Abstract
Mutagenesis of protein-encoding sequences occurs ubiquitously; it enables evolution, accumulates during aging, and is associated with disease. Many biotechnological methods exploit random mutations to evolve novel proteins. To quantitate protein tolerance to random change, it is vital to understand the probability that a random amino acid replacement will lead to a protein's functional inactivation. We define this probability as the “x factor.” Here, we develop a broadly applicable approach to calculate x factors and demonstrate this method using the human DNA repair enzyme 3-methyladenine DNA glycosylase (AAG). Three gene-wide mutagenesis libraries were created, each with 105 diversity and averaging 2.2, 4.6, and 6.2 random amino acid changes per mutant. After determining the percentage of functional mutants in each library using high-stringency selection (>19,000-fold), the x factor was found to be 34% ± 6%. Remarkably, reanalysis of data from studies of diverse proteins reveals similar inactivation probabilities. To delineate the nature of tolerated amino acid substitutions, we sequenced 244 surviving AAG mutants. The 920 tolerated substitutions were characterized by substitutability index and mapped onto the AAG primary, secondary, and known tertiary structures. Evolutionarily conserved residues show low substitutability indices. In AAG, β strands are on average less substitutable than α helices; and surface loops that are not involved in DNA binding are the most substitutable. Our results are relevant to such diverse topics as applied molecular evolution, the rate of introduction of deleterious alleles into genomes in evolutionary history, and organisms' tolerance of mutational burden.
Afundamental aspect of evolution is that mutations generate novel alleles that are then favored by selection. However, new coding mutations can be deleterious, neutral, or beneficial. Mutations can result from environmental and endogenous damage to DNA and from errors during DNA synthetic processes. In humans, random mutations produce inherited diseases and accumulate with aging and cancer (1). Conversely, targeted hypermutagenesis by immune defenses helps to generate antibody diversity and was recently shown to inactivate retroviral genomes (2). John Maynard Smith (3) proposed more than 30 years ago that the occurrence of functional mutant proteins that differ from wild type by one residue is likely frequent for evolution to be possible. Since then, numerous evolutionary and mutagenesis studies have led to the assertion that proteins are highly plastic in tolerating amino acid changes (4, 5). However, to date, we lack a quantitative measure of the degree of proteins' tolerance for random amino acid changes that occur at a random position in the protein. If a rigorous measure of proteins' degree of tolerance of random amino acid changes can be defined, then such fundamental calculations as the steepness of protein fitness landscapes or the rate of introduction of deleterious mutations into coding genomes can be more clearly delineated. Further understanding of the nature of tolerated amino acid substitutions can also lend insight into protein folding and design.
Here, we develop the concept of the probability of inactivating a protein with a random codon replacement producing amino acid change at a random location along its sequence. For conciseness, this concept is named the x factor. We describe an analytical method for calculating the x factor of proteins from randomly mutated libraries and demonstrate the method using the human DNA repair enzyme 3-methyladenine DNA glycosylase [AAG, methyl purine DNA glycosylase (MPG), and alkyl purine DNA glycosylase (ANPG)]. Nine hundred and twenty tolerated amino acid substitutions in active mutant enzymes were identified and substitutions were mapped to the available x-ray crystal structure of AAG. We examine the applicability of the x factor concept to diverse proteins by reanalyzing results from prior studies. These findings reveal a similar range of inactivation probabilities.
Materials and Methods
Escherichia coli strain MV1932 (ada alkA1) was previously derived from strain AB1157 (6). Chemicals were from Sigma–Aldrich, enzymes were from NEB (Beverly, MA), and DNA oligonucleotides were purchased from IDT (Coralville, IA), unless otherwise indicated.
Construction of PCR Mutagenesis Libraries. The low, medium, and highly mutated AAG libraries were generated by using a previously undescribed PCR mutagenesis protocol that produces similar mutational frequencies at G:C and A:T base pairs. Briefly, PCR mutagenesis was carried out sequentially with Mutazyme in the genemorph kit (Stratagene), which preferentially mutates at G:C sites, and with TaqDNA polymerase with 0.5 mM Mn++ and dNTP bias, which prefers A:T (7). Libraries were cloned into a pUC-based plasmid and transformed into MV1932 for genetic complementation. Mutants from each library were sequenced before methyl methanesulfonate (MMS) selection. For detailed PCR mutagenesis and cloning methods, refer to Supporting Methods and Tables 4, 5, 6, which are published as supporting information on the PNAS web site.
Genetic Selection for Active Enzymes. MV1932 cells were transformed with pGRFP2-AAG, low, medium, and high libraries and with empty pGRFP2 vector and grown to confluence, diluted 1:100, and grown to midlogarithmic phase in LB–carbenicillin (LB-carb) at 37°C. Cultures were treated with 0.2% MMS for 1 hr, and the drug was washed away. Pretreated and posttreated cultures were serially diluted and plated on LB-carb in triplicate to calculate survival means and standard deviations. The fractions of surviving clones in libraries were normalized to wild-type survival. The dose of MMS used was within a range of drug in which the library and control populations were proportionally affected. MMS sensitivity assays were also performed at 0.15% and 0.25% MMS; and the percent library survivals relative to controls and each other were similar at these MMS concentrations (data not shown).
Supporting Methods present methods used for (i) PCR mutagenesis, (ii) DNA sequencing, (iii) AAG protein activity assay, and (iv) x factor calculation and protein substitutability visualization.
Results and Discussion
Calculating Protein Tolerance to Random Amino Acid Substitutions. The probability of protein inactivation with one random amino acid substitution, the x factor (xsub), can be calculated from the fractions of mutants (amino acid mutation load frequencies, fn) with (n) number of amino acid changes within a gene-wide randomly mutated library, and from the proportion of mutants that survive functional selection (S). For example, f0 denotes the fraction of the unselected library with 0-aa change, f1 denotes the fraction with 1-aa change, and so on:
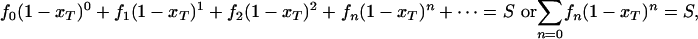 |
[1] |
where xT is the total protein inactivation probability with random amino acid change, including frameshifts (indels, i). xT can be solved after experimental determination of the fn, S, and i values. Indels are found at low percentages in random mutagenesis libraries, but invariably produce protein inactivation. To determine the true x factor (xsub) resulting only from a random codon substitution (missense or nonsense mutation), the indel fraction in the total mutational pool (i) is subtracted from xT to obtain xsub.
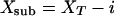 |
[2] |
To measure the probability of inactivation by random amino acid substitutions, we used the gene encoding the human AAG. AAG protects cells against DNA alkylation damage by excising alkylated base lesions including 3-methyladenine, 7-methylguanine, and 1,N6-ethenoadenine (εA) (8). The 894-bp AAG cDNA encodes a 298-aa 33-kDa monomeric protein that complements the DNA alkylation repair-deficient strain MV1932 (ada alkA1) (6) against toxicity induced by the alkylating drug MMS (9). Under MMS challenge, MV1932 cells expressing AAG from our pUC based vector exhibit >19,000-fold survival advantage over non-AAG-expressing MV1932 controls (Table 1), thus providing a stringent and specific selection for active mutant AAG enzymes.
Table 1. Calculating the x factor.
| % of library with (n) number of amino acid changes (fn × 100)
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Average mutation frequency | Library size | % Indels (i × 100) | % Survival (S × 100) | x factor (xsub) | |
| WT-AAG | 100 | — | — | — | — | — | — | — | — | — | — | — | — | 100 ± 8.6 | ||||
| Low | 5 | 20 | 40 | 20 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.2 | 2 × 105 | 6.1 | 32.7 ± 3.5 | 0.39 ± 0.038 |
| Medium | 5.6 | 5.6 | 5.6 | 11.1 | 5.6 | 33.3 | 22.2 | 5.6 | 0 | 5.6 | 0 | 0 | 0 | 4.6 | 1 × 105 | 9.9 | 18.2 ± 3.3 | 0.30 ± 0.052 |
| High | 0 | 3.6 | 7.1 | 10.7 | 3.6 | 10.7 | 14.3 | 10.7 | 17.9 | 14.3 | 3.6 | 3.6 | 0 | 6.2 | 0.9 × 105 | 5.5 | 10.7 ± 2.3 | 0.33 ± 0.034 |
| Vector only | — | — | — | — | — | — | — | — | — | — | — | — | — | 0.0051 ± 0.00017 | ||||
| Average x factor | 0.34 × 0.06 | |||||||||||||||||
Distribution of amino acid mutation load frequencies (fn), library survival (S), and x factors (xsub) in the low, medium, and highly mutated libraries. Indels (i) are expected to produce nearly 100% inactivation and are thus subtracted from the unadjusted x factors (xT) to yield x factors due to amino acid substitutions (xsub). As expected, increasing average mutation load results in lower percentage of active enzymes.
The crystal structure of the catalytically competent ΔN79 (residues 80–298) AAG protein complexed with an 1,N6-ethenoadenine substrate oligo reveals that the enzyme binds to DNA via a flat positively charged face. A β-hairpin extends into the DNA minor groove and flips the targeted nucleotide into the enzyme active site (10, 11). A water molecule is deprotonated by Glu-125 to form a hydroxyl nucleophile that cleaves the glycosylic bond between the damaged base and the sugar. The resulting abasic site is later cleaved and replaced with a normal nucleotide by the subsequent actions of an endonuclease, a DNA polymerase, and a DNA ligase (8).
We used PCR mutagenesis to generate low, medium, and highly mutated AAG cDNA libraries averaging 2.2-, 4.6-, and 6.2-aa changes per gene (a change is defined as a missense, nonsense, or indel). Sequencing of 20, 18, and 28 mutants from each unselected library revealed the fn and i values of each library (Table 1). Expression of AAG and AAG mutant libraries protected MV1932 cells against MMS-induced cell death. The fractional survival of each library relative to wild-type yielded the S values (Table 1). Solving for xsub using Eqs. 1 and 2 yielded the x factors (xsub) of the low, medium, and high libraries at: 39% ± 4%, 30% ± 5%, and 33% ± 3% (mean ± standard deviation), respectively. The x factors from the three libraries are within the 95% confidence interval of each other. The average x factor is 34% ± 6%. Thus, the overall probability of inactivating AAG with a single random amino acid change occurring randomly in the protein is ≈34%, or one-third (Table 1).
The x Factor and the Substitutability of Proteins. Using three different libraries, we obtained a consistent value for the probability that a random amino acid change will inactivate AAG. Our findings beg the question of whether a similar x factor is seen in other proteins. It may be argued that the wide range of protein functions should demand drastically different mutabilities of various proteins. On the other hand, proteins face essentially similar requirements, such as the need to properly fold into soluble globular structures necessary for function (12). General types of changes leading to unfolding would inactivate various proteins. To address these questions, we reanalyzed data from diverse published studies and calculated inactivation probabilities. First, we examined random oligonucleotide mutagenesis studies in which mutations were targeted to the catalytic center of enzymes, and from which fn and S data are available (13–18). We reasoned that these critical segments are expected to tolerate few substitutions. The results from human, bacterial, and viral enzymes are shown in Table 2. Despite the different enzymes and selection systems used, inactivation probabilities within these sensitive regions range from 44% to as high as 81%, averaging ≈60%, thus supporting our hypothesis. Second, Markiewicz and coworkers (19) examined 12 or 13 different amino acid substitutions at each residue across 90% of the 360-aa E. coli lac repressor protein using amber codon suppressor strains, which often corresponded to two or three nucleotide changes per codon. In our reanalysis of their data, we counted close to 1,380 single mutants that were inactive, ≈20% of which were temperature sensitive, of a total of 4,049 examined. This yielded an x factor for the lac repressor gene of 34%, which correlates well with our results for human AAG. Third, the x factor of a protein is conceptually similar to the proportion of new deleterious alleles that arise during the evolution of the source organism. Eyre-Walker and Keightley (20) calculated the percentage of deleterious substitution mutations that were eliminated from the human lineage by purifying selection. They examined synonymous and nonsynonymous substitution rates from coding regions of 46 homologous proteins from humans and chimpanzees. Interestingly, they conclude that at least 38% of spontaneous mutations in the human lineage were sufficiently deleterious to have been eliminated by selection (20). Together, these findings from multiple and independent experimental approaches suggest a range of similar x factors over the length of diverse proteins.
Table 2. x values calculated from active-site targeted cassette mutagenesis studies.
| Protein | Organism | Protein region (amino acid no.) | Average substitution frequency | Survival fraction (S) | x value | Ref. |
|---|---|---|---|---|---|---|
| DNA polymerase η | Homo sapiens | 52—73 | 3.8 | 0.02 | 0.81 | 13 |
| DNA polymerase I | Thermus aquaticus | 605—617 (Motif A) | 3.8 | 0.04 | 0.59 | 14 |
| Thymidylate synthase | H. sapiens | 196—199, 204—212 | 4.2 | 0.1 | 0.6 | 15 |
| Reverse transcriptase | HIV | 67—78 (β3 β4 loop) | 4.1 | 0.11 | 0.59 | 16 |
| DNA polymerase I | Thermus aquaticus | 659—671 (O helix) | 2.7 | 0.11 | 0.8 | 17 |
| Thymidine kinase | Herpes Simplex-1 | 155, 161—165 | 2.4 | 0.32 | 0.44 | 18 |
Available fn and S values were used to derive the inactivation probabilities. Although the various complementation systems may require differing levels of minimal enzyme activity, nevertheless the x values were >34% due to the concentration of mutations near the enzyme active sites.
Enzyme inactivation can result from indirect structure disrupting mutations or from direct alterations of the catalytic mechanism. The AAG functional assay is sensitive to both modalities. Minimal AAG activity necessary for complementation was assessed by measuring initial reaction rates under saturating substrate conditions in lysates of 10 random surviving clones. The results indicated that ≈5–10% of wild-type activity is necessary for survival at the MMS dose used (data not shown). Hydrophobic/hydrophilic properties appear to be crucial overall determinants of protein structure (5, 12). The buried core is sensitive to nonhydrophobic changes and those that disrupt packing, whereas the solvent-accessible surface is generally more tolerant of change. Residue size, charge, hydrogen-bonding characteristics, and bond angle flexibility are other folding factors that may be perturbed by random substitutions.
AAG is a simple monomeric protein. Larger proteins with multiple functional domains and multiple interacting partners may exhibit more complex inactivation dynamics. The x factor calculation assumes that the effects of multiple mutations are independent, in that the effects of mutations on protein function are largely additive. This is supported by findings on the λ repressor (21). However, at higher mutational loads effects of mutations may interact in more complex ways, with increased possibility of compensatory or synergistic effects. These results with AAG may slightly underestimate the x factor, because the N-terminal 79 aa of AAG are not required for enzymatic activity. Tolerated substitutions are slightly elevated in this N-terminal region. Nevertheless, protein-folding principles apply, and mutations in this region that cause overall misfolding or aggregation will produce inactivation.
There likely are variations in the substitutability of different proteins. The hydrophobic core is generally less tolerant of change than the solvent accessible exterior (5). Therefore, x factors may also be influenced by protein sizes and surface-to- volume ratios. Axe and coworkers found that 5% of single amino acid substitutions lead to an inactivated barnase enzyme (22). Rennell et al. (23) found that ≈16% of amino acid substitutions in T4 lysozyme caused inactivation. The differences from the above findings may be attributed to barnase and T4 lysozyme small sizes, which are 110 and 164 aa, respectively. Highly conserved proteins such as histones are likely to be relatively intolerant to mutation, whereas protein domains such as Fv regions of antibodies may exhibit increased tolerance against misfolding. Residues that are posttranslationally modified are also expected to be intolerant of change.
The x factor is calculated for amino acid replacements and can include the generation of stop codons. The frequencies of stop codons in the low, medium, and high libraries are 4%, 9%, and 7.5%, respectively. The x factor can be converted for single-nucleotide substitutions. Largely due to degeneracy at the third position, the nucleotide x factor is expected to be less than the amino acid x factor. Multiplying the amino acid x factor of ≈34% by the probability of nonsynonymous codon change accessible by one nucleotide (415/549) yields the nucleotide x factor of ≈26%.
Our mutagenesis scheme of creating predominantly random single nucleotide substitutions mimics the generation of natural diversity. Three naturally occurring human single-nucleotide polymorphisms arose in our database of tolerated AAG substitutions: P64L, T199A, and A258V. These variations did not exhibit appreciable effects on MV1932 complementation when individually assayed (data not shown).
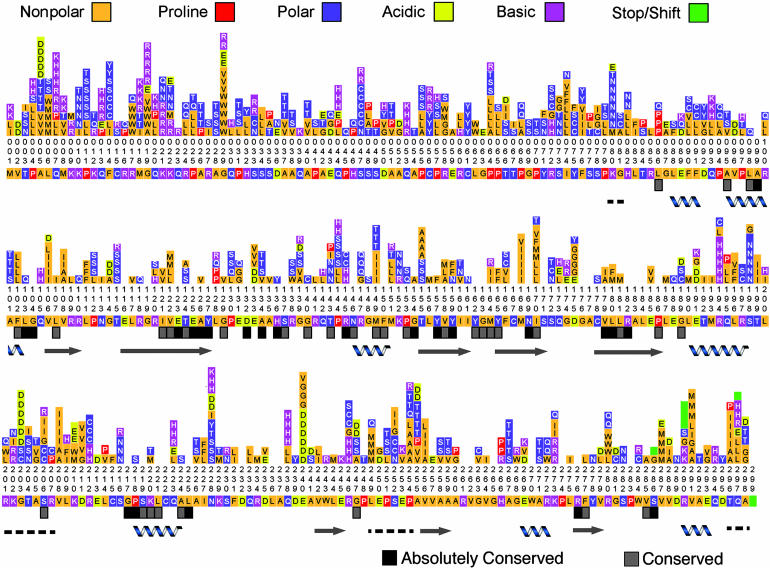
Substitutability and Structure. Previously, we have focused on the probability of amino acid changes being inactivating. We have also examined situations in which amino acid substitutions are tolerated. To analyze the nature of tolerated substitutions, we sequenced 244 mutant AAG cDNAs from the highly mutated library that complemented MV1932. This yielded a total of 920 tolerated amino acid changes. Fig. 1 maps the mutations along the AAG primary sequence. The types of tolerated amino acid substitutions at each position are indicated. Residues without bars reflect zero identified substitutions.
Fig. 1.
Tolerated amino acid changes along the AAG primary sequence, shown with evolutionary conservation and secondary structures. Two hundred forty-four active AAG mutants were sequenced, and observed amino acid substitutions are shown above the wild-type sequence. Colored bars indicate general categories of amino acids. Numbers are read vertically and indicate residue position. Below the wild-type sequence, evolutionarily invariant residues are marked by black boxes and conserved residues by gray boxes. α helices (helix), β strands (arrows), and disordered regions (dashed lines) are indicated. Homologous sequences (from human, mouse, rat, Borrelia burgdorferi, Bacillus subtilis, Arabidopsis thaliana, and Mycobacterium tuberculosis) were identified with psi-blast (10), and secondary structure calling was performed with molecular operating environment (moe, CCG, Montreal, Canada).
A residue's “substitutability index” is defined as the percent sequenced clones with a substitution at that residue. Many positions that are evolutionarily conserved are also essential for activity (10, 11) and did not tolerate changes in our assay. Examples include Glu-125, Arg-182, and Val-262, each of which interacts with the activated water molecule that hydrolyzes the sugar-base glycosylic bond. Other nonsubstituted amino acid residues include Tyr-162, which projects from a surface β hairpin and acts as a “nucleotide flipper.” Met-164 and Tyr-165 assist in this base-flipping mechanism by destabilizing the base pair adjacent to the flipped nucleotide. Y162A, M164A, and Y165A single substitution mutants were generated by Lau et al. (11) and assayed by using a genetic complementation system. The Y162A mutant exhibited large impairment of glycosylase activity, whereas M164A and Y165A showed only moderate impairment (11). Correspondingly, in our study, no substitutions were observed at Tyr-162, whereas positions Met-164 and Tyr-165 showed moderate substitutability, allowing Ile, Arg, and Phe substitutions, respectively (Fig. 1). Within the substrate-binding pocket, the flipped-out base stacks between the aromatic side-chains of Tyr-127, His-136, and Tyr-159. Y127F, H136Q, and Y159F mutants were also generated previously (11). Y127F exhibited the most profound decrease in activity, whereas Y159F was the least affected (11). In our data set, Tyr-127 was concordantly unsubstituted, and His-136 tolerated only one Tyr replacement. Tyr-159 was substituted by both Phe and Asn.
There are positions in AAG that are not evolutionarily conserved but did not exhibit any tolerated changes. The individual spatial arrangements of these interactions are likely unique to AAG. Although some of these positions may display substitutions if even more mutants are sequenced, the structural basis for lack of substitutions at many of these positions highlights three general mechanisms: specific hydrogen-bonding interactions, unique hydrophobic packing, and ion binding. For example, specific hydrogen-bonding requirements are emphasized by Glu-116's interaction with Arg-118, which, in turn, interacts with Glu-188 and Glu-245 in a three-way interaction. Arg-261 provides a hydrogen pair partner to the evolutionary conserved and unsubstituted Asp-132. This pair packs adjacent to Tyr-127, which forms part of the active site pocket. Hydrophobic packing constraints are observed at Gly-119, which is at the core of a β strand, <4.5 Å away from Leu-184. No other side chains can fit in this tight space. Similar packing constraints are observed at Leu-184, which is <4.5 Å from the unsubstituted Leu-225. Cys-167 is buried <4.5 Å from Ile-227 and close to the Cα of Cys-222. Mutations of buried residues may require concomitant mutations of other closely packed residues to maintain optimal packing. Interestingly, at least one mutant in our study appears to demonstrate this principle. It contains I170V and L181M substitutions that pack adjacently in the hydrophobic core. The conversion of Leu-181 to the slightly bulkier methionine is found to coexist with the conversion of Ile-170 to the smaller valine. Last, lack of substitutions at Ser-171 highlights the role of ion binding. Ser-171's side-chain oxygen binds to a Na+ ion, which has been postulated to enhance the structural stability of the active site floor (11).
In contrast, certain regions in AAG appear highly substitutable. Examples include the first 79 N-terminal residues that have been shown previously to be unnecessary for in vitro enzyme activity and DNA-binding specificity (24). Residues 80–81, 200–207, 249–254, and 296–298 are also highly substitutable (Fig. 1). In accord, they display low electron density in x-ray crystallography and were inferred to be disordered loops (10).
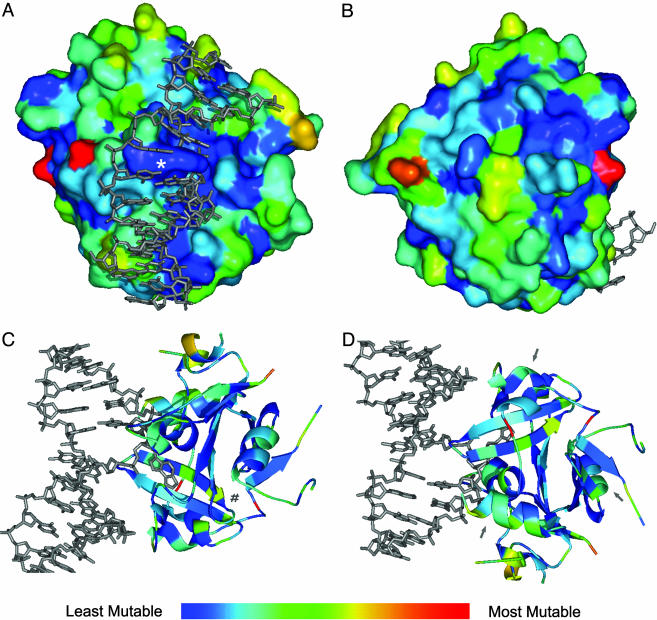
In Fig. 2, the relative substitutability indices of residues are mapped onto the available crystal structures of the N79Δ AAG mutant. Dark-blue residues are the least substitutable, and red residues are the most tolerant of change. Fig. 2 A and B shows surface residues, and Fig. 2 C and D facilitates views into the protein core. One striking feature is the general immutability of the DNA-interacting face and specifically, the nucleotide-flipper Tyr-162 (Fig. 2A). A surface region distant from the DNA-binding face (Fig. 2B) was also observed to have low substitutability scores; Glu-188, Arg-118, Glu-245, Glu-116, and Arg-110 participate in a network of charged contacts that likely contribute to protein stability. In the protein interior, a conspicuous pattern of alternating unsubstituted and substitutable sites is seen in the β4 (165–171) strand (Figs. 1 and 2 C and D). Cys-167, Asn-169, and Ser-171 are relatively unsubstituted, because their side chains face toward the active site and are involved in substrate recognition or Na+ binding (11). In contrast, Met-168 and Ile-170 tolerate hydrophobic substitutions, because their side chains face the opposite direction and pack into the hydrophobic core. Solvent-accessible surfaces generally exhibit higher substitutability compared with buried residues. This is evident in Fig. 2 C and D, where the exposed exterior sides of severalα helices exhibit greater substitutability than their interiorfacing sides.
Fig. 2.
Substitutability of AAG amino acid residues and structure. Individual residues' substitutability scores are indicated by their color in the spectrum, with red being the most substitutable and dark blue the least. (A) The DNA interacting face of AAG. The DNA-binding face and intercalating Tyr-162 (*) are largely intolerant of substitution, whereas distant loops are generally tolerant of change. (B) Rotation of A by 180°, showing the opposite side of the AAG surface. (C and D) The substitutability of AAG resides shown by secondary and tertiary structure representation. Views are rotated 180° relative to each other. Residues near the active site of AAG, adjacent to the extrahelical and 1,N6-ethenoadenine DNA lesion, are generally intolerant of change. The β4 (165–171) strand is indicated by #. Arrows point toward α helices with solvent accessible faces that exhibit greater substitutability than their buried sides.
Averages of substitutability indices in different structural motifs are presented in Table 3. In AAG, evolutionarily conserved and catalytically crucial residues are significantly less substitutable than the rest of the protein. Nonconserved residues adjacent to conserved residues in the primary sequence are generally less substitutable than other nonconserved residues, reflecting their involvement in functionally important regions. This observation suggests they may also be fruitful targets for directed evolution studies. β strand residues, as a group, are less tolerant of substitution than are α helices. This may be explained in part by the fact that in this α-β protein, the β-sheets are generally less solvent accessible and therefore possess fewer surface residues that are more likely to tolerate substitutions. Loops and turns, expectedly, are the most substitutable.
Table 3. Mean substitutability indices of AAG motifs.
| Motif residue substitutability
|
Nonmotif residues
|
t test
|
|
|---|---|---|---|
| Mean ± SD | Mean ± SD | P value | |
| Entire protein | 1.38 ± 1.10 | ||
| Evolutionarily conserved | 0.75 ± 0.85 | 1.53 ± 1.10 | 6.37E-08 |
| Nonconserved adjacent to conserved | 1.25 ± 0.84 | 1.60 ± 1.16 | 0.017 |
| α-helices | 1.19 ± 0.97 | 1.41 ± 1.12 | 0.19 |
| β-strands | 0.73 ± 0.76 | 1.52 ± 1.12 | 1.01E-08 |
| Turns and loops | 1.57 ± 1.12 | 0.93 ± 0.89 | 3.46E-07 |
| Functionally important residues | 0.56 ± 0.60 | 1.41 ± 1.11 | 1.04E-04 |
The substitutability index of individual residues (number of observed amino acid changes/number of active mutants sequenced at that position) is expressed as a percentage (×100), categorized by motifs, and averaged. The t test is performed against indices of motif nonmembers for differences in mean substitutability indices, indicative of differing importance to enzyme function.
Some Implications of the x Factor. We observed that various residues of a protein are differentially sensitive to substitutions, and that tolerance of the entire protein to random change can be defined by the x factor. The x factor is a description of an intrinsic property of individual proteins and protein motifs and can be a guiding parameter in the study of natural and artificial evolutionary processes. For example, using the estimated inactivation probability of ≈34% and assuming mutually independent effects on inactivation probability by multiple mutations, the isolation of active mutants harboring many mutations from large random mutagenesis libraries (>105) is not surprising (25). In contrast, a single, non-3-bp indel event almost certainly leads to inactivation (x≈1). Therefore, indel frequencies should be minimized in efforts to evolve novel proteins from high mutation load libraries. Retroviruses, such as HIV, may be susceptible to increased mutational burden, and lethal mutagenesis of viral genomes by introducing mutations through the use of nucleoside and ribonucleotide analogs has been proposed (26). Given our findings, such efforts may be further enhanced by the use of analogs that efficiently induce frameshift mutations. Viral genomes that encode multiple proteins as different reading frames of the same genetic sequence may be particularly sensitive to agents that generate frameshifts.
It is estimated that the human mutation rate per coding diploid genome per generation is 3.2, including base substitutions, indels, and larger changes (27). Multiplying this number by the general x factor of ≈34%, the rate of introducing deleterious coding alleles by random substitution is ≈1.0 per diploid genome per sexual generation. This is likely an underestimate, because indels inactivate coding regions much more efficiently than base substitution mutations. Dominant negative mutations may also more efficiently produce a deleterious phenotype, although the frequency of mutations that act in a dominant negative manner is largely unknown. Interestingly, our deleterious coding allele rate calculation of 1.0 is congruent with the estimate of 1.6 independently calculated by Eyre-Walker and Keightley (20), which was based on the assumption of 60,000 genes in the human genome.
Overall, our method of gene-wide random mutagenesis and sequencing highlights the relative importance of specific residues to enzyme structure and function through the numbers and types of tolerated substitutions. This work validates and extends from previous structural studies. Interestingly, the substitutability indices of individual residues can be obtained independently of conservation or structural information and are generally consistent with both. The extensive database of tolerated amino acid substitutions is obtained from a more expedient form of gene-wide study than previous techniques, such as alanine scanning. This database can provide a valuable resource for predicting the effects of mutations on protein function, which has been a focus of recent investigations (28, 29).
We advance the concept of the x factor as a measure of protein tolerance to random substitutions. The x factor may also be useful in measuring genomic robustness against mutations. It has been hypothesized that evolvability, or the ability to generate heritable variation, may be favored in certain environments (30). Genomes experiencing high mutational burden may face selective pressure to evolve proteins that are tolerant of change, in which case the observed x factors are expected to be less than x factors of homologous proteins from more faithfully propagated genomes. It may be of particular interest to examine x factors from various protein families and diverse organisms.
Supplementary Material
Acknowledgments
We are indebted to Dr. Elinor Adman for structural discussions and modeling; Drs. Greg Ireton and Django Sussman for help in generating Fig. 2; Drs. Steve Henikoff, David Baker, and Michael Fry for discussions; Drs. John Davidson, Ann Blank, and Raymond Monnat for critical reading of the manuscript; and Dan Shen for x-calculator web site programming. This work was supported by National Institutes of Health Grants CA78885 and CA80993. H.H.G. and J.C. are also supported by the Medical Scientist Training Program of the University of Washington.
Abbreviations: AAG, human 3-methyladenine DNA glycosylase; MMS, methyl methanesulfonate.
References
- 1.Loeb, L. A., Loeb, K. R. & Anderson, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris, R. S., Sheehy, A. M., Craig, H. M., Malim, M. H. & Neuberger, M. S. (2003) Nat. Immunol. 4, 641-643. [DOI] [PubMed] [Google Scholar]
- 3.Smith, J. M. (1970) Nature 225, 563-564. [DOI] [PubMed] [Google Scholar]
- 4.Creighton, T. E. (1993) Proteins (Freeman, New York).
- 5.Bowie, J. U., Reidhaar-Olson, J. F., Lim, W. A. & Sauer, R. T. (1990) Science 247, 1306-1310. [DOI] [PubMed] [Google Scholar]
- 6.Posnick, L. M. & Samson, L. D. (1999) J. Bacteriol. 181, 6763-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawate, H., Landis, D. M. & Loeb, L. A. (2002) J. Biol. Chem. 277, 36304-36311. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt, M. D., Allan, J. M., Lau, A. Y., Ellenberger, T. E. & Samson, L. D. (1999) BioEssays 21, 668-676. [DOI] [PubMed] [Google Scholar]
- 9.Samson, L., Derfler, B., Boosalis, M. & Call, K. (1991) Proc. Natl. Acad. Sci. USA 88, 9127-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau, A. Y., Scharer, O. D., Samson, L., Verdine, G. L. & Ellenberger, T. (1998) Cell 95, 249-258. [DOI] [PubMed] [Google Scholar]
- 11.Lau, A. Y., Wyatt, M. D., Glassner, B. J., Samson, L. D. & Ellenberger, T. (2000) Proc. Natl. Acad. Sci. USA 97, 13573-13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beasley, J. R. & Hecht, M. H. (1997) J. Biol. Chem. 272, 2031-2034. [DOI] [PubMed] [Google Scholar]
- 13.Glick, E., Vigna, K. L. & Loeb, L. A. (2001) EMBO J. 20, 7303-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel, P. H. & Loeb, L. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landis, D. M. & Loeb, L. A. (1998) J. Biol. Chem. 273, 25809-25817. [DOI] [PubMed] [Google Scholar]
- 16.Kim, B., Hathaway, T. R. & Loeb, L. A. (1996) J. Biol. Chem. 271, 4872-4878. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki, M., Baskin, D., Hood, L. & Loeb, L. A. (1996) Proc. Natl. Acad. Sci. USA 93, 9670-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black, M. E. & Loeb, L. A. (1993) Biochemistry 32, 11618-11626. [DOI] [PubMed] [Google Scholar]
- 19.Markiewicz, P., Kleina, L. G., Cruz, C., Ehret, S. & Miller, J. H. (1994) J. Mol. Biol. 240, 421-433. [DOI] [PubMed] [Google Scholar]
- 20.Eyre-Walker, A. & Keightley, P. D. (1999) Nature 397, 344-347. [DOI] [PubMed] [Google Scholar]
- 21.Gregoret, L. M. & Sauer, R. T. (1993) Proc. Natl. Acad. Sci. USA 90, 4246-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axe, D. D., Foster, N. W. & Fersht, A. R. (1998) Biochemistry 37, 7157-7166. [DOI] [PubMed] [Google Scholar]
- 23.Rennell, D., Bouvier, S. E., Hardy, L. W. & Poteete, A. R. (1991) J. Mol. Biol. 222, 67-88. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor, T. R. (1993) Nucleic Acids Res. 21, 5561-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaccolo, M. & Gherardi, E. (1999) J. Mol. Biol. 285, 775-783. [DOI] [PubMed] [Google Scholar]
- 26.Loeb, L. A., Essigmann, J. M., Kazazi, F., Zhang, J., Rose, K. D. & Mullins, J. I. (1999) Proc. Natl. Acad. Sci. USA 96, 1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drake, J. W., Charlesworth, B., Charlesworth, D. & Crow, J. F. (1998) Genetics 148, 1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng, P. C. & Henikoff, S. (2001) Genome Res. 11, 863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders, C. T. & Baker, D. (2002) J. Mol. Biol. 322, 891-901. [DOI] [PubMed] [Google Scholar]
- 30.Radman, M., Matic, I. & Taddei, F. (1999) Ann. N. Y. Acad Sci. 870, 146-155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.