Abstract
Eukaryotic translation elongation factors 1 alpha, eEF1A1 and eEF1A2, are not only translation factors but also pleiotropic proteins that are highly expressed in human tumors, including breast cancer, ovarian cancer, and lung cancer. eEF1A1 modulates cytoskeleton, exhibits chaperone-like activity and also controls cell proliferation and cell death. In contrast, eEF1A2 protein favors oncogenesis as shown by the fact that overexpression of eEF1A2 leads to cellular transformation and gives rise to tumors in nude mice. The eEF1A2 protein stimulates the phospholipid signaling and activates the Akt-dependent cell migration and actin remodeling that ultimately favors tumorigenesis. In contrast, inactivation of eEF1A proteins leads to immunodeficiency, neural and muscular defects, and favors apoptosis. Finally, eEF1A proteins interact with several viral proteins resulting in enhanced viral replication, decreased apoptosis, and increased cellular transformation. This review summarizes the recent findings on eEF1A proteins indicating that eEF1A proteins play a critical role in numerous human diseases through enhancement of oncogenesis, blockade of apoptosis, and increased viral pathogenesis.
Keywords: eEF1A, cancer, apoptosis, virus, HIV
Introduction
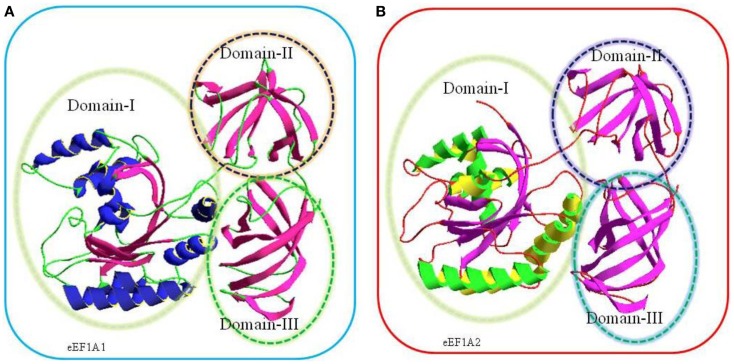
Translation is the central event leading to protein synthesis and translation factors are key actors involved in the translation process (1–3). Recently, it has been suggested that translation factors could represent a new class of potential oncoproteins that could favor cellular transformation through protein translation infidelity, association with cytoskeleton alterations, and modulation of signaling pathways (3–5). The eukaryotic translation elongation factors 1 alpha, eEF1A1 and eEF1A2, are the second most abundant protein (1–3% of total protein content) after actin and an important component of translation machinery. In its GTP bound form, eEF1A1 and eEF1A2 deliver the aminoacylated-tRNA to the A site of the ribosome for decoding of mRNA by codon–anticodon interactions (6). Following the hydrolysis of GTP, eEF1A (eEF1A1 and eEF1A2), GDP is released from the ribosome (6). Thus, eEF1A1 and eEF1A2 are GTP-binding proteins and consist of three domains namely domain I, domain II, and domain III. Domain I spans over 1–240 residues, which made up of a Rossmann-fold topology. Domain II (241–336 amino acids) and domain III (residues 337–443) consist of beta strands and each domain contains two beta sheets that form the beta barrel (7–10) (Figure 1).
Figure 1.
Comparative three-dimensional (3-D) model of eEF1A1 (A) and eEF1A2 (B) on the basis of crystal structure of homologous eEF1A from yeast. The target sequences used were eEF1A1 (Swiss-Prot Accession No: P68104) and eEF1A2 (Swiss-Prot Accession No: Q05639). The amino acids sequences of each protein were submitted to SWISS-MODEL server to build a 3D model (10).The highest resolved structure, 1.67-Å X-ray-derived eEF1A protein structure from yeast (Saccharomyces cerevisiae) with PDB ID: 1f60 and E-value 0.0 (sequence identity: 80.371%) was used as a template for modeling. Structurally, each model consists of three domains, domain I, domain II, and domain III as shown in the above cartoon. Domain I (residues 1–240) is made up of Rossmann-fold topology. Domain II (residues 241–336) and domain III (residues 337–443) are made up of entirely from beta-strands; each domain contains two beta sheets that form a beta barrel (7).
There are two known isoforms of protein eEF1A, i.e., eEF1A1 and eEF1A2. The cellular expression of eEF1A is divided into three classes. First are the majority of cell types that express only eEF1A1. Second, neurons and muscle cells that express only eEF1A2. Third class belongs to certain tumor cell types and cell lines that express both eEF1A isoforms (11–13). eEF1A1 protein has been mapped on chromosomes 6q14. eEF1A1 protein shares homology with eEF1A2 protein at the nucleotide level (75%) and amino acid level (96%). eEF1A2 does not bind GDP and GTP with the same relative affinity as eEF1A1. The GDP dissociation rate constant is seven times higher for eEF1A1 than for eEF1A2. In addition, the nucleotide preference ratio (GDP/GTP) for eEF1A1 is 0.82 and for eEF1A2 is 1.50 (14–17). Furthermore, eEF1A has been shown to be a novel component of the nuclear export machinery in mammalian cells and is involved in the nuclear export of specific proteins such as VHL van Hippel-Lindau (VHL) tumor suppressor and poly(A)-binding protein (PABP1) (18).
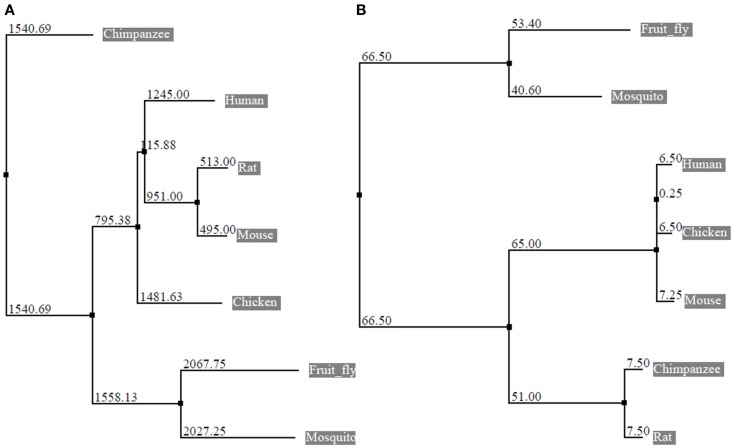
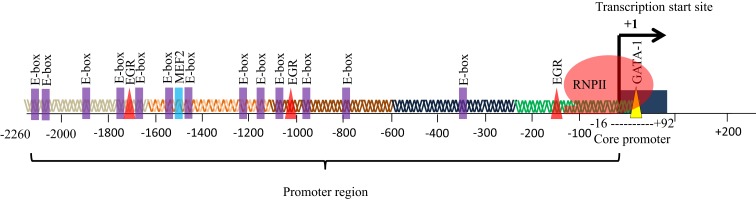
The eEF1A2 gene is present in the common ancestor of eukaryotes (Table 1; Figure 2). Human eEF1A2 gene spans approximately 12 kb human genome sequence, which consists of eight exons and seven introns plus 2 kb upstream promoter region, and has been mapped on chromosome 20q13.3. Analysis of the region −2064 to +220 reveals that the promoter region contains the binding sites for several important cis-regulatory elements (E-boxes, EGR family proteins, GATA motif, and MEF binding site) with no TATA elements (Figure 3). The core region of the promoter is mapped from position −16 to +92 (9, 19). Beyond its central role in translation machinery, eEF1A2 plays an important role in cell cycle regulation, heat-shock response, aging, posttranslational modifications, and signal transduction (20–23).
Table 1.
Orthologs for eEF1A2 gene.
| Organism | Similarity with human |
|
|---|---|---|
| Nucleotide (%) | Amino acid (%) | |
| Chimpanzee | 78.31 | 92.41 |
| Rat | 89.27 | 99.78 |
| Mouse | 89.13 | 99.78 |
| Chicken | 88.91 | 99.57 |
| Mosquito | 78.99 | 85.42 |
| Fruit fly | 77.68 | 83.91 |
Figure 2.
Phylogenetic tree constructed from the alignment of nucleotide and protein sequences of eEF1A2. The horizontal lines are the branches and suggest the amount of evolutionary changes over time. (A) Phylogenetic tree based on nucleotide sequences of eEF1A2 (Human: NM_001958.3, Chimpanzee: XM_003954094.1, Rat: NM_012660.2, Mouse: NM_007906.2, Fruit fly: NM_079872.4, Chicken: NM_001032398.2, and Mosquito: XM_003436467.1) using neighbor-joining distance method. The numbers indicate the evolutionary distances. (B) Phylogenetic tree based on amino acid sequences of eEF1A2 (Human: NP_001949.1, Chimpanzee: XP_003954143.1, Rat: NP_036792.2, Mouse: NP_031932.1, Fruit fly: NP_524611.1, Chicken: NP_001027570.1, and Mosquito: XP_003436515.1 using neighbor-joining (PAM250). The Jalview program was used for tree construction.
Figure 3.
Promoter of eEF1A2. The promoter of eEF1A2 spans approximately 2.26 kb. Analysis of promoter sequence reveals 13 E-boxes, 3 EGR binding sites, and 1 MEF2 binding site. The core promoter region spans from position −16 to +92 bp.
Protein synthesis is one of the most sophisticated biochemical processes occurring in the cell, which requires hundred of proteins, including eEF1A proteins, eEF1A1 and eEF1A2. Besides the role of eEF1A proteins in the translational process, an increasing series of data are presently emerging about the non-canonical roles of these proteins in oncogenesis, modulation of apoptosis and viral pathogenesis (1, 20).
Cancer
Breast cancer
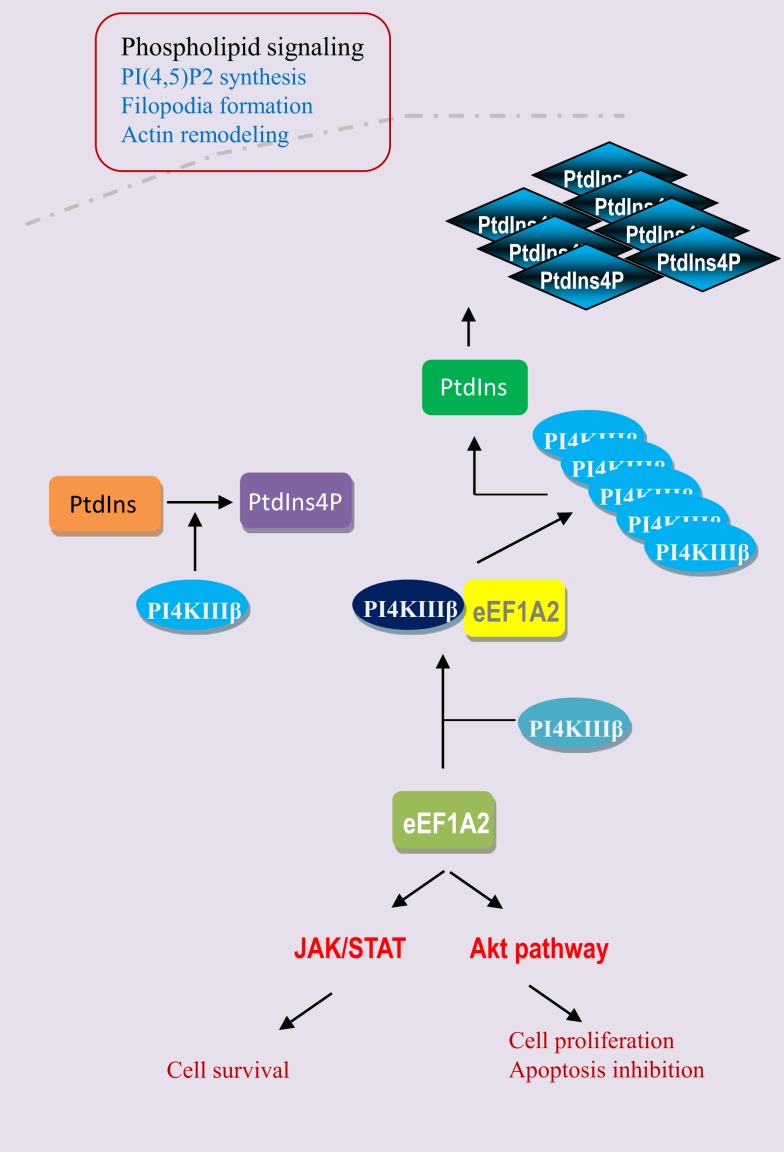
Breast cancer is the most common cancer in women both in the developing and developed world; there are an estimated 1 million new cases per year. The use of gene signature or the identification of changes in gene expression in breast tumors relative to normal surrounding tissue is of great importance in terms of prognostic indicators and therapeutic targets (24). The eEF1A2 is hardly detectable in normal human breast tissue but the expression of eEF1A2 is strongly upregulated in most of breast tumors (25, 26). High levels of eEF1A2 proteins are detected in 60% of primary breast tumors and metastases, but not in normal epithelium (Table 2). The expression of eEF1A2 is sufficient to stimulate the formation of filopodia in BT549 human breast cancer cells and non-transformed Rat2 cells. Moreover, its expression is sufficient to activate Akt in a PI3K-dependent fashion, as down-regulation of eEF1A2 by siRNA reduces Akt activity. In breast cancer cell line BT549, eEF1A2 expression stimulates cell migration and invasion in a largely PI3K and Akt-dependent manner, suggesting eEF1A2 regulates oncogenesis through Akt and PI3K-dependent cytoskeleton remodeling (27–29). In fact, eEF1A2 participates in the regulation of the phospholipids signaling pathway (Figure 4). Phosphatidylinositols are negatively charged membrane bound phospholipids that participate in the pathways that regulate the cell proliferation, survival, cytoskeleton organization, vesicular trafficking, and oncogenesis (30, 31). Phosphoinositols are composed of an inositol ring in which one or more −OH groups at the 3-, 4-, and 5-position of inositol ring are esterified with a phosphate group in all possible combinations. Specific kinase families (PI3K, PI4K, and PI5K) are responsible for phosphorylation at these sites (32, 33). Overexpression of eEF1A2 protein up-regulates overall PI4K activity and cellular phosphatidylinositol 4-phosphate (PI4P) generation in human cells. Furthermore, eEF1A2 directly interacts with and activates phosphatidylinositol-4 kinase III β (a subfamily of PI4K), an enzyme that converts phosphatidylinositol to PI4P. Knockdown of eEF1A2 using eEF1A2 siRNA results in down-regulation of phosphatidylinositol-4-kinase activity indicating that eEF1A2 is a physiological regulator of PI4KIIIβ signaling (34, 35). In addition, eEF1A2 expression up-regulates the generation of phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] in the cytoplasm and at the plasma membrane. The subsequent increase in PI(4,5)P2 at the plasma membrane stimulates the eEF1A2-induced filopodia formation through binding and activation of PI4KIII β. Therefore, eEF1A2 is involved in phosphatidylinositol signaling and actin remodeling (34, 36). Moreover, the gene expression profiling of primary mouse B cell lineage showed the high expression of eEF1A2 in plasmacytomas (PCT), which results in progression of plasma cell neoplasms in both mouse and human (29). Finally, the knockdown of eEF1A2 expression delays or impairs the IL-6-induced activation of STAT3 and Akt signaling pathways suggesting that activation of STAT3 and Akt through eEF1A2 involvement that favors cell proliferation, cell cycle progression, and inhibition of apoptosis (29, 37, 38). Altogether, eEF1A2 protein activates the PIK-Akt-STAT3 pathways that have been extensively shown to favor cellular transformation and oncogenesis (39–44) (Figure 4).
Table 2.
Expression of eEF1A2 in different human cancers.
| Type of cancer | Methods of detection | Relevant findings | Reference |
|---|---|---|---|
| Breast cancer | Real-time reverse transcription-PCR/Tissue array/Immunohistochemistry/western blot | eEF1A2 mRNA/protein is highly expressed in 50–60% in primary human breast cancer | (25, 39) |
| Ovarian cancer | Tissue-microarray/Immunohistochemistry | eEF1A2 is highly expressed in 30% of primary ovarian tumors | (40, 56) |
| Lung cancer | Comparative genomic hybridization | Positive Ki-67 expression associated with positive eEF1A2 and KCIP-1 | (41, 42) |
| Hepatocellular carcinoma | Immunohistochemistry | eEF1A2 is highly expressed in half of hepatocellular carcinoma | (43, 44) |
Figure 4.
eEF1A2 activates the phospholipid, JAK/STAT, and Akt pathways. eEF1A2 is a physiological regulator of phosphatidylinositol-4-kinase (PI4K). It directly interacts with phosphatidyloinositol-4 kinase III β (PI4KIIIβ) and enhances its lipid kinase activity by converting the phosphatidylinositol (PtdIns) into phosphatidylinositol-4-phosphate (PtdIns4P). Then PI4KIIIβ and PtdIns4P increase the phosphatidylinositol-4,5 bisphosphate generation at the plasma membrane level, which results in the filopodia formation and actin remodeling. eEF1A2 also directly or indirectly regulates the JAK/STAT and Akt signaling pathways.
In breast cancer, the genes in 20q13 are frequently amplified and have been shown by using comparative genomic hybridization and fluorescence in situ hybridization (45). The differential screening of cDNA libraries from metastatic and non-metastatic cell lines derived from the same parental rat mammary adenocarcinoma showed a 1.5-fold overexpression of eEF1A in the metastatic compared to the non-metastatic cells (46). Studying cancer cell lines derived from breast, lung, prostate, and skin, eEF1A2 gene expression exhibited the highest alteration in the cancer cell lines compared to normal controls using a profiling array (47–50).
Ovarian cancer
Ovarian cancer represents 4% of all female cancer. It has the highest fatality to case ratio of all gynecological cancers because the majority of cases are diagnosed in the late stage. Despite of significant efforts to improve the early detection and advances in chemotherapy, metastasis remains a major challenge in clinical management of ovarian cancer (51–54). The eEF1A2 gene is not expressed in normal ovary but highly expressed in ovarian cancer (55). eEF1A2 expression enhances the ovarian cell growth in vitro and favors the tumorsphere formation (56), suggesting that eEF1A2 could favor the development of ovarian primary tumor formation. High levels of eEF1A2 expression was observed in 30% all the primary ovarian tumors, 50% of serous tumors, 30% of endometrioid tumors, 19% of mucinous tumors, and 8% of clear-cell tumors (Table 2) (56). Furthermore, the eEF1A2 protein and RNA expression levels are upregulated in clear-cell ovarian carcinoma by 75 and 33% respectively. The eEF1A2 gene is unmethylated both in normal and tumor cells, suggesting that up-regulation of eEF1A2 gene expression is not dependent on epigenetic modifications (at least for the methylation status), but instead that the inappropriate expression of trans-acting factor(s) could be involved (55). The enhanced expression of eEF1A2 protein in ovarian cancers correlates with poor prognosis (57).
The oncogenic properties of eEF1A2 have also been studied in different ovarian tumors and established cell lines. In rodent fibroblasts, eEF1A2 protein favors anchorage-independent growth and results in decreased doubling time during cellular proliferation. Furthermore, the induced expression of eEF1A2 in NIH3T3 cells makes them tumorigenic and increases the growth rate of ES-2 ovarian carcinoma cells xenografted in nude mice (40). The transcription factor ZNF217 and eEF1A2, both located on chromosome 20q13, are frequently amplified in ovarian epithelial carcinomas. The stable preneoplastic ovarian cell lines that over-express eEF1A2 provides the evidence that up-regulation of eEF1A2 expression contributes to the neoplastic properties of precursor cells of ovarian carcinomas mediated through ZNF217 (58).
Resveratrol, a phytoalexin produced naturally by several fruit plants, has been extensively studied for its chemopreventive and chemotherapeutic effects in cancer and animal models. Resveratrol blocks the angiogenesis, induces the autophagocytosis and apoptosis in proliferating cells, and is a well-known sirtuin 1 activator (59, 60). The expression of eEF1A2 is increased in the PA-1 ovarian cell line after serum or insulin stimulation. Resveratrol up-regulates the caspase-3 level in PA-1 cells by down regulating the expression of eEF1A2 via the blockade of upstream Akt pathway. Additionally, resveratrol suppresses growth of human ovarian cancer cells in culture and in a murine xenograft model with reduced expression of proliferating cell nuclear antigen and increased TUNEL staining (61). All together resveratrol down-regulates eEF1A2 in ovarian cancer cells and thereby favors apoptosis.
Lung cancer
Lung cancer is the most common cause of cancer related death both in men and women. It accounts for 1.3 million deaths worldwide annually. In spite of continuous efforts and clinical trials to develop new diagnostic and prognostic markers, a better understanding of the molecular pathways involved in lung cancer is essential for developing new therapeutic targets (62, 63). Four lung adenocarcinoma cell lines (HKULC 1–4) were established from patients with different clinical characteristics. Comprehensive studies of these cell lines show that eEF1A2 is a putative oncogene that is highly expressed in all the HKULC cell lines (64). In addition, high-resolution analysis of genomic aberration by metaphase and comparative genomic hybridization array identify the involvement of the 20q region, suggesting the potential role of eEF1A2 as a candidate tumor gene in lung cancer cell lines (41). In another study, 183 genes with increment in both genomic copy number and transcript in six lung adenocarcinoma were analyzed. Forty-two proteins were overexpressed in these cell lines as compared to the normal cells. Comparing the 183 genes with the 42 proteins, the expression of four candidates, namely, PRDX1, eEF1A2, CALR, and KCIP-1 was correlated with increased DNA copy number and transcript levels. Furthermore, expression of siRNA targeting eEF1A2 and KCIP-1 in these cell lines suppressed cellular proliferation and triggered apoptosis. Therefore, the overexpression of eEF1A2 and KCIP-1 in lung tumor samples strongly suggests that both proteins could be involved in lung adenocarcinoma and could be potential therapeutic targets in lung cancer (42).
Liver cancer
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in humans (65, 66). HCC cell lines, HepG2, HuH7, and JHH6 show increased expression of eEF1A2 as compare to normal liver tissue, but the mRNA of eEF1A2 is markedly increased only in JHH6 cells (43). In addition, the MDM4 and eEF1A2 proteins show increased expression in cryptogenic HCC (44). eEF1A2 gene silencing reduces cell viability, proliferation, and increases the apoptosis rates in HCC cell lines (44). Furthermore, eEF1A2 is overexpressed in 43% of HCC and activates the Akt pathway as observed in 40–60% of primary HCCs (Table 2) (44, 67). Using array based comparative genomic hybridization, frequent amplification and gain of DNA copy number at 1q, 6p, 8q, 17q, and 20q were observed in HCCs, suggesting that these genetic aberrations might facilitate the malignant transformation (68).
Pancreatic cancer
Pancreatic cancer is one of the unsolved health problems associated with aggressive malignancies and therefore there is need to develop improved early diagnosis tools (69, 70). The expression of eEF1A2 gene, located on chromosome 20q13 is significantly upregulated in pancreatic cancer. Although little or no eEF1A2 expression is present in normal pancreatic tissue, 83% of pancreatic cancers display increased expression of eEF1A2, suggesting that eEF1A2 plays an important role in pancreatic carcinogenesis (71–73).
Apoptosis
Cell proliferation and cell death are highly regulated and coordinated during the normal development, and both are crucial for the maintenance of tissue homeostasis. Impaired apoptosis can lead to autoimmunity or malignancy (74, 75). An increasing number of evidence suggests the involvement of eEF1A2 at the onset of cell transformation. Peroxiredoxin-1 (Prx-1) is a protein that is ubiquitously expressed in all mammalian cells and protects the cells from oxidative stress by reducing the range of reactive oxygen species. eEF1A2 interacts directly with Prx-1 and protects the cells from stress-induced apoptosis by the down-regulation of caspase-3 and caspase-8 activation parallel to increased expression of the pro-survival factor Akt (76, 77). Additionally, the eEF1A expression varies in cells treated with various concentrations of hydrogen peroxide, a strong apoptotic inducer associated with decreased mitochondrial respiration and an inhibitor of Prx-1 (78, 79). Finally, eEF1A provides protection against all endoplasmic reticulum (ER) stress-mediated cell death. FL5.12 cells are non-transformed murine B-cells that require IL-3 for growth, survival, and proliferation. Enforced expression of eEF1A in these cells protects them from IL-3 withdrawal without cellular transformation (80).
Wasted mouse is a spontaneous autosomal recessive mutation associated with neurological defects, immune system abnormalities, and defective response to DNA damage repair in immune cells. Thymus and spleen from wasted mice show extensive apoptosis. The loss of activity in wasted mice and immunological abnormalities are solely linked to the presence of a deletion in the eef1a2 gene (81–84). Furthermore, eEF1A2 is highly expressed in terminally differentiated cells such as neurons, cardiomyocytes, and myofibers. During skeletal muscle development, undifferentiated myoblasts are susceptible to serum deprived caspase 3-mediated apoptosis but become resistant to apoptosis after differentiating into myotubes. In muscle cells, the eEF1A1 is expressed during embryonic development of myoblast but is replaced by eEF1A2 after 2 weeks of birth indicating that the eEF1A1 isoform plays a pro-apoptotic role while the eEF1A2 isoform displays anti-apoptotic properties (85, 86).
Viral Infections
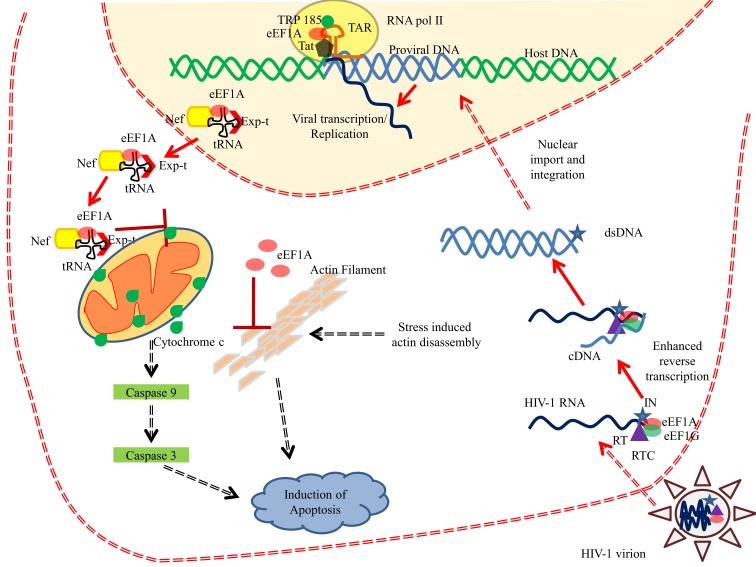
The eEF1A proteins play a critical role in several viral infections at distinct stages of the viral cycle [reviewed in Ref. (87)] (Table 3). Among the most studied viruses, human immunodeficiency virus (HIV) interacts with numerous cellular proteins, including eEF1A (88). First, eEF1A has been detected as part of the HIV virions as an actin-binding protein (89). Second, eEF1A has been reported to plays a critical role at early stages of HIV replication (Figure 5). eEF1A and also EF1G are part of HIV reverse transcription complex (RTC) (90). Both eEF1A and EF1G proteins coimmunoprecipitated with the p51 subunit of reverse transcriptase (RT) and integrase using an endogenous reverse transcription assay (90) (Figure 5). The depletion of eEF1A and EF1G using siRNAs decreased reverse transcription parallel to reduced levels of RTC in treated cells (90). Since integrase is also an RT binding protein, a tight interplay between RT, integrase, and eEF1A could be involved in several stages of HIV replication. Actually, integrase has been shown to interact with eEF1A in vitro (91, 92). eEF1A also interacts with other viral proteins involved in early stages of HIV replication such as Nef and Tat. Several reports indicate that HIV Nef interacts with eEF1A (77, 93) (Figure 5). Abbas and colleagues have shown that the Nef-eEF1A interaction favors the nucleo-cytoplasmic shuttling of eEF1A and ultimately inhibits stress-mediated apoptosis in monocyte-derived macrophages (77). Additionally, Nef interaction with actin impairs human podocyte actin cytoskeletal integrity and eEF1A could play a role in the development of podocyte phenotype in HIV-1 associated nephropathy (93). The trans-activation response (TAR) element is critical for the activation of HIV-1 transcription. eEF1A stimulates the RNA polymerase II and TRP-185 binding to TAR RNA, which in turn regulates the HIV-1 gene expression (94). eEF1A has been described to interact with the HIV Gag protein (88). Finally, eEF1A is responsible for the selection and binding of cognate aminoacyl-tRNAs to the A site of the ribosome. HIV Nef interacts with two components of the 40S small ribosomal subunit, the RPS10 protein, and the 18S rRNA, and also to tRNAs, and has been reported to decrease translation using an in vitro translation assay (95). We cannot exclude that eEF1A that binds to both Nef and tRNA participates in the control of translation in HIV-infected cells. Interestingly, EF1-delta has been reported to interact with the second coding exon of HIV Tat, and to result in reduced efficiency of the translation of cellular proteins, but not of viral mRNAs (96).
Table 3.
Virus-eEF1A interplay.
| Virus | Description of eEF1A/viral protein interaction | Reference |
|---|---|---|
| HIV | eEF1A part of HIV-1 virion and reverse transcriptase complex | (77, 89, 90, 93) |
| eEF1A also interacts with Nef and modulate apoptosis in MDMs | ||
| HBV | HBx interacts with eEF1A1 resulting in blockade of actin bundling | (97) |
| HDV | HDV genome also interacts with eEF1A1 | (98) |
| HPV 38 | E7 protein interacts with both eEF1A1 and eEF1A2. The interaction is associated with cellular immortalization and transformation of primary keratinocytes | (99) |
| WNV | eEF1A interacts with the 3′stem–loop region of the viral genomic RNA and favors replication | (100, 101) |
| DENV | Viral genomic RNA sequesters eEF1A and hence decreases the concentration of Sphk1, thereby governing cell survival | (102) |
| TGEV | eEF1A interacts with the nucleocapsid of the virus and favors virus replication | (103) |
| VSV | eEF1A is found within the virion | (104) |
| Vaccinia virus | eEF1A as a part of mature virion | (105, 106) |
| CMV | eEF1A found to be as a part of virion proteome | (107, 108) |
| SARS-CoV | eEF1A found in mature virion | (109) |
HIV, human immunodeficiency virus; MDMs, monocyte-derived macrophages; HBV, hepatitis B virus; HDV, hepatitis delta virus; HPV, human papillomavirus; WNV, West Nile virus; DENV, dengue virus; VSV, vesicular stomatitis virus; CMV, cytomegalovirus; TGEV, transmissible gastroenteritis coronavirus; SARS, severe acute respiratory syndrome-associated coronavirus (SARS-CoV).
Figure 5.
eEF1A interferes with HIV-1 replication. eEF1A plays important role in various phases of HIV-1 life cycle. eEF1A has been reported in the mature HIV-1 virion and is also found to be a part of reverse transcription complex (RTC). Binding of eEF1A/EF1G to the RTC resulted in enhanced reverse transcription. In addition, eEF1A also help in the recruitment of RNA polymerase II and TRP-185 to the TAR RNA, which in turn regulates the viral transcription from 5′LTR. Moreover, HIV-1 Nef has been shown to interact with eEF1A and resulted in nucleo-cytoplasmic shuttling of eEF1A and ultimately in the inhibition of stress-induced apoptosis. Role of eEF1A in inhibiting the actin filament disassembly has been also proposed (77). Abbreviations: eEF1A, eukaryotic translation elongation factor 1 alpha; Exp-t, exportin-t; dsDNA, double stranded DNA; cDNA, complementary DNA; IN, integrase; RT, reverse transcriptase; RTC, reverse transcription complex; tRNA, transfer RNA; TAR, trans-activation response element; RNA pol II, RNA polymerase II; Tat, transactivator protein.
Since overexpression of eEF1A has been reported in several cancers, we cannot exclude a link between oncoviruses and eEF1A. Among oncoviruses, hepatitis B virus expresses the HBx protein that has been previously involved in liver cancer, especially HCC (110). HBx protein has been reported to interact with eEF1A1 in Huh-7 hepatoma cells infected with recombinant adenovirus expressing HBx protein (97). Interaction of HBx protein with eEF1A1 blocks filamentous actin bundling (97). Interestingly, the hepatitis delta virus (HDV) that can propagate only in the presence of HBV has a RNA genome that also interacts with eEF1A1 (98). A dozen of human papillomavirus (HPV) types including HPV16 and HPV18 are well-known oncoviruses that express two oncoproteins E6 and E7, and favor cellular transformation, oncogenesis, and the appearance of cervical cancers (111). E7 protein of HPV38 has been shown to interact with both eEF1A1 and eEF1A2 proteins leading to cellular immortalization and transformation of primary keratinocytes probably through disruption of actin stress fiber formation, a critical event linked to tumor formation (99). eEF1A facilitates virus replication complex (RPC) assembly and favors replication of West Nile virus and dengue virus (100–102). Similarly, eEF1A interacts with the nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) and favors TGEV replication (103). Since eEF1A is found in highly purified virions of numerous RNA and DNA viruses including vesicular stomatitis virus (104), vaccinia virus (105, 106), cytomegalovirus (107, 108), severe acute respiratory syndrome coronavirus (SARS-CoV) (109), and HIV-1 (88, 112), its role in viral pathogenesis needs to be further investigated.
Conclusion
Recent findings in the field of cellular and molecular biology reveal that the eEF1A proteins are not only involved in the translational process but display also non-canonical functions. The specific up-regulation of eEF1A2 in numerous tumors and its transforming properties indicate that it could play a significant role in tumorigenesis. Furthermore, eEF1A2 activates the phospholipid and Akt signaling pathways favoring cell survival. Additionally, eEF1A proteins block apoptosis and favors viral replication. All together, the non-canonical functions of eEF1A proteins are involved at the crossroads of oncogenesis, blockade of apoptosis, and viral pathogenesis. Therefore, the eEF1A proteins might play an important role in the pathophysiology of tumors and apoptosis, especially in response to stress and viral infections. Future studies need to be done to further highlight the role of eEF1A proteins in human diseases.
Author Contributions
WA and AK were responsible for drafting and revising the manuscript. GH was involved in critical reading of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the University of Franche-Comté, the Région Franche-Comté (RECH-FON12-000013), the Agence Nationale de Recherche sur le SIDA (ANRS, no. 13543 and 13544), and HIVERA 2013 (EURECA project) (to GH), by grants from Higher Education Commission (HEC) of Pakistan (to WA). AK is a recipient of a postdoctoral grant of the Agence Nationale de Recherche sur le SIDA (ANRS, no. 13543 and 13544) and HIVERA 2013 (EURECA project).
References
- 1.Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med (Berl) (2003) 81(9):536–48. 10.1007/s00109-003-0461-8 [DOI] [PubMed] [Google Scholar]
- 2.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature (2011) 475(7356):308–15. 10.1038/nature10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garner AL, Janda KD. Protein-protein interactions and cancer: targeting the central dogma. Curr Top Med Chem (2011) 11(3):258–80. 10.2174/156802611794072614 [DOI] [PubMed] [Google Scholar]
- 4.Caraglia M, Budillon A, Vitale G, Lupoli G, Tagliaferri P, Abbruzzese A. Modulation of molecular mechanisms involved in protein synthesis machinery as a new tool for the control of cell proliferation. Eur J Biochem (2000) 267(13):3919–36. 10.1046/j.1432-1327.2000.01465.x [DOI] [PubMed] [Google Scholar]
- 5.Clemens MJ. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene (2004) 23(18):3180–8. 10.1038/sj.onc.1207544 [DOI] [PubMed] [Google Scholar]
- 6.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem (2002) 269(22):5360–8. 10.1046/j.1432-1033.2002.03290.x [DOI] [PubMed] [Google Scholar]
- 7.Soares DC, Barlow PN, Newbery HJ, Porteous DJ, Abbott CM. Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PLoS One (2009) 4(7):e6315. 10.1371/journal.pone.0006315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panwar D, Rawal L, Ali S. Molecular docking uncovers TSPY binds more efficiently with eEF1A2 compared to eEF1A1. J Biomol Struct Dyn (2014) 9:1–12. 10.1080/07391102.2014.952664 [DOI] [PubMed] [Google Scholar]
- 9.Yaremchuk A, Shalak VF, Novosylna OV, Negrutskii BS, Crepin T, El’skaya AV, et al. Purification, crystallization and preliminary X-ray crystallographic analysis of mammalian translation elongation factor eEF1A2. Acta Crystallogr Sect F Struct Biol Cryst Commun (2012) 68(Pt 3):295–7. 10.1107/S1744309112000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res (2003) 31(13):3381–5. 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen SM, Frydenberg J, Clark BF, Leffers H. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur J Biochem (1993) 215(3):549–54. 10.1111/j.1432-1033.1993.tb18064.x [DOI] [PubMed] [Google Scholar]
- 12.Sasikumar AN, Perez WB, Kinzy TG. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA. (2012) 3(4):543–55. 10.1002/wrna.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doig J, Griffiths LA, Peberdy D, Dharmasaroja P, Vera M, Davies FJ, et al. In vivo characterization of the role of tissue-specific translation elongation factor 1A2 in protein synthesis reveals insights into muscle atrophy. FEBS J (2013) 280(24):6528–40. 10.1111/febs.12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahns S, Lund A, Kristensen P, Knudsen CR, Clark BF, Cavallius J, et al. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res (1998) 26(8):1884–90. 10.1093/nar/26.8.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund A, Knudsen SM, Vissing H, Clark B, Tommerup N. Assignment of human elongation factor 1alpha genes: eEF1A maps to chromosome 6q14 and eEF1A2 to 20q13.3. Genomics (1996) 36(2):359–61. 10.1006/geno.1996.0475 [DOI] [PubMed] [Google Scholar]
- 16.Soares DC, Abbott CM. Highly homologous eEF1A1 and eEF1A2 exhibit differential post-translational modification with significant enrichment around localised sites of sequence variation. Biol Direct (2013) 8:29. 10.1186/1745-6150-8-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanges C, Scheuermann C, Zahedi RP, Sickmann A, Lamberti A, Migliaccio N, et al. Raf kinases mediate the phosphorylation of eukaryotic translation elongation factor 1A and regulate its stability in eukaryotic cells. Cell Death Dis (2012) 3:e276. 10.1038/cddis.2012.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khacho M, Mekhail K, Pilon-Larose K, Pause A, Cote J, Lee S. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol Biol Cell (2008) 19(12):5296–308. 10.1091/mbc.E08-06-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischoff C, Kahns S, Lund A, Jorgensen HF, Praestegaard M, Clark BF, et al. The human elongation factor 1 A-2 gene (EEF1A2): complete sequence and characterization of gene structure and promoter activity. Genomics (2000) 68(1):63–70. 10.1006/geno.2000.6271 [DOI] [PubMed] [Google Scholar]
- 20.Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids (2004) 26(4):443–8. 10.1007/s00726-004-0088-2 [DOI] [PubMed] [Google Scholar]
- 21.Gandin V, Gutierrez GJ, Brill LM, Varsano T, Feng Y, Aza-Blanc P, et al. Degradation of newly synthesized polypeptides by ribosome-associated RACK1/c-Jun N-terminal kinase/eukaryotic elongation factor 1A2 complex. Mol Cell Biol (2013) 33(13):2510–26. 10.1128/MCB.01362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veeramah KR, Johnstone L, Karafet TM, Wolf D, Sprissler R, Salogiannis J, et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia (2013) 54(7):1270–81. 10.1111/epi.12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura P, Coriati A, Belanger G, De RY, Lee J, Kothary R, et al. The utrophin A 5’-UTR drives cap-independent translation exclusively in skeletal muscles of transgenic mice and interacts with eEF1A2. Hum Mol Genet (2010) 19(7):1211–20. 10.1093/hmg/ddp591 [DOI] [PubMed] [Google Scholar]
- 24.Hinestrosa MC, Dickersin K, Klein P, Mayer M, Noss K, Slamon D, et al. Shaping the future of biomarker research in breast cancer to ensure clinical relevance. Nat Rev Cancer (2007) 7(4):309–15. 10.1038/nrc2144 [DOI] [PubMed] [Google Scholar]
- 25.Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, et al. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer (2005) 5:113. 10.1186/1471-2407-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao N, Chen CY, Wu CY, Motonishi K, Kung HJ, Lam KS. Novel flavonoids with antiproliferative activities against breast cancer cells. J Med Chem (2011) 54(13):4339–49. 10.1021/jm101440r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amiri A, Noei F, Jeganathan S, Kulkarni G, Pinke DE, Lee JM. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene (2007) 26(21):3027–40. 10.1038/sj.onc.1210101 [DOI] [PubMed] [Google Scholar]
- 28.Pecorari L, Marin O, Silvestri C, Candini O, Rossi E, Guerzoni C, et al. Elongation factor 1 alpha interacts with phospho-Akt in breast cancer cells and regulates their proliferation, survival and motility. Mol Cancer (2009) 8:58. 10.1186/1476-4598-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Qi CF, Shin DM, Zingone A, Newbery HJ, Kovalchuk AL, et al. eEF1A2 promotes cell growth, inhibits apoptosis and activates JAK/STAT and AKT signaling in mouse plasmacytomas. PLoS One (2010) 5(5):e10755. 10.1371/journal.pone.0010755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem (1998) 67:481–507. 10.1146/annurev.biochem.67.1.481 [DOI] [PubMed] [Google Scholar]
- 31.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer (2010) 10(5):342–52. 10.1038/nrc2842 [DOI] [PubMed] [Google Scholar]
- 32.Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol (2006) 16(7):351–61. 10.1016/j.tcb.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 33.Carpenter CL, Cantley LC. Phosphoinositide kinases. Curr Opin Cell Biol (1996) 8(2):153–8. 10.1016/S0955-0674(96)80060-3 [DOI] [PubMed] [Google Scholar]
- 34.Jeganathan S, Lee JM. Binding of elongation factor eEF1A2 to phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and phosphatidylinositol 4-phosphate generation. J Biol Chem (2007) 282(1):372–80. 10.1074/jbc.M602955200 [DOI] [PubMed] [Google Scholar]
- 35.Pinke DE, Lee JM. The lipid kinase PI4KIIIbeta and the eEF1A2 oncogene co-operate to disrupt three-dimensional in vitro acinar morphogenesis. Exp Cell Res (2011) 317(17):2503–11. 10.1016/j.yexcr.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 36.Jeganathan S, Morrow A, Amiri A, Lee JM. Eukaryotic elongation factor 1A2 cooperates with phosphatidylinositol-4 kinase III beta to stimulate production of filopodia through increased phosphatidylinositol-4,5 bisphosphate generation. Mol Cell Biol (2008) 28(14):4549–61. 10.1128/MCB.00150-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vislovukh A, Kratassiouk G, Porto E, Gralievska N, Beldiman C, Pinna G, et al. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer (2013) 108(11):2304–11. 10.1038/bjc.2013.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MH, Choi BY, Cho YY, Lee SY, Huang Z, Kundu JK, et al. Tumor suppressor p16(INK4a) inhibits cancer cell growth by downregulating eEF1A2 through a direct interaction. J Cell Sci (2013) 126(Pt 8):1744–52. 10.1242/jcs.113613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkarni G, Turbin DA, Amiri A, Jeganathan S, Andrade-Navarro MA, Wu TD, et al. Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Res Treat (2007) 102(1):31–41. 10.1007/s10549-006-9315-8 [DOI] [PubMed] [Google Scholar]
- 40.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, et al. Protein elongation factor eEF1A2 is a putative oncogene in ovarian cancer. Nat Genet (2002) 31(3):301–5. 10.1038/ng904 [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, Lam DC, Han KC, Tin VP, Suen WS, Wang E, et al. High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Lett (2007) 245(1–2):303–14. 10.1016/j.canlet.2006.01.020 [DOI] [PubMed] [Google Scholar]
- 42.Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, et al. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene (2006) 25(18):2628–35. 10.1038/sj.onc.1209289 [DOI] [PubMed] [Google Scholar]
- 43.Grassi G, Scaggiante B, Farra R, Dapas B, Agostini F, Baiz D, et al. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Biochimie (2007) 89(12):1544–52. 10.1016/j.biochi.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 44.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology (2008) 47(2):511–20. 10.1002/hep.22033 [DOI] [PubMed] [Google Scholar]
- 45.Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, et al. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res (1995) 1(12):1455–61. [PubMed] [Google Scholar]
- 46.Edmonds BT, Wyckoff J, Yeung YG, Wang Y, Stanley ER, Jones J, et al. Elongation factor-1 alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J Cell Sci (1996) 109(Pt 11):2705–14. [DOI] [PubMed] [Google Scholar]
- 47.Joseph P, O’Kernick CM, Othumpangat S, Lei YX, Yuan BZ, Ong TM. Expression profile of eukaryotic translation factors in human cancer tissues and cell lines. Mol Carcinog (2004) 40(3):171–9. 10.1002/mc.20033 [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Du C, Wang B, Zhang Y, Liu X, Ren G. Up-regulation of eEF1A2 promotes proliferation and inhibits apoptosis in prostate cancer. Biochem Biophys Res Commun (2014) 450(1):1–6. 10.1016/j.bbrc.2014.05.045 [DOI] [PubMed] [Google Scholar]
- 49.Rehman I, Evans CA, Glen A, Cross SS, Eaton CL, Down J, et al. iTRAQ identification of candidate serum biomarkers associated with metastatic progression of human prostate cancer. PLoS One (2012) 7(2):e30885. 10.1371/journal.pone.0030885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatsuka M, Mitsui H, Wada M, Nagata A, Nojima H, Okayama H. Elongation factor-1 alpha gene determines susceptibility to transformation. Nature (1992) 359(6393):333–6. 10.1038/359333a0 [DOI] [PubMed] [Google Scholar]
- 51.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer (2003) 3(7):502–16. 10.1038/nrc1123 [DOI] [PubMed] [Google Scholar]
- 52.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer (2005) 5(5):355–66. 10.1038/nrc1611 [DOI] [PubMed] [Google Scholar]
- 53.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet (2009) 374(9698):1371–82 10.1016/S0140-6736(09)61338-6 [DOI] [PubMed] [Google Scholar]
- 54.Kim A, Ueda Y, Naka T, Enomoto T. Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res (2012) 31:14. 10.1186/1756-9966-31-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomlinson VA, Newbery HJ, Bergmann JH, Boyd J, Scott D, Wray NR, et al. Expression of eEF1A2 is associated with clear cell histology in ovarian carcinomas: overexpression of the gene is not dependent on modifications at the eEF1A2 locus. Br J Cancer (2007) 96(10):1613–20. 10.1038/sj.bjc.6603748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinke DE, Kalloger SE, Francetic T, Huntsman DG, Lee JM. The prognostic significance of elongation factor eEF1A2 in ovarian cancer. Gynecol Oncol (2008) 108(3):561–8. 10.1016/j.ygyno.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 57.Lee MH, Surh YJ. eEF1A2 as a putative oncogene. Ann N Y Acad Sci (2009) 1171:87–93 10.1111/j.1749-6632.2009.04909.x [DOI] [PubMed] [Google Scholar]
- 58.Sun Y, Wong N, Guan Y, Salamanca CM, Cheng JC, Lee JM, et al. The eukaryotic translation elongation factor eEF1A2 induces neoplastic properties and mediates tumorigenic effects of ZNF217 in precursor cells of human ovarian carcinomas. Int J Cancer (2008) 123(8):1761–9. 10.1002/ijc.23708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kundu JK, Surh YJ. Cancer chemopreventive and therapeutic potential of resveratrol: mechanistic perspectives. Cancer Lett (2008) 269(2):243–61. 10.1016/j.canlet.2008.03.057 [DOI] [PubMed] [Google Scholar]
- 60.Wendling D, Abbas W, Godfrin-Valnet M, Guillot X, Khan KA, Cedoz JP, et al. Resveratrol, a sirtuin 1 activator, increases IL-6 production by peripheral blood mononuclear cells of patients with knee osteoarthritis. Clin Epigenetics (2013) 5(1):10. 10.1186/1868-7083-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee MH, Choi BY, Kundu JK, Shin YK, Na HK, Surh YJ. Resveratrol suppresses growth of human ovarian cancer cells in culture and in a murine xenograft model: eukaryotic elongation factor 1A2 as a potential target. Cancer Res (2009) 69(18):7449–58. 10.1158/0008-5472.CAN-09-1266 [DOI] [PubMed] [Google Scholar]
- 62.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin (2010) 60(5):277–300 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- 63.Marugame T, Hirabayashi Y. Comparison of time trends in lung cancer mortality (1990-2006) in the world, from the WHO mortality database. Jpn J Clin Oncol (2009) 39(10):696–7 10.1093/jjco/hyp138 [DOI] [PubMed] [Google Scholar]
- 64.Lam DC, Girard L, Suen WS, Chung LP, Tin VP, Lam WK, et al. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol (2006) 1(9):932–42. 10.1097/01243894-200611000-00003 [DOI] [PubMed] [Google Scholar]
- 65.Effendi K, Sakamoto M. Molecular pathology in early hepatocarcinogenesis. Oncology (2010) 78(2):157–60 10.1159/000312658 [DOI] [PubMed] [Google Scholar]
- 66.Ayyappan AP, Jhaveri KS. CT and MRI of hepatocellular carcinoma: an update. Expert Rev Anticancer Ther (2010) 10(4):507–19. 10.1586/era.10.24 [DOI] [PubMed] [Google Scholar]
- 67.Pellegrino R, Calvisi DF, Neumann O, Kolluru V, Wesely J, Chen X, et al. eEF1A2 inactivates p53 by way of PI3K/AKT/mTOR-dependent stabilization of MDM4 in hepatocellular carcinoma. Hepatology (2014) 59(5):1886–99. 10.1002/hep.26954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer (2006) 6(9):674–87. 10.1038/nrc1934 [DOI] [PubMed] [Google Scholar]
- 69.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet (2004) 363(9414):1049–57. 10.1016/S0140-6736(04)15841-8 [DOI] [PubMed] [Google Scholar]
- 70.Shaib YH, Davila JA, El-Serag HB. The epidemiology of pancreatic cancer in the United States: changes below the surface. Aliment Pharmacol Ther (2006) 24(1):87–94. 10.1111/j.1365-2036.2006.02961.x [DOI] [PubMed] [Google Scholar]
- 71.Cao H, Zhu Q, Huang J, Li B, Zhang S, Yao W, et al. Regulation and functional role of eEF1A2 in pancreatic carcinoma. Biochem Biophys Res Commun (2009) 380(1):11–6. 10.1016/j.bbrc.2008.12.171 [DOI] [PubMed] [Google Scholar]
- 72.Xu C, Hu DM, Zhu Q. eEF1A2 promotes cell migration, invasion and metastasis in pancreatic cancer by upregulating MMP-9 expression through Akt activation. Clin Exp Metastasis (2013) 30(7):933–44. 10.1007/s10585-013-9593-6 [DOI] [PubMed] [Google Scholar]
- 73.Duanmin H, Chao X, Qi Z. eEF1A2 protein expression correlates with lymph node metastasis and decreased survival in pancreatic ductal adenocarcinoma. Hepatogastroenterology (2013) 60(124):870–5. 10.5754/hge12869 [DOI] [PubMed] [Google Scholar]
- 74.Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The cell is dead. Long live the cell! Trends Cell Biol (2008) 18(10):467–73. 10.1016/j.tcb.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams JM, Cory S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol (2007) 19(5):488–96. 10.1016/j.coi.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang R, Wang E. Mouse translation elongation factor eEF1A-2 interacts with Prdx-I to protect cells against apoptotic death induced by oxidative stress. J Cell Biochem (2007) 100(2):267–78. 10.1002/jcb.20969 [DOI] [PubMed] [Google Scholar]
- 77.Abbas W, Khan KA, Kumar A, Tripathy MK, Dichamp I, Keita M, et al. Blockade of BFA-mediated apoptosis in macrophages by the HIV-1 Nef protein. Cell Death Dis (2014) 5:e1080. 10.1038/cddis.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen E, Proestou G, Bourbeau D, Wang E. Rapid up-regulation of peptide elongation factor EF-1alpha protein levels is an immediate early event during oxidative stress-induced apoptosis. Exp Cell Res (2000) 259(1):140–8. 10.1006/excr.2000.4952 [DOI] [PubMed] [Google Scholar]
- 79.Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell (2012) 45(3):398–408. 10.1016/j.molcel.2011.11.027 [DOI] [PubMed] [Google Scholar]
- 80.Talapatra S, Wagner JD, Thompson CB. Elongation factor-1 alpha is a selective regulator of growth factor withdrawal and ER stress-induced apoptosis. Cell Death Differ (2002) 9(8):856–61. 10.1038/sj.cdd.4401078 [DOI] [PubMed] [Google Scholar]
- 81.Newbery HJ, Loh DH, O’Donoghue JE, Tomlinson VA, Chau YY, Boyd JA, et al. Translation elongation factor eEF1A2 is essential for post-weaning survival in mice. J Biol Chem (2007) 282(39):28951–9. 10.1074/jbc.M703962200 [DOI] [PubMed] [Google Scholar]
- 82.Chambers DM, Peters J, Abbott CM. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc Natl Acad Sci U S A (1998) 95(8):4463–8. 10.1073/pnas.95.8.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Potter M, Bernstein A, Lee JM. The wst gene regulates multiple forms of thymocyte apoptosis. Cell Immunol (1998) 188(2):111–7. 10.1006/cimm.1998.1346 [DOI] [PubMed] [Google Scholar]
- 84.Nakajima J, Okamoto N, Tohyama J, Kato M, Arai H, Funahashi O, et al. De novo eEF1A2 mutations in patients with characteristic facial features, intellectual disability, autistic behaviors and epilepsy. Clin Genet (2015) 87(4):356–61. 10.1111/cge.12394 [DOI] [PubMed] [Google Scholar]
- 85.Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem (2002) 277(7):5418–25. 10.1074/jbc.M110685200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abbott CM, Newbery HJ, Squires CE, Brownstein D, Griffiths LA, Soares DC. eEF1A2 and neuronal degeneration. Biochem Soc Trans (2009) 37(Pt 6):1293–7 10.1042/BST0371293 [DOI] [PubMed] [Google Scholar]
- 87.Li D, Wei T, Abbott CM, Harrich D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol Mol Biol Rev (2013) 77(2):253–66. 10.1128/MMBR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol (1999) 73(7):5388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ott DE, Coren LV, Johnson DG, Kane BP, Sowder RC, Kim YD, et al. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology (2000) 266(1):42–51. 10.1006/viro.1999.0075 [DOI] [PubMed] [Google Scholar]
- 90.Warren K, Wei T, Li D, Qin F, Warrilow D, Lin MH, et al. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc Natl Acad Sci U S A (2012) 109(24):9587–92. 10.1073/pnas.1204673109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allouch A, Cereseto A. Identification of cellular factors binding to acetylated HIV-1 integrase. Amino Acids (2011) 41(5):1137–45. 10.1007/s00726-009-0444-3 [DOI] [PubMed] [Google Scholar]
- 92.Parissi V, Calmels C, De Soultrait VR, Caumont A, Fournier M, Chaignepain S, et al. Functional interactions of human immunodeficiency virus type 1 integrase with human and yeast HSP60. J Virol (2001) 75(23):11344–53. 10.1128/JVI.75.23.11344-11353.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan R, Patni H, Tandon P, Luan L, Sharma B, Salhan D, et al. Nef interaction with actin compromises human podocyte actin cytoskeletal integrity. Exp Mol Pathol (2013) 94(1):51–7. 10.1016/j.yexmp.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu-Baer F, Lane WS, Gaynor RB. Identification of a group of cellular cofactors that stimulate the binding of RNA polymerase II and TRP-185 to human immunodeficiency virus 1 TAR RNA. J Biol Chem (1996) 271(8):4201–8. 10.1074/jbc.271.8.4201 [DOI] [PubMed] [Google Scholar]
- 95.Abbas W, Dichamp I, Herbein G. The HIV-1 Nef protein interacts with two components of the 40S small ribosomal subunit, the RPS10 protein and the 18S rRNA. Virol J (2012) 9:103. 10.1186/1743-422X-9-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao H, Neuveut C, Benkirane M, Jeang KT. Interaction of the second coding exon of Tat with human EF-1 delta delineates a mechanism for HIV-1-mediated shut-off of host mRNA translation. Biochem Biophys Res Commun (1998) 244(2):384–9. 10.1006/bbrc.1998.8274 [DOI] [PubMed] [Google Scholar]
- 97.Lin WS, Jiao BY, Wu YL, Chen WN, Lin X. Hepatitis B virus X protein blocks filamentous actin bundles by interaction with eukaryotic translation elongation factor 1 alpha 1. J Med Virol (2012) 84(6):871–7. 10.1002/jmv.23283 [DOI] [PubMed] [Google Scholar]
- 98.Sikora D, Greco-Stewart VS, Miron P, Pelchat M. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology (2009) 390(1):71–8. 10.1016/j.virol.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 99.Yue J, Shukla R, Accardi R, Zanella-Cleon I, Siouda M, Cros MP, et al. Cutaneous human papillomavirus type 38 E7 regulates actin cytoskeleton structure for increasing cell proliferation through CK2 and the eukaryotic elongation factor 1A. J Virol (2011) 85(17):8477–94. 10.1128/JVI.02561-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3’ stem-loop region of West Nile virus genomic RNA. J Virol (1997) 71(9):6433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davis WG, Blackwell JL, Shi PY, Brinton MA. Interaction between the cellular protein eEF1A and the 3’-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol (2007) 81(18):10172–87. 10.1128/JVI.00531-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carr JM, Kua T, Clarke JN, Calvert JK, Zebol JR, Beard MR, et al. Reduced sphingosine kinase 1 activity in dengue virus type-2 infected cells can be mediated by the 3’ untranslated region of dengue virus type-2 RNA. J Gen Virol (2013) 94(Pt 11):2437–48. 10.1099/vir.0.055616-0 [DOI] [PubMed] [Google Scholar]
- 103.Zhang X, Shi H, Chen J, Shi D, Li C, Feng L. EF1A interacting with nucleocapsid protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Vet Microbiol (2014) 172(3–4):443–8. 10.1016/j.vetmic.2014.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moerdyk-Schauwecker M, Hwang SI, Grdzelishvili VZ. Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol J (2009) 6:166. 10.1186/1743-422X-6-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J Virol (2006) 80(5):2127–40. 10.1128/JVI.80.5.2127-2140.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. Protein composition of the vaccinia virus mature virion. Virology (2007) 358(1):233–47. 10.1016/j.virol.2006.08.025 [DOI] [PubMed] [Google Scholar]
- 107.Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, et al. Identification of proteins associated with murine cytomegalovirus virions. J Virol (2004) 78(20):11187–97. 10.1128/JVI.78.20.11187-11197.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol (2004) 78(20):10960–6. 10.1128/JVI.78.23.13395.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol (2008) 82(11):5279–94. 10.1128/JVI.02631-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci (2006) 97(10):977–83. 10.1111/j.1349-7006.2006.00299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes (2010) 40(1):1–13. 10.1007/s11262-009-0412-8 [DOI] [PubMed] [Google Scholar]
- 112.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol (2006) 80(18):9039–52. 10.1128/JVI.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]