Abstract
The process of bacterial DNA replication generates chromosomal topological constraints that are further confounded by simultaneous transcription. Topoisomerases play a key role in ensuring orderly replication and partition of DNA in the face of a continuously changing DNA tertiary structure. In addition to topological constraints, the cellular position of the replication origin is strictly controlled during the cell cycle. In Caulobacter crescentus, the origin of DNA replication is located at the cell pole. Upon initiation of DNA replication, one copy of the duplicated origin sequence rapidly appears at the opposite cell pole. To determine whether the maintenance of DNA topology contributes to the dynamic positioning of a specific DNA region within the cell, we examined origin localization in cells that express temperature-sensitive forms of either the ParC or ParE subunit of topoisomerase (Topo) IV. We found that in the absence of active Topo IV, replication initiation can occur but a significant percent of replication origins are either no longer moved to or maintained at the cell poles. During the replication process, the ParC subunit colocalizes with the replisome, whereas the ParE subunit is dispersed throughout the cell. However, an active ParE subunit is required for ParC localization to the replisome as it moves from the cell pole to the division plane during chromosome replication. We propose that the maintenance of DNA topology throughout the cell cycle contributes to the dynamic positioning of the origin sequence within the cell.
Recent work using cytological methods has shown that the circular bacterial chromosome has a specific spatial orientation within the cell, with the origin of replication and terminus at opposite edges of the nucleoid and DNA regions in the middle arranged linearly between them (1–3). In Escherichia coli and Bacillus subtilis, the origin migrates to midcell before DNA replication initiation; however, upon initiation, both copies of the duplicated origin move toward opposite cell poles (3–6). The mechanism that carries out origin movement to the cell poles is not known. However, based on the characterization of proteins, such as RacA and Spo0J of B. subtilis (4, 7–11) and ParA and ParB from Caulobacter crescentus (12), the evidence increasingly suggests that a mitotic-like apparatus for the segregation of chromosomes exists in bacteria. RacA is located at the cell poles and is required for anchoring the origin region at the pole (7). Spo0J binds to eight sites in the origin-proximal region of the chromosome and is positioned at polar regions of the nucleoid (4, 8, 9, 11). Null mutations in Spo0J cause a chromosome partitioning defect (8, 10). The Caulobacter homolog of Spo0J, ParB, binds to the origin-proximal regions of the chromosome and both ParA and ParB localize to the cell poles. Overexpression of ParA and ParB disrupts the polar localization of these two proteins, resulting in defects in cell division and chromosome partitioning (12).
The bacterium C. crescentus is a useful model for studying chromosome replication and segregation because DNA replication occurs once and only once per cell-division cycle, and the replication origin is always positioned at a cell pole (13). Each cell division is asymmetric, producing two distinct cell types: a motile swarmer cell and a stalked cell (Fig. 1A). In the swarmer cell, which is unable to initiate DNA replication, the origin of replication is located at the flagellated pole (1). DNA replication initiates when the swarmer cell differentiates into a stalked cell and replisome components assemble onto the replication origin at the stalked cell pole (14). A copy of the replicated origin moves rapidly to the pole opposite the stalk in what is thought to be an active process [see the companion article by Viollier et al. (15) in this issue of PNAS], and as DNA replication proceeds, the replisome progresses from the stalked pole to the division plane (14). Once replication is complete, the replisome disassembles, only to assemble anew at the polar replication origin of the progeny stalked cell (14).
Fig. 1.
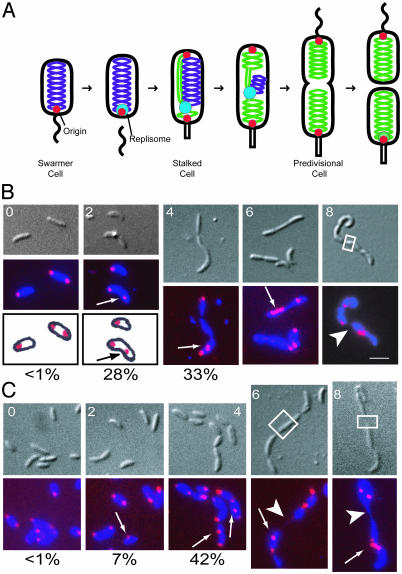
ParC and ParE are required for positioning the replication origin at the cell pole. (A) Schematic of the C. crescentus cell cycle. Unreplicated chromosomal DNA is labeled purple, newly replicated chromosomal DNA is in green, and the replisome is in blue. The replication origins are shown as red dots. Fluorescence in situ hybridization (FISH) of the origins of replication (1) in parC ts (B) and parE ts (C) strains. Cells were grown in minimal media (M2G) at 28°C to log phase and shifted to 37°C. Samples were taken at 0, 2, 4, 6, and 8 h after the temperature shift, fixed, and hybridized with a Cy3-labeled origin probe. (Upper) Nomarski differential interference contrast (DIC) microscopy images. (Lower) Overlays of the 4′,6-diamidino-2-phenylindole (a DNA-binding dye) and the Cy3 signal. The lower images are a schematic diagram of the intracellular location of the origins for 0 and 2 h after the temperature shift. Origins of replication are pseudocolored red, and DNA stained with 4′,6-diamidino-2-phenylindole is blue. Arrows indicate mislocalized origins. Arrowheads indicate a DNA-partitioning defect. White boxes indicate regions that lack DNA. (White scale bar, 2 μm.) The percentage of cells that show one polar origin and a second origin not at the pole at 0, 2, and 4 h is indicated below the FISH overlays.
To determine whether the maintenance of DNA topology contributes to the directed movement of the origin, we examined the localization of replication origins in cells carrying temperature-sensitive forms of either the ParC or ParE subunit of topoisomerase (Topo) IV. Here we present evidence that Topo IV, an enzyme required for the decatenation and segregation of daughter chromosomes (16, 17), mediates the intracellular positioning of the replication origin. When temperature-sensitive mutants of either ParC or ParE Topo IV subunits are shifted to the restrictive temperature, DNA replication continues to initiate, but the origins mislocalize. The active form of Topo IV consists of a heterotetramer of the ParC and ParE proteins (ParC2ParE2) (18). The ParC protein contains the DNA binding, cleavage, and religation domains, whereas the ParE protein contains the ATPase domain (18). We demonstrate that the ParC subunit colocalizes with the DnaB helicase component of the replisome and ParE is dispersed throughout the cell. However, a functional ParE subunit is essential for ParC association with the replisome, which in turn is required for origin placement at the cell pole, suggesting that continuous maintenance of DNA topology contributes to the cellular positioning of a specific region of the replicating chromosome.
Materials and Methods
Strains and Growth Conditions. The C. crescentus parC and parE temperature-sensitive (ts) strains, in a CB15 genetic background, were obtained from A. Newton (Department of Molecular Biology, Princeton University, Princeton) (19, 20). All other Caulobacter strains were derived from CB15N. Synchronization was performed as described (21). All strains were grown at 28°C or 37°C in M2-glucose minimal medium (22) but using 8.7 g/liter Na2HPO4, 5.3 g/liter KH2PO4, 0.2% glucose, and 0.5 mM MgSO4 in place of 0.5 mM MgCl2. When indicated, strains were induced for 2 h with 0.3% xylose. Transductions were performed with ΦCr30 (22).
The ParC-GFP strain was constructed as follows. The parC gene was amplified by using primers ParC-1 (5′-GGAATTCCATATGAACAAGCCTGTCCTTCC-3′) and ParC-22 (5′-TATGAATTCAGTTTGGGACGGAACCGCTTG-3′). The NdeI- and EcoRI-digested PCR products and the EcoRI–NotI gfp fragment from pEGFP-N3 (Clontech) were cloned into the integration vector pXGFP4 (gift from M. R. K. Alley, Anacor Pharmaceuticals, Palo Alto, CA). The resulting plasmid (pSCW449) was integrated into the chromosome of CB15N at the xylose locus by a single crossover event to produce LS3744. The GFP-ParE strain was constructed as follows. The parE gene was amplified by using primers ParE-31 (5′-TTAACTAGTGTGGAGCCGCGCGTCGAGC-3′) and ParE-41 (5′-TATAGTTAACCTACAAATCCAGATCCGCCGACGC-3′). The SpeI- and HpaI-digested PCR products were cloned into the integration vector pXGFP4-C1 (gift from M. R. K. Alley). The resulting plasmid (pSCW537) was integrated into the chromosome of CB15N at the xylose locus by a single crossover event to produce LS3745. Western blotting was performed on both ParC-GFP and GFP-ParE strains after induction with 0.3% xylose for 2 h to confirm integration at the xylose locus. GFP fusion proteins of expected sizes were expressed.
The ParC-CFP construct was constructed as follows: Asp718- and HpaI-digested pSCW449 was ligated with Asp718- and XmnI-digested PJC2 to make pSCW540. pSCW540 was then digested with AflII, the ends blunted, digested with NheI, and then ligated with SpeI- and HpaI-digested pXGFP7 to produce pSCW546. pSCW546 was then integrated into the CB15N chromosome at the xylose locus to produce LS3746.
For strains, see Table 1, which is published as supporting information on the PNAS web site.
Immunoblots. Western blot analysis was performed as described (23). Samples were normalized so that approximately equal amounts of total protein were loaded into each lane. Rabbit polyclonal anti-ParC and anti-ParE antibodies (24) were used at a dilution of 1:5,000. Horseradish peroxidase-conjugated donkey anti-rabbit antibodies (Jackson ImmunoResearch) were used at a 2:25,000 dilution. The Perkin–Elmer Western Lightning Chemiluminescence Reagent Plus Kit was used for detection.
Live Cell Microscopy. Cells were imaged as described (14) with the following modifications. Cells were immobilized on a thin layer of agarose containing M2-glucose medium (M2G) or M2G including 0.3% xylose when appropriate.
Results
Topo IV Subunits ParC and ParE Are Required for Positioning the Replication Origin at the Cell Pole. To determine the relationship between DNA topology and the cellular positioning of chromosomal regions, we used FISH to observe the cellular location of the origin in parC ts and parE ts strains (19, 20) after a shift from the permissive to the nonpermissive temperature (Fig. 1 B and C).
Temperature-sensitive mutants of parC (PC8861) and parE (PC8830) display a late-stage cell division defect at the nonpermissive temperature (37°C), resulting in the formation of filamentous chains of cells (19, 20). Sequence analysis of these strains (data not shown) revealed that the parC ts allele contains a point mutation at residue 680 that converts isoleucine to serine. The parE ts allele contains a point mutation at residue 404 that converts leucine to serine. Both parE and parC are essential genes (19). On shift to nonpermissive temperature, the parC ts and parE ts mutant strains exhibit a gradual decrease in colony forming units (data not shown). Flow cytometry analysis of the parC and parE ts mutant strains after a shift to the nonpermissive temperature shows that these strains continue to initiate DNA replication while unable to complete cell division (data not shown).
At the time of the shift to the restrictive temperature (0 h), both parC ts and parE ts cells exhibit a wild-type pattern of origin localization, with either a unipolar origin (nonreplicating swarmer cells) or bipolar origins (cells that have initiated DNA replication). Two hours after the shift, of cells containing only two origins, 28% of parC ts and 7% of parE ts cells have one origin at the cell pole with the other origin not at the pole (Fig. 1 B and C), whereas <1% of cells in both strains have two nonpolar origins (n > 100 cells). At 4 h after the shift, the percentage of cells with one polar and one nonpolar origin increases to 33% in the parC ts strain and 42% in the parE ts strain, whereas ≈4% of cells in both strains have two nonpolar origins (n > 25 cells). In wild-type cells shifted to the restrictive temperature for 4 h, the percentage of cells with one polar and one nonpolar origin is ≈5%, whereas no cells with two nonpolar origins were observed (n > 100 cells). These results imply that in ts mutants of either the ParC or ParE subunits of Topo IV, the newly duplicated origin does not complete the journey to the opposite cell pole at the restrictive temperature.
At 6 and 8 h after the shift to restrictive temperature, chains of cells with multiple nonpolar origins are commonly observed, indicating that the cells are able to initiate DNA replication despite being unable to complete cell division (Fig. 1 B and C). After 8 h at the restrictive temperature, 82% of parC ts cells and 94% of parE ts cells have nonpolar origins. In addition to origin mislocalization, both the parC and parE ts mutants exhibit a DNA partitioning defect at the restrictive temperature, where DNA is unevenly distributed throughout the cell or chains of cells (Fig. 1 B and C, boxed regions). Consistent with previous results (19), we observed that ≈4% of both parC ts and parE ts mutant cells have a partitioning defect. These results indicate that ParC and ParE Topo IV subunits are necessary for origin localization and chromosome partitioning.
The ParC Subunit of Topo IV Colocalizes with the DnaB Helicase Component of the Replisome, but the ParE Subunit Is Dispersed Throughout the Cell. To determine whether the ParC and ParE proteins are localized in Caulobacter, we constructed both gfp and cfp fusions to the C terminus of the parC gene and a gfp fusion to the N terminus of parE, all under the control of a xylose-inducible promoter (25). The constructs were integrated at the chromosomal xylose locus so that their transcription is controlled by the xylose promoter, whereas the wild-type copy of each gene is present at its normal chromosomal site. To determine whether the GFP fusions are functional, the parC-gfp and gfp-parE fusion constructs were also integrated at the chromosomal xylose locus in the parC ts and parE ts mutant strains (19), respectively. The GFP fusions complemented the temperaturesensitive defects in their respective strains (data not shown), indicating that the ParC-GFP and GFP-ParE derivatives are fully functional.
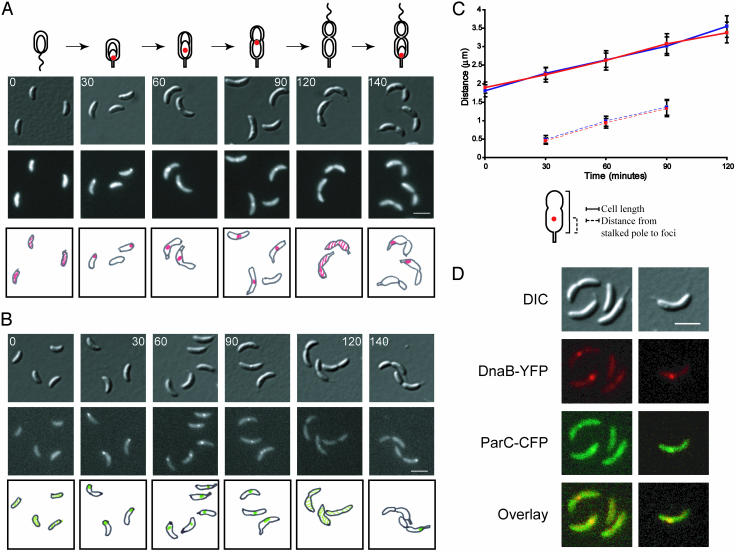
To observe the intracellular position of ParC-GFP as a function of the cell cycle, strain LS3744 containing a wild-type copy of parC and a copy of parC-gfp under the control of the xylose promoter was incubated in the presence of 0.3% xylose for 2 h to induce expression of the ParC-GFP protein. Swarmer cells were isolated and allowed to progress synchronously through the cell cycle (Fig. 2A). ParC-GFP is initially dispersed throughout the swarmer cell (0 min). At the swarmer-to-stalked cell transition, when DNA replication is initiated, a fluorescent focus appears at the stalked cell pole (30 min). As the cells proceed to the predivisional stage, the focus gradually moves toward the division plane (60 and 90 min). At the late-predivisional stage (120 min), when DNA replication is completed, the focus disperses and then reappears at the stalked cell pole (140 min). The cell-cycle localization pattern of ParC-GFP foci parallels that of the replisome components as observed previously (14). As cells progressed through the cell cycle, we observed an increased number of cells with ParC-GFP foci. At the swarmer-to-stalked cell transition, 21% of the cells had visible ParC-GFP foci. At the stalked cell stage of the cell cycle, 31% of the cells had visible foci, and at the early predivisional stage, 44% of the cells had foci. The low percentage of initial focus formation could be attributed to the fact that the wild-type copy of ParC is still present in the cell. However, when ParC-GFP was expressed in a parC ts strain that was incubated for 4 h at the nonpermissive temperature, a similar low initial percentage of foci formation still occurred in early stalked cells (data not shown).
Fig. 2.
ParC colocalizes with DnaB component of the replisome. Intracellular localization of ParC-GFP in LS3744(A) and DnaB-YFP in LS3587 (B) during the cell cycle. LS3744 and LS3587 swarmer cells were isolated and allowed to progress synchronously through the cell cycle. Images were collected at the indicated times (in minutes). LS3744 was grown in minimal media and induced with 0.3% xylose for 2 h to induce expression of ParC-GFP before synchronization. (Top) DIC images. (Middle) Either GFP or YFP fluorescence. (Bottom) Schematic diagrams of ParC-GFP fluorescence in red and DnaB-YFP fluorescence in green. (White scale bars, 2 μm.) (C) Measurements of average cell length (solid lines) for LS3744 (blue) and for LS3587 (red). The distance of ParC-GFP foci (blue) and DnaB-YFP foci (red) from the stalked pole at successive time points in a synchronized cell cycle are shown as dashed lines. At least 50 cells were measured for each time point. (D) Colocalization of DnaB helicase and ParC. LS3733 containing parC-cfp at the xyl locus and dnaB-yfp at its endogenous chromosomal locus was grown in minimal media containing 0.3% xylose for 2 h to induce expression of ParC-CFP. Samples of cells from a mixed culture were imaged by fluorescence microscopy. Top row of images show DIC images, second row shows DnaB-YFP foci (pseudocolored red), third row shows ParC-CFP foci (pseudocolored green), and bottom row shows overlays of the YFP and CFP channels. The overlap of red and green signal gives yellow. (White scale bar, 2 μm.)
An early step in the commitment to chromosome replication and the formation of the replisome in E. coli is the recruitment of the DnaB helicase to the unwound AT-rich region of the origin (26). We previously showed that in Caulobacter, the DnaB helicase tracks with replisome components, the β-clamp loaders HolB and HolC, during DNA replication (14). The cell-cycle pattern of DnaB-YFP foci in strain LS3587, containing a dnaB-yfp fusion replacing its respective wild-type copy under the control of its endogenous promoter, is shown in Fig. 2B. DnaB-YFP is dispersed in swarmer cells (0 min). At the swarmer-to-stalked cell transition (30 min), a DnaB-YFP focus appears at the stalked cell pole. The DnaB-YFP focus gradually moves toward the division plane as cells proceed to the predivisional stage (60 and 90 min). At the late-predivisional stage (120 min), the DnaB-YFP focus disperses and then reappears at the stalked cell pole (140 min). Approximately 80–90% of cells have visible DnaB-YFP foci at time points where DNA replication takes place.
The cell length and the distance from the stalked pole to the middle of the DnaB-YFP or ParC-GFP foci were measured as a function of the cell cycle for both LS3587 and LS3744, respectively (Fig. 2C). The ParC-GFP and DnaB-YFP foci tracked together throughout the cell cycle, indicating colocalization with the replisome. To confirm this, we examined Caulobacter strain LS3733 in which parC-cfp was integrated at the xylose locus and a dnaB-yfp gene fusion replaced its respective wild-type copy as the only chromosomal copy under the control of its endogenous promoter. In this strain, DnaB-YFP colocalized with the ParC-CFP foci in all cells in which both foci were visible (Fig. 2D), suggesting that ParC is part of the replisome complex.
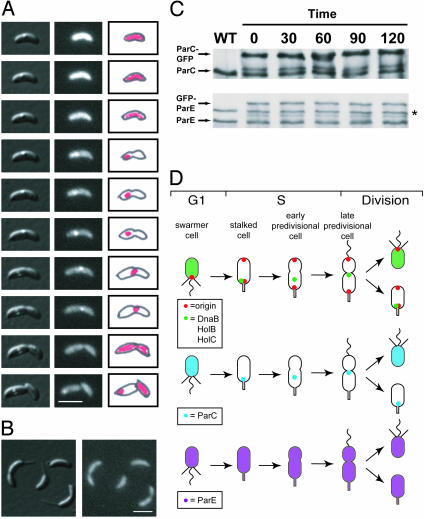
We performed time-lapse microscopy of living cells expressing ParC-GFP (LS3744) progressing through the cell cycle. Images were collected every 30 min over a 330-min cell cycle (Fig. 3A; see Movie 1, which is published as supporting information on the PNAS web site). The central dot observed in the cells shown in the DIC images (Left) is an aggregate of what is most likely β-hydroxybutyric acid, which is normally observed in cultures of C. crescentus. In the fluorescent images (Center), ParC-GFP is initially dispersed in the swarmer cell but forms a focus at the stalked pole at the swarmer-to-stalked cell transition. Swarmer cells appear brighter in the time-lapse images because cells are progressively bleached at each time point. As the cell cycle proceeds, ParC-GFP moves closer to the division plane, until it disperses in the late-predivisional cell, close to the time of cell division. ParC-GFP then reappears at the stalked pole, as was observed with other replisome components (14).
Fig. 3.
ParC localizes with the replisome whereas ParE is dispersed intracellularly. (A) Time-lapse microscopy of ParC-GFP in strain LS3744. Cells were grown in minimal media with 0.3% xylose for 2 h to induce expression of ParC-GFP. Swarmer cells were isolated and placed on an agarose pad containing xylose and images of the same cells were acquired every 30 min as the cells progressed through the cell cycle. (Left) DIC images. (Center) GFP fluorescence. (Right) Schematic diagrams of the observed fluorescence in red. (White scale bar, 2 μm.) (B) Intracellular localization of GFP-ParE. Cells from a mixed culture of strain LS3745 were grown in 0.3% xylose for 2 h to induce expression of GFP-ParE before being placed on a xylose-containing agarose pad and examined by fluorescence microscopy. (Left) DIC image. (Right) GFP fluorescence. (White scale bar, 2 μm.) (C) Immunoblots of ParC-GFP and GFP-ParE throughout the cell cycle by using antibodies to ParC or ParE (24). Samples were taken from a wild-type (WT) control and from synchronized populations of strains LS3744 and LS3745 at the indicated times (in minutes), after induction with 0.3% xylose for 2 h. Approximately equal amounts of cells were loaded into each lane. Asterisk indicates a cross-reacting band. (D) Schematic of the cell cycle localization patterns of the origin (red), Topo IV ParC (blue) and ParE (purple) subunits, and the replisome components (green) DnaB helicase, and the clamp loaders HolB and HolC (14).
In contrast to ParC, the GFP-ParE subunit is dispersed throughout the cell at all times during the cell cycle (Fig. 3B). To ascertain that GFP-ParE is dispersed in vivo and not being dispersed because of competition from wild-type ParE protein, the gfp-parE fusion construct was integrated into the chromosomal locus in the parE ts mutant strain. GFP-ParE was dispersed throughout the cell even after 4 h at the restrictive temperature in the mutant background (data not shown).
Immunoblots of ParC-GFP and GFP-ParE during the cell cycle show that both proteins are present throughout the cell cycle and are expressed at approximately equivalent amounts to the wild-type protein (Fig. 3C). A summary of the positions of the replication origin, ParC, ParE, and DnaB as a function of the cell cycle is shown in Fig. 3D.
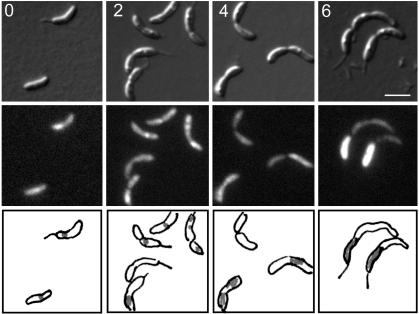
To determine whether ParC localization to the replisome requires active ParE, we transduced the Pxyl-parC-gfp construct into the parE ts strain (LS3775). When strain LS3775 is shifted to the nonpermissive temperature, the percentage of cells with ParC-GFP foci begins to decrease at 2 h, and by 4 h ParC is dispersed with ParC-GFP predominantly sequestered into the stalked compartment of the dividing cells (Fig. 4). Thus, active Topo IV is required for the ParC subunit to localize to the replisome.
Fig. 4.
Intracellular localization of ParC-GFP requires functional ParE. (A) LS3775 containing parC-gfp in the parE ts strain, was grown at 28°C to log phase, induced with 0.3% xylose for 2 h, and shifted to 37°C. Images were taken at 0, 2, 4, and 6 h after shift to 37°C. (Top) DIC images. (Middle) GFP fluorescence. (Bottom) Schematic diagram of ParC-GFP fluorescence in gray. (White scale bar, 2 μm.)
Discussion
Caulobacter swarmer cells are unable to initiate DNA replication until they differentiate into stalked cells (Fig. 1 A). The origin of replication is always located at a cell pole, but the replisome assembles on the origin only in the stalked cell (1, 14). As soon as the origin region is replicated, a copy of the origin is found at the opposite pole where it is retained throughout the rest of the cell cycle (1). As DNA replication proceeds, the replisome moves to the division plane, where it disassembles on completion of replication (14). DNA condensation mediated by structural maintenance of chromosomes (1) and likely other factors, such as gyrase and Topo IV, work in concert to condense, decatenate, and segregate the chromosomes. Here, we have questioned whether Topo IV activity is required for the placement of the origin at the cell poles and we have examined the cellular position of the Topo IV subunits ParC and ParE as a function of the cell cycle.
Two hours after a shift to the restrictive temperature, a significant number of cells expressing temperature-sensitive mutants of either Topo IV subunit had one polar and one nonpolar origin. These results suggest that active Topo IV subunits are required to allow the placement of a copy of the duplicated origin at the opposite cell pole. We cannot exclude the possibility that Topo IV is involved in retaining origins at the pole. However, very few cells with two nonpolar origins were observed, arguing for the former possibility. During Caulobacter DNA replication, duplicated origin regions exhibit a rapid and directed movement to the cell poles (15). This directed movement suggests the existence of a motor that rapidly drives the origin to the cell pole. In this scheme, a tangled chromosome, possibly present in the Caulobacter Topo IV ts mutants, may activate a checkpoint that inhibits the motor that drives the origins to the cell poles. In a simpler model, origin regions may fail to reach the opposite cell pole in the ts mutants because of topological constraints that cannot be resolved by Topo IV during the replication process.
Continued incubation of the parC and parE ts mutants at the restrictive temperature results in the formation of filamentous chains of cells followed by cell death. Although cell division is blocked under these conditions, perhaps because of the inability to separate the chromosomes, new rounds of replication continued to initiate; FISH analysis showed multiple origin regions (Fig. 1 B and C) and fluorescence-activated cell-sorter measurements revealed continued replication initiation (data not shown). The replication origins in these filamentous ts cells were positioned randomly in a majority of the cells. Thus, continued replication initiation in the absence of Topo IV activity appears to disrupt the cellular position of the replicated origins. Hence, during replication, we propose that inherent control of DNA topology, directly or indirectly mediated by Topo IV, contributes to the intracellular positioning of critical chromosomal regions, such as replication origins.
To determine the cellular position of the Topo IV ParC and ParE subunits during the cell cycle, we constructed gfp fusions to the parC and parE genes. The ParC subunit of Topo IV was found to colocalize with the DnaB helicase component of the replisome and to dissociate on completion of DNA replication. The ParE subunit, on the other hand, was found throughout the cytoplasm at all times. The localization patterns observed are consistent with that found in other bacterial systems. In B. subtilis, ParC localizes to the cell poles, whereas ParE is dispersed in the cytoplasm of the cell (27). In E. coli, ParC colocalizes with the replisome, and ParE accumulates in the nucleoid-free regions of the cell (28). The conservation of the disparate cellular positions of the two subunits of Topo IV among widely differing bacteria suggests that their cellular locations may serve a functional role in the regulation of Topo IV activity. Similar to B. subtilis (27), we observed that functional ParE subunits are necessary to localize ParC to the Caulobacter replisome, suggesting that an active Topo IV complex is necessary for the localization process.
The dissimilar localization pattern of two subunits of the same enzyme suggests two methods in which Topo IV activity could be regulated. (i) Release of ParC from the replisome on completion of DNA replication allows it to associate with free ParE in the cytoplasm to form an active complex at the end of replication, which coincides with the time when it is required to decatenate daughter chromosomes. (ii) ParC at the replication fork interacts with ParE to form an active Topo IV complex even while associated with the replisome. Espeli et al. (28) observed that in E. coli the majority of Topo IV activity was restricted to late in the cell cycle, and they suggested the former method of regulating Topo IV activity. However, this method assumes that whenever ParC is delocalized, high levels of Topo IV activity exist in the cell. This assumption may not be the case in Caulobacter. In swarmer cells, where DNA replication is repressed and, presumably, little need exists for Topo IV activity, both ParC and ParE are dispersed intracellularly. Thus, the second method of Topo IV regulation may also be operative in Caulobacter, while still ensuring high levels of Topo IV activity at the completion of DNA replication. During DNA replication, DNA gyrase and Topo IV are required to resolve the positive supercoils produced ahead of and behind the fork (precatenanes), respectively, during the replication process for the replication fork to proceed (29, 30). In stalked cells, at the start of DNA replication, we observed relatively low numbers of ParC-GFP foci. Although ParC-GFP was more dispersed at the start of replication, increased levels of ParC were found associated with the replisome as replication proceeded. On completion of DNA replication, ParC would then be concentrated at the cell-division plane where it is needed to decatenate replicated chromosomes and to resolve the precatenanes that accumulate behind the replication fork. This temporal and spatial regulation of the ParC subunit allows effective concentrations of Topo IV to be available at the right time and at the right place in the cell for replication to proceed, for chromosomes to be decatenated, and, consequently, for cell division to take place.
FtsK, an essential cell-division protein, localizes to the division plane in E. coli, and the C terminus has been implicated in chromosome segregation (31, 32). Recently, it has been shown that in E. coli, ParC and FtsK physically interact and that FtsK stimulates the decatenation activity of Topo IV (33). If Caulobacter FtsK localizes to the division plane, it is possible that Topo IV and FtsK may work together at the division plane to clear chromosomes from the division site just before cell division. This may explain the block in cell separation observed in Caulobacter Topo IV mutants because abnormal segregation of the chromosomes may cause a cell-cycle checkpoint to be activated that blocks the completion of cell division. A comparable phenotype has been observed in Caulobacter in the case of smc deletion mutants; these mutants exhibit abnormal chromosome segregation and cells arrest at the predivisional stage (1). Thus, we propose that Topo IV activity and its intracellular position contributes to cell cycle progression by controlling chromosome topology, segregation, and the cellular position of replication origins in the cell.
Supplementary Material
Acknowledgments
We thank Noriko Ohta and Austin Newton for the parC ts and parE ts mutant strains and for antibodies to the Topo IV ParC and ParE subunits; Patrick Viollier and Joseph Chen for stimulating scientific discussions and valuable advice; and other members of the Shapiro laboratory and Rasmus Jensen for critical reading of the manuscript. This work was supported by National Institutes of Health Grants GM32506 and GM51426. S.C.W. was supported by a predoctoral fellowship from the National Science Foundation.
Abbreviations: DIC, differential interference contrast; ts, temperature-sensitive; Topo, topoisomerase; FISH, fluorescence in situ hybridization.
References
- 1.Jensen, R. B. & Shapiro, L. (1999) Proc. Natl. Acad. Sci. USA 96, 10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teleman, A. A., Graumann, P. L., Lin, D. C., Grossman, A. D. & Losick, R. (1998) Curr. Biol. 8, 1102-1109. [DOI] [PubMed] [Google Scholar]
- 3.Niki, H., Yamaichi, Y. & Hiraga, S. (2000) Genes Dev. 14, 212-223. [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis, P. J. & Errington, J. (1997) Mol. Microbiol. 25, 945-954. [DOI] [PubMed] [Google Scholar]
- 5.Webb, C. D., Teleman, A., Gordon, S., Straight, A., Belmont, A., Lin, D. C., Grossman, A. D., Wright, A. & Losick, R. (1997) Cell 88, 667-674. [DOI] [PubMed] [Google Scholar]
- 6.Webb, C. D., Graumann, P. L., Kahana, J. A., Teleman, A. A., Silver, P. A. & Losick, R. (1998) Mol. Microbiol. 28, 883-892. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., Rudner, D. Z. & Losick, R. (2003) Science 299, 532-536. [DOI] [PubMed] [Google Scholar]
- 8.Lin, D. C., Levin, P. A. & Grossman, A. D. (1997) Proc. Natl. Acad. Sci. USA 94, 4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, D. C. & Grossman, A. D. (1998) Cell 92, 675-685. [DOI] [PubMed] [Google Scholar]
- 10.Ireton, K., Gunther, N. W. t. & Grossman, A. D. (1994) J. Bacteriol. 176, 5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser, P., Sharpe, M. E., Raether, B., Perego, M., Ohlsen, K. & Errington, J. (1997) Genes Dev. 11, 1160-1168. [DOI] [PubMed] [Google Scholar]
- 12.Mohl, D. A. & Gober, J. W. (1997) Cell 88, 675-684. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, R. B., Wang, S. C. & Shapiro, L. (2002) Nat. Rev. Mol. Cell Biol. 3, 167-176. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, R. B., Wang, S. C. & Shapiro, L. (2001) EMBO J. 20, 4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viollier, P. H., Thanbichler, M., McGrath, P. T., West, L., Meewan, M., McAdams, H. H. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng, H. & Marians, K. J. (1993) Proc. Natl. Acad. Sci. USA 90, 8571-8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams, D. E., Shekhtman, E. M., Zechiedrich, E. L., Schmid, M. B. & Cozzarelli, N. R. (1992) Cell 71, 277-288. [DOI] [PubMed] [Google Scholar]
- 18.Peng, H. & Marians, K. J. (1993) J. Biol. Chem. 268, 24481-24490. [PubMed] [Google Scholar]
- 19.Ward, D. & Newton, A. (1997) Mol. Microbiol. 26, 897-910. [DOI] [PubMed] [Google Scholar]
- 20.Ohta, N., Ninfa, A. J., Allaire, A., Kulick, L. & Newton, A. (1997) J. Bacteriol. 179, 2169-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evinger, M. & Agabian, N. (1977) J. Bacteriol. 132, 294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ely, B. (1991) Methods Enzymol. 204, 372-384. [DOI] [PubMed] [Google Scholar]
- 23.Domian, I. J., Quon, K. C. & Shapiro, L. (1997) Cell 90, 415-424. [DOI] [PubMed] [Google Scholar]
- 24.Ward, D. V. & Newton, A. (1999) J. Bacteriol. 181, 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meisenzahl, A. C., Shapiro, L. & Jenal, U. (1997) J. Bacteriol. 179, 592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York).
- 27.Huang, W. M., Libbey, J. L., van der Hoeven, P. & Yu, S. X. (1998) Proc. Natl. Acad. Sci. USA 95, 4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espeli, O., Levine, C., Hassing, H. & Marians, K. J. (2003) Mol. Cell 11, 189-201. [DOI] [PubMed] [Google Scholar]
- 29.Khodursky, A. B., Peter, B. J., Schmid, M. B., DeRisi, J., Botstein, D., Brown, P. O. & Cozzarelli, N. R. (2000) Proc. Natl. Acad. Sci. USA 97, 9419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postow, L., Crisona, N. J., Peter, B. J., Hardy, C. D. & Cozzarelli, N. R. (2001) Proc. Natl. Acad. Sci. USA 98, 8219-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, G., Draper, G. C. & Donachie, W. D. (1998) Mol. Microbiol. 29, 893-903. [DOI] [PubMed] [Google Scholar]
- 32.Yu, X. C., Weihe, E. K. & Margolin, W. (1998) J. Bacteriol. 180, 6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espeli, O., Lee, C. & Marians, K. J. (2003) J. Biol. Chem. 278, 44639-44644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.