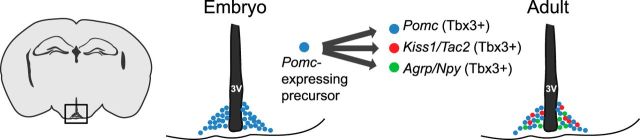
Abstract
Hypothalamic neuronal populations are central regulators of energy homeostasis and reproductive function. However, the ontogeny of these critical hypothalamic neuronal populations is largely unknown. We developed a novel approach to examine the developmental pathways that link specific subtypes of neurons by combining embryonic and adult ribosome-tagging strategies in mice. This new method shows that Pomc-expressing precursors not only differentiate into discrete neuronal populations that mediate energy balance (POMC and AgRP neurons), but also into neurons critical for puberty onset and the regulation of reproductive function (Kiss1 neurons). These results demonstrate a developmental link between nutrient-sensing and reproductive neuropeptide synthesizing neuronal populations and suggest a potential pathway that could link maternal nutrition to reproductive development in the offspring.
Keywords: gene expression, hypothalamus, metabolism, reproduction
Introduction
Proopiomelanocortin (POMC) neurons located in arcuate nucleus (ARC) of the hypothalamus have a prominent role in the regulation of feeding, metabolism, and body weight (Cone, 2005). These neurons, together with the functionally antagonistic agouti-related peptide (AgRP) neurons, are first-order neurons in the melanocortin system and tightly regulate energy homeostasis by integrating signals from the periphery (leptin, ghrelin, insulin) and from other neurons in the hypothalamus and brain stem (Varela and Horvath, 2012; Sohn et al., 2013). Perturbations in energy homeostasis are known to have profound effects on reproductive function, and POMC neurons, among other neuronal populations in the ARC such as Kiss1 neurons and AgRP neurons, are thought to mediate the link between energy balance and reproduction (Hill et al., 2008; Castellano et al., 2010; Xu et al., 2011; Wu et al., 2012). Although the role of these neuronal populations as central regulators of energy homeostasis and reproduction in the adult has been investigated extensively, studies on the embryonic development and differentiation of these cells are just emerging (Padilla et al., 2010; Shimogori et al., 2010; Pelling et al., 2011). Previous evidence has shown that ARC progenitors expressing the Pomc gene during the specification and differentiation period are highly sensitive to the maternal metabolic status (Wattez et al., 2013), and only a fraction of these progenitors adopt a POMC terminal fate in the adult. Some of the remaining Pomc-expressing progenitors give rise to the orexigenic AgRP/NPY neurons (Padilla et al., 2010). We implemented a novel technique using genomic and viral-mediated ribosome tagging to characterize these Pomc-expressing progenitors and determine whether they also give rise to other neurons involved in metabolism and reproduction. Our results show that Pomc-expressing progenitors also differentiate into ARC localized Kiss1 neurons and that all of the Pomc progeny continue to express the Tbx3 transcription factor gene and are subject to nutritional regulation.
Materials and Methods
Animals.
Mice were housed in a temperature and humidity controlled facility with a 12 h light/dark cycle. All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Washington. RiboTag mice were generated as described previously (Sanz et al., 2009). Pomc-Cre mice bacterial artificial chromosome (BAC) transgene line 1 (Balthasar et al., 2004), BAC transgene line 2 (McHugh et al., 2007), and Kiss1-CreEGFP mice (Gottsch et al., 2011) were described previously. All mice used were adults (3 to 4 months old) and on a C57BL/6 background, except for embryonic expression studies, where embryonic day 12.5 (E12.5) and E15.5 embryos on a Swiss–Webster background were examined. Unless stated otherwise, equal numbers of male and female mice were used in the experiments.
Generation of Agrp-CreEGFP mice.
The Agrp-CreEGFP targeting construct was prepared by inserting a cassette containing (1) a Cre-EGFP fusion protein with an myc tag and nuclear localization signal at the N terminus and an SV40 polyA at the 3′ end followed by (2) an frt-flanked SV-NeoR gene into the Pml1 site of an Agrp plasmid that has an ∼1 kb 5′ arm and ∼10 kb 3′ arm flanked by Pgk-DTa and HSV-TK genes for negative selection. The construct was electroporated in G4 hybrid (129/Sv × C57BL/6) ES cells. Correct targeting was observed in 31 of 83 clones analyzed by Southern blot after digesting DNA with BamH1 and using a probe 5′ of the short arm. Several clones were expanded and injected into blastocysts, and one of them that gave high-percentage chimeras was bred with C57BL/6 mice carrying Gt(Rosa)26 Sor–FLP recombinase to remove the SV-NeoR gene. Agrp-CreEGFP offspring were continuously backcrossed to C57BL/6 thereafter.
Adeno-associated viral vector generation and delivery.
We generated an adeno-associated virus (AAV) that would initiate Cre-inducible RiboTag expression using a double floxed inverted open reading frame (DIO) approach. An NheI/XhoI fragment containing a construct coding for the mouse ribosomal protein Rpl22 gene fused to three hemagglutinin (HA) sequences (Rpl22 · 3HA) (Sanz et al., 2009) was first cloned in NheI/XhoI-digested pIRES2-GFP. Subsequently, Rpl22 · 3HA-IRES-GFP was excised using NcoI, digesting 5′ of Rpl22 and at the starting codon (ATG) of the GFP cassette (Rpl22 · 3HA-IRES). Finally, this fragment was cloned into a NcoI-digested pAAV-EF1a-DIO-YFP-WPRE-hGH polyA plasmid to obtain a pAAV1-EF1a-DIO Rpl22 · 3HA-IRES-YFP-WPRE-hGH polyA construct (AAV-DIO-RiboTag). An AAV (AAV1 serotype) vector was produced and CsCl-gradient purified as described previously (Quintana et al., 2012a). Pomc-Cre mice were injected bilaterally with a total of 1 μl of the RiboTag viral vector (7 × 1011 viral genomes/ml; 0.5 μl per side) as described previously (Quintana et al., 2012b) using the following coordinates: ±0.27 mm mediolateral (ML), −1.14 mm anteroposterior (AP), and −5.22 mm dorsoventral (DV) from bregma. Mice were killed 3 to 4 weeks after injections.
RiboTag assays.
Punches containing the mediobasal hypothalamus of two male and two female mice were pooled and homogenized in 1 ml of buffer as described previously (Sanz et al., 2009). After centrifugation, 4 μl of anti-HA antibody (MMS-101R, 2–3 mg/ml; Covance) was added to 800 μl of the cleared lysate and incubated for 4 h at 4°C. Remaining lysate was saved as input sample. After incubation, 300 μl of protein A/G magnetic beads (Thermo Scientific) was added and incubated overnight at 4°C with rotation. Immunoprecipitates (IPs) were washed in high salt buffer and RNA from inputs and IPs extracted as described in the original protocol (Sanz et al., 2009). In experiments to examine the Kiss1 neurons in the anteroventral periventricular nucleus/periventricular nucleus (AVPV/PeN), that anterior region of the hypothalamus was first isolated from a coronal slice 2 mm rostral to the supraoptic decussation, and then punches of tissue surrounding the third ventricle were isolated for RiboTag analysis as above.
qRT-PCR analysis.
For qRT-PCR analysis, equal amounts of RNA were assayed using the Brilliant II SYBR Green qRT-PCR 1-Step Master Mix (Agilent Technologies) or the Brilliant II qRT-PCR 1-Step Master Mix (Agilent Technologies), depending on the system used (SYBR or Taqman). Relative expression values were obtained using the standard curve method and normalized to Actb levels. Amplification efficiencies were calculated using MxPro software (Stratagene) and were within accepted parameters (80–120%). Kiss1 and Tbx3 mRNAs were determined using specific Taqman assays (Mm03058560_m1 for Kiss1 and Mm01195726_m1 for Tbx3; Applied Biosystems), and sequences for the different primer sets used in SYBR assays were obtained from Primerbank (Spandidos et al., 2010) and were as follows: Pomc, forward, 5′-ACCTCACCACGGAGAGCAAC-3′; reverse, 5′-GCGAGAGGTCGAGTTTGCA-3′; Agrp, forward 5′-TAGATCCACAGAACCGCGAGT-3′; reverse, 5′-GAAGCGGCAGTAGCACGTA-3′; Npy, forward, 5′-CTCCGCTCTGCGACACTAC-3′; reverse, 5′-AGGGTCTTCAAGCCTTGTTCT-3′; Tac2, forward, 5′-TTCCACAGAAACGTGACATGC-3′; reverse, 5′-GGGGGTGTTCTCTTCAACCAC-3′.
Microarray analysis.
RNA (10 ng) was amplified using the Ovation Pico SL WTA system (NuGEN). Fidelity of amplification was confirmed by qPCR analysis of the resulting cDNA using the QuantiTect kit (Qiagen) before biotinylation according to the EncoreIL biotinilation kit (NuGEN). Biotinilated cDNA was quantified and product size distribution was analyzed using the 2100 Bioanalyzer system with the RNA 6000 Nano chips (Agilent Technologies). Biotinylated cDNA (750 ng) was hybridized at 48°C to MouseRef-8 v2 expression beadchips (Illumina) for 16 h before washing and analyzing according to the manufacturer's directions. Signal was detected using a BeadArray Reader (Illumina), and data were analyzed for differential expression using the GenomeStudio data analysis software (Illumina). Average normalization, the Illumina custom error model, and multiple testing corrections using the Benjamini and Hochberg false discovery rate were applied to the analysis. Only transcripts with a differential score of >13 (p < 0.05) were considered. Normalized and raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE56917).
Tissue processing.
For double-labeling in situ hybridization (ISH)/immunofluorescence (IF) experiments, ovariectomized adult Pomc-Cre:RiboTag females were killed by Beuthanasia-D administration (86 mg/kg pentobarbital sodium, 11 mg/kg phenytoin sodium) followed by cardiac perfusion with 4% paraformaldehyde in PBS, pH 7.4. Brains were dissected and postfixed at 4°C overnight and cryoprotected in 30% sucrose. Tissue was frozen in dry ice and stored at −80°C until sectioning. Coronal sections (15 μm) containing the anterior to posterior extent of the arcuate nucleus were used for analysis. For embryonic tissue, embryos (E12.5 and E15.5) were dissected in PBS at 4°C and fixed in 4% paraformaldehyde for 3 h before cryoprotection with 30% sucrose. Whole heads were embedded in Tissue-Tek OCT compound (Sakura), frozen, and sectioned as described for adult brains.
In situ hybridization.
For probe synthesis, plasmids for Kiss1 (Gottsch et al., 2004) and Pomc (Thornton et al., 1997) were linearized with HindIII before transcription in vitro and digoxigenin labeling using the DIG RNA labeling kit (Roche) with T7 or SP6 RNA polymerase, respectively. For single-labeled ISH, sections were acetylated with triethanolamine/acetic anhydride and permeabilized in PBS with 1% Triton X-100 before hybridizing at 68°C overnight. The following day, signal was detected using the TSA plus Cyanine 3 (Cy3) system (PerkinElmer). Sheep anti-digoxigenin-HRP antibody (11207733910; Roche) was used at a 1:200 dilution. Double-labeled ISH/IF was accomplished by simultaneous incubation of the anti-digoxigenin-HRP antibody with a rabbit anti-HA antibody (71-5500; Zymed) before detection using the TSA plus Cy3 System and an anti-rabbit Alexa Fluor 488-conjugated secondary antibody.
Immunofluorescence.
Brains from adult Pomc-Cre:RiboTag (BAC1 and BAC2) mice, Pomc-Cre mice injected with the RiboTag viral vector (AAV-DIO-RiboTag), Kiss1-CreEGFP mice, and Agrp-CreEGFP mice were processed as described previously, and staining was accomplished as described previously (Gottsch et al., 2011). At least three animals were used in each analysis unless noted otherwise in the figure legends. Rabbit anti-GFP antibody (A-11122; Invitrogen), rabbit anti-Kisspeptin antibody (AB9754; Millipore), Alexa Fluor 488-conjugated mouse anti-HA antibody (A488-101L; Covance), and rabbit anti-HA antibody (71-5500; Zymed) were used at 1:1000 dilutions. Goat anti-Tbx3 (sc-31657; Santa Cruz Biotechnology) was used at a 1:100 dilution. Rabbit anti-HA antibody and Alexa Fluor 488-conjugated mouse anti-HA antibodies were used for HA single staining and Kisspeptin/ HA and Tbx3/HA double staining. For Kisspeptin labeling, colchicine (30 μg) was administered into the third ventricle, and mice were killed by Beuthanasia-D administration followed by cardiac perfusion with 4% paraformaldehyde 24 h after injection.
Results
To characterize the cellular fate of the Pomc-expressing precursors, we HA tagged the polysomes of the Pomc-derived lineage and of adult POMC neurons and determined their differential polysome-associated mRNA profile (translatome) by an affinity purification method followed by gene array analysis (RiboTag).
To obtain mice with a ribosomal HA tag on the Pomc-derived lineage, a BAC-transgenic line of Pomc-Cre(BAC1) mice (Balthasar et al., 2004) was crossed to RiboTag mice (Sanz et al., 2009; Fig. 1A). In these mice, developmental expression of Cre recombinase driven by the Pomc promoter activates the expression of the epitope-tagged ribosomal protein, RPL22-HA, in Pomc-expressing progenitors, leading to HA-tagged polysomes in all cells that expressed Pomc during development (Fig. 1B). Thus, if Pomc-Cre was expressed in a precursor population that subsequently gave rise to mature POMC neurons as well as other neuronal types that no longer expressed Pomc, tagged ribosomes would be found not only in POMC neurons, but in the other progeny as well. To label ribosomes in the progeny of Pomc-expressing neurons that turn off expression of Pomc as neurons differentiate, it is critical to obtain robust expression of Cre recombinase as soon as the Pomc gene turns on to allow recombination of the RiboTag locus before subsequent inactivation of Pomc in progeny that will become non-POMC neurons.
Figure 1.
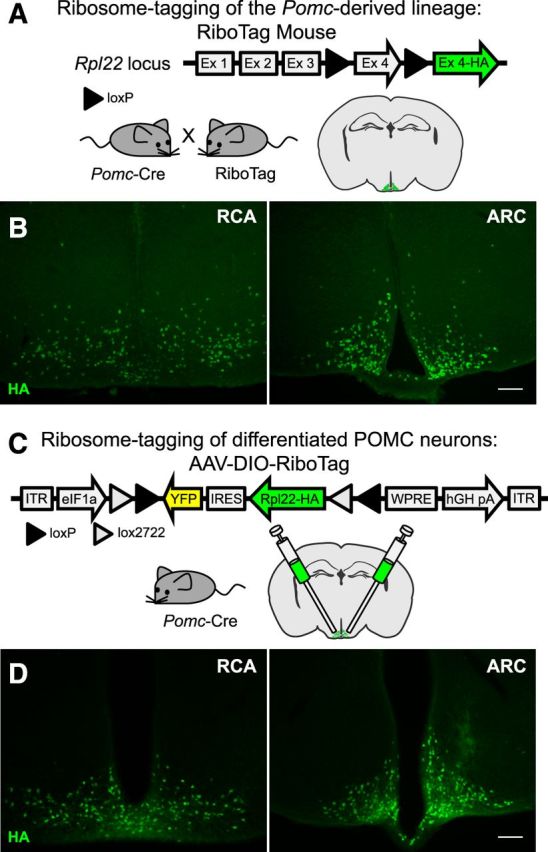
Ribosome tagging of the POMC-derived lineage and adult POMC neurons. A, Locus of the ribosomal protein Rpl22 in the RiboTag mouse and breeding strategy used to obtain Pomc-Cre:RiboTag mice. B, Immunofluorescence showing HA (green) staining in the RCA and ARC of Pomc-Cre(BAC1):RiboTag mice. Sections were cut at 30 μm. C, RiboTag viral vector (AAV-DIO-RiboTag) construct and strategy used to express the RiboTag viral vector in POMC neurons of adult mice. D, Immunofluorescence analysis using anti-HA antibodies in hypothalamic sections containing the RCA and the ARC of Pomc-Cre(BAC1) mice injected bilaterally into the ARC with the RiboTag viral vector (AAV-DIO-RiboTag). Sections were cut at 60 μm. Scale bars: 100 μm. Images show sections obtained from the same animal.
To HA tag ribosomes only in adult differentiated POMC neurons, a Cre-dependent RiboTag adenoassociated viral vector (AAV-DIO-RiboTag) was generated and injected into the ARCs of adult Pomc-Cre(BAC1) mice (Fig. 1C). This viral vector was created by inserting the cDNA of an HA-tagged Rpl22 into a double-floxed inverse open reading frame (DIO) AAV construct. HA staining on hypothalamic sections of adult Pomc-Cre mice injected with AAV-DIO-RiboTag was performed to show continuous and restricted signal from the retrochiasmatic area (RCA) to the ARC, where adult Pomc-expressing neurons are located (Fig. 1D). With this approach, only those neurons that express Pomc-Cre in the adult will have labeled ribosomes. One potential problem is that the viral approach depends on dispersion of the virus after injection and may not transduce all the Pomc-expressing neurons. However, incomplete transduction of all Pomc neurons will not compromise the results as long as the population transduced is representative. For that reason we injected relatively large volumes of virus (0.5 μl) bilaterally and pooled hypothalamic punches from four animals for each assay.
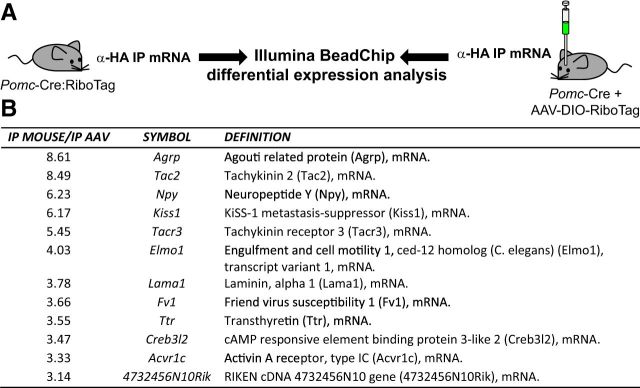
The characterization of the Pomc-derived lineage was accomplished by comparing the RiboTag results from Pomc-Cre(BAC1):RiboTag mice with Pomc-Cre(BAC1) mice injected with AAV-DIO-RiboTag (Fig. 2A). Differential microarray analysis revealed a much higher abundance of specific transcripts in the immunoprecipitates from Pomc-Cre(BAC1):RiboTag mice compared to those from Pomc-Cre(BAC1) mice injected with AAV-DIO-RiboTag (Fig. 2B). The most differentially expressed transcripts were Agrp, Tac2, Npy, and Kiss1. Quantitative RT-PCR analysis of the immunoprecipitates from Pomc-Cre(BAC1):RiboTag and Pomc-Cre(BAC1):AAV-DIO-RiboTag hypothalamic punches (Fig. 3A) showed that both immunoprecipitates were highly enriched in Pomc transcripts when compared to their respective inputs (Fig. 3B) and confirmed the microarray results (Fig. 3C–F). Transcripts for Agrp, Npy, Kiss1, and Tac2 were all enriched compared to input in the immunoprecipitates from the RiboTag mouse and de-enriched compared to the input when cell-specific polyribosomes were isolated using the AAV-DIO-RiboTag viral injection in adult Pomc-Cre(BAC1) mice (Fig. 3C–F).
Figure 2.
Ribosomal tagging reveals the molecular identity of the Pomc-derived lineage. A, Illustration depicting the samples compared by differential gene expression analysis for the identification of the Pomc-derived lineage. α-HA IP mRNA, mRNA from anti-Hemagglutinin immunoprecipitates. B, Microarray analysis data reveal the mRNA transcripts enriched threefold or higher in the immunoprecipitates of Pomc-Cre(BAC1):RiboTag mice (IP mouse) compared to the immunoprecipitates of Pomc-Cre(BAC1) mice injected with the RiboTag adenoassociated viral vector (IP AAV).
Figure 3.
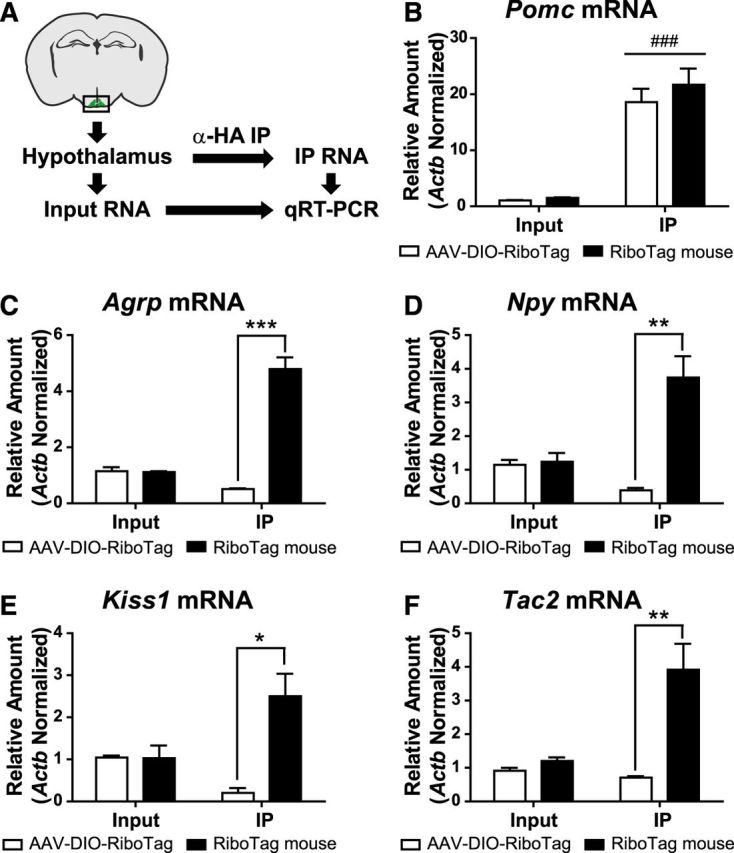
qRT-PCR confirmation of microarray data. A, Diagram showing the RiboTag immunoprecipitation procedure and the method used for the analysis of the resulting samples. B–F, qRT-PCR analysis for Pomc (B), Agrp (C), Npy (D), Kiss1 (E), and Tac2 (F) transcripts in inputs and IPs of Pomc-Cre(BAC1):RiboTag mice (RiboTag mouse) or Pomc-Cre(BAC1) mice injected with the RiboTag viral vector (AAV-DIO-RiboTag). Graphs show the mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-tests. *p < 0.05; **p < 0.01; ***p < 0.001.
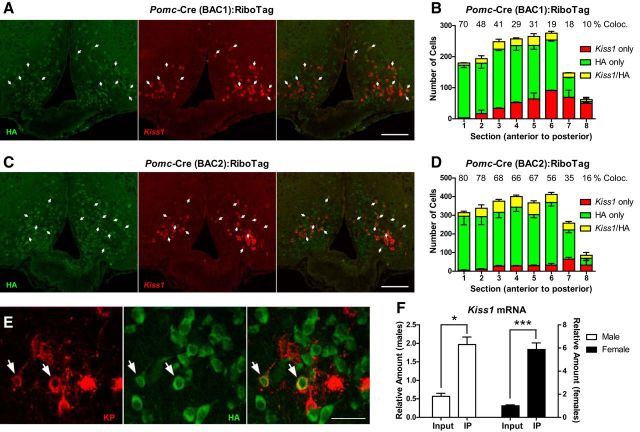
The presence of Agrp and Npy mRNAs in the Pomc lineage confirmed previous results (Padilla et al., 2010). Unexpectedly, our approach also revealed the presence of transcripts including Kiss1 and Tac2 in the Pomc-Cre(BAC1) marked lineage. Kiss1 and Tac2 are known to be coexpressed in Kiss1 neurons in the ARC (Navarro et al., 2009), suggesting that Pomc-expressing progenitors also give rise to Kiss1 neurons. To confirm this, we performed double ISH/IF analysis for Kiss1 mRNA and RPL22-HA in female ovarectomized Pomc-Cre:RiboTag mice using two different BAC transgenic lines of Pomc-Cre mice [BAC1 (Balthasar et al., 2004) and BAC2 (McHugh et al., 2007); Fig. 4A–D]. Ovariectomies were performed to maximize Kiss1 mRNA signal in the ARC. Double ISH/IF showed the presence of Kiss1 mRNA in HA-tagged cells in both Pomc-Cre lines. However, Pomc-Cre(BAC1):RiboTag mice showed a reduced number of HA-positive cells in the ARC compared to Pomc-Cre(BAC2):RiboTag mice (Fig. 4A–D). The percentage of Kiss1 neurons that also express Rpl22-HA is shown in Figure 4, B and D, for each section. Cell counts showed that an average of 27% of Kiss1-positive neurons expressed the HA tag in Pomc-Cre(BAC1):RiboTag mice, whereas in Pomc-Cre(BAC2):RiboTag mice, 59% of the total Kiss1 neurons were Rpl22-HA positive. The Pomc-Cre(BAC2) also elicits a more efficient recombination in the hypothalamus (Fig. 4A–D) and other areas where Pomc-expressing cells are known to be localized (McHugh et al., 2007). This suggests that the transgenic integration site of the Pomc-Cre(BAC2) allows for more robust expression of the Cre recombinase protein. Kisspeptin also colocalized with HA at the protein level (Fig. 4E), and Kiss1 mRNA showed enrichment in the immunoprecipitates of intact Pomc-Cre(BAC1):RiboTag mice in both males and females (Fig. 4F). We examined the timing and anatomical localization of both Pomc mRNA and Kiss1 mRNA in the embryonic hypothalamus and observed that Pomc expression in the presumptive ARC of the mouse embryo precedes Kiss1 expression, and that both localize to a similar region of the ARC by E15.5 (Fig. 5A).
Figure 4.
Kiss1 neurons arise from Pomc-expressing progenitors. A, Double in situ hybridization/immunofluorescence analysis for Kiss1 mRNA (red) and HA (green) in female ovarectomized Pomc-Cre(BAC1):RiboTag mice. Arrows indicate double-positive cells. B, Quantification of the total number of non-HA-tagged Kiss1-expressing cells (red), HA-tagged non-Kiss1-expressing cells (green), and Kiss1-expressing HA-tagged cells (yellow) in different ARC sections (every 150 μm, −1.34 to −2.54 mm from Bregma) from female ovarectomized Pomc-Cre(BAC1):RiboTag mice. C, Double in situ hybridization/immunofluorescence analysis for Kiss1 mRNA (red) and HA (green) in female ovarectomized Pomc-Cre(BAC2):RiboTag mice. Arrows indicate double-positive cells. D, Quantification of the total number of non-HA-tagged Kiss1-expressing cells (red), HA-tagged non-Kiss1-expressing cells (green), and Kiss1-expressing HA-tagged cells (yellow) in different ARC sections (every 150 μm, −1.34 to −2.54 mm from Bregma) from female ovarectomized Pomc-Cre(BAC2):RiboTag mice. E, Confocal images of Kisspeptin (red) and HA (green) immunofluorescent staining in arcuate sections of ovarectomized and colchicine-treated female Pomc-Cre(BAC1):RiboTag mice. Arrows indicate double-positive cells. F, qRT-PCR analysis for Kiss1 mRNA in inputs and IPs of intact male and female Pomc-Cre(BAC1):RiboTag mice. Each sample was obtained from a pool of four hypothalamic punches. Graphs show the mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-tests. *p < 0.05; ***p < 0.001. Scale bars: A, C, 100 μm; E, 30 μm.
Figure 5.
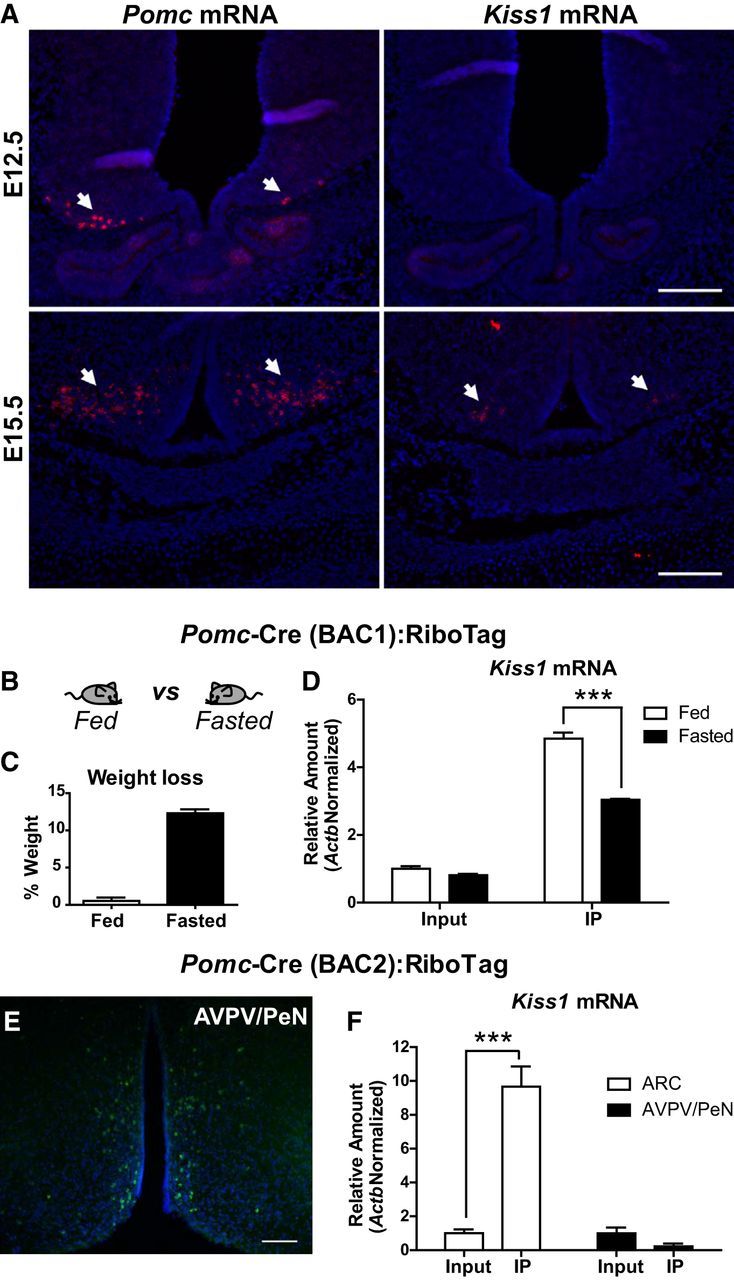
Developmental and nutritional regulation of Kiss1 neurons. A, In situ hybridization analysis for Pomc and Kiss1 mRNA in embryonic hypothalamic sections (E12.5 and E15.5). Arrows indicate Pomc- or Kiss1-expressing cells. B, Illustration showing the different treatments compared in C and D. C, Graph showing the percentage of weight loss after a 24 h fasting in Pomc-Cre(BAC1):RiboTag mice. D, qRT-PCR analysis for Kiss1 mRNA in inputs and IPs of Pomc-Cre(BAC1):RiboTag mice fed or fasted for 24 h. E, Immunofluorescence for HA staining in the AVPV/PeN of Pomc-Cre(BAC2):RiboTag mice. F, qRT-PCR analysis for Kiss1 mRNA in inputs and IPs of sections containing the AVPV/PeN or the ARC in Pomc-Cre(BAC2):RiboTag mice. Scale bars: 100 μm. Graphs show the mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-tests. ***p < 0.001.
The expression of the neuropeptides Agrp, Npy, and Pomc is regulated in AgRP and POMC neurons by the nutritional status of the animal (Hahn et al., 1998; Mizuno et al., 1998). To explore whether the Pomc-derived Kiss1 neurons are also nutritionally regulated, we performed the RiboTag assay in Pomc-Cre(BAC1):RiboTag mice fasted for 24 h and assessed Kiss1 expression in inputs and IPs by qRT-PCR analysis (Fig. 5B–D). Results showed that Kiss1 mRNA was significantly reduced in Pomc-derived Kiss1 neurons (IPs), suggesting that the fertility-regulating neuropeptide Kisspeptin is modified by the nutritional status in these cells.
The other major population of fertility-regulating Kiss1 neurons is present in the AVPV/PeN in the anterior hypothalamus of the female mouse, and female Pomc-Cre(BAC2):RiboTag mice also show specific RiboTag activation in the area (Fig. 5E). To define whether this other population of Kiss1 neurons in the AVPV/PeN is also derived from Pomc-expressing progenitors, we performed the RiboTag assay in hypothalamic punches containing the AVPV/PeN or the ARC of Pomc-Cre(BAC2):RiboTag mice and assessed the relative amount of Kiss1 mRNA in these cells (IPs; Fig. 5F). IPs from ARC RiboTag assays showed a significant enrichment for Kiss1 mRNA compared to the input RNA, as expected. However, RiboTag assays performed on AVPV/PeN punches did not show any enrichment in the IP, suggesting that the Pomc-driven HA-positive cells in the AVPV/PeN were not Kiss1 neurons. Enrichment for Kiss1 in the ARC using Pomc-Cre(BAC2):RiboTag mice was higher than when the Pomc-Cre(BAC1) line was used (compare Figs. 4F, 5F), which is consistent with the significantly larger number of Kiss1/HA-positive cells obtained with the Pomc-Cre(BAC2) mouse line (Fig. 4D).
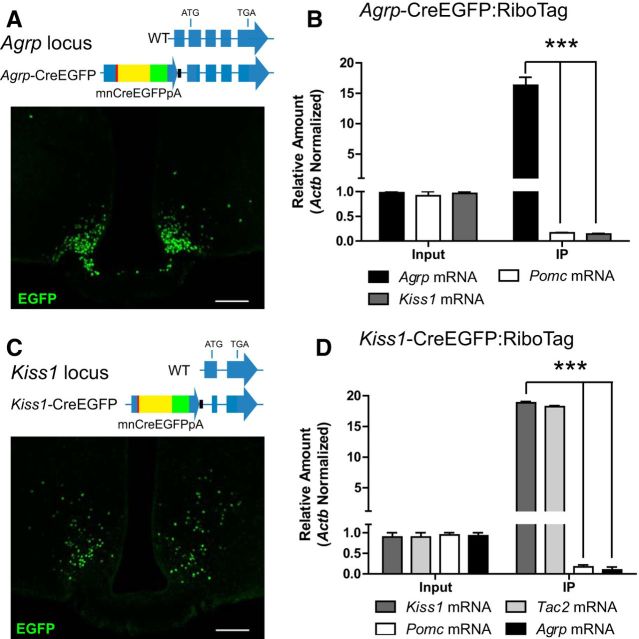
We next asked whether the adult AgRP and Kiss1 ARC subpopulations originate from different Pomc-expressing precursors or are sequentially generated. To test whether Agrp-expressing cells were precursors of Kiss1 neurons, a line of mice expressing a Cre-EGFP recombinase fusion protein under the control of the endogenous Agrp promoter was generated (Agrp-CreEGFP; Fig. 6A) and crossed to RiboTag mice. Quantitative RT-PCR analysis following RiboTag assays in Agrp-CreEGFP:RiboTag mice confirmed the enrichment for Agrp mRNA, but Kiss1 and Pomc mRNAs were depleted in the immunoprecipitates, indicating that Agrp-expressing neurons do not differentiate into Pomc- or Kiss1-expressing neurons (Fig. 6B). A similar genetic lineage tracing strategy was performed using mice expressing Cre recombinase (fused to EGFP) regulated by the endogenous Kiss1 promoter (Gottsch et al., 2011; Fig. 6C) crossed to RiboTag mice (Kiss1-CreEGFP:RiboTag; Fig. 6D). RiboTag activation in Kiss1 neurons in the ARC was confirmed by enrichment for Kiss1 and Tac2 in the immunoprecipitates of the RiboTag assays (Fig. 6D). However, Agrp and Pomc transcripts were depleted in the immunoprecipitates of these mice, suggesting that AgRP neurons are not derived from Kiss1-expressing cells.
Figure 6.
Kiss1/Tac2- and Agrp/Npy-expressing neurons are derived from independent subpopulations of Pomc-expressing progenitors. A, Agrp locus in wild-type (WT) and Agrp-CreEGFP mice and immunofluorescent analysis for EGFP in ARC sections of Agrp-CreEGFP mice. Mice were fasted for 30 h to induce CreEGFP expression by activating the Agrp promoter. B, qRT-PCR analysis for Agrp, Pomc, and Kiss1 transcripts in inputs and IPs of Agrp-CreEGFP:RiboTag mice. C, Kiss1 locus in WT and Kiss1-CreEGFP mice and immunofluorescence for EGFP in ARC sections of Kiss1-CreEGFP mice. Mice were gonadectomized to induce CreEGFP expression by activating the Kiss1 promoter. D, qRT-PCR analysis of Kiss1, Tac2, Pomc, and Agrp transcripts in inputs and IPs of Kiss1-CreEGFP:RiboTag mice. Each sample was obtained from a pool of four hypothalamic punches. Graphs show the mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-tests. *** p < 0.001. Scale bars: 100 μm.
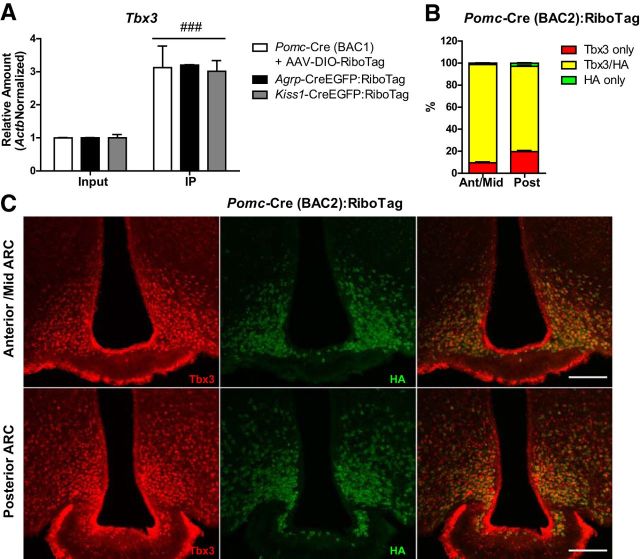
Whether these Pomc-derived cell populations share a molecular signature after differentiation it is not known. The expression of the transcription factor gene, Tbx3, is restricted to the ARC in the mediobasal hypothalamus of the mouse brain (Eriksson and Mignot, 2009), and mutations in this transcription factor are known to result in obesity and affect fertility (Schinzel et al., 1987; Bamshad et al., 1997). Therefore, we asked whether Tbx3 expression might be a common signature of the Pomc-derived neuronal populations. To test this, we analyzed the expression of Tbx3 mRNA in RiboTag assays of Pomc-Cre mice injected with AAV-DIO-RiboTag, Agrp-CreEGFP:Ribotag mice, and Kiss1-CreEGFP:RiboTag mice (Fig. 7A) and observed that the immunoprecipitates of all three groups were enriched for Tbx3 transcripts. This suggests that the Tbx3 transcription factor is quite specific to POMC, AgRP, and Kiss1 neurons when compared to all the other cell types in the dissected area. To confirm that these Pomc-derived neuronal populations specifically express Tbx3, staining for Tbx3 and HA was performed in Pomc-Cre(BAC2):RiboTag mice (Fig. 7B,C) to obtain the highest percentage of recombination of the RiboTag allele in Pomc-expressing progenitors. Results showed that almost all HA-positive cells across the ARC have Tbx3-stained nuclei. However, a small fraction of Tbx3-positive nuclei (10% in the anterior/middle ARC and 20% in the posterior ARC) did not show HA staining (Fig. 7B). Representative images are shown in Figure 7C. This may be due to either incomplete recombination of the RiboTag allele in Pomc-expressing progenitors or expression of Tbx3 in specific cell types other than the Pomc lineage.
Figure 7.
Pomc-derived ARC neurons express the transcription factor Tbx3. A, qRT-PCR analysis for Tbx3 in inputs and IPs of Pomc-Cre(BAC1) mice injected with AAV-DIO-RiboTag and Agrp-CreEGFP:RiboTag and Kiss1-CreEGFP:RiboTag mice. Graph shows the mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-tests. ###p < 0.001. B, Quantification of immunofluorescence analysis for Tbx3 and HA in the anterior/middle (−1.34 to −1.94 mm from Bregma) and posterior ARC (−1.94 to −2.46 mm from Bregma) of Pomc-Cre(BAC2):RiboTag mice. Data are presented as the average percentage of Tbx3-only (red), HA-only (green), and Tbx3/HA double-stained (yellow) cells in 30-μm-thick sections every 180 μm. Graphs show the mean ± SEM of the combined results from two mice. C, Representative images showing the presence of Tbx3-positive nuclei in HA-labeled cells in the anterior/mid and posterior ARC of Pomc-Cre(BAC2):RiboTag mice. Scale bars: 100 μm.
Discussion
Our results reveal that the progenitor neuron population in the ARC that expresses Pomc early in development subsequently differentiates into neurons that express Pomc, or Agrp/Npy or Kiss1/Tac2 (Fig. 8). Direct counting of Kiss1 neurons that were labeled during development in Pomc-Cre:RiboTag mice showed different results depending on the BAC transgene Pomc-Cre line used. The BAC2 transgenic line gave a higher percentage of Kiss1 neurons with tagged ribosomes, indicating a more robust Pomc-driven Cre recombinase expression in this line compared to the BAC1 line. Thus, the results obtained using the BAC1 line probably underestimate the total number of Pomc-derived Kiss1 neurons due to a less robust expression of Cre recombinase in the Pomc-expressing precursors and the stochastic nature of Cre-mediated excision of loxP-flanked sites (Nagy, 2000). The promoter activity of BAC transgenics is influenced by the genomic environment at the random site of integration of the transgene, and the level of expression of Cre recombinase determines how rapidly loxP targeted recombination takes place. For these reasons, we are confident that at least 59% of the Kiss1 neurons in the ARC are derived from Pomc-expressing progenitors, and it is possible that all Kiss1 neurons share the same progenitor lineage. However, our results do not rule out the possibility that a fraction of Kiss1 neurons in the ARC derive from a non-Pomc-expressing precursor. The Kiss1 neurons that reside in the AVPV/PeN, however, are clearly not differentiated from the same Pomc-expressing precursors that are the ancestors of the ARC Kiss1 neurons. Our RiboTag analysis demonstrated that the AVPV/PeN Pomc-Cre-labeled neurons did not express Kiss1 mRNA.
Figure 8.
Cellular fate of Pomc-expressing progenitors in the ARC. Pomc-expressing progenitors give rise to three independent neuronal populations (Pomc-, Kiss1/Tac2-, and Agrp/Npy-expressing neurons) in the adult mouse. 3V, Third ventricle.
Our data demonstrate that the ARC Pomc-derived lineage is characterized by a selective expression of the transcription factor Tbx3. In the mouse brain, Tbx3 expression is restricted to the ARC, the tuberomamillary nucleus, and the NTS of the dorsal medulla (Eriksson and Mignot, 2009). Mutations in this gene in humans result in ulnar-mammary syndrome (UMS), which is characterized by multiple phenotypes, including obesity, hypogenitalism, and delayed puberty (Schinzel et al., 1987; Bamshad et al., 1997). Although the function of Tbx3 in the ARC is not known, it is thought that reduced activity of this transcription factor in this hypothalamic nucleus may be responsible for the obesity and reproductive phenotypes in this syndrome (Eriksson and Mignot, 2009). Our findings showing Tbx3 expression as a molecular signature of the Pomc-derived lineage identifies the cell types potentially responsible for these phenotypes. POMC and AgRP/NPY neurons are well-known regulators of food intake; therefore, Tbx3 mutations in these neurons may disrupt their expression profile and affect their energy-balancing role. In addition, reduced activity of Tbx3 in Kiss1/Tac2 neurons may explain the reproductive phenotype in UMS patients, which is linked to gonadotropin deficiency (Sasaki et al., 2002).
There is considerable evidence suggesting a strong influence of prenatal factors on hypothalamic development. Maternal nutritional challenges have profound effects on the development of the hypothalamic melanocortin system and may lead to disturbances in energy homeostasis in the adult (Wattez et al., 2013). In addition, reproductive function and puberty onset are also sensitive to nutritional influences during embryonic development (Dupont et al., 2012). Our results demonstrate that the Pomc-derived lineage includes cells with a critical role in the regulation of reproductive function, suggesting that disturbances in the embryonic Pomc system may also have life-long effects on the reproductive capacity of the adult mouse. The effect of prenatal nutritional perturbations on postnatal Kiss1 neurons and reproductive function has already been established (Iwasa et al., 2010); however, further experiments are required to determine whether alterations in Kiss1 neurons are due to the nutritional programming of their Pomc-expressing precursors.
The Cre-dependent viral RiboTag technique that we developed overcomes the limitations associated with developmental activation of Cre recombinase transgenes and allows direct mRNA profiling of adult neurons. Other Cre-dependent mouse lines designed for ribosome tagging and translatome analysis (Stanley et al., 2013; Zhou et al., 2013; Hupe et al., 2014) also share the same limitation imposed by developmental expression as does the RiboTag mouse line (Sanz et al., 2009). The novel viral vector dependent approach described here will overcome this limitation and allow for cell-specific mRNA profiling of adult neuronal cells.
Footnotes
This work was supported by the National Institutes of Health Grants MH086386 and GM032875 (G.S.M.) and R01-DA24908 (R.D.P.). E.S. and A.Q. were recipients of a postdoctoral mobility program Fellowship from the Spanish Ministry of Innovation and Science. We acknowledge Dr. Simina Popa for giving expert advice and performing ovariectomies, Dr. Anna La Torre for supplying mouse embryos, Dr. Karl Diesseroth for sharing the DIO-AAV construct, Dr. William Mahoney and Ambika Gunaje for kindly helping with the Illumina Beadchip microarray, and Drs. Paul Amieux and Stephanie Padilla for providing invaluable advice and critical reading of this manuscript.
The authors declare no competing financial interests.
References
- Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC, Jr, Elmquist JK, Lowell BB. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Bentsen AH, Mikkelsen JD, Tena-Sempere M. Kisspeptins: bridging energy homeostasis and reproduction. Brain Res. 2010;1364:129–138. doi: 10.1016/j.brainres.2010.08.057. [DOI] [PubMed] [Google Scholar]
- Cone RD. Anatomy and regulation of the central melanocortin system. Nat Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- Dupont C, Cordier AG, Junien C, Mandon-Pépin B, Levy R, Chavatte-Palmer P. Maternal environment and the reproductive function of the offspring. Theriogenology. 2012;78:1405–1414. doi: 10.1016/j.theriogenology.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Eriksson KS, Mignot E. T-box 3 is expressed in the adult mouse hypothalamus and medulla. Brain Res. 2009;1302:233–239. doi: 10.1016/j.brainres.2009.08.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Popa SM, Lawhorn JK, Qiu J, Tonsfeldt KJ, Bosch MA, Kelly MJ, Rønnekleiv OK, Sanz E, McKnight GS, Clifton DK, Palmiter RD, Steiner RA. Molecular properties of Kiss1 neurons in the arcuate nucleus of the mouse. Endocrinology. 2011;152:4298–4309. doi: 10.1210/en.2011-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- Hill JW, Elmquist JK, Elias CF. Hypothalamic pathways linking energy balance and reproduction. Am J Physiol Endocrinol Metab. 2008;294:E827–E832. doi: 10.1152/ajpendo.00670.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 2014;42:e14. doi: 10.1093/nar/gkt995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa T, Matsuzaki T, Murakami M, Fujisawa S, Kinouchi R, Gereltsetseg G, Kuwahara A, Yasui T, Irahara M. Effects of intrauterine undernutrition on hypothalamic Kiss1 expression and the timing of puberty in female rats. J Physiol. 2010;588:821–829. doi: 10.1113/jphysiol.2009.183558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- Mizuno TM, Kleopoulos SP, Bergen HT, Roberts JL, Priest CA, Mobbs CV. Hypothalamic pro-opiomelanocortin mRNA is reduced by fasting and [corrected] in ob/ob and db/db mice, but is stimulated by leptin. Diabetes. 1998;47:294–297. doi: 10.2337/diab.47.2.294. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci. 2009;29:11859–11866. doi: 10.1523/JNEUROSCI.1569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla SL, Carmody JS, Zeltser LM. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med. 2010;16:403–405. doi: 10.1038/nm.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling M, Anthwal N, McNay D, Gradwohl G, Leiter AB, Guillemot F, Ang SL. Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev Biol. 2011;349:406–416. doi: 10.1016/j.ydbio.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Quintana A, Zanella S, Koch H, Kruse SE, Lee D, Ramirez JM, Palmiter RD. Fatal breathing dysfunction in a mouse model of Leigh syndrome. J Clin Invest. 2012a;122:2359–2368. doi: 10.1172/JCI62923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A, Sanz E, Wang W, Storey GP, Güler AD, Wanat MJ, Roller BA, La Torre A, Amieux PS, McKnight GS, Bamford NS, Palmiter RD. Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue-dependent behaviors. Nat Neurosci. 2012b;15:1547–1555. doi: 10.1038/nn.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki G, Ogata T, Ishii T, Hasegawa T, Sato S, Matsuo N. Novel mutation of TBX3 in a Japanese family with ulnar-mammary syndrome: implication for impaired sex development. Am J Med Genet. 2002;110:365–369. doi: 10.1002/ajmg.10447. [DOI] [PubMed] [Google Scholar]
- Schinzel A, Illig R, Prader A. The ulnar-mammary syndrome: an autosomal dominant pleiotropic gene. Clin Genet. 1987;32:160–168. doi: 10.1111/j.1399-0004.1987.tb03347.x. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn JW, Elmquist JK, Williams KW. Neuronal circuits that regulate feeding behavior and metabolism. Trends Neurosci. 2013;36:504–512. doi: 10.1016/j.tins.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S, Domingos AI, Kelly L, Garfield A, Damanpour S, Heisler L, Friedman J. Profiling of glucose-sensing neurons reveals that GHRH neurons are activated by hypoglycemia. Cell Metab. 2013;18:596–607. doi: 10.1016/j.cmet.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Cheung CC, Clifton DK, Steiner RA. Regulation of hypothalamic proopiomelanocortin mRNA by leptin in ob/ob mice. Endocrinology. 1997;138:5063–5066. doi: 10.1210/endo.138.11.5651. [DOI] [PubMed] [Google Scholar]
- Varela L, Horvath TL. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012;13:1079–1086. doi: 10.1038/embor.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattez JS, Delahaye F, Lukaszewski MA, Risold PY, Eberlé D, Vieau D, Breton C. Perinatal nutrition programs the hypothalamic melanocortin system in offspring. Horm Metab Res. 2013;45:980–990. doi: 10.1055/s-0033-1357182. [DOI] [PubMed] [Google Scholar]
- Wu Q, Whiddon BB, Palmiter RD. Ablation of neurons expressing agouti-related protein, but not melanin concentrating hormone, in leptin-deficient mice restores metabolic functions and fertility. Proc Natl Acad Sci U S A. 2012;109:3155–3160. doi: 10.1073/pnas.1120501109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Faulkner LD, Hill JW. Cross-talk between metabolism and reproduction: the role of POMC and SF1 neurons. Front Endocrinol (Lausanne) 2011;2:98. doi: 10.3389/fendo.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, Pu WT. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci U S A. 2013;110:15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]