SUMMARY
Aerobic glycolysis (AG), i.e., non-oxidative metabolism of glucose despite the presence of abundant oxygen, accounts for 10–12% of glucose used by the adult human brain. AG varies regionally in the resting state. Brain AG may support synaptic growth and remodeling; however, data supporting this hypothesis are sparse. Here, we report on investigations on the role of AG in the human brain. Meta-analysis of prior brain glucose and oxygen metabolism studies demonstrates that AG increases during childhood, precisely when synaptic growth rates are highest. In resting adult humans, AG correlates with persistence of gene expression typical of infancy (transcriptional neoteny). In brain regions with the highest AG, we find increased gene expression related to synapse formation and growth. In contrast, regions high in oxidative glucose metabolism express genes related to mitochondria and synaptic transmission. Our results suggest that brain AG supports developmental processes, particularly those required for synapse formation and growth.
INTRODUCTION
The adult human brain, though a mere 2–3% of total body weight, consumes nearly 20% of the human body’s basal metabolic rate (Clarke and Sokoloff, 1999). Building a human brain is even more expensive. Conservative estimates suggest that an infant’s brain consumes more than 40% of the body’s basal metabolic rate (Durnin, 1981). This figure is particularly impressive given that the basal metabolic rate per square meter of surface area is larger in children than it is in adults (Durnin, 1981).
Glucose normally supplies the vast majority of calories consumed by the adult brain. Most of this glucose is oxidized to supply the large amounts of ATP required to maintain membrane ion gradients and other cellular processes related to synaptic transmission (Attwell and Laughlin, 2001; Lennie, 2003; Raichle and Mintun, 2006; Sibson et al., 1997; Sibson et al., 1998). Yet, quantitative measurements of brain metabolism reveal that approximately 10–12% of the total glucose consumed by a normal adult brain (CMRglc) is in excess of oxygen consumption (CMRO2) (Boyle et al., 1994; Madsen et al., 1995; Powers et al., 2007; Raichle et al., 1970; Vaishnavi et al., 2010). The metabolic origin of this non-oxidative metabolism of glucose is debated. In normal, awake, resting adult humans, lactate efflux likely accounts for ~20% of this excess glucose consumption (Madsen et al., 1995; Raichle et al., 1970), though estimates vary (Dalsgaard et al., 2004). We refer to this total excess brain glucose consumption as ‘aerobic glycolysis’ (AG) based on a similar, well-described phenomenon found in cancer cells (Lunt and Vander Heiden, 2011; Vaishnavi et al., 2010).
In the resting adult human brain, AG varies regionally: nearly 25% of resting glucose consumption is non-oxidative in the medial prefrontal gyrus, while AG comprises only 2% of glucose consumption in the cerebellum (Figure 1) (Vaishnavi et al., 2010). A recent meta-analysis of studies on human brain oxygen and glucose consumption identified a similar regional variance of AG (Hyder et al., 2013). AG correlates with centrality as defined by large scale, structural and resting state function connectivity studies of the human brain (Bullmore and Sporns, 2012; Vaishnavi et al., 2010). Further, AG identifies regions most vulnerable to amyloid-beta deposition in Alzheimer’s disease (Vlassenko et al., 2010). Uncoupling of glucose metabolism from oxygen consumption is characteristic of the brain’s response to imposed tasks (‘activation’) (Fox et al., 1988). Curiously, AG persists for at least 40 minutes following cognitive demanding task performance, despite the return of blood flow and lactate production to control levels (Fox et al., 1988; Madsen et al., 1995).
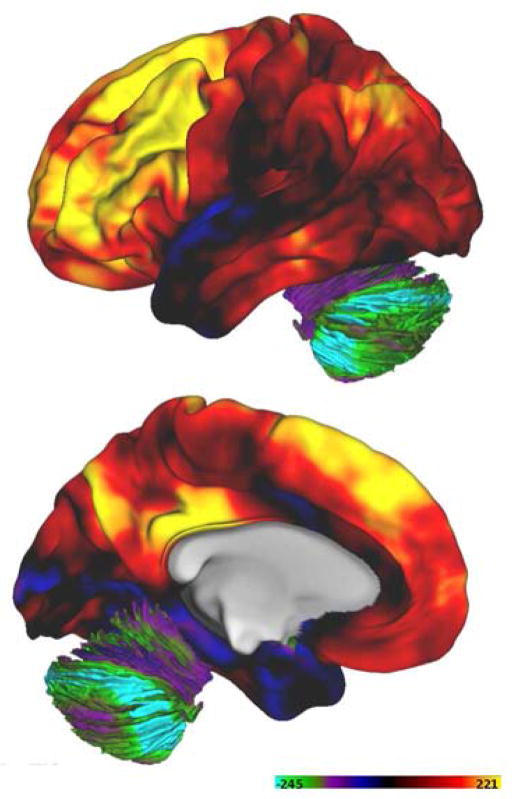
Figure 1. Human brain AG varies regionally in healthy adults.
AG was measured as previously described (Vaishnavi et al. 2010) in 33 normal young adults. AG, expressed as the glycolytic index, is illustrated here on the lateral and medial brain surface. Computed AG has been geodesically (parallel to the cortical surface) smoothed. The highest levels of AG occur in the medial frontal gyrus, precuneus, and posterior cingulate cortex, as previously reported.
AG is a prominent feature of cancer cell metabolism; its main role is thought to be support of biosynthetic pathways required for cellular proliferation (Locasale and Cantley, 2011; Lunt and Vander Heiden, 2011). Might AG serve a similar role in the brain, i.e., biosynthetic support of synapse and neurite formation? To explore this hypothesis, we performed a series of analyses, starting with a literature-based meta-analysis of prior investigations of glucose and oxygen consumption across the lifespan.
We also compared in vivo, regional adult brain AG measurements, previously obtained by our laboratory with PET imaging, to post-mortem gene expression data obtained in 16 different brain regions in over 50 brains acquired across the human lifespan (BrainSpan Study [BSS]) (Brainspan; Kang et al., 2011). To improve the external validity of our results, we used a second, independent source of adult human brain gene expression (Allen Human Brain Atlas [AHBA]) (ABHA; Hawrylycz et al., 2012) to identify genes that are most highly expressed in brain regions with high AG. Finally, we contrasted the relationship between gene expression and AG to gene expression relationships with other metabolic parameters, such as CMRglc. The observed patterns of gene expression support our hypothesis that brain AG supports the biosynthetic requirements of synaptic growth and remodeling.
RESULTS
Meta-analysis of prior studies of brain glucose and oxygen consumption
We first present a meta-analysis of prior studies of whole brain glucose and oxygen consumption across the human lifespan. We identified several studies of human subjects dating back to 1953 that obtained quantitative measurements of whole brain or cerebral glucose and oxygen consumption during various stages of life, including premature neonates, growing normal children, young adults, and the elderly (Altman et al., 1993; Burns and Tyrrell, 1992; Chugani et al., 1987; Frewen et al., 1991; Himwich et al., 1959; Kennedy and Sokoloff, 1957; Kinnala et al., 1996; Kuhl et al., 1982; Leenders et al., 1990; Pantano et al., 1984; Perlmutter et al., 1987; Petit-Taboue et al., 1998; Scheinberg et al., 1953; Takahashi et al., 1999; Yoxall and Weindling, 1998). We combined the reported values of whole brain or cerebral glucose and oxygen consumption from these studies to create a summary representation of these metabolic variables as a function of age across the human lifespan (Table S1). Glucose consumption is plotted in standard units across the human lifespan, and for comparison, oxygen consumption is plotted in terms of the amount of glucose consumption to be expected if all the glucose consumed were ultimately metabolized via oxidative phosphorylation (Figure 2a).
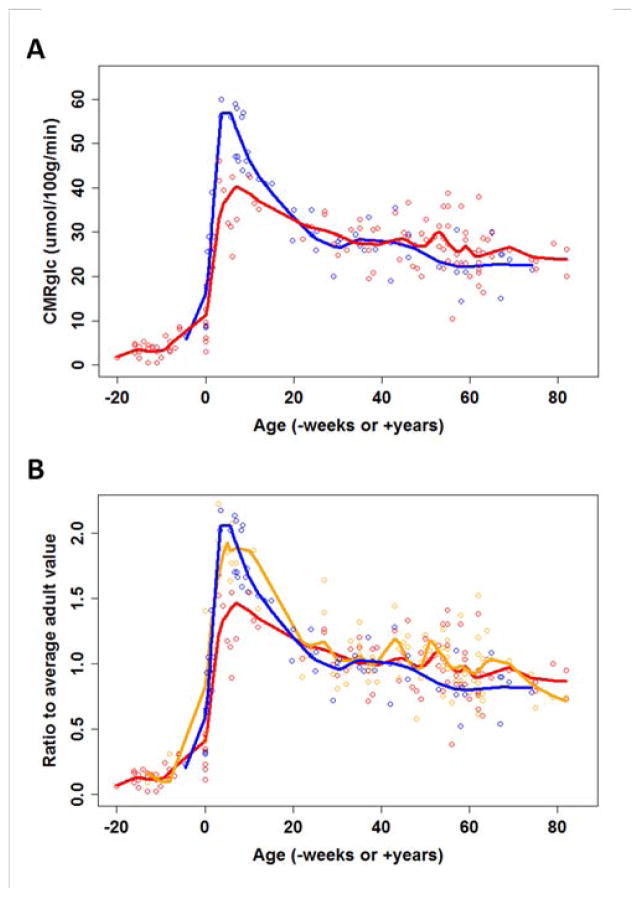
Figure 2. see also Table S1. Brain metabolism across the lifespan.
As discussed in the main text, data were gathered from 15 different studies dating back to 1953. (A) Whole brain or cerebral total glucose consumption rates (blue circles) and a fit obtained using loessR (blue line, see Experimental Procedures for details) demonstrate an approximate doubling of CMRglc during early childhood. The expected glucose consumption based on measured oxygen consumption rates is also plotted (red circles and line). This also increases during early childhood, though less than the measured changes in CMRglc, suggesting that approximately 30% of the CMRglc during childhood is in excess of oxygen consumption (i.e., aerobic glycolysis). (B) CMRglc (blue), CMRO2 (red), and CBF (orange) were plotted across the lifespan as normalized proportions of average adult values. This analysis again shows an approximate 2-fold rise in CMRglc and 1.5-fold rise in CMRO2 during early childhood. Interestingly, CBF also increases 2-fold during early childhood, matching the changes in CMRglc, but then appears to more closely follow changes in CMRO2 during adulthood. The raw and normalized data are shown in Table S1.
During much of adult life, brain glucose consumption is only slightly greater than that expected based on brain oxygen consumption, and whole brain AG appears to virtually disappear in the elderly (Figure 2a). However, AG increases dramatically during childhood, and accounts for approximately one-third of total glucose consumption at its peak around age 5 years, when total glucose consumption exceeds adult levels by a factor of two. To compare glucose and oxygen consumption to relative changes in cerebral blood flow, we replotted the data in terms of a ratio relative to adult values (Figure 2b). Interestingly, resting cerebral blood flow tracks glucose utilization during childhood, but then tracks oxygen consumption during adulthood.
We averaged reported metabolic data across the lifespan and then calculated an oxygen-glucose index (OGI) 1 for each developmental stage. We find that OGI is approximately 5 during the fetal period, decreases to less than 4 during early childhood, progressively rises to 5.67 in young and mid-age adults, and ultimately exceeds 6 in older adults (Table S1). The decrease in AG from young adulthood to a mean age in the 70’s has been described previously (Dastur, 1985); in that study, the whole brain OGI was 4.7 in normal subjects with mean age 21 as compared to an OGI of 5.9 in older subjects with mean age of 71. Thus, the available data demonstrate that brain AG peaks (OGI is at a minimum) precisely when synaptic development peaks in the human brain, i.e., during early childhood.
Comparison of regional AG to transcriptional neoteny
We reasoned that, if AG is increased in the developing brain, then regionally high AG in the adult human brain may reflect persistent developmental processes. Persistence of developmental features and processes into adulthood is referred to as ‘neoteny’ (Bufill et al., 2011). A classic example is that adult human faces tend to better resemble juvenile chimpanzee faces than their adult counterparts. Neoteny traditionally has been assessed in terms of cross-species comparisons (Gould, 2000). More recently, the concept has been broadened to encompass within-species delayed maturation of specific features (Petanjek et al., 2011).
To measure regional transcriptional neoteny in the human brain, we used the Brainspan Study (BSS), a publicly available, whole-genome, gene expression microarray dataset representing 16 different brain regions over 50 human brains across the fetal – adulthood lifespan (Brainspan; Kang et al., 2011). We assessed transcriptional neoteny using a measure previously constructed to demonstrate neoteny in the dorsolateral prefrontal cortex and superior frontal gyrus in humans compared to chimpanzees and macaques (Somel et al., 2009); incidentally, both regions exhibit high AG in normal human adults (Vaishnavi et al., 2010). Specifically, we defined the regional neoteny index as the number of neotenous genes within each of the 16 regions assessed in the BSS, relative to the cerebellum; the cerebellum was chosen as the ‘reference region’ in this analysis because of its lowest aerobic glycolysis relative to any other of the 16 regions.
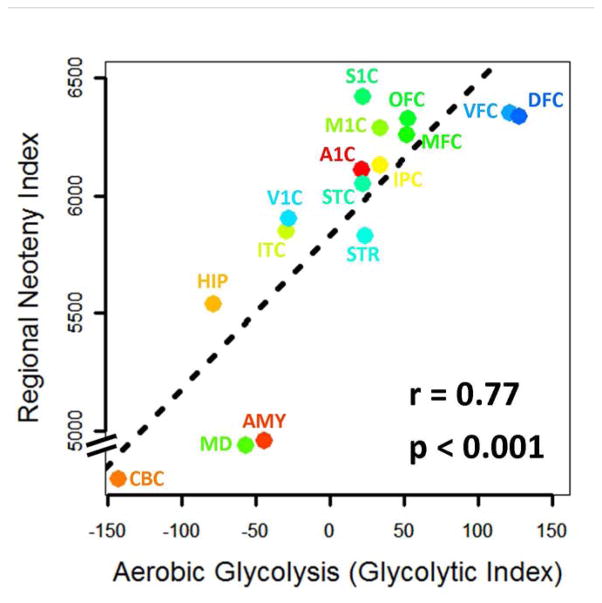
To investigate the relationship between this regional neoteny index and AG, we assigned each of the 16 regions an AG value using regional PET imaging data previously obtained by our laboratory in resting, healthy young adults (Vaishnavi et al., 2010). For example, AG in the cerebellum, as measured using the glycolytic index, was −143, while AG in the dorsolateral frontal cortex (including Brodmann areas 9 and 46) was 127 (Vaishnavi et al., 2010). Across these 16 regions, the neoteny index was significantly correlated with AG (Pearson correlation = 0.77, r-squared = 0.59, p=0.0008, 95% CI r = 0.43–0.92; Figure 3). A related measure of regional transcriptional neoteny—the median ‘age-shift’ (Somel et al., 2009) —revealed very similar results (Pearson correlation = 0.71, r-squared = 0.51, p = 0.002, 95% CI r = 0.33–0.89). Thus, gene expression data strongly suggest that the regional dependence of AG in the adult brain reflects a persistent developmental character, specifically, neoteny.
Figure 3. Transcriptional neoteny regionally correlates with aerobic glycolysis.
We assessed the transcriptional neoteny in each of 15 regions from the BSS as compared to the cerebellum. A gene was defined as neotenous in a particular region if its expression demonstrated delayed, prolonged, or potentiated expression as compared to its expression in the cerebellum. Thus, by definition, there are zero neotenous genes in the cerebellum (CBC) as compared to itself. The number of neotenous genes in each region, that is, the regional neoteny index, increases in relation to aerobic glycolysis (r = 0.77, Pearson correlation, p=0.0008, 95% CI 0.43–0.92). The median age-shift across all genes for each region as compared to the cerebellum also correlates with aerobic glycolysis (r = 0.71, Pearson correlation, p=0.003, 95% CI 0.31–0.90). Regions are named according to the BSS (Kang et al., 2011): CBC = cerebellum, MD = thalamus, AMY = amygdala, HIP = hippocampus, ITC = inferior temporal cortex, STR = striatum, V1C = primary visual cortex, STC = superior temporal cortex, A1C = auditory cortex, IPC = inferior parietal cortex, M1C = primary motor cortex, S1C = primary somatosensory cortex, MFC = medial frontal cortex, OFC = orbital frontal cortex, DFC = dorsal frontal cortex, VFC = ventral frontal cortex.
AG is related to spatially specific gene expression
Next, we sought to identify specific genes whose expression spatially correlates with AG in the adult brain. To maximize the integrity of this analysis, we limited the comparison to 5 age-matched BSS brains, each including 1 or 2 samples from 16 different regions. We further included another regional brain gene expression data set from the AHBA, which was acquired independently from the BSS and included whole-genome microarray measurements from approximately 900 different brain regions.
For each of the 16 regions sampled in the 5 BSS brains, we assigned an AG value in the same manner as described above. We assigned an AG value to each of the ~900 brain regions sampled in the AHBA brain by measuring the glycolytic index in our PET data at the MNI coordinate provided for each of the AHBA regions. AG was then correlated to the expression of each gene, and the top 1000 correlations were identified for each of the 6 brains (5 BSS and 1 AHBA brains, cumulative FDR for top 1000 correlations across all 6 brains = 0.045). Finally, to maximize the external validity of this analysis, we isolated genes consistently found in the top 1000 for all 6 brains (given cumulative FDR < 0.05, the chance that the same false positive gene would occur in all 6 brains is exceedingly low).
The number of genes satisfying these stringent criteria was 116 (Table S2, see also Table 1). To further assess the statistical significance of this finding, we randomly shuffled aerobic glycolysis values among the brain regions and repeated the analysis. Only ~2% of the random permutations reproduced an equal or greater number of consistent genes (Figure S1). Thus, aerobic glycolysis in the adult human brain is significantly associated with a consistent pattern of gene expression as identified by these 116 genes. In contrast, performing the same analysis to identify genes associated with CMRglc revealed only 38 genes.
Table 1. see also Figure S1 and Tables S2–S4. Gene ontology results for the top 116 genes correlating with AG.
The top 116 genes (Table S2) associated with adult human brain AG were identified as those that consistently occurred among the top 1,000 out of 17,205 genes whose expression correlates with AG in the AHBA and 5 matched BSS brains. DAVID Bioinformatics Resources (v6.7) was used to functionally characterize these genes (Huang da et al., 2009). Top functional terms with a group enrichment score >1.0 are listed, and in general include axons, dendrites, and the regulation of their development (see Table S3 for the full list). Similar results are obtained using other gene ontology tools including GeneMANIA.org and userListEnrichment (Table S4) (Miller et al., 2011; Warde-Farley et al., 2010). A permutation analysis showed that the discovery of the 116 genes is statistically unlikely by chance alone (Figure S1).
| Functional Term | Group Enrichment Score |
|---|---|
| Neuronal projections; dendrites; axons | 2.66 |
| Ion channels | 2.25 |
| Synapses; dendritic spines | 1.57 |
| Regulation of secretion and transport | 1.40 |
| Neuronal development; axonogenesis | 1.18 |
| Cytoplasmic vesicles | 1.18 |
| Neuronal differentiation; neuron projection development | 1.11 |
| Regulation of cell development and differentiation | 1.04 |
Gene EPHB6 (ephrin type-B receptor 6) emerged as the gene with the strongest association with aerobic glycolysis. This gene was among the top 40 in all 6 brains, which is extremely unlikely by chance alone (p<10−8, Bonferroni corrected). Ephrins and ephrin receptors are known to support a variety of development and remodeling functions, including regulation of axon guidance and development of dendritic filopodia (Klein, 2009). Other “top 116” associations with aerobic glycolysis included potassium ion channels, genes implicated in synaptic transmission and plasticity, neuropeptides and neuropeptide receptors, and NRGN (neurogranin), a gene implicated in schizophrenia (Stefansson et al., 2009).
We used DAVID Bioinformatics Resources (v6.7) to characterize the functional relevance of these 116 genes (Huang da et al., 2009). These AG related genes are enriched for genes associated with axons, dendrites, and their development (Tables 1 and S3). In contrast, the 38 genes associated with CMRglc are only enriched for genes related to regulation of transcription and other DNA binding activities (group enrichment score 1.36). For the 116 AG-related genes, an independent gene ontology tool, GeneMANIA.org, confirmed that these genes are associated with neuronal projections and axonal and dendritic development (Table S4a) (Warde-Farley et al., 2010). Using userListEnrichmentR, a program in the weighted gene co-expression network analysis (WGCNA) package (Langfelder and Horvath, 2008) that identifies gene list enrichment in brain specific datasets, we found that the 116 AG-related genes are also enriched for neuron specific markers, genes downregulated in Alzheimer disease and aging, genes associated with autism, and proteins found in the human post-synaptic density (p<0.05, Bonferroni corrected, Table S4b). There was no significant enrichment of genes related to glycolysis or genes regulated by agonal stress (Langfelder and Horvath, 2008; Li et al., 2007).
Having identified 116 genes that are consistently expressed in glycolytic regions of adult human brains, we next explored their temporal expression by measuring their average gene expression in the BSS across the human lifespan. We found that the 116 glycolytic genes collectively exhibit a unique expression pattern across the lifespan (Figure 4a) that strikingly varies across the brain. During the early fetal period, there is little differential regional expression. Postnatally, cerebellar expression (the area in the adult brain with the lowest level of AG, see Figure 1) falls while cortical expression continues to rise. Within the cortex, the rise during postnatal development and subsequent persistence during adulthood is highest within association areas that, as a whole, include the brain regions with the highest AG (Figure 4b).
Figure 4. Expression of the 116 glycolytic genes consistently expressed in glycolytic regions of the adult human brain progressively rises and persists in the cortex, particularly neocortical areas.
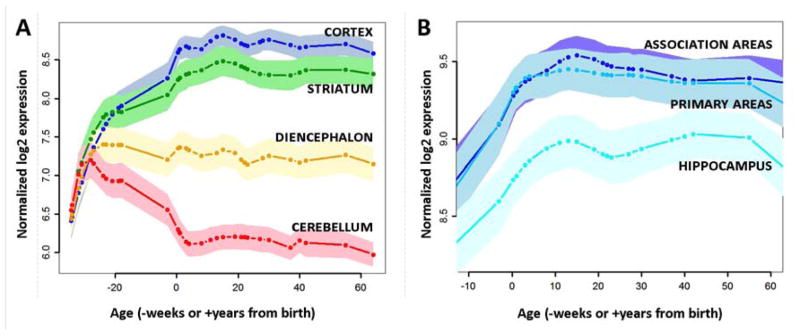
(A) The expression of the 116 genes most consistently correlating with aerobic glycolysis in adults is plotted across the lifespan in the BrainSpan Study. Lines are smoothed and normalized group averages. Shaded regions represent 95% confidence intervals of the mean. The first few months of post-conception fetal life are associated with parallel increases in expression across the brain. However, a dramatic divergence occurs at mid-gestation, with expression failing in the cerebellum (red line), stabilizing in the diencephalon (orange), and continuing to increase in the striatum (green) and, even more so, in the cerebral cortex (blue). (B) Within the postnatal cerebral cortex, the 116 genes are most highly expressed in neocortex, particularly in association areas (dark blue line), slightly (but not significantly) less so in primary sensorimotor regions (teal line), and significantly less so in the hippocampus and amygdala (skyblue). The overall relationship among these regions suggests that these genes are more likely to persist at higher levels during adulthood in regions associated with high aerobic glycolysis.
It is noteworthy that genes coding for enzymes directly associated with glycolysis were not included among the 116 genes consistently correlating with aerobic glycolysis. This is not surprising, since glycolysis occurs equally in regions with high oxidative metabolism. Using a less stringent statistical test, we found only two glycolytic isoenzymes that significantly correlated with aerobic glycolysis in all 6 brains (PDK3—pyruvate dehydrogenase kinase 3, and PPM2C—pyruvate dehydrogenase phosphatase catalytic subunit 1, p<0.05, Holm-Bonferroni corrected across glycolysis isoenzymes). Both PDK3 and PPM2C directly regulate pyruvate metabolism in that PDK3 potently inhibits pyruvate dehydrogenase activity and PPM2C contributes to pyruvate dehydrogenase activation. The two genes thus act as ‘switches’ that determines whether the end product of glycolysis is directed towards oxidative phosphorylation or alternative pathways. Indeed, upregulation of PDK3 acts as a switch for aerobic glycolysis in certain cancer cell lines (Lu et al., 2008).
Comparison of AG related brain gene expression to CMRglc and other metabolic measures
A critical question remains: are the above-identified transcriptional patterns specific for AG compared to other metabolic parameters? As a preliminary step, we applied multidimensional scaling to the AHBA data, which places each of the ~900 microarray samples in a two-dimensional coordinate space based on their relation to the first two principle components of the dataset. Regions with high AG were found to cluster independently from regions with low AG along the y-axis (principle coordinate 2) in the resulting plot while, for example, high versus low OEF regions separated along the x-axis (principle coordinate 1) (Figure S2a). This result demonstrates that metabolic differences among brain regions are represented in their transcriptional differences.
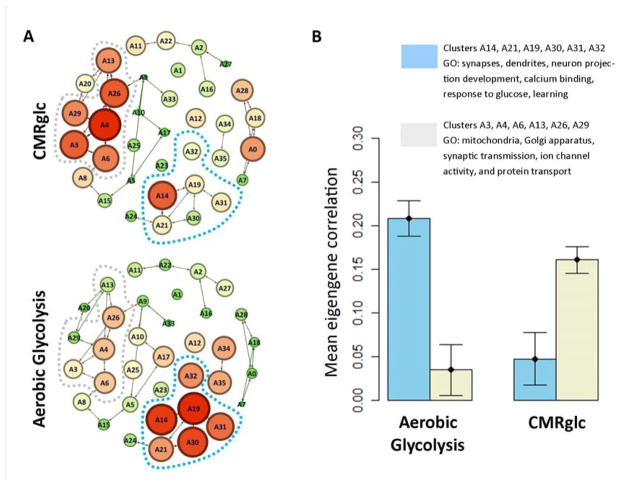
To further analyze these differences, we used weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008). WGCNA analyzes gene co-expression graphs, identifies clusters within the graph and then calculates the first principle component (eigengene) within each cluster. Thus, WGCNA reduces the dimensionality of a very large data set (~30,000 × 30,000 co-expressed genes) to ~30 eigengenes across ~900 brain regions. We found distinct correlations between AG and AHBA eigengenes (Figure 5). AG most highly correlated with clusters associated with neurite growth, synaptic growth and development (cluster eigengenes A14, A19, and A30). In contrast, CMRglc best correlated with clusters related to mitochondria and synaptic transmission (A3, A4, and A26), which result is consistent with the principle that synaptic activity drives demand for ATP (Figure 5 and Table S5). Other metabolic parameters, including CMRO2, CBF, CBV, and OEF also showed specific and distinct relationships to the AHBA eigengenes (Figure S2b).
Figure 5. see also Figure S2 and Tables S5-S6. Aerobic glycolysis is associated with a distinct spatio-temporal transcriptional profile compared to CMRglc.
(A) Genes in the AHBA were clustered using WGCNA (see text). Node connections represent inter-nodal eigengene correlations. Spatial grouping in the figure represents gene co-expression. Correlations between each eigengene and total glucose consumption (CMRglc) or aerobic glycolysis were assessed. Node size and color (green = low, red = high) represent eigengene correlation with either CMRglc (top) or aerobic glycolysis (bottom). Aerobic glycolysis is associated with gene clusters surrounding A19. In contrast, CMRglc is associated gene clusters near A4. (B) Mean correlations between CMRglc vs. AG and eigengenes representing gene clusters surrounding A4 and A19. According to DAVID gene ontology analysis (Table S5), A4 and adjacent (A3, A6, A13, A26, and A29) gene clusters are associated with mitochondria, Golgi apparatus, synaptic transmission, and protein transport. In contrast, gene clusters surrounding A19 (A14, A21, A30, A31, and A32) are associated with synapses and dendrites, neuron projection development, calcium binding, response to glucose, and “learning.” Error bars represent standard error (S.E.M.) in the correlations for the five clusters in each group.
DISCUSSION
In all life forms, there is a fundamental trade-off between the efficient production of energy and increases in biomass. Bacteria evolve according to this trade-off to occupy niches based on either high ATP yield or rapid growth (Pfeiffer et al., 2001; Schuetz et al., 2012). Cancer cells, defined by their rapid, uncontrolled proliferation, amplify the latter by shifting glucose consumption towards biosynthetic reactions, that is, towards AG (Locasale and Cantley, 2011; Lunt and Vander Heiden, 2011). It is not surprising, then, that the rapidly growing human child brain is also highly engaged in AG. Further, the most neotenous regions of the adult brain retain relatively increased levels of AG. Taken together, these results support the hypothesis that brain AG has a similar role to that seen in cancer cells, namely, to support the biosynthesis required for brain growth. More specifically, we hypothesize that AG supports synaptic and neurite formation, as well as turnover, which likely persists throughout the lifespan (Goyal and Raichle, 2013; Marder and Goaillard, 2006).
It is intriguing to speculate upon the metabolic mechanisms by which AG may support brain development and synaptic turnover. In astrocytes, AG is an obligatory step in the metabolism of glutamate released at active synapses (Pellerin and Magistretti, 1994). Some fraction of the resulting lactate may be taken up by neurons; this process is known as the ‘astrocyte-neuron lactate shuttle’ (Kasischke et al., 2004). The ultimate fate of this lactate is uncertain. Much of it may be converted to pyruvate and then metabolized through oxidative phosphorylation. Lactate has metabolic signaling functions (Barros, 2013). It also stimulates gene expression related to long-term memory (Barros, 2013; Suzuki et al., 2011). Lactate affects the redox state of neurons, thereby directing glycolytic intermediates towards biosynthetic pathways as well as increasing neuronal excitability (Cerdan et al., 2006; Ido et al., 2004; Mintun et al., 2004). In neurons, lactate may spare some glucose from oxidative phosphorylation; this glucose then may contribute three-carbon fragments to anabolic pathways used for creating or turning over synapses (Pavlides et al., 2010; Vaishnavi et al., 2010). Measured AG may also represent lactate efflux generated by glycolysis, or a glycogen shunt, to rapidly supply ATP at intensely active synapses (Shulman et al., 2001). Accordingly, it may not be coincidental that neuronal bursting plays a role in shaping synaptic connections during development (Katz and Shatz, 1996). These alternative accounts of AG are not mutually exclusive; experimental investigation is needed to sort through the various possibilities.
Interestingly, in the adult brain, the highest levels of resting state AG are found in the default mode network (DMN), a constellation of brain regions originally identified as most active in the absence of attention-demanding, goal-directed task performance (Raichle et al., 2001). More recently, the DMN has emerged as among the most functionally robust networks in the brain as evidenced by its correlated intrinsic activity (Fox and Raichle, 2007), a property now widely referred to as functional connectivity (Bullmore and Sporns, 2012). The DMN also constitutes a central hub of structural (axonal) connectivity (Bullmore and Sporns, 2012; Vaishnavi et al., 2010). An attractive hypothesis is that the reason these centrally connected regions in the human brain are high in AG is that their function requires high levels of new synaptic growth and remodeling.
Similarly, brain AG was previously found to be highest in regions most vulnerable to amyloid deposition (Vlassenko et al., 2010). Given the present findings, it is intriguing to consider the possibility that amyloid deposition is linked to high rates of synaptic turnover (Bufill et al., 2011; Nikolaev et al., 2009). Further studies investigating the evolution of brain AG in neurodegenerative diseases, as well as neurodevelopmental abnormalities such as schizophrenia, are warranted.
The existence of topographic gene expression databases (e.g., AHBA, BSS) raises the possibility of using imaging to inform the significance of gene expression patterns, and vice versa. However, few studies along these lines have been published. As far as we are aware, this study is the first example of such an analysis resulting in a strong correspondence between the regional variance of an imaging parameter, in this case AG, and specific gene expression.
Our study has several limitations. The meta-analysis of human brain AG (Figure 2) is based on incomplete data obtained using different methods across studies and age groups. These results should be confirmed. Unfortunately, obtaining additional quantitative metabolic data during the first two decades of life using PET in normal humans cannot be justified because of radiation safety concerns. Efforts to obtain data of this type in developing non-human primates should be considered. Another caveat is that gene expression levels may only modestly correlate with protein and enzyme levels because of post-transcriptional modifications. Finally, our results do not address causation. Hence, further studies will be needed to explore whether AG is induced by the biosynthetic requirements of synapse and neurite formation. Thus, our analyses link AG to synaptic and neurite growth and formation but do not elucidate the specific mechanisms by which this occurs. Possible functions of AG include providing carbon-enriched substrates, providing ATP and NAD+ rapidly, and reducing reactive oxygen species. All of these functions are believed to be important in proliferating cells, but some may be relatively more important in the brain.
In summary, AG is potentiated during childhood, persists in neotenous adult brain regions and spatially correlates with gene expression related to synapse formation and neurite growth. These observations suggest that a primary role of AG is biosynthetic support of the aforementioned cellular processes. Accordingly, in vivo measurement of AG may be informative regarding synaptic growth and turnover in health and disease. Thus, impaired brain AG may represent a useful biomarker of pathology in neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Meta-analysis of brain metabolism across the lifespan
The meta-analysis reported in Figure 2 was based on in-depth review of the literature, using Pubmed, Google Scholar, and reference lists in several papers identifying studies that measured glucose or oxygen metabolism over the whole brain or cerebral cortex. We counted measurements obtained with either the (modified) Kety-Schmidt method or PET imaging. In some cases, reported values were extracted from charts and/or tables. Metabolic data were expressed as glucose μmol/100g/min and oxygen mL/100g/min. Oxygen metabolism was also expressed in terms of ‘expected’ glucose consumption that would be required to fully account for oxygen consumed, assuming a 6:1 stoichiometric relationship between oxygen and glucose consumption. Loess curves were fit to the data using the built-in R script, loessR. Data were also normalized to average adult values to compare oxygen and glucose consumption to cerebral blood flow across the human lifespan. Finally, the oxygen-glucose index (OGI) was computed as the molar ratio of oxygen consumption to glucose consumption: OGI = CMRO2 (mL/100g/min) × 44.7 μmol/mL / CMRglc (μmol/100g/min).
Brain PET data analysis and regional alignment to gene expression data
The remaining analyses comparing gene-expression to PET imaging data, were based on regional measurements of CMRglc, CMRO2, CBF, and CBV acquired in our laboratory in 33 normal young adults. A prior report discusses the regional dependence of these parameters, as well as AG calculated via the glycolytic index—a measure of glucose consumption in excess of that expected based on CMRO2. AG and the oxygen-glucose index (OGI) are nearly perfectly inversely correlated (r = −0.98) (Vaishnavi et al., 2010). The regional PET data for each metabolic parameter, including AG, were aligned to individual regions acquired in the AHBA via Montreal Neurological Institute (MNI) coordinates, as provided by the Allen Brain Institute and via atlas registration of the PET data. The regional PET data were aligned to the BrainSpan data via regional and Brodmann area measurements provided in the original report of this data (Vaishnavi et al., 2010). We further compared these measurements to a similar set of PET measurements in FreeSurfer-derived regions matched to the BSS regional nomenclature, and found only slight differences.
Preprocessing of brain gene expression data
Methods describing production and initial analysis of adult human brain aerobic glycolysis, adult human brain gene expression, and human brain gene expression across the lifespan have been previously published (ABHA; Brainspan; Hawrylycz et al., 2012; Kang et al., 2011; Vaishnavi et al., 2010). Donor brains with post-mortem intervals >24 hours or with >25% of microarray samples with an RNA integrity index (RIN) less than 7.5 were excluded to maximize the integrity of our analysis. Hence, data from only one of the AHBA donors was used, since only this brain met our stringent RNA integrity and post-mortem interval criteria. The AHBA data were corrected for batch effects and normalized to the 75% quantile, as previously described (Hawrylycz et al., 2012). BSS data also were quantile normalized.
Calculation of regional transcriptional neoteny
The regional neoteny index was calculated as the number of genes (effectively, the percentage of genes) demonstrating significant neoteny based on the method of Somel and colleagues (Somel et al., 2009). A gene was defined as neotenous in a particular region if its expression exhibited prolonged, delayed, or increased expression in that region relative to its expression in the cerebellum (p<0.01 cut-off). Thus, by definition, there are zero neotenous genes in the cerebellum (CBC) as compared to itself. The cerebellum was used as the ‘reference species’ in these calculations, since the cerebellum shows the lowest AG amongst all brain regions assessed in the BSS. Median age-shift was also used as an alternative measure of regional transcriptional neoteny, again calculated using the method of Somel and colleagues (Somel et al., 2009). The regional neoteny index and median age-shift calculations then were compared to AG using region-wise Pearson correlation.
Regional comparative analysis of the brain PET and gene expression data
Genes in common between the AHBA and BSS (n = 17,205) were rank-ordered according to their correlation with AG. Similarity among rank ordered gene lists was computed using OrderedListR (Lottaz et al., 2006). We searched for genes consistently found in the top 1,000 out of the 17,205 in each of the AHBA and 5 BSS age- and RIN-matched adult brains; this search identified the 116 genes described in the main text. To evaluate the significance of this finding, the same analysis was performed after repeatedly shuffling the AG values among the 16 BSS regions to measure the percentage of instances that produced an equivalent number of genes as compared to the actual AG values (AHBA was excluded from this permutation analysis given the difficulty in relating shuffled BSS AG values to the AHBA regions). The results of the permutation analysis are reported in Figure S1. The 116 AG-associated genes were analyzed using three gene ontology methods including GeneMANIA (Warde-Farley et al., 2010), DAVID v6.7 (Huang da et al., 2009), and userListEnrichmentR (Miller et al., 2011), as described in the text and figure legends.
Multidimensional scaling (MDS) was performed using cmdscaleR, a built-in function in the open-source R statistical programming language (v2.13.2), on the AHBA data across regions. The first two principal components derived from the MDS analysis are shown in Figure S2a. PET-derived metabolic measurements are superimposed on the MDS results. Regions with high (> 1 SD above the mean) or low (> 1 SD below the mean) metabolic values are color-coded (red vs. blue, respectively).
Weighted Gene Co-expression Network Analysis was performed using the WGCNA (v1.18-2) R package. Metabolic parameters were compared to each cluster of genes in the AHBA using Pearson correlation between the spatial pattern of metabolism with each cluster eigengene (Langfelder and Horvath, 2008). Clusters of genes in the BSS were compared to each metabolic parameter by calculating the correlation of gene significance for metabolism with its membership in each cluster. Gene ontology analysis of each cluster was performed using DAVID v6.7 (Huang da et al., 2009). Gephi was used to construct Fruchterman-Reingold representations of the AHBA network, sized and colored according to their correlation with aerobic glycolysis and CMRglc.
The R scripts and pre-processed data used by these scripts are available for free download at ftp://imaging.wustl.edu/pub/goyal/.
Supplementary Material
Highlights.
Brain aerobic glycolysis regionally relates to synaptic growth gene expression.
Oxidative glycolysis instead relates to mitochondria and synaptic transmission.
In human, whole brain aerobic glycolysis peaks during childhood.
Neotenous regions of the adult brain maintain relatively high aerobic glycolysis.
Acknowledgments
We thank Matthew Glasser for providing a geodesically smoothed representation of human brain aerobic glycolysis on the brain surface. We are also grateful for the many thoughtful discussions that we had with Anish Mitra, Ben Shannon, and Ed Lein, as well as the thoughtful comments provided by our anonymous reviewers. We thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement and support. The Allen Human Brain Atlas was supported in part by award numbers 1C76HF15069-01-00 and 1C76HF19619-01-00 from the Department of Health and Human Services Health Resources and Services Administration. This work was supported by National Institutes of Health (P50NS006833 to M.E.R. and A.Z.S.) and the National Institute of Neurologic Disorders and Stroke (P30 NS048056 to A.Z.S.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Department of Health and Human Services.
Footnotes
Six moles of oxygen are consumed for each mole of glucose oxidized to carbon dioxide and water; this ratio is referred to as the oxygen-glucose index or OGI; an OGI less than 6 indicates AG, whereas an OGI more than 6 indicates that the brain is using an alternative fuel for energy, e.g., ketone bodies (Dastur, 1985).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ABHA Allen Brain Human Atlas [Internet] Seattle (WA): Allen Institute for Brain Science; ©2009. Available from: http://www.brain-map.org. [Google Scholar]
- Altman DI, Perlman JM, Volpe JJ, Powers WJ. Cerebral oxygen metabolism in newborns. Pediatrics. 1993;92:99–104. [PubMed] [Google Scholar]
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Barros LF. Metabolic signaling by lactate in the brain. Trends in neurosciences. 2013;36:396–404. doi: 10.1016/j.tins.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Boyle PJ, Scott JC, Krentz AJ, Nagy RJ, Comstock E, Hoffman C. Diminished brain glucose metabolism is a significant determinant for falling rates of systemic glucose utilization during sleep in normal humans. The Journal of clinical investigation. 1994;93:529–535. doi: 10.1172/JCI117003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainspan BrainSpan: Atlas of the Developing Human Brain [Internet] Funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01, and 1RC2MH089929-01. © 2011 Available from: http://developinghumanbrain.org.
- Bufill E, Agusti J, Blesa R. Human neoteny revisited: The case of synaptic plasticity. American journal of human biology : the official journal of the Human Biology Council. 2011;23:729–739. doi: 10.1002/ajhb.21225. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nature reviews Neuroscience. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Burns A, Tyrrell P. Association of age with regional cerebral oxygen utilization: a positron emission tomography study. Age and ageing. 1992;21:316–320. doi: 10.1093/ageing/21.5.316. [DOI] [PubMed] [Google Scholar]
- Cerdan S, Rodrigues TB, Sierra A, Benito M, Fonseca LL, Fonseca CP, Garcia-Martin ML. The redox switch/redox coupling hypothesis. Neurochemistry international. 2006;48:523–530. doi: 10.1016/j.neuint.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Chugani HT, Phelps ME, Mazziotta JC. Positron emission tomography study of human brain functional development. Ann Neurol. 1987;22:487–497. doi: 10.1002/ana.410220408. [DOI] [PubMed] [Google Scholar]
- Clarke DD, Sokoloff L. Circulation and energy metabolism of the brain. In: Agranoff BW, Siegel GJ, editors. Basic Neurochemistry Molecular, Cellular and Medical Aspects. Philadelphia: Lippincott-Raven; 1999. pp. 637–670. [Google Scholar]
- Dalsgaard MK, Quistorff B, Danielsen ER, Selmer C, Vogelsang T, Secher NH. A reduced cerebral metabolic ratio in exercise reflects metabolism and not accumulation of lactate within the human brain. The Journal of physiology. 2004;554:571–578. doi: 10.1113/jphysiol.2003.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastur DK. Cerebral blood flow and metabolism in normal human aging, pathological aging, and senile dementia. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1985;5:1–9. doi: 10.1038/jcbfm.1985.1. [DOI] [PubMed] [Google Scholar]
- Durnin JVGA. Basal metabolic rate in man. Joint FAO/WHO/UNU Expert Consultation on Energy and Protein Requirements; Rome. 1981. [Google Scholar]
- Fox MD, Raichle M. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nature Reviews Neuroscience. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME, Mintun MA, Dence C. Nonoxidative glucose consumption during focal physiologic neural activity. Science. 1988;241:462–464. doi: 10.1126/science.3260686. [DOI] [PubMed] [Google Scholar]
- Frewen TC, Kissoon N, Kronick J, Fox M, Lee R, Bradwin N, Chance G. Cerebral blood flow, cross-brain oxygen extraction, and fontanelle pressure after hypoxic-ischemic injury in newborn infants. The Journal of pediatrics. 1991;118:265–271. doi: 10.1016/s0022-3476(05)80500-1. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Of coiled oysters and big brains: how to rescue the terminology of heterochrony, now gone astray. Evolution & development. 2000;2:241–248. doi: 10.1046/j.1525-142x.2000.00067.x. [DOI] [PubMed] [Google Scholar]
- Goyal MS, Raichle ME. Gene expression-based modeling of human cortical synaptic density. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6571–6576. doi: 10.1073/pnas.1303453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himwich WA, Benaron HB, Tucker BE, Babuna C, Stripe MC. Metabolic studies on perinatal human brain. J Appl Physiol. 1959;14:873–877. doi: 10.1152/jappl.1959.14.6.873. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hyder F, Fulbright RK, Shulman RG, Rothman DL. Glutamatergic function in the resting awake human brain is supported by uniformly high oxidative energy. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2013;33:339–347. doi: 10.1038/jcbfm.2012.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido Y, Chang K, Williamson JR. NADH augments blood flow in physiologically activated retina and visual cortex. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:653–658. doi: 10.1073/pnas.0307458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kennedy C, Sokoloff L. An adaptation of the nitrous oxide method to the study of the cerebral circulation in children; normal values for cerebral blood flow and cerebral metabolic rate in childhood. The Journal of clinical investigation. 1957;36:1130–1137. doi: 10.1172/JCI103509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnala A, Suhonen-Polvi H, Aarimaa T, Kero P, Korvenranta H, Ruotsalainen U, Bergman J, Haaparanta M, Solin O, Nuutila P, et al. Cerebral metabolic rate for glucose during the first six months of life: an FDG positron emission tomography study. Archives of disease in childhood Fetal and neonatal edition. 1996;74:F153–157. doi: 10.1136/fn.74.3.f153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nature neuroscience. 2009;12:15–20. doi: 10.1038/nn.2231. [DOI] [PubMed] [Google Scholar]
- Kuhl DE, Metter EJ, Riege WH, Phelps ME. Effects of human aging on patterns of local cerebral glucose utilization determined by the [18F]fluorodeoxyglucose method. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1982;2:163–171. doi: 10.1038/jcbfm.1982.15. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenders KL, Perani D, Lammertsma AA, Heather JD, Buckingham P, Healy MJ, Gibbs JM, Wise RJ, Hatazawa J, Herold S, et al. Cerebral blood flow, blood volume and oxygen utilization. Normal values and effect of age. Brain : a journal of neurology. 1990;113(Pt 1):27–47. doi: 10.1093/brain/113.1.27. [DOI] [PubMed] [Google Scholar]
- Lennie P. The cost of cortical computation. Curr Biol. 2003;13:493–497. doi: 10.1016/s0960-9822(03)00135-0. [DOI] [PubMed] [Google Scholar]
- Li JZ, Meng F, Tsavaler L, Evans SJ, Choudary PV, Tomita H, Vawter MP, Walsh D, Shokoohi V, Chung T, et al. Sample matching by inferred agonal stress in gene expression analyses of the brain. BMC genomics. 2007;8:336. doi: 10.1186/1471-2164-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell metabolism. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottaz C, Yang X, Scheid S, Spang R. OrderedList--a bioconductor package for detecting similarity in ordered gene lists. Bioinformatics. 2006;22:2315–2316. doi: 10.1093/bioinformatics/btl385. [DOI] [PubMed] [Google Scholar]
- Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. The Journal of biological chemistry. 2008;283:28106–28114. doi: 10.1074/jbc.M803508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Madsen PL, Hasselbalch SG, Hagemann LP, Olsen KS, Bulow J, Holm S, Wildschiodtz G, Paulson OB, Lassen NA. Persistent resetting of the cerebral oxygen/glucose uptake ratio by brain activation: evidence obtained with the Kety-Schmidt technique. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1995;15:485–491. doi: 10.1038/jcbfm.1995.60. [DOI] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nature reviews Neuroscience. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S. Strategies for aggregating gene expression data: the collapseRows R function. BMC bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun MA, Vlassenko AG, Rundle MM, Raichle ME. Increased lactate/pyruvate ratio augments blood flow in physiologically activated human brain. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:659–664. doi: 10.1073/pnas.0307457100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pantano P, Baron JC, Lebrun-Grandie P, Duquesnoy N, Bousser MG, Comar D. Regional cerebral blood flow and oxygen consumption in human aging. Stroke; a journal of cerebral circulation. 1984;15:635–641. doi: 10.1161/01.str.15.4.635. [DOI] [PubMed] [Google Scholar]
- Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, Wang C, Pestell RG, Martinez-Outschoorn UE, Howell A, et al. Transcriptional evidence for the “Reverse Warburg Effect” in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer’s disease, and “Neuron-Glia Metabolic Coupling”. Aging. 2010;2:185–199. doi: 10.18632/aging.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter JS, Powers WJ, Herscovitch P, Fox PT, Raichle ME. Regional asymmetries of cerebral blood flow, blood volume, and oxygen utilization and extraction in normal subjects. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1987;7:64–67. doi: 10.1038/jcbfm.1987.9. [DOI] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit-Taboue MC, Landeau B, Desson JF, Desgranges B, Baron JC. Effects of healthy aging on the regional cerebral metabolic rate of glucose assessed with statistical parametric mapping. NeuroImage. 1998;7:176–184. doi: 10.1006/nimg.1997.0318. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, Perlmutter JS. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Mintun MA. Brain work and brain imaging. Annual review of neuroscience. 2006;29:449–476. doi: 10.1146/annurev.neuro.29.051605.112819. [DOI] [PubMed] [Google Scholar]
- Raichle ME, Posner JB, Plum F. Cerebral blood flow during and after hyperventilation. Archives of neurology. 1970;23:394–403. doi: 10.1001/archneur.1970.00480290014002. [DOI] [PubMed] [Google Scholar]
- Scheinberg P, Blackburn I, Rich M, Saslaw M. Effects of aging on cerebral circulation and metabolism. AMA archives of neurology and psychiatry. 1953;70:77–85. doi: 10.1001/archneurpsyc.1953.02320310083007. [DOI] [PubMed] [Google Scholar]
- Schuetz R, Zamboni N, Zampieri M, Heinemann M, Sauer U. Multidimensional optimality of microbial metabolism. Science. 2012;336:601–604. doi: 10.1126/science.1216882. [DOI] [PubMed] [Google Scholar]
- Shulman RG, Hyder F, Rothman DL. Cerebral energetics and the glycogen shunt: neurochemical basis of functional imaging. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6417–6422. doi: 10.1073/pnas.101129298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibson NR, Dhankhar A, Mason GF, Behar KL, Rothman DL, Shulman RG. In vivo 13C NMR measurements of cerebral glutamine synthesis as evidence for glutamate-glutamine cycling. Proc Natl Acad Sci U S A. 1997;94:2699–2704. doi: 10.1073/pnas.94.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibson NR, Dhankhar A, Mason GF, Rothman DL, Behar KL, Shulman RG. Stoichiometric coupling of brain glucose metabolism and glutamatergic neuronal activity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:316–321. doi: 10.1073/pnas.95.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. Transcriptional neoteny in the human brain. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, Alberini CM. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell. 2011;144:810–823. doi: 10.1016/j.cell.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Shirane R, Sato S, Yoshimoto T. Developmental changes of cerebral blood flow and oxygen metabolism in children. AJNR Am J Neuroradiol. 1999;20:917–922. [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassenko AG, Vaishnavi SN, Couture L, Sacco D, Shannon BJ, Mach RH, Morris JC, Raichle ME, Mintun MA. Spatial correlation between brain aerobic glycolysis and amyloid-beta (Abeta ) deposition. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17763–17767. doi: 10.1073/pnas.1010461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoxall CW, Weindling AM. Measurement of cerebral oxygen consumption in the human neonate using near infrared spectroscopy: cerebral oxygen consumption increases with advancing gestational age. Pediatric research. 1998;44:283–290. doi: 10.1203/00006450-199809000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.