Abstract
One of the most exciting developments in the field of bacterial pathogenesis in recent years is the discovery that many pathogens utilized complex nanomachines to deliver bacterially encoded effector proteins into target eukaryotic cells. These effector proteins modulate a variety of cellular functions for the pathogen’s benefit. One of these protein-delivery machines is the type III secretion system (T3SS). T3SSs are widespread in nature and are encoded not only by bacteria pathogenic to vertebrates or plants, but also by bacteria that are symbiotic to plants or insects. A central component of T3SSs is the needle complex, a supramolecular structure that mediates the passage of the secreted proteins across the bacterial envelope. Working in conjunction with several cytoplasmic components, the needle complex engages specific substrates in sequential order, moves them across the bacterial envelope, and ultimately delivers them into eukaryotic cells. The central role of T3SSs in pathogenesis makes them great targets for novel antimicrobial strategies.
Introduction
Many bacteria have evolved specialized machines to deliver “effector” proteins into target eukaryotic cells with the capacity to modulate a variety of cellular activities (7; 60; 95). Type III protein secretion systems (T3SS) are arguably the best characterized of these protein injection machines (20; 32; 63). In the “pre-genomic era”, these systems were most often identified in the context of searches for genes involved in intimate host/pathogen interactions (61; 69; 113). However, in the “post-genomic era”, it is now clear that T3SS are widespread in nature, playing important roles not only in pathogenic but also in symbiotic interactions in the context of vertebrate, plant or insect hosts (21; 34; 130). Although the secretion machine is highly conserved across bacterial species, the effector proteins that they deliver are specific for each individual pathogen or symbiont that encode them (60). In this article, we will describe the main components of these fascinating nanomachines and will discuss what is known about their function. We will not attempt to comprehensively review the literature. Rather, we will describe what in our view are the most important aspects of the structure and function of these machines. This article will focus on T3SS involved in protein delivery into eukaryotic cells, which we view as substantially different in many important aspects from related systems involved in motility (54). In addition, we will not cover the diverse activities mediated by the effector proteins delivered by these machines. Finally, we recognize that the nomenclature of the genes involved in T3SS in different bacteria can be confusing and can hamper comparisons between different systems. Therefore to facilitate the reading of this article we provide a Table (Table 1) listing the nomenclature of homologs in the most studied systems as well as a previously introduced unifying nomenclature (71). When necessary, we will provide gene or protein names, with emphasis on homologs from T3SS of Salmonella, Shigella, and Yersinia, which are arguably the most studied.
Table 1.
Nomenclature of type III secretion components in different bacteria
| Yersinia spp. | Shigella spp | Salmonella enterica | Pseudomonas syringae | Sct common nomenclature1 | Flagellar apparatus | Proposed Function | |
|---|---|---|---|---|---|---|---|
| SPI-1 | SPI-2 | ||||||
| YscC | MxiD | InvG | SsaC | HrcC | SctC | - | Needle complex outer rings |
| YscD | MxiG | PrgH | SsaD | HrpQ | SctD | - | Needle complex inner rings |
| YscJ | MxiJ | PrgK | SsaJ | HrcJ | SctJ | - | Needle complex inner rings |
| YscR | Spa24 | SpaP | SsaR | HrcR | SctR | FliP | Export apparatus (protein channel?) |
| YscS | Spa9 | SpaQ | SsaS | HrcS | SctS | FliQ | Export apparatus (protein channel?) |
| YscT | Spa29 | SpaR | SsaT | HrcT | SctT | FliR | Export apparatus (protein channel?) |
| YscU | Spa40 | SpaS | SsaU | HrcU | SctU | FlhB | Export apparatus; substrate switching |
| YscV | MxiA | InvA | SsaV | HrcV | SctV | FlhA | Export apparatus; (proton channel?) |
| YscK | MxiK | OrgA | - | HrpD | SctK | - | Cytoplasmic sorting platform |
| YscQ | Spa33 | SpaO | SsaQ | HrcQA+B | SctQ | FliM + FliN | Cytoplasmic sorting platform |
| YscL | MxiN | OrgB | SsaK | HrpE | SctL | FliH | Links ATPase to sorting platform (?) |
| YscN | Spa47 | InvC | SsaN | HrcN | SctN | FliI | ATPase |
| YscO | Spa13 | InvI | SsaO | HrpO | SctO | - | Cytoplasmic component; function unknown |
| YscF | MxiH | PrgI | SsaG | HrpA | SctF | - | Needle filament component |
| YscI | MxiI | PrgJ | SsaI | HrpB | SctI | - | Inner rod component |
| YscP | Spa32 | InvJ | SsaP | HrpP | SctP | FliK | Inner rod assembly; substrate switching |
| LcrV | IpaD | SipD | - | - | - | Tip complex; translocase deployment | |
| YopB | IpaB | SipB | SseC | HrpK | - | Effector translocase | |
| YopD | IpaC | SipC | SseD | - | - | Effector translocase | |
| YscW | MxiM | InvH | - | - | - | Pilotin (assembly of outer rings) | |
| YopN | MxiC | InvE | SsaL | HrpJ | SctW | - | Controls translocase secretion |
Previously proposed common nomenclature (71).
Evolutionary origins of T3SS
Recent phylogenetic analyses support the notion that T3SSs are an evolutionary exaptation of the flagellar apparatus (115) in a process that may have proceeded in two steps (1). The first step, the descendants of which can still be detected in Myxococcales (81), led to a structure that is competent for protein secretion although it is no longer able to carry out motility functions. The second step, which may have occurred more than once, involved the recruitment of “secretins”, a family of outer membrane proteins that are involved in phage release or protein secretion. Composed of more than 20 proteins, T3SSs are among the most complex protein secretion systems known. Such complexity may have emerged out of the need to modulate complex cellular processes requiring the delivery of several bacterial proteins to the same eukaryotic cell. The delivery of multiple proteins to the same cell in a coordinated fashion demands adaptations much more complex than those that have evolved, for example, to deliver a single exotoxin to a target cell.
The needle complexand associated elements
The core component of the T3SS is the needle complex, a 3.5 MD multi-ring structure that spans the bacterial envelope (83) (Fig. 1). Originally identified in Salmonella Typhimurium (83), this structure has been subsequently visualized in many other bacteria (17; 38; 129) showing a highly conserved architecture. In recent years, high-resolution cryo-electron microscopy combined with the “molecular docking” of the atomic structures of some of its components has provided a remarkable high-resolution view of this structure (59; 96; 99; 100; 127; 128; 133) (Fig. 1). In addition to the structure embedded in the bacterial envelope, there are several cytoplasmic components that in a dynamic fashion associate with the needle complex to mediate specific steps of the secretion process. Finally, passage of effector proteins through the target eukaryotic cell membrane requires the assembly of a protein channel (the “translocon”), composed of proteins deployed by the T3SS itself.
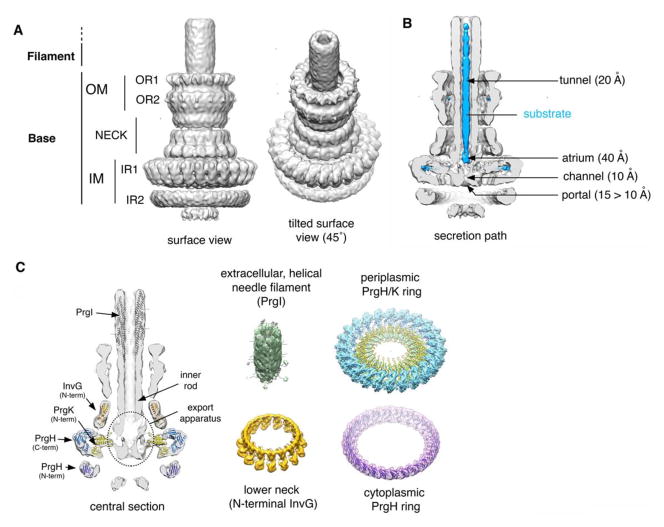
Fig. 1.
Needle complex structure from Salmonella typhimurium. A. Surface views of the 3D reconstruction of the cryo EM map of the S. typhimurium needle complex. The different substructures are noted. B. Surface view of a half-sectioned needle complex containing a trapped substrate within the central tunnel (119). Relevant structural details and dimensions are noted. C. Docking of the atomic structures of the different needle complex components onto the 3D cryo-EM map.
a) General architecture of the needle complex
The needle complex is composed of a “base” substructure embedded in the bacterial envelope and a needle-shaped extension that protrudes from the bacterial surface (24; 82; 96; 99; 100; 119; 149). The base itself is approximately 25 nm wide and 30 nm long and is composed of two rings associated with the inner membrane, inner ring 1 (IR1) and 2 (IR2), which connect to two outer membrane rings, outer ring 1 (OR1) and 2 (OR2), through a “neck” (Fig. 1). The needle itself is approximately 50 nm in length and is linked to the base through the “inner rod”, which docks into a “socket” like structure within the base. The entire structure is traversed by a channel ~20Å in diameter that serves as a conduit for proteins traveling this pathway (119). In addition to the core needle complex, there are other sub-structures often lost during its biochemical isolation. At the tip of the needle there is the “tip complex”, presumably involved in sensing the target cell and in the deployment of the protein translocases (see below) (53; 111). A group of inner membrane proteins (the “export apparatus”) located at the center of the inner rings presumably serve as a channel that mediate the passage of type III secreted proteins through the inner membrane (140). Finally, several cytoplasmic proteins are linked to the cytoplasmic side of the needle complex, presumably forming a defined structure as recently suggested by electron tomographic visualization of the needle complex in situ (2; 77; 85).
b) Structural Organization of the needle complex
The base
Despite its architectural complexity, the core needle complex is composed of a relatively small number of proteins (83). The lower rings (IR1 and IR2) are composed of two proteins, [PrgK/YscJ/MxiJ and PrgH/YscD/MxiG in Salmonella, Yersinia and Shigella, respectively, (Table 1)], while the outer rings (OR1 and OR2) and neck are composed of just one protein, (InvG/YscC/MxiD), a member of the secretin family of outer membrane proteins (Fig. 1). In Salmonella, the needle complex exhibits a three-fold symmetry in which 15 subunits of the outer ring and neck connect to 24 subunits of the IM ring (128). This observation implies that there is a local symmetry mismatch between the neck and inner ring subunits. A different stoichiometry has been proposed for needle complexes from Shigella (70), although it is unclear whether this represents real differences in the organization of needle complexes from different bacteria. PrgH and PrgK are assembled into two concentric rings 27 and 18 nm in diameter, respectively (127; 133) (Fig. 1). PrgK is composed of a larger N-terminal lipid-anchored periplasmic domain separated from a short (and in some homologs absent) cytoplasmic domain by a transmembrane segment. PrgH shares a similar architecture of two domains separated by a single transmembrane segment but with an inverted topology. However, its cytoplasmic domain is much larger than PrgK’s and forms the IR2 ring of the needle complex. The secretin that forms the outer rings of the needle complex posses a long periplasmic domain that makes the neck of the base substructure and directly contactsthe inner rings (127).
The atomic structures of the soluble domains of the base components revealed that these three proteins share a small domain with a αββαβ configuration (133). The fact that all these three proteins organize in a ring-like fashion has led to the proposal that this domain may be responsible for ring formation. This hypothesis was suggested by the observation that one of these protein homologs, EscJ (homolog of PrgK), crystalized as a superhelical structure that, when collapsed in its axis, results in a ring of dimensions compatible with those predicted for the smaller ring within IR2 (127; 133). However, this domain is present in proteins that do not form rings and even in proteins that form oligomeric rings, this domain has been shown to be dispensable for ring formation (2). Therefore the relationship between the presence of this domain and ring formation is still unclear.
The needle
The needle substructure is assembled from multiple (~100) copies of a single ~80 residue subunit, PrgI/YscF/MxiH, arranged in a helical fashion (31; 64; 84; 96) (Fig. 1). In its native form, the length of the needle ranges from 30 to 70 nm and its width ranges from 10 to 13 nm. The crystal structures of needle protomers from different T3SSs show a conserved organization consisting of an α-helical hairpin made of two α-helices of roughly the same size linked by a short segment most often containing two Prolines separated by two amino acids (the so called PXXP motif) (40; 118; 153). The structure of the in vitro assembled PrgI needle filament from Salmonella obtained by solid state NMR spectroscopy and Rossetta modeling has provided a high-resolution view of this substructure. The NMR structure shows a ~80Å wide filament with a ~25Å diameter lumen with a right-handed helical organization consisting of ~5.7 subunits per turn and a helical pitch of ~24Å (96). The assembled subunits show a short (5 amino acids) N-terminal extended domain followed by an α-helix, the PXXP loop (pointing towards the tip), and a C-terminal α-helix. The subunits are stabilized by multiple inter and intra subunit contacts resulting in a rather rigid structure. Of note is the presence of a small kink (residues Val20-Asn22 in PrgI) that interrupts the N-terminal α-helix that is not observed in the crystal structure of the soluble protomer. It is tempting to hypothesize that this structure may be involved in signal transduction upon activation of the secretion machine (see below). The solid state NMR structure shows that the N-terminal domain of PrgI faces the exterior of the needle filament while the C-terminus faces the lumen. Residues that line the lumen of the channel are highly conserved and mostly polar, and analysis of their electrostatic potential reveals alternating positive and negative charge regions. Although the significance of this observation remains to be determined, it is intriguing to hypothesize that such alternating charge distribution may be important for substrate progression within the channel. A different organization has been proposed for the needle structure of Shigella (58). However, a recent solid state NMR study has confirmed the same organization for the Shigella and Salmonella needles (43).
The inner rod
The inner rod is also built from a single ~90 amino acid subunit, PrgJ/YscG/MxiI in Salmonella/Yersinia/Shigella, respectively (99). The atomic structure solved by NMR of the PrgJ monomer from Salmonella is available but in its soluble form this protein is largely unfolded (154). However, PrgJ is predicted to have a similar structure to the needle subunit PrgI and in silico modeling has shown that these two proteins share a similar α-helical hairpin shape flanked by flexible regions. In fact, the two monomeric structures align very well around critical domains required for filament assembly (92). The modeled structure of the inner rod (based on the needle filament) suggests that, because of significant divergence at its amino terminus, the inner rod may not be able to elongate beyond two turns of the helix (~11 subunits). However, this structure has not been visualized at high resolution and its actual length is currently unknown.
Inner membrane export apparatus
All T3SSs contain five highly conserved inner membrane proteins that are essential for their function (InvA, SpaP, SpaQ, SpaR and SpaS in Salmonella) (6; 58; 62; 66; 67; 117). Recent cryo EM studies have correlated the presence of a defined density in the lumen of the inner rings of the needle complex with the presence of the inner membrane export apparatus (140). These results indicate that at least a subset of these inner membrane proteins are located within the needle complex, presumably serving as a protein channel to facilitate the export of target proteins through the inner membrane. Although these proteins are usually considered as a group, it is likely that they perform specialized functions. For example, one of these membrane proteins (InvA in Salmonella) has a large cytoplasmic domain that, as shown by structural studies, can form a circular nonamer (2). Indeed, the presence of this protein has been correlated with the presence of a toroidal shape density immediately below the cytoplasmic IR2 ring of the needle complex. Although the functional significance of this observation is unknown, it is tempting to hypothesize that this ring-structure may aid the preparation of substrates before their translocation through the inner membrane channel. Another member of this group of membrane proteins (SpaS in Salmonella) has a unique C-terminal domain, which functions as a protease for its own autocatalytic processing (49; 57; 89; 106). This processing event has been linked to altered secretion therefore this protein has been postulated to play a role in the establishment of the secretion hierarchy (see below).
Cytosolic components
There are several cytosolic proteins that are essential for secretion and are conserved across all T3SSs (29; 30; 51; 58; 110; 146) (Fig. 2). Although interactions among some of these components have been detected (74; 88; 132), the organization of these proteins within the bacterial cytoplasm remains poorly understood. In the flagellar apparatus, some of these components form a defined structure known as the “C ring”, which is involved in switching the direction of flagellar rotation (48; 79). However, there is no definitive evidence of the existence of stable C ring equivalent in T3SSs and lack of a Cring-like structure has been recently observed in tomographic reconstructions of T3SSs from different bacteria in situ (77; 85; 119). In fact, homologs of the flagellar C ring component (FliG) that anchors this structure to the flagellar body are absent from T3SSs. Nevertheless, it is likely that the homologs of C ring components form a complex that may dynamically associate with the needle complex. In fact, evidence for such a complex or “platform” has been obtained (88) (see below). Although some densities seen in tomograms of needle complexes in situ have been assigned to these components (2; 77; 85), there is still no direct demonstration of how these complexes are organized in the three dimensional space. Another highly conserved cytosolic component is an ATPase with structural similarity to F0F1 ATPases (51; 146), which is thought to be involved in substrate recognition and unfolding (4) (see below). Low-resolution tomograms of T3SS needle complex and associated structures in situ have correlated the presence of a density beneath the needle complex with the ATPase (2; 77; 85). Although it is certainly expected that the ATPase associates with the needle complex at least at some point during its functional cycle, the data available are not sufficient to establish its precise subcellular location. Indeed, at least in the flagellar system evidence is accumulating that the location of some of the cytoplasmic components is dynamic (42; 93) and therefore it is possible that the location of the ATPase may change during its functional cycle. An attractive model is that a linker protein (OrgB in Salmonella) recruits the ATPase to the sorting platform (SpaO/OrgA in Salmonella), which may bring it in close proximity to the needle complex and export apparatus. In support of this model, interactions among the relevant proteins have been demonstrated (46; 74; 88; 132). An alternative hypothesis implicates another cytoplasmic component (InvI in Salmonella), which would work in manner analogous to the “stalk” that links F0F1 ATPases to the plasma membrane. This proposal is largely (but not solely) based on the observation that the crystal structure of FliJ (the homolog in flagella) exhibits structural similarity with the “stalk” protein (73). However, the structural feature that led to this proposal is in essence a coiled-coil domain, a feature not distinctive enough to extend functional analogy to these proteins. Indeed, the coiled-coil domain of FliJ shows higher similarity to coiled coil domains of functionally unrelated proteins. Therefore more studies are needed to ascertain how much the analogy to ATP synthases can be extended to the T3SSs components.
Fig. 2.
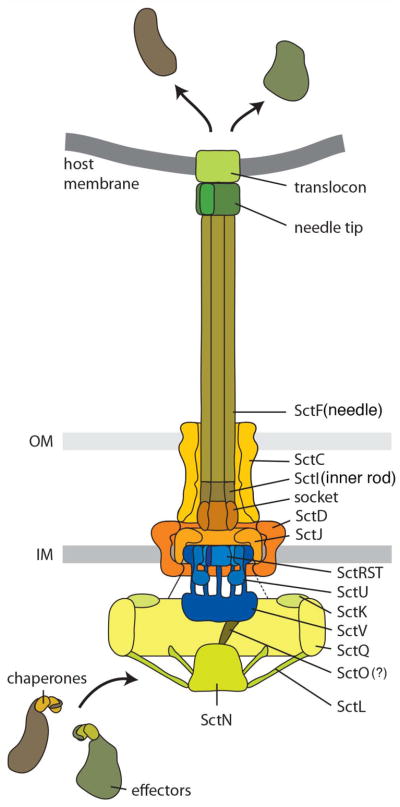
Diagram of the needle complex and associated structures. A previously suggested common nomenclature (71) was used to indicate the potential localization of the different components and facilitate comparison across different systems.
The needle tip structure and needle extension
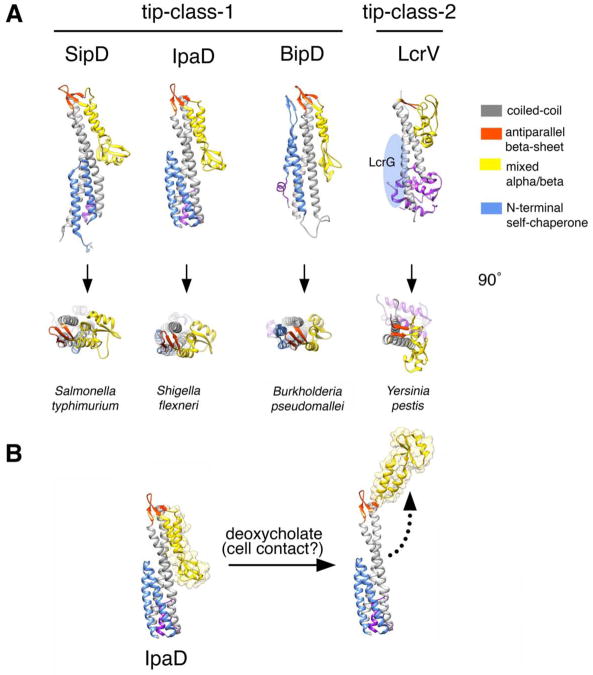
The needle filament is either “capped” at its tip by a single protein (53; 111) or it is extended by yet another filament that is longer in length than the needle itself (80). Based on their structure similarities the “tip” proteins can be classified in two related groups: the SipD/IpaD (from Salmonella and Shigella) and the LcrV/PcrV (from Yersinia and Pseudomonas) families (Fig. 3). The SipD/IpaD family displays a distinct domain organization: an N-terminal α-helical hairpin, a central coiled-coil domain, and a C-terminal region composed of mixed structural elements (55; 75). The LcrV family has the conserved central coiled-coil domain but lacks the N-terminal α-helical hairpin and has and extended globular domain at is amino terminus that is absent from the SipD/IpaD family (44). Specific functions have been proposed for some of these defined structural elements. For example, it is thought that the highly conserved central coil-coiled links the tip proteins to the end of the needle filaments presumably through interactions that in many ways may resemble those that link the needle protomers in the needle filament (97; 120; 152). The α-helical hairpin has been proposed to function as a self-chaperone preventing the self-oligomerization of SipD/IpaD within the bacterial cytoplasm (75). In the LcrV/PcrV family, which lacks this domain, the chaperone function is thought to be carried out by a cytoplasmic protein (LcrG/PcrG) (41; 101), which is absent in bacteria encoding members of the IpaD/SipD family. Under low-resolution EM the LcrV needle tip shows a “head, neck and base” configuration (19). In contrast, the IpaD tip complex visualized by low-resolution cryo EM exhibits a 5-fold symmetric “scepter”-like structure with a diameter of 78Å at its widest point (53). Docking of the crystal structure of the IpaD monomer was possible only if a large conformation change of the C-terminal domain was introduced into the model, suggesting that the tip protein may undergo significant conformational changes upon assembly into the tip complex.
Fig. 3.
The tip complex of type III secretion systems. A. Crystal structures of tip proteins from different bacteria. Relevant structural features are noted. B. Conformational changes in the IpaD tip protein induced by the binding of deoxycholate, which is thought to mimic the activation event that occurs upon contact with target cells.
A substantial variation in the structure at the tip of the needle filament occurs in T3SSs from some pathogenic strains of E. coli. In these T3SSs the needle extends into a long filament that presumably serves as a link between the bacteria and its target cell (80). In E. coli the filament is made of a single protein, EspA, which is structurally similar to FliC and assembles into a helical structure similar to the needle filament (150). However, in contrast to the rigid needle, the helical filament extension appears to be flexible since despite having a fixed twist of 5.6 subunits per turn, its axial rise varies substantially from 3.6 Å to 5.6 Å (142). The functional significance of this observation is currently unknown.
The translocon
The last step in the T3SS-mediated delivery of effector proteins into eukaryotic cells is their passage through the target host-cell membrane (see below). This process is mediated by the protein translocases (SipB/SipC, IpaB/IpaC, and YopB/YopD in Salmonella, Shigella and Yersinia, respectively) (28; 122). After secretion by the T3SS, the translocases insert into the target host cell membrane where they presumably form a protein channel (16; 112). Although the protein translocases are not well conserved at the primary amino acid sequence level, they are all α helical proteins with transmembrane helixes (72; 109; 124). The crystal structures of the amino terminus of IpaB and SipB revealed the presence of a trimeric coiled-coil domain formed by three antiparallel α helixes, an organization reminiscent of other membrane-active proteins such as colicins or viral envelope glycoproteins (13). The mechanism by which the translocases insert in the membrane is not understood although it is likely that is orchestrated by the tip complex. Indeed, in the absence of the tip protein, the translocases cannot insert into the host-target membrane although they can be efficiently secreted (28; 68; 98; 139).
Needle complex assembly
Assembly of the needle complex and associated structures occurs in a step-wise fashion (46; 47; 136; 140) (Fig. 4). The sec machinery mediates the export or membrane insertion of all the base and inner membrane export apparatus components prior to their assembly into the final base substructure. Assembly starts at the inner membrane with the formation of a complex of a subset of the export apparatus components (SpaP and SpaR in Salmonella) (140), followed by the incorporation of additional export apparatus components, which finally template the assembly of the lower rings, first the inner ring of IR1 (made of PrgK in Salmonella) and then the outer rings of IR1 and IR2 made of PrgH (see Fig. 4). Several observations support this model. Although assembly of the needle complex can occur in the absence of the inner membrane export apparatus, the efficiency of assembly is drastically reduced. Furthermore, at least 4 of the 5 inner membrane apparatus components (SpaP, SpaQ, SpaR and SpaS in Salmonella) cannot be incorporated into previously assembled bases indicating that functional needle complexes cannot be assembled without the prior deployment of the export apparatus (140). The outer rings and neck are formed by a single protein of the secretin family (InvG in Salmonella), a process that in most (but not all) systems requires the assistance of an accessory lipoprotein of the “pilotin” family (InvH in Salmonella). The outer and inner rings of the base substructure can assemble independently although their stability in the absence of one another is compromised (46). Therefore, it is likely that these two structures are assembled independently but that are rapidly linked to one another to form a stable structure.
Fig. 4.
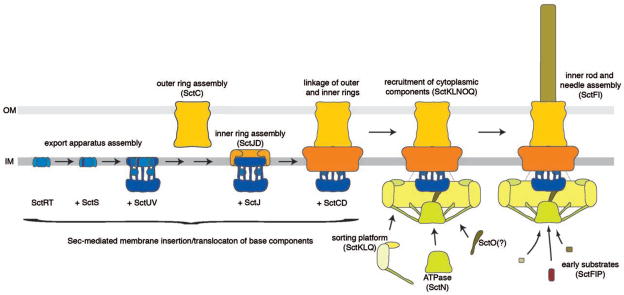
Model for the assembly of the type III secretion needle complex and associated structures. To facilitate comparison of the assembly pathway in different type III secretion systems, a previously suggested nomenclature of the different components was used (71) (see Table 1). See text for details.
Once the base substructure is assembled, several cytoplasmic proteins must be recruited so that the base can become competent for type III secretion. Although this process is poorly understood the available evidence suggests that some of these cytoplasmic factors may be pre-assembled into a complex prior to their recruitment to the base. For example, it is likely that the components of the sorting platform (OrgA, and SpaO) may exist in the cytoplasm in a pre-assembled estate (88). Whether the ATPase is part of that pre-assembled complex is unclear but it is known that the recruitment of the ATPase to the needle complex requires the components of the sorting platform (2; 46). How the sorting platform is recruited to the needle complex or even what specific interactions bring this cytoplasmic complex to that site is not known. Although it has been proposed that the cytoplasmic domain of the InvA family of proteins is involved in the recruitment of cytoplasmic components (108), recent electron tomography studies suggest otherwise (2). Most likely components of the needle complex itself may serve as an anchor for this recruitment since in its absence there is no recruitment of cytoplasmic components to the membrane.
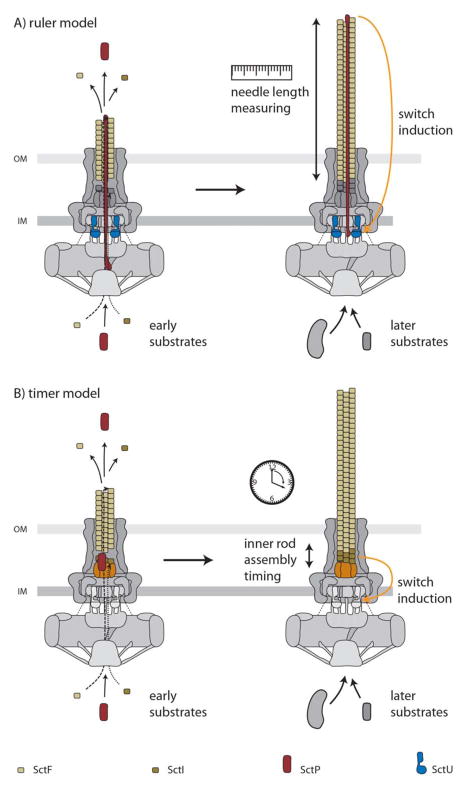
Once the cytoplasmic components are recruited, the base substructure can function as a limited-specificity type III protein secretion machine that can only recognize the inner rod and needle protomers as well as an accessory protein required for proper needle complex assembly (InvJ/YscP in Salmonella and Yersinia). Like flagella, assembly of the needle substructure occurs by sequential addition of subunits at the growing tip (37; 118). Unlike flagella, however, assembly of the T3SS needle does not require a “capping” protein that facilitates addition of the subunits at the tip. Assembly of the inner rod is less well understood but, unlike the needle substructure, it requires the function of an accessory protein (InvJ in Salmonella), which like the flagellar “capping” proteins, does not form part of the final structure and is discarded in the culture supernatant (92; 99). In the absence of InvJ, assembly of the inner rod does not take place although needles of improper length (see below) can efficiently assemble. At some point during the assembly process the type III secretion machine switches substrate specificity so that it can no longer recognize the early substrates (needle and inner rod protomers) and becomes competent for the secretion of middle (i. e. the tip protein and translocases) and late substrates (i. e. effector proteins). The mechanisms by which the secretion machine is reprogramed are incompletely understood although the accessory protein InvJ/YscP is required for this process. In its absence, the needle complex assembles abnormally long needles because it is unable to switch substrates (84). The function of this accessory protein is incompletely understood and the subject of some controversy. For example, YscP has been proposed to function as a molecular ruler, measuring the length of the needle and triggering substrate switching once an appropriate length of the needle is achieved (76) (Fig. 5). How this protein would measure the length is not understood but it has been proposed that the fully extended form of YscP located within the lumen of the secretion channel performs the “measuring” by interacting with proteins at the tip and at the base of the needle complex (YscU in Yersinia, see below) triggering substrate switching. Support for this model comes from the observation that alterations in the length of YscP result in needles of different length (76). A significant caveat for these experiments is that measurements were not made with isolated needle complexes but on “shed’ needles, which can be subject to artifacts. In addition, artificial elongation of YscP could result in partial loss of function that could lead to longer needles (see below), which can further complicate the interpretation of these results. Finally, this model is not compatible with the fact that needle length in wild type needle complexes varies substantially following a rather broad length distribution (99). This distribution would not be expected if a molecular ruler mechanism was involved in length determination and, instead, it suggests a stochastic process. An alternative model has been proposed based on the observation that InvJ is required for the assembly of the inner rod, a process that leads to the firm anchoring of the needle filament to the base (92; 99) (Fig. 5). Anchoring of the needle results in substantial conformational changes on the cytoplasmic face of the needle complex (99; 100), which is hypothesized to, directly or indirectly, trigger substrate switching. Therefore in this model the termination of the assembly of the inner rod is the critical event that determines substrate switching. In this case the role of InvJ in needle determination is indirect through its role in inner rod assembly. It has been proposed that SpaS/YscU, a component of the export apparatus, may also be involved in the mechanisms of substrate switching (18; 49; 121; 148). As discussed above, SpaS/YscU protein family possesses a long cytoplasmic domain with autocatalytic protease activity (57). Mutations in the catalytic site result in a strain that is competent for secretion of effectors but unable to secrete the protein translocases (49). It has been proposed that the autoproteolitic cleavage of SpaS, which is hypothesized to be triggered by its putative interaction with InvJ/YscP, may determine substrate switching. Although there is no question that the catalytic mutant of SpaS exhibits altered secretion, needle length control is unaffected in this mutant, which is inconsistent with its proposed role in needle length determination.
Fig. 5.
Proposed models for the mechanism of substrate switching and needle length control in the assembly pathway of the needle complex. See text for details.
Substrate recognition by the type III protein secretion machine
Proteins destined to travel the type III secretion pathway are targeted to the secretion machine by a set of secretion signals that ensure specificity (11). One of the secretion signals encompasses the first 20–25 amino acids (105; 131). This signal is highly variable in sequence and can often tolerate significant changes without affecting function (10; 123), an observation that led to the proposition that the 5′ mRNA of some type III secreted proteins was responsible for their targeting (8; 9). However, it is now believed that the amino acid sequence acts as the targeting element and that the tolerance for mutagenesis stems from the fact that the targeting signal is indeed unstructured. Furthermore, bioinformatic analyses have identified different common features in this amino acid sequence (102). These features include enrichment in serine and threonine and depletion of charge and hydrophobic residues such as leucine. A second signal, at least in some type III secreted proteins, serves as binding site for specific chaperones and spans from residues ~25–100 (26; 131; 143; 144). Unlike other chaperones, such as the GroEL and DnaK/Hsp70 protein family, T3SS-associated chaperones lack nucleotide-binding or hydrolysis activities. T3SS chaperones exhibit limited primary amino-acid sequence similarity to one another although they share structural similarity as well as some physical properties, such a small size and an acidic pI (135). Based on their tertiary structure and specificity of binding, these chaperones have been classified in two general groups referred to as class I and class II (33; 116; 137). An additional distinction is usually made among class I chaperones between those that bind a single protein (unicargo) vs those that bind several (multicargo). T3SS chaperones usually form homodimers along a helical interface and exhibit extended hydrophobic patches and a large hydrophobic groove that accommodates the amino terminal region of their cognate effectors (14; 134). A notable feature of this interaction is that the chaperone-bound effector domain is completely non-globular although it retains significant amounts of secondary structure. Since substrates are transported in at least a partially unfolded state (119), it is believed that this configuration aids the secretion process. Although the primary sequence of chaperone-binding domains varies greatly, some common motifs have been identified. For example, a β-strand motifs is present at the amino-terminal region of many effectors and a conserved chaperone-binding domain consensus sequence [(LMIF)1XXX(IV)5XX(IV)8XN10] that overlaps with the β-motif has also been identified (35; 94). Multi cargo chaperones exhibit a similar interacting interface with their cognate effectors although usually burying less surface (94). In the absence of chaperones, particularly those with a single cargo, the cognate effector proteins are degraded within the bacterial cytoplasm (103; 144). This is often not the case for effectors chaperoned by multi cargo chaperones (50; 90). In this case it is possible that the stability of these effectors in the absence of cognate chaperone may be necessary to facilitate complex assembly since they are usually encoded away from their cognate chaperones. This is most often not the case for single cargo chaperones, which are not only encoded in the immediate vicinity of their cognate cargo but also, in some instances, have specific translation regulatory mechanisms in place to coordinate their synthesis with that of their effector (22). It is now well accepted that the main function of these chaperones is to target their cognate effectors to the secretion pathway. Absence of these chaperones results in lack of secretion of their corresponding effectors. Furthermore, removal of the chaperone-binding domain also prevents secretion through their cognate type III secretion pathway (147) (125), although mistargeting to the flagellar secretion pathway (by the amino terminal secretion sequence) can occur (91). Class II chaperones usually interact with protein translocases and, in some T3SS, with protomers of the needle or of the extended appendages such as EspA (36; 103). The structural feature that characterizes this type of chaperones is the presence of tetratricopeptide repeats (23). Although it is clear that these chaperones stabilize or prevent detrimental interactions of their cognate target proteins, their role in secretion is still unclear.
The mechanisms by which substrates of the type III secretion systems are ultimately recognized are poorly understood. The chaperone-effector complexes are most likely recognized and targeted to the secretion machinery by a group of cytoplasmic proteins that are associated with the needle complex such as the “sorting platform” (see above). It has also been shown that the ATPase interacts with chaperone/effector complexes (65) and that it is able to dissociate them (4), a necessary step before secretion since the chaperones remain in the cytoplasm after secretion of the cognate effector. Furthermore, chaperones are essential for the recruitment of the secreted proteins to the sorting platform (88). It is therefore possible that the chaperones may be involved in the establishment of the secretion hierarchy perhaps by exhibiting different affinity to the relevant sorting platform components. Subsequent to their recognition by the secretion machine, proteins must be at least partially unfolded prior to or simultaneously with their delivery to the secretion channel, which is too narrow to accommodate folded proteins. The ATPase is capable of unfolding effector proteins in vitro so it is possible that it plays an equivalent role in vivo (4).
Energizing the type III secretion system
Although there are no reliable measurements of the speed at which proteins are moved through individual type III secretion machines, the available estimates suggest that the process must be fast (126). Furthermore, the type III secretion machine is able to deliver proteins that have been engaged on the bacterial cytoplasm directly into the host target cell. Undoubtedly these activities must demand a significant amount of energy. There are at least two possible sources of energy for this system. One source is likely derived from the hydrolysis of ATP by the conserved T3SS-associated ATPase since it is known that its catalytic activity is essential for secretion (52; 146). How ATP hydrolysis could be coupled to the secretion process is incompletely understood. However, since at least in vitro these ATPases can unfold the effector proteins (3), it is possible that the energy “stored” in the unfolded proteins may contribute to the progression of substrates through the secretion channel. Several pieces of evidence indicate that the proton motive force is also required for type III secretion (145). However, how the PMF is potentially coupled to the secretion process is unknown. It has been proposed that a conserved component of the inner membrane export apparatus in the flagellar system can function as a proton-protein antiporter that uses the two components of proton motive force, Δψ and ΔpH, for protein export (107). However, definitive demonstration of this hypothesis awaits further investigation. Recently a radically different mechanism has been proposed to explain the movement of flagellar subunits within the flagellar channel (56). This model proposes a pulling mechanism derived from the crystallization of subunits at the growing flagellar tip that would harness the entropic energy of the unfolded subunits. In this model, subunits would be linked in a head-to-tail configuration within the flagellar channel so that the crystallization of subunits at the tip would “pull” all the subunits in the channel. Although this model could potentially account for the movement of the subunits that are destined to form the needle substructure of the needle complex, it is unclear how this model could explain the movement of effector proteins. Effector proteins do not crystalize at the tip and their diversity makes the proposed head-to-tail arrangement within the secretion channel hard to accommodate. Furthermore, this model is at odds with previous thermodynamic calculations, which concluded that diffusion could account for the movement of flagellar subunits within the flagellar channel (138; 151). Therefore, more experiments will be required to explore the universality and validity of this new proposal.
Sensing and firing: type III secretion machines in action
Type III secretion machines require an activating signal before they can be competent for protein secretion and delivery. Although the activation process is poorly understood, there is compelling evidence that activation occurs upon contact with target cells (104; 155). Such a mechanism presumably ensures that effector proteins are delivered directly to target cells and not to the extracellular space, where they would be functionally irrelevant. How cell contact activates the secretion machine is unknown but most likely the tip complex is involved in the sensing process (15). In fact, some compounds that bind the tip complex such as bile salts or congo red, can stimulate type III secretion (12; 114) presumably by introducing specific conformational changes in the tip protein that probably resemble those induced by cell contact (25; 45; 141) (Fig. 3). The signal presumably sensed by the tip complex must be transduced to the cytoplasmic side of the secretion machine, a process most likely mediated by conformational changes in the needle and inner rod substructures of the needle complex (15). In support of this hypothesis, several mutations in the needle and inner rod proteins have been identified, which result in de-repressed and/or otherwise altered patterns of protein secretion (27; 39; 78; 92; 139). Activation of the T3SS ultimately leads to the deployment of the translocases on the target cell membrane, which in turn will mediate the passage of effectors through the target cell plasma membrane. In this model, the secretion machine must engage the translocases prior to the effectors, a mechanism that most likely involves a cytoplasmic sorting platform. Consistent with this model, prior to activation of the secretion machine, only the translocases are found on the sorting platform and it is only in the absence of the translocases that effector proteins can be detected at this location (88). Deployment of the translocases leads to intimate attachment of the bacteria to the target host cell, which presumably aids the translocation process (87). An alternative “two-step model” has been proposed in which effector proteins are first delivered to the bacterial surface and a second step, akin to the mechanisms of AB toxins, in which the effectors are moved through the plasma membrane by the protein translocases (5). This model was proposed based on the observation that, under certain conditions, effectors are seen on the surface of Yersinia and that effector proteins artificially deposited on the bacterial surface can be translocated into eukaryotic cells. However, in other bacteria no effectors are seen on the bacterial surface prior to target cell contact (86; 104; 155). Furthermore, it is possible that the observed translocation of exogenously applied effector proteins (5) is the result of the artificial capture of translocation intermediates on the bacterial surface. More experiments will be required to support this model, which is inconsistent with a substantially amount ofavailable data.
Outstanding questions
Although a great deal is known about the structure of the type III secretion machine, there are still protein densities observed in the high-resolution cryo-electron microscopic map that are unaccounted for. Some of these densities are likely to represent transmembrane domains of export apparatus components, whose crystal structures are not yet available. Short of solving the atomic structure of the entire needle complex, the solution of the atomic structures of these membrane proteins is clearly one of the major challenges for the future. The actual mechanism of secretion is still poorly understood and there are many outstanding questions that are likely to guide and inspire future research. How is the host cell sensed and how is the signal transduced to the secretion machinery, in particular to its cytoplasmic components? If the needle and inner rod are indeed the signal transducers, how do they accomplish this function? Are they as rigid as their structures suggest or do they transduce signals by yet undetected conformational changes? How does the sorting platform select its substrates and deliver them to the secretion channel? Are there differences in the recognition mechanisms of translocases and effectors? How do the substrates move through the secretion channel and what is the source of energy that drives the secretion process? How do the effectors traverse the target cell plasma membrane? The last 10 years have seen remarkable progress in the understanding of the structure and organization of the type III secretion machine. However this knowledge has been largely focus on snap shots of the machine with limited studies focusing on the dynamic aspects of the secretion machine. With the availability of powerful live imaging tools, it is expected that during the next few years we will be able to catch it in action. Finally, the central role of T3SS in the pathogenesis of several bacterial pathogens of great public health importance has stimulated efforts to develop novel therapeutic strategies targeted to this machine. It is expected that during the next 10 years these efforts may begin to translate into effective therapeutics.
Acknowledgments
We apologize to our colleagues that due to the strict limits imposed on the length of the manuscript and the limited number of references that can be cited, we have not been able to discuss or cite important papers. Research in the Galán laboratory related to work discussed in this article is supported by Grants AI030492 and AI055472 from the National Institutes of Health.
References cited
- 1.Abby S, Rocha EPC. The non-flagellar type III secretion system evolved form the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genetics. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby M, Hendrixon D, et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akeda Y, Galan JE. Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J Bacteriol. 2004;186:2402–12. doi: 10.1128/JB.186.8.2402-2412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–5. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 5.Akopyan K, Edgren T, HW-E, Rosqvist R, Fahlgren A, et al. Translocation of suface-localizd effectors in type III secretion. Proc Natl Acad Sci USA. 2011;108:1639–44. doi: 10.1073/pnas.1013888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allaoui A, Woestyn S, Sluiters C, Cornelis GR. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–42. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Martinez C, Christie P. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DM, Fouts DE, Collmer A, Schneewind O. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc Natl Acad Sci U S A. 1999;96:12839–43. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson DM, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–3. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 10.Anderson DM, Schneewind O. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol. 1999;31:1139–48. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 11.Arnold R, Jehl A, Rattei T. Targetting effectors: the molecular recognition of type III secreted proteins. Microbes Infect. 2010;12:346–58. doi: 10.1016/j.micinf.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Bahrani FK, Sansonetti PJ, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infection & Immunity. 1997;65:4005–10. doi: 10.1128/iai.65.10.4005-4010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barta ML, Dickenson NE, Patil M, Keightley A, Wyckoff GJ, et al. The structures of coiled-coil domains from type III secretion system translocators reveal homology to pore-forming toxins. J Mol Biol. 2012;417:395–405. doi: 10.1016/j.jmb.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–80. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- 15.Blocker A, Deane J, Veenendaal A, Roversi P, Hodkinson J, et al. What’s the point of the type III secretion needle? Proc Natl Acad Sci USA. 2008;105:6507–13. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–93. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, et al. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–63. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 18.Botteaux A, Kayath C, Page A, Jouihri N, Sani M, et al. The 33 carboxyl-terminal reisdues of Spa40 orchestrate the multi-step assembly process of the type III secretion needle complex in Shigella flexneri. Microbiology. 2010;156:2807–17. doi: 10.1099/mic.0.039651-0. [DOI] [PubMed] [Google Scholar]
- 19.Broz P, Mueller CA, Muller SA, Philippsen A, Sorg I, et al. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol. 2007;65:1311–20. doi: 10.1111/j.1365-2958.2007.05871.x. [DOI] [PubMed] [Google Scholar]
- 20.Buttner D. Protein export according to schedule: architecture, assembly, and regulatin of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buttner D, He S. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–64. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Button JE, Galan JE. Regulation of chaperone/effector complex synthesis in a bacterial type III secretion system. Mol Microbiol. 2011;81:1474–83. doi: 10.1111/j.1365-2958.2011.07784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, et al. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun. 2013;81:629–35. doi: 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee S, Battaile K, Lovell S, Plano G, De Guzman R. Structure and biophysics of type III secretion in bacteria. Biochemistry. 2013;52:2508–17. doi: 10.1021/bi400160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee S, Zhong D, Nordhues B, Battaile K, Lovell S, De Guzman RN. The crystal structure of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Portein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng LW, Anderson DM, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Molecular Microbiology. 1997;24:757–65. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 27.Cherradi Y, Schiavolin L, Moussa S, Megharoui A, Meksem A, et al. Interplay between predicted inner-rod and gatekeeper in controlling substrate speciicity of the type III secretion system. Mol Microbiol. 2013;87:1183–99. doi: 10.1111/mmi.12158. [DOI] [PubMed] [Google Scholar]
- 28.Collazo C, Galán JE. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–56. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 29.Collazo CM, Galan JE. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–31. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collazo CM, Zierler MK, Galan JE. Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 31.Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003 doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 32.Cornelis G. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–25. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 33.Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–74. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 34.Correa V, Majerczak D, Ammar e-D, Merighi M, Pratt R, et al. The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl Environ Microbiol. 2012;78:6327–36. doi: 10.1128/AEM.00892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa S, Schmitz A, Jahufar F, Boyd J, Cho M, et al. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. MBio. 2012;3:e00243–11. doi: 10.1128/mBio.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Creasey E, Friedberg D, Shaw R, Umanski T, Knutton S, et al. CesAB is an enteropathogenic Escherichia coli chaperone for the type III translocator EspA and EspB. Microbiology. 2003;149:469–77. doi: 10.1099/mic.0.26735-0. [DOI] [PubMed] [Google Scholar]
- 37.Crepin V, Shaw R, Abe C, Knutton S, Frankel G. Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J Bacteriol. 2005;187:2881–9. doi: 10.1128/JB.187.8.2881-2889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniell S, Takahashi N, Wilson R, Friedberg D, Rosenshine I, et al. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:865–71. doi: 10.1046/j.1462-5822.2001.00168.x. [DOI] [PubMed] [Google Scholar]
- 39.Davis A, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specfically abrogate effector translocation into host cellls. J Bacteriol. 2010;189:83–97. doi: 10.1128/JB.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deane JE, Roversi P, Cordes F. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci USA. 2006;103:12529–33. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBord K, Lee V, Schneewind O. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J Bacteriol. 2001;183:4588–98. doi: 10.1128/JB.183.15.4588-4598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delalez N, Wadhams G, Rosser G, Xue Q, Brown M, et al. Signal-dependent turnover of teh bacterial flagellar swith protein FliM. Proc Natl Acad Sci USA. 2010;107:11347–51. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demers J, Sgourakis N, Gupta R, Loquet A, Giller K, et al. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog. 2013 Mar;9(3):e1003245. doi: 10.1371/journal.ppat.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derewenda U, Mateja A, Devedjiev Y, Routzahn KM, Evdokimov AG, et al. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–6. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Dickenson NE, Zhang L, Epier C, Adam P, Picking W, Picking WD. Cpnformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry. 2011;50:172–80. doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis G. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–40. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diepold A, Wiesand U, Cornelis G. The assembly of the export apparatus (YscR,-S,-T,-U,-V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol Microbiol. 2011;82:502–14. doi: 10.1111/j.1365-2958.2011.07830.x. [DOI] [PubMed] [Google Scholar]
- 48.Driks A, DeRosier D. Additional structures associated with bacterial flagellar basal body. J Mol Biol. 1990;211:669–72. doi: 10.1016/0022-2836(90)90063-R. [DOI] [PubMed] [Google Scholar]
- 49.Edqvist P, Olsson J, Lavander M, Sundberg L, Forsberg A, et al. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–66. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehrbar K, Friebel A, Miller S, Hardt W. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmoella effector proteins encoded outsite of SPI-1. J Bacteriol. 2003;185:7279–84. doi: 10.1128/JB.185.23.6950-6967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichelberg K, Ginocchio C, Galán JE. Molecularandfunctional characterization of the Salmonella typhimurium invasion genes invB and invC: Homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–10. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eichelberg K, Ginocchio CC, Galan JE. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–10. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epier C, Dickenson N, Bullitt E, Picking W. Ultrastructural analysis of IpaDa the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol. 2012;420:29–39. doi: 10.1016/j.jmb.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erhardt M, Namba K, Hughes K. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Wong Fat Sang N, et al. High Resolution Structure of BipD: An InvasionProtein Associated with the Type III Secretion System of Burkholderia pseudomallei. J Mol Biol. 2006;363:125–36. doi: 10.1016/j.jmb.2006.07.069. [DOI] [PubMed] [Google Scholar]
- 56.Evans L, Poulter S, Terentjev E, Hughes C, Fraser G. A chain mechanism for flagellar growth. Nature. 2013;504:287–90. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferris H, Furukawa Y, Minamino T, Kroetz M, Kihara M, et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–42. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 58.Fields KA, Plano GV, Straley SC. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–79. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuji T, Cheung M, Blanco A, Kato T, Blocker A, Namba K. Structure of a type III secretion needle at 7 A resolution privdes insight into its assembly and signaling mechanisms. Proc Natl Acad Sci USA. 2012;109:4461–6. doi: 10.1073/pnas.1116126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galán J. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–9. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galán JE, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sc USA. 1989;86:6383–7. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galán JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: Homology of InvA to members of a new protein family. J Bacteriol. 1992;17:4338–49. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–73. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 64.Galkin V, Schmied W, Schriadt O, Marlovits T, Egelman E. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J Mol Biol. 2010;396:1392–7. doi: 10.1016/j.jmb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gauthier A, Finlay BB. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J Bacteriol. 2003;185:6747–55. doi: 10.1128/JB.185.23.6747-6755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ginocchio CC, Galan JE. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect Immun. 1995;63:729–32. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groisman EA, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–87. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harmon D, Murphy J, Davis A, Mecsas J. A mutant with aberrant extracellular LcrV-YscF interactions fails to form pores and translocate Yop effector proteins but retains the ability to trigger Yop secretion in response to cell contact. J Bacteriol. 2013;195:2244–54. doi: 10.1128/JB.02011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–3. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 70.Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16:477–85. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hume PJ, McGhie EJ, Hayward RD, Koronakis V. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol Microbiol. 2003;49:425–39. doi: 10.1046/j.1365-2958.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 73.Ibuki T, Imada K, Minamino T, Kato T, Miyata T, Namba K. Common architecture of the flagellar type III protein export apparatus and F- and V- type ATPases. Nat Struct Mol Biol. 2011;18:277–82. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]
- 74.Jackson M, Plano G. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol Lett. 2000;186:85–90. doi: 10.1111/j.1574-6968.2000.tb09086.x. [DOI] [PubMed] [Google Scholar]
- 75.Johnson S, Roversi P, Espina M, Olive A, Deane JE, et al. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–44. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–60. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 77.Kawamoto A, Morimoto Y, Miyata T, Minamino T, Hughes K, et al. Common and distinct structural featues of Salmonella injectisome and flagellar basal body. Sci Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kenjale R, Wilson J, Zenk S, Saurya S, Picking W, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–37. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 79.Khan I, Reese T, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–60. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO Journal. 1998;17:2166–76. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Konovalova A, Petters T, Sagaard-Andersen L. Extracellular biology of Mycococcus xanthus. MS Microbiol Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 82.Kosarewicz A, Konigsmaier L, Marlovits T. The blueprint of the type 3 injectisome. Philos Trans R Soc London B Biol Sci. 2012;367:1140–54. doi: 10.1098/rstb.2011.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 84.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A. 2000;97:10225–30. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kudryashev M, Stenta M, Schmelz S, Amstutz M, Wiesand U, et al. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2013;2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–42. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lara-Tejero M, Galan JE. The Salmonella Typhimurium SPI-1 type III secretion translocases mediate intimate attachment to non-phagocytic cells. Infect Immun. 2009;77:2635–42. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–91. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lavander M, Sundberg L, Edqvist P, Lloyd S, Wolf-Watz H, Forsberg A. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–9. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee SH, Galan JE. InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE. J Bacteriol. 2003;185:7279–84. doi: 10.1128/JB.185.24.7279-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–95. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 92.Lefevre M, Galan J. The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1319698111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lele P, Branch R, Nathan V, Berg H. Mechanism for adaptive remodeling of teh bacterial flagellar switch. Proc Natl Acad Sci USA. 2012;109:20018–22. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lilic M, Vujanac M, Stebbins C. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell. 2006;21:653–64. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 95.Llosa M, Roy C, Dehio C. Bacterial type IV secretion systems in human disease. Mol Microbiol. 2009;73:141–51. doi: 10.1111/j.1365-2958.2009.06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loquet A, Sgourakis N, Gupta R, Giller K, Riedel D, et al. Atomic model of the type III secretion system needle. Nature. 2012;486:276–9. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI-SipD: Insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marenne M, Journet L, Mota L, Cornelis G. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersnia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog. 2003;35:243–58. doi: 10.1016/s0882-4010(03)00154-2. [DOI] [PubMed] [Google Scholar]
- 99.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–40. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 100.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–2. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matson JS, Nilles ML. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J Bacteriol. 2001;183:5082–91. doi: 10.1128/JB.183.17.5082-5091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McDermott J, Corrigan A, Peterson E, Oehmen C, Niemann G, et al. Computational Prediction of Type III and IV Secreted Effectors in Gram-Negative Bacteria. Infect Immun. 2011;79:23–32. doi: 10.1128/IAI.00537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Menard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;4:515–25. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 104.Ménard R, Sansonetti PJC. The secretion of the Shigella flexneri Ipa invasins is induced by the epithelial cell and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Michiels T, Cornelis GR. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–85. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Minamino T, Macnab R. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol. 2000;182:4906–14. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Minamino T, Morimoto Y, Hara N, Namba K. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun. 2011;2:475–83. doi: 10.1038/ncomms1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Minamino T, Shimada M, Okabe M, Saijo-Hamano Y, Imada K, et al. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagella type III protein export. J Bacteriol. 2010;192:1929–36. doi: 10.1128/JB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Montagner C, Arquint C, Cornelis GR. Translocators YopB and YopD from Yersinia enterocolitica form a multimeric integral membrane complex in eukaryotic cell membranes. J Bacteriol. 2011;193:6923–8. doi: 10.1128/JB.05555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281:599–60. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 111.Mueller C, Broz P, Muller S, Ringler P, Erne-Brand F, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–6. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 112.Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: Requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–81. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 113.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification Of a Pathogenicity Island Required For Salmonella Survival In Host Cells. Proc Natl Acad Sc USA. 1996;93:7800–4. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Olive A, Kenjale R, Espina M, Moore D, Picking W, Picking W. Bile salts stimulate recruitment of IpaB to the Shigella flexnei surface, where it colocalizes with IpaD at the tip fo the type III secretion needle. Infect Immun. 2007;75:2626–9. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pallen M, Beatson S, Bailey C. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol Rev. 2005;29:201–29. doi: 10.1016/j.femsre.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 116.Parsot C, Hamiaux C, ALP The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;56:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 117.Plano GV, Barve SS, Straley SC. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, et al. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol. 2010;17:788–92. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- 119.Radics J, Konigsmaier L, Marlovits T. Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2722. [DOI] [PubMed] [Google Scholar]
- 120.Rathinavelan T, Tang C, De Guzman RN. Characterization of the Interaction between the Salmonella Type III Secretion System Tip Protein SipD and the Needle Protein PrgI by Paramagnetic Relaxation Enhancement. J Biol Chem. 2011;286:4922–30. doi: 10.1074/jbc.M110.159434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008;68:1485–501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rosqvist R, Persson C, Hakansson S, Nordfeldt R, Wolf-Watz H. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contributions to Microbiology & Immunology. 1995;13:230–4. [PubMed] [Google Scholar]
- 123.Russmann H, Kubori T, Sauer J, Galan J. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol Microbiol. 2002;46:769–79. doi: 10.1046/j.1365-2958.2002.03196.x. [DOI] [PubMed] [Google Scholar]
- 124.Ryndak MB, Chung H, London E, Bliska JB. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect Immun. 2005;73:2433–43. doi: 10.1128/IAI.73.4.2433-2443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schesser K, Frithz-Lindsten E, Wolf-Watz H. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J Bacteriol. 1996;178:7227–33. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schlumberger M, Muller A, Ehrbar K, Winnen B, Duss I, et al. Real time imaging of type III secretion: Salmonella SipA injection into host cells. Proc Natl Acad Sci USA. 2005;102:12548–53. doi: 10.1073/pnas.0503407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schraidt O, Lefebre M, Brunner M, Schmied W, Schmidt A, et al. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010;6:e1000824. doi: 10.1371/journal.ppat.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schraidt O, Marlovits T. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–5. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 129.Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci U S A. 2001;98:11638–43. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Silver A, Kikuchi Y, Fadl A, Sha J, Chopra A, Graf J. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci. 2007;104:9481–6. doi: 10.1073/pnas.0700286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sory M-P, Boland A, Lambermount I, Cornelis G. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sc USA. 1995;92:11998–2002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spaeth K, Chen Y, Valdivia R. The Chlamydia type III secretion system Cring engages a chaperone-effector protein complex. PLoS Pathog. 2009;5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Spreter T, Yip CK, Sanowar S, Andre I, Kimbrough TG, et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol. 2009;16:468–76. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- 135.Stebbins CE, Galan JE. Priming virulence factors for delivery into the host. Nat Rev Mol Cell Biol. 2003;4:738–43. doi: 10.1038/nrm1201. [DOI] [PubMed] [Google Scholar]
- 136.Sukhan A, Kubori T, Wilson J, Galan JE. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol. 2001;183:1159–67. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thomas N, Ma I, Preasad M, Rafuse C. Expanded roles for multicargo and class 1B effector chaperones in type III secretion. J Bacteriol. 2012;194:3767–3. doi: 10.1128/JB.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Turner L, Stern A, Berg H. Growth of flagellar filaments of Escherichia coli is independent of filament lenght. J Bacteriol. 2012;194:2437–42. doi: 10.1128/JB.06735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Veenendaal A, Hodkinson J, Schwarzer L, Stabat D, Zenk S, Blocker A. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol. 2007;63:1719–30. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 140.Wagner S, Königsmaier L, Lara-Tejero M, Lefebre M, Marlovits T, Galán J. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci USA. 2010;107:17745–50. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y, Nordhues B, Zhong D, De Guzman R. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry. 2010;49:4220–6. doi: 10.1021/bi100335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Y, Yu X, Yip C, Strynadka N, Egelman E. Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved apporach to helical reconstruction. Structure. 2006;14:75–81. doi: 10.1016/j.str.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 143.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis GR. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sc U S A. 1994;91:10493–7. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wattiau P, Cornelis GR. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–31. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 145.Wilharm G, Lehmann V, Krauss K, Lehnert B, Richter S, et al. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect Immun. 2004;72:4004–9. doi: 10.1128/IAI.72.7.4004-4009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Woestyn S, Allaoui A, Wattiau P, Cornelis GR. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–9. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Woestyn S, Sory MP, Boland A, Lequenne O, Cornelis GR. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Molecular Microbiology. 1996;20:1261–71. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 148.Wood S, Jin J, Lloyd S. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol. 2008;190:4252–62. doi: 10.1128/JB.00328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Worral L, Lameignere E, Strynadka N. Structural overview of the bacterial injectisome. Curr Opin Microbiol. 2011;14:3–8. doi: 10.1016/j.mib.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 150.Yip C, Fnlay B, Strynadka N. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 151.Yuan J, Berg H. Thermal and solvent-isotope effects on the flaellar rotary motor near zero load. Biophys J. 2010;98:21212126. doi: 10.1016/j.bpj.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang L, Wang Y, Olive AJ, Smith ND, Picking WD, et al. Identification of the MxiH needle protein residues responsible for anchoring invasion plasmid antigen D to the type III secretion needle tip. J Biol Chem. 2007;282:32144–51. doi: 10.1074/jbc.M703403200. [DOI] [PubMed] [Google Scholar]
- 153.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–30. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 154.Zhong D, Lefebre M, Kaur K, McDowell MA, Gdowski C, et al. The Salmonella Type III Secretion System Inner Rod Protein PrgJ is Partially Folded. J Biol Chem. 2012 doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zierler MK, Galan JE. Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect Immun. 1995;63:4024–8. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]