SUMMARY
Loss-of-function (LOF) (i.e., nonsense, splice site, and frameshift) variants that lead to disruption of gene function are likely to contribute to the etiology of neuropsychiatric disorders. Here, we perform a systematic investigation of the role of both de novo and inherited LOF variants in schizophrenia using exome sequencing data from 231 case and 34 control trios. We identify two de novo LOF variants in the SETD1A gene, which encodes a subunit of his-tone methyltransferase, a finding unlikely to have occurred by chance, and provide evidence for a more general role of chromatin regulators in schizophrenia risk. Transmission pattern analyses reveal that LOF variants are more likely to be transmitted to affected individuals than controls. This is especially true for private LOF variants in genes intolerant to functional genetic variation. These findings highlight the contribution of LOF mutations to the genetic architecture of schizophrenia and provide important insights into disease pathogenesis.
INTRODUCTION
Schizophrenia (SCZ) is a severe, common psychiatric disorder with a strong genetic component (Rodriguez-Murillo et al., 2012). Elucidating the genomic architecture of SCZ and identifying specific risk genes and affected pathways can inform the underlying disease pathophysiology and lead to identification of novel treatment targets.
Recent findings make increasingly clear that rare de novo copy number variants (CNVs) and de novo protein-altering mutations (defined as point substitutions or single-nucleotide variants [SNVs] and small insertions/deletions [indels]) contribute to the risk of SCZ and a number of other brain disorders, such as autism and intellectual disability (Karayiorgou et al., 1995; Kirov et al., 2012; Malhotra et al., 2011; Xu et al., 2008; de Ligt et al., 2012; Girard et al., 2011; Gulsuner et al., 2013; Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012b; Rauch et al., 2012; Sanders et al., 2012; Xu et al., 2011, 2012). De novo mutations are often transmitted by relatively asymptomatic carriers in afflicted families (Karayiorgou et al., 2012; Rodriguez-Murillo et al., 2012), thus contributing to the heritable component of these disorders. The relative contribution of de novo or inherited mutations to each disorder remains to be determined, but it is expected to correlate with the impact of the disease on fitness and fecundity. Thus, investigation of both variant types should be particularly important for diseases with partial reduction in fecundity, such as SCZ. Among de novo and inherited variants, ones that lead to loss-of-function (LOF) by disrupting protein-coding genes have a high probability of being deleterious and are of great interest in disease etiology (Veltman and Brunner, 2012).
This study was designed to evaluate the role that de novo and inherited LOF variants play in conferring SCZ risk. First, our investigation of the impact of de novo variation confirmed an excess of de novo LOFs in SCZ patients and led to the identification of a candidate risk gene (SETD1A) harboring two de novo LOF indels in two individuals with SCZ. Second, our investigation of the impact of transmitted variation revealed a relative increase in the transmission of private and rare LOF variants to SCZ probands compared to controls, especially in genes that are more intolerant to functional genetic variation. Collectively, these findings highlight the contribution of LOF mutations to the genetic architecture of SCZ and provide important insights into disease pathogenesis.
RESULTS
Systematic Search for De Novo LOF Indels Identifies Two Mutations in SETD1A
Recently, we sequenced a total of 795 exomes from 231 parent-proband case trios enriched for sporadic SCZ cases, as well as 34 control trios (Xu et al., 2012), and using GATK (McKenna et al., 2010) and Dindel (Albers et al., 2011) we identified 14 indels (ten frameshift and four in-frame variants) with a validation rate of ~20%. The low accuracy of variant calls for de novo indels suggested that we might have missed a considerable number of indels, including LOF frameshift indels. Here, we undertake a more extensive analysis of de novo LOF indels, using two recently developed software packages specifically designed to detect indels from high-throughput sequencing data by applying split-read-based methods, PRISM (Jiang et al., 2012) and Pindel (Ye et al., 2009). Initial variant calling using PRISM and Pindel produced 24 and 36 candidate de novo LOF indels, respectively. Of these, 16 candidates overlapped (for a total of 44 unique indels using the two programs), and all of the ten LOF indels validated in our previous study (nine in cases and one in controls) (Xu et al., 2012) were included among them (Figure S1 available online). The remaining 34 candidates for previously unidentified de novo LOF indels (29 in cases and five in controls) were manually inspected using the Integrative Genomics Viewer (Robinson et al., 2011) for supporting reads in the proband but not in either of the parents. Twenty candidates in cases and all five candidates in controls were excluded by this step due to lack of reads supporting the indel in the proband or existence of multiple reads supporting the indel in either of the parents. We tested the remaining nine candidates by Sanger sequencing, and five de novo LOF indels (all in cases) were validated (Table 1). (A complete list of de novo LOF mutations in our cohort is shown in Table S1.) The total number of individuals carrying de novo LOF variants (nonsense, splice site, and frameshift variants) in our case and control data sets were, respectively, 25 and 1, resulting in a 3.7-fold enrichment of de novo LOF mutation carriers in SCZ subjects (25/231; 10.8% in cases and 1/34; 2.9% in controls), although the enrichment is not statistically significant. We also asked whether the load of de novo LOF variants correlates with any of four available clinical variables (severity of the disease [based on clinical course and functional outcome, see Supplemental Experimental Procedures for detailed information], age at disease onset, history of childhood learning disabilities, and comorbidity of mental retardation). We observed a significant correlation between history of childhood learning difficulties and the number of de novo LOF variants per individual (p = 0.0006, Spearman's rank correlation r = 0.26), indicating a higher incidence of childhood learning difficulties in patients with de novo LOF variants.
Table 1.
List of the Newly Identified De Novo Loss-of-Function Variants in 265 Trios
| Position (hg19) | Reference / Variant Allele | Type | Effect | Property | Gene | Pop. | Diagnosis |
|---|---|---|---|---|---|---|---|
| chr1:3753245-3753259 | CAGGGACTCTGCTG / - | D | Canonical splice site + Inframe | c.1120-3_1131 del, p.A374_L377 del | CEP104 | U.S. | SCZAFF |
| chr12: 26816636-26816638 | CTG / AGGTCAGTGTC | D+ I | Frameshift | c.1693_1695 del3ins11, p.Q565fs | ITPR2 | Afr. | SCZ |
| chr12: 57843366 | A/ - | D | Frameshift | c.619del1, p.Q207fs | INHBC | Afr. | SCZAFF |
| chr16: 30976335 | C / - | D | Frameshift | c.1272 del1, p.D424fs | SETD1A | Afr. | SCZ |
| chr16: 30992058-30992059 | AG / - | D | Canonical splice site | c.4582-2_4582-1 del2 | SETD1A | U.S. | SCZ |
D, deletion; I, insertion, Afr., Afrikaner; SCZ, schizophrenia; SCZAFF, schizoaffective disorder.
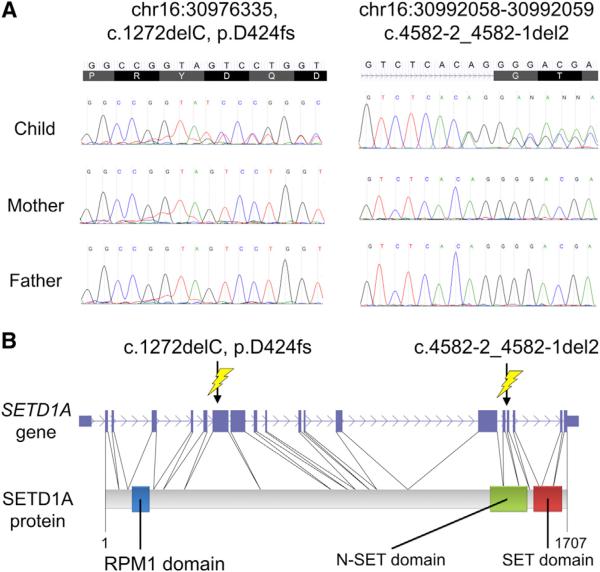
Notably, two of the five validated LOF indels were found in the same gene, SETD1A (Figure 1), with two case probands each carrying a frameshift de novo indel and a de novo indel changing the canonical splice acceptor site sequence from AG to GG. The other three validated de novo LOF indels were located within the CEP104, ITPR2, and INHBC genes (Table 1 and Figure S2). SETD1A encodes for a component of a histone methyltransferase complex that produces mono-, di-, and trimethylated histone H3 at Lysine 4 (H3K4) (Miller et al., 2001; Roguev et al., 2001). The de novo frameshift indel (D424fs) in the first proband creates an early stop codon and leads to a predicted protein truncation. The de novo indel changing the canonical splice acceptor site (c.4582-2_4582-1 del2 variant) in the second proband is predicted to lead to loss of exon 16 and disruption of N-SET domain that plays an important role for H3K4 methyltransferase activity (Dehé et al., 2006; Schlichter and Cairns, 2005). We used a rather conservative approach to assess the statistical significance of observing at least two de novo LOF mutations in the same gene that takes into account the length of the gene and the gene-specific GC content. Based on the number of de novo LOFs found in our cases (a total of 25: 18 coding LOFs [nonsense SNVs and frameshift indels] and 7 splice site LOFs [canonical splice site SNVs and indels affecting canonical splice site]), we determined that two or more de novo LOF mutations in a single gene were unlikely to occur by chance (p = 0.035). As mentioned above, because increased rates of de novo LOFs in patients with psychiatric disorders have now been reported in most of the published exome studies in psychiatric and neurodevelopmental disorders, this is a conservative analysis since it conditions on the number of observed de novo LOF mutations in cases (Girard et al., 2011; Iossifov et al., 2012; O'Roak et al., 2012b; Sanders et al., 2012; Xu et al., 2011, 2012). We used an additional method to evaluate the significance of observing at least two LOF de novo mutations in SETD1A, which results in a p value that is specific to SETD1A. Specifically, we first estimated the probability of an LOF de novo mutation per chromosome for SETD1A and then used a Poisson model for the probability of observing two or more LOF de novo events in this gene. The key element in this calculation is specifying the mutation rate for SETD1A. To achieve this, we made use of the recently developed transmission and de novo association (TADA) software (He et al., 2013), which estimates the mutation rate per gene based on its exonic length and its nucleotide content (Sanders et al., 2012). For SETD1A, we obtained an LOF mutation rate per chromosome of 4.75 × 10−6. The resulting probability of two or more LOF mutations is 2.4 × 10−6. Assuming that there are 20,000 genes, the genome-wide p value is 0.048, very similar to our first analysis.
Figure 1. Two De Novo LOF Indels in SETD1A.
(A) Two de novo LOF indels in the SETD1A gene confirmed by Sanger sequencing. (B) Structure of the SETD1A gene and the SETD1A protein along with the positions of the two LOF indels. The de novo frameshift indel variant (D424fs) (left) creates an early stop codon and leads to protein truncation. The de novo indel variant c.4582-2_4582-1 del2 variant (right) changes the canonical splice acceptor site sequence adjacent to exon 16 from AG to GG.
Considering the phenotypic presentation of the mutation carriers, it is notable that both SCZ cases carrying de novo LOF mutations in SETD1A also manifest symptoms consistent with a secondary diagnosis of obsessive-compulsive disorder (OCD). Detailed description of the clinical presentation of these two individuals is available in Supplemental Information. For both, SCZ is the primary lifetime diagnosis and the one associated with their long-term disability. Overall, clinical information for obsessive-compulsive symptoms is available for 205 of the 231 probands in our SCZ cohort, with 17 probands meeting full diagnostic criteria for secondary, comorbid OCD. The proportion of subjects with OCD in our SCZ cohort (17/205, 8.3%) is within the range reported in previous studies (7.8%–26%; Bottas et al., 2005). Given the number of subjects with comorbid OCD in our SCZ cohort, occurrence of de novo LOF mutations in SETD1A was significantly associated with SCZ comorbid with OCD (p = 0.002, Barnard's test). Whether this highly similar phenotypic outcome suggests that patients with LOF mutations in SETD1A could define a new clinical subtype of SCZ comorbid with OCD symptoms (Bottas et al., 2005) remains to be determined by additional sequencing studies of larger cohorts of both SCZ and OCD patients well-phenotyped for psychotic and obsessive-compulsive symptoms.
Damaging De Novo Variants Highlight a Role of Chromatin Regulators in SCZ
Further analysis aiming at prioritizing genes and variants for their likely pathogenicity provided additional evidence for a role of both de novo damaging mutations and chromatin regulators. Specifically, application of a recently established scoring system that makes use of human polymorphism data and reflects the intolerance of each gene against functional genetic variation in the general human population (residual variation intolerance score [RVIS]) (Petrovski et al., 2013), to all de novo mutations observed in individuals with SCZ in our and other studies (Girard et al., 2011; Gulsuner et al., 2013; Xu et al., 2012), as well as in control individuals from family-based whole-exome sequencing (WES) studies for SCZ, autism spectrum disorders (ASDs), and intellectual disability (ID) (Gulsuner et al., 2013; Iossifov et al., 2012; O'Roak et al., 2012b; Rauch et al., 2012; Sanders et al., 2012; Xu et al., 2012), revealed significant enrichment of genes intolerant to variation among the genes hit by de novo LOF and missense variants (but not by de novo silent variants) in cases (Figure S3A). No significant enrichment of any type of de novo variant was observed within the control samples. Two-dimensional (2D) plotting analyses that used the RVIS percentiles for genes (y axis) and PolyPhen-2 quantitative scores (Adzhubei et al., 2010) for missense mutations (x axis) revealed that de novo missense variants located within genes intolerant to variation and predicted to be damaging by PolyPhen-2 (quantitative score ≥ 0.95) are enriched in cases and therefore are more likely to contribute to disease risk (Figure S3B). A similar pattern of excess of genes intolerant to variation among genes hit by functional de novo mutations was also reported in cohorts ascertained for epilepsy, ID, and ASD (Petrovski et al., 2013), suggesting that intolerant genes carrying functional variants, including LOF ones, are more likely to be associated with disease.
Based on these observations, in the combined data set of family-based WES studies of SCZ, we selected a total of 62 genes thatare intolerant to variation and harbordenovoLOF ordamaging missense (PolyPhen-2 scores ≥ 0.95) variants: 38 genes from our studies (36 from the Xu et al., 2012 study plus SETD1A and ITPR2 identified in the current analysis) as well as 25 genes from two other studies (Girard et al., 2011; Gulsuner et al., 2013) (de novo damaging variants in KIAA1109 were found across data sets). Gene set enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009) revealed “chromatin regulator” as the most significantly enriched term among these 62 genes (Figure S4A and Table S2, also see Supplemental Information). Direct and indirect interactions among chromatin regulator genes harboring de novo damaging variants, including SETD1A, RBBP5, TRRAP, UBR5, KDM2B, and KDM5C, are shown in Figure S4B.
The Inherited Component of SCZ Risk Is Enriched in Rare LOF Variants
Recent WES studies in ASD provided evidence that inherited LOF mutations were enriched in affected individuals (Lim et al., 2013; Yu et al., 2013). Therefore, detailed analysis of rare inherited variants promises to provide important insights toward our understanding of the familial component of SCZ risk. To evaluate evidence for such etiologies in SCZ, we studied the transmission patterns of common and rare SNVs and indel variants identified from our WES data.
Because recent whole-genome and exome studies suggested that many deleterious and disease-related variants, especially LOF variants, exist at low frequency in the human population (Lim et al., 2013; MacArthur et al., 2012; Abecasis et al., 2012; Zhu et al., 2011), we first partitioned the variants into disjoint classes according to their minor allele frequencies (MAF) in the parents as follows: private variants, observed only once in the parents; rare variants, observed more than once and MAF ≤ 0.05; common variants, MAF > 0.05.
For the functional annotation and classification of variants, we used the SnpEff software (Cingolani et al., 2012). By following the definition of functionality of variants provided in this software, we identified three classes: nonsense variants, variants disrupting start and stop codons, canonical splice site variants, and frame-shift variants were classified as “LOF” variants; missense variants and in-frame indels were classified as “MODERATE”-effect variants; and silent variants, variants generating start codons in untranslated regions, and nonsynonymous variants generating alternative start codons were classified as “LOW”-effect variants.
When we analyzed the transmission (number of transmissions from parents to offspring) to untransmission (number of untran-smissions) ratio (T:U ratio) of LOF variants, we observed a tendency toward undertransmission in both cases and controls, i.e., T:U < 1 (Table 2; private LOFs: 0.86, rare LOFs: 0.83 and common LOFs: 0.95 in cases; private LOFs: 0.58, rare LOFs: 0.63 and common LOFs: 0.88 in controls). A T:U ratio of 1 is expected for neutral loci and it is what we observe for the common synonymous (LOW-effect) variants (T:U = ~1) (Table 2). There are several possible explanations for the observed distortion in the transmission ratios of the LOF variants. One possibility is that this is the result of selection acting against these LOF variants, a mutation load-reducing mechanism that facilitates elimination of deleterious mutations per generation and maintains higher mean fitness. For example, if some of these variants are associated with embryonic lethality or early onset of deleterious developmental defects (Edmonds et al., 1982), some of the carriers could be eliminated immediately in the next generation. Selection may also be acting prior to fertilization in the form of higher failure rate in gametogenesis, lower chance for fertilization (Otto and Hastings, 1998), or in the form of cellular processes related to selfish spermatogonial selection (Goriely et al., 2013). In addition, the combination of LOF variants from the two parents in the offspring may lead to more deleterious effects (Crow, 2000). Finally, as we generally collected subjects without severe comorbid diseases or developmental disorders for both controls and cases, LOF variants linked to such maladies should be represented less frequently among the transmitted alleles.
Table 2.
Numbers of Transmitted and Untransmitted Alleles in Cases and Controls
| Cohort | Transmitted | Untransmitted | T:U ratio | RTR (Case/Control) | p Value |
|---|---|---|---|---|---|
| Loss-of-Function Variants (Nonsense, Canonical Splice Site, and Frameshift Variants) | |||||
| Private (hit once in parental population) | |||||
| Cases | 1,682 | 1,945 | 0.86 | 1.49 | 0.0086 |
| Controls | 159 | 274 | 0.58 | - | - |
| Rare (nonprivate and frequency ≤ 0.05) | |||||
| Cases | 5,948 | 7,168 | 0.83 | 1.31 | 0.0051 |
| Controls | 902 | 1,425 | 0.63 | - | - |
| Common (frequency > 0.05) | |||||
| Cases | 26,350 | 27,724 | 0.95 | 1.08 | 0.037 |
| Controls | 3,732 | 4,231 | 0.88 | - | - |
| MODERATE-Effect Variants | |||||
| Private | |||||
| Cases | 26,558 | 25,250 | 1.05 | 0.99 | 0.607 |
| Controls | 2,286 | 2,155 | 1.06 | - | - |
| Rare | |||||
| Cases | 146,052 | 158,361 | 0.92 | 1.10 | 0.022 |
| Controls | 20,603 | 24,663 | 0.84 | - | - |
| Common | |||||
| Cases | 920,641 | 958,085 | 0.96 | 1.02 | 0.023 |
| Controls | 127,551 | 135,706 | 0.94 | - | - |
| LOW-Effect Variants | |||||
| Private | |||||
| Cases | 16,622 | 15,249 | 1.09 | 0.99 | 0.361 |
| Controls | 1,464 | 1,335 | 1.10 | - | - |
| Rare | |||||
| Cases | 129,196 | 136,780 | 0.94 | 1.05 | 0.176 |
| Controls | 17,610 | 19,562 | 0.90 | - | - |
| Common | |||||
| Cases | 1,148,993 | 1,177,446 | 0.98 | 1.01 | 0.058 |
| Controls | 159,769 | 165,749 | 0.96 | - | - |
T:U ratio, transmitted to untransmitted ratio; RTR, relative transmission ratio (T:U ratio in cases/T:U ratio in controls). Detailed definition for MODERATE-and LOW-effect variants are described in the Supplemental Experimental Procedures. p values were calculated by one-sided permutations.
Although we observed global undertransmission of LOFs in both cases and controls, the deviation from the 1:1 ratio was more attenuated in cases than in controls. Therefore, we then compared the T:U ratio in cases and controls to calculate the relative transmission ratio (RTR = T:U ratio in cases/T:U ratio in controls). Since sequencing and genotyping errors are expected to affect cases and controls equally, contrasting T:U ratios in cases versus controls should provide a robust result. We observed significantly higher RTRs than expected for all allele frequency classes of LOF variants (Table 2; private LOFs: RTR = 1.49, p = 0.0086; rare LOFs: RTR = 1.31, p = 0.0051; common LOFs: RTR = 1.08, p = 0.037, one-sided permutation test). These results suggest that LOF variants identified in cases, especially the private and rare ones, are enriched in disease-causing variants.
To further confirm that our observation was not due to errors in genotyping calls, we analyzed “MODERATE”-effect variants and “LOW”-effect variants. Unlike LOF variants, the T:U ratios for MODERATE- and LOW-effect variants were close to 1:1 for all allele frequency classes (Table 2). Therefore, the global under-transmission of LOF variants that we observed is unlikely due to sequencing errors but rather to selection acting against the LOF variants. When we calculated RTRs, we observed significantly higher RTRs for the rare and common MODERATE-effect variants, but the effect sizes observed were more modest than LOFs (Table 2; private MODERATE: RTR = 0.99, p = 0.607; rare MODERATE: RTR = 1.10, p = 0.022; common MODERATE: RTR = 1.02, p = 0.023, one-sided permutation test). For the variants with LOW effect, we did not obtain any significant results (Table 2). These findings indicate less contribution from variants classified as MODERATE- or LOW-effect to the disease etiology than from LOF variants. We also performed an analysis excluding all European American case trios and confirmed that the observed results were not influenced by the combination of two population groups (Table S3, and Supplemental Information).
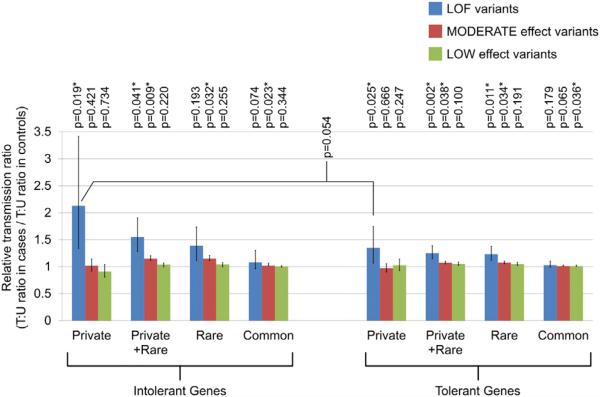
Application of RVIS provided further evidence for a role of selection pressure in shaping the transmission patterns of inherited variants. We observed further increase in RTR for private LOF variants in genes intolerant to functional variation (Figure 2; RTR = 2.13, p = 0.0193, one-sided permutation test) compared to private LOFs in all genes (RTR = 1.49). RTR for private LOF variants in tolerant genes was 1.35 (p = 0.025). Difference of RTRs between private LOF variants in intolerant and tolerant genes showed marginal significance (p = 0.054). By contrast, a difference of RTRs between intolerant and tolerant genes was not apparent for other classes of variants (rare and common LOFs, all categories of missense and silent variants). These results suggest that private LOFs in intolerant genes in cases are more likely to contribute to disease etiology than other variants. It should be also noted that global undertransmission of private and rare LOF variants was prominent for intolerant genes both in cases and controls (private LOF variants: T:U = 0.81 in cases and 0.38 in controls, rare LOF variants: T:U = 0.57 in cases and 0.41 in controls), further indicating that the observed global undertransmission of LOF variants is not due to sequencing errors.
Figure 2. Prominent Overtransmission of Private LOF Variants in Intolerant Genes.
Relative transmission ratios (RTRs) are displayed for variants categorized by their functionality (blue: loss-of-function [LOF] variants; red: MODERATE-effect variants; and green: LOW-effect variants), minor allele frequencies (private, rare, or common), and gene intolerance. Detailed definition for LOF, MODERATE-, and LOW-effect variants is provided in the Supplemental Experimental Procedures. *p < 0.05. Error bars indicate 95% confidential intervals. p values for RTRs were calculated by one-sided permutation analysis with random shuffling of the case-control status.
We also asked whether the load of inherited LOF variants correlates with four clinical variables (see above, also described in Supplemental Experimental Procedures) and observed a significant correlation between severity scores of the disease and per-individual number of private LOF variants (p = 2.77 × 10−5, Spearman's rank correlation r = 0.29). This finding highlights the importance of private inherited LOFs for disease manifestation. Thus, both analysis of transmission patterns and phenotypic correlations suggest that the inherited component of SCZ risk is enriched in private LOF variants, especially ones in genes intolerant to functional variation. A gene set enrichment analysis showed that despite high genetic heterogeneity, there is an enrichment of a few biological processes and/or functions among such genes, most notably enrichment of terms related to ATPase activity (see Supplemental Information and Table S4).
Finally, to explore individual promising candidate genes further, we cross-compared the list of intolerant genes harboring at least one private LOF variant in SCZ with lists of candidate genes from previously published literature (see Supplemental Information and Table S5). We found several genes in common that represent promising candidate genes worthy of follow-up analysis. In particular, when we cross-compared the list of intolerant genes harboring private LOFs with previously published literature on CNVs associated with neuropsychiatric and neuro-developmental diseases (Malhotra and Sebat, 2012), we found two genes (LZTR1 and RFC2) in common. LZTR1, encoding a protein exclusively localized to the Golgi network helping stabilize the Golgi complex (Nacak et al., 2006), is located within the 3-Mb 22q11.2 deletion locus, a finding consistent with recent evidence establishing a strong link between the Golgi apparatus and neuronal phenotypes associated with the 22q11.2 microdeletion (Mukai et al., 2008; Xu et al., 2013). RFC2 is located on the chromosome 7q11.2 Williams-Beuren syndrome (WBS) critical locus, encodes for a protein playing a role in response to DNA damage, and may be involved in microcephaly and growth retardation observed in WBS patients (Merla et al., 2010). This finding is also consistent with recent findings implicating DNA repair mechanisms in the pathogenesis of SCZ and ASD (Ionita-Laza et al., 2014).
DISCUSSION
In this study, we performed a systematic investigation of the role of both de novo and inherited LOF variants in order to elucidate the role that gene-disruptive events play in the genetic architecture of SCZ.
We confirmed an excess of de novo LOFs in SCZ patients and identified a single gene (SETD1A) harboring two de novo LOF indels in two individuals with SCZ. SETD1A encodes a catalytic subunit of the histone methyltransferase protein complex, named Set/COMPASS (complex protein associated with Set1). This complex mediates mono-, di-, and trimethylation of H3K4, a mark of active gene transcription (Miller et al., 2001; Roguev et al., 2001). In humans, Set/COMPASS is comprised of one of the six catalytic subunits (SETD1A, SETD1B, KMT2A [also known as MLL], KMT2D [MLL2], KMT2C [MLL3], and KMT2B [MLL4]), four common subunits (WDR5, RBBP5, ASH2L, and DPY30), and several subtype-specific proteins (Mohan et al., 2010; Shilatifard, 2012). Interestingly, de novo mutations in other subunits comprising Set/COMPASS have been reported in SCZ and ASD cases: a missense variant in RBBP5 in SCZ (Gulsuner et al., 2013), as well as a nonsense variant in KMT2C and a missense variant in KMT2A in ASD (Neale et al., 2012; O'Roak et al., 2012b). It is also noteworthy that the ATP-dependent chromatin helicase CHD8, in which recurrent de novo LOF mutations have been identified in ASD patients (O'Roak et al., 2012a, 2012b), directly interacts with common subunits of Set/COMPASS and plays a role in the regulation of H3K4 trimethylation (Yates et al., 2010).
Implication of SETD1A involvement, as well as the observed enrichment of chromatin regulators among genes hit by de novo damaging variants in SCZ, are in line with accumulating data indicating a role of chromatin-related genes in various psychiatric and neurodevelopmental disorders (Ben-David and Shifman, 2013; Ronan et al., 2013). Involvement of chromatin modification pathways in SCZ was also suggested from analysis integrating data from de novo CNVs, de novo SNVs, and genome-wide association studies (GWASs) (Gilman et al., 2012). Although there was no statistically significant enrichment of chromatin-related pathways among genes at loci identified in the largest GWAS for SCZ so far, some genes involved in chromatin regulation, such as PBRM1 and SETD8, were found in the proximity of SNPs with genome-wide significant association (Ripke et al., 2013).
H3K4 trimethylation, in particular, is known to be essential for neuronal differentiation (Wynder et al., 2005) and fear memory formation (Gupta et al., 2010). The H3K4 trimethylation status in prefrontal cortex neurons is dynamically regulated from the late prenatal period to early adulthood (Shulha et al., 2013). Interestingly, there are reports of decreased H3K4 trimethylation at the GAD1 promoter region in the prefrontal cortex of female SCZ patients (Huang et al., 2007), as well as of upregulation of Gad1 H3K4 trimethylation induced by clozapine but not by haloperidol (Huang et al., 2007). It is also noteworthy that a de novo LOF variant in KDM5C, encoding JARID1C that mediates demethylation of H3K4, was found in another SCZ subject in our cohort (Xu et al., 2012). Mutations in KDM5C have also been linked to nonsyndromic X-linked mental retardation (Jensen et al., 2005).
Application of RVIS identified SETD1A within the first percentile of the most intolerant genes against functional variants including LOF and missense variants (Petrovski et al., 2013). When we calculated in the same manner intolerance scores specifically against LOF variants using data from Exome Variant Server (http://evs.gs.washington.edu/EVS), SETD1A was included in the fourth percentile of the genes most intolerant against LOF variants. In addition, there is no LOF variant disrupting SETD1A in the 1000 Genomes Project database (Abecasis et al., 2012), another large-scale data source of human genetic variation. The high intolerance of SETD1A specifically against LOF variants further supports that the identification of two de novo LOF mutations in our SCZ cohort is unlikely to have occurred by chance.
Our current analysis using PRISM and Pindel revealed five previously unidentified de novo LOF indels. The fact that these LOFs were not identified in our previous analysis using Dindel and its default filtering parameters is primarily due to the low coverage of these variant sites in relation to their high GC content or their complex structure. Specifically, for four of these LOFs (CEP104, INHBC, and the two SETD1A indels), the coverage of their genomic position was 5×–13×, with 56–70 GC% content in the surrounding 100 bp regions (50 bp upstream and 50 bp downstream). For the fifth LOF (ITPR2 indel), the genomic position was well covered (42× coverage, GC% content = 51), but the variant was probably missed in our previous analysis because of its complexity and relatively long length (a combination of an 11 bp insertion and a 3 bp deletion). Therefore, our current analysis using two different software and custom filtering criteria may be advantageous for identifying indels with complex structures or at low coverage regions. It is also notable that the SETD1A frameshift indel and the CEP104 indel were solely identified by PRISM and Pindel, respectively. This indicates that the sensitivity of detecting de novo indels could be increased substantially by using multiple software.
Identification of relatively increased transmission of private and rare inherited LOF variants in SCZ probands highlighted the significance of such events as risk factors for SCZ, while correlation of mutational load with disease severity implicated them in clinical outcome. Similar to the de novo CNVs and SNVs, these mutations are distributed across many different genes and therefore would have been missed through previous association or linkage studies. Results from our WES family-based analysis are consistent with recent case-control studies performing targeted resequencing of candidate genes, which also reported excess of rare LOF variants in SCZ patients (Hu et al., 2013; Kenny et al., 2013).
Results from the current study and the findings in other studies for de novo SNVs and indels (Girard et al., 2011; Xu et al., 2011, 2012) collectively highlight the significance of LOF mutations in the overall genetic etiology of SCZ. In addition, LOF events provide further collective insight into the heritable component of SCZ, which has not yet been accounted for by de novo CNVs and SNVs. Finally, identification of SETD1A as one of the first genes recurrently hit by de novo LOF mutations in SCZ, strongly suggests that further studies using large sample sets enriched in truly sporadic cases as well as intensive analysis of currently available data sets focusing on LOF variants will lead to the discovery of additional recurrently affected genes, a key step toward elucidating the neural mechanisms underlying susceptibility to SCZ and designing novel therapies (Karayiorgou et al., 2012).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the families who participated in this research. We thank S.L. Lundy for valuable assistance with clinical database maintenance. We also thank S. Levy and the HudsonAlpha Genomics Services Laboratory for sequencing. This work was partially supported by National Institute of Mental Health (NIMH) grants MH061399 (to M.K.) and MH097879 (to J.A.G.) and the Lieber Center for Schizophrenia Research at Columbia University. A.T. was supported by the JSPS Postdoctoral Fellowship for Research Abroad. B.X. was partially supported by a National Alliance for Research in Schizophrenia and Depression (NARSAD) Young Investigator Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Text, Supplemental Experimental Procedures, four figures, and fives tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2014.04.043.
REFERENCES
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA, 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol. Psychiatry. 2013;18:1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- Bottas A, Cooke RG, Richter MA. Comorbidity and patho-physiology of obsessive-compulsive disorder in schizophrenia: is there evidence for a schizo-obsessive subtype of schizophrenia? J. Psychiatry Neurosci. 2005;30:187–193. [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodríguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- Edmonds DK, Lindsay KS, Miller JF, Williamson E, Wood PJ. Early embryonic mortality in women. Fertil. Steril. 1982;38:447–453. [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Goriely A, McGrath JJ, Hultman CM, Wilkie AO, Malaspina D. “Selfish spermatogonial selection”: a novel mechanism for the association between advanced paternal age and neurodevelopmental disorders. Am. J. Psychiatry. 2013;170:599–608. doi: 10.1176/appi.ajp.2013.12101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, et al. Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J. Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang B, Liu W, Paciga S, He W, Lanz TA, Kleiman R, Dougherty B, Hall SK, McIntosh AM, et al. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol. Psychiatry. 2013 doi: 10.1038/mp.2013.131. Published online October 15, 2013. http://dx.doi.org/10.1038/ mp.2013.131. [DOI] [PMC free article] [PubMed]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J. Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I, Xu B, Makarov V, Buxbaum JD, Roos JL, Gogos JA, Karayiorgou M. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc. Natl. Acad. Sci. USA. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang Y, Brudno M. PRISM: pair-read informed split-read mapping for base-pair level detection of insertion, deletion and structural variants. Bioinformatics. 2012;28:2576–2583. doi: 10.1093/bioinformatics/bts484. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al. Schizophrenia susceptibility associated with interstitial deletions of chromo-some 22q11. Proc. Natl. Acad. Sci. USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Flint J, Gogos JA, Malenka RC, Genetic and Neural Complexity in Psychiatry 2011 Working Group The best of times, the worst of times for psychiatric disease. Nat. Neurosci. 2012;15:811–812. doi: 10.1038/nn.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C, Kelleher E, Ennis S, Tropea D, Anney R, et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol. Psychiatry. 2013 doi: 10.1038/mp.2013.127. Published online October 15, 2013. http://dx.doi.org/10.1038/mp.2013.127. [DOI] [PubMed]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, et al. NHLBI Exome Sequencing Project Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. 1000 Genomes Project Consortium A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams-Beuren syndrome 7q11.23 region. Hum. Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat. Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacak TG, Leptien K, Fellner D, Augustin HG, Kroll J. The BTB-kelch protein LZTR-1 is a novel Golgi protein that is degraded upon induction of apoptosis. J. Biol. Chem. 2006;281:5065–5071. doi: 10.1074/jbc.M509073200. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Hastings IM. Mutation and selection within the individual. Genetica. 1998;102-103:507–524. [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kaähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, et al. Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Gogos JA, Karayiorgou M. The genetic architecture of schizophrenia: new mutations and emerging paradigms. Annu. Rev. Med. 2012;63:63–80. doi: 10.1146/annurev-med-072010-091100. [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 2005;7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JA, Menon T, Thompson BA, Bochar DA. Regulation of HOXA2 gene expression by the ATP-dependent chromatin remodeling enzyme CHD8. FEBS Lett. 2010;584:689–693. doi: 10.1016/j.febslet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am. J. Hum. Genet. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.