Abstract
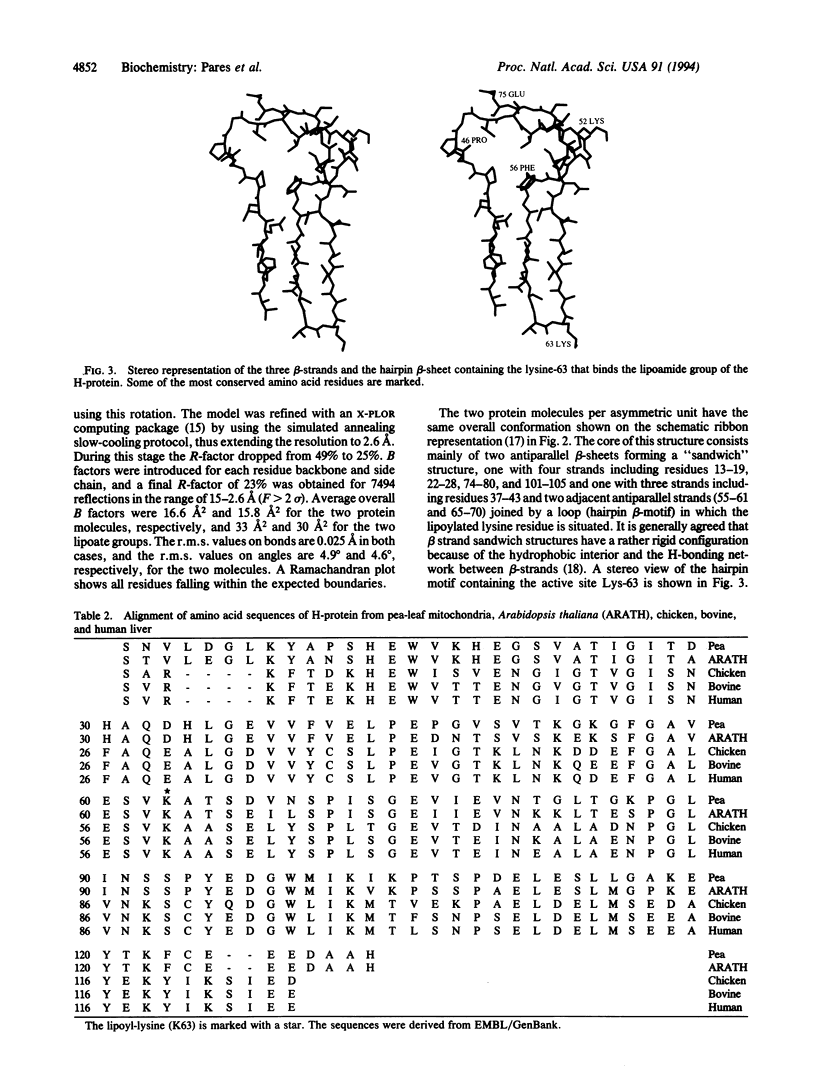
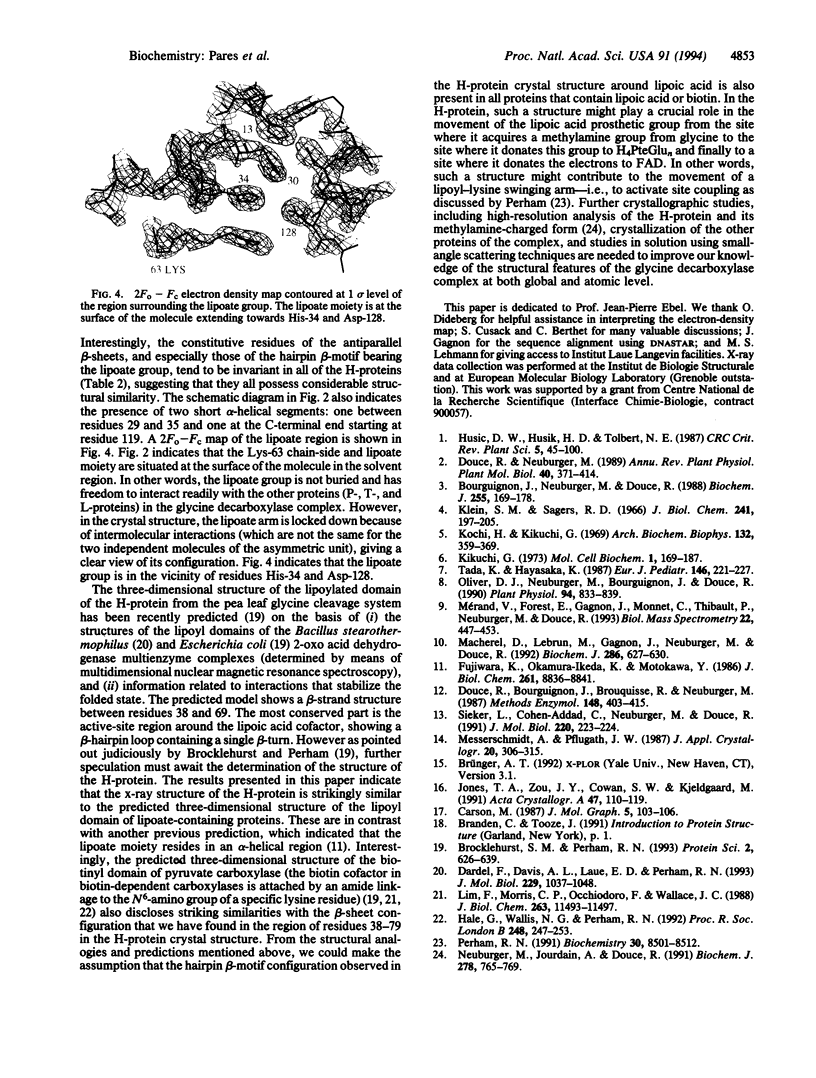
H-protein, a lipoic acid-containing protein of the glycine decarboxylase (EC 1.4.4.2) complex from pea (Pisum sativum) was crystallized from ammonium sulfate solution at pH 5.2 in space group P3(1)21. The x-ray crystal structure was determined to 2.6-A resolution by multiple isomorphous replacement techniques. The structure was refined to an R value of 23% for reflections between 15- and 2.6-A resolution (F > 2 sigma), including the lipoate moiety and 50 water molecules, for the two protein molecules of the asymmetric unit. The 131-amino acid residues form seven beta-strands arranged into two antiparallel beta-sheets forming a "sandwich" structure. One alpha-helix is observed at the C-terminal end. The lipoate cofactor attached to Lys-63 is located in the loop of a hairpin configuration. The lipoate moiety points toward the residues His-34 and Asp-128 and is situated at the surface of the H-protein. This allows the flexibility of the lipoate arm. This is the first x-ray determination of a lipoic acid-containing protein, and the present results are in agreement with previous theoretical predictions and NMR studies of the catalytic domains of lipoic acid- and biotin-containing proteins.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourguignon J., Neuburger M., Douce R. Resolution and characterization of the glycine-cleavage reaction in pea leaf mitochondria. Properties of the forward reaction catalysed by glycine decarboxylase and serine hydroxymethyltransferase. Biochem J. 1988 Oct 1;255(1):169–178. doi: 10.1042/bj2550169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst S. M., Perham R. N. Prediction of the three-dimensional structures of the biotinylated domain from yeast pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: a new automated method for the prediction of protein tertiary structure. Protein Sci. 1993 Apr;2(4):626–639. doi: 10.1002/pro.5560020413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardel F., Davis A. L., Laue E. D., Perham R. N. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. J Mol Biol. 1993 Feb 20;229(4):1037–1048. doi: 10.1006/jmbi.1993.1103. [DOI] [PubMed] [Google Scholar]
- Fujiwara K., Okamura-Ikeda K., Motokawa Y. Chicken liver H-protein, a component of the glycine cleavage system. Amino acid sequence and identification of the N epsilon-lipoyllysine residue. J Biol Chem. 1986 Jul 5;261(19):8836–8841. [PubMed] [Google Scholar]
- Hale G., Wallis N. G., Perham R. N. Interaction of avidin with the lipoyl domains in the pyruvate dehydrogenase multienzyme complex: three-dimensional location and similarity to biotinyl domains in carboxylases. Proc Biol Sci. 1992 Jun 22;248(1323):247–253. doi: 10.1098/rspb.1992.0069. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973 Jun 27;1(2):169–187. doi: 10.1007/BF01659328. [DOI] [PubMed] [Google Scholar]
- Klein S. M., Sagers R. D. Glycine metabolism. I. Properties of the system catalyzing the exchange of bicarbonate with the carboxyl group of glycine in Peptococcus glycinophilus. J Biol Chem. 1966 Jan 10;241(1):197–205. [PubMed] [Google Scholar]
- Kochi H., Kikuchi G. Reactions of glycine synthesis and glycine cleavage catalyzed by extracts of Arthrobacter globiformis grown on glycine. Arch Biochem Biophys. 1969 Jul;132(2):359–369. doi: 10.1016/0003-9861(69)90377-4. [DOI] [PubMed] [Google Scholar]
- Lim F., Morris C. P., Occhiodoro F., Wallace J. C. Sequence and domain structure of yeast pyruvate carboxylase. J Biol Chem. 1988 Aug 15;263(23):11493–11497. [PubMed] [Google Scholar]
- Macherel D., Bourguignon J., Douce R. Cloning of the gene (gdcH) encoding H-protein, a component of the glycine decarboxylase complex of pea (Pisum sativum L.). Biochem J. 1992 Sep 1;286(Pt 2):627–630. doi: 10.1042/bj2860627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérand V., Forest E., Gagnon J., Monnet C., Thibault P., Neuburger M., Douce R. Characterization of the primary structure of H-protein from Pisum sativum and location of a lipoic acid residue by combined liquid chromatography/mass spectrometry and liquid chromatography/tandem mass spectrometry. Biol Mass Spectrom. 1993 Aug;22(8):447–456. doi: 10.1002/bms.1200220805. [DOI] [PubMed] [Google Scholar]
- Neuburger M., Jourdain A., Douce R. Isolation of H-protein loaded with methylamine as a transient species in glycine decarboxylase reactions. Biochem J. 1991 Sep 15;278(Pt 3):765–769. doi: 10.1042/bj2780765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. J., Neuburger M., Bourguignon J., Douce R. Interaction between the Component Enzymes of the Glycine Decarboxylase Multienzyme Complex. Plant Physiol. 1990 Oct;94(2):833–839. doi: 10.1104/pp.94.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham R. N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991 Sep 3;30(35):8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Sieker L., Cohen-Addad C., Neuburger M., Douce R. Crystallographic data for H-protein from the glycine decarboxylase complex. J Mol Biol. 1991 Jul 20;220(2):223–224. doi: 10.1016/0022-2836(91)90007-s. [DOI] [PubMed] [Google Scholar]
- Tada K., Hayasaka K. Non-ketotic hyperglycinaemia: clinical and biochemical aspects. Eur J Pediatr. 1987 May;146(3):221–227. doi: 10.1007/BF00716464. [DOI] [PubMed] [Google Scholar]