. 2013).
Here, we demonstrate that miR-182 acts as a tumor suppressor by controlling the expression and activity of oncogenes deregulated in GBM; i.e., Bcl2L12, c-Met, and hypoxia-inducible factor 2α (HIF2A). Treatment with miR-182 increased apoptotic cell death in response to chemotherapeutic treatments such as TMZ and RTK inhibitors (RTKIs) in a Bcl2L12-dependent manner and influenced sphere formation, expansion, and differentiation capacities of patient-derived glioma-initiating cells (GICs) by repressing stem cell-associated mRNA signatures and phenotypes controlled by c-Met and HIF2A. To further evaluate antitumor properties of miR-182 in GBM, we used SNAs functionalized with mature miR-182 sequences (182-SNAs) that penetrate transformed glioma cells and GICs in the absence of auxiliary transfection agents. Upon systemic, intravenous administration to glioma-bearing mice, 182SNAs disseminated throughout extravascular tumor parenchyma that resulted in a reduction of tumor burden and an increase in animal subject survival in vivo in the absence of significant adverse side effects. In summary, our studies suggest that miR-182 expression represents a novel therapeutic avenue against GBM tumors and point to the SNAs as a platform for miRNA-based biotherapeutic gene silencing for the treatment of GBM.
Results
Identification of miR-182 as a p53-controlled proapoptotic factor in GBM that targets Bcl2L12
Bcl2L12 is a critical inhibitor of therapy-induced apoptosis with overexpression in nearly all GBM (Stegh et al. 2007, 2008a,b, 2010; Stegh and DePinho 2011). We aimed to identify miRNAs that control the expression of Bcl2L12 in GBM, as such miRNAs represent potent tumor suppressors with chemosensitizing activity and are thus ideal candidates for subsequent design and preclinical evaluation of SNA therapeutics.
As a first step, in silico studies of GBM samples of the multidimensional Cancer Genome Atlas (TCGA) data set (http://cancergenome.nih.gov/dataportal; Cancer Genome Atlas Research Network 2008) were conducted to discover miRNAs with expression levels that negatively correlate with Bcl2L12 mRNA abundance. This oncogenomic analysis identified miR-182 that negatively correlated with Bcl2L12 mRNA, especially in proneural tumor specimens (R=−0.32, P=0.01) (Supplemental Fig. S1A), which are characterized by frequent p53 mutation (Verhaak et al. 2010). Negative correlation between Bcl2L12 and miR-182 expression levels were subsequently confirmed by RT-qPCR in a collection of frozen primary GBM specimens (n=45, R=−0.61, P=8.65×10−6) (Supplemental Fig. S1B). Here, GBM tumors carrying p53 mutations showed elevated levels of miR-182 compared with those with wild-type p53 (Supplemental Fig. S2A).
Next, we determined molecular mechanisms that regulate miR-182 expression. In silico promoter analysis (Transcriptional Regulatory Element Database [TRED]) identified a total of five putative p53-binding sites, three of which are located in the promoter proximal to the miR-182 transcriptional start site (termed proximal p53 response elements [p53REproximal]), and two found in an upstream promoter region that controls the expression of the entire miR-183-96-182 cluster (p53-REdistal) (Supplemental Fig. S2B). Luciferase assays revealed that wild-type but not dominant-negative p53 repressed reporter activity of both proximal and distal promoters (Supplemental Fig. S2C,D). Induction of a luciferase reporter containing multimerized p53 consensus sites is shown as a positive control to confirm that ectopically expressed p53 is functional (Supplemental Fig. S2E). Similarly, endogenous miR-182 levels decreased upon treatment of U87MG and transformed astrocytes with doxorubicin, a well-described DNA damage-inducing agent and p53 activator (Supplemental Fig. S2F). Together, these data suggest that miR182 expression in GBM is controlled by p53, and that elevated miR-182 levels in proneural GBM tumors negatively correlate with Bcl2L12 mRNA abundance.
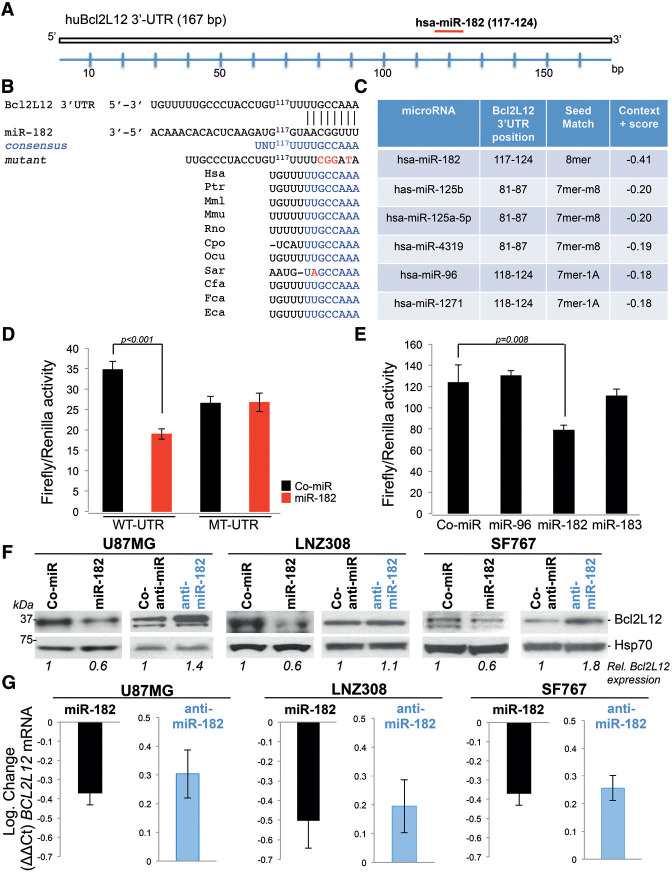
To functionally validate these in silico analysis results and further characterize a potential Bcl2L12–miR-182 axis, we engineered a luciferase reporter construct harboring the full-length (167-base-pair [bp]) human Bcl2L12 3′ untranslated region (UTR) (Fig. 1A), including an evolutionarily conserved miR-182-binding site identified by TargetScan (position 117–124 of the Bcl2L12 3′ UTR) (Fig. 1B,C). Cotransfection with synthetic miR-182 but not a nontargeting control miR (Co-miR) resulted in 50% reduction in luciferase levels (Fig. 1D). miR-96 and miR-183, members of the 183-96-182 miR cluster, failed to repress Bcl2L12 3′ UTR reporter activity (Fig. 1E). To further confirm that miR-182 binds the Bcl2L12 3′ UTR, we generated a luciferase reporter with a mutated miR182-binding site. Disrupting the binding site completely prevented repression by the synthetic miR-182 (Fig. 1D). Subsequently, Co-miR or miR-182 sequences were ectopically expressed in three glioma cell lines (i.e., U87MG, LNZ308, and SF767) (see Supplemental Table S1 for p53 mutational status of glioma cell lines used in this study). Western blot and RT-qPCR analyses confirmed robust down-regulation of Bcl2L12 on mRNA and protein levels upon miR-182 expression and also showed increased Bcl2L12 protein and mRNA levels upon expression of a miR-182 inhibitor (anti-miR-182) (Fig. 1F,G). These studies indicate that miR-182, but not other members of the miR-183-96-182 cluster transcribed from the same polycistronic transcript, directly binds to the 3′ UTR of Bcl2L12 and represses its mRNA and protein expression.
Figure 1.
miR-182 negatively regulates Bcl2L12 expression in GBM. (A) Schematic representation of the 3′ UTR of the Bcl2L12 gene, including the miR-182 target site identified by TargetScan. (B) Position of the miR-182-binding site within the 3′ UTR of Bcl2L12 and alignment of miR182-binding sites among different species. (C ) Ranking of miRNAs that bind to the Bcl2L12 3′ UTR sequence. (D) Luciferase activity measured in 293T cells 24 h after transfection of wild-type or mutant Bcl2L12-3′ UTR-pGL3 reporter vectors in combination with synthetic, premature miR-182 or Co-miR sequences at a concentration of 200 nM. (E) Luciferase activity measured 24 h after transfection of 293T cells transiently expressing a Bcl2L12-3′ UTR-pGL3 construct in combination with 200 nM miR-96, miR-182, or miR-183. (F,G) U87MG, LNZ308, and SF767 cells were transfected with 15 nM Co-miR or miR-182 and 100 nM Co-anti-miR or anti-miR-182 for 48 h, and the effects on Bcl2L12 protein and mRNA levels were assessed by Western blotting (F ) and RTqPCR (G). Results are presented as log2 expression. In all cases, Hsp70 is shown as a loading control. Histograms depict mean values±standard deviations.
miR-182 sensitizes glioma cells toward chemotherapy via down-regulation of Bcl2L12
Next, we determined whether miR-182 regulates oncogenic properties of Bcl2L12, in particular its ability to neutralize effector caspases in cells and tumors. To assess the functional consequences of miR-182-mediated knockdown of Bcl2L12, we examined effector caspase activation, activity, and overall apoptosis rates upon ectopic miR-182 expression in three glioma cell lines (U87MG, LNZ308, and LN229). miR-182 transfectants treated with the pankinase inhibitor staurosporine (STS) displayed significantly higher levels of active caspase-3 and caspase-7, and, correspondingly, enforced expression of an antimiR-182 sequence blocked effector caspase activation and activity (Fig. 2A–D). Increased effector caspase activation upon miR-182 expression translated into a significant increase in Annexin V positivity (Fig. 2E). To determine whether proapoptotic effects of miR-182 are mediated by its capacity to neutralize Bcl2L12 expression, we expressed anti-miR-182 and Bcl2L12-targeting siRNAs as single reagents or in combination and monitored effector caspase activation and activity. Cotransfection with Bcl2L12-specific siRNAs negated the caspase inhibitory effect of anti-miR-182 (Fig. 2C,D), suggesting that Bcl2L12 is a critical anti-apoptotic miR-182 target. Next, we investigated whether miR-182, by inhibiting Bcl2L12, sensitized glioma cells to TMZ and small ATP mimetics targeting the activity of RTKs; i.e., EGFR-targeting erlotinib, the c-Met inhibitor SU11274, an inhibitor of insulin growth factor receptor (AG1204), and imatinib, a small molecule inhibitor of platelet-derived growth factor receptor (PDGFR). Glioma cells transfected with miR-182 or anti-miR-182 sequences and subsequently treated with TMZ and RTK inhibitor exhibited increased or decreased levels of caspase-3 and caspase-7 maturation, respectively (Fig. 2F,G). Importantly, and similar to our results from analyzing STS-mediated apoptosis (Fig. 2A–D), coexpression of Bcl2L12-specific siRNAs prevented caspase inhibitory effects of anti-miR-182 sequences (Fig. 2G). Thus, the broad chemo-sensitizing effects of miR-182 in response to chemotherapy and RTK inhibition are largely driven by repression of Bcl2L12 expression.
Figure 2.
miR-182 sensitizes glioma cells to therapy-induced apoptosis. (A–D) LN229 and U87MG cells were transfected with 15 nM each Co-miR and miR-182, 50 nM each Co-anti-miR and anti-miR-182, and 100 nM each scramble siRNA or siRNA targeting Bcl2L12 alone and in combination with 100 nM anti-miR-182 (in C,D). After 48 h, cultures were treated with 0.5 µM STS for the indicated periods of time. Activation of caspase-3 (aC-3) and caspase-7 (aC-7) was measured by Western blotting (A,C ), and caspase activity was quantified by a fluorometric DEVDase activity assay (B,D). (E) Overall apoptosis was quantified by FACS-based Annexin V staining in U87MG, LN229, and LN308 cells. (F,G) LNZ308 cells were transfected with 15 nM each Co-miR and miR-182, 50 nM each Co-anti-miR and anti-miR-182, and a combination of siRNA and anti-miR-182 sequences for 24 h (in G) and then treated with 100 µM TMZ for 48 h (F ) and with 5 µM erlotinib (Erl), 5 µM SU11274 (SU), 5 µM AG1024 (AG), and 5 µM imatinib (Im) (G). Levels of cleaved caspase-3 and caspase-7 were measured by Western blotting. In all cases, Hsp70 is shown as a loading control, and histograms depict mean values±standard deviations. (LS) Large subunit; (LS+N) large subunit plus N-peptide.
miR-182 controls a stem cell-associated signature in GBM
Consensus clustering of miRNA expression profiles in 261 TCGA high-grade gliomas identified five clinically and genetically distinct subclasses of GBM resembling major stages of neural cell development; i.e., radial glia, oligoneural, neural, neuroepithelial/neural crest, and astrocyte precursors (Kim et al. 2011). miR-182 is expressed in the oligoneural GBM subtype, which is characterized by reduced mRNA expression of embryonic and neural stem cell signatures and is associated with significantly increased patient survival (Kim et al. 2011). In support of a role for miR-182 in regulating growth and differentiation processes, expression of miR-182 in GICs decreased neurosphere size and expansion upon low-density seeding (Supplemental Fig. S3). In addition, miR-182 reduced cellular growth in response to mitogenic factors, such as transforming growth factor β1 (TGFβ1) (Supplemental Fig. S4), a critical driver of GBM progression with pleiotropic effects on tumor cell growth, invasion, self-renewal, and neo-angiogenesis (Golestaneh and Mishra 2005). Furthermore, miR-182 expression was decreased in CD133-positive, patient-derived GICs in comparison with CD133-negative GICs (Fig. 3A). Upon ectopic expression, miR-182 promoted a more differentiated phenotype, as evidenced by decreased expression of stem cells markers (i.e., Oct4, CD44, Nanog, and Sox2) (Fig. 3B–D), increased numbers of adherent cells (Supplemental Fig. S3C), and elevated levels of MAP2 and GFAP cell positivity (Fig. 3E,F).
Figure 3.
miR-182 regulates GIC stemness. (A) RT-qPCR analysis of miR-182 in patient-derived CD133-positive and CD133negative GICs; shown is the percentage change±SD. (B–D) RT-qPCR analysis of stem cell marker expression in GIC-20, GIC-23, and LNZ308 cells expressing miR182 relative to Co-miR-expressing cultures, shown as log2 fold change±SD. (E,F ) Immunofluorescence staining and quantification of GFAPand MAP2-positive cells in Co-miR-overexpressing or miR-182-overexpressing GICs attached on poly-D-lysine/ laminin-coated coverslips. Bar, 50 µm. (G) In silico overlap analysis of the miR-182 downregulated transcriptome in GICs with the mRNA signatures of seven stages of neural cell differentiation. Numbers next to each celltyperepresent thenumberofoverlapping genes. (ESCs) Embryonic stem cells; (HSCs) hematopoietic stem cells; (NSCs) neural stem cells. (H) Western blot analysis of HIF2A expression levels in GIC-20, GIC-16, LNZ308, and U87MG cells overexpressing Co-miR or miR-182. For the analysis of HIF2A protein levels, cultures were placed in a hypoxic chamber (1.5% O2) for 24 h. (I ) RT-qPCR analysis of HIF2A mRNA levels in GICs overexpressing Co-miR or miR-182. (J) Luciferase activity measured in 293T cells 24–48 h after transfection of HIF2A-3′ UTRpGL3 reporter vectors in combination with miR-182 or Co-miR sequences at a concentration of 200 nM. (K ) LNZ308 cells were transfected with 100 nM siCo, 100 nM siRNA targeting HIF2A, and 100 nM antimiR-182 alone and in combination with 100 nM siHIF2A and, 24 h later, were placed in a hypoxic chamber (1.5% O2) for 24 h. The levels of CD44, Nestin, CCNB1, and Sox2 were measured by Western blotting. (L) Densitometric analysis of Western blots in K.
To investigate the molecular basis of miR-182-dependent growth and differentiation, we subjected patient-derived GICs, following modification with lentivirus for expression of miR-182 or Co-miR, to whole-genome expression profiling (a heat map and a complete list of significantly affected genes are shown in Supplemental Fig. S5; Supplemental Table S2). A comparative analysis of the transcripts repressed by miR-182 and mRNA signatures up-regulated in seven stages of neural cell differentiation (i.e., mRNA expression profiles derived from differentiated neural cells [astrocytes, oligodendrocytes, and neurons] and stem cells [embryonic, hematopoetic, and neural stem cells]) revealed that, similar to miR-182-expressing oligoneural GBM tumors, the miR-182-repressed transcriptome in GICs is enriched for stem cell-related genes (Fig. 3G; see Supplemental Table S3 for a full list of overlapping genes). Together, these comparative genomic studies and functional experiments point to miR-182 as a transcriptional regulator of GIC stemness.
To further define molecular signatures repressed by miR-182, the expression profiling data were used for sequence-based prediction of miR-182 interactions using the TargetScan algorithm. Gene lists were integrated using Ingenuity Pathway analysis, which revealed a miR182-controlled oncogenic signature that includes CDK1, CCNB1, AURKB, and PLK1 as indirect targets (Supplemental Fig. S6A) and c-Met, WNT5a, ZIC2, and HIF2A as putative direct miR-182 target genes (Supplemental Fig. S6B), with TGFβ1 as a top upstream regulator (overlap P = 2.74×10−5, Z score of activation=−2.53 for the direct miR-182 target genes; Z score of activation = −4.63, overlap P = 2.45×10−12 for the all-182 target genes) (Supplemental Fig. S6C). Gene ontology analysis revealed enrichment of genes with annotated functions in cell death, survival, and (embryonic) development (Supplemental Fig. S7), further supporting miR-182 as playing a critical role in the regulation of cell death, differentiation, and cell cycle progression. RT-qPCR and Western blot analyses in GICs as well as in established glioma cell lines confirmed that miR-182 regulates the expression of this gene signature (Supplemental Fig. S8).
miR-182 controls HIF2A-dependent cell dedifferentiation in GBM
Next, we determined molecular mechanisms of miR-182 enhanced cell differentiation. We found that miR-182 recognized an evolutionarily conserved site within the 3′ UTR of HIF2A (Supplemental Fig. S9A). UTR binding was confirmed by reporter assay demonstrating significant reduction in luciferase activity in the presence of miR-182 as compared with Co-miR (Fig. 3J). Furthermore, miR-182 could reduce HIF2A mRNA and protein levels under normoxic and hypoxic conditions (Fig. 3H,I). As HIF2A is a well-annotated mediator of hypoxic stress in the cellular environment that is coexpressed with cancer stem cell markers and promotes self-renewal, cell survival, and the tumor-initiating potential of GICs (Li et al. 2009), we determined whether miR-182, by downregulating HIF2A, impacts stem cell marker expression. Anti-miR-182 expression triggered an increase in stem cell and proliferation marker expression (e.g., Nestin, Sox2, CD44, and CCNB1), which was negated by concomitant siRNA-mediated silencing of HIF2A, indicating that HIF2A knockdown contributes to the regulation of stem cell and proliferation marker expression in response to miR-182 expression (Fig. 3K, see L for densitometric quantification of Western blot data).
miR-182 regulates c-Met-controlled glioma cell growth
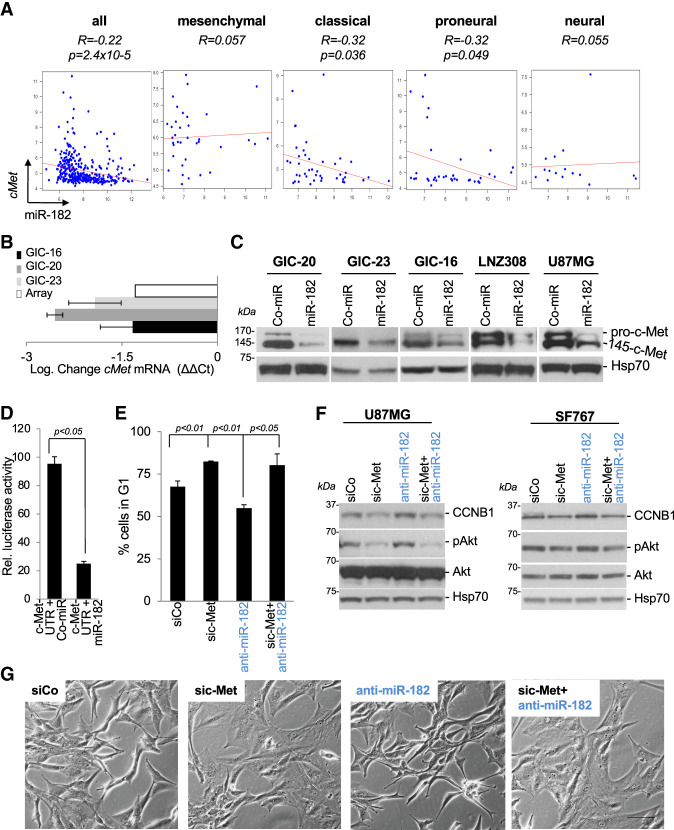
Besides hypoxia-driven, HIF2A-controlled differentiation processes, we found that miR-182 also regulates glioma cell growth by targeting the RTK c-Met, which represents a functional glioma stem cell marker and promoter of GIC self-renewal (Boccaccio and Comoglio 2013). A survey of the TCGA data set showed a prominent negative correlation between miR-182 and c-Met mRNA expression, predominantly in classical and proneural GBM tumors (Fig. 4A). A series of RT-qPCR (Fig. 4B), Western blot (Fig. 4C), and luciferase reporter assays (Fig. 4D) in GICs and transformed glioma cell lines revealed that miR-182, by binding to an evolutionarily conserved 8-mer 3′ UTR sequence (Supplemental Fig. S9B), down-regulates c-Met mRNA and protein. c-Met knockdown contributes to the cell cycle inhibitory activity of miR-182 (Fig. 4E; Supplemental Fig. S4), as transfection of anti-miR-182 sequences provoked a decrease in G1-phase content, which was antagonized by c-Met-targeting siRNAs (Fig. 4E). Similarly, analyses of cell morphology and cell cycle marker expression revealed that c-Met knockdown mediated a morphological transition into a flattened cell shape and antagonized anti-miR-182-driven Akt activation and CCNB1 induction (Fig. 4F,G). Thus, c-Met and HIF2A are critical miR-182 target genes that regulate GIC growth and differentiation in GBM.
Figure 4.
miR-182 regulates cell cycle progression through c-Met. (A) In silico correlation analysis of c-Met and miR-182 mRNA levels using the TCGA data set. (B) RT-qPCR analysis of c-Met mRNA levels in different GICs overexpressing ComiR or miR-182, shown as log2 fold change ± SD. (C ) Western blot analysis of c-Met protein levels in GICs and adherent cell lines overexpressing Co-miR or miR-182. Hsp70 served as loading control. (D) Luciferase activity measured in 293T cells 24–48 h after transfection of c-Met-3′ UTRpGL3 reporter vectors in combination with miR-182 or Co-miR sequences at a concentration of 200 nM. (E) Cell cycle analysis of U87MG cells transfected with 100 nM siCo, 100 nM sic-Met, 100 nM anti-miR-182, or a combination of sic-Met and anti-miR-182 for 48 h. (F ) Western blot analysis of c-Met downstream targets in U87MG and SF767 cells transfected with siCo, sic-Met, or anti-miR-182 as single agents or in combination for 48 h. (G) Bright-field images of U87MG cells transfected with 100 nM siCo, 100 nM sic-Met, or 100 nM anti-miR-182 alone or in combination with 100 nM sic-Met for 48 h. Bar, 60 µm.
miR-182-based SNAs reduce expression of miR-182 target genes and show anti-tumor activity in vitro
Inherent barriers of miRNA-based functional studies and miRNA-directed cancer therapy include the lack of delivery tools that enable robust, nontoxic, nonimmunogenic penetration of miRNAs into tumor cells and tissue. Delivery is in particularly challenging for brain tumors, as miRNA–carrier complexes have to cross the BBB and BTB and distribute throughout extravascular areas of the glioma parenchyma. Here, we designed a series of functional and efficacy studies to test the hypothesis that conjugation of gold nanoparticles bearing a monolayer of alkylthiol-modified miR-182 duplexes (Fig. 5A–C) could provide an approach for delivering proapoptotic and anti-proliferative miR-182 into GBM cells and tumors. First, we investigated the uptake of Cy5.5-labeled 182-SNAs into glioma lineages. In contrast to nonconjugated linear Cy5.5miR-182 oligoucleotides (Fig. 5D, inset), 182-SNAs robustly penetrated >90% of glioma cells and patient-derived GICs (Fig. 5D), significantly decreased Bcl2L12 and c-Met protein levels compared with Co-SNA-treated cultures (Fig. 5E), increased apoptosis in association with STS treatment (Fig. 5F), and diminished proliferation of glioma cells, as indicated by reduced BrdU incorporation (Fig. 5G).
Figure 5.
miR-182-based SNAs penetrate glioma cells, robustly down-regulate miR182 target genes, and phenocopy cellular effects of lipoplex-delivered miR-182 sequences. (A) miR-182 or Co-miR–RNA duplexes were hybridized to citrate stabilized gold nanoparticles (AuNPs) via thiolgold bond and passivated with polyethylene glycol-Thiol (mPEG-SH). (B) Sequence of miR-182 and Co-miR duplexes. (C ) Physico–chemical characterization of SNAs as outlined in the Materials and Methods. (D) Confocal images of U87MG and GIC-20 treated with Cy5.5-labeled SNAs or free miR-182 sequences (inset) and counterstained with Hoechst dye to visualize the nuclei. Bar, 50 µm. (E) U87MG cells were treated with 10 nM Co-SNA or 182-SNAs for 48 h, and protein levels of Bcl2L12 and c-Met were assessed by Western blotting. (F ) Western blot analysis for active caspase-3 and caspase-7 in U87MG cells that were transfected with 10 nM Co-SNAs or 182-SNAs for 48 h and subsequently treated with 0.5 µM STS for the indicated periods of time. (G) BrdU incorporation assays in the presence of 10% FBS or 10 ng/mL TGF-β1 in U87MG and LNZ308 treated with 10 nM Co-SNAs or 10 nM 182-SNAs for 24 h.
miR-182 displays significant organ retention and a low-toxicity profile in vivo
To evaluate the miR-SNA platform in vivo, we assessed 182-SNA pharmacokinetics, distribution, toxicity, cytokine induction, and intratumoral accumulation upon intravenous administration. Pharmacokinetics of 182SNAs follow a two-compartment model, as shown by nonlinear regression analysis, suggesting that transport between the central and peripheral compartments and the elimination of SNA conjugates follow first-order kinetics (Fig. 6A). 182-SNA half-life for distribution (αt1/2) was ∼27 min (Fig. 6A), and SNA retention was significant, as indicated by a slow elimination phase (half-life for elimination, βt1/2, ∼12.6 h) (Fig. 6B) and low levels of SNAs in the blood throughout the 72-h course of the study. Tissue distribution in healthy animals was primarily in the liver and spleen, with small accumulations in the lungs, kidneys, heart, brain, and intestines (Supplemental Fig. S10).
Figure 6.
SNAs effectively penetrate the BBB/BTB in xenograft mouse models of GBM. (A) Pharmacokinetics of 182-SNAs follow a two-compartment model, as shown by nonlinear regression analysis. (B) Pharmacokinetic parameters of SNA distribution and elimination. (C ) Histopathological survey of major organ systems of Sprague-Dawley treated with 10 mg/kg 182-SNAs for 14 d. Representative H&E stainings are shown for the brain, heart, lung, liver, kidney, muscle, spleen, and small intestine. (D) U87MG cells wereintracranially implanted into SCID mice and intravenously injected with 182-SNA-Cy5.5 or saline. 182-SNA content in the tumor was evaluated by in vivo imaging system (IVIS) analysis of brains 48 h after injection. Sham surgery was used as control. Dorsal brain images demonstrate accumulation of SNAs within the tumor xenografts, as indicated by increased fluorescence. (E) Inductively coupled plasma mass spectrometry (ICP-MS) analysis to quantify gold content in tumor tissue of two independent GIC-derived xenograft models. (F ) High-magnification images of coronal brainsections of mice harboring GIC-20 (F ) and U87MG-derived tumor xenografts (G) and treated with CoSNAs or 182-SNAs. Spherical nucleic acids were visualized by silver staining. Bars: F, 50 µm; G, left panel, 100 µm; G, middle panel, 50 µm; G, right panel, 200 µm.
To determine potential adverse side effects of SNAs, 182-SNAs (10 mg of RNA per kilogram of animal weight) or an equivalent volume of saline were administered as a single dose via intravenous injection into SpragueDawley rats, with the effects on body weight, blood chemistry, blood counts, and major organ histopathology determined. Importantly, we did not observe 182-SNA-related clinical signs or difference in body or organ weight at 1 or 14 d after SNA administration. Complete blood counts (CBCs/hematology) and blood chemistry panels were within normal range (Supplemental Table S4), confirming the absence of early or late toxic responses at therapeutic dosages. Histopathologic evaluation of major organs failed to detect malignant lesions or signs of abnormal processes and indicated organ structures that were consistent with clinically normal, ageand strain-matched rats (Fig. 6C). To address whether there was any indication of induced immune response, a screen for inflammatory cytokine induction was performed using blood samples from SCID mice inoculated with U87MG cells and treated with Co-SNAs, 182-SNAs, or saline every other day. After receiving seven treatments and ∼10 mg/kg SNAs, no significant differences were noticed between the groups that would support an inflammatory response (Supplemental Table S5).
We next determined intraglioma SNA accumulation upon systemic administration to elucidate whether 182SNAs crossed the BBB/BTB. In orthotopic GIC-derived GBM xenografts systemically injected with Cy5.5-labeled 182-SNAs, we demonstrated by in vivo imaging system (IVIS) (Fig. 6D), inductively coupled plasma mass spectrometry (ICP-MS) (Fig. 6E), and silver staining of coronal sections (Fig. 6F,G) that 182-SNAs penetrated glioma cell line and GIC-derived tumor grafts, with robust accumulation of Co-SNAs and 182-SNAs in glioma-associated vasculature and parenchyma (Fig. 6F,G).
miR-182 is associated with increased survival and a less aggressive glioma phenotype in vivo
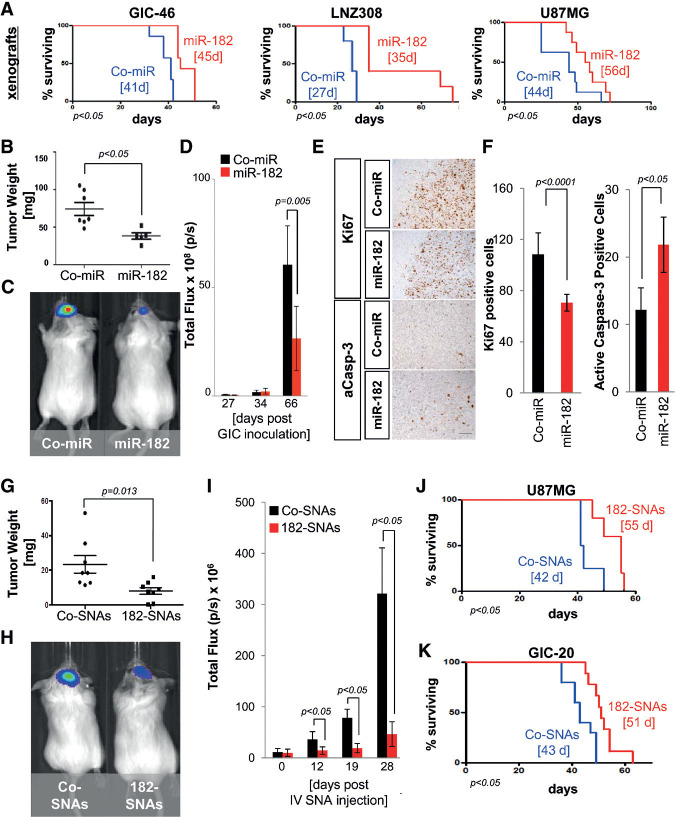
To further evaluate the tumor-suppressive functions of miR-182 and 182-SNAs, we analyzed TCGA patient survival as a function of miR-182 expression levels. miR-182 expression levels in TCGA GBM tumors, in particular in the proneural subclass, positively correlated with patient survival (Supplemental Fig. S11; Celiku et al. 2014). Studies in orthotopic xenograft models using transformed glioma cells and GICs ectopically expressing miR-182 showed that the presence of miR-182 increased glioma-free survival (Fig. 7A) and reduced tumor burden, as determined by measurement of tumor weight (Fig. 7B) and bioluminescence imaging of luciferase-labeled GICs (Fig. 7C,D). Histopathological analysis of Co-miR-expressing and miR-182-expressing tumors revealed that the presence of miR-182 decreased the number of proliferating and increased the number of apoptotic tumor cells, as determined by Ki67 and caspase-3 immunohistochemistry (IHC), respectively (Fig. 7E,F).
Figure 7.
182-SNAs reduce tumor growth in vivo. (A) Orthotopic xenograft survival analysis with glioma cells and GICs engineered to stably express miR-182 revealed that miR-182 expression increases survival of animal subjects. Median survival is indicated. (B–F) Analysis of tumor burden by weight (B) and bioluminescence imaging (C,D). (E) Ki67 and caspase-3 IHC in coronal brain sections of Co-miR-expressing and miR-182-expressing GIC-derived xenografts. Bar, 100 µm. (F) Quantification of Ki67 and caspase-3 in xenograft specimens. (G) Weight of U87MG-derived xenografted tumors extracted from SCID mice 21 d after intravenous treatment with Co-SNAs or 182-SNAs. (H) Bioluminescence imaging of xenograft tumors derived from GIC-20 12 d after intravenous treatment with CoSNAs or 182-SNAs. (I) Quantification of bioluminescence signal up to 28 d after treatment with Co-SNAs or 182-SNAs. (J, K) Kaplan-Meyer survival curves of SCID mice carrying xenografted glioma tumors (U87MG and GIC-20), which were treated with intravenously administered Co-SNAs or 182-SNAs. Median survival is indicated.
To assess the impact of 182-SNAs on GBM progression in vivo, we administered 182-SNAs and Co-SNAs through tail vein injection to SCID mice that were inoculated with U87MG cells and GIC-20. We found that robust BBB/BTB penetration and intratumoral accumulation of 182-SNAs resulted in significant reduction in tumor burden, as assessed by weighing of tumor tissue (Fig. 7G), bioluminscence imaging of xenografts from luciferase-labeled GICs (Fig. 7H,I), and prolonged animal subject survival (Fig. 7J,K). Thus, efficient BBB/BTB penetration and robust intraglioma dissemination of 182-SNAs upon systemic administration resulted in decreased tumor burden and increased xenograft survival in the absence of adverse side effects.
Discussion
In this study, we report the identification and functional characterization of miR-182 as a regulator of growth, differentiation, chemotherapeutic susceptibility, and tumor progression in GBM by targeting an oncogenic signature that includes Bcl2L12, c-Met, and HIF2A via direct binding to evolutionarily conserved, 3′ UTR-localized sequences. Using miR-182-functionalized SNAs, we demonstrate that miR-182 sequences effectively reduce tumor burden, thereby providing the first proof of concept that systemically administered miRNAs can cross the BBB/ BTB, infiltrate glioma parenchyma, and inhibit GBM progression. miR-182 has been the topic of extensive research in developmental and cancer biology. Together with its family members, miR-96 and miR-183, miR-182 was first described in mouse neurosensory cells, specifically in the retina, inner ear, and dorsal root ganglia (Kloosterman and Plasterk 2006; Xu et al. 2007; Weston et al. 2011). During the embryonic stages of development, miR-182 levels are low but significantly increase postnatally, suggesting a role for miR-182 in terminal differentiation of retinal progenitor cells and maintenance of mature retinal neuron function (Xu et al. 2007). Several other recent studies have suggested that miR-182 promotes cellular differentiation by regulating the expression of the transcription factor SNAI2 and induces mesenchymal-to-epithelial transition (Qu et al. 2013). Consistent with these findings, we demonstrate that miR-182 overexpression in GICs reduces sphere size and expansion and promotes differentiation along neuronal and astroglial axes at least in part via targeting of c-Met and HIF2A signaling. We further show that the miR-182-repressed transcriptome of GICs was enriched for stem cell signatures.
The miR-182 target gene c-Met is hyperactivated in GBM (Huang et al. 2007; Stommel et al. 2007; Cancer Genome Atlas Research Network 2008); represents a critical driver of GBM growth, invasion, hypoxia-induced dedifferentiation, and tumor progression (Boccaccio and Comoglio 2006, 2013); and is critical for adult stem cells to transition between a quiescent G0 and an “alert” G phase. Such transition primes stem cells for cell cycle entry and enables them to effectively mobilize their tissue-regenerative function (Rodgers et al. 2014). Similarly, HIF2A is an important effector of hypoxia-driven stem cell maintenance that promotes GIC self-renewal, growth, and tumorigenicity, and its expression negatively correlates with glioma patient survival (Li et al. 2009).
Further demonstrating prodifferentiation activity of miR-182, GBM tumors, according to TCGA-based miRNA clustering, are classified into five clinically and genetically distinct groups that each relates to a different neural precursor cell type (Kim et al. 2011). In particular, the oligoneural tumors, which are characterized by elevated miR-182 levels, exhibit significantly longer survival than patients with radial glial, neural, astrocytic, or neuromesenchymal tumors. On a genetic level, oligoneural tumors show repression of neural and embryonic stem cellrelated mRNA signatures, suggesting a more differentiated, less stem-like state.
We propose that miR-182 expression in GICs promotes a more differentiated, less aggressive phenotype and that this activity is at least in part due to transcriptional repression of c-Met and HIF2A. While our mRNA, protein, and 3′ UTR reporter studies point to direct transcriptional regulation of c-Met and HIF2A by miR-182, only c-Met but not HIF2A mRNA expression is negatively correlated with miR-182 levels in TCGA GBM. This suggests that HIF2A is regulated through additional mechanisms in GBM tumors besides transcriptional repression, including repression of mRNA translation or modulation of HIF2A protein expression/stability.
In addition to the regulation of GIC and glioma cell plasticity, we identified miR-182 as a chemosensitizer that promotes apoptosis induced by conventional and targeted therapies through down-regulation of Bcl2L12, a validated oncogene with high-level expression in GBM tumors (Stegh et al. 2007; Jensen et al. 2013). Bcl2L12 is significantly expressed in the majority of primary GBM tumor specimens and is distantly related to canonical Bcl-2 proteins. Mechanistically, our previous studies determined that, unlike prototypic Bcl-2 family members, Bcl2L12 did not safeguard mitochondrial membrane integrity but instead potently inhibited apoptosis at the level of post-mitochondrial effector caspase-3 and caspase-7 activation. A combination of enforced expression, RNAimediated extinction, colocalization, and protein-interaction studies revealed that Bcl2L12 inhibits caspase-3 and caspase-7 via distinct mechanisms. Direct physical interaction underlies Bcl2L12's inhibition of caspase-7 processing, whereas Bcl2L12-induced transcriptional upregulation of the small heat-shock protein αB-crystallin is instrumental to the neutralization of caspase-3 activation (Stegh et al. 2007, 2008a,b; Stegh and DePinho 2011). By down-regulating Bcl2L12, miR-182 sensitizes cells toward extant therapies primarily by increasing effector caspase activity.
Similar to other miRNAs, miR-182 has been reported to have cancer-promoting or inhibitory activity, which largely depends on cancer type and genetic context. For instance, it has been shown that miR-182 has oncogenic properties in medulloblastoma and cervical, breast, colon, prostate, and bladder cancer and tumor-suppressive activity in lung cancer, gastric adenocarcinoma, oropharyngeal carcinoma, and uveal melanoma by regulating oncogenes such as Bcl-2, Cyclin D2, MITF, and, via indirect mechanisms, c-Met. In addition, miR-182 acts as a tumor suppressor in renal cell carcinoma by targeting the FLOT1/ AKT/FOXO3/CyclinD1 pathway (Sarver et al. 2009; Segura et al. 2009; Cekaite et al. 2012; Hirata et al. 2012, 2013; Kong et al. 2012; Liu et al. 2012, 2013; Poell et al. 2012; Weeraratne et al. 2012; Yan et al. 2012; Hui et al. 2013; Li et al. 2014; Tang et al. 2014; Xu et al. 2014; Yang et al. 2014). This is particularly relevant to GBM, as a recent study suggested that miR-182 can trigger TGF-β1-dependent NF-κB induction in glioma cells and provoke a more aggressive glioma phenotype by targeting the deubuiquitinase CYLB (Song et al. 2012). These data suggest that miR-182 activity is highly context-dependent, pointing to important roles of the tumor microenvironment and the cooperating cancer (subtype)-specific genetic landscape in shaping miR-182 responses.
With regard to molecular mechanisms controlling miR182 expression, we propose that p53 negatively regulates miR-182 transcription. p53 is an important regulator of cancer stem cell differentiation, as loss of p53 promotes neurosphere formation and a stem cell-like phenotype (Puzio-Kuter and Levine 2009). Thus, we hypothesize that in tumors with compromised p53 function, miR182 levels are elevated and compensate for p53 loss of function by promoting a more differentiated GIC state.
Drug delivery to intracranial tumor sites represents a major challenge for the development of GBM therapeutics. In particular, the BBB/BTB represents a major physiological barrier that often prevents the accumulation of therapeutic drug concentrations within intracerebral lesions. Delivery of RNA-based therapeutics is particularly challenging, since RNA can be unstable, often entrapped in endosomes, and incapable of infiltrating tumor parenchyma beyond perivascular regions (Blakeley 2008; Allhenn et al. 2012). In a recent study, we characterized SNAs as a novel RNAi-based nanoconjugate using siRNAs designed to target Bcl2L12 (Jensen et al. 2013). SNAs exhibit a number of unique properties—i.e., high cellular uptake (>90% of cell populations), including patient-derived GICs without the use of toxic, auxiliary transfection agents or viral delivery platforms; extraordinary stability in physiological environments; robust resistance to nuclease degradation (Giljohann et al. 2009; Seferos et al. 2009); no acute or long-term toxicity at high and therapeutic doses in animal models; and low activation of the innate immune response (Massich et al. 2009). siRNA-based SNAs crossed the BBB/BTB and displayed preferential accumulation within intracerebral glioma due to the enhanced permeability and retention (EPR) effect; i.e., the increased accumulation of nanomaterials in tumors due to the abnormal form and architecture of tumor blood vessels. Thus, SNAs represent a powerful platform for RNAi-mediated biotherapeutic gene silencing, allowing us to combine potent knockdown of GBM oncogenes with efficient BBB/BTB penetration and robust intraglioma dissemination upon systemic administration. Here, we developed and characterized a novel construct of SNAs functionalized with mature miR-182 sequences to harness the tumor-suppressive activities of miR-182. 182-SNAs effectively crossed the BBB/BTB upon systemic intravenous administration and disseminated throughout glioma parenchyma, as evidenced by IVIS-based imaging, ICP-MS, and silver staining on serial brain sections. SNAs therefore have potential as a tractable, nontoxic platform for biotherapeutic gene silencing via passive tumor targeting driven by EPR of the tumor-associated vasculature. While SNA conjugates were readily detectable in non-tumor-bearing mice by IVIS, intracranial levels were significantly higher in glioma-bearing mice, suggesting that a compromised BBB/BTB together with EPR significantly contributes to accumulation of 182SNAs in tumor versus normal brain elements. Validation experiments in glioma cells revealed robust cellular uptake of 182-SNAs that resulted in potent down-regulation of Bcl2L12 and c-Met proteins, sensitization toward druginduced apoptosis, and reduction in cellular proliferation. Mirroring proapoptotic and growth inhibitory effects in cells in vitro, systemic delivery of 182-SNAs reduced tumor burden in an orthotopic GBM xenograft model. Together with toxicity studies confirming the absence of significant adverse side effects and inflammatory responses associated with systemic introduction of miR-182, we propose that 182-SNAs represent a tool for discovery science to characterize miR function in cells and animals in the absence of viral or lipoplex delivery agents and a novel therapeutic paradigm for the treatment of GBM and possibly other genetic malignancies and disorders of the CNS. Thus, the potent tumor-suppressive activity of miR-182 resulting from repression of critical GBM oncogenes— primarily Bcl2L12, c-Met, and HIF2A—together with the capacity of the SNA platform to effectively infiltrate GBM tumors points to SNA-mediated miR-182 transfer as an attractive strategy to target multiple oncogenes.
Materials and methods
Reagents
The pan-specific protein kinase inhibitor STS (0.5 µM) (Sigma) and the following RTK inhibitors at 5 µM each were used to treat glioma cell lines: EGFR inhibitor erlotinib (Sigma), c-Met inhibitor SU11274 (Sigma), PDGFR inhibitor imatinib mesylate (Selleck), insulin-like growth factor-1, and receptor inhibitor AG1024 (Calbiochem-EMD). The alkylating agent TMZ (Sigma) was used at a final concentration of 100 µM.
Cell culture and human patient samples
Glioma cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco). Patient-derived tumor neurospheres (GIC) were a gift from Dr. Kenneth Aldap (University of Texas M.D. Anderson Cancer Center) and were cultivated under neurosphere conditions according to established protocols (Bao et al. 2006). Cells were transduced with lentivirus stably expressing firefly luciferase to assess xenograft tumor growth in vivo by bioluminescence imaging (IVIS Spectrum, PerkinElmer) (Dinca et al. 2007). Tumor tissue from GBM patients (n=45) was obtained after surgery at Northwestern Memorial Hospital in compliance with the Northwestern Memorial Hospital Institutional Review Board.
Cell differentiation
GICs were plated at 50,000 cells per milliliter on poly-D-lysine/ laminin-coated coverslips in GIC medium with 1 ng/mL EGF and bFGF (Invitrogen). Immunocytochemistry was performed as previously described (Srikanth et al. 2013) using the following antibodies: rabbit anti-GFAP (1:1000; DakoCytomation) and mouse anti-MAP2 (1:500; BD Pharmingen). Cells were imaged using a Leica SP-5 confocal microscope, and quantification was performed using NIH ImageJ software.
Cell cycle analysis
U87MG cells were plated in six-well plates and, at ∼50% confluency, were transfected with different siRNA or miRNA constructs for 24 h at low serum. The next day, media were changed to 10% FBS for 48 h. Next, the cells were starved for 3 h and lysed by Nicolleti buffer containing propidium iodide (Stegh et al. 2007). Cells were analyzed by flow cytometry (30,000 counts; LSRFortessa).
Comparative in silico analysis using TCGA
Pearson's correlation coefficient (PCC) was used to measure the correlation between miR-182 and Bcl2L12 mRNA levels in TCGA GBM samples. PCCs were calculated for correlations in mesenchymal, classical, proneural, and neural samples individually and all tumor samples collectively.
SNA synthesis
Gold nanoparticles (13 nm±1 nm) were prepared using the Frens method (Frens 1973). The particles were treated with DEPC (0.1% v/v) and autoclaved before being functionalized with miRNA duplexes. The miRNA-conjugated SNAs were purified by three successive centrifugations steps, supernatant removal, and addition of PBS at 4°C or by tangential flow filtration. Each 182-SNA contained ∼30 antisense strands per 13-nm gold nanoparticle. For the generation of fluorochrome-labeled SNAs, Cy5.5 phosphoramidite (Glen Research) was manually coupled to the 5′ end of either Co-SNA or 182-SNA sense strands prior to hybridization. For further information on the functionalization and characterization of 182-SNA and Co-SNA, see the Supplemental Material.
Tumor xenograft model and in vivo SNA administration
All animals used were under an approved protocol of the Institutional Animal Care and Use Committee of Northwestern University. Female SCID mice (6–8 wk) were intracranially injected with U87MG or GIC-20 cells to form tumors according to published protocols (Jensen et al. 2013). To demonstrate the therapeutic efficacy of the 182-SNAs, 500 nM 182-SNAs or Co-SNAs was administered via the tail vein at a dose of 1.4 mg of RNA per kilogram of mouse weight per injection, with injections performed every other day. The mice received seven injections of SNAs.
Statistical analysis
All experimental data are presented as mean±SD (n≥3) unless otherwise stated. Tumor weights were analyzed by Mantel-Cox and Gehan-Breslow-Wilcoxon test. Control and treatment groups in in vitro experiments were compared by two-tailed Student's t-test. A P value of ≤0.05 was considered statistically significant.
Competing interest statement
D.A.G., C.A.M., W.L.D., and A.H.S. have interest in AuraSense Therapeutics, which develops spherical nucleic acid-based technologies.
Acknowledgments
We thank the members of the Stegh laboratory as well as Dr. Kazuo Fushimi and Dr. Paolo Ceppi for constructive discussions and help, and Kurt Harris for his help with the IncuCyte ZOOM instrument. This research was supported by the Center for Cancer Nanotechnology Excellence (CCNE) initiative of the National Institutes of Health (NIH) (U54 CA151880 to C.A.M. and A.H.S.), National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)/NIH grants (R01AR060810 and R21AR062898 to C.A.M.), the Dixon Translational Research Grants Initiative of the Northwestern Memorial Foundation (to C.A.M. and A.H.S.), a Research Scholar award from the American Cancer Society (to A.H.S.), the James S. MacDonnell 21st Century Initiative, and the Coffman Charitable Trust (to A.H.S.). L.C. was supported by funding from the NIH (P01 CA095616, U01 CA141508, and U24 CA143845) and the Ben and Catherine Ivy Foundation. The research was also supported by the NIH National Cancer Institute (NCI) under award number F32CA171949 to E.S.D. E.S.D. and T.J.M. were supported by the Northwestern University International Institute of Nanotechnology. Core facilities at Northwestern University (i.e., the Center for Advanced Molecular Imaging, the Cellular Imaging Facility, the Developmental Therapeutics Core, and the Mouse Histology and Phenotyping Laboratory) are all supported by NCI Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, and the Quantitative Bioelemental Imaging Center is supported by NASA Ames Research Center (NNA06CB93G). This work was also supported in part by the Defense Advanced Projects Agency (DARPA) Cooperative agreement number HR0011-13-20018.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.257394.114.
References
- Allhenn D, Boushehri MA, Lamprecht A. 2012. Drug delivery strategies for the treatment of malignant gliomas. Int J Pharm 436: 299–310. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760. [DOI] [PubMed] [Google Scholar]
- Blakeley J. 2008. Drug delivery to brain tumors. Curr Neurol Neurosci Rep 8: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccio C, Comoglio PM. 2006. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer 6: 637–645. [DOI] [PubMed] [Google Scholar]
- Boccaccio C, Comoglio PM. 2013. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res 73: 3193–3199. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekaite L, Rantala JK, Bruun J, Guriby M, Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe RA, et al. 2012. MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia 14: 868– 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiku O, Johnson S, Zhao S, Camphausen K, Shankavaram U. 2014. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PLoS One 9: e101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley RW, Pouratian N, Sheehan JP. 2006. Gamma knife surgery for glioblastoma multiforme. Neurosurg Focus 20: E17. [DOI] [PubMed] [Google Scholar]
- Dinca EB, Sarkaria JN, Schroeder MA, Carlson BL, Voicu R, Gupta N, Berger MS, James CD. 2007. Bioluminescence monitoring of intracranial glioblastoma xenograft: response to primary and salvage temozolomide therapy. J Neurosurg 107: 610–616. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. 2012. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev 26: 756–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frens G. 1973. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat Phys Sci 241: 20–22. [Google Scholar]
- Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. 2009. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc 131: 2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestaneh N, Mishra B. 2005. TGF-β, neuronal stem cells and glioblastoma. Oncogene 24: 5722–5730. [DOI] [PubMed] [Google Scholar]
- Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. 2012. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One 7: e51056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. 2013. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One 8: e55502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci 104: 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui AB, Lin A, Xu W, Waldron L, Perez-Ordonez B, Weinreb I, Shi W, Bruce J, Huang SH, O'Sullivan B, et al. 2013. Potentially prognostic miRNAs in HPV-associated oropharyngeal carcinoma. Clin Cancer Res 19: 2154–2162. [DOI] [PubMed] [Google Scholar]
- Huse JT, Holland EC. 2009. Yin and yang: cancer-implicated miRNAs that have it both ways. Cell Cycle 8: 3611–3612. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. 2009. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27: 5848–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. 2012. Causes and consequences of microRNA dysregulation. Cancer J 18: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al. 2013. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 5: 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385. [DOI] [PubMed] [Google Scholar]
- Kim TM, Huang W, Park R, Park PJ, Johnson MD. 2011. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 71: 3387–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. 2006. The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450. [DOI] [PubMed] [Google Scholar]
- Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, Tang H. 2012. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J 279: 1252–1260. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. 2009. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 15: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Sheng C, Huang L, Zhang H, Cheng Z, Zhu Q. 2014. MiR183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res 16: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, Hernando E, Wei JJ. 2012. MiR-182 overexpression in tumourigenesis of highgrade serous ovarian carcinoma. J Pathol 228: 204–215. [DOI] [PubMed] [Google Scholar]
- Liu R, Li J, Teng Z, Zhang Z, Xu Y. 2013. Overexpressed microRNA-182 promotes proliferation and invasion in prostate cancer PC-3 cells by down-regulating N-myc downstream regulated gene 1 (NDRG1). PLoS One 8: e68982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. 2009. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol Pharm 6: 1934–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. 2013. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93: 98–104. [DOI] [PubMed] [Google Scholar]
- Poell JB, van Haastert RJ, de Gunst T, Schultz IJ, Gommans WM, Verheul M, Cerisoli F, van Noort PI, Prevost GP, Schaapveld RQ, et al. 2012. A functional screen identifies specific microRNAs capable of inhibiting human melanoma cell viability. PLoS One 7: e43569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzio-Kuter AM, Levine AJ. 2009. Stem cell biology meets p53. Nat Biotechnol 27: 914–915. [DOI] [PubMed] [Google Scholar]
- Qu Y, Li WC, Hellem MR, Rostad K, Popa M, McCormack E, Oyan AM, Kalland KH, Ke XS. 2013. MiR-182 and miR-203 induce mesenchymal to epithelial transition and self-sufficiency of growth signals via repressing SNAI2 in prostate cells. Int J Cancer 133: 544–555. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al. 2014. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM, et al. 2009. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 9: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. 2009. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett 9: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, AlvarezDiaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, et al. 2009. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci 106: 1814–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Helmy K, Khan AA, Silber J, Arvey A, Neezen F, Agius P, Huse JT, Holland EC, Leslie CS. 2012. Inferring transcriptional and microRNA-mediated regulatory programs in glioblastoma. Mol Syst Biol 8: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C, Wu J, Hu B, Cheng SY, Li M, et al. 2012. TGF-β induces miR-182 to sustain NF-κB activation in glioma subsets. J Clin Invest 122: 3563–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth M, Das S, Berns EJ, Kim J, Stupp SI, Kessler JA. 2013. Nanofiber-mediated inhibition of focal adhesion kinase sensitizes glioma stemlike cells to epidermal growth factor receptor inhibition. Neuro Oncol 15: 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH, DePinho RA. 2011. Beyond effector caspase inhibition: Bcl2L12 neutralizes p53 signaling in glioblastoma. Cell Cycle 10: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H, Park K, Hannon GJ, Yuan J, Louis DN, et al. 2007. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev 21: 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH, Chin L, Louis DN, DePinho RA. 2008a. What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell Cycle 7: 2833–2839. [DOI] [PubMed] [Google Scholar]
- Stegh AH, Kesari S, Mahoney JE, Jenq HT, Forloney KL, Protopopov A, Louis DN, Chin L, Depinho RA. 2008b. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci 105: 10703– 10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP, Protopopov A, Chin L, Depinho RA. 2010. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev 24: 2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. 2007. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318: 287–290. [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10: 459–466. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang Z, Liu Q, Liu X, Wu M, Li G. 2014. Disturbing miR182 and -381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PLoS One 9: e84146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, et al. 2012. Pleiotropic effects of miR-183∼96∼182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol 123: 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, RochaSanchez S, Beisel KW, Soukup GA. 2011. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn 240: 808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. 2007. Micro-RNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem 282: 25053–25066. [DOI] [PubMed] [Google Scholar]
- Xu X, Wu J, Li S, Hu Z, Zhu Y, Liang Z, Wang X, Lin Y, Mao Y, Chen H, et al. 2014. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol Cancer 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Dong XD, Chen X, Yao S, Wang L, Wang C, Hu DN, Qu J, Tu L. 2012. Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One 7: e40967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Tang Y, Guo W, Du Y, Wang Y, Li P, Zang W, Yin X, Wang H, Chu H, et al. 2014. Up-regulation of microRNA138 induce radiosensitization in lung cancer cells. Tumour Biol 35: 6557–6565. [DOI] [PubMed] [Google Scholar]