Abstract
Complex diseases are caused by a combination of genetic and environmental factors, creating a difficult challenge for diagnosis and defining subtypes. This review article describes how distinct disease subtypes can be identified through integration and analysis of clinical and multi-omics data. A broad shift toward molecular subtyping of disease using genetic and omics data has yielded successful results in cancer and other complex diseases. To determine molecular subtypes, patients are first classified by applying clustering methods to different types of omics data, then these results are integrated with clinical data to characterize distinct disease subtypes. An example of this molecular-data-first approach is in research on Autism Spectrum Disorder (ASD), a spectrum of social communication disorders marked by tremendous etiological and phenotypic heterogeneity. In the case of ASD, omics data such as exome sequences and gene and protein expression data are combined with clinical data such as psychometric testing and imaging to enable subtype identification. Novel ASD subtypes have been proposed, such as CHD8, using this molecular subtyping approach. Broader use of molecular subtyping in complex disease research is impeded by data heterogeneity, diversity of standards, and ineffective analysis tools. The future of molecular subtyping for ASD and other complex diseases calls for an integrated resource to identify disease mechanisms, classify new patients, and inform effective treatment options. This in turn will empower and accelerate precision medicine and personalized healthcare.
Introduction
Complex diseases are caused by a combination of genetic, biological, and environmental factors. The determination of disease etiology necessitates the alignment of clinical phenotypes with underlying biomolecular mechanisms. Consequently, researchers have traditionally first identified distinct clinical phenotypes and then identified and compared biomolecular factors that may explain differences in disease manifestation. Biomolecular comparisons across clinical phenotype have been successful in a variety of complex diseases, such as cancer (Kehoe et al., 1999; Ringman et al., 2014; Wong et al., 2015). However, the rapidly expanding availability of genetic and biomolecular expression data from new high-throughput technologies is beginning to shift this traditional phenotype-first approach to a genetics or molecular data-first approach. A molecular data-first approach identifies recurrent genetic variants or expression patterns in order to reduce heterogeneity prior to phenotypic profiling.
Molecular subtyping through gene sequencing, gene expression, and other epigenetic and omics data has been used with great success in cancer to classify subtypes for more effective treatment, understanding prognosis, and identifying disease mechanisms. For example, major molecular subtypes in breast cancer showed marked difference in their clinical features, treatment response, and outcomes (Bertucci et al., 2012; Dvorkin-Gheva and Hassell, 2014; Engstrøm et al., 2013; Schnitt, 2010). Molecular subtypes of lung cancer determined by genetic aberrations were associated with specific tests to assign the subtype and potentially relevant therapies (West et al., 2012; Yauch et al., 2005). Three subtypes of pancreatic cancer identified by transcriptomics analysis showed differences in clinical outcomes and therapeutic response (Collisson et al., 2011). The molecular subtypes of colorectal cancer showed marked differences in survival times (Marisa et al., 2013; Phipps et al., 2015). Molecular subtypes of classical Hodgkin's disease correlated with response to therapy and clinical outcome (Devilard et al., 2002). Three subtypes of gastric cancer established from gene expression data correlated with differences in patients' responses to therapy (Tan et al., 2011). Molecular subtyping of this type is being adapted for other complex diseases such as ASD.
This approach has recently been applied to the study of neurodevelopmental disorders, such as Autism Spectrum Disorders (ASD). ASD is characterized by deficits in social communication and the presence of repetitive, restricted patterns of behavior and interests (American Psychiatric Association, 2013). This neurodevelopmental disorder affects 1 in 68 children, impacting more males than females (Baio, 2012). While individuals with ASD share a core set of features, their genetic etiology and phenotypic presentation are heterogeneous and complex (Betancur, 2011; O'Roak et al., 2012a; Sanders et al., 2012; Stessman et al., 2014). Recently, ASD was redefined in the Diagnostic and Statistical Manual for Mental Disorders, version 5 (DSM-5), to encompass previously distinct classifications of Autistic Disorder, Asperger's Syndrome, Childhood Disintegrative Disorder, and Pervasive Developmental Disorder-Not Otherwise Specified (American Psychiatric Association, 2013). In addition to concerns about reliability and validity of the previous diagnostic criteria (Lord et al., 2012; Sharma et al., 2012) and a lack of behaviorally-defined subtype specific treatments, growing genetic advances failed to find causal differences between these behaviorally-defined subtypes, suggesting instead that a general continuum of autism spectrum disorders with varying levels of severity was more appropriate (King et al., 2014).
These recent developments are prompting efforts to define the etiology of ASD more clearly by shifting research from the predominantly phenotypic classification of the disorder to a genetics-first, and ultimately a molecular data-first approach. With this broader initiative toward identifying disease subtypes using molecular data, known as molecular subtyping, distinct subtypes of ASD are being explored through genetic testing and omics data (genomics, transcriptomics, proteomics). The use of molecular subtyping and the identification of genes and other omics molecules that affect common functional networks shows promise as a means to reduce heterogeneity and explore similarities and differences between interacting genotypes (Iossifov et al., 2014; Jeste and Geschwind, 2014; O'Roak et al., 2012a; Stessman et al., 2014).
The clinical relevance of molecular subtyping relies on its ability to connect underlying disease mechanisms with clinical and phenotypic data. This is exemplified in ASD research: following the identification of recurrent gene disruptions associated with ASD, neurological and behavioral mapping is taking place through imaging and psychometric testing, identifying distinct phenotypic features that accompany a targeted genotype (Bernier et al., 2014; Frazier et al., 2014; Vandeweyer et al., 2014; van Bon et al., 2015).
The function and pathogenicity of many of the genetic mutations found in individuals with ASD are still unknown. Continued gene discovery requires large sample collections, substantial data infrastructure, multidisciplinary collaboration across research sites, and the ability to work iteratively with families to characterize the clinical presentation adequately (Stessman et al., 2014). This publication is a review of the current state of genetics, omics, imaging, psychometric, and clinical data methods as they relate to ASD subtyping. In addition, a broadly applicable approach to statistical methodology is presented, along with ASD-specific examples of the generation of biologically and clinically significant molecular subtypes. Lastly, this review discusses the need for the integration of genotypic and phenotypic data to inform personalized outcomes and treatment options for patients and their families.
Data in Molecular Subtyping
Molecular data
A broad array of genetics and omics data is being used to carry out molecular subtyping of complex diseases. Molecular subtyping is most often based upon the identification of common genetic mutations and copy number variants, as well as the patterns of gene and protein expression. Regulators of gene and protein expression and activity such as miRNA, DNA methylation, and protein phosphorylation are also increasingly being used for molecular characterization of complex diseases.
Candidate gene discovery and the identification of putative causal copy number variations mark the first steps in a genetics-first approach to subtyping ASD. Exome sequencing projects are well underway and have already identified de novo likely gene disrupting mutations (LGD) associated with ASD (Iossifov et al., 2014; O'Roak et al., 2012a, 2012b; Sanders et al., 2012). Using whole exome sequencing, Iossifov and colleagues (2014) determined that de novo mutations, including copy number variants, account for 30% of simplex autism cases (one affected individual in a family). Of the LGDs identified thus far in individuals with ASD, many belong to shared networks, indicating common biological pathways are at play (Hormozdiari et al., 2015; Iossifov et al., 2014; O'Roak et al., 2012a, 2012b). The use of targeted sequencing, such as molecular-inversion probes (MIP), provides an efficient, cost-effective way to resequence recurrent genetic events in larger populations (O'Roak et al., 2012a; Turner et al., 2009).
However, individual sample collections often lack the size to reach statistical significance; large samples of affected and control subjects are needed. This has prompted multi-site collaborations, such as the Autism Sequencing Consortium, which involves researchers who have agreed to share data in order to determine genetic markers more quickly (Buxbaum et al., 2012; Stessman et al., 2014). Patient–clinician–researcher networks, such as that of the Simons VIP Consortium (Simons VIP Consortium, 2012), have been developed for recurrent copy number variations and single gene disrupting mutations associated with developmental disorders (e.g., 16p11.2) in order to determine clinical profiles through comprehensive behavioral phenotyping and neuroimaging. It is the hope that similar research networks can be developed for specific candidate genes as pathogenicity is confirmed.
The utility of identified ASD-associated gene disrupting mutations remains incomplete without further understanding of how these gene disruptions impact transcription, protein expression, and biological pathway modulators. Understanding the molecular mechanisms involved in ASD is the subject of transcriptomic and proteomic research, both of which have indicated abnormal neuronal development and inflammation (Broek et al., 2014). Transcriptomic studies, which use cDNA microarray and RNA sequencing, have identified dysregulated hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors in individuals with ASD, which have known functions in synaptic plasticity (Isaac et al., 2007; Purcell et al., 2001; van Spronsen and Hoogenraad, 2010). Disruptions to the gamma-aminobutyric acid (GABA) receptor system, which is involved in signal transduction and cellular adhesion, have also been found in ASD samples (Broek et al., 2014; Voineagu, 2012; Voineagu et al., 2011).
Proteomic studies have investigated protein expression in serum, plasma, and postmortem brains of individuals with ASD using techniques such as mass spectrometry and immunoassay (Broek et al., 2014; Schwarz et al., 2011; Taurines et al., 2010). Studies have found increased levels of proteins involved in synapse formation, such as brain-derived neurotrophic factor (BDNF) and glial fibrillary acidic protein (GFAP) in ASD samples (Chauhan et al., 2011; Correia et al., 2010; Fatemi et al., 2002; Riikonen, 2003; Schwarz et al., 2011; Thanseem et al., 2012). Proteomic analyses have also found altered levels of immune system-regulating proteins, such as apolipoprotein in the cerebrospinal fluid and blood of individuals with ASD compared to controls (Corbett et al., 2007; Molloy et al., 2006; Woods et al., 2012; Zimmerman et al., 2005). Proteins show great promise as useful biomarkers for ASD, but large-scale replications are needed. Importantly, despite group differences between individuals with ASD and typically developing counterparts, significant variability is observed within the ASD group underscoring the relevance of using this variance to define subpopulations within ASD.
Clinical data
Molecular subtyping via genetics and omics data gains clinical significance and utility when associated with phenotypic and clinical features. Genetic sequencing provides a “sieve” for the vast heterogeneity of complex diseases such as ASD, sorting individuals at the molecular level prior to phenotypic profiling, thus reducing heterogeneity and simplifying the phenotypic subtyping process (Stessman et al., 2014). While secondary in a genetics-first approach, comprehensive standardized psychometric testing gathers critical information on cognitive, adaptive, behavioral, motor, and neuropsychological levels of functioning. Once recurrent genetic loci have been identified through genetic testing, individuals with ASD and an identified genetic event should be evaluated clinically in order to determine whether phenotypic characteristics suggest a unique ASD subtype (e.g., CHD8) (Bernier et al., 2014). Previous associations have been found between genes associated with ASD and intellectual disability (Kaufman et al., 2010), and already specific gene disrupting mutations identified through molecular subtyping show co-morbidity with significant cognitive deficits (e.g., ADNP, PTEN) (Frazier et al, 2014; Helsmoortel et al, 2014; Vandeweyer et al, 2014).
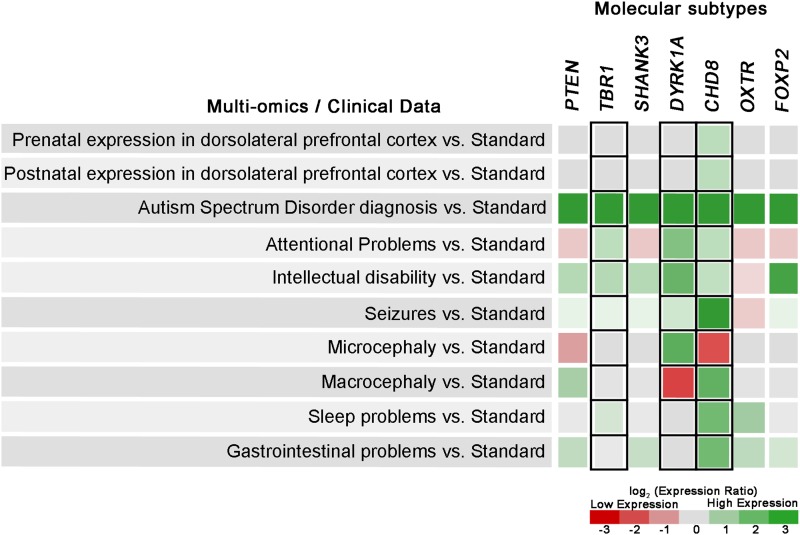
Clinical phenotyping should also include imaging and physical examination, with an emphasis on head circumference and physical dysmorphology, as these physical measurements are easily, consistently, and reliably collected in clinical settings (Stessman et al., 2014). These latter observations are often part of patient medical records, underscoring the utility of integration of electronic medical records with omics data to parse the heterogeneity of neurodevelopmental disorders such as ASD. Individuals with recurrent disrupted genes belonging to a beta catenin/Wnt signaling-associated protein–protein interaction network show variations in head circumference by subset; macrocephaly is found in individuals with PTEN and CHD8 while microcephaly is predominant in individuals with DYRK1A mutations (Bernier et al., 2014; Frazier et al., 2014; O'Roak et al., 2012a).
While the extent of known genotype–phenotype connections are still limited to small samples sizes, strong associations between identified ASD-associated genes and distinct cognitive and physical phenotypes indicate successful first steps in molecular subtyping. Longitudinal data is needed to better understand the genetic contributions to phenotypic presentation over the course of development. Understanding long-term impact of gene and protein disruption due to disruptive mutations is essential to identifying additional medical and psychiatric risks and informing treatment plans across the lifespan.
Imaging data
Over the past decade, imaging studies of children and adults with ASD continue to investigate whether there is common convergence within the structural and/or functional brain, despite the known heterogeneity of ASD (Anagnostou and Taylor, 2011). A variety of technologies are used, including functional and structural magnetic resonance imaging (fMRI; sMRI), electroencephalography (EEG), and functional near-infrared spectroscopy (fNIRS). The primary goal of ASD imaging research has been to identify biomarkers that are strong diagnostic indicators (Ruggeri et al., 2014). Several studies have implicated possible robust biomarkers, including hypoactive social and language brain areas (Carter et al., 2012; Pelphrey et al., 2011; Williams et al., 2013), and atypical EEG rhythms and components (Bernier et al., 2007; Maxwell et al., 2013; Oberman et al., 2013; Webb et al., 2012). However, many imaging studies report mixed findings, such as conflicting reports of long-range brain connections that suggest under-connectivity (Just et al., 2004) versus reports of over-connectivity (McFadden and Minshew, 2013).
To address these potential inconsistencies, recent work is focusing on how molecular and genetic subtypes may impact patterns of brain activation. In one study using sMRI (Qureshi et al., 2014), a mirror phenotype of brain volume was observed for individuals with ASD and a copy number variation within the 16p11.2 locus, such that compared to controls, deletion carriers exhibited increases and duplication carriers exhibited decreases in brain size. Other work has associated autism risk genes to structural and functional brain connectivity (e.g., CNTNAP2) (Dennis et al., 2011; Rudie et al., 2012). These studies highlight the importance of specifying ASD subtypes when investigating neural biomarkers of ASD, considering the differences that are likely derived from the etiology of subgroups of ASD.
In addition to characterizing atypical neural patterns of individuals with ASD, other recent imaging initiatives seek to detect neuroendophenotypes in individuals with a typical clinical phenotype but also a genetic vulnerability for developing ASD (e.g., unaffected relatives). Neuroendophenotypes are heritable indicators that persist in unaffected individuals, regardless of whether the pathology developed (Gottesman and Gould, 2003). In other words, this approach provides an opportunity to observe the neurobiological mechanisms by which high-risk individuals (e.g., siblings of children with autism) overcome genetic susceptibility (Constantino et al., 2010). For instance, unaffected children who have a sibling with ASD exhibit increased activation within key social perception brain regions, such as the superior temporal sulcus and ventromedial prefrontal cortex, above and beyond typically developing children (Kaiser et al., 2010). Similarly, other studies have targeted other social brain regions, such as the amygdala, as being functionally and structurally distinct for unaffected siblings (Dalton et al., 2007; Segovia et al., 2014; Spencer et al., 2011). These compensatory mechanisms may highlight areas of strength for unaffected relatives that can be used for targeted treatment of children with ASD and may better elucidate molecular etiologies in tandem with state-like biomarkers of ASD.
Data resources
An increasing array of publically available genetics and omics data is helping to greatly expand the use of molecular subtyping in complex diseases. This is most readily apparent in cancer where the Cancer Genome Atlas (TCGA) (McLendon et al., 2008; TCGA Network, 2011, 2012) maintains a repository of omics data including sequencing, gene and protein expression, SNPs, miRNA, and methylation for thousands of tumors across dozens of types of cancer. GEO and Array Express maintain huge repositories of gene expression data (Barrett et al., 2010; Kolesnikov et al., 2015) and other repositories are now storing raw proteomics data (Farrah et al., 2014; Vizcaíno et al., 2013). Other resources such as the Multi-Omics Profiling Expression Database (MOPED) are also processing these data to present more accessible and standardized views of gene and protein expression data (Higdon et al., 2014; Kolker et al., 2012; Montague et al., 2015, 2014).
In the field of autism research, large databases such as the National Database for Autism Research (NDAR, ndar.nih.gov) have been developed to house multidisciplinary biomolecular data, including exome sequencing, brain imaging, and clinical diagnostic data. A large-scale genome mapping study, AUT10K (funded by Autism Speaks) is using Google Cloud to manage, analyze, and disseminate its data, which provides new opportunities for broader access. However, the utility of these resources is significantly limited due to the lack of detailed clinical and phenotypic data being explicitly linked to the molecular data.
Summary of data in molecular subtyping
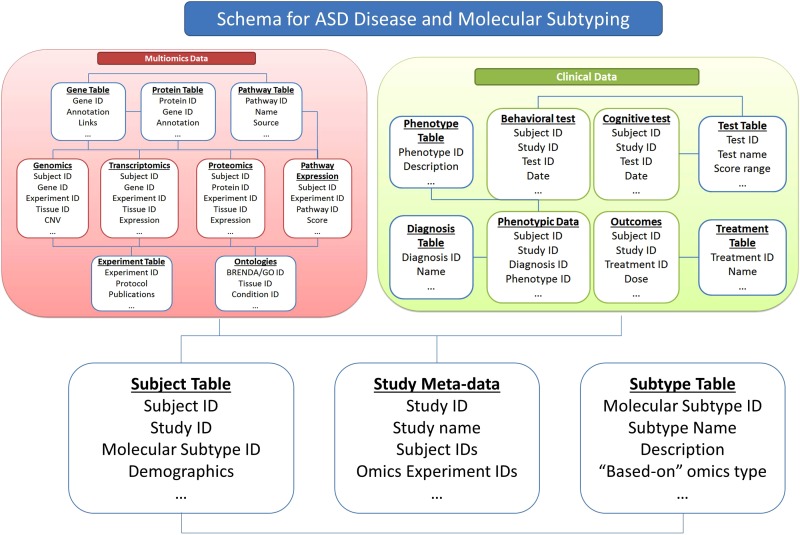
With rapid advances in genetics, omics, imaging, and clinical research on ASD, the need for collaborative data sharing and integration becomes imperative for building a comprehensive understanding of ASD etiology. In order to initiate effective avenues for data integration, a clear understanding of the commonly used methods and resources for data sharing and analysis in molecular subtyping is needed, as they relate to subtype identification and patient classification (Dumbill and Kolker, 2013; Field et al., 2009; Higdon et al., 2013; Kolker and Stewart, 2014). Figure 1 shows a sample database schema for integrating ASD data that can be used for the generation of molecular subtypes.
FIG. 1.
A model schema showing data used for molecular and disease subtyping in ASD.
Data Analysis and Integration for Molecular Subtyping
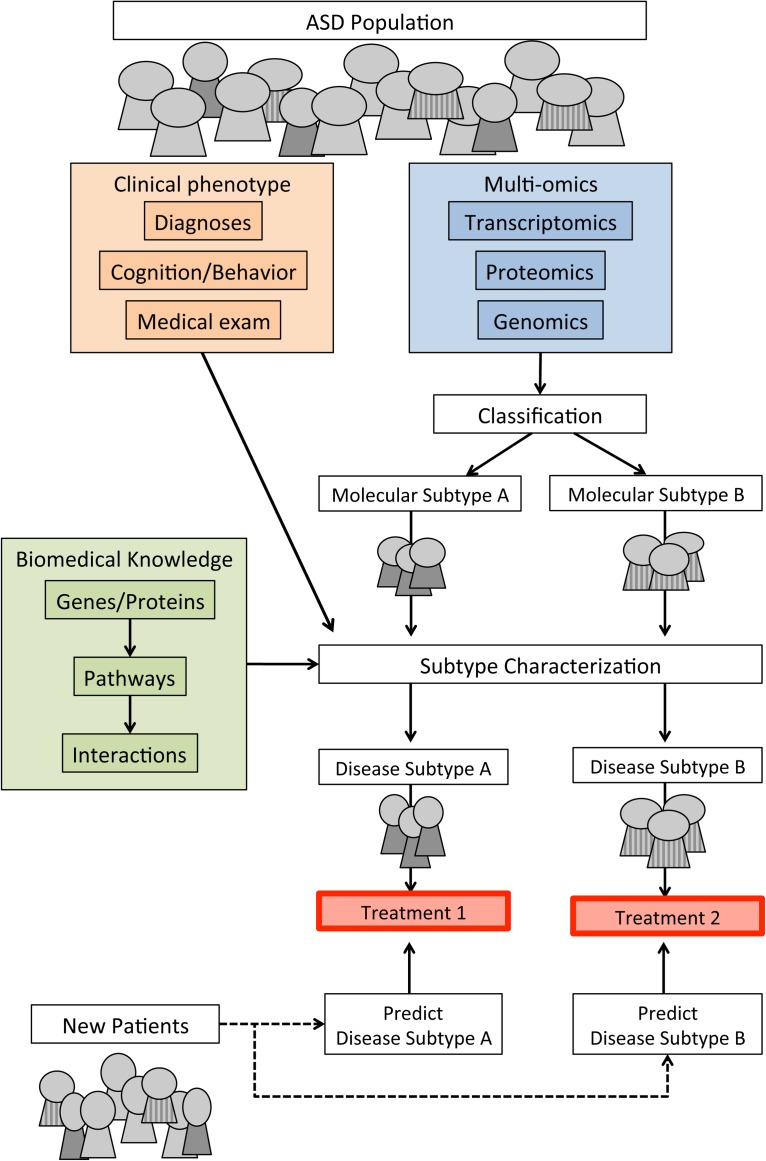
Figure 2 outlines an analysis approach to generating molecular and disease subtypes. This subtype and analysis approach can be used to characterize disease mechanisms, relate molecular data to disease phenotypes through clinical and environmental data, classify new patients, and identify optimal and potential new treatments for different subtypes.
FIG. 2.
Approach to molecular and disease subtyping in ASD.
Classifying patients into subtypes
Following molecular data collection, the methods used to classify patient genetic data into molecular subtypes vary greatly depending on type and amount of data. When subtyping by identified genetic mutations or CNVs, which are often sparsely distributed across patient populations and rarely occur with high multiplicity in any individual patients, the mere presence of these mutations is enough to generate molecular subtypes. However, differential gene expression and common biological pathways can make phenotypic manifestations difficult to differentiate at a clinical subtyping level (Stessman et al., 2014). In the case of ASD, it is anticipated that while some gene mutations may be highly penetrant and possess distinct clinical features indicative of syndromic subsets of ASD, other mutations are linked to common pathways such that phenotypic profiles are highly interconnected and more difficult to tease apart (Stessman et al., 2014).
Due to this variability in genetic background, gene expression data obtained from microarrays or RNA-Seq provide a more quantitative basis for generating molecular subtypes. The use of clustering methods with gene expression data is well established and widespread (de Souto et al., 2008; Eisen et al., 1998). The simplest and most commonly used approach is hierarchical clustering where patients are iteratively grouped by using a distance metric based upon expression values. This approach has been used in many previous molecular subtyping studies (Prat et al., 2010; Rouzier et al., 2005; Sørlie et al., 2001). Iossifov and colleagues (2014) clustered functional classes to determine enrichment of LGDs in individuals with ASD and their siblings in the following functional domains: Fragile-X mental retardation protein (FMRP) target genes, chromatin modifying genes, genes encoding postsynaptic density proteins, and those expressed in embryonic development.
There are many other clustering and unsupervised learning approaches, two of the most frequently used for molecular subtyping are k-means and self-organizing maps (SOM) (Borkowska et al., 2014; Hartigan and Wong, 1979; Kohonen, 1989). These approaches can often create more definitive and interpretable clusters than hierarchical methods. An approach that has become popular in molecular subtyping for cancer is consensus clustering. In this approach, a consensus is created from repeated clustering using multiple subsets of data and different numbers of clusters (Monti et al., 2003).
Typically, gene expression datasets contain data on thousands of genes, which create a level of noise and complexity that can make clustering algorithms inefficient and results hard to interpret. This has led to the use of data reduction methods such as principal components analysis (PCA) (Yeung and Ruzzo, 2001) and partial least squares (PLS) (Nguyen and Rocke, 2002) to reduce the dimension of data. Other methods limit the number of feature or genes used for clustering, creating both tighter and easier to interpret clusters; among these are sparse clustering, gene shaving, and dendogram sharpening (Hastie et al., 2000; Stanberry et al., 2013, 2003; Witten and Tibshirani, 2010).
The methods used to analyze gene expression have been adapted for other types of data platforms as well, such as miRNA, methylation, SNP array, proteomics, and metabolomics. Difficulties arise when the generation of subtypes needs to be based upon multiple platforms. Clustering can be applied individually to different sets of data and then the clusters can be merged using various ad hoc approaches. Alternatively, clustering can be applied jointly to different sets; indeed, several approaches have been developed to do that (Shen et al., 2010). All of these approaches will benefit for consistently processed, normalized, and analyzed expression data (Holzman and Kolker, 2004; Kolker et al., 2011).
Characterizing shared pathways and mechanisms in molecular subtyping
The use of clustering algorithms applied to molecular data can easily generate subgroups, but these are of little value if they do not help characterize the underlying disease mechanisms. Once molecular subtypes are established, they are most often characterized by describing their common genetic mutations and molecular features or by the over or under expression of specific molecules (genes, proteins, or other omics).
As mentioned previously, in the case of ASD, further disease mechanism characterization is performed by linking different genes according to their involvement in biological pathways or interaction networks (Hormozdiari et al., 2015; Iossifov et al., 2014). Many public (e.g., Reactome, Panther, BioCyc, KEGG) (Caspi et al., 2014; Croft et al., 2014; Kanehisa et al., 2014; Mi et al., 2013) and commercial pathway resources (e.g. Ingenuity) are available, as well as interaction databases (e.g., String, Intact) (Franceschini et al., 2013; Kerrien et al., 2011). Some methods take advantage of these resources to jointly combine pathway information with clustering in order to generate subtypes that are more directly interpretable and connected with existing pathway knowledge (Milone et al., 2014). Joint clustering of genes and patients may be able to identify sets of genes operating in concert outside of known pathways (Shen et al., 2010).
When subtypes are based on expression data, conventional analysis tools can be applied to molecular subtypes in order to characterize them. This includes identifying genes (or other molecules) that are highly differentially expressed across subtypes. Popular models for differential expression analysis include linear models for microarray analysis (LIMMA) and significance analysis of microarrays (SAM) (Smyth, 2004; Tusher et al., 2001). For example, a study by Zeidan-Chulia and colleagues (2014) used LIMMA modeling to identify altered expression of Alzheimer's-related genes in the NOTCH and Wnt signaling cascades in a sample of individuals with ASD, particularly the downregulation of mitochondria-regulating genes.
In addition, gene set approaches (Subramanian et al., 2005; Wu et al., 2010; Wu and Smyth, 2012) can be used to identify important pathways or networks that differ across subtypes in their expression pattern. Improvements to these approaches incorporate pathway structure to help identify important sub-pathways as is done in the Differential Expression of Pathways (DEAP) method (Haynes et al., 2013). It is important to note that if the same data are used for characterizing subtypes as was used for generating subtypes, then measures of statistical significance (p-values, etc.) will be highly biased.
Statistical models for connecting molecular subtypes with clinical phenotype
In order to understand and treat a complex disease such as ASD, molecular characteristics must be linked with the clinical manifestation of the disease. Standard models can be used to compare clinical data across established molecular subtypes, including linear (for continuous data), generalized linear (categorical, dichotomous, ordinal or count data), or Cox (for survival or time to event data) models (Cox and Oakes, 1984; McCullagh and Nelder, 1989). These models can be adjusted for the interaction of molecular data with environmental data (race, gender, parental factors, exposure during pregnancy) by adding terms to the model.
An important aspect of complex diseases such as ASD is progression of the disorder over time. Models that can incorporate longitudinal data, such as linear or generalized linear mixed models, are important for identifying mechanisms related to progression of the disorder or response to treatment (Breslow and Clayton, 1993; Laird and Ware, 1982). Longitudinal models incorporating molecular data have begun in studies on the progression of ASD in early development, such as the work of Glatt and colleagues (2012), who associated mRNA biomarkers in infants who showed early signs of ASD between 12 and 36 months of age. This ability to detect ASD subtypes during early infancy will accelerate opportunities for immediate and personalized treatment decisions. Other methods have been developed for complex diseases that combine the generation of subtypes with analysis of clinical data, such as survival, allowing for analysis when subtypes are not pre-defined (Bair and Tibshirani, 2004).
New patient classification and treatment identification
Molecular subtyping can greatly enhance the classification of new patients to disease subtypes, a crucial step to understanding a prognosis and appropriately targeting treatments. Since new patients will often have more limited data than the set patients used to generate the molecular subtypes, new classifications need to be based on simpler, more easily obtained data. In some cancer subtyping, small panels of biomarkers are used to classify patients to subtypes (Bastien et al., 2012; Choudhury et al., 2015; Taylor et al., 2012). Previous analyses can be used to help identify sets of potential predictor variables. The data can be randomly divided into training and test sets to help build and validate the model. If the amount of available data is small, cross-validation approaches can be used to evaluate the model. A wide range of supervised learning models are available to build a classifier such as logistic discriminant analysis, support vector machines, nearest neighbor and Bayes classifiers (Hastie et al., 2009). Software such as WEKA (Hall et al., 2009) can implement a wide range of models, so both the predictors and the type model can be easily evaluated. Additionally, electronic medical records can be a simple, more easily obtained resource that can be mined for the validation of subtype classification.
Two of the key objectives for molecular subtyping are early identification of patients (i.e., early infancy) to target the most effective treatment for individuals and to identify new treatment targets. Genes and pathways identified in the characterization of subtypes can be tied to different pharmacological databases such as PharmGkb and DrugBank (Law et al., 2014; Whirl-Carrillo et al., 2012). These resources can be used to identify compounds that inhibit or induce specific gene or protein expression or that block or stimulate specific pathways. Clinical data regarding response to treatment can be related back to molecular subtypes to identify those with the best responses to particular treatments. Linking longitudinal data to etiologically-derived subtypes can reduce the variability in phenotypic presentation, unmasking common behaviors or biomarkers that aid in diagnosis and predict prognosis.
Genetics has already informed pharmacological treatment exploration for ASD (Jeste and Geschwind, 2014). For example, CNTNAP2 variants have been associated with ASD and other neurodevelopmental disorders (Alarcon et al., 2008; Arking et al., 2008); this variant has been shown to have increased expression in frontostriatal circuits of the brain (Abrahams et al., 2007). CNTNAP2-mutant mouse models, which present ASD-like symptoms, have shown alleviated repetitive behaviors, but no change in social deficits when treated with risperidone, a dopamine antagonist (Penagarikano et al., 2011; Penagarikano and Geschwind, 2012). As molecular subtyping continues in the field of ASD research, it is expected that personalized treatment will emerge for distinct molecular subtypes, better equipping medical professionals to address symptoms of ASD, as well as co-morbid conditions.
Emerging Molecular Subtypes in Autism
Even in its early stages, molecular subtyping with multiple integrated data methods has been shown to be successful in ASD research (Bernier et al., 2014; Frazier et al., 2014; Vandeweyer et al., 2014). Using exome and targeted sequencing technology, recurrent de novo disruptive mutations, such as CHD8, ADNP, DYRK1A, and PTEN have been found in individuals with ASD (Iossifov et al., 2014; O'Roak et al., 2012a, 2012b). Comprehensive phenotyping of a growing number of individuals with these disruptive mutations indicates a high likelihood of autism and unique medical, psychiatric, and morphological characteristics that suggest specific genetic subtypes for ASD (Bernier et al., 2014; Frazier et al., 2014; van Bon et al., 2015). Autism, intellectual disability, and dysmorphic features have been found in individuals with a disruptive mutation to ADNP, along with multiple reports of visual and cardiac defects (Vandeweyer et al, 2014; Helsmoortel et al, 2014).
Individuals with mutations to CHD8 have enriched instances of chronic gastrointestinal complications, distinct facial dysmorphology, and macrocephalic head size (Bernier et al, 2014). In contrast, those with DYRK1A mutations have greater likelihood of microcephalic head size and early growth difficulties (van Bon et al, in press). In a recent study by Frazier and colleagues (2014), individuals with PTEN mutations showed abnormal white matter brain volume in addition to autism symptoms. These data, while still preliminary due to small sample sizes, suggest that the genetic heterogeneity of autism can be successfully reduced to smaller subsets of recurrent disruptive mutations in biologically related networks (Stessman et al., 2014). Figure 3 shows an example comparative analysis of gene expression across different clinical features and phenotypes for molecular subtypes of ASD.
FIG. 3.
Comparison of expression levels across clinical features and phenotypes of mutated genes characterizing different molecular subtypes of ASD.
Future Directions
The potential of molecular subtyping for advancing research, guiding personalized treatments, and providing a clear understanding of disease prognosis and progression has clearly been shown in diseases such as cancer and is emerging in ASD. Further, the emergence of family groups and communities centered on etiologically driven subgroups of neurodevelopmental disorders, such as Simons VIP Connect (www.simonsvipconnect.org) (Simons VIP Consortium, 2012), allows for improved quality of life for families. However, a broader use of molecular subtyping is impeded by data heterogeneity, diversity of standards, ineffective analysis tools, and a lack of rich clinical phenotypic and clinical data linked to molecular data. This limits the reproducibility and usage of subtypes across patients, experiments, and diseases.
Challenges arise when trying to generate robust and reproducible molecular subtypes from different experiments and datasets with widely varying data types and experimental protocols. This issue has impacted the generation of molecular subtypes for diseases such as ASD (Stessman et al., 2014). In addition, as new data and methods become available, the need to update subtypes must be addressed.
Achieving reproducible and robust molecular subtyping will require resources and technology that provide data integration using community standards, proper normalization, and sufficient meta-data across different omics (Chain et al., 2009; Dumbill and Kolker, 2013; Field et al., 2009; Galperin and Kolker, 2006; Garrity et al., 2008; Higdon et al., 2013, 2008; Hogan et al., 2006; Holzman and Kolker, 2004; Kolker and Stewart, 2014). These resources should enable molecular subtyping based upon only the highest quality data, experiments, and standards, use only the most robust models and analysis methods, and facilitate validation on sets of very well characterized subjects (Hather et al., 2010; Higdon et al., 2004; 2007; 2011; Higdon and Kolker, 2006; Kolker et al., 2011). Robust models need to be available to accurately predict the subtypes of new patients. Finally, molecular subtypes need to be connected to rich, but easily attainable, clinical and phenotypic data so that molecular subtyping can be used to create personalized treatments for patients. The development of such resources to assist the molecular subtyping of ASD has the potential to accelerate both the classification of new patients and the development of treatment regimens tailored to the specific presentation of a given subtype. As a result, these resources will empower and accelerate precision medicine and personalized healthcare and will ultimately serve families in more proactive and comprehensive ways.
Acknowledgments
We thank Maggie Lackey for her critical reading. This article was supported by National Science Foundation under the Division of Biological Infrastructure [0969929]; The National Institutes of Health R01-HL-060666-10A1; The Robert B. McMillen Foundation; Seattle Children's Research Institute-Northeastern University [to EK]. Additional support was provided by the National Institutes of Health R01 MH101221, R01 NIMH MH 092367, R01 MH100028, and R01 MH100047 [to RB].
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, and Geschwind DH. (2007). Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci USA 104, 17849–17854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, et al. (2008). Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 82, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatry Association. (2013). The Diagnostic and Statistical Manual of Mental Disorders: DSM 5 bookpointUS [Google Scholar]
- Anagnostou E, and Taylor MJ. (2011). Review of neuroimaging in autism spectrum disorders: What have we learned and where we go from here. Mol Autism 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, et al. (2008). A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet 82, 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baio J. (2012). Prevalence of autism spectrum disorders: Autism and developmental disabilities monitoring network, 14 Sites, United States, (2007). Morbidity and Mortality Weekly Report. Surveillance Summaries. Volume 61, Number 3 Centers for Disease Control and Prevention; [PubMed] [Google Scholar]
- Bair E, and Tibshirani R. (2004). Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, et al. (2010). NCBI GEO: Archive for functional genomics data sets–10 years on. Nucleic Acids Res 39, D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien RRL, Rodríguez-Lescure Á, Ebbert MTW, et al. (2012). PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Dawson G, Webb S, and Murias M. (2007). EEG mu rhythm and imitation impairments in individuals with autism spectrum disorder. Brain Cogn 64, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, and Birnbaum D. (2012). Basal breast cancer: A complex and deadly molecular subtype. Curr Mol Med 12, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. (2011). Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res 1380, 42–77 [DOI] [PubMed] [Google Scholar]
- Borkowska EM, Kruk A, Jedrzejczyk A, et al. (2014). Molecular subtyping of bladder cancer using Kohonen self-organizing maps. Cancer Med 3, 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow NE, and Clayton DG. (1993). Approximate inference in generalized linear mixed models. J Am Stat Assoc 88, 9–25 [Google Scholar]
- Broek JA, Brombacher E, Stelzhammer V, Guest PC, Rahmoune H, and Bahn S. (2014). The need for a comprehensive molecular characterization of autism spectrum disorders. Int J Neuropsychopharmacol 17, 651–673 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, and State MW. (2012). The autism sequencing consortium: Large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 76, 1052–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EJ, Williams DL, Minshew NJ, and Lehman JF. (2012). Is he being bad? Social and language brain networks during social judgment in children with autism. PLoS One 7, e47241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42, D459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PSG, Grafham DV, Fulton RS, et al. (2009). Genomics. Genome project standards in a new era of sequencing. Science 326, 236–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, and Chauhan V. (2011). Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem 117, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury Y, Wei X, Chu Y-H, et al. (2015). A multigene assay identifying distinct prognostic subtypes of clear cell renal cell carcinoma with differential response to tyrosine kinase inhibition. Eur Urol 67, 17–20 [DOI] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, et al. (2011). Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17, 500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Zhang Y, Frazier T, Abbacchi AM, and Law P. (2010). Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry 167, 1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett BA, Kantor AB, Schulman H, et al. (2007). A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry 12, 292–306 [DOI] [PubMed] [Google Scholar]
- Correia CT, Coutinho AM, Sequeira AF, et al. (2010). Increased BDNF levels and NTRK2 gene association suggest a disruption of BDNF/TrkB signaling in autism. Genes Brain Behav 9, 841–848 [DOI] [PubMed] [Google Scholar]
- Cox DR, and Oakes D. (1984). Analysis of Survival Data. CRC Press, Boca Raton, FL [Google Scholar]
- Croft D, Mundo AF, Haw R, et al. (2014). The Reactome pathway knowledgebase. Nucleic Acids Res 42, D472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton KM, Nacewicz BM, Alexander AL, and Davidson RJ. (2007). Gaze-fixation, brain activation, and amygdala volume in unaffected siblings of individuals with autism. Biol Psychiatry 61, 512–520 [DOI] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, Rudie JD, et al. (2011). Altered structural brain connectivity in healthy carriers of the autism risk gene, CNTNAP2. Brain Connect 1, 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souto MCP, Costa IG, de Araujo DSA, Ludermir TB, and Schliep A. (2008). Clustering cancer gene expression data: A comparative study. BMC Bioinformatics 9, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilard E, Bertucci F, Trempat P, et al. (2002). Gene expression profiling defines molecular subtypes of classical Hodgkin's disease. Oncogene 21, 3095–3102 [DOI] [PubMed] [Google Scholar]
- Dumbill E, and Kolker E. (2013). Introducing a metadata checklist for omics data. Big Data 1, 195–195 [DOI] [PubMed] [Google Scholar]
- Dvorkin-Gheva A, and Hassell JA. (2014). Identification of a novel luminal molecular subtype of breast cancer. PloS One 9, e103514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, and Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrøm MJ, Opdahl S, Hagen AI, et al. (2013). Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat 140, 463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrah T, Deutsch EW, Omenn GS, et al. (2014). State of the human proteome in 2013 as viewed through PeptideAtlas: Comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven Human Proteome Project. J. Proteome Res 13, 60–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, and Realmuto GR. (2002). Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol Psychiatry 52, 805–810 [DOI] [PubMed] [Google Scholar]
- Field D, Sansone S-A, Collis A, et al. (2009).’Omics data sharing. Science 326, 234–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, et al. (2013). STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, D808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, and Eng C. (2014). Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, and Kolker E. (2006). New metrics for comparative genomics. Curr Opin Biotechnol 17, 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Field D, Kyrpides N, et al. (2008). Toward a standards-compliant genomic and metagenomic publication record. OMICS J Integr Biol 12, 157–160 [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Tsuang MT, Winn M, et al. (2012). Blood-based gene expression signatures of infants and toddlers with autism. J Am Acad Child Adolesc Psychiatry 51, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman I, and Gould TD. (2003). The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry 160, 636–645 [DOI] [PubMed] [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, and Witten IH. (2009). The WEKA Data Mining Software: An Update. SIGKDD Explor Newsl 11, 10–18 [Google Scholar]
- Hartigan JA, and Wong MA. (1979). Algorithm AS 136: A K-means clustering algorithm. J R Stat Soc Ser C Appl Stat 28, 100–108 [Google Scholar]
- Hastie T, Tibshirani R, Eisen MB, et al. (2000). “Gene shaving” as a method for identifying distinct sets of genes with similar expression patterns. Genome Biol. 1, research0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, and Friedman J. (2009). The Elements of Statistical Learning, 2nd ed. Springer, New York [Google Scholar]
- Hather GJ, Haynes W, Higdon R, et al. (2010). The United States of America and Scientific Research. PLoS ONE 5, e12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WA, Higdon R, Stanberry L, Collins D, and Kolker E. (2013). Differential expression analysis for pathways. PLoS Comput Biol 9, e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsmoortel C, Vulto-van Silfhout AT, Coe BP, et al. (2014). A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet 46, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon R, Haynes W, Stanberry L, et al. (2013). Unraveling the complexities of life sciences data. Big Data 1, 42–50 [DOI] [PubMed] [Google Scholar]
- Higdon R, Hogan JM, Kolker N, van Belle G, and Kolker E. (2007). Experiment-specific estimation of peptide identification probabilities using a randomized database. OMICS 11, 351–366 [DOI] [PubMed] [Google Scholar]
- Higdon R, and Kolker E. (2006). A predictive model for identifying proteins by a single peptide match. Bioinformatics 23, 277–280 [DOI] [PubMed] [Google Scholar]
- Higdon R, Kolker N, Picone A, van Belle G, and Kolker E. (2004). LIP index for peptide classification using MS/MS and SEQUEST search via logistic regression. OMICS 8, 357–369 [DOI] [PubMed] [Google Scholar]
- Higdon R, Reiter L, Hather G, et al. (2011). IPM: An integrated protein model for false discovery rate estimation and identification in high-throughput proteomics. J. Proteomics 75, 116–121 [DOI] [PubMed] [Google Scholar]
- Higdon R, Stewart E, Stanberry L, et al. (2014). MOPED enables discoveries through consistently processed proteomics data. J Proteome Res 13, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon R, van Belle G, and Kolker E. (2008). A note on the false discovery rate and inconsistent comparisons between experiments. Bioinformatics 24, 1225–1228 [DOI] [PubMed] [Google Scholar]
- Hogan JM, Higdon R, and Kolker E. (2006). Experimental standards for high-throughput proteomics. OMICS 10, 152–157 [DOI] [PubMed] [Google Scholar]
- Holzman T, and Kolker E. (2004). Statistical analysis of global gene expression data: Some practical considerations. Curr Opin Biotechnol 15, 52–57 [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, Penn O, Borenstein E, and Eichler EE. (2015). The discovery of integrated gene networks for autism and related disorders. Genome Res 25, 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac JT, Ashby MC, and McBain CJ. (2007). The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54, 859–871 [DOI] [PubMed] [Google Scholar]
- Jeste SS, and Geschwind DH. (2014). Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol 10, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, and Minshew NJ. (2004). Cortical activation and synchronization during sentence comprehension in high-functioning autism: Evidence of underconnectivity. Brain 127, 1811–1821 [DOI] [PubMed] [Google Scholar]
- Kaiser MD, Hudac CM, Shultz S, et al. (2010). Neural signatures of autism. Proc Natl Acad Sci USA 107, 21223–21228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M. (2014). Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res 42, D199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Ayub M., and Vincent JB. (2010). The genetic basis of non-syndromic intellectual disability: A review. J Neurodev Disord 2, 182–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, et al. (1999). A full genome scan for late onset Alzheimer's disease. Hum Mol Genet 8, 237–245 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, et al. (2011). The IntAct molecular interaction database in 2012. Nucleic Acids Res 40, D84100D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BH, Navot N, Bernier R, and Webb SJ. (2014). Update on diagnostic classification in autism. Curr Opin Psychiatry 27, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen PT. (1989). Self-organizing feature maps In: Self-Organization and Associative Memory, Springer Series in Information Sciences. Springer Berlin Heidelberg, pp. 119–157 [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, et al. (2015). ArrayExpress update—Simplifying data submissions. Nucleic Acids Res 43, D1113–D1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E, Higdon R, Haynes W, et al. (2012). MOPED: Model Organism Protein Expression Database. Nucleic Acids Res 40, D1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E, Higdon R, Welch D, Bauman A, Stewart E, Haynes W, Broomall W, and Kolker N. (2011). SPIRE: Systematic Protein Investigative Research Environment. J Proteomics 75, 122–126 [DOI] [PubMed] [Google Scholar]
- Kolker E, and Stewart E. (2014). OMICS studies: How about metadata checklist and data publications? J Proteome Res. 13, 1783–1784 [DOI] [PubMed] [Google Scholar]
- Laird NM, and Ware JH. (1982). Random-effects models for longitudinal data. Biometrics 38, 963–974 [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, et al. (2014). DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res 42, D1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Petkova E, Hus V, et al. (2012). A multisite study of the clinical diagnosis of different autism spectrum disorders. Arch Gen Psychiatry 69, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisa L, de Reyniès A, Duval A, et al. (2013). Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med 10, e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CR, Villalobos ME, Schultz RT, Herpertz-Dahlmann B, Konrad K, and Kohls G. (2015). Atypical laterality of resting gamma oscillations in autism spectrum disorders. J Autism Dev Disord 45, 292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, and Nelder JA. (1989). Generalized Linear Models, Second Edition, 2 edition. ed. Chapman and Hall/CRC, Boca Raton [Google Scholar]
- McFadden K, and Minshew NJ. (2013). Evidence for dysregulation of axonal growth and guidance in the etiology of ASD. Front Hum Neurosci 7, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, et al. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, and Thomas PD. (2013). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41, D377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone DH, Stegmayer G, López M, Kamenetzky L, and Carrari F. (2014). Improving clustering with metabolic pathway data. BMC Bioinformatics 15, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy CA, Morrow AL, Meinzen-Derr J, et al. (2006). Elevated cytokine levels in children with autism spectrum disorder. J Neuroimmunol 172, 198–205 [DOI] [PubMed] [Google Scholar]
- Montague E, Janko I, Stanberry L, et al. (2015). Beyond protein expression, MOPED goes multi-omics. Nucleic Acids Res. 43, D1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague E, Stanberry L, Higdon R, et al. (2014). MOPED 2.5. An integrated multi-omics resource: Multi-Omics Profiling Expression Database now includes transcriptomics data. OMICS 18, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, and Golub T. (2003). Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52, 91–118 [Google Scholar]
- Nguyen DV, and Rocke DM. (2002). Tumor classification by partial least squares using microarray gene expression data. Bioinformatics 18, 39–50 [DOI] [PubMed] [Google Scholar]
- Oberman LM, McCleery JP, Hubbard EM, Bernier R, Wiersema JR, Raymaekers R, and Pineda JA. (2013). Developmental changes in mu suppression to observed and executed actions in autism spectrum disorders. Soc Cogn Affect Neurosci 8, 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, et al. (2012a). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, et al. (2012b). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelphrey KA, Shultz S, Hudac CM, and Vander Wyk BC. (2011). Research review: Constraining heterogeneity: The social brain and its development in autism spectrum disorder. J Child Psychol Psychiatry 52, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, et al. (2011). Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, and Geschwind DH. (2012). What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med 18, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Limburg PJ, Baron JA, et al. (2015). Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 148, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, et al. (2010). Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res BCR 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell AE, Jeon OH, Zimmerman AW, Blue ME, and Pevsner J. (2001). Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology 57, 1618–1628 [DOI] [PubMed] [Google Scholar]
- Qureshi AY, Mueller S, Snyder AZ, et al. (2014). Opposing brain differences in 16p11.2 deletion and duplication carriers. J Neurosci 34, 11199–111211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reporting Checklist For Life Sciences Articles [www Document], (2013). Nat. Publ. Group. URL http://www.nature.com/authors/policies/checklist.pdf ; accessed Jly 23, 2013
- Riikonen R. (2003). Neurotrophic factors in the pathogenesis of Rett syndrome. J Child Neurol 18, 693–697 [DOI] [PubMed] [Google Scholar]
- Ringman JM, Goate A, Masters CL, et al. (2014). Genetic heterogeneity in Alzheimer disease and implications for treatment strategies. Curr Neurol Neurosci Rep 14, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier R, Perou CM, Symmans WF, et al. (2005). Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11, 5678–5685 [DOI] [PubMed] [Google Scholar]
- Rudie JD, Hernandez LM, Brown JA, et al. (2012). Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron 75, 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri B, Sarkans U, Schumann G, and Persico AM. (2014). Biomarkers in autism spectrum disorder: The old and the new. Psychopharmacol Berl 231, 1201–1216 [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitt SJ. (2010). Classification and prognosis of invasive breast cancer: From morphology to molecular taxonomy. Mod Pathol 23, S60–S64 [DOI] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Rahmoune H, et al. (2011). Sex-specific serum biomarker patterns in adults with Asperger's syndrome. Mol Psychiatry 16, 1213–1220 [DOI] [PubMed] [Google Scholar]
- Segovia F, Holt R, Spencer M, et al. (2014). Identifying endophenotypes of autism: A multivariate approach. Front Comput Neurosci 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Woolfson LM, and Hunter SC. (2012). Confusion and inconsistency in diagnosis of Asperger syndrome: A review of studies from 1981 to 2010. Autism 16, 465–486 [DOI] [PubMed] [Google Scholar]
- Shen R, Olshen AB, and Ladanyi M. (2010). Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 26, 292–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons VIP Consortium, (2012). Simons Variation in Individuals Project (Simons VIP): A genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron 73, 1063–1067 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article3. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MD, Holt RJ, Chura LR, Suckling J, Calder AJ, Bullmore ET, and Baron-Cohen S. (2011). A novel functional brain imaging endophenotype of autism: The neural response to facial expression of emotion. Transl Psychiatry 1, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanberry L, Mias GI, Haynes W, Higdon R, Snyder M, and Kolker E. (2013). Integrative analysis of longitudinal metabolomics data from a personal multi-omics profile. Metabolites 3, 741–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanberry L, Nandy R, and Cordes D. (2003). Cluster analysis of fMRI data using dendrogram sharpening. Hum Brain Mapp 20, 201–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Bernier R, and Eichler EE. (2014). A genotype-first approach to defining the subtypes of a complex disease. Cell 156, 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IB, Ivanova T, Lim KH, et al. (2011). Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 141, 476–485, 485.e1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurines R, Dudley E, Conner AC, et al. (2010). Serum protein profiling and proteomics in autistic spectrum disorder using magnetic bead-assisted mass spectrometry. Eur Arch Psychiatry Clin Neurosci 260, 249–255 [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, et al. (2012). Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol (Berl) 123, 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Network (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, Anitha A, Nakamura K, et al. (2012). Elevated transcription factor specificity protein 1 in autistic brains alters the expression of autism candidate genes. Biol Psychiatry 71, 410–418 [DOI] [PubMed] [Google Scholar]
- Turner EH, Lee C, Ng SB, Nickerson DA, and Shendure J. (2009). Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods 6, 315–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, and Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bon B, Coe B, Bernier R, et al. (2015). Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol Psychiatry. Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Helsmoortel C, Van Dijck A, et al. (2014). The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am J Med Genet C Semin Med Genet 166c, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Spronsen M, and Hoogenraad CC. (2010). Synapse pathology in psychiatric and neurologic disease. Curr Neurol Neurosci Rep 10, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I. (2012). Gene expression studies in autism: Moving from the genome to the transcriptome and beyond. Neurobiol Dis 45, 69–75 [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, et al. (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SJ, Merkle K, Murias M, Richards T, Aylward E, and Dawson G. (2012). ERP responses differentiate inverted but not upright face processing in adults with ASD. Soc Cogn Affect Neurosci 7, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West L, Vidwans SJ, Campbell NP, et al. (2012). A novel classification of lung cancer into molecular subtypes. PLoS ONE 7, e31906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. (2012). Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DL, Cherkassky VL, Mason RA, Keller TA, Minshew NJ, and Just MA. (2013). Brain function differences in language processing in children and adults with autism. Autism Res 6, 288–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten DM, and Tibshirani R. (2010). A framework for feature selection in clustering. J Am Stat Assoc 105, 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SQ, Behren A, Mar VJ, et al. (2015). Whole exome sequencing identifies a recurrent RQCD1 P131L mutation in cutaneous melanoma. Oncotarget 6, 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods AG, Sokolowska I, and Darie CC. (2012). Identification of consistent alkylation of cysteine-less peptides in a proteomics experiment. Biochem Biophys Res Commun 419, 305–308 [DOI] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, and Smyth GK. (2010). ROAST: Rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, and Smyth GK. (2012). Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 40, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Januario T, Eberhard DA, et al. (2005). Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res Off J Am Assoc Cancer Res 11, 8686–8698 [DOI] [PubMed] [Google Scholar]
- Yeung KY, and Ruzzo WL. (2001). Principal component analysis for clustering gene expression data. Bioinforma Oxf Engl 17, 763–774 [DOI] [PubMed] [Google Scholar]
- Zeidan-Chulia F, de Oliveira BH, Salmina AB, et al. (2014). Altered expression of Alzheimer's disease-related genes in the cerebellum of autistic patients: A model for disrupted brain connectome and therapy. Cell Death Dis 5, e1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AW, Jyonouchi H, Comi AM, Connors SL, Milstien S, Varsou A, and Heyes MP. (2005). Cerebrospinal fluid and serum markers of inflammation in autism. Pediatr Neurol 33, 195–201 [DOI] [PubMed] [Google Scholar]