Abstract
The MED1 subunit of the Mediator transcriptional coregulator complex coactivates GATA1 and induces erythropoiesis. Here, we show the dual mechanism of GATA1- and MED1-mediated transcription. MED1 expression levels in K562 erythroleukemia cells paralleled the levels of GATA1-targeted gene transcription and erythroid differentiation. An N-terminal fragment of MED1, MED1(1–602), which is incapable of interacting with GATA1, enhanced GATA1-targeted gene transcription and erythroid differentiation, and introduction of MED1(1–602) into Med1−/− mouse embryonic fibroblasts (MEFs) partially rescued GATA1-mediated transcription. The C-terminal zinc-finger domain of GATA1 interacts with the MED1(1–602)-interacting coactivator CCAR1, CoCoA, and MED1(681–715). CCAR1 and CoCoA synergistically enhanced GATA1-mediated transcription from the γ-globin promoter in MEFs. Recombinant GATA1, CCAR1, CoCoA, and MED1(1–602) formed a complex in vitro, and GATA1, CCAR1, CoCoA, and MED1 were recruited to the γ-globin promoter in K562 cells during erythroid differentiation. Therefore, in addition to the direct interaction between GATA1 and MED1, CoCoA and CCAR1 appear to relay the GATA1 signal to MED1, and multiple modes of the GATA1-MED1 axis may help to fine-tune GATA1 function during GATA1-mediated homeostasis events.
INTRODUCTION
A number of nuclear factors that activate or repress lineage-specific gene expression specifically regulate hematopoietic cell differentiation (reviewed in Cantor & Orkin 2002; Kerenyi & Orkin 2010; Doré & Crispino 2011). GATA1, an erythroid and megakaryocytic lineage-specific transcription factor, is essential in erythropoiesis and megakaryopoiesis; GATA1 knockout mice show a lethal phenotype due to a complete block of erythromegakaryocytic precursor cell differentiation (Pevny et al. 1991; Shivdasani et al. 1997). GATA1 activates the transcription of genes involved in erythroid differentiation and proliferation and represses myeloid differentiation through its interaction with multiple activators and coregulators (reviewed in Cantor & Orkin 2002; Kerenyi & Orkin 2010; Doré & Crispino 2011). Among the GATA1-interacting activators involved in erythroid differentiation are FOG1 (Tsang et al. 1997), KLF-1 (Crossley et al. 1994), Sp1 (Merika & Orkin 1995) and RUNX1 (Elagib et al. 2003). GATA1 also interacts with LMO2 and forms a pentameric complex with SCL/Tal-1, E2A, LDB1, and LMO2, which activates transcription both from the GATA-binding site and from the E-box that neighbors GATA-binding site (Wadman et al. 1997). GATA1 and PU.1, a myeloid-specific activator that binds GATA1, are mutually antagonistic, and repression of PU.1 by GATA1 blocks myelomonocytic differentiation of erythroid precursors (Rekhtman et al. 1999; Zhang et al. 1999). Coactivators that act downstream of GATA1 include the histone acetyltransferases CBP/p300 (Blobel et al. 1998). These coactivators relax the chromatin structure through histone modification, and enable the preinitiation complex (PIC) to form on the GATA1-bound promoter.
The Mediator transcriptional coregulator is an approximately 2-MDa multi-subunit polypeptide complex that integrates a variety of intracellular signals by physically and specifically forming a bridge between DNA-bound transcription factors and RNA polymerase II (Ito et al. 2002; Zhang et al. 2005; reviewed in Malik & Roeder 2010). The Mediator subunit MED1 was originally identified as a ligand-dependent coactivator of thyroid hormone receptor α (TRα) (Yuan et al. 1998). MED1 acts as specific coactivator of nuclear hormone receptors (NRs) (reviewed in Ito & Roeder 2001; Chen & Roeder 2011), C/EBPβ (Li et al. 2008), BRCA1 (Wada et al. 2004), and GATA family activators (Crawford et al. 2002; Stumpf et al. 2006), and plays an important role in the differentiation of distinct lineages of cells, including non-hematopoietic cells such as adipocytes (Ge et al. 2002; Ge et al. 2008) and mammary epithelial cells (Jiang et al. 2010; Hasegawa et al. 2012). We previously showed that MED1 plays a crucial role in the ligand-dependent myelomonocytic differentiation of human promyelocytic leukemia HL-60 cells, which differentiate into monocytes in the presence of 1,25-dihydroxyvitamin D3 treatment, and differentiate into granulocytes in the presence of all-trans retinoic acid (ATRA) (Urahama et al. 2005). Med1-knockout mice are early embryonic lethal (Ito et al. 2000; Zhu et al. 2000), and analysis of Med1-knockout and Med1-conditional knockout mice showed that MED1 is crucial for normal erythroid differentiation during both the embryonic and adult stages (Stumpf et al. 2006; Stumpf et al. 2010). MED1 binds directly to GATA1 and activates the transcription of GATA1-targeted genes. However, the detailed mechanism of the nuclear signaling pathway is still unknown. Furthermore, a recent study of CD4-positive cell-specific Med1 conditional knockout mice suggested the significance of MED1 in intrathymic development of invariant natural killer T (iNKT) cells, although the precise underlying mechanism is not yet known (Yue et al. 2011).
Cell cycle and apoptosis regulator 1 (CCAR1; also known as CARP-1), originally identified as a mediator of apoptosis signaling by retinoid CD437 (Rishi et al. 2003) and recently shown to be a coiled-coil coactivator (CoCoA)/calcium binding and coiled coil domain 1 (Calcoco1) C-terminal activation domain binding protein, activates estrogen receptor (ER)- and glucocorticoid receptor (GR)-mediated transcription through simultaneous interaction with NRs and the N-terminus of MED1 (Kim et al. 2008). Therefore, CCAR1 constitutes an important bypass for Mediator recruitment to ER and GR targeted genes. CCAR1 and CoCoA also interact with p53 and activate p53-mediated transcription, probably by bypassing the interaction between p53 and MED1 (Kim et al. 2008). Furthermore, CCAR1 has been reported to be required for transcriptional activation by Wnt, β-catenin (Ou et al. 2009), and Ngn3 (Lu et al. 2012).
This study demonstrates the role of, and mechanism underlying, MED1 action in GATA1-mediated transcription using K562 erythroleukemia cells, which were originally isolated from a patient with chronic myeloid leukemia (Lozzio & Lozzio 1975). We confirm that MED1 is essential for optimal erythroid differentiation of erythroleukemic cells. We show, unexpectedly, that the direct interaction between GATA1 and MED1 is dispensable for GATA1-mediated transcription and erythroid differentiation, and we identified CCAR1 and CoCoA as a bypass between GATA1 and the MED1 subunit of Mediator, and suggest that the multiple modes of GATA1 signaling might serve as a fine tuning mechanism for GATA1 action during homeostatic events that most probably include erythropoiesis.
RESULTS
The Mediator subunit MED1 directs GATA1-induced transcription and erythroid differentiation in K562 cells
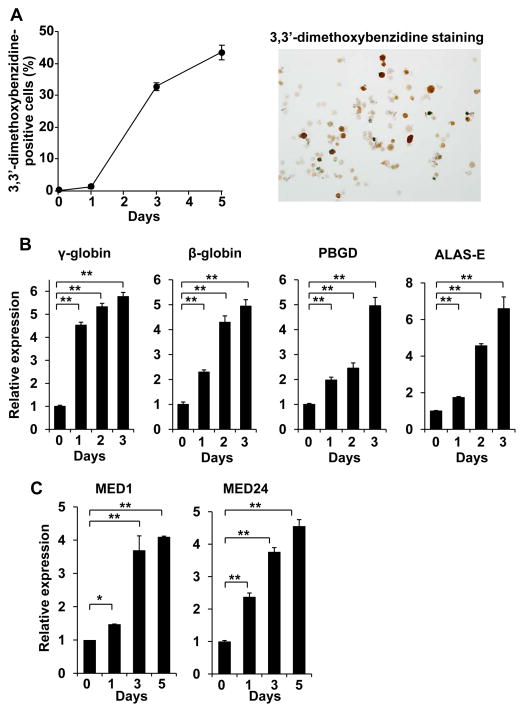
The Mediator subunit MED1 acts as a GATA1-specific coactivator and promotes normal erythropoiesis (Stumpf et al. 2006). To analyze the mechanism of MED1 action in GATA1-mediated transcription, we used the hemin-induced and GATA1-dependent erythroid differentiation system of K562 erythroleukemia cells. When K562 cells were exposed to hemin, the cells differentiated toward the erythroid lineage, as evidenced by positive 3,3′-dimethoxybenzidine staining (Fig. 1A). During the first 3 days, transcription of GATA1-targeted erythroid differentiation marker genes, including genes encoding β-globin, γ-globin, porphobilinogen deaminase (PBGD), and 5-aminolevulinate synthase (ALAS-E), was significantly activated (Fig. 1B). Unexpectedly, expression of Mediator subunits, including MED1 and MED24, were also notably (over 4-fold) induced (Fig. 1C). In contrast, expression of GATA1 and known GATA1-associated activators (KLF1, SCL, LMO2 and FOG1) was unchanged (GATA1, SCL, LMO2) or only slightly (up to 1.5-fold) increased (KLF1, FOG1) (Fig. S1). Because induced gene products often have an important physiological role, the Mediator complex could play a major role in GATA1-mediated erythroid differentiation.
Figure 1. MED1 requirement for GATA1-targeted gene transcription and erythroid differentiation in K562 cells.
(A) Erythroid differentiation of 3,3′-dimethoxybenzidine-treated cells. Approximately 45% of cells differentiated after 5 days of treatment. Values (mean ± SD) from a representative experiment performed in triplicate are shown.
(B) Induction of GATA1-targeted gene transcription. Values (mean ± SD) from a representative experiment performed in quadruplicate are shown (**p < .01). Representative GATA1-targeted genes were induced during cell differentiation.
(C) Results of quantitative RT-PCR of MED1 (left panel) and MED24 (right panel) are shown. Values (mean ± SD) of a representative experiment performed in duplicate are plotted (*p < .05, **p < .01). Transcription of the genes encoding the Mediator subunits MED1 and MED24 are induced during differentiation of cells.
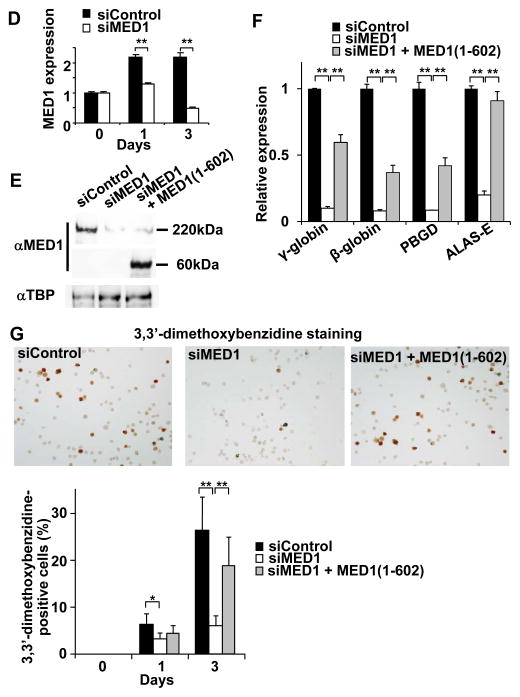
(D) Results of quantitative RT-PCR of MED1 after transfection with MED1 or control siRNA are shown. Values (mean ± SD) of a representative experiment performed in duplicate are shown (**p < .01). si, siRNA.
(E) Results of western blot analyses of MED1, and TBP as a control, 2 days after transfection with MED1 or control siRNA, and siRNA-resistant hMED1(1–602) expression vector are shown. α, anti.
(F) Reduced GATA1-targeted gene transcription 3 days after transfection with MED1 siRNA. Simultaneous expression of siRNA-resistant hMED1(1–602) partially rescues the GATA1-targeted gene transcription. Values (mean ± SD) of a representative experiment performed in quadruplicate are shown (**p < .01).
(G) Attenuated erythroid differentiation after transfection with MED1 siRNA. The siRNA-resistant MED1(1–602) rescues the attenuated erythroid differentiation. Control siRNA was used as a control. Values (mean ± SD) of a representative experiment performed in quadruplicate are shown (*p < .05, **p < .01).
The results were reproducible in three independent experiments.
Since the MED1 subunit, as well as other Mediator subunits, was reported to physically interact with GATA1 and coactivate GATA1-mediated transcription (Stumpf et al. 2006), depletion of MED1 in K562 cells might impair GATA1 function and erythroid differentiation. Indeed, when MED1 expression was suppressed by RNA interference (Fig. 1D, E), hemin-induced transcriptional activation of the GATA1-targeted genes encoding β-globin, γ-globin, PBGD, and ALAS-E was strongly attenuated 3 days after hemin treatment (Fig. 1F). As a result, the number of erythroid-differentiated cells that were positive for 3,3′-dimethoxybenzidine staining was significantly attenuated during 3 days of hemin treatment (Fig. 1G). In contrast, expression of GATA1 and associated activators was unchanged (KLF1, LMO2, FOG1) or rather slightly (up to 1.5-fold) increased (GATA1, SCL) (Fig. S2). When MED1 was knocked down in hemin-untreated K562 cells, expression of the GATA1-targeted genes was also strongly reduced (Fig. S3). These results indicate that MED1 plays a major role in GATA1-mediated transcription and erythroid differentiation in K562 cells.
Human MED1(681–715) domain interacts with GATA1
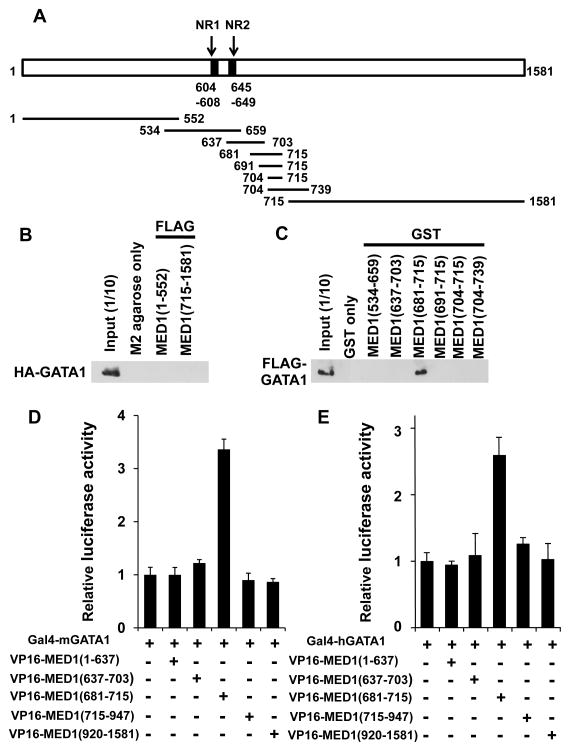
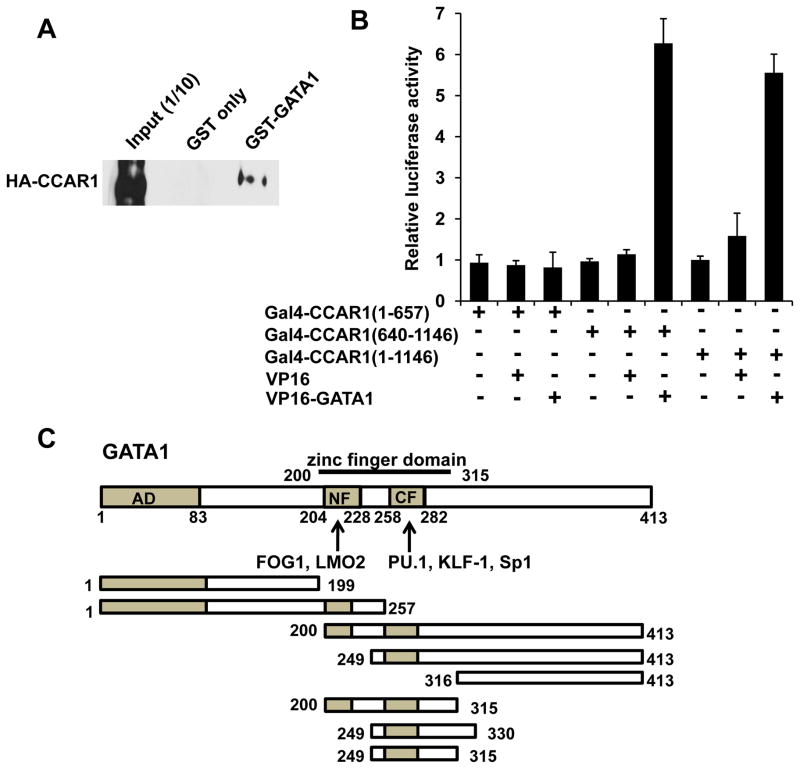
It was previously reported that the middle portion of mouse (m) MED1 (amino acids 622–701), mMED1(622–701), interacts with GATA1 (Stumpf et al. 2006). To narrow down the region of human (h) MED1 that interacts with GATA1, and to further analyze this interaction at the mechanistic level, serial binding assays were performed. HA-tagged recombinant mGATA1 was not pulled down by recombinant FLAG-tagged N-terminal and C-terminal hMED1 fragments [hMED1(1–552) and hMED1(715–1,581)] (Fig. S4A) immobilized to M2-agarose, which was in agreement with a previous report (Fig. 2A, B) (Stumpf et al. 2006). Next, a series of hMED1 GST-fusion fragments (Fig. S4B) were tested for their interaction with recombinant HA-tagged mGATA1, and hMED1(681–715) was the shortest fragment tested that was found to significantly interact with mGATA1 (Fig. 2C). To confirm this interaction in a more physiological setting within mammalian cells, we next assessed it in a mammalian two-hybrid assay using Med1−/− MEFs, and the interaction between hMED1(681–715) and (both mouse and human) GATA1 was confirmed (Fig. 2D, E).
Figure 2. GATA1 physically interacts with MED1(681–715).
(A) Diagram of MED1, showing 2 nuclear receptor recognition motifs (NR boxes; NR1 and NR2) and the fragments used in this study.
(B) Fragments of FLAG-tagged MED1 were incubated with HA-tagged GATA1 and immunoprecipitated with M2-agarose. HA-GATA1 was visualized with an anti-HA antibody.
(C) GST-pull down assays using GST-fused to various MED1 fragments and FLAG-tagged GATA1. An anti-FLAG antibody was used to visualize FLAG-GATA1.
(D, E) Mammalian two-hybrid assays. Luciferase activity of the reporter with 5× Gal4-binding sites was measured. Gal4-fused mGATA1 (D) and Gal4-fused hGATA1 (E) interact with VP16-fused hMED1(681–715). Values (mean ± SD) of a representative experiment performed in duplicate (D) or triplicate (E) are shown.
hMED1(681–715) is dispensable for GATA1-mediated transcription and erythroid differentiation
We next assessed the role of the GATA1-interacting region of hMED1(681–715) in GATA1 function and erythroid differentiation. We first tested the effect of overexpressing various hMED1 fragments (Yuan et al. 1998; Sumitomo et al. 2010) by transfection with mammalian expression vectors into K562 cells (Fig. 3A). MED1 is a substoichiometric component of Mediator, and Mediator that is associated with RNA polymerase II core complex is enriched in the Mediator fraction containing the activator-bound MED1 (Zhang et al. 2005; reviewed in Malik & Roeder 2010). The free Mediator subunit is destabilized and subject to an enhanced turnover (Ito et al. 2002). When the FLAG-tagged mutant MED1 transfected in HeLa cells is immunopurified with anti-FLAG antibody beads, the isolated proteins contain stoichiometric amounts of mutant MED1 and other representative Mediator subunits (Ge et al. 2002). Hence, overexpression of MED1 corresponds to saturation of MED1 in the Mediator complex and may lead to enhanced GATA1-induced transcription.
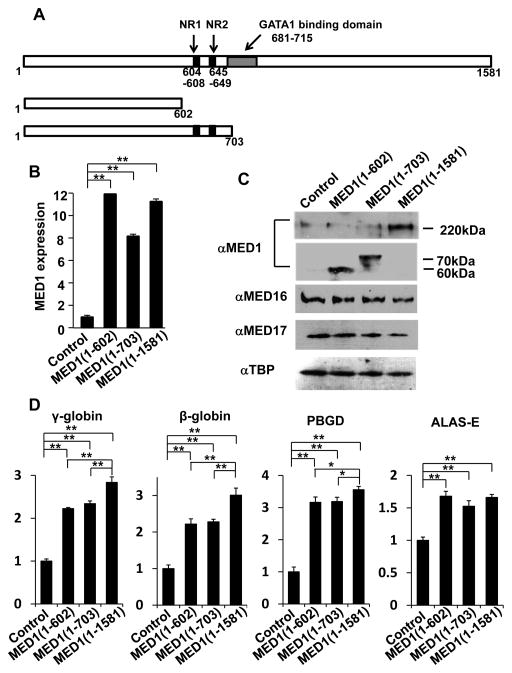
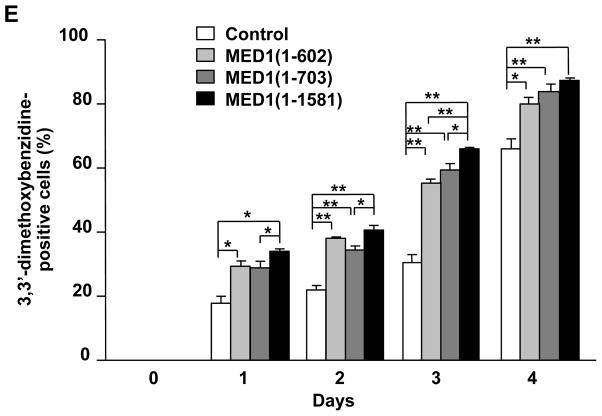
Figure 3. MED1(1–602) and MED1(1–703) induce GATA1-mediated transcription and erythroid differentiation in K562 cells.
(A) Diagram of MED1, showing the domains of 2 NR boxes (NR1 and NR2) and the GATA1 binding domain and the MED1 fragments used in these experiments.
(B, C) Results of quantitative RT-PCR for MED1 (B) and western blot analyses for Mediator subunits and TBP as a control (C) 3 days after transfection with expression vectors with various fragments of MED1 or an empty expression vector as a control in hemin-treated K562 cells. Values (mean ± SD) of a representative experiment performed in duplicate are shown (**p < .01) (B).
(D) Enhanced GATA1-targeted gene transcription after overexpression of various fragments of MED1 or an empty expression vector as a control in hemin-treated cells. Values (mean ± SD) of a representative experiment performed in quadruplicate are shown (*p < .05, **p < .01).
(E) Enhanced erythroid differentiation after overexpression of various MED1 fragments in hemin-treated cells. An empty expression vector was used as a control. Values (mean ± SD) of a representative experiment performed in triplicate are plotted (*p < .05, **p < .01).
The results were reproducible in three independent experiments.
When full-length hMED1 [hMED1(1–1,581)] was overexpressed in K562 cells (Fig. 3B, C), transcription of GATA1-targeted genes was prominently activated 3 days after transfection (Fig. 3D), and the number of differentiated cells stained with 3,3′-dimethoxybenzidine increased for 4 days after transfection (Fig. 3E). MED1 has 2 closely located LxxLL NR recognition motifs (NR boxes) (Yuan et al. 1998). We then overexpressed 2 N-terminal hMED1 fragments, hMED1(1–602), which lacks the NR boxes, and hMED1(1–703), which contains the NR boxes. Both of these mutants lack the binding domain for, hence the capacity of binding to, GATA1 (Fig. 3A). The exogenously overexpressed MED1 truncations and the endogenous MED1 were incorporated in the Mediator competitively according to their molar ratios (Fig. 3C). Unexpectedly, overexpression of either of these mutant MED1 fragments also enhanced transcriptional activation of GATA1-targeted genes to levels similar to, but possibly not as prominent as, those following overexpression of full-length hMED1 (Fig. 3D). Erythroid differentiation, as assessed by the number of cells stained with 3,3′-dimethoxybenzidine, was also enhanced by overexpression of hMED1(1–602) and hMED1(1–703), but the enhancement may not have been as prominent as when full-length hMED1 was overexpressed (Fig. 3E). Of note, introduction of siRNA-resistant MED1(1–602) to MED1-knockdown K562 cells rescued the attenuated GATA1-targeted gene transcription and erythroid differentiation (Fig. 1F, G).
Meanwhile, hMED1(1–602) overexpression caused expression of GATA1 and associated activators unchanged (SCL, LMO2, FOG1) or minimally changed (GATA1, KLF1) (Fig. S5). In contrast, in the absence of hemin, expression of GATA1-targeted genes did not change (γ-globin, β-globin, PBGD) or only slightly elevated (ALAS-E) by MED1(1–602) overexpression. Expression of GATA1 and associated activators also did not change (KLF1, FOG1) or marginally elevated (SCL, LMO2, GATA1) (Fig. S6). These results suggest that direct interaction between GATA1 and MED1 is dispensable, and that the N-terminal hMED1(1–602) domain is sufficient for hemin-induced erythroid differentiation and associated coactivation of GATA1, but that the C-terminal domain of MED1 that contains the GATA1-binding motif might be necessary for full activation of GATA1-mediated erythroid differentiation.
MED1-mediated transcription initiation of the γ-globin promoter
The human γ-globin promoter includes 2 consensus sequences for GATA1 (−175 to −171 and −189 to −185); when GATA1 binds to these sequences, γ-globin transcription is strongly induced (Martin et al. 1989). To assess the mechanism underlying MED1 function in GATA1-mediated transcription, luciferase reporter assays using the human γ-globin promoter (–299 to +37) were employed.
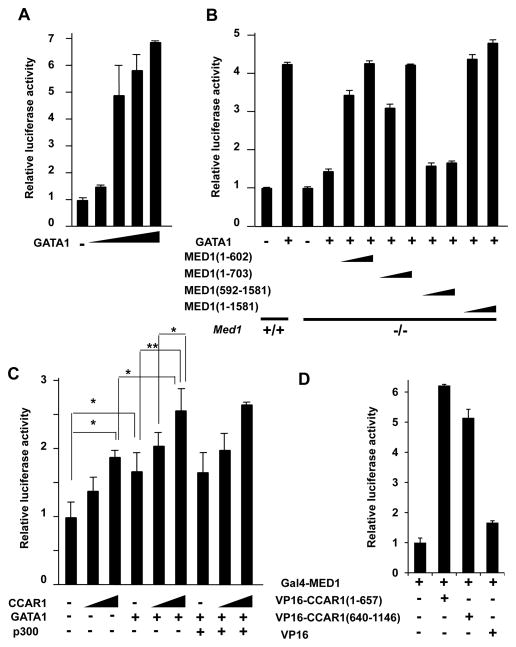
The reporter assays showed that transcription in Med1+/+ MEFs was strongly activated (up to 7-fold) in the presence of mGATA1 in a dose-dependent manner (Fig. 4A). When a fixed amount of mGATA1 (100 ng) was added, transcription was 4-fold higher in the Med1+/+ MEFs than in the absence of mGATA1; however, transcription was only modestly increased in Med1−/− MEFs (1.5-fold) compared to transcription in these cells in the absence of mGATA1, even though residual activation might occur in these cells (Fig. 4B). Therefore, MED1 appeared to be necessary for optimal GATA1-mediated transcription, even though a redundant mechanism that does not require MED1 might exist. Introduction of exogenous full-length hMED1(1–1,581) into Med1−/− MEFs rescued GATA1-dependent transactivation to wild-type levels (Fig. 4B). Next, various hMED1 mutants were cotransfected to determine the MED1 domain(s) responsible for GATA1-dependent activation (Fig. 4B). Both MED1(1–602) and MED1(1–703) significantly restored GATA1-mediated transcriptional activation to similar levels in a dose-dependent manner; however, transcription did not appear to be fully restored compared to transcription levels when full-length MED1(1–1,581) was introduced (Fig. 4B). This difference might reflect the contribution of the direct interaction between MED1(681–715) and GATA1. In contrast, MED1(592–1,581), which is incapable of complex formation (Ge et al. 2008), was unable to restore any transcriptional activity (Fig. 4B).
Figure 4. N-terminal MED1 and CCAR1 drive GATA1-mediated transcription from the γ-globin promoter. Luciferase reporter assays of the human γ-globin promoter in MEFs are shown.
(A) GATA1-dependent transcriptional activation of the γ-globin promoter in Med1+/+ MEFs in the presence of 10, 50, 100, and 200 ng of GATA1.
(B) GATA1-dependent and MED1-mediated transcriptional activation. The amounts of the expression vectors added are as follows: 100 ng of GATA1 and 10 or 100 ng of various MED1 fragments.
(C) CCAR1-enhanced activation of GATA1-dependent transcription in Med1+/+ MEFs. The amounts of expression vectors added are as follows: 20 ng of GATA1, 20 or 100 ng of CCAR1, and 100 ng of p300 (*p < .05, **p < .01).
(D) Mammalian two-hybrid assays. Luciferase activities of the reporter with 5× Gal4-binding sites are measured. Gal4-fused MED1 interacts with VP16-fused CCAR1(1–657) and CCAR1(640–1,146).
Values (mean ± SD) of a representative experiment performed in duplicate are shown (A–D). The results were reproducible in three independent experiments.
These data might suggest that both MED1(1–602) and MED1(603–1,581) contribute to γ-globin transcription. MED1(1–602) appears to stimulate GATA1-mediated transactivation independent of direct interaction with GATA1, while MED1(603–1,581) possibly contributes to GATA1-mediated transactivation through the direct interaction of GATA1 and MED1(681–715). The results also indicate that the MED1 NR boxes are dispensable for GATA1 function.
CCAR1 enhances GATA1-mediated transactivation
Given that MED1(1–602) enhanced GATA1-initiated transactivation, a GATA1 signaling mechanism that bypasses its interaction with MED1 appears to exist. In the case of NRs, the bypass molecule CCAR1 reportedly simultaneously interacts with both the NRs (ERs and GR) and with the N-terminus of MED1(1–670), and mediates a bypass pathway that does not require the direct interaction between the NRs and the MED1 NR boxes (Kim et al. 2008). Based on this information, we hypothesized that CCAR1 might also mediate a bypass pathway between GATA1 and MED1, and tested this hypothesis.
Luciferase reporter assays using the human γ-globin promoter (−299 to +37) in Med1+/+ MEFs showed that CCAR1 alone enhanced reporter activity in a dose-dependent manner. When GATA1 was coexpressed with CCAR1, reporter activity was further enhanced (Fig. 4C). It is known that GATA1 interacts with the chromatin-modifying coactivator p300 (Blobel et al. 1998) and that p300 interacts with CoCoA (Kim et al. 2006), a partner activator of CCAR1. Therefore, p300 could indirectly recruit CCAR1 and MED1 to GATA1 for transactivation. However, the addition of p300 did not further enhance reporter activity in this system (Fig. 4C). Therefore, CCAR1 might act as a coactivator of not only NRs, but also GATA1, possibly through a p300-independent mechanism.
MED1 interacts with both the N- and C-terminal halves of CCAR1
Since a previous report suggested that the N-terminus of MED1 interacts with CCAR1 (Kim et al. 2008), we attempted to further analyze this interaction. A mammalian two-hybrid assay was used to identify which parts of CCAR1 interact with MED1. Indeed, Gal4-MED1 specifically interacted with both VP16-CCAR1(1–657) and VP16-CCAR1(640–1,146) (Fig. 4D). Therefore, MED1 appears to interact with both the N- and C-terminal halves of CCAR1.
The C-terminal zinc finger (CF) domain of GATA1 is necessary for the direct interaction with the C-terminus of CCAR1 and MED1
Since CCAR1 enhanced GATA1-mediated transcription irrespective of p300, we asked if CCAR1 could directly convey the GATA1 signal to MED1, analogous to the case with NRs. Indeed, a GST-pull down assay showed that GATA1 directly interacts with CCAR1 in vitro (Fig. 5A). A mammalian two-hybrid assay in Med1−/− MEFs confirmed that this interaction takes place within mammalian cells, and showed that this intranuclear interaction required the C-terminal half of CCAR1 [CCAR1(640–1,146)] (Fig. 5B).
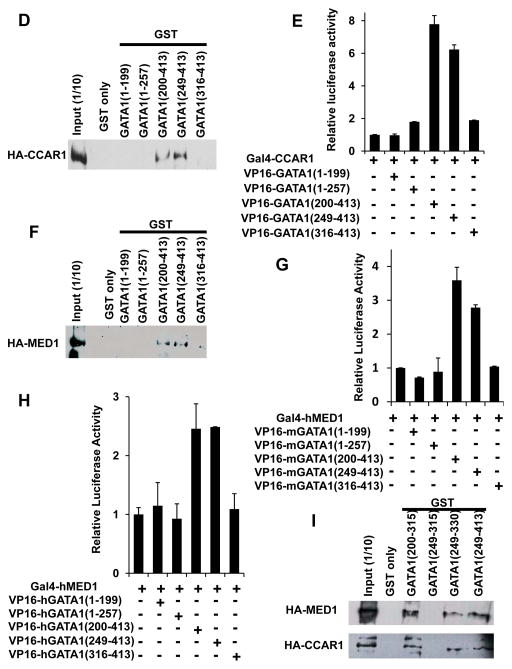
Figure 5. The CF domain of GATA1 is necessary for the direct interaction with CCAR1(640–1,146) and MED1.
(A, D, F, I) GST-pull down assays. HA-CCAR1 (A, D, I) or HA-MED1 (F, I) is pulled down by various GST-fusion fragments of GATA1. HA-CCAR1 and HA-MED1 were visualized with an anti-HA antibody.
(B, E, G, H) Mammalian two-hybrid assays. Luciferase activities of a reporter with 5×Gal4-binding sites were measured. The interaction between various Gal4-fusion fragments of mCCAR1 (B, E) or hMED1 (G, H) and various VP16-fusion fragments of mGATA1 (B, E, G) or hGATA1 (H) were tested. Values (mean ± SD) of a representative experiment performed in duplicate (B, E, G) or triplicate (H) are shown. The results were reproducible in three independent experiments.
(C) Diagram of mGATA1, showing the 2 zinc-finger domains, NF and CF, and the transcription factors that interact with these zinc finger domains, as well as the fragments used in these experiments.
To analyze the mechanism of GATA1 action in CCAR1-mediated transcription, we next asked which domain of GATA1 interacts with CCAR1. GATA1 contains 2 zinc finger domains, an N-terminal zinc finger (NF) and a C-terminal zinc finger (CF), which bind DNA and various activators and cofactors, including FOG1 and LMO2 (the NF domain), and PU.1, EKLF, and Sp1 (the CF domain) (Wilkinson-White et al. 2011; Rekhtman et al. 1999; Merika & Orkin 1995) (Fig. 5C). Serial GST-pull down assays and mammalian two-hybrid assays in Med1−/− MEFs, determined that CCAR1 interacted with GATA1(249–413) but not with GATA1(316–413) (Fig. 5D, E). Therefore, GATA1(249–315), the CF domain of GATA1, was necessary for the interaction with CCAR1. However, GATA1(249–315) was not enough for the interaction, and either the N-terminal [GATA1(200–315)] or C-terminal [GATA1(249–330)] extension outside of GATA1(249–315) was necessary (Fig. 5I).
Regarding the direct interaction between GATA1 and MED1, a previous report suggested that MED1 interacts with the GATA1 NF domain (Stumpf et al. 2006). We also assessed their interaction using GST-pull down assays and mammalian two-hybrid assays, as were used for the analyses of CCAR1 interaction (above). Unexpectedly, in our relatively stringent system, neither mGATA1(1–257) nor hGATA1(1–257) interacted with MED1. Indeed, both mouse and human GATA1(249–413) interacted with hMED1, but neither mouse nor human GATA1(316–413) did (Fig. 5F–H). Therefore, GATA1(249–315), which contains the CF domain, was required for specific interaction with MED1. However, as was the case for CCAR1, GATA1(249–315) was not enough for this interaction, and either N-terminal [GATA1(200–315)] or C-terminal [GATA1(249–330)] extension was necessary (Fig. 5I). The reason for this discrepancy might be the differences in the experimental procedures; however, the physical interaction of the GATA1 containing its CF domain with both CCAR1 (above) and MED1 appears to be relevant in vivo.
The AD2 domain of CoCoA, a partner of CCAR1, interacts with the GATA1 CF domain
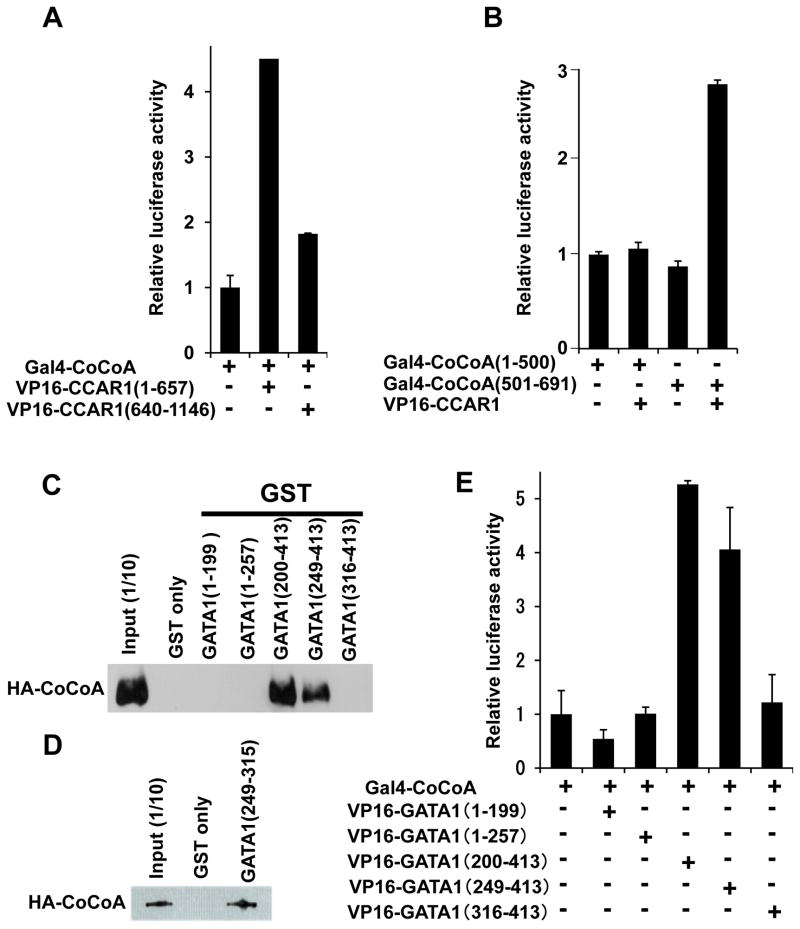
CoCoA is a known dimerization partner of CCAR1 (Kim et al. 2008) that acts as a transcriptional coactivator for some activators, including NRs (Kim et al. 2003), β-catenin (Yang et al. 2006a; Yang et al. 2006b), aryl hydrocarbon receptor (Kim et al. 2004), and p53 (Kim et al. 2008). Indeed, during hemin-induced differentiation of K562 cells, CoCoA expression was prominently (4-fold) induced after 2 days, while expression of CCAR1 only minimally changed (Fig. S1). Expression of CoCoA was modestly induced when MED1 was knocked down (Fig. S2), and when MED1(1–530) was overexpressed both in the presence and absence of hemin (Fig. S5, S6). Therefore, CoCoA might have an important role in MED1-mediated transcriptional control, and we next assessed the role of CoCoA in GATA1-mediated activation. First, the interaction between CCAR1 and CoCoA was analyzed by mammalian two-hybrid assays in Med1−/− MEFs. Consistent with a previous study that utilized GST-pull down assays (Kim et al. 2008), Gal4-CoCoA interacted with VP16-CCAR1(1–657) in Med1−/− MEFs, and VP16-CCAR1 interacted with CoCoA(501–691) (Fig. 6A, B). Therefore, we confirmed that the N-terminal half of CCAR1 interacts specifically with the C-terminus of CoCoA.
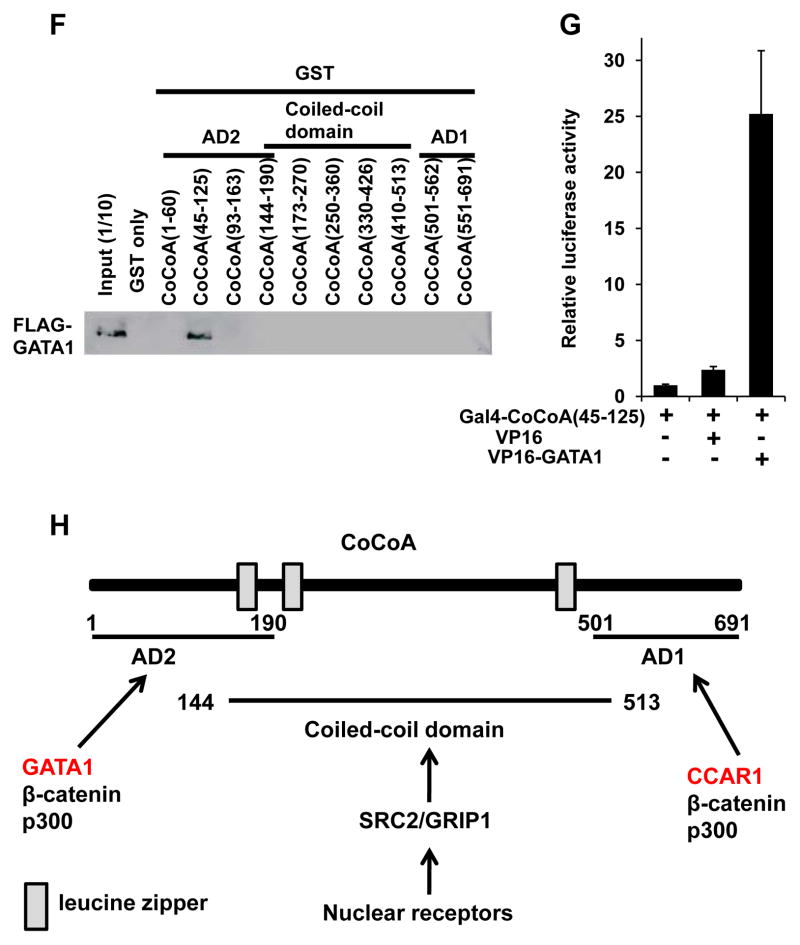
Figure 6. The AD1 domain of CoCoA interacts with CCAR1(501–691), while the AD2 domain of CoCoA interacts with the GATA1 CF domain.
(A, B, E, G) Mammalian two-hybrid assays. The luciferase activity of the reporter with 5×Gal4-binding sites was measured. The interaction between various Gal4-fusion fragments of CoCoA (A, B, E, G) and various VP16-fusion fragments of CCAR1 (A, B) or GATA1 (E, G) were tested. Values (mean ± SD) of a representative experiment performed in duplicate are shown. The results were reproducible in three independent experiments.
(C, D, F) GST-pull down assays. HA-CoCoA (C, D) or FLAG-GATA1 (F) was pulled down by various GST-fusion fragments of GATA1 (C, D) or CoCoA (F). HA-CoCoA and FLAG-GATA1 were visualized using an anti-HA or -FLAG antibody, respectively.
(H) Diagram of CoCoA showing the activation domains AD1 and AD2, the coiled-coil domain, and the leucine zippers. Transcription factors that interact with these domains are also shown.
CoCoA is composed of 3 domains, CoCoA(1–190), an N-terminal activation domain 2 (AD2), CoCoA(144–513), a coiled-coil domain, and CoCoA(501–691), a C-terminal AD1 (Yang et al. 2008) (Fig. 6H). NR signaling may be conveyed through the CoCoA coiled-coil domain via SRC2/GRIP1 (Kim et al. 2003), while β-catenin and p300 interact with both the AD2 and AD1 of CoCoA (Kim et al. 2006; Yang et al. 2006a; Yang et al. 2006b). We now add CCAR1 to the list of specific CoCoA AD1-interacting proteins (Fig. 6G).
We next tested if CoCoA could interact with GATA1. Intriguingly, serial GST-pull down assays and mammalian two-hybrid assays (as described above) demonstrated a specific and significant interaction between CoCoA and the GATA1(249–315) that corresponds to the CF domain (Fig. 6C–E). Then, serial GST-pull down assays were performed to determine which domain of CoCoA interacted with CCAR1. CoCoA(45–125), which corresponds to activation domain 2 (AD2) of CoCoA, was found to bind specifically to GATA1 (Fig. 6E). Mammalian two-hybrid assays confirmed the intracellular interaction between GATA1 and CoCoA(45–125) (Fig. 6F). These results indicate that CoCoA AD2 may interact with GATA1, CoCoA AD1 interacts with CCAR1 (Fig. 6G), and that the GATA1 CF domain serves as a docking surface for multiple coactivators, including CoCoA, CCAR1, and MED1.
GATA1, MED1, CCAR1, and CoCoA are recruited to the γ-globin promoter during erythroid differentiation
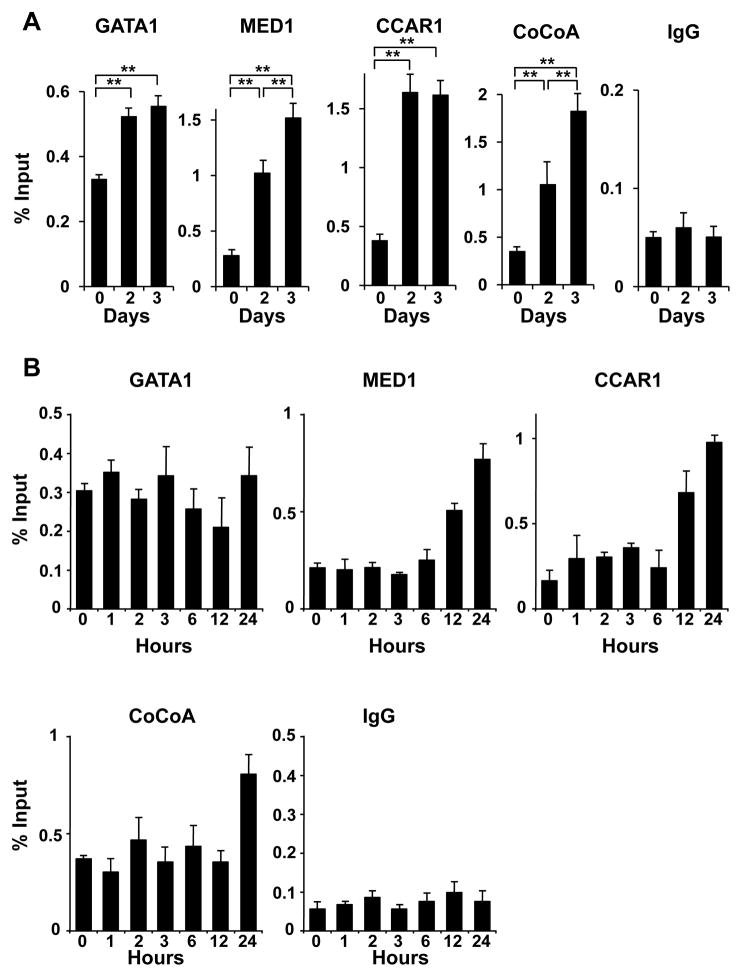
Since CCAR1 appeared to be involved in GATA1- and MED1-mediated transcriptional activation and erythroid differentiation independent of the direct interaction between GATA1 and MED1, and since CoCoA might be a GATA1- and CCAR1-interacting molecule, we next asked if these molecules were actually recruited to the γ-globin promoter during erythroid differentiation of K562 cells. When K562 cells were tested using chromatin immunoprecipitation (ChIP) assays before and after treatment with hemin, GATA1, MED1, CCAR1, and CoCoA were all recruited onto the γ-globin promoter at the GATA1-binding sites after 2 and 3 days during erythroid differentiation (Fig. 7A). Therefore, the intrinsic GATA1 binding sites were occupied by GATA1, CCAR1/CoCoA, and Mediator during the transcriptional activation of the γ-globin promoter. Next, the occupancies of GATA1, MED1, CCAR1 and CoCoA were assessed chronologically for 24 h so that the order of the occupancies may be more apparent.
Figure 7. GATA1, MED1, CCAR1 and CoCoA are recruited onto the γ-globin promoter during erythroid differentiation of K562 cells.
(A, B) ChIP assays showing the occupancies of GATA1, MED1, CCAR1, and CoCoA on the γ-globin promoter during 3 days (A) and 24 h (B) of erythroid differentiation of hemin-treated K562 cells.
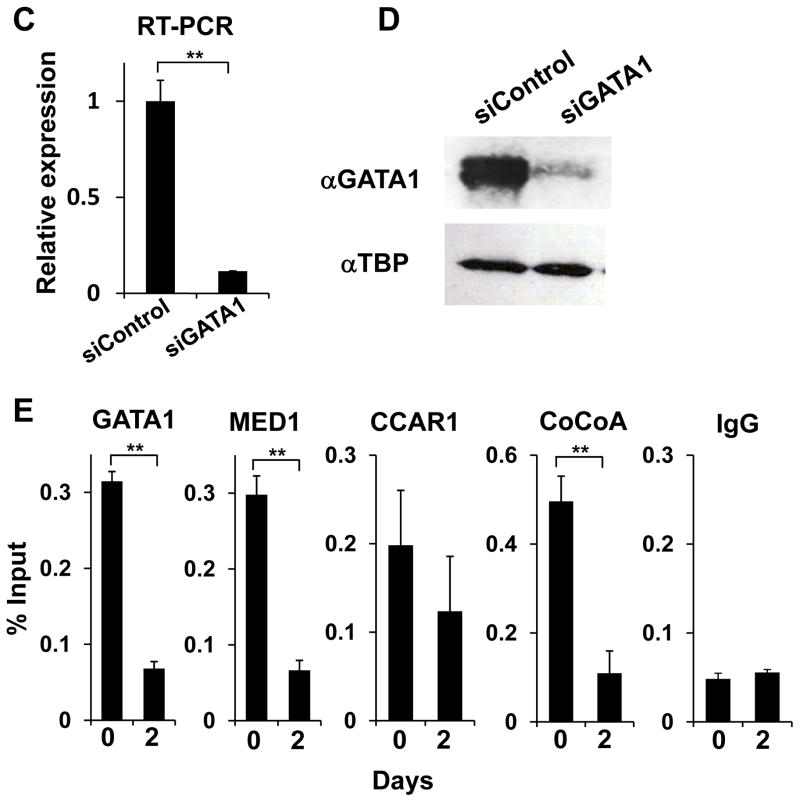
(C, D) Results of quantitative RT-PCR (C) and western blot analyses (D) of GATA1 2 days after transfection with GATA1 siRNA in K562 cells are shown. Values (mean ± SD) of a representative experiment performed in triplicate are shown (**p < .01).
(E) ChIP assays of hemin-treated K562 cells showing the occupancies of indicated cofactors on the γ-globin promoter before and 3 days after transfection with GATA1 siRNA.
Unimmunized rabbit IgG is used as a negative control. Values (mean ± SD from a representative experiment performed in triplicate) are plotted as percentages of the value of the input (A, B, E). The results were reproducible in three independent experiments.
After 12 h, the occupancy of CCAR1 and MED1 began to increase, followed by CoCoA recruitment at 24 h. The occupancy of GATA1 appeared unchanged during this period (Fig. 7B). Thus, during the first day, GATA1 already associated with the promoter appeared to be used for recruitment of CCAR1 and MED1. After 2 days, together with the enhanced recruitment of GATA1, the occupancy of all of these cofactors increased. While the occupancy of CCAR1 appeared to reach the maximum after 2 days, more CoCoA and MED1 were recruited after 3 days (Fig. 7A), concurrently with the induced expression of CoCoA and MED1 (Fig. 1C, Fig. S1).
To prove that GATA1 is the platform for recruitment of these cofactors, knockdown of GATA1 was performed followed by ChIP assays. Indeed, 3 days after GATA1 was knocked down in hemin-treated K562 cells (Fig. 7C, D), the occupancy of CoCoA and MED1 was reduced and CCAR1 did not increase (Fig. 7E), even though expression of these cofactors was upregulated (Fig. S7). Hence, DNA-bound GATA1 appeared to be the interface for recruitment of these cofactors.
GATA1, CCAR1/CoCoA, and MED1(1–602) simultaneously associate
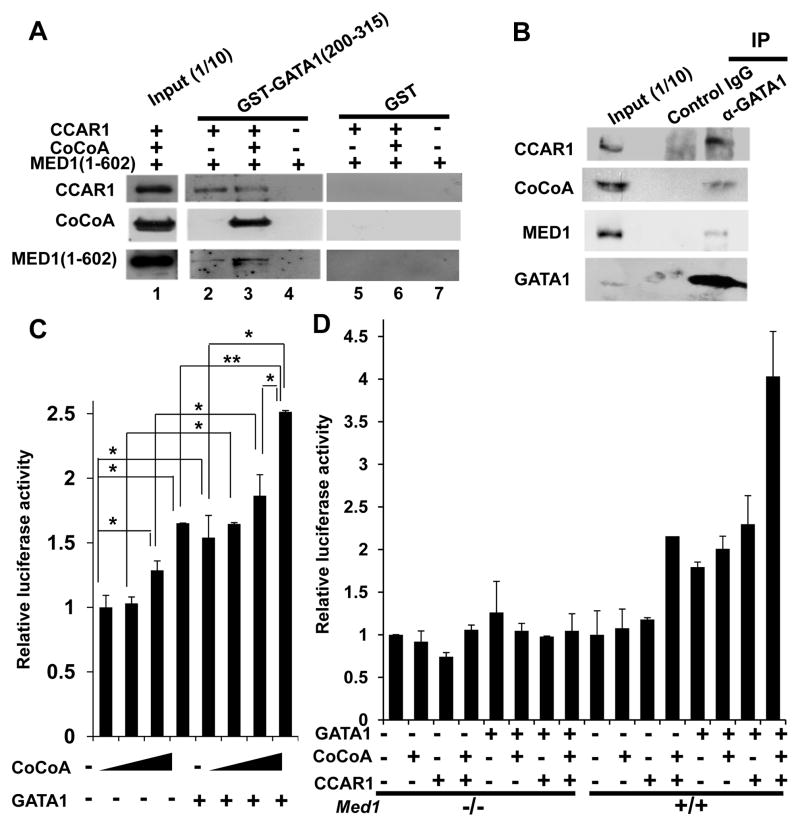
Since GATA1, CCAR1/CoCoA, and MED1 interact with one another both in vitro and in vivo and since they are recruited to endogenous GATA1-binding sites during transcription, we next asked if these molecules could associate. We prepared recombinant CCAR1, CoCoA, and MED1(1–602) and performed GST-pull down assays using recombinant GST-GATA1(200–315), which contains the zinc finger domains. Although GST-GATA1(200–315) did not interact with MED1(1–602) alone (Fig. 8A, lane 4), it did interact with CCAR1 and MED1(1–602) (Fig. 8A, lane 2), indicating that CCAR1 can simultaneously interact with MED1 and GATA1. When CoCoA was also added, it efficiently co-purified with CCAR1 and MED1(1–602) (Fig. 8A, lane 3).
Figure 8. CCAR1 and CoCoA cooperatively enhance GATA1- and MED1-mediated transcriptional activation of the γ-globin promoter.
(A) GST-pull down assay. The interaction between baculovirus expressed CCAR1, CoCoA, and/or MED1(1–602), and bacterially expressed GST-GATA1(200–315) (lanes 2–4) or GST (lanes 5–7) was tested. GATA1(200–315), CCAR1, and CoCoA form a ternary complex (lane 2), and the addition of CoCoA enables formation of a tetramer complex (lane 3).
(B) Coimmunoprecipitation of K562 cells treated with hemin for 2 days. Endogenous CCAR1 and CoCoA are co-immunoprecipitated together with endogenous GATA1. Anti-CCAR1, anti-CoCoA, anti-MED1, or anti-GATA1 antibody was used (A, B).
(C, D) Luciferase reporter assays of the human γ-globin promoter in Med1+/+ (C, D) and Med1−/− (D) MEFs. (C) CoCoA dose-dependently enhances GATA1-mediated transcriptional activation. The amounts of each expression vector added were as follows: 20, 50, or 100 ng of CoCoA and 20 ng of GATA1. (D) CCAR1 and CoCoA synergistically enhance GATA1-mediated transcriptional activation in a MED1-dependent manner. The amounts of each expression vector added were as follows: 20 ng of GATA1, 20 ng of CoCoA, and 20 ng of CCAR1.
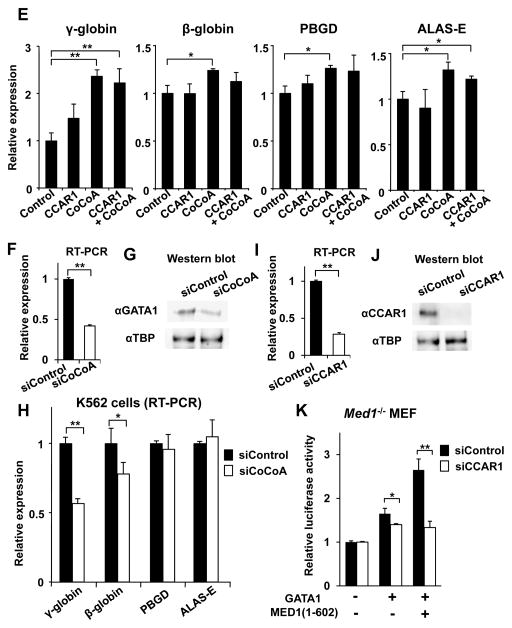
(E) Overexpression of CCAR1 and/or CoCoA in hemin-treated K562 cells. The mRNA expression of GATA1-targeted genes 3 days after transfection with the CCAR1 and/or CoCoA expression vectors is shown. An empty expression vector was used as a control.
(F, G) Results of quantitative RT-PCR (F) and western blot analyses (G) of GATA1 2 days after transfection with hCoCoA siRNA in K562 cells are shown. Anti-TBP antibody was used as a control (G).
(H) GATA1-targeted gene transcription 3 days after transfection with hCoCoA siRNA in hemin-treated K562 cells.
(I, J) Results of quantitative RT-PCR (I) and western blot analyses (J) of GATA1 2 days after transfection with mCCAR1 siRNA in Med1−/− MEFs are shown. Anti-TBP antibody was used as a control (J).
(K) Luciferase reporter assays of the human γ-globin promoter in Med1−/− MEFs. Knockdown of CCAR1 prominently reduces GATA1- and MED1(1–602)-mediated transcriptional activation.
Values (mean ± SD) of a representative experiment performed in duplicate (C, D) or triplicate (E, F, I, H, K) are shown (*p < .05, **p < .01). The results were reproducible in three independent experiments.
It is known that GATA1 interacts with endogenous Mediator through MED1 subunit in vivo (Stumpf et al. 2006). We next asked if endogenous GATA1 and CCAR1/CoCoA associate during transcription. When total cell lyates of hemin-treated K562 cells were used for immunoprecipitation with the anti-GATA1 antibody, both CCAR1 and CoCoA were co-immunoprecipitated together with GATA1 (Fig. 8B). Therefore, endogenous GATA1 and CCAR1/CoCoA associate during GATA1-mediated transcription and erythroid differentiation.
These results indicate that GATA1, CCAR1/CoCoA, and MED1 can form a complex, at least transiently, and may contribute to GATA1- and MED1-mediated transcriptional activation independent of the direct interaction of GATA1 and MED1.
GATA1, CCAR1, and CoCoA cooperatively activate transcription at the γ-globin promoter
Since CoCoA might be involved in GATA1-mediated transcriptional activation, we next tested the role of CoCoA at the γ-globin promoter using luciferase reporter assays in Med1+/+ MEFs. CoCoA modestly enhanced γ-globin promoter activity in a dose-dependent manner (up to 1.7 times the basal level). When a small amount of GATA1 (20 ng) was added, transcriptional activation was enhanced in a CoCoA dose-dependent manner (up to 2.5 times) (Fig. 8C). When small amounts of GATA1 (20 ng), CCAR1 (20 ng), and/or CoCoA (20 ng) were added to Med1+/+ MEFs in various combinations, any combination of 2 components significantly activated transcription (up to 2 times the basal level). When all 3 components were added altogether, transcriptional activation was further enhanced (4 times the basal level) (Fig. 8D). However, GATA1- and CCAR1/CoCoA-mediated transactivation was not induced in Med1−/− MEFs, indicating that the action of CCAR1/CoCoA is dependent on MED1 (Fig. 8D). These results indicate that the CCAR1/CoCoA pair can cooperatively enhance GATA1- and MED1-mediated transcriptional activation.
We next assessed the requirement of CCAR1 and CoCoA for GATA1-dependent gene transcription during K562 cell differentiation by overexpression of these genes (Fig. S8). Indeed, CoCoA overexpression did enhance expression of GATA1-targeted and erythroid marker genes, among which γ-globin expression was prominent, as well as some GATA1-associated regulators (FOG1, KLF1, SCL) (Fig. 8E, Fig. S9). However, overexpression of CCAR1 did not enhance transcription of these genes, possibly indicating, consistent with the fact that CCAR1 was not induced during differentiation of K562 cells (Fig. S1), that endogenous CCAR1 was sufficient for this function, while CoCoA might be a limiting factor in this process. Indeed, when CoCoA was knocked down in K562 cells (Fig. 8F, G), expression of GATA1-targeted genes including γ-globin and β-globin was significantly reduced during erythroid differentiation (Fig. 8H). Since hCCAR1 siRNA that efficiently attenuates endogenous CCAR1 in K562 cells was unavailable, we tested human γ-globin promoter in Med1−/− MEFs. When endogenous CCAR1 was knocked down in these cells (Fig. 8I, J), GATA1 function was attenuated. While the addition of MED1(1–602) prominently enhanced the reporter activity in control cells, the activity was unchanged when CCAR1 was downregulated (Fig. 8K). These results further confirm that CCAR1 and CoCoA have an important role in GATA1- and MED1-mediated transcription.
DISCUSSION
In the process of analyzing the role of MED1 in GATA1-mediated transcriptional activation in K562 cells, we gathered evidence that the direct interaction between MED1 and GATA1 is partially redundant, and that the CCAR1/CoCoA pair can bypass the need for this interaction in GATA1- and MED1-mediated transcriptional activation (Fig. 9). This bypass pathway constitutes a novel post-GATA1 intranuclear signaling pathway that might be of biological significance.
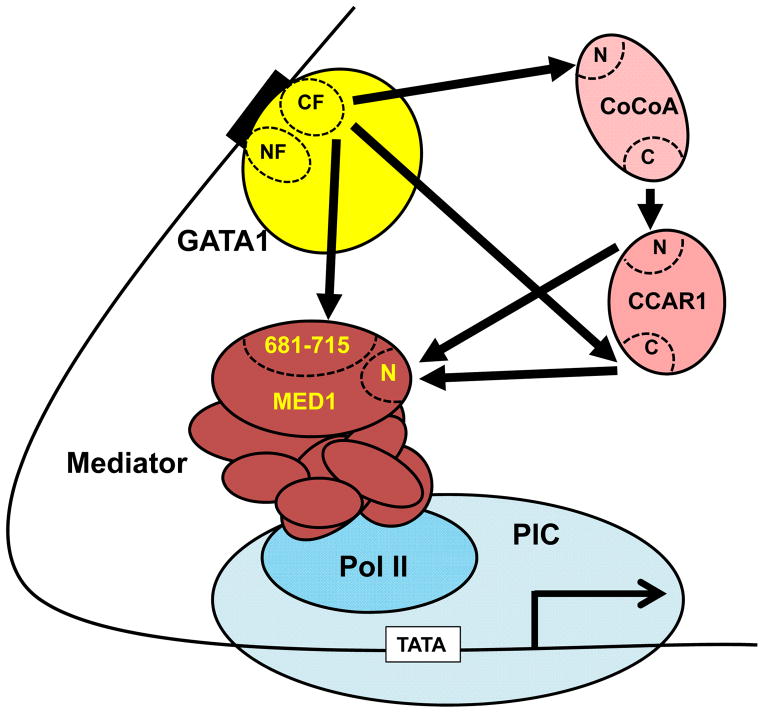
Figure 9.
Proposed model for the role of CCAR/CoCoA in the bypass mechanism of GATA1- and MED1-mediated transcriptional activation. CCAR1, CoCoA, and MED1(681–715) interact with the CF domain either competitively or simultaneously. CCAR1 and CoCoA interact with the CF domain of GATA1 and the N-terminal domain of MED1 simultaneously via multiple interfaces. MED1-led Mediator then recruits RNA polymerase II, eventually forming a preinitiation complex for transcription initiation. The multiple pathways that underlie the GATA1-MED1 axis may help to fine-tune the coupling between GATA1 and the basal transcriptional machinery.
The transcriptional bypass CCAR1/CoCoA coactivator as a secondary coactivation system
Previous studies indicated that CoCoA, identified as a NR-specific coactivator, interacts with both SRC2/GRIP1 and p300 (Kim et al. 2003; Kim et al. 2006). Although the precise mechanisms underlying CoCoA action in transcriptional activation are not yet known, CoCoA might serve as an intermediary, connecting activator signaling to histone-modifying coactivators (Kim et al. 2006). CCAR1, identified as partner of CoCoA and as a bypass molecule for ER and GR, conveys intranuclear post-receptor signal to Mediator via MED1 (Kim et al. 2008). Thus, the CCAR1 and CoCoA pair appears to function cooperatively, and in multiple, distinct, and supplementary ways, to coactivate NR functions. It is conceivable that transactivation induced by p53, β-catenin, and Ngn-3 is similarly mediated by the CCAR1 and CoCoA pair, where CCAR1 and CoCoA play either a supplementary or major role in coactivation.
This study re-identifies GATA1 as a novel activator that is driven by the CCAR1/CoCoA pair. In addition to the role of CCAR1 (and CoCoA) as a molecular bridge between GATA1 and MED1 during recruitment of RNA polymerase II holoenzyme to the promoter and PIC formation, its interacting partner CoCoA might simultaneously recruit chromatin remodeling complex(es), which may contribute to relaxation of nucleosome structures and enable recruitment of the basal transcriptional machinery. Therefore, the CCAR1/CoCoA pair might constitute a secondary coactivation machinery beyond primary coactivators, which include chromatin modifying coactivators (p160 coactivators, histone acetyltransferases [e.g., CBP and p300], and histone methyltransferases [e.g., CARM1 and PRMT1]) and Mediator. The multiple layers in the coactivation systems proposed here are probably used for efficient transactivation in vivo not only by GATA1, but also by other activators, including NRs and p53.
Model for GATA1-induced transcriptional activation
Besides its association with MED1 and CCAR1/CoCoA (Kim et al. 2008; this study), GATA1 is known to interact with acetyltransferase coactivators (e.g., CBP/p300) (Blobel et al. 1998). This interaction is reminiscent of the one with NRs, and the multistep activation model of GATA1 function can be proposed analogously to that for NR function (Ito & Roeder 2001). In this model, GATA1-induced activation might start by dissociation of PU.1 (a suppressor of GATA1 function) from GATA1, and subsequent interaction of DNA-bound GATA1 with either CBP/p300 or Mediator. Destabilization of the nucleosome by CBP/p300 may be prerequisite for Mediator-facilitated recruitment of RNA polymerase II and formation of the PIC. In this model, CCAR1/CoCoA might contribute to the increased mechanistic complexity, enabling multiple interfaces for GATA1 to access to both CBP/p300 and MED1 (or Mediator).
GATA1 also interacts with several DNA-bound and unbound activators and coactivators [e.g., FOG1 (Tsang et al. 1997), KLF-1 (Crossley et al. 1994), Sp1 (Merika & Orkin 1995), RUNX1 (Elagib et al. 2003), and LMO2 (Wadman et al. 1007)]. Each of these factors, and a tetrameric complex of SCL/Tal-1, E2A, LDB1, and LMO2, may, either singly or in combination, contribute to Mediator recruitment and formation of the PIC, irrespective of MED1. The residual erythropoiesis in Med1−/− embryos (Ito et al. 2000; Stumpf et al. 2006) and residual GATA1-mediated activation of the γ-globin promoter in Med1−/− MEFs (Fig. 4B) indicate that GATA1-mediated, MED1-independent Mediator recruitment and transcriptional activation occur. The contribution of these activators/coactivators might add, in parallel, complexity to GATA1-mediated activation, and might modify the multistep model described above in a trans-acting manner. It is known that acetylation of GATA1 at lysine residues adjacent to the zinc finger domains is required for association with bromodomain protein Brd3 (Lamonica et al. 2011). Modification(s) (e.g., acetylation) of GATA1 might also alter affinities for CCAR1/CoCoA or the abovementioned activators/coactivators, enabling and facilitating a switch of the factors that associate with GATA1. Changes in accessibility of CCAR1/CoCoA and MED1 to DNA-bound GATA1 12 to 24 h after induction of erythroid differentiation (Fig. 7A, B) might reflect changes in GATA1 modification.
Redundancy and the requirement for direct interaction between GATA1 and MED1
Our study indicates that GATA1- and MED1-mediated transcriptional activation occurs via both GATA1-MED1 direct interaction-dependent and -independent mechanisms, the latter of which is mediated by CCAR1/CoCoA; however, the underlying mechanism may not be restricted to CCAR1/CoCoA. Multiple GATA1-MED1 pathways may have important mechanistic and physiological implications.
First, while the GATA1 CF domain serves as a molecular dock for MED1(681–715), CCAR1, CoCoA (Fig. 9), and several activators (Fig. 5C), we do not know whether a single GATA1 molecule interacts with all of these simultaneously. Studies of the three-dimensional structure suggest that a DNA-bound GATA1 NF domain can bind to FOG1 (Liew et al. 2005), and that the GATA1 NF domain binds FOG1 and LMO2 simultaneously (Wilkinson-White et al. 2011), suggesting that the GATA1 CF domain might contain a binding interface for multiple molecules. If this is the case, then GATA1 CF domain-bound activators/coactivators might contribute to the interaction with Mediator via either MED1 or other subunit(s) of Mediator. Another possibility is that multiple GATA1 molecules bound to multiple GATA-binding sites of a promoter (e.g., the γ-globin promoter has 2 GATA-binding sites.) might bind to different activators/coactivators, and eventually the multiplicity of activators/coactivators on the same promoter would contribute to a stronger interaction with, and recruitment of, Mediator, and thus might synergize the activator/coactivator signals, eventually facilitating PIC formation on the promoter.
Second, the CCAR1/CoCoA pair has multiple functions in GATA1-MED1 interaction, namely, (i) direct binding of GATA1 to MED1(681–715), (ii) binding of GATA1 to MED1(1–602) via the C-terminus of CCAR1, and (iii) binding of GATA1 to MED1(1–602) via CoCoA and subsequently CCAR1. Although the contribution of CCAR1/CoCoA appears to be significant, it does not exclude the possibility that other cofactor(s) might also play a role in this bypass process. The multiple and redundant GATA1-MED1 pathways are reminiscent of the PPARγ2-dependent, and MED1 NR box-independent, adipocyte differentiation in cultured cells (Ge et al. 2008), and the almost normal development and homeostasis of MED1 NR box-mutant knock-in mice (Jiang et al. 2010). However, NR box-dependent activation is specifically necessary in some circumstances, as shown by the retarded ERα-dependent pubertal growth of mammary epithelial cells in NR box-mutant knock-in mice (Jiang et al. 2010).
The physiological significance of the multiple modes for the nuclear GATA1-MED1 pathway is not yet known. However, because the intact MED1 is required for maximal transactivation and erythroid differentiation (this study), MED1 is probably required for a full GATA1 function in a living animal. Full GATA1 activity is presumably required, in erythropoiesis for instance, in some special and extreme circumstances, such as to meet physiological needs of erythropoiesis during development and pregnancy, and in pathological conditions such as hemolysis and bleeding, and recovery from myelosuppression due to chemotherapeutic agents. It would be intriguing to determine if there is disordered function(s) in one of the GATA1-mediating molecules in megaloblastosis and/or neoplastic diseases such as myelodysplastic syndromes. Disordered function(s) in one of these molecules may also result from some disease conditions including some types to leukemia where alterations of GATA1 are involved (Shimizu et al. 2008). GATA1-mediated homeostatic events other than erythropoiesis would also be governed in a likewise manner. The answer to these questions would require studies of CCAR1 and/or CoCoA mouse knockouts.
EXPERIMENTAL PROCEDURES
Plasmids
Mammalian expression vectors pIRESneo-hMED1 (full-length), pIRESneo-hMED1(1–602), pIRESneo-hMED1(1–703), and pIRESneo-hMED1(592–1,581) (Sumitomo et al. 2010), and CMV-p300 (Gu et al. 1997) have been described. The cDNAs for mGATA1 and hGATA1 kindly provided by M. Yamamoto, and for mCCAR1 and mCoCoA prepared by reverse transcriptase-PCR (RT-PCR) using the ReverTra Ace qPCR RT Kit and KOD FX (Toyobo, Japan), were cloned in either pIRESneo (Clontech) or pcDNA3.1(+) (Invitrogen) to create pIRESneo-mGATA1, pIRESneo-mCCAR1, and pcDNA3.1-mCoCoA, respectively. For the expression of glutathione S-transferase (GST)-fusion proteins, cDNAs encoding hMED1(534–659), hMED1(637–703), hMED1(681–715), hMED1(691–715), hMED1(704–715), hMED1(704–739), mGATA1 (full-length), mGATA1(1–199), mGATA1(1–257), mGATA1(200–413), mGATA1(249–413), mGATA1(316–413), mGATA1(200–315), mGATA1(249–315), mGATA1(249–330), mCoCoA(1–60), mCoCoA(45–125), mCoCoA(93–163), mCoCoA(144–190), mCoCoA(173–270), mCoCoA(250–360), mCoCoA(330–426), mCoCoA(410–513), mCoCoA(501–562), and mCoCoA(551–691) were amplified by PCR and subcloned into pGEX4T-3 (GE Healthcare). mGATA1 was HA- and FLAG-tagged, and subcloned into pVL1392 (BD Biosciences) to generate pVL-HA-mGATA1 and pVL-FLAG-mGATA1, respectively. mCCAR1, mCoCoA, hMED1 (full-length), hMED1(1–552), hMED1(715–1,581), and hMED1(1–602) were HA- or FLAG-tagged, and subcloned into either pVL1392 or pVL1393 to create pVL-HA-mCCAR1, pVL-HA-mCoCoA, pVL-HA-hMED1, pVL-FLAG-hMED(1–552), pVL-FLAG-hMED1(715–1,581) and pVL-HA-hMED1(1–602), respectively. For mammalian two-hybrid assays, Gal4-hMED1 in pCDM8 (Invitrogen) (pGal4-hMED1) was used, which has been described previously (Sumitomo et al. 2010). cDNAs encoding mCCAR1(1–1,146), mCCAR1(1–657), mCCAR1(640–1,146), mCoCoA(1–691), mCoCoA(1–500), mCoCoA(501–691), and mCoCoA(45–125) were fused to Gal4 and subcloned into pCDM8 to generate pGal4-mCCAR1(1–1,146), pGal4-mCCAR1(1–657), pGal4-mCCAR1(640–1,146), pGal4-mCoCoA(1–691), pGal4-mCoCoA(1–500), pGal4-mCoCoA(501–691), and pGal4-mCoCoA(45–125), respectively. cDNAs encoding mCCAR1(1–657), mCCAR1(640–1,146), mGATA1 (full-length), mGATA1(1–199), mGATA1(1–257), mGATA1(200–413), mGATA1(249–413), mGATA1(316–413), mCCAR1 (full-length), mCCAR1(1–657), and mCCAR1(640–1,146) were fused to VP16 and subcloned into pcDNA3.1(+) (Invitrogen) to generate pVP16-mCCAR1(1–657), pVP16-mCCAR1(640–1,146), pVP16-mGATA1, pVP16-mGATA1(1–199), pVP16-mGATA1(1–257), pVP16-mGATA1(200–413), pVP16-mGATA1(249–413), pVP16-mGATA1(316–413), pVP16-mCCAR1, pVP16-mCCAR1(1–657), and pVP16-mCCAR1(640–1,146), respectively. Expression vectors for Gal4-fused full-length hGATA1 and for VP16-fused various truncations of hGATA1 were likewise prepared. For luciferase reporters, the human γ-globin promoter (–299 to +37) was amplified from genomic DNA using KOD FX (Toyobo, Japan) and cloned into the firefly luciferase reporter plasmid pGL4.10 to create γ-globin-LUC. The reporter containing 5 Gal4 binding sites (5×Gal4-LUC) has been described (Sumitomo et al. 2010).
Cell culture
K562 cells (Lozzio & Lozzio 1975), obtained from the Japanese Collection of Research Bioresources (Osaka, Japan), were cultured in RPMI1640 supplemented with 10% fetal bovine serum (FBS) at 37°C. Erythroid differentiation of K562 cells was induced by the addition of 50 μM hemin, and quantified by counting the number of cells that were positive for 3,3′-dimethoxybenzidine (MP Biomedicals, Inc.) staining (Lam et al. 2010). Stable lines of Med1+/+ p53−/− and Med1−/− p53−/− mouse embryonic fibroblasts (MEFs), established from embryonic day 10.0 (E10.0) embryos derived from a single crossing of Med1+/− p53+/− male and Med1+/− p53+/− female mice in a C57BL6 background, have been described (Sumitomo et al. 2010). These Med1+/+ and Med1−/− MEFs (in p53−/−) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS at 37°C.
Transient expression and RNA interference
For transient expression, 1.0 × 106 K562 cells in a 6-well dish were transfected with 5 μg of pIRESneo-hMED1, pIRESneo-hMED1(1–602), pIRESneo-hMED1(1–703), pIRESneo-mCCAR1 and/or pcDNA3.1-mCoCoA, or empty pIRESneo as a control, using Lipofectamine™ LTX Reagent (Invitrogen) or Cell Line Solution V with Nucleofector II device (Lonza). Hemin was added after 6 h.
For RNA interference, hMED1 siRNA (Custom siRNA, Qiagen; sense, GGCUCUCAAAGUAACAUCUdTdT; antisense, AGAUGUUACUUUGAGAGCCdTdT), siRNA for hCoCoA or hGATA1 (Silencer select predesigned siRNA; Applied Biosystems), or control siRNA (Invitrogen) (5 nM in 24-well plates), was transfected using either Lipofectamine RNAiMAX (Invitrogen) or Cell Line Solution V with Nucleofector II devise (Lonza) according to the manufacturer’s protocol. Cotransfection of hMED1 siRNA together with siRNA-resistant pIRES-MED1(1–602) (Hasegawa et al. 2012) was performed by using Cell Line Solution V with Nucleofector II device (Lonza).
Luciferase reporter assay and mammalian two-hybrid assay
For luciferase reporter assays, MEFs (2 × 104) in 24-well plates were transfected with pIRESneo-hMED1, pIRESneo-hMED1(1–602), pIRESneo-hMED1(1–703), pIRESneo-hMED1(592–1,581), pIRESneo-mGATA1, pIRESneo-mCCAR1 and/or pcDNA3.1-mCoCoA, together with γ-globin-LUC (50 ng) and the Renilla control luciferase vector (5 ng) using Lipofectamine (Invitrogen). Cotransfection of mCCAR1 siRNA (Silencer select predesigned siRNA; Applied Biosystems) together with γ-globin-LUC, pIRESneo-mGATA1, and/or pIRESneo-hMED1(1–602) was performed by using Lipofectamine RNAiMAX to introduce siRNA followed by using Lipofectamine LTX the next day to introduce plasmids (Invitrogen). After 48 h, reporter activity was measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to the activity of the control Renilla luciferase (Sumitomo et al. 2010).
Mammalian two-hybrid assays were similarly performed by transfection with 10 ng of Gal4-fused expression vector [pGal4-hMED1, pGal4-m/hGATA1, pGal4-mCCAR1(1–1,146), pGal4-mCCAR1(1–657), pGal4-mCCAR1(640–1,146), pGal4-mCoCoA(1–691), pGal4-mCoCoA(1–500), pGal4-mCoCoA(501–691), or pGal4-mCoCoA(45–125)], 200 ng of VP16-fused expression vector [pVP16-hMED1(1–637), pVP16-hMED1(637–703), pVP16-hMED1(681–715), pVP16-hMED1(715–947), pVP16-hMED1(920–1,581), pVP16-mCCAR1(1–657), pVP16-mCCAR1(640–1,146), pVP16-mGATA1, pVP16-m/hGATA1(1–199), pVP16-m/hGATA1(1–257), pVP16-m/hGATA1(200–413), pVP16-m/hGATA1(249–413), pVP16-m/hGATA1(316–413), pVP16-mCCAR1, pVP16-mCCAR1(1–657) or pVP16-mCCAR1(640–1,146)], and 5×Gal4-LUC (100 ng), together with the Renilla control luciferase vector (5 ng).
Quantification of mRNA expression
For quantitative PCR (qPCR), total RNA (1 μg), extracted with ISOGEN (NIPPON GENE, Japan), were used to prepare cDNAs with the ReverTra Ace qPCR RT kit (Toyobo, Japan). The expression of various genes was identified by qPCR (StepOnePl™ real-time PCR system; Applied Biosystems). Values were normalized to the values of human or mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) measured as a reference marker. The sequences of the primers and the PCR conditions used for amplification are available upon request.
Co-immunoprecipitation and western blot analysis
For the co-immunoprecipitstion, anti-GATA1 rat polyclonal antibody (N6; Santa Cruz Biotechnology) or control IgG were pretreated with Dynabeads Protein G (Life Technologies) for 10 min at room temperature. K562 cells were treated with hemin for 2 days, and total cell lysates in BC150 buffer with 0.1% NP40 and 1 mM phenylmethylsulfonyl fluoride were incubated with the protein G-bound antibodies for 30 min at room temperature, and the bound proteins were isolated.
For the western blot analysis, proteins were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with polyclonal antibodies (Sumitomo et al. 2010).
In vitro protein-protein interaction analysis
For bacterial expression, GST-fused proteins were expressed in and purified from Escherichia coli strain JM109. For baculovirus-mediated expression, pVL-HA-mCCAR1, pVL-HA-mCoCoA, pVL-HA-hMED1, pVL-FLAG-hMED(1–552), pVL-FLAG-hMED1(715–1,581), and pVL-HA-hMED1(1–602) were cotransfected into Sf9 cells using BacVector-2000 (Novagen). Whole cell lysates of Sf9 cells expressing these recombinant proteins were used in the analyses.
For GST-pull down assays, immobilized GST or GST-fusion protein (1.5 μg) and 3 μL of Sf9 cell lysate were used. For co-immunoprecipitation assays, FLAG-tagged hMED1(1–552) or hMED1(715–1,581) (1 μg) immobilized to M2 agarose (Sigma) and 3 μL of Sf9 cell lysate containing HA-tagged GATA1 were used. These were incubated in BC150 buffer with 0.1% NP40 and 1 mM β-mercaptoethanol at 4°C for 1 h. Then the beads were washed extensively with the binding buffer and bound proteins were eluted in 0.3% sarkosyl. Samples were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with either anti-HA (12CA5; Boehringer) or anti-FLAG (M5; Sigma) antibody.
ChIP assay
For ChIP assays, 1.0 × 107 K562 cells (before or after treatment with hemin) were fixed with 1% formalin for 10 min and used to prepare chromatin by sonication according to the ChIP-IT Express manual (Active Motif). Samples were immunoprecipitated with anti-GATA1, anti-MED1 (M255; Santa Cruz Biotechnology), anti-CCAR1 (ab70244; Abcam), and anti-CoCoA (Bethyl Laboratories) antibodies and control IgG (Sigma), and used to amplify the γ-globin promoter region, from −299 to +37, by quantitative PCR. The sequences of the primers and the PCR conditions used for amplification are available upon request.
Statistical analysis
The significance of differences between independent means was assessed by Student’s t test. We considered a P value of <0.05 to be statistically significant.
Supplementary Material
Acknowledgments
We thank M. Yamamoto for the mGATA1 and hGATA1 plasmids, S. Asano, H. Matsuoka and members in our laboratories for helpful discussion. This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to M.I. and N.U.), the Global Center for Excellence Program ‘Global Center of Excellence for Education and Research on Signal Transduction Medicine in the Coming Generation’ from MEXT, Japan Brain Foundation, Takeda Science Foundation and Sagawa Foundation for Promotion of Cancer Research (to M.I.), and from a grant from the NIH (to R.G.R.).
References
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Chen W, Roeder RG. Mediator-dependent nuclear receptor functions. Semin Cell Dev Biol. 2011;22:749–758. doi: 10.1016/j.semcdb.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- Crossley M, Tsang AP, Bieker JJ, Orkin SH. Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. J Biol Chem. 1994;269:15440–15444. [PubMed] [Google Scholar]
- Doré LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118:231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- Hasegawa N, Sumitomo A, Fujita A, Aritome N, Mizuta S, Matsui K, Ishino R, Inoue K, Urahama N, Nose J, Mukohara T, Kamoshida S, Roeder RG, Ito M. Mediator Subunits MED1 and MED24 Cooperatively Contribute to Pubertal Mammary Gland Development and Growth of Breast Carcinoma Cells. Mol Cell Biol. 2012;32:1483–1495. doi: 10.1128/MCB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002;21:3464–3475. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- Jiang P, Hu Q, Ito M, Meyer S, Waltz S, Khan S, Roeder RG, Zhang X. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc Natl Acad Sci USA. 2010;107:6765–6770. doi: 10.1073/pnas.1001814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi MA, Orkin SH. Networking erythropoiesis. J Exp Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. J Biol Chem. 2004;279:49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yang CK, Stallcup MR. Downstream signaling mechanism of the C-terminal activation domain of transcriptional coactivator CoCoA. Nucleic Acids Res. 2006;34:2736–2750. doi: 10.1093/nar/gkl361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Ronchini C, Norton J, Capobianco AJ, Bresnick EH. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by notch-1. J Biol Chem. 2000;275:19676–19684. doi: 10.1074/jbc.M002866200. [DOI] [PubMed] [Google Scholar]
- Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, Blobel GA. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108:E159–168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gade P, Nallar SC, Raha A, Roy SK, Karra S, Reddy JK, Reddy SP, Kalvakolanu DV. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-γ. J Biol Chem. 2008;283:13077–13086. doi: 10.1074/jbc.M800604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CK, Simpson RJ, Kwan AH, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci USA. 2005;102:583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–324. [PubMed] [Google Scholar]
- Lu CK, Lai YC, Lin YF, Chen HR, Chiang MK. CCAR1 is required for Ngn3-mediated endocrine differentiation. Biochem Biophys Res Commun. 2012;418:307–312. doi: 10.1016/j.bbrc.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Tsai SF, Orkin SH. Increased γ-globin expression in a nondeletion HPFH mediated by an erythroid-specific DNA-binding factor. Nature. 1989;338:435–438. doi: 10.1038/338435a0. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Kim JH, Yang CK, Stallcup MR. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and β-catenin and for anchorage-independent growth of human colon carcinoma cells. J Biol Chem. 2009;284:20629–20637. doi: 10.1074/jbc.M109.014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi AK, Zhang L, Boyanapalli M, Wali A, Mohammad RM, Yu Y, Fontana JA, Hatfield JS, Dawson MI, Majumdar AP, Reichert U. Identification and characterization of a cell-cycle and apoptosis regulatory protein-1 as a novel mediator of apoptosis signaling by retinoid CD437. J Biol Chem. 2003;278:33422–33435. doi: 10.1074/jbc.M303173200. [DOI] [PubMed] [Google Scholar]
- Shimizu R, Engel JD, Yamamoto M. GATA1-related leukaemias. Nat Rev Cancer. 2008;8:279–287. doi: 10.1038/nrc2348. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M, Waskow C, Krötschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci USA. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc Natl Acad Sci USA. 2010;107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo A, Ishino R, Urahama N, Inoue K, Yonezawa K, Hasegawa N, Horie O, Matsuoka H, Kondo T, Roeder RG, Ito M. The transcriptional mediator subunit MED1/TRAP220 in stromal cells is involved in hematopoietic stem/progenitor cell support through osteopontin expression. Mol Cell Biol. 2010;30:4818–4827. doi: 10.1128/MCB.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Urahama N, Ito M, Sada A, Yakushijin K, Yamamoto K, Okamura A, Minagawa K, Hato A, Chihara K, Roeder RG, Matsui T. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells. 2005;10:1127–1137. doi: 10.1111/j.1365-2443.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Wada O, Oishi H, Takada I, Yanagisawa J, Yano T, Kato S. BRCA1 function mediates a TRAP/DRIP complex through direct interaction with TRAP220. Oncogene. 2004;23:6000–6005. doi: 10.1038/sj.onc.1207786. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci USA. 2011;108:14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CK, Kim JH, Ann DK, Stallcup MR. Differential regulation of the two transcriptional activation domains of the coiled-coil coactivator CoCoA by sumoylation. BMC Mol Biol. 2008;9:12. doi: 10.1186/1471-2199-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CK, Kim JH, Li H, Stallcup MR. Differential use of functional domains by coiled-coil coactivator in its synergistic coactivator function with β-catenin or GRIP1. J Biol Chem. 2006;281:3389–3397. doi: 10.1074/jbc.M510403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CK, Kim JH, Stallcup MR. Role of the N-terminal activation domain of the coiled-coil coactivator in mediating transcriptional activation by β-catenin. Mol Endocrinol. 2006;20:3251–3262. doi: 10.1210/me.2006-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C-X, Ito M, Fondell JD, Fu Z-Y, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Izcue A, Borggrefe T. Essential role of Mediator subunit Med1 in invariant natural killer T-cell development. Proc Natl Acad Sci USA. 2011;108:17105–17110. doi: 10.1073/pnas.1109095108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J Biol Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.