Abstract
The chromosomal origin and terminus of replication are precisely localized in bacterial cells. We examined the cellular position of 112 individual loci that are dispersed over the circular Caulobacter crescentus chromosome and found that in living cells each locus has a specific subcellular address and that these loci are arrayed in linear order along the long axis of the cell. Time-lapse microscopy of the location of the chromosomal origin and 10 selected loci in the origin-proximal half of the chromosome showed that during DNA replication, as the replisome sequentially copies each locus, the newly replicated DNA segments are moved in chronological order to their final subcellular destination in the nascent half of the predivisional cell. Thus, the remarkable organization of the chromosome is being established while DNA replication is still in progress. The fact that the movement of these 10 loci is, like that of the origin, directed and rapid, and occurs at a similar rate, suggests that the same molecular machinery serves to partition and place many, if not most, chromosomal loci at defined subcellular sites.
Bacterial chromosomes are not static structures. They undergo dynamic topological changes during DNA replication, segregation, and transcription (1–5). For bacteria with circular chromosomes, replication initiates at a single origin of replication (ori) and proceeds bidirectionally toward the terminus of replication (ter) (6). During replication, the DNA double helix is unwound locally, introducing compensatory superhelicity and entanglements that are relieved by the action of topoisomerases (1). Before newly replicated DNA can be segregated, the two sister chromosomes are unlinked by topoisomerases, resolvases, and recombinases (2, 7, 8). After the completion of replication and segregation, each half of the predivisional cell contains one sister chromosome. The mechanism whereby the chromosomes are moved, positioned, and finally restructured before cell division is poorly understood.
Fluorescence in situ hybridization (FISH) has been used to visualize distinct chromosomal loci in fixed bacterial cells (9). In addition, a technique to label chromosomal loci in live cells has been developed by using a lac repressor GFP hybrid protein (LacI-GFP) that binds to arrays of lac operator (lacO) sequences inserted at specific sites on the chromosome (10–12). By using this labeling technique along with time-lapse fluorescence microscopy (FM), both the Escherichia coli and the Bacillus subtilis ori have been shown to move rapidly toward the cell poles once DNA replication has initiated (13–15).
Chromosome replication and segregation is coordinated with other events during the cell cycle, such as polar morphogenesis and cell division. In Caulobacter, DNA replication initiates once and only once per cell cycle and proceeds bidirectionally from a single origin (16, 17). Caulobacter cell division is asymmetric, yielding a replicative stalked cell and a nonreplicative swarmer cell with polar pili and a polar flagellum (18–20). DNA replication is blocked in the swarmer cell, as in the eukaryotic G1 phase cell. Chromosome replication can only initiate once the swarmer cell has differentiated into a stalked cell. This transition is thus analogous to the eukaryotic G1-to-S transition. Swarmer cells are separable from stalked cells by density gradient centrifugation and, once separated out and resuspended in fresh medium, progress synchronously through the cell cycle. In swarmer cells, the ori is located at the pole that bears the pili and the flagellum (Fig. 1D). After the initiation of DNA replication, a second copy of ori appears at the opposite cell pole (21). At the end of the cell cycle, as DNA replication ceases, the two copies of ter are observed near the division plane. In support of this observation, immunocytological experiments in which newly replicated DNA was localized by using antibodies to the thymine analog BrdUrd showed that DNA replicated early in the cell cycle was found near the cell poles. As the cell cycle progressed, newly replicated DNA was located at increasing distance from the cell poles, progressively closer to the plane of division, where the DNA replicated last was observed (22).
Fig. 1.
The positions of four distinct chromosomal loci in Caulobacter swarmer cells. (A) Schematic of the circular Caulobacter chromosome showing the location of the pilA, pleC, and podJ loci relative to that of ori and ter.(B and C) Histograms showing the fraction of cells with ori, pilA, pleC, and podJ at a given position in the cell as determined by FROS (B) or FISH (C). The positions of ori, pilA, pleC, and podJ are shown in green, blue, magenta, and yellow, respectively. (D) Diagram of the average subcellular positions of the four loci. Wavy line, flagellum; straight lines, pili. (E) Average subcellular positions of the pilA, pleC, and podJ loci plotted as a function of their distance from ori on the Caulobacter chromosome as determined by FISH (blue line) and FROS (magenta line).
Whereas the positioning of the ori and ter has been extensively studied in E. coli, B. subtilis, and Caulobacter, little is known about the movement and deposition of specific intervening chromosomal loci during the cell cycle. Two loci between the ori and ter have been shown to occupy distinct subcellular positions in live B. subtilis cells (23). Analysis of 22 chromosomal loci by FISH in fixed E. coli cells has similarly provided evidence for the orderly arrangement of the chromosome (24). Here, we describe a comprehensive analysis of the arrangement of >100 chromosomal sites in live cells and explore the arrangement of a subset of these sites in relation to one another as cells progress through the cell cycle. We demonstrate that, like the ori, newly replicated loci rapidly move in an orderly fashion to their specific address in the incipient daughter cell. Thus, the remarkable organization of the chromosome is established while DNA replication is ongoing.
Experimental Procedures
Bacterial Strains and Growth Conditions. Caulobacter crescentus CB15N and derivatives were grown in M2G minimal or peptoneyeast extract containing 0.02% glucose complex medium. The composition of M2G and peptone-yeast extract, protocols for bacteriophage ΦCr30-mediated generalized transductions, plasmid mobilization from E. coli to Caulobacter by conjugation, and synchronization by Percoll density gradient centrifugation have been described (25, 26). To prepare electrocompetent Caulobacter cells, cultures were grown in 2× peptone-yeast extract to an OD600 of 1.0 and processed according to Ely (25). For Caulobacter, liquid medium was supplemented with kanamycin (5 μg/ml) or spectinomycin (25 μg/ml) and solid medium was supplemented with kanamycin (20 μg/ml), gentamycin (2.5 μg/ml), hygromycin (100 μg/ml), or a mixture of spectinomycin (30 μg/ml) and streptomycin (5 μg/ml) when required. Standard antibiotic concentrations were used for E. coli (27–29). E. coli strains DH10B/λ pir and S17–1/λ pir (Lucy Shapiro, Laboratory collection) were used for construction and mobilization of the mariner plasmids pHPV499 and pHPV560, respectively. E. coli EC100D™ pir-116 (Epicentre Technologies, Madison, WI) was used as host when mapping the site of the mariner insertion on the Caulobacter chromosome. E. coli TOP10 or STBL2 (Invitrogen) were used as hosts for all other clonings.
FM. FISH and immunofluorescence microscopy (IFM) were performed as described (21). The ori, pilA, pleC, and podJ FISH probes were 5- to 10-kb fragments liberated from plasmids pGM1342 (G. Marczynski, unpublished work), pJS2 (30), pSCW400 (31), and pHPV58 (see Supporting Experimental Procedures, which is published as supporting information on the PNAS web site), respectively, by restriction enzyme digestion. For time-lapse FM experiments, 10 strains, each carrying (lacO)n at a different site in the origin-proximal half of the chromosome and the repressor expression construct (pHPV472) integrated at the xylX locus, were cultivated in M2G minimal medium containing kanamycin and spectinomycin at 30°C to the midexponential phase. Synthesis of LacI-cyan fluorescent protein (CFP) was induced by the addition of 0.03% xylose. At the same time, 50 μM isopropyl β-d-thiogalactoside (IPTG) was added to the culture, which was then further cultivated for 75 min at 30°C. Subsequently, swarmer cells were isolated as described (26) with the exception that 50 μM IPTG was added to the washing buffer. After resuspension in M2G medium containing 50 μM IPTG, the cells were transferred onto a pad of 1% agarose prepared in M2G medium containing 50 μMIPTG(t = 0 min). The pad was covered with a coverslide, was sealed, and was incubated at room temperature (25°C) until the start of the experiment. Phase contrast and fluorescence images were taken every 2 min. For fluorescence images, the illumination time was 3 sec.
The mariner strains carrying both (tetO)n and (lacO)n were grown at room temperature to the midexponential phase in peptone-yeast extract containing 0.02% glucose complex medium. At that time, synthesis of LacI-CFP and tet repressoryellow fluorescent protein (TetR-YFP) was induced with 0.2 mM xylose for 60–90 min, and swarmer (G1) cells were isolated as described (26). Cells were spotted on a 1% agarose pad, and phase contrast and fluorescence (YFP and CFP channels) images were acquired.
Strain and Plasmid Constructions, Transposon Mapping, Image and Data Processing. Detailed descriptions for these procedures are published in Supporting Experimental Procedures.
Results and Discussion
Determination of the Subcellular Position of Four Distinct Chromosomal Loci by FISH and the Fluorescent Repressor-Operator System (FROS) in Swarmer Cells. The intracellular position of chromosomal loci was visualized in live Caulobacter cells by using a reporter system based on the binding of a LacI-CFP chimera to tandem arrays of lac operator sites [(lacO)n] inserted at specific locations on the chromosome. This FROS has previously been used to investigate chromosome localization in living cells (10, 11). To confirm that FROS accurately identifies the position of chromosomal loci within live cells, we compared the subcellular position of four different chromosomal loci as determined by both the FROS and FISH techniques (9, 21). These four loci lie near the ori, the pilA, the pleC, and the podJ genes. They are located in sequential order on the 4.01 Mb circular Caulobacter chromosome (32), close to the 12, 9, 8, and 7 o'clock positions, respectively (Fig. 1 A).
FROS was adapted to Caulobacter to simultaneously visualize the position of two distinct loci, each bound by a different reporter protein: a LacI-CFP chimera bound to (lacO)n and a TetR-YFP chimera bound to arrays of tet operator sites [(tetO)n] (27). We created strains with (tetO)n inserted 4 kb to the right of ori and (lacO)n inserted at the pilA, pleC, or podJ locus, as described in Experimental Procedures. An artificial operon containing the coding sequences for LacI-CFP and TetR-YFP under the control of the Caulobacter xylX promoter (PxylX) was first integrated into the xylX locus of the wild-type strain by homologous recombination and subsequently transferred into the doubly marked ori::(tetO)n pilA::(lacO)n, ori::(tetO)n pleC::(lacO)n,or ori:: (tetO)n podJ::(lacO)n strains by generalized transduction. In each of the resulting strains, the two differentially labeled, operator-bearing loci appear as distinct subcellullar fluorescent foci in vivo after inducing the expression of LacI-CFP and TetR-YFP by the addition of xylose (2 mM; Fig. 1C and E). In Caulobacter swarmer cells, the single chromosome is in the nonreplicative G1 state (20). Hence, there is only one copy of each locus per cell. The two fluorescent foci that were observed in each doubly marked swarmer cell are: a TetR-YFP-derived ori focus at the flagellated pole and a LacI-CFP-derived focus at the position of either the pilA, pleC, or podJ locus. In each of these strains, the position of the foci along the long cell axis can be measured relative to the (“old”) flagellated cell pole. [Similar experiments in E. coli and B. subtilis are hampered by the lack of polar markers to distinguish the “old” from the “new” cell pole (23, 24)].
To facilitate the visualization of the loci and the determination of their position in a large number of cells (typically n = 500), we developed an automated image processing program (see Experimental Procedures). This program determines the outline of individual cells from phase contrast microscopy images and then identifies the point of maximal fluorescence emission for each focus within each cell from the FM images. For each cell in an image, the length of the long axis as well as the distances of the foci from both cell poles were then automatically measured, taking into account the curvature of the crescent-shaped Caulobacter cells. From these values, the ori-proximal (flagellated) pole was identified as well as the absolute and fractional distances of the pilA-, pleC-, or podJ-derived foci from the flagellated pole. The histograms in Fig. 1 B and C represent distributions of measured foci positions for FROS and FISH measurements, respectively, expressed as fractional cell lengths from the flagellated pole (the absolute swarmer cell length ranges from 1.8 to 2.2 μm). The histograms show that our measurements of the position of the peak fluorescence intensity in each individual cell have an SD of ≈0.1 relative cell lengths. Using FROS, we found that on average the ori, pilA, pleC, and podJ foci are located 0.11, 0.45, 0.59, and 0.76 relative cell lengths from the flagellated cell pole, respectively.
The corresponding analysis using FISH was performed on fixed swarmer cells using ori, pilA, pleC,or podJ DNA probes (see Experimental Procedures). After the FISH treatment, IFM was used to visualize the McpA chemoreceptor, a molecular marker for the flagellated cell pole (21, 33). The images were analyzed and the subcellular position of ori, pilA, pleC,or podJ and McpA was determined by using FM (Fig. 1C), as described for the FROS samples. The FISH results showed the ori, pilA, pleC, and podJ loci (on average) to be located at 0.17, 0.42, 0.58, and 0.76 relative cell lengths, respectively. The fact that the independent FISH and FROS techniques yielded similar results confirms that the latter reliably determines the subcellular position of chromosomal loci in living cells.
The FISH and FROS experiments indicated an approximately linear relationship between the chromosomal location of these three sites and their longitudinal position in live swarmer cells (Fig. 1 D and E). Similar ordering was also observed for replicating B. subtilis cells harboring (lacO)n at the 0, 3, 6, or 9 o'clock position of the circular chromosome (23).
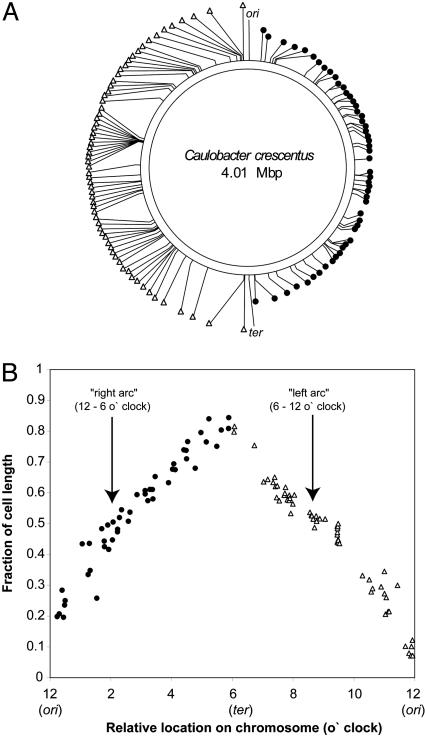
Chromosome Organization in Live G1 Cells Determined by FROS Analysis of 112 Distinct Loci. To determine whether the linear relationship between the positions of the genes on the chromosome and their location within live G1 cells applies throughout the chromosome, we analyzed 112 different loci by using FROS. A mariner (Himar1)-based transposition strategy (34) was used for the random delivery of either (tetO)n or (lacO)n into the chromosome of wild-type Caulobacter, followed by the transduction of the arrays into strains containing the LacI-CFP and TetR-YFP expression construct at the xylX locus and either (lacO)n or (tetO)n at ori (strains MT15 and MT16, respectively). MT15- or MT16-derived strains harboring the mariner (lacO)n or (tetO)n transposon were isolated and the sites of the transposon insertions were mapped (see Experimental Procedures). Fig. 2A shows the location of 83 mariner insertions and 29 additional insertions constructed by homologous recombination (see Experimental Procedures, and Fig. 5, which is published as supporting information on the PNAS web site) into MT16. Swarmer cells were isolated from these strains (henceforth referred to as mariner strains) and the position of the two fluorescent foci, the focus at ori marking the flagellated pole, and the other focus derived from the mariner insertion, were determined by FROS. Image data were processed and analyzed as described above, generating histograms of the position of each locus in several hundred cells. From the histogram for each mariner strain, the average subcellular position of the focus derived from the mariner insertion relative to the flagellated pole was calculated (see Table 1, which is published as supporting information on the PNAS web site). Fig. 2B shows the resulting average position of the focus for each mariner strain plotted versus the location of the operator insertions on the Caulobacter chromosome. The plot shows that for insertions located on both the “right” and “left” arcs of the chromosome (12–6 o'clock and 6–12 o'clock regions, respectively), there is a linear correlation between the physical position of a given insertion on the chromosome and its position along the long cell axis. The ori and ter, located at 12 and 6 o'clock on the chromosome (Fig. 1 A), are located at 0.1 and 0.9 relative cell lengths from the flagellated cell pole, respectively. A similar, but less detailed study of the organization of the replicating E. coli chromosome by using FISH (fixed cells), suggested the existence of two macrodomains of ≈920 kb, encompassing ori and ter, that behave as distinct units with respect to their subcellular positioning and their segregation during the replication cycle (24). Our results do not show any such exceptional regions for Caulobacter.
Fig. 2.
Positions of 112 distinct chromosomal loci in Caulobacter swarmer cells determined by FROS. (A) Positions of the 112 loci tagged either with (lacO)n or (tetO)n (see Fig. 5 for detailed information) relative to ori and ter (12 and 6 o'clock, respectively) on the chromosome. (B) The average subcellular position (see Table 1) of the 112 loci in A plotted as a function of their relative location on the chromosome. In both A and B, insertions on the right (12–6 o'clock; •) and left (6–12 o'clock; ▵) arc of the chromosome are depicted.
Our analysis locates chromosomal regions comprising of as little as 125–250 kb to distinct subcellular positions. Thus, loci separated by at least 250 kb are positioned at discrete places in the cell. Because the order of the genes on the chromosome likely reflects their intracellular position, transposing genes from their native location to ectopic sites on the chromosome should alter their position inside the cell. Indeed, when the B. subtilis spoIIAB regulatory gene is moved from its native originproximal site to the origin-distal end of the chromosome, it becomes trapped in the wrong cellular compartment during sporulation (35).
Rapid Movement of the Newly Replicated Copy of ori to the Opposite Pole of Living Cells. Previous experiments using the FISH technique showed that, shortly after the onset of DNA replication, there is a switch from a unipolar to a bipolar pattern of ori localization (8). To directly observe the movement of the origin sequence, we used FROS and time-lapse FM of a synchronized MT15 cell population. In this strain, (lacO)n is located 4 kb from ori. MT15 swarmer cells were isolated by density gradient centrifugation, spotted onto an agarose pad containing nutrients on a microscope slide, and were imaged at 2-min intervals. Fig. 3A shows representative images of such a time course study and Fig. 3B shows the ori movement (averaged over 22 cells) as a function of time. The rate of the ori movement far exceeds the rate of cell elongation: after 10 min, the new ori has moved >1 μm from one pole of the cell to the other, whereas the cells have elongated by <0.2 μm during this period. This finding suggests there is an active transport mechanism for ori with an average speed of ≈0.27 μm/min (Fig. 4C). The changing slope of the curve may be due to a first phase of rapid and active ori movement, followed by a slower process associated with ori capture at the cell pole. If so, it would suggest that different gene products may be involved in each process. It is also possible that the slower movement of ori is associated with the compaction of this region of the chromosome during the terminal stages of segregation. In ≈10% of MT15 cells, but not in the strains described below where (lacO)n is located at least 43 kb from ori, the movement of the focus appeared to be discontinuous and even transiently reversed before the consistent movement to the pole resumed. These cells were excluded from the set used to calculate the average rate of movement. The erratic movement of the foci in some MT15 cells could be an artifact caused, directly or indirectly, by the interference of repetitive lacO sequences and/or of a large region of protein-bound DNA (LacI-CFP bound to lacO) in the vicinity of ori with the segregation process.
Fig. 3.
Rapid and directed movement of (lacO)n/LacI-CFP-tagged ori during the early stages of the DNA replication cycle. (A) Swarmer cells were isolated and spotted on an agarose pad containing nutrients. Phase contrast and fluorescence images were acquired at 2-min intervals immediately before and during foci duplication and movement to the opposite cell pole. Each image represents an overlay of a phase contrast and a fluorescence image. The number in the upper left corner of each image indicates the time (min) relative to the starting point (0 min) at which the image was taken. Diagrams illustrating the movement of the locus are shown above and below the image frames. (Bar, 1 μm.) (B) Movement of the ori focus within the cell (average of 22 cells) plotted as a function of time. The green line shows the increase in cell length. The blue and magenta lines mark the distance of the “old” and the “new” focus from the stalked cell pole over time.
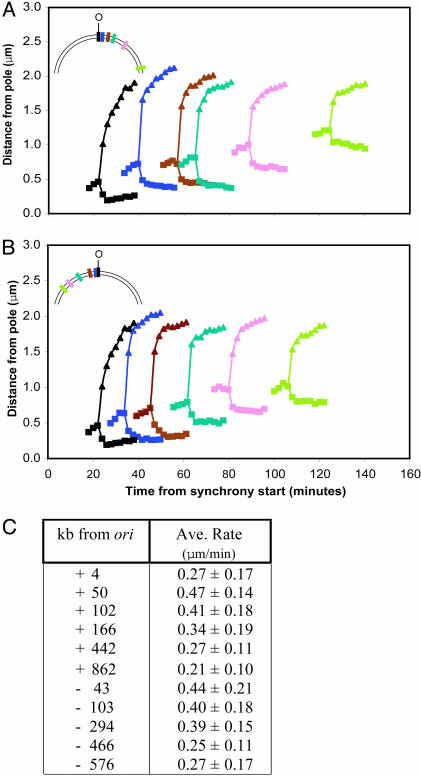
Fig. 4.
Rapid, directed, and sequential movement of different (lacO)n/LacI-CFP-tagged loci during the DNA replication cycle. (A and B) The average movement of the fluorescent foci from at least 20 cells was analyzed as in Fig. 3 for 10 strains, each harboring a (lacO)n insertion at a distinct chromosomal position. The result of the time lapse experiment shown in Fig. 3B by using the strain that carries (lacO)n 4 kb from ori are reproduced in A and B (black line) as a reference for the results obtained with the 10 additional strains. (Insets) The relative position of the insertions in the ori-proximal half of the chromosome. The movement of five (lacO)n/LacI-CFP-tagged loci located 50 (blue), 102 (brown), 166 (turquoise), 442 (pink), and 864 kb (green) to the “right” and five (lacO)n/LacI-CFP-tagged loci located 43 (blue), 103 (brown), 294 (turquoise), 466 (pink), and 576 kb (green) to the “left” of ori was plotted on the same graph in A and B, respectively. Synchronized swarmer cells were isolated from each strain, spotted on an agarose pad containing nutrients (time point 0 min) and were allowed to proceed through the cell cycle until the movement of the loci began. Images were then acquired at 2-min intervals to record the movement. With increasing distance from ori, the movement of the focus occurred progressively later in the cell cycle. O, ori. (C) The average rates of movement of (lacO)n/LacI-CFP-tagged chromosomal loci. The strain used in Fig. 3 containing (lacO)n 4 kb from ori, as well as 10 other strains described were analyzed.
To identify factors that mediate ori movement to the cell pole, we and others have examined several Caulobacter mutant strains that have impaired ori localization. These include strains with mutations in SMC, topoisomerase IV, ParB, and the actin-like protein MreB (21, 36–38). Because experiments with these strains were performed by using FISH, we could not determine whether these mutations affect ori movement or polar capture. SMC, topoismerase IV, and MreB could, directly or indirectly, be involved in mediating ori movement. ParB, on the other hand, may play a role in capturing ori at the cell pole, because the ParB protein colocalizes with ori at the cell poles and belongs to a family of proteins that bind centromere-like sequences, most of which are located in close proximity to the origin of replication (39, 40). In B. subtilis, a protein, RacA, has been identified that is localized to the cell poles of sporulating B. subtilis cells and, through its interaction with origin-proximal binding sites, tethers the origin of replication to the pole (41). In the absence of RacA, the polar positioning of the origins is impaired: they are not located at the cell pole, but remain in its vicinity, suggesting that RacA is not involved in mediating the poleward migration of the origin.
Rapid, Directed, and Sequential Segregation of 10 Chromosomal Loci into the Incipient Daughter Cell During DNA Replication. We performed time-lapse FM of 10 synchronized (lacO)n-bearing strains by using FROS to determine the rate of segregation of chromosomal loci other than ori and to analyze the chronology and dynamics of the chromosome segregation process. It should be noted that we observed heterogeneity in the compactness of foci during movement: in some cells the focus moving into the swarmer cell half of the predivisional cell appeared blurred compared to the other foci, especially in the early stages of the segregation process. Although this observation could reflect different states of DNA compaction as the newly replicated locus travels through the cell, we cannot exclude the possibility that the binding ability of LacI-CFP or TetR-YFP is compromised during certain stages of chromosome segregation. Of the 10 strains harboring ori-proximal (lacO)n insertions that were analyzed, five carried insertions to the right side of ori (Fig. 4A Inset) and five to the left side of ori (Fig. 4B Inset). With increasing distance from ori, the intracellular displacement of foci and, therefore, the spatial and temporal resolution of the experiment decreases (compare the absolute distance traveled by foci derived from ori-proximal and -distal (lacO)n insertions in Fig. 4A). As a result, only strains with insertions <900 kb from ori were analyzed. To minimize exposure of cells to UV light, images were acquired only during the period of the cell cycle when foci duplication and separation occurred. Because synchronized swarmer cells were the starting point for each experiment, we could determine the time in the cell cycle when the locus is moved across the cell. Fig. 4 A and B show the movement of these individual loci as the cells progressed through the cell cycle. For each set of insertions, the five loci are moved one after the other to their target positions in each incipient daughter cell. Each of the loci moves in a similar manner, at a rate comparable with that observed for ori (Fig. 4C). This result suggests that the same mechanisms may be involved in segregating ori and sites up to 900 kb away from ori into the daughter cell half of the predivisional cell. The average time in the cell cycle when each locus is moved correlates with the relative physical distance of each insertion from ori. DNA replication is still ongoing at the time the most distal of these insertions is segregated. In other experiments, under comparable experimental conditions (22), chromosome replication (determined by the presence of an assembled replisome) lasted from the 30-min to the 200-min time point of a 240-min cell cycle. Thus, our results show that replication and segregation occur concurrently, as would be predicted from the extrusion and capture model for chromosome partitioning in which the energy released by the DNA replication factory is thought to provide, at least in part, the force required for segregating newly replicated DNA (42). However, other factors, such as MreB, SMC, RNA polymerase, or topoisomerase IV, may also be involved in the segregation process by actively exerting pulling and/or pushing forces on the newly replicated DNA.
Concluding Remarks. The two principal findings of this study are: (i) each chromosomal locus and the genes within that locus consistently occupy specific subcellular positions, and the position of a given gene inside the cell correlates linearly with its relative position on the chromosome, and (ii) the movement of newly replicated chromosomal loci to a specific position in the incipient daughter cell is rapid, directed, and sequential, occurring with exquisite precision and at a similar rate. Based on these observations, we conclude that a highly organized genome is maintained in bacterial cells at the most fundamental level to facilitate replication and segregation. The next challenge is to identify the machinery used for active movement of chromosomal loci and to determine whether and how the site-specific location of genes inside the cell has been integrated into cellular regulatory systems.
Supplementary Material
Acknowledgments
We thank Michael Laub (Bauer Center for Genomic Research, Harvard University, Cambridge, MA) and Craig Stephens (Biology Department, Santa Clara University, Santa Clara, CA) for providing the ordered cosmid library; M. R. K. Alley (Imperial College London, London) for providing pXGFP4 and pXGFP7C1; Heidi Kaplan (Department of Microbiology and Molecular Genetics, University of Texas Medical School, Houston) for providing pMiniHimar-LacZ; David Sherratt (Department of Biochemistry, University of Oxford, Oxford) for providing pLAU43, pLAU44, and pLAU53 before publication; Ellen Judd for advice with matlab; and Charles Yanofsky and members of the Shapiro and McAdams laboratories for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM 51426 (to L.S.), and Defense Advanced Research Planning Agency Grant MDA972-00-0032 and Department of Energy DE-FG03-01ER63219 (to H.H.M.). M.T. is funded by a long-term fellowship from the European Molecular Biology Organization.
Abbreviations: CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; FISH, fluorescence in situ hybridization; FROS, fluorescent repressor-operator system; FM, fluorescence microscopy; IFM, immunofluorescence microscopy; LacI, lac repressor; TetR, tet repressor.
See Commentary on page 9175.
References
- 1.Postow, L., Crisona, N. J., Peter, B. J., Hardy, C. D. & Cozzarelli, N. R. (2001) Proc. Natl. Acad. Sci. USA 98, 8219-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherratt, D. J. (2003) Science 301, 780-785. [DOI] [PubMed] [Google Scholar]
- 3.Deng, S., Stein, R. A. & Higgins, N. P. (2004) Proc. Natl. Acad. Sci. USA 101, 3398-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera, J. E. & Jin, D. J. (2003) Mol. Microbiol. 50, 1493-1505. [DOI] [PubMed] [Google Scholar]
- 5.Dworkin, J. & Losick, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14089-14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York).
- 7.Draper, G. C. & Gober, J. W. (2002) Annu. Rev. Microbiol. 56, 567-597. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, G. S. & Wright, A. (2000) Annu. Rev. Microbiol. 54, 681-708. [DOI] [PubMed] [Google Scholar]
- 9.Niki, H. & Hiraga, S. (1997) Cell 90, 951-957. [DOI] [PubMed] [Google Scholar]
- 10.Robinett, C. C., Straight, A., Li, G., Willhelm, C., Sudlow, G., Murray, A. & Belmont, A. S. (1996) J. Cell Biol. 135, 1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straight, A. F., Belmont, A. S., Robinett, C. C. & Murray, A. W. (1996) Curr. Biol. 6, 1599-1608. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, G. S., Sitnikov, D., Webb, C. D., Teleman, A., Straight, A., Losick, R., Murray, A. W. & Wright, A. (1997) Cell 90, 1113-1121. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, P., Sharpe, M. E., Raether, B., Perego, M., Ohlsen, K. & Errington, J. (1997) Genes Dev. 11, 1160-1168. [DOI] [PubMed] [Google Scholar]
- 14.Webb, C. D., Teleman, A., Gordon, S., Straight, A., Belmont, A., Lin, D. C., Grossman, A. D., Wright, A. & Losick, R. (1997) Cell 88, 667-674. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, S., Rech, J., Lane, D. & Wright, A. (2004) Mol. Microbiol. 51, 461-469. [DOI] [PubMed] [Google Scholar]
- 16.Marczynski, G. T. & Shapiro, L. (2002) Annu. Rev. Microbiol. 56, 625-656. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall, A. & Shapiro, L. (1989) Proc. Natl. Acad. Sci. USA 86, 119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, R. B., Wang, S. C. & Shapiro, L. (2002) Nat. Rev. Mol. Cell Biol. 3, 167-176. [DOI] [PubMed] [Google Scholar]
- 19.Ryan, K. R. & Shapiro, L. (2003) Annu. Rev. Biochem. 72, 367-394. [DOI] [PubMed] [Google Scholar]
- 20.Degnen, S. T. & Newton, A. (1972) J. Mol. Biol. 64, 671-680. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, R. B. & Shapiro, L. (1999) Proc. Natl. Acad. Sci. USA 96, 10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, R. B., Wang, S. C. & Shapiro, L. (2001) EMBO J. 20, 4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teleman, A. A., Graumann, P. L., Lin, D. C., Grossman, A. D. & Losick, R. (1998) Curr. Biol. 8, 1102-1109. [DOI] [PubMed] [Google Scholar]
- 24.Niki, H., Yamaichi, Y. & Hiraga, S. (2000) Genes Dev. 14, 212-223. [PMC free article] [PubMed] [Google Scholar]
- 25.Ely, B. (1991) Methods Enzymol. 204, 372-384. [DOI] [PubMed] [Google Scholar]
- 26.Alley, M. R. (2001) Mol. Microbiol. 40, 1335-1343. [DOI] [PubMed] [Google Scholar]
- 27.Lau, I. F., Filipe, S. R., Soballe, B., Okstad, O. A., Barre, F. X. & Sherratt, D. J. (2003) Mol. Microbiol. 49, 731-743. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 29.Blondelet-Rouault, M. H., Weiser, J., Lebrihi, A., Branny, P. & Pernodet, J. L. (1997) Gene 190, 315-317. [DOI] [PubMed] [Google Scholar]
- 30.Skerker, J. M. & Shapiro, L. (2000) EMBO J. 19, 3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. P., Sharma, P. L., Schoenlein, P. V. & Ely, B. (1993) Proc. Natl. Acad. Sci. USA 90, 630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J., Heidelberg, J. F., Alley, M. R., Ohta, N., Maddock, J. R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alley, M. R., Maddock, J. R. & Shapiro, L. (1992) Genes Dev. 6, 825-836. [DOI] [PubMed] [Google Scholar]
- 34.Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dworkin, J. & Losick, R. (2001) Cell 107, 339-346. [DOI] [PubMed] [Google Scholar]
- 36.Figge, R. M., Easter, J. & Gober, J. W. (2003) Mol. Microbiol. 47, 1225-1237. [DOI] [PubMed] [Google Scholar]
- 37.Wang, S. C. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gitai, Z., Dye, N. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohl, D. A. & Gober, J. W. (1997) Cell 88, 675-684. [DOI] [PubMed] [Google Scholar]
- 40.Lin, D. C. & Grossman, A. D. (1998) Cell 92, 675-685. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Yehuda, S., Rudner, D. Z. & Losick, R. (2003) Science 299, 532-536. [DOI] [PubMed] [Google Scholar]
- 42.Lemon, K. P. & Grossman, A. D. (2001) Genes Dev. 15, 2031-2041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.