Abstract
Herpes simplex virus 1 encodes at least four genes whose functions include blocking apoptosis induced by exogenous agents (e.g., sorbitol, Fas ligand, and BAD protein) or replication-incompetent mutants (e.g., the d120 mutant lacking both copies of the α4 gene). US3, one of these four genes, encodes a serine-threonine kinase that has been demonstrated to block apoptosis induced by proapoptotic cellular proteins or by the d120 mutant. The amino acid context of serine-threonine phosphorylated by US3 is similar to that of the cAMP-dependent protein kinase PKA. We report that (i) the pattern of proteins phosphorylated by US3 in transduced cells or in cells infected with WT virus overlaps that of phosphoproteins targeted by PKA, (ii) activation of PKA blocks apoptosis induced by d120 mutant or by BAD protein independently of US3, (iii) US3 protein kinase phosphorylates peptides containing the serine or threonine targeted by PKA including that present in the regulatory type IIα subunit of PKA, and (iv) in WT virus-infected cells the regulatory type IIα subunit is phosphorylated in a US3-dependent manner. We conclude that a major determinant of the antiapoptotic activity of the US3 protein kinase is the phosphorylation of PKA substrates by either or both enzymes.
Keywords: replication-defective viruses, BAD protein, forskolin, RIIα subunit of PKA
Herpes simplex virus 1 (HSV-1) blocks apoptosis induced by exogenous agents such as FAS ligand (1–4), sorbitol (1, 5–7), and activated proapoptotic cellular proteins (8–12), or by replication-incompetent mutants such as those lacking the viral regulatory genes encoding ICP4 (1, 13–15) or ICP27 (16). Our laboratory reported that viral glycoprotein D, glycoprotein J, and the protein kinase encoded by the US3 gene each blocks apoptosis induced by replication-incompetent mutants or by proapoptotic cellular proteins such as BAD (1, 5, 8, 9, 14, 15, 17). Other studies added viral ribonucleotide reductase to the list of antiapoptotic viral proteins (18). These studies were confirmed and extended in numerous subsequent studies (2, 3, 7, 10–12). Inasmuch as US3 protein kinase can block apoptosis induced by proapoptotic cellular proteins or replication-incompetent d120 mutant, which lacks both copies of the α4 gene in the absence of other viral proteins, it could be predicted that US3 protein exerts its function through phosphorylation of specific cellular substrates. The identity of these substrate(s) has so far remained elusive. In vitro studies on synthetic peptides have characterized aUS3 optimal consensus sequence as (R)nX(S/T)YY, where n = 3, X can be Arg, Ala, Val, Pro, or Ser, and Y can be any of these residues except that an acidic residue is unacceptable in those positions (19). This sequence closely resembles the target sequence of the cellular cAMP-dependent protein kinase PKA (20). Interestingly, extensive literature indicates that PKA is a key enzyme important in the regulation of metabolism, survival, and proliferation of eukaryotic cells, and it mediates most of the biological effects of the second messenger cAMP (20–25). Structurally, the enzyme is an inactive tetramer composed of two regulatory (R) and two catalytic subunits. Each R subunit binds two molecules of cAMP, causing the holoenzyme to dissociate and the catalytic subunits to be released and become available to phosphorylate target proteins. Four types of R subunits have been identified, Iα, Iβ, IIα, and IIβ. They differ in molecular mass, physiochemical properties, and tissue distribution. One important difference is that RII but not RI subunits can be phosphorylated by the catalytic subunits, and this event enhances net dissociation and activation of the catalytic subunits (25, 26). Further regulation of the PKA pathway is provided by anchoring proteins (AKAPs) that sequester PKA to a specific subcellular location by binding to the R subunits (25).
In this article we report that the US3 protein kinase may work by activating PKA and by phosphorylating proteins targeted by activated PKA.
Materials and Methods
Cells and Viruses. SK-N-SH and HEp-2 cell lines obtained from the American Type Culture Collection were grown in DMEM containing 5% (HEp-2) or 10% (SK-N-SH) newborn calf serum. Rabbit skin cells were originally obtained by J. McLaren and grown in DMEM supplemented with 5% newborn calf serum. Insect cell line Sf9 (Spodoptera frugiperda) was obtained from Pharmingen. HSV-1(F) is the prototype HSV-1 strain used in the laboratory. The HSV-1(KOS) d120 mutant, a kind gift from N. DeLuca (University of Pittsburgh, Pittsburgh), lacks both copies of the α4 gene and was grown in a Vero-derived cell line (E5) expressing the α4 gene (13). The recombinant virus R7041, lacking the US3 gene, is described in ref. 27.
Antibodies. Rabbit polyclonal antibody against phosphorylated (Ser/Thr) PKA substrates (Cell Signaling Technology, Beverly, MA) was used at a dilution of 1:1,000. Rabbit polyclonal antibodies against PKA RIIα subunit and poly(ADP-ribosyl) synthetase (PARP) (Santa Cruz Biotechnology) were used at a dilution of 1:200 and 1:700, respectively. The polyclonal antibody to ICP22 is described in ref. 28. Monoclonal antibodies to ICP0 and ICP27 were purchased from the Goodwin Institute (Plantation, FL).
Peptides, Proteins, and Reagents. CREBtide was obtained from Santa Cruz Biotechnology, and KEMPtide was obtained from Promega. The PKA RIIα(81–99) autophosphorylation peptide and the casein kinase II substrate peptide were obtained from Biomol (Plymouth Meeting, PA), and histone H1 was obtained from Roche. Forskolin was obtained from Sigma. The myristoylated peptide PKI 14–22 amide (p-PKI) was obtained from Calbiochem. p-PKI is a cell-permeable, active fragment derived from the endogenous heat-stable protein kinase inhibitor (PKI), a potent and specific inhibitor of PKA (29).
Baculoviruses. Construction of transfer plasmids encoding US3 and gstBAD3S/A proteins under the control of the immediate-early cytomegalovirus promoter is described in refs. 11 and 16. The baculoviruses expressing US3 or gstBAD3S/A were constructed by cotransfecting the respective transfer plasmids with BaculoGold DNA (Pharmingen).
Purification of GST-US3 Protein. The US3 coding sequence was transferred from US3-MTS-1 into a suitable transfer plasmid. GST-US3- and GST-expressing baculoviruses were constructed by cotransfecting the respective transfer plasmids with BaculoGold DNA. Infected Sf9 cultures were harvested, rinsed twice with PBS (0.14 M NaCl/3 mM KCl/10 mM Na2HPO4/1.5 mM KH2PO4) and lysed in radioimmunoprecipitation assay buffer (1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS in PBS) in the presence of protease inhibitors (Complete, Roche) as recommended by the manufacturer. Lysed cells were held on ice for 10 min before brief sonication and centrifugation at 14,000 rpm for 10 min in an Eppendorf 5415 C centrifuge. The GST-chimeric proteins were bound to glutathione Sepharose beads (Amersham Biosciences), rinsed four times with PBS, and stored at 4°C.
Immunoblot Assays. Cells grown in 25-cm2 flasks were either mock-infected or exposed to 10 pfu of virus per cell for 1 h and 30 min at 37°C in medium 199V [mixture 199 (Sigma) supplemented with 1% calf serum] and then maintained in DMEM supplemented with the proper amount of serum. Cells were harvested at the indicated times after infection, rinsed three times with PBS, and solubilized in radioimmunoprecipitation assay buffer in the presence of phosphatase inhibitors (10 mM NaF/10 mM β-glycerophosphate/0.1 mM sodium vanadate) and protease inhibitors (Complete). Lysed cells were stored on ice for 10 min before centrifugation at 14,000 rpm for 10 min. The protein concentration of the supernatant fluids was determined with the aid of a Bio-Rad protein assay. Protein samples denatured in disruption buffer (50 mM Tris, pH 7.0/2.75% sucrose/5% 2-mercaptoethanol/2% SDS) were heated at 95°C for 5 min, electrophoretically separated in denaturing polyacrylamide gels, electrically transferred to a nitrocellulose sheet, blocked, and reacted with primary antibody followed by appropriate secondary antibody conjugated to alkaline phosphatase (Bio-Rad). Protein bands were visualized with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (Denville Scientific, Metuchen, NJ).
DEVDase Activity Assay. Caspase 3 activity was assayed by using a tetrapeptide conjugated to phenylnitraniline (DEVD-pNA) (Biomol) (5, 8, 9, 15). Cells were harvested at the indicated times after infection with 10 pfu of virus per cell, rinsed three times with PBS, resuspended in 75 μl of lysis solution A {0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate/50 mM Hepes, pH 7.4/1 mM DTT/0.1 mM EDTA}, held on ice for 10 min, and centrifuged at 14,000 rpm for 10 min at 4°C. Equal amounts of protein in supernatant fluids were tested for DEVDase activity according to the manufacturer's instructions (Biomol).
Kinase Assays. One microgram of GST or GST-US3 attached to glutathione Sepharose beads was reacted with either 2 or 4 μg of the indicated substrates, in 50 μl of kinase buffer (20 mM Tris·HCl, pH 7.5/50 mM KCl/10 mM MgCl2/10 mM 2-mercaptoethanol), supplemented with 100 μM ATP and 20 μCi (1 Ci = 37 GBq) of [γ-32P]ATP. The samples were reacted at 30°C for 30 min. The beads were pelleted by low-speed centrifugation, and 35 μl of the supernatant was spotted onto cellulose phosphate paper filters (Whatman P81). The filters were rinsed four times with 0.5% H3PO4 for 5 min each time and rinsed with ethanol, and the amount of bound radioactivity was measured with a Beckman LS 8000 scintillation counter. Alternatively, 4 μg of histone H1 was reacted with the kinase as described above. The reactions were terminated by the addition of gel-loading buffer, heated to 95°C for 5 min, resolved by PAGE, transferred to nitrocellulose membrane, and analyzed by autoradiography. Quantification of 32P phosphorylation of the substrates was done with the aid of a Molecular Dynamics Storm 860 PhosphorImager.
Results
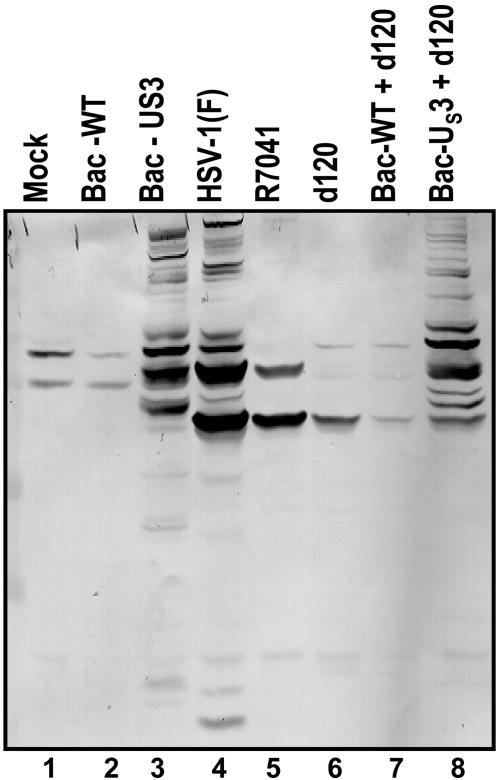
The Profile of Phosphorylated Serines and Threonines in the Context of the Sequence Arg-X-X-Thr*-X-X or Arg-Arg-X-Ser*-X-X in Cells Transduced with the US3 Protein Kinase Is Similar to That of Cells Infected with WT HSV-1. At 18 h after mock infection or transduction with WT baculovirus (Bac-WT), a baculovirus expressing the US3 protein kinase (Bac-US3), or infection with WT HSV-1(F), or with mutants lacking the US3 ORF (R7041) or the α4 genes (d120), SK-N-SH cells were harvested, solubilized, and subjected to electrophoresis on denaturing gels. The electrophoretically separated proteins reacted with an antibody specific for phosphorylated serines and threonines in the context of the sequences Arg-X-X-Thr*-X-X or Arg-Arg-X-Ser*-X-X. This antibody is used to identify the proteins phosphorylated by PKA. As shown in Fig. 1, the antibody reacted with a small number of bands in lysates of cells that were either mock-infected or transduced with Bac-WT or infected with R7041(ΔUS3) or d120 (Δα4) mutants (lanes 1, 2, 5, and 6). In contrast, the antibody reacted with a large number of bands in lysates of cells infected with HSV-1(F) or exposed to a Bac-US3 (lanes 3 and 4). Of particular interest are the patterns of phosphoproteins detected in cells doubly infected with d120 mutant and a baculovirus expressing the US3 protein kinase (lanes 6–8). Many of the numerous phosphoprotein bands reactive with the antibody appeared to comigrate with phosphoprotein bands in cells infected with a Bac-US3 or HSV-1(F). Comparable results were obtained after ectopic expression of US3 in HEp-2 cells or in rabbit skin cells (data not shown). We conclude from these studies that US3 protein kinase mediates the phosphorylation of PKA target sequences Arg-X-X-Thr-X-X or Arg-Arg-X-Ser-X-X.
Fig. 1.
Profiles of electrophoretically separated proteins reacted with antibody specific for phosphorylated serines or threonines in the context of sequences recognized by PKA. SK-N-SH cells were harvested 18 h after mock infection or infection with HSV-1(F), the recombinant virus R7041(ΔUS3) or d120(Δα4), or baculovirus infection. The electrophoretically separated proteins in a denaturing polyacrylamide gel were transferred to a nitrocellulose sheet and reacted with the antibody as described in Materials and Methods.
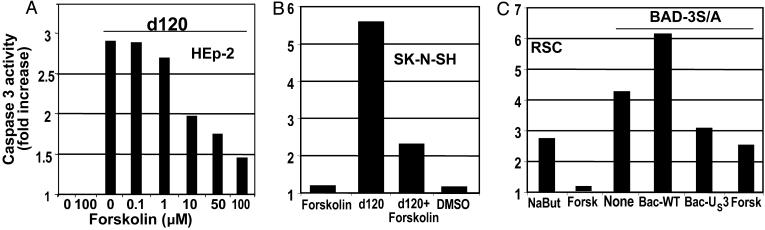
Forskolin Blocks Apoptosis Induced by Infection of Cells with d120 Mutant or Transduction with BAD. The results shown in Fig. 1 suggest that either the US3 protein kinase has a substrate similar to that of PKA or the US3 protein kinase activates PKA, and this, in turn, activates an antiapoptotic pathway. Two series of experiments were done to test the hypothesis that activated PKA can block apoptosis induced by the d120 mutant or BAD. In the first, replicate cultures of HEp-2 cells infected with d120 mutant were exposed to an increased amount of forskolin, a PKA activator. As shown in Fig. 2A, caspase 3 activity induced by the d120 infection decreased as a function of forskolin concentration in the medium. Light microscopic examination revealed complete destruction of the cells in untreated cultures or cultures exposed to low doses of forskolin and a virtually intact cell monolayer in infected cells treated with the highest dose of forskolin. Forskolin (100 μM) also blocked the activation of caspase 3 in rabbit skin cells transduced with a form of GST-BAD in which all of the serines subject to inhibitory phosphorylation had been replaced with alanines as reported in ref. 9 and as shown in Fig. 2C. Finally, forskolin blocked activation of caspase 3 in SK-N-SH cells exposed to the d120 mutant (Fig. 2B).
Fig. 2.
The effect of forskolin on the DEVDase activity of cells infected with the HSV-1 d120 mutant or transduced with a baculovirus encoding gstBAD3S/A chimeric protein. (A) HEp-2 cells were either mock-infected or infected with d120 mutant (10 pfu per cell) in the presence of increasing concentrations of forskolin. Cells were harvested 18 h after infection, and the cell lysates were assayed for DEVDase activity. (B) SK-N-SH cells were either mock-infected or infected with d120 mutant (10 pfu per cell) in the presence of 100 μM forskolin. Cells were harvested 18 h after the infection and assayed for DEVDase activity. (C) Rabbit skin cells were either mock-infected or transduced with a baculovirus expressing gstBAD3S/A (10 pfu per cell), in the presence or absence of 100 μM forskolin. Cells were harvested 18 h after infection and assayed for DEVDase activity. In lanes 4 and 5, cells were exposed to either Bac-WT or Bac-US3 for 6 h, superinfected with Bac-gstBAD3S/A for 18 h, lysed, and assayed for DEVDase activity.
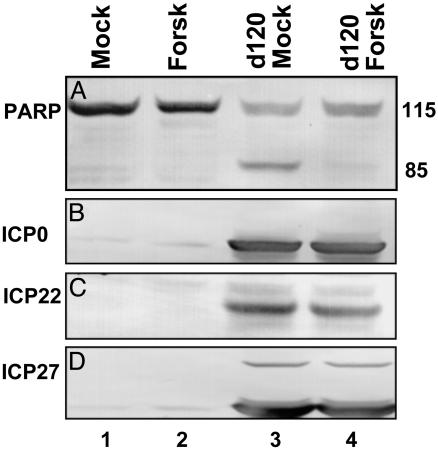
In the second series of experiments replicate cultures of mock-infected or d120-mutant-infected cells were exposed to forskolin (100 μM). At 18 h after infection the cells were harvested, processed as described in the legend to Fig. 3, and reacted with antibody to PARP. The results (Fig. 3A, lanes 1–4) showed that forskolin blocked the caspase 3-dependent cleavage of PARP that is characteristic of d120-mutant-infected cells (lane 3) (5). To determine whether forskolin blocks the expression of viral genes in d120-mutant-infected cells, aliquots of the lysates probed for the status of PARP were probed with antibodies to α proteins known to be produced in cells infected with this mutant. As shown in Fig. 3 B–D, the accumulation of the α proteins ICP0, ICP22, or ICP27 in forskolin-treated cells (lane 4) could not be differentiated from those present in untreated cells (lane 3). Hence, forskolin did not act by blocking the expression of replication-defective mutant. Other studies have shown that foskolin did not enable the d120 mutant to express the US3 gene (data not shown).
Fig. 3.
The effect of forskolin on the cleavage of PARP and the synthesis of viral proteins in cells infected with the d120 mutant. HEp-2 cells mock-infected or infected with the d120 mutant (10 pfu per cell) in the presence or absence of 100 μM forskolin were harvested 18 h after infection, solubilized, subjected to electrophoresis in a denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with anti-PARP antibody (A) and antibody to infected-cell protein (ICP) no. 0 (B), ICP22 (C), or ICP27 (D).
We conclude from this series of experiments that forskolin blocks apoptosis induced either by d120 mutant or BAD in a manner analogous to that of US3 protein kinase.
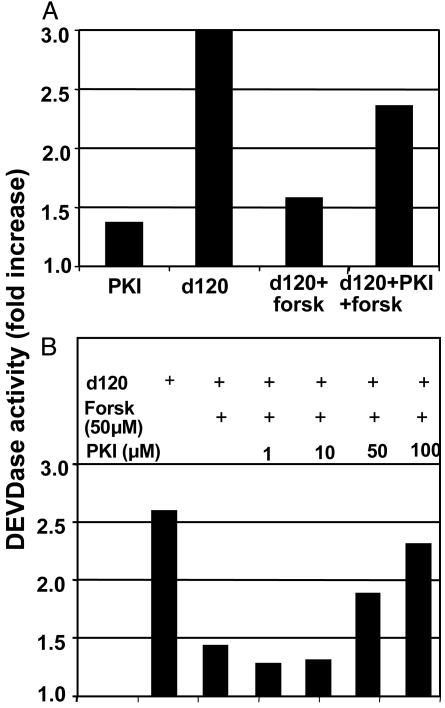
Forskolin Acts by Activating PKA. As noted in the Introduction, PKA is activated by cAMP or phosphorylation of the regulatory type II subunits and is blocked by the interaction of the catalytic subunit with an inhibitory peptide (p-PKI) (29). Forskolin has been shown to activate PKA by increasing the levels of cAMP in infected cells. To test whether the antiapoptotic activity of forskolin is exerted by activation of PKA, we exposed cells infected with d120 mutant to forskolin and the p-PKI peptide. The expectation was that if forskolin acts by activating PKA, p-PKI would inhibit PKA and block the antiapoptotic activity exerted by forskolin. If forskolin acts by another mechanism, p-PKI would not block the antiapoptotic effect. We report two series of experiments. In the first, replicate cultures of HEp2 cells were exposed to p-PKI (100 μM), d120 mutant (10 pfu per cell), d120 mutant and forskolin (100 μM), or d120 mutant, p-PKI, and forskolin. As shown in Fig. 4A, forskolin decreased the level of caspase 3 induced by the d120 mutant. This decrease was reversed in cells exposed to p-PKI.
Fig. 4.
DEVDase activity of cells infected with the HSV-1 d120 mutant in the presence or absence of forskolin or PKI. (A) Replicate cultures of HEp-2 cells were incubated for 90 min in medium containing 100 μM PKI. The cells were then either mock-infected, exposed to the d120 mutant (10 pfu per cell), or infected with the d120 mutant and treated with 100 μM forskolin. (B) Replicate cultures of HEp-2 cells were incubated for 90 min in the presence of increasing concentrations of PKI. The cells were then either mock-infected, infected with d120, or infected with d120 and treated with forskolin. In both sets of experiments, the cells were harvested 18 h after the infection, and the cell lysates were assayed for DEVDase activity.
The results of the second series of experiments are illustrated in Fig. 4B. In these experiments cells exposed to d120 mutant (10 pfu per cell) alone or in combination with forskolin (50 μM) were exposed to increasing amounts of p-PKI. The cells were harvested 18 h after infection and tested for caspase activity as described in Materials and Methods. The results show that forskolin decreased the activation of caspase 3 activity characteristic of d120-mutant-infected cells and that p-PKI caused a dose-dependent increase in caspase 3 activity.
We conclude from this series of experiments that forskolin blocks the proapoptotic functions induced by the d120 mutant by activating PKA.
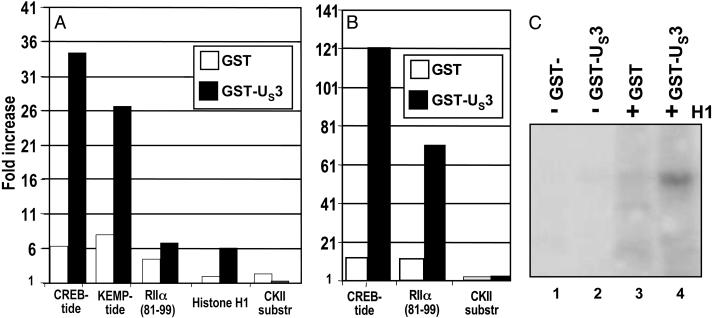
US3 Protein Kinase Phosphorylates PKA Substrates. The objective of this series of experiments was to determine whether US3 protein kinase phosphorylates PKA substrates. Purified GST-US3 chimeric protein was reacted with a panel of PKA substrate peptides and proteins. The procedures for the purification of GST-US3 protein and for measuring the phosphorylation of the substrates are described in Materials and Methods. We used GST-US3 because it is readily purified and is active in phosphorylating known substrates. The results of three separate experiments are shown in Fig. 5 A–C. The results presented here show that GST-US3 phosphorylates in vitro both peptides known to be substrates of PKA (CREB-tide, KEMP-tide, RIIα residues 81–99) and intact proteins (e.g., histone H1) known to be targets of PKA phosphorylation (20). A peptide containing the target site of phosphorylation by casein kinase II served as a negative control and was not phosphorylated by GST-US3.
Fig. 5.
Phosphorylation of known PKA substrates by GST-US3 kinase. Two micrograms (A)or4 μg(B) of the indicated peptides were reacted with 1 μg of purified GST or GST-Us3 attached to glutathione Sepharose beads, as described in the text. Phosphorylated substrates were adsorbed onto cellulose phosphate paper filters, and radioactivity was measured with a scintillation counter. The data show the increase in cpm over a mock reaction carried out with no substrate. (C) Four micrograms of histone H1 were reacted with 1 μg of purified GST or GST-US3 attached to glutathione Sepharose beads, as described in the text. Samples were subjected to polyacrylamide gel electrophoresis, nitrocellulose membrane transfer, and autoradiography.
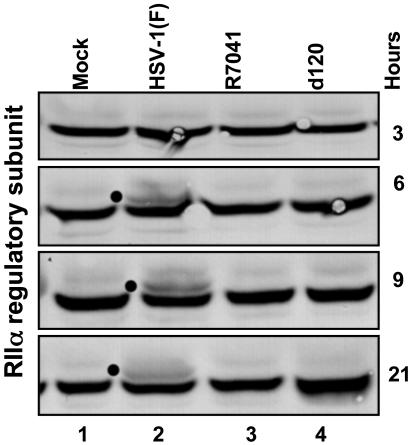
The US3 Protein Kinase Mediates the Posttranslational Modification of the RIIα Catalytic Subunit of PKA in WT HSV-1-Infected Cells. The experiments described above indicated that peptides derived from RIIα catalytic subunit of PKA were phosphorylated in vitro by the US3 protein kinase. The experiments described here were conducted to determine whether the RIIα subunit of PKA was posttranslationally modified in a US3-kinase-dependent manner. HEp-2 cells were mock-infected or infected with HSV-1(F), or the mutant viruses R7041 and d120. At different times after infection, the cells were harvested, lysed, and immunoblotted for the RIIα subunit. As shown in Fig. 6, an additional band appears 6 h after infection with HSV-1 but not after infection with the mutants that do not express US3. A similar modification was observed in a similar experiment done in SK-N-SH cells. As predicted, forskolin treatment of either cell line did not cause the posttranslational modification seen in WT-virus-infected cells. Additionally, the RIIβ subunit was not detected in HEp-2 cells. Whereas the RIIβ subunit was detected in SK-N-SH cells, it was not posttranslationally modified after ectopic expression of the US3 protein kinase (data not shown). These results are consistent with the phosphorylation of the RIIα peptide reported in the preceding section and indicate that RIIα is also modified in a US3-dependent fashion in infected cells.
Fig. 6.
Posttranslational modification of PKA RIIα in infected cells requires the presence of US3 protein kinase. HEp-2 cells were either mock-infected or infected with HSV-1(F) or the R7041 or d120 mutant viruses (10 pfu per cell). The cells were harvested at 3, 6, 9, or 21 h after infection. Cell lysates were separated on an 8% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with antibody against the RIIα subunit of PKA. The dots identify the appearance of posttranslationally modified forms in infected cells.
Discussion
An impressive feature of WT HSV-1-infected cells is that the virus blocks apoptosis induced by a variety of exogenous agents (1–6). Moreover, whereas cells infected by WT virus do not undergo apoptosis, at least three replication-incompetent deletion mutants, that is, mutants lacking the essential genes α4 (13–15), α27 (16), or glycoprotein D (17), induce apoptosis. The apoptosis induced by mutants lacking glycoprotein D is blocked by ectopic expression of glycoproteins D or J (17), whereas that induced by Δα4 mutant is alleviated by the ectopic expression of the US3 protein kinase (14, 15). The viral protein kinase also blocks the proapoptotic activity of ectopically expressed BAD protein and that of other proapoptotic proteins (8–12). Although it has been assumed that US3 protein kinase acts by phosphorylating specific target proteins, the targets themselves remained elusive. In light of the many apparent proapoptotic proteins whose function is blocked by the US3 protein, we sought to determine whether US3 could have a cellular counterpart that could be involved in blocking proapoptotic events, which could interfere with viral replication. The context of the serines or threonines phosphorylated by US3 protein kinase pointed to the cAMP-dependent protein kinase PKA (20, 25). In this study we report several findings that alter the fundamental perception of the function of the US3 protein kinase. Specifically we show the following.
(i) The fundamental hypothesis of these studies is that US3 activates PKA by phosphorylating one of its regulatory subunits, and that apoptosis is blocked by the concerted action of both activated PKA and US3 protein kinase. To activate PKA we used forskolin, a drug commonly used for this purpose. The evidence that forskolin acted by activating PKA is based on the observation that p-PKI, a specific inhibitor of the catalytic subunit, reversed the effect of forskolin.
(ii) Activation of PKA blocked apoptosis induced by the replication defective Δα4 mutant d120 in the absence of ectopic expression of the US3 protein kinase. This mutant induces classical apoptosis in a wide variety of cell lines (1, 5), and earlier studies have shown that apoptosis induced by the d120 mutant is blocked by ectopic expression of the US3 protein kinase (14, 15). In infected cells, the d120 mutant expresses only the α or immediate early genes, and most likely it is the expression of these genes that induces apoptosis. As shown in Fig. 3, forskolin did not block apoptosis by blocking the synthesis of α proteins.
(iii) In earlier studies we used an ectopically expressed BAD protein as a model for the study of apoptosis by activation of a proapoptotic protein. In these studies, the US3 protein kinase blocked apoptosis induced by BAD (8, 9). To enhance its activity, the coding sequence of BAD protein was modified by substitution of three serines, which, when phosphorylated, preclude the protein from inducing apoptosis (9). Activation of PKA also blocked apoptosis induced by the modified BAD3S/A protein in the absence of US3 protein kinase.
Relevant to the results and conclusions of these studies are the following.
(i) Extensive literature indicates that PKA plays an important role in regulating apoptosis in eukaryotic cells (21–24). The data presented in this report indicates that in the experimental context used in these studies, activated PKA performed the same functions as the US3 protein kinase in blocking apoptosis induced by either a replication deficient virus or a proapoptotic cellular protein.
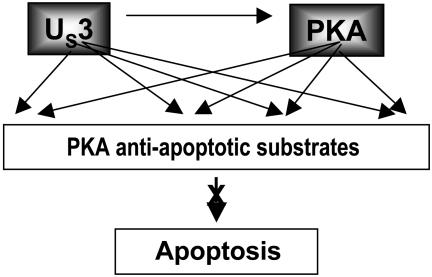
(ii) The model we propose is that the substrate specificity of the US3 protein kinase is similar to that of PKA, that PKA can be activated, and that either US3 alone or both enzymes phosphorylate specific PKA substrates to block apoptosis (Fig. 7). In the few examples presented in this report, we showed that US3 phosphorylated several peptides known to contain PKA phosphorylation sites. One of these peptides contained the autophosphorylation site for the RIIα subunit of PKA. In another experiment we showed that PKA RIIα was phosphorylated in infected cells in a US3-protein-kinase-dependent manner. At this time it is not clear whether the actual block of apoptosis is due to activation of PKA or the function of both PKA and US3 enzymes. Inasmuch as US3 mediated the posttranslational modification of the RIIα but not the RIIβ subunit, the question arises whether tetramers containing both RIIα and RIIβ can be activated by US3 protein kinase.
Fig. 7.
A model for the antiapoptotic interplay between US3 and PKA. Earlier studies have shown that the US3 protein kinase blocks apoptosis induced by d120, a replication-incompetent virus, and by ectopically expressed BAD protein. This report shows that activated PKA blocks apoptosis induced by either d120 mutant or BAD protein, and that US3 can phosphorylate PKA substrates including the autophosphorylation site of the RIIα subunit of PKA. The model predicts that the US3 protein kinase activates PKA, and that either PKA or both PKA and US3 enzymes block apoptosis.
(iii) A well documented antiapoptotic function of activated PKA is the phosphorylation of one of the regulatory residues of BAD, Ser-155 (20, 21). This phosphorylation prevents the binding of BAD to antiapoptotic members of the Bcl-2 family and therefore results in inactivation of BAD itself. However, in the studies reported here, activation of PKA by forskolin was sufficient to block apoptosis induced by a mutant of BAD in which the regulatory phosphorylation sites were mutated. This result is of particular interest because it suggests that the activation of PKA can produce a redundancy of antiapoptotic events. Such a redundancy may explain why US3 has been implicated in protection against apoptosis induced by many and different unrelated stimuli (3, 7–12, 14, 15). Simultaneous activation of several antiapoptotic pathways by a single viral protein would be highly beneficial to the virus.
Lastly, it is of interest to note that activation of PKA by the US3 protein kinase conforms with and extends our understanding of the fundamental strategy by which HSV-1 takes control of the infected cell. Earlier studies have shown that HSV scavenges the cell for useful proteins and diverts them to serve the needs of the virus (30–33). Although our data suggest that US3 protein kinase interacts with and activates PKA, the data also indicate that the enzyme mimics and usurps the functions of activated PKA.
Acknowledgments
This work was supported by National Cancer Institute Grants CA78766, CA71933, CA83939, CA87661, and CA88860 and a grant from the Public Health Service.
Abbreviations: Bac-US3, baculovirus expressing the US3 protein kinase; HSV-1, herpes simplex virus 1; PARP, poly(ADP-ribosyl) synthetase; PKI, protein kinase inhibitor; p-PKI, myristoylated peptide PKI 14–22 amide; R, regulatory.
References
- 1.Galvan, V. & Roizman, B. (1998) Proc. Natl. Acad. Sci. USA 95, 3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jerome, K. R., Chen, Z., Lang, R., Torres, M. R., Hofmeister, J., Smith, S., Fox, R., Froelich, C. J. & Corey, L. (2001) J. Immunol. 167, 3928-3935. [DOI] [PubMed] [Google Scholar]
- 3.Jerome, K. R., Fox, R., Chen, Z., Sears, A. E., Lee, H.-Y. & Corey, L. (1999) J. Virol. 73, 8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieg, S., Yildirim, Z., Smith, D., Kayagaki, N., Yagita, H., Huang, Y. & Kaplan, D. (1996) J. Virol. 70, 8747-8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galvan, V., Brandimarti, R. & Roizman, B. (1999) J. Virol. 73, 3219-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koyama, A. H. & Miwa, Y. (1997) J. Virol. 71, 2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murata, T., Goshima, F., Yamauchi, Y., Koshizuka, T., Takakuwa, H. & Nishiyama, Y. (2002) Microbes Infect. 4, 707-712. [DOI] [PubMed] [Google Scholar]
- 8.Munger, J. & Roizman, B. (2001) Proc. Natl. Acad. Sci. USA 98, 10410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benetti, L., Munger, J. & Roizman, B. (2003) J. Virol. 77, 6567-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogg, P. D., McDonell, P. J., Ryckman, B. J., Knudson, C. M. & Roller, R. J. (2004) Virology 319, 212-224. [DOI] [PubMed] [Google Scholar]
- 11.Cartier, A., Broberg, E., Komai, T., Henriksson, M. & Masucci, M. G. (2003) Cell Death Differ. 10, 1320-1328. [DOI] [PubMed] [Google Scholar]
- 12.Cartier, A., Komai, T. & Masucci, M. G. (2003) Exp. Cell Res. 291, 242-250. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca, N. A., McCarth, M. & Schaffer, P. A. (1985) J. Virol. 56, 558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leopardi, R., Van Sant, C. & Roizman, B. (1997) Proc. Natl. Acad. Sci. USA 94, 7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munger, J., Chee, A. V. & Roizman, B. (2001) J. Virol. 75, 5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubert, M. & Blaho, J. A. (1999) J. Virol. 73, 2803-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, G., Galvan, V., Campadelli-Fiume, G. & Roizman, B. (2000) J. Virol. 74, 11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins, D., Pereira, E. F. R., Gober, M., Yarowsky, P. J. & Aurelian, L. (2002) J. Virol. 76, 1435-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purves, F. C., Deana, A. D., Marchiori, F., Leader, D. P. & Pinna, L. A. (1986) Biochim. Biophys. Acta 889, 208-215. [DOI] [PubMed] [Google Scholar]
- 20.Shabb, J. B. (2001) Chem. Rev. (Washington, D.C.) 101, 2381-2411. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, X.-M., Liu, Y., Payne, G., Lutz, R. J. & Chittenden, T. (2000) J. Biol. Chem. 275, 25046-25051. [DOI] [PubMed] [Google Scholar]
- 22.Manna, P. P. & Frazier, W. A. (2004) Cancer Res. 64, 1026-1036. [DOI] [PubMed] [Google Scholar]
- 23.Fladmark, K. E., Gjertsen, B. T., Doskeland, S. O. & Vintermyr, O. K. (1997) Biochem. Biophys. Res. Commun. 232, 20-25. [DOI] [PubMed] [Google Scholar]
- 24.Yusta, B., Estall, J. & Drucker, D. J. (2002) J. Biol. Chem. 277, 24896-24906. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, D. A., Akamine, P., Radzio-Andzelm, E., Madhusudan, M. & Taylor, S. S. (2001) Chem. Rev. (Washington, D.C.) 101, 2243-2270. [DOI] [PubMed] [Google Scholar]
- 26.Granot, J., Mildvan, A. S., Hiyama, K., Kondo, H. & Kaiser, T. (1980) J. Biol. Chem. 255, 4569-4573. [PubMed] [Google Scholar]
- 27.Purves, F. C., Spector, D. & Roizman, B. (1991) J. Virol. 65, 5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackermann, M., Sarmiento, M. & Roizman, B. (1985) J. Virol. 56, 207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, D. A., Ashby, C. D., Gonzales, C., Calkins, D., Fischer, E. H. & Krebs, E. G. (1971) J. Biol. Chem. 246, 1977-1985. [PubMed] [Google Scholar]
- 30.He, B., Gross, M. & Roizman, B. (1997) Proc. Natl. Acad. Sci. USA 94, 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaguchi, Y., Van Sant, C. & Roizman, B. (1997) J. Virol. 71, 7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Sant, C., Lopez, P., Advani, S. J. & Roizman, B. (2001) J. Virol. 75, 1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Advani, S. J., Brandimarti, R., Weichselbaum, R. R. & Roizman, B. (2000) J. Virol. 74, 8-15. [PMC free article] [PubMed] [Google Scholar]