Organization is a hallmark of life (1). As observational methods have improved, the extent of organization that is revealed has grown continuously. It was once thought that the bacterial nucleoid might be the last refuge of entropy because of its amorphous appearance in micrographs. This view was undermined by the demonstration that a few specific sequences, such as the origin and terminus of replication, were at least roughly localized within the cell (2–4). Several other regions of the chromosome were also found to reside between the origin and terminus, suggesting that the arrangement of chromosomal loci in the cell might correspond to their order along the DNA (5, 6). Now, in a technical tour de force from the Shapiro and McAdams laboratories at Stanford presented in this issue of PNAS, Viollier et al. (7) studied 112 sites across the genome of Caulobacter crescentus and found that every one has a specific location within the nucleoid. They accomplished this feat by using the fluorescent repressor–operator system (FROS), in which an array of transcriptional operators inserted into the chromosome is illuminated by means of a fluorescent derivative of the cognate repressor (2, 8, 9).
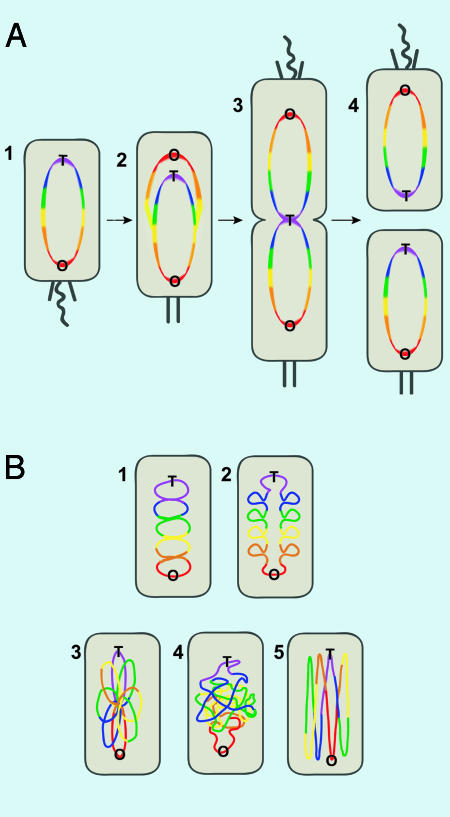
The pattern is perfectly simple (Fig. 1A1). In resting G1 cells, the cellular position of any gene correlates linearly with how far it is along the chromosome from the replication origin. Thus, genes close to the origin localize near the origin at one cell pole, those genetically near the terminus are near the opposite cell pole, and the rest are ordered in between. Because replication proceeds bidirectionally from the origin, the pattern also reflects replication timing, with origin-proximal loci being replicated earliest. Equally dramatic live-cell imaging demonstrated that, after replication, loci segregate actively to mirror-symmetric positions (Fig. 1 A2). The two origins move to opposite cell poles, and genes increasingly distant from the origin are located closer and closer to the cell center (Fig. 1 A3). Cell division then reproduces the G1 organization (Fig. 1 A4).
Fig. 1.
Organization and replication dynamics of the Caulobacter chromosome. Color indicates distance from the origin along the length of the chromosome; the order is red, orange, yellow, green, blue, and purple. O and T indicate the origin and terminus of replication. The flagellum and pili are shown as curved and slanted black lines, respectively, and the cell stalk is represented by two vertical black segments. (A1) In the G1 phase, the replication origin and nearby sequences (red) are close to the flagellar pole of the cell. (A2) After initiation, one copy of the origin moves to the other end of the cell and the other moves a little closer to the flagellar pole. (A3) As regions increasingly distant from the origin are copied, they too are segregated. (A4) After division, the chromosomes in each daughter cell are positioned as they were in the parent before replication. (B) The chromosome must adopt a folded conformation to fit within the cell volume. (B 1 and 2) Depicted is a series of stacked loops orthogonal to the cell axis; either model is consistent with the data of Viollier et al. (7). The two schemes differ as to whether the left and right halves of the chromosome occupy the same region (B1) or are kept apart (B2). Other possible organizations—a rosette in which loops emanate in various directions (B3), a random coil (B4), and loops along the cell axis (B5)—are ruled out by the data of Viollier et al.
How can all these different sequences be guided to their proper addresses? When only a few sequences were known to reside at specific positions, it was possible to imagine that each was attached to structural components with a fixed address via specific adapter proteins. Although this scenario appears to be the case for the origin region in several bacteria (4, 9, 10), it is absurd to think that >100 different positioning proteins bind specifically to an equal number of fixed sites. Rather, we suggest that an underlying structure of the chromosome maintains a linear register, positioning genes according to their distance from the origin and therefore their time of replication. The overall orientation with respect to the cell is set by pinning the origin and perhaps the terminus to a specific location. We find an analogy in the Grand Canyon. The linear order of strata results simply from the time of deposition. In the nucleoid, of course, there is more flexibility than in layers of granite, particularly within the fundamental structural unit of the chromosome, the topological domain (12, 13). Because recent work shows that these domains have sizes of only ≈10 kb in Escherichia coli (14), the deviation from linearity that occurs within domains would be smaller than the resolution limit of the Viollier et al. (7) experiments (SD = 0.1 cell length). To prevent diffusion of supercoils between domains, the DNA must be tightly constrained. It is reasonable to think that these constraints may also be involved in ordering the domains, thereby preventing large-scale diffusion of the DNA; this may be accomplished through connections to the cytoskeleton (15).
Viollier et al. (7) observed that the ≈12-kb array of illuminated operators forms a discrete focus very soon after replication and concluded that newly replicated DNA segments are condensed right after replication—thank goodness, because a longer delay would impede partitioning instead of promoting it (16). Once each locus has been copied, it moves directly to its final destination and remains there until the next S phase, implying that it is incorporated immediately into a nucleoid structure that persists for a whole cell cycle. In this view, there are connections between domains, and, in follow-the-leader fashion, the domains are placed in linear order according to their time of replication.
These results, together with our understanding of topological domains, argue that the chromosome is organized in a series of loops orthogonal to the cell axis. Future studies should elucidate whether the two arms of the chromosome occupy the same region (Fig, 1B1) or, much more likely, are kept separate (Fig. 1B2). The current results did not have the resolution necessary to distinguish between these alternatives. The results do, however, rule out several other organizational schemes, such as a rosette structure (Fig. 1B3) in which loops emanate randomly from the central core; a random coil model (Fig. 1B4); and a model in which the chromosome is looped parallel rather than perpendicular to the cell axis (Fig. 1B5). None of these three models would order genes linearly.
What provides the driving force for the movement of DNA during active segregation? The force of ejection from a replisome (11) cannot accomplish this task, because nascent DNA, not yet deposited into an ordered structure, would collapse into a flexible coil rather than shooting directly from the polymerase to the cell pole. Cotranscriptional translation of membrane proteins (17), which attaches various points on the chromosome to the slowly growing membrane, also cannot account for the rapidity and specificity with which the Caulobacter chromosome moves. Thus, a fast motor is needed. DNA translocases (18), such as FtsK from E. coli, can move a load many times larger than a single domain at high speed; for FtsK, this figure is 5 kb/s (O. Levy, P. J. Pease, and N.R.C., unpublished results). The Bacillus subtilis translocase, SpoIIIE, pumps the bulk of the chromosome into the small volume of a developing spore (19).
The Shapiro laboratory also demonstrated roles for Smc (structural maintenance of chromosomes), MreB, and topoisomerase IV in chromosomal organization in three other recent reports in PNAS (20–22). The widely conserved Smc has been shown in various systems to condense DNA and aid in successful segregation (23). Approximately 10–15% of the Caulobacter smc mutants fail to segregate oriC to opposite cell poles (24). Depletion or overexpression of MreB, a bacterial actin homolog, resulted in mislocalization of the origin as well as a segregation defect in Caulobacter (20). The MreB homolog ParM forms filaments that physically push plasmids to opposite ends of a cell (15), suggesting a direct role for MreB in chromosome segregation. Finally, loss of function of topoisomerase IV (topo IV), a widely conserved, essential bacterial protein that decatenates and unknots DNA and relaxes positive supercoils (25, 26), prevents segregation of origins in approximately one-third of cells (22). This finding indicates that topo IV disentangles DNA throughout S phase in Caulobacter in addition to decatenating chromosomes at the end of replication.
The dynamic movement and segregation of the Caulobacter chromosome provide further support for the factory model for DNA replication advanced most convincingly by the Grossman laboratory (27). In this model, the two replisomes responsible for copying the left and right halves of the chromosome are colocalized and move very little compared with the length of DNA replicated. The Caulobacter replisome does move through the cell during S phase (28), but the speed is only ≈0.3 μm/hour as determined by where each locus duplicates. Consistent with the factory model, Viollier et al. (7) show that DNA moves to the replisome before replication and moves away afterward hundreds of times more quickly than the movement of the replisome through space.
Harvesting these results required a massive scale-up of FROS. At the heart of the article is the ability to measure accurately the position of 112 different chromosomal loci; each locus was measured in 500 cells. The two key innovations were (i) the use of a transposon carrying the operator array to generate strains marked at many different locations around the chromosome and (ii) the analysis of the resulting micrographs. In all, over 50,000 images were analyzed. These analyses were done not by locking graduate students in a room with a pile of images; instead, Viollier et al. (7) constructed image analysis software that allowed high-throughput measurement of focus position relative to the cell pole. These advances were essential to establish the reliability of the data, given that there was variability in focus position among cells.
Is this lovely pattern of organization specific to Caulobacter crescentus, an aquatic member of the α-proteobacterial class? Caulobacter does change shape in a way most other bacteria do not, but it is hard to see that the choreography of gene movement has anything to do with its development. Similar studies in other organisms will have to be done to be sure, but there is already lower resolution evidence in E. coli and B. subtilis that there is a global positioning of genes, directed movement, and partitioning during replication (5, 6).
Acknowledgments
We particularly thank Pat Higgins, Alan Grossman, Dave Sherratt, and Andrew Wright for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM31655 (to N.R.C.) and a Howard Hughes Medical Institute predoctoral fellowship (to A.M.B.).
See companion article on page 9257.
References
- 1.Schrodinger, E. (1967) What Is Life? The Physical Aspect of the Living Cell & Mind and Matter (Cambridge Univ. Press, Cambridge, U.K.).
- 2.Lau, I. F., Filipe, S. R., Soballe, B., Okstad, O. A., Barre, F. X. & Sherratt, D. J. (2003) Mol. Microbiol. 49, 731-743. [DOI] [PubMed] [Google Scholar]
- 3.Webb, C. D., Teleman, A., Gordon, S., Straight, A., Belmont, A., Lin, D. C., Grossman, A. D., Wright, A. & Losick, R. (1997) Cell 88, 667-674. [DOI] [PubMed] [Google Scholar]
- 4.Niki, H. & Hiraga, S. (1998) Genes Dev. 12, 1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niki, H., Yamaichi, Y. & Hiraga, S. (2000) Genes Dev. 14, 212-223. [PMC free article] [PubMed] [Google Scholar]
- 6.Teleman, A. A., Graumann, P. L., Lin, D. C., Grossman, A. D. & Losick, R. (1998) Curr. Biol. 8, 1102-1109. [DOI] [PubMed] [Google Scholar]
- 7.Viollier, P. H., Thanbichler, M., McGrath, P. T., West, L., Meewan, M., McAdams, H. H. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinett, C. C., Straight, A., Li, G., Willhelm, C., Sudlow, G., Murray, A. & Belmont, A. S. (1996) J. Cell Biol. 135, 1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon, G. S., Sitnikov, D., Webb, C. D., Teleman, A., Straight, A., Losick, R., Murray, A. W. & Wright, A. (1997) Cell 90, 1113-1121. [DOI] [PubMed] [Google Scholar]
- 10.Glaser, P., Sharpe, M. E., Raether, B., Perego, M., Ohlsen, K. & Errington, J. (1997) Genes Dev. 11, 1160-1168. [DOI] [PubMed] [Google Scholar]
- 11.Lemon, K. P. & Grossman, A. D. (2001) Genes Dev. 15, 2031-2041. [DOI] [PubMed] [Google Scholar]
- 12.Pettijohn, D. E. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, D.C.), Vol. 1, pp. 158-166. [Google Scholar]
- 13.Higgins, N. P. (1999) in Organization of the Prokaryotic Genome, ed. Charlebois, R. L. (Am. Soc. Microbiol., Washington, D.C.), Vol. 1, pp. 189-202. [Google Scholar]
- 14.Postow, L. A., Hardy, C. D., Arsuaga, J. & Cozzarelli, N. R. (2004) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 15.Gerdes, K., Moller-Jensen, J., Ebersbach, G., Kruse, T. & Nordstrom, K. (2004) Cell 116, 359-366. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, C. D., Crisona, N. J., Stone, M. D. & Cozzarelli, N. R. (2004) Philos. Trans. R. Soc. Lond. B 359, 39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woldringh, C. L. (2002) Mol. Microbiol. 45, 17-29. [DOI] [PubMed] [Google Scholar]
- 18.Donachie, W. D. (2002) Mol. Cell 9, 206-207. [DOI] [PubMed] [Google Scholar]
- 19.Bath, J., Wu, L. J., Errington, J. & Wang, J. C. (2000) Science 290, 995-997. [DOI] [PubMed] [Google Scholar]
- 20.Gitai, Z., Dye, N. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA. 101, 8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan, K., Huntwork, S. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 7415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, S. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA. 101, 9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haering, C. H. & Nasmyth, K. (2003) BioEssays 25, 1178-1191. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, R. B. & Shapiro, L. (1999) Proc. Natl. Acad. Sci. USA 96, 10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crisona, N. J., Strick, T. R., Bensimon, D., Croquette, V. & Cozzarelli, N. R. (2000) Genes Dev. 14, 2881-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams, D. E., Shekhtman, E. M., Zechiedrich, E. L., Schmid, M. B. & Cozzarelli, N. R. (1992) Cell 71, 277-288. [DOI] [PubMed] [Google Scholar]
- 27.Lemon, K. P. & Grossman, A. D. (2000) Mol. Cell 6, 1321-1330. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, R. B., Wang, S. C. & Shapiro, L. (2001) EMBO J. 20, 4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]