Abstract
Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, remains one of the most prevalent human pathogens and a major cause of mortality worldwide. Metabolic network is a central mediator and defining feature of the pathogenicity of Mtb. Increasing evidence suggests that lysine succinylation dynamically regulates enzymes in carbon metabolism in both bacteria and human cells; however, its extent and function in Mtb remain unexplored. Here, we performed a global succinylome analysis of the virulent Mtb strain H37Rv by using high accuracy nano-LC-MS/MS in combination with the enrichment of succinylated peptides from digested cell lysates and subsequent peptide identification. In total, 1545 lysine succinylation sites on 626 proteins were identified in this pathogen. The identified succinylated proteins are involved in various biological processes and a large proportion of the succinylation sites are present on proteins in the central metabolism pathway. Site-specific mutations showed that succinylation is a negative regulatory modification on the enzymatic activity of acetyl-CoA synthetase. Molecular dynamics simulations demonstrated that succinylation affects the conformational stability of acetyl-CoA synthetase, which is critical for its enzymatic activity. Further functional studies showed that CobB, a sirtuin-like deacetylase in Mtb, functions as a desuccinylase of acetyl-CoA synthetase in in vitro assays. Together, our findings reveal widespread roles for lysine succinylation in regulating metabolism and diverse processes in Mtb. Our data provide a rich resource for functional analyses of lysine succinylation and facilitate the dissection of metabolic networks in this life-threatening pathogen.
Post-translational modifications (PTMs)1 are complex and fundamental mechanisms modulating diverse protein properties and functions, and have been associated with almost all known cellular pathways and disease processes (1, 2). Among the hundreds of different PTMs, acylations at lysine residues, such as acetylation (3–6), malonylation (7, 8), crotonylation (9, 10), propionylation (11–13), butyrylation (11, 13), and succinylation (7, 14–16) are crucial for functional regulations of many prokaryotic and eukaryotic proteins. Because these lysine PTMs depend on the acyl-CoA metabolic intermediates, such as acetyl-CoA (Ac-CoA), succinyl-CoA, and malonyl-CoA, lysine acylation could provide a mechanism to respond to changes in the energy status of the cell and regulate energy metabolism and the key metabolic pathways in diverse organisms (17, 18).
Among these lysine PTMs, lysine succinylation is a highly dynamic and regulated PTM defined as transfer of a succinyl group (-CO-CH2-CH2-CO-) to a lysine residue of a protein molecule (8). It was recently identified and comprehensively validated in both bacterial and mammalian cells (8, 14, 16). It was also identified in core histones, suggesting that lysine succinylation may regulate the functions of histones and affect chromatin structure and gene expression (7). Accumulating evidence suggests that lysine succinylation is a widespread and important PTM in both eukaryotes and prokaryotes and regulates diverse cellular processes (16). The system-wide studies involving lysine-succinylated peptide immunoprecipitation and liquid chromatography-mass spectrometry (LC-MS/MS) have been employed to analyze the bacteria (E. coli) (14, 16), yeast (S. cerevisiae), human (HeLa) cells, and mouse embryonic fibroblasts and liver cells (16, 19). These succinylome studies have generated large data sets of lysine-succinylated proteins in both eukaryotes and prokaryotes and demonstrated the diverse cellular functions of this PTM. Notably, lysine succinylation is widespread among diverse mitochondrial metabolic enzymes that are involved in fatty acid metabolism, amino acid degradation, and the tricarboxylic acid cycle (19, 20). Thus, lysine succinylation is reported as a functional PTM with the potential to impact mitochondrial metabolism and coordinate different metabolic pathways in human cells and bacteria (14, 19–22).
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is a major cause of mortality worldwide and claims more human lives annually than any other bacterial pathogen (23). About one third of the world's population is infected with Mtb, which leads to nearly 1.3 million deaths and 8.6 million new cases of TB in 2012 worldwide (24). Mtb remains a major threat to global health, especially in the developing countries. Emergence of multidrug resistant (MDR) and extensively drug-resistant (XDR) Mtb, and also the emergence of co-infection between TB and HIV have further worsened the situation (25–27). Among bacterial pathogens, Mtb has a distinctive life cycle spanning different environments and developmental stages (28). Especially, Mtb can exist in dormant or active states in the host, leading to asymptomatic latent TB infection or active TB disease (29). To achieve these different physiologic states, Mtb developed a mechanism to sense diverse signals from the host and to coordinately regulate multiple cellular processes and pathways (30, 31). Mtb has evolved its metabolic network to both maintain and propagate its survival as a species within humans (32–35). It is well accepted that metabolic network is a central mediator and defining feature of the pathogenicity of Mtb (23, 36–38). Knowledge of the regulation of metabolic pathways used by Mtb during infection is therefore important for understanding its pathogenicity, and can also guide the development of novel drug therapies (39). On the other hand, increasing evidence suggests that lysine succinylation dynamically regulates enzymes in carbon metabolism in both bacteria and human cells (14, 19–22). It is tempting to speculate that lysine succinylation may play an important regulatory role in metabolic processes in Mtb. However, to the best of our knowledge, no succinylated protein in Mtb has been identified, presenting a major obstacle to understand the regulatory roles of lysine succinylation in this life-threatening pathogen.
In order to fill this gap in our knowledge, we have initiated a systematic study of the identities and functional roles of the succinylated protein in Mtb. Because Mtb H37Rv is the first sequenced Mtb strain (40) and has been extensively used for studies in dissecting the roles of individual genes in pathogenesis (41), it was selected as a test case. We analyzed the succinylome of Mtb H37Rv by using high accuracy nano-LC-MS/MS in combination with the enrichment of succinylated peptides from digested cell lysates and subsequent peptide identification. In total, 1545 lysine succinylation sites on 626 proteins were identified in this pathogen. The identified succinylated proteins are involved in various biological processes and render particular enrichment to metabolic process. A large proportion of the succinylation sites are present on proteins in the central metabolism pathway. We further dissected the regulatory role of succinylation on acetyl-CoA synthetase (Acs) via site-specific mutagenesis analysis and molecular dynamics (MD) simulations showed that reversible lysine succinylation could inhibit the activity of Acs. Further functional studies showed that CobB, a sirtuin-like deacetylase in Mtb, functions as a deacetylase and as a desuccinylase of Acs in in vitro assays. Together, our findings provide significant insights into the range of functions regulated by lysine succinylation in Mtb.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The M. tuberculosis strain H37Rv (ATCC 27294) was obtained from the National Center for Medical Culture Collections (CMCC, Beijing, China). The seed culture was grown to log phase (OD600 ≈ 1.2) in Middlebrook 7H9 liquid medium (Difco) supplemented with ADC (Albumin-dextrose-catalase) at 37 °C. After 7 days of growth of log phase, the cultures were pellet, washed twice with PBS (0.1 m Na2HPO4, 0.15 m NaCl, pH 7.5), and resuspended in fresh liquid medium followed by incubation for 2–3 weeks to a final OD600 ≈ 0.6–0.8. For carbon source utilization studies, Mtb seed culture was resuspended in fresh complex growth media containing additional carbon sources, such as 0.2% pyruvate, 0.2% glucose, 0.2% glycerol, 0.2% succinate, or 0.2% acetate, respectively.
Preparation of Protein Lysate and In-solution Trypsin Digestion
To identify as many succinylation events as possible, we have performed a series of preliminary experiments using different amount of Mtb proteins to find the best protocol. Following is the optimized experimental procedures based on these preliminary experiments. In brief, bacterial cells (250 ml) were harvested by centrifugation at 2500 × g for 10 min at 4 °C. The pellet was washed twice with ice-cold PBS, resuspended in ice-cold PBS containing 1× protease inhibitor mixture (Roche Diagnostics Ltd, Mannheim, Germany) and 1 mm PMSF (Beyotime Institute of Biotechnology, Jiangsu, China). The mixture was placed in Lysing Matrix B bead beater vials (pre-filled with 0.1 mm silica; MP Biomedicals, Santa Ana, CA) and homogenized using the FastPrep-24 sample preparation system (MP Biomedicals, CA). The cells were lysed by bead-beating for five cycles of 35 s at a setting of 6.5 m/s. Between each cycle of bead beating, the samples were incubated on ice for 3 min. Cell lysates were cleared of silica and cellular debris via centrifugation at 12,000 × g for 15 min at 4 °C. Supernatant was then filtered through the 0.2 μm syringe filters twice to sterilize lysate. The clarified lysate was precipitated by using 10% trichloroacetic acid and 1% sodium deoxycholate, and then washed twice with ice cold acetone. The precipitated proteins (2 mg) were redissolved in 50 mm ammonium bicarbonate, then in-solution digested by trypsin according to previously described (14, 42). The protein concentration was determined with the Bradford assay (Bio-Rad, Hercules, CA).
Immunoaffinity Enrichment of Lysine Succinylated Peptides
Succinylated peptides were enriched using agarose-conjugated anti-succinyl lysine antibody (PTM Biolabs Inc., Chicago, IL) as previously described (14). Briefly, tryptic peptides were redissolved in NETN buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40), and then incubated with anti-succinyllysine antibody conjugated protein A agarose beads at 4 °C for 6h with gentle rotation. The supernatant was removed and the beads were washed three times with NETN buffer, twice with ETN buffer (50 mm Tris-Cl, pH 8.0, 100 mm NaCl, 1 mm EDTA), followed by three times wash with water. The bound peptides were eluted by washing three times with 1% trifluoroacetic acid. The eluates were combined and dried in a SpeedVac. The resulting succinylated peptides were loaded onto self-packed C18 STAGE tips according to the manufacturer's instructions to desalt the sample, prior to nano-HPLC-MS/MS analysis.
Nano-HPLC-MS/MS Analysis
The enriched peptides were dissolved in the HPLC buffer A (0.1% (v/v) formic acid in water), and analyzed by online nanoflow LC-MS/MS using an easy nLC-1000 system (Thermo Scientific) connected to a Q-Exactive (Thermo Scientific) mass spectrometer. Briefly, samples were injected onto the analytical C18-nanocapillary LC column (5 μm particle size, 100 Å pore diameter) and eluted at a flow rate of 300 nl/min with a 45 min gradient from 5% solvent B (90% ACN/0.1% formic acid, v/v) to 80% solvent B. The peptides were then directly ionized and sprayed into a Q-Exactive mass spectrometer by a nanospray ion source. Mass spectrometer was operated in a data-dependent mode with an automatic switch between MS and MS/MS acquisition. Full MS spectra from m/z 350 to 1600 were acquired with a resolution of 70,000 at m/z = 200 in profile mode. Following every survey scan, up to 15 most intense precursor ions were picked for MS/MS fragmentation by higher energy C-trap dissociation (HCD) with normalized collision energy of 35%. Lock mass at m/z 445.12003 was enabled for full MS scan. The dynamic exclusion duration was set to be 5 s with a repeat count of one and ± 10 ppm exclusion window.
Protein Sequence Database Search
All acquired raw data were processed with the MaxQuant software (version 1.3.0.5) (43). The peak lists were searched against the Mtb H37Rv protein database from NCBI ftp site (http://www.ncbi.nlm.nih.gov/) containing 4018 protein sequences. Two missed cleavages were allowed for trypsin. The precursor and fragment ion mass tolerances were 10 ppm and 0.02 Da, respectively. Carbamidomethylation (Cys) was set as a fixed modification, whereas oxidation (Met), deamidation (Asn/Gln), succinylation (Lys), and acetylayion (Protein N-terminal) were set as variable modifications. Minimum peptide length was set at six. The estimated false discovery rate (FDR) thresholds for modification site, peptide and protein were specified at maximum 1%. All MS/MS spectra for the identified peptides with succinylation modifications were manually inspected using criteria as previously reported (44, 45). Furthermore, to improve the reliability of result, all succinylation sites assigned to the peptide C-terminal were removed, prior to bioinformatics analysis.
Bioinformatics Analysis
The identified succinylated proteins were grouped into biological process and molecular function class based on the Gene Ontology (GO) terms by Blast2GO software (46). The subcellular localization of succinylated proteins was analyzed with PSORTb program (47). To gain information on the over-representations, GO enrichment analysis was performed using Cytoscape plugin BiNGO (48, 49) with the parameters as previously described (50). The reference GO ontology in Cytoscape ontology format was created using whole Mtb H37Rv proteome that extracted from MTBbase (http://www.ark.in-berlin.de/Site/MTBbase.html). Pathway and Pfam functional domain enrichment analyses were performed using the DAVID bioinformatics resource (51, 52). Functional interaction network analysis was performed using interaction data as previously reported (53) and the interaction network was visualized by Cytoscape (version 3.0.2) (48). Amino acid sequence motifs were analyzed using motif-X (54) and an in-house script as previously reported (42). The position-specific heatmap was generated by plotting the log10 of the ratio of frequencies. The secondary structure types were determined for positions with succinylated lysine using NetSurfP tool (55). In our data, the corresponding p value < 0.05 was considered statistically significant.
Production of Specific Antibodies against Mtb Proteins
The generations of polyclonal antibodies against the specific Mtb proteins were carried out by ABclonal Inc. (Wuhan, Hubei, China). Briefly, to produce antibodies against the Mtb proteins, i.e. ATP synthase alpha chain (AtpA), ATP synthase beta chain (AtpF), 3-oxoacyl-[acyl-carrier protein] synthase 1 (KasA), acyl-[acyl-carrier protein] desaturase (DesA1), acyl-CoA ligase (FadD31), F420-dependent glucose-6-phosphate dehydrogenase (Fgd1), malate synthase G (GlcB), heat shock protein (HspX), two component system transcriptional regulator (MtrA) and polyketide synthase (Pks2), the full-length of the genes were cloned into the expression vector pGEX-4T (Pharmacia, Piscataway, NJ), and then each expression vector was transformed into E. coli strain BL21 (DE3) for overexpression. For expression, 1 mm isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added into the logarithmically growing bacterial culture and cells were harvested after 4 h of induction at 30 °C. Following purification of these antigens, immunization and sampling of the anti-sera from rabbit were performed by ABclonal Inc. The specificity of the generated antibodies was determined by the manufacturer using ELISA and Western blotting.
Immunoprecipitation and Western blotting
To validate succinylation of identified proteins, we performed immunoprecipitation (IP) experiments to analysis the succinylated proteins in vivo using the generated specific protein antibodies. For IP, the protein specific antibody was conjugated to the protein G Dynabeads (Invitrogen AS, Oslo, Norway) in PBS by gentle rocking for 4 h at 4 °C. The conjugated beads were washed three times with PBS and incubated with the whole cell lysates overnight at 4 °C. The beads were then washed three times with PBS to remove the unbound proteins. Bound proteins were boiled in SDS loading buffer for 5 min, then subjected to 15% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. For Western blotting, the membrane was blocked overnight at 4 °C or 37 °C for 2 h in TBS buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl) containing 5% bovine serum albumin (BSA) and incubated with either the specific Mtb protein antibody or the anti-succinyl lysine antibody (PTM Biolabs Inc., Chicago, IL) (1:2000, in TBS/5% BSA) overnight at 4 °C. After washing three times with TBST buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Tween20), the membrane was incubated with horseradish peroxidase- conjugated goat anti-rabbit antibody (1:5000 dilutions) for 1 h at 37 °C. The membrane was then washed with TBST buffer and visualized with enhanced chemiluminescence (ECL) immunoblotting detection reagents (Advansta Inc., Menlo Park, CA). The density of each band was determined with a fluorescence scanner (ImageQuant TL, GE Healthcare). Densitometry analysis was performed via the ImageJ suite (http://rsbweb.nih.gov/ij/).
Site-directed Mutagenesis and Purification of Acs
The deacetylase (CobB) and Acs expression vectors (pET32a-cobB and pET20b-acs, kindly provided by Professor Jiao-Yu Deng, Wuhan Institute of Virology, CAS) were transformed into E. coli BL21 (DE3) for protein expression. The site-directed mutation of acs gene was introduced into the selected sites by PCR reaction. The PCR product was transformed into E. coli BL21 (DE3), and subsequently the mutagenesis was completely sequenced to confirm the presence of the site-directed mutation. Mutagenic primers were given below with the mutated base triplets underlined: K193R-sense: 5′-CGCGGCAGACCATCGCCCCTCA AGGCGGCC-3′, K193R-antisense: 5′-CGATGGTCTGCCGCGCCGAAACTGCCCGTC-3′, K366R-sense: 5′-TTTATGAGATGGGGCCGTGAGATCCCCGAC-3′, K366R-antisense: 5′-GCCCCATCTCATAAACATCCGGATGAGGGT-3′. The wild type E. coli BL21 (DE3)/pET32a-cobB, E. coli BL21 (DE3)/pET20b-acs and the site-directed mutants of acs gene were grown in 5 ml Luria-Bertani (LB) medium containing ampicillin (50 μg/ml) overnight at 37 °C, and then the cultures were transformed into 500 ml fresh LB medium with ampicillin (50 μg/ml) at 37 °C in shaking flasks to optical density at 600 nm of 0.4∼0.6. Cells were induced with 0.2 mm of IPTG at 18 °C for 12h. The cultured cells were harvested by centrifugation and washed twice with ice-cold PBS, resuspended in binding buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl and 5 mm imidazole). The mixture was disrupted by high pressure cell disrupter with an output of 1500 W (1500 bar, JN-02C, JNBIO, Guangzhou, China). Cellular debris was removed by centrifugation at 8000 × g for 30 min at 4 °C, and the resulting supernatants were loaded onto an affinity Ni2+ column (Qiagen Inc., Chatsworth, CA) pre-equilibrated with the binding buffer. After the column was washed with binding buffer, followed by washing buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl and 60 mm imidazole or 80 mm imidazole), the protein was eluted with elution buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl and 250 mm imidazole). The elution was collected and concentrated using a 30,000 MWCO concentrator (Millipore) in storage buffer (20 mm Tris-HCl, pH 8.0, 50 mm NaCl). The protein concentration was determined with the Bradford assay (Bio-Rad).
Acs Activity Assay
The activity of purified Acs and site-directed mutants of K193R, K366R, K193R/K366R proteins were measured as described (56, 57). Briefly, each reaction contained 50 mm Tris-HCl (pH 8.0), 600 mm hydroxylamine-HCl (pre-neutralized with KOH), 10 mm MgCl2, 2 mm dl-Dithiothreitol (DTT), 1 mm Coenzyme A (CoA), 30 mm ATP, and 5 mm potassium acetate. Reactions were pre-incubated at 37 °C for 5 min before the addition of purified proteins (1 μm). Samples without protein served as a blank, and formation of acetyl-hydroxamate by acetyl-phosphate (AcP) served as a standard. After 10 min incubation, the reactions were quenched by adding an equal volume of stop solution [2.5% (w/v) FeCl3 in 2 m HCl, 10% (w/v) trichloroacetic acid]. The reaction products were centrifuged to remove turbidity, and then measured at 540 nm.
In Vitro Desuccinylation Activity Assay
The purified recombinant CobB and Acs were incubated in 50 μl of reaction buffer (50 mm Tris-HCl, pH 8.5, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2) with or without 500 μm NAD+ and in the presence or absence of 2 mm Nicotinamide (NAM). The mixture was incubated at 25 °C for 10 h. The reaction products were analyzed by SDS-PAGE and Western blotting.
Molecular Dynamics Simulations
The initial model of the Acs module was prepared from the available crystal structure (PDB code 2P20, 50.56% sequence identity) via structural modeling using CPHmodels 3.2 Server (http://www.cbs.dtu.dk/services/CPHmodels). All simulations were based on the initial model and a total of seven systems were prepared for simulation using GROMACS 4.5.5 (58) in conjunction with the OPLS-AA/L all-atom force field (59). The protein was then solvated in simple point charge (SPC) water molecules in a cubic box (60), with the box edges ∼1.0 nm from any atom of the protein, and eighteen additional Na+ ions were added to neutralize the charge of each system. Each system was then energy minimized using a steepest descents integrator either until the maximum force was less than 1000 kJ/mol/nm on any atom or until additional steps resulted in a potential energy change of less than 1 kJ/mol. Next, the simulations were performed under a constant temperature of 300 K and the V-rescale algorithm (61) was used with a temperature coupling time constant of 0.1 ps. All bond lengths were constrained using the linear constraint solver (LINCS) algorithm (62). Van der Waals interactions used a simple cut off at 1.4 nm and long-range electrostatic interactions were handled using the particle mesh Ewald (PME) method (63, 64) with a fourth-order spline interpolation and a 0.1 nm Fourier grid spacing. Once each system was sufficiently equilibrated around the target temperature, isotropic was used for pressure coupling and the constant pressure was set to 1.0 bar in all directions and a pressure coupling time constant of 1.0 ps. Finally, each system was subjected to 10 ns of molecular dynamics (MD) simulation and the time step used in the simulations was 2 fs. All analyses were performed using the GROMACS suite of tools and a secondary structure recognition algorithm (DSSP) (65), which was implemented in GROMACS. The PyMOL Molecular Graphics System (version 1.7.2, http://www.pymol.org) was employed to present the structural results of this study.
RESULTS
Detection of Protein Lysine Succinylation in Mtb
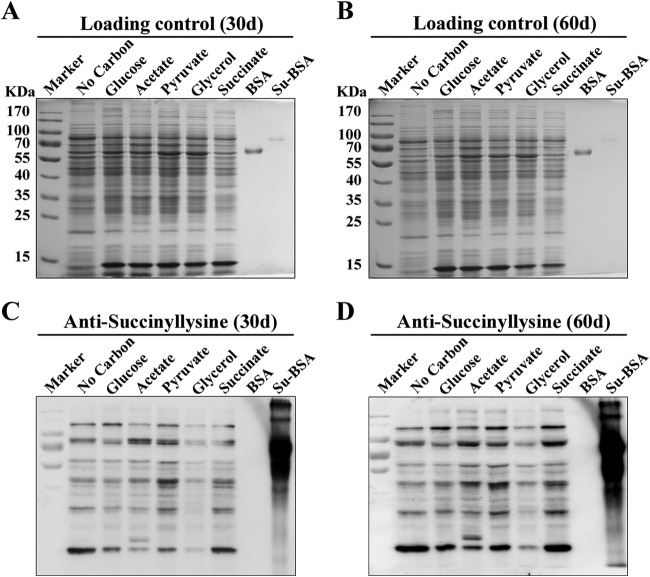
To examine the diversity and relative abundance of lysine succinylation proteins from different growth conditions of Mtb, Western blotting analysis was performed using with a polyclonal anti-succinyl lysine antibody (Fig. 1). The virulent Mtb strain H37Rv were grown in minimal medium, either without an additional carbon source or treated with pyruvate, glucose, glycerol, succinate, or acetate and harvested at 30 day or 60 day. Strong immunoblot signals were observed at both 30 day and 60 day (Fig. 1), indicating that lysine succinylation was abundant in H37Rv under different culture conditions. Immunoblots with succinylated or non succinylated BSA confirmed that the protein bands detected were specific (Fig. 1). However, succinylation signals were stronger when H37Rv were grown in medium containing succinate or pyruvate as a sole carbon source than those in no carbon medium (Fig. 1), suggesting that different growth mediums may alter the lysine succinylation profile. As expected, Mtb exhibited different growth rate in media containing different carbon source (supplemental Fig. S1). These results implied that the lysine succinylation is likely linked with energy metabolism in Mtb. Moreover, as shown in supplemental Fig. S2, we obtained a similar result as previously reported on lysine acetylation (66) when the strain was grown on different carbon source.
Fig. 1.
Anti-lysine succinylation immunoblots showing that different carbon sources altered succinylation profiles of Mtb H37Rv. Coomassie blue staining of protein lysates from A 30 days and B 60 days of cultures. Coomassie blue staining was used for the loading control. Western blotting analysis of lysine succinylation level in protein lysates from C 30 days and D 60 days of cultures with a pan anti-succinyllysine antibody. Specificity of succinylation signals was validated by succinylated or non succinylated BSA assay.
Enrichment and Identification of Lysine Succinylation Sites in Mtb
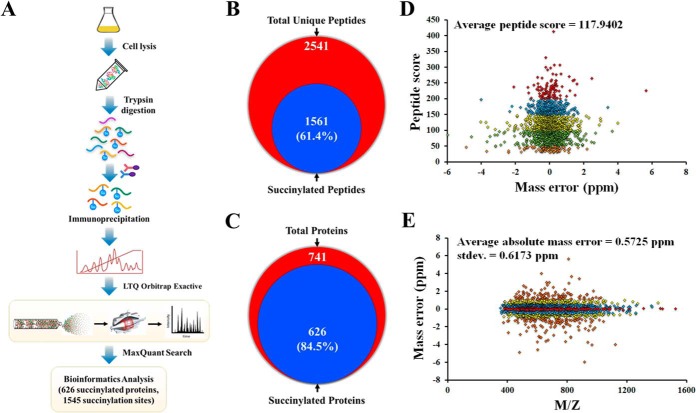
To identify the lysine succinylation sites in Mtb, we used a similar procedure as previously described (7, 14, 16) (Fig. 2A). In total, we identified 1561 unique modified peptides encompassing 1545 succinylation sites (class I) from 626 proteins with a FDR below 1% for modified peptides. The proteomic data showed that 61.4% of identified unique peptides (2541) across 84.5% of the identified proteins (741) were immune-enriched, indicating the highly efficient enrichment procedure were used in this study (Fig. 2B and 2C). All spectra containing succinylation were manually inspected to ensure diagnostic b- or y-ion series as previously described (44, 45). Detailed information of all identified succinylated peptides are provided in the supplemental Table S1. The raw data has been uploaded onto a public database PeptideAtlas (http://www.peptideatlas.org) and can be accessed through the following link: http://www.peptideatlas.org/PASS/PASS00518. Annotated peptide spectra for all succinylated peptides were deposited in PeptideAtlas and supplied as supplementary Dataset. The average peptide score was 117.9402 (Fig. 2D) and the overall absolute peptide mass accuracy was 0.5752 parts per million (ppm) (standard deviation, 0.6173 ppm) (Fig. 2E), confirming the high accuracy of modified peptide data obtained from MS. Furthermore, to assess the distribution of the average modified peptide length and succinylation sites, we calculated the number of amino acids per peptide and identified modification sites per protein. Supplemental Fig. S3A showed that the average peptide length was 15 amino acids and the average site coverage per identified protein was three lysine succinylation sites (supplemental Fig. S3B). We reasoned that multiple succinylation sites on one protein may play an important role in stabilization/destabilization of protein conformation and fulfill the delicate regulatory function.
Fig. 2.
Profiling lysine succinylome in Mtb H37Rv. A, Workflow for succinylome analysis of Mtb H37Rv. B, Venn diagram showing the number of identified succinylated peptides. C, Venn diagram showing the number of identified succinylated proteins. D, Distribution of peptide score. E, Distribution of precursor mass deviations.
Conservation of Succinylated Proteins
To explore the evolutionary conservation of all identified lysine succinylation sites in this study, we searched the orthologs of 626 succinylated proteins of Mtb against the following previously reported succinylomes: Escherichia coli DH10B (14), Escherichia coli BW25113 (16), Saccharomyces cerevisiae S288c (16), Homo sapiens (16), Mus musculus (16), and Drosophila melanogaster (7). The orthologs that showed the highest homology to each other in both directions were counted and reported as the percentage of the total number of proteins in all analyzed species. The percentage of Mtb homologs of succinylated proteins in diverse organisms is showed in Table I. As a result of a global homology search using the amino acid sequence of target proteins, orthologs of 205 (∼32.7%) Mtb succinylated proteins can be detected in the succinylomes of other species (supplemental Table S2). Moreover, we found that 164 and 107 Mtb succinylated proteins have succinylated homologs in bacterial and eukaryotic cell, respectively (supplemental Fig. S4). Interestingly, most succinylated proteins are involved in central carbon metabolism and lipids metabolism. These conserved succinylation events suggest that succinylation may play an important role in essential biological processes in this pathogen.
Table I. Comparison of Mtb H37Rv succinylome with other published succinylomes. “su” = succinylation.
| Species | Genome size (Mb) | Proteins | No. of su-sites | No. of su-proteins | Homologue with H37Rv | Ref |
|---|---|---|---|---|---|---|
| E. coli DH10B | 4.47 | 4,124 | 2,580 | 670 | 128 | (14) |
| E. coli BW25113 | 4.64 | 4,141 | 2,572 | 990 | 153 | (16) |
| S. cerevisiae S288c | 12.16 | 5,906 | 1,345 | 474 | 60 | |
| H. sapiens (HeLa) | 3,209.29 | 67,778 | 2,004 | 738 | 24 | |
| M. musculus (liver) | 2,798.79 | 73,462 | 2,140 | 750 | 58 | |
| D. melanogaster | 139.485 | 17,526 | - | 104 | 6 | (7) |
| M. musculus (MEFs) | 2,798.79 | 73,462 | 2,565 | 779 | 75 | (19) |
| M. musculus (liver) | 2,798.79 | 73,462 | 1,190 | 252 | 45 | (20) |
| M. tuberculosis H37Rv | 4.41 | 4,018 | 1,545 | 626 | - | In this study |
Functional Annotation and Cellular Localization of Succinylated Proteins
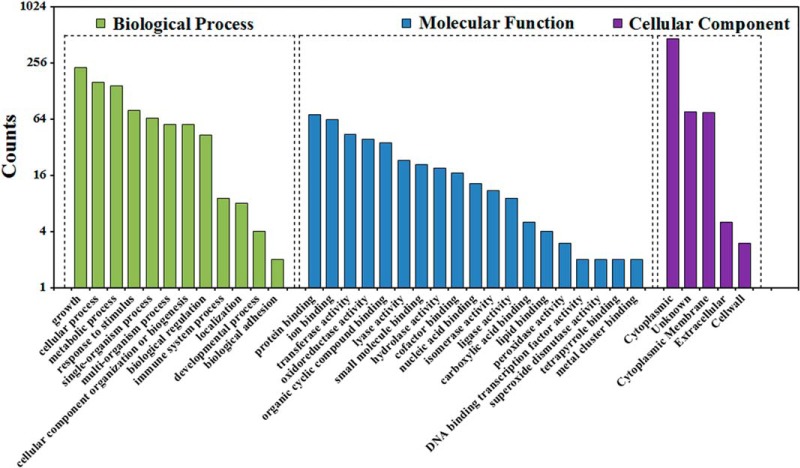
To better understand the lysine succinylome in Mtb, we performed functional annotation analysis using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Pfam database. Firstly, we investigated the GO functional classification of all the succinylated proteins based on their biological process and molecular function (Fig. 3). The classification results for biological process showed that the largest protein group of succinylated proteins was related to growth (26.60%), followed by cellular process (18.44%), metabolic process (17.04%) and response to stimulus (9.33%). Interestingly, 1.05% and 0.12% of the total succinylated proteins were involved in immune system process and signaling, respectively, which could be an important mechanism for the survival of Mtb. In the molecular function classification, most succinylated proteins were related to the binding of various targets and enzyme activity. Approximately half of the lysine succinylated proteins (50.76%) were categorized as catalytic activity-related proteins. Another large succinylated protein group is binding to targets (45.45%) (supplemental Table S3). Within the cluster of cellular localization, a total of 74.44% of the succinylated proteins was in the cytoplasmic category by the bacterial protein subcellular localization prediction program PSORTb (47). This result was also in agreement with the biological process analysis that most succinylated proteins were involved in cellular metabolism. Importantly, we also found that a sizable portion of succinylation proteins were located on cytoplasmic membrane (11.98%), cell wall (0.48%), and extracellular (0.8%). These proteins could play a role in regulating the envelope-related proteins to impact the virulence of mycobacteria.
Fig. 3.
Histogram representations of the distribution of identified succinylated proteins according to their biological processes, molecular functions and cellular localization.
To further elucidate the biological functions of succinylated proteins, we performed enrichment analysis using Cytoscape plugin BinGO. As shown in supplemental Figs. S5 and S6, the succinylated proteins were mostly enriched in growth (p = 9.86 × 10−15), and also statistically enriched in cellular metabolism process with specific enrichment in cellular ketone metabolic process (p = 3.6 × 10−9), carboxylic acid metabolic process (p = 3.86 × 10−9), oxoacid metabolic process (p = 3.86 × 10−9), and organic acid metabolic process (p = 3.86 × 10−9) (supplemental Fig. S5 and supplemental Table S4A). Consistently, the GO enrichment analysis of molecular functions further demonstrated that many functions were enriched in our set, including catalytic activity and binding (supplemental Fig. S6 and supplemental Table S4A). We next performed cellular compartment analysis of all succinylated proteins and obtained the similar results. As expected, a significant portion of succinylated proteins were significantly enriched in membrane and cell wall (supplemental Fig. S6 and supplemental Table S4A). Moreover, proteins involved in ribosome, citrate cycle (TCA cycle), fatty acid metabolism, glycolysis/gluconeogenesis, pyruvate metabolism, oxidative phosphorylation, glyoxylate, and dicarboxylate metabolism were also enriched, as determined through the KEGG pathway analysis (supplemental Fig. S7 and supplemental Table S4B), indicating that lysine succinylation substrates were associated with the ribosome and cellular metabolism events. The Pfam domain analysis illustrated that most succinylated substrates were enriched with the acyl-CoA dehydrogenase domain, which catalyzed the first step in the β-oxidation of fatty acids and catabolism of some amino acids (supplemental Fig. S7 and supplemental Table S4B). Taken together, these findings suggested that the identified succinylated proteins have a wide distribution of functions and cellular localizations and render particular enrichment to metabolic process in Mtb.
Sequence Recognition Motifs and Local Structural Properties
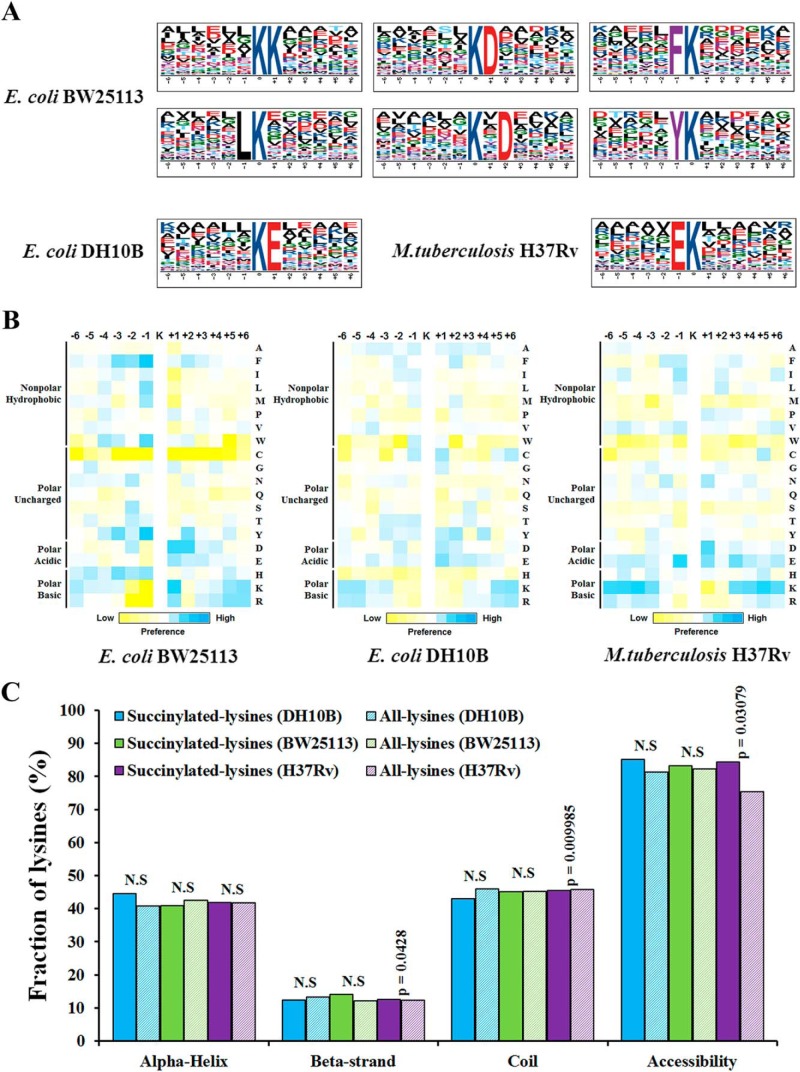
We further compared the position-specific amino acid frequencies of the surrounding sequences (six amino acids to both termini) of succinylated lysine residues with those of all lysine residues that occurred in the Mtb proteome. We found that negatively charged amino acids (glutamic acids) was drastically overrepresented at position −1 using motif-X (Fig. 4A and supplemental Table S5A). Then, we compared our data sets to the reported succinylome of E. coli, showing that the succinylation pattern found in E. coli BW25113 were quite different from Mtb site-specific succinylation motifs. Interestingly, the same amino acid (glutamic acids) was preferred at position +1 in E. coli DH10B. To further assess if there is significant enrichment or depletion of specific amino acids with respect to the general amino acid composition of the entire proteome, a position-specific intensity map was generated as previously described (42). The result showed a preference for polar acidic and polar basic amino acids (Fig. 4B). It can be seen that aspartic acid was most commonly found at +1 and glutamic acid was most commonly found at −1, +2. As well, we observed a significant preference for lysine at the −6 to −4, and +5 positions. These results indicated that succinylation preferentially occurred in lysine-rich regions and polar acidic/basic amino acid regions of proteins. We next compared these data with the patterns generated from other succinylomes. Although we did not detect a strong bias for a specific succinylation site motif in E. coli DH10B, there were modest biases for flanking amino acid residues. The different preference of amino acid residues surrounding lysine sites suggested the unique substrate preferences in Mtb. Next, we predicted the structural features of lysine succinylation sites in identified proteins with NetSurfP algorithm (55). As expected, succinylation sites were enriched on the protein surface (84.4%) as compared with 75.5% of the all lysine residue (p = 3.079 × 10−2). In accordance with previous report (16), the succinylated lysine was more frequently found in structured regions. Overall, 54.4% of succinylated lysines were predicted to be localized in α helix and β sheet structures in comparison to 45.5% of all lysines (Fig. 4C and supplemental Table S5B). Therefore, lysine succinylation may have an effect on the surface properties of modified proteins similar as other PTMs, such as acetylation and phosphorylation.
Fig. 4.
Bioinformational analysis of succinylation sites. A, Sequence Logo representation of significant motifs identified by Motif-X software. The motifs with significance of p < 0.000001 are shown. B, Position-specific under- or over-representation of amino acids flanking the succinylation sites. Colors were plotted by using intensity map and represent the log10 of the ratio of frequencies within succinyl-13-mers versus nonsuccinyl-13-mers (blue shows enrichment, yellow shows depletion). C, Distribution of succinylated and nonsuccinylated lysines in protein secondary structures. Probabilities for different secondary structures (α helix, beta-strand and coil) of succinylated lysine were compared with the secondary structure probabilities of nonsuccinylated lysine on all proteins identified in this study. Significance was calculated by Wilcoxon test.
Functional Interaction Networks of Succinylated Proteins in Mtb
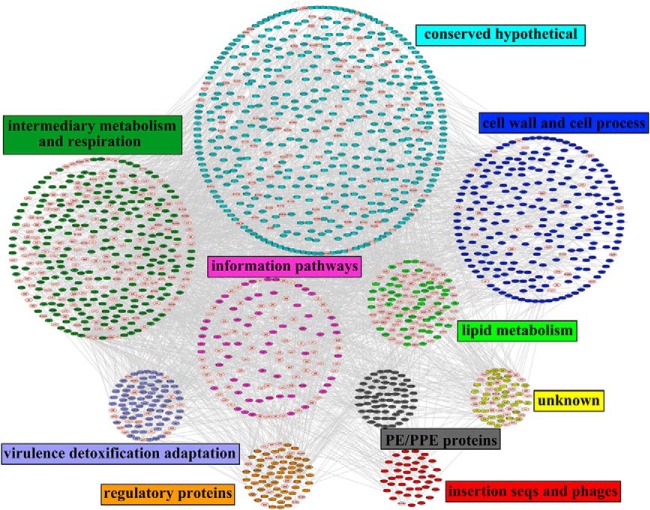
We generated a protein interaction network of all succinylated proteins using protein interaction information. The protein interaction map was visualized using Cytoscape (48) and consisted of a large network covering 1672 proteins among which there were 462 identified succinylated proteins (supplemental Table S6). Although further functional experiments were needed to validate the potential interactions, such bioinformatic analysis should be useful for formulating testable hypotheses to understand the function of the identified succinylated proteins in Mtb. Then, the functional category was used to group the identified proteins (supplemental Table S7). As shown in Fig. 5, several complexes and cellular functions formed prominent and highly connected clusters, such as cell wall and cell processes, intermediary metabolism and respiration, lipid metabolism, and virulence detoxification adaptation. The interaction networks of succinylated proteins involved in the ESX systems as well as virulence, detoxification and adaptation function in Mtb are illustrated in supplemental Fig. S8. We speculate that the physiological interaction among these protein clusters might contribute to the cooperation and/or coordination of their functions in the control of diverse signaling and regulatory pathways in Mtb.
Fig. 5.
The complete interaction network of identified succinylated proteins and their interacting proteins. The succinylated proteins were grouped using functional annotation and interaction network was visualized with Cytoscape. The red node border represents the identified succinylated proteins.
Succinylation of Enzymes Involved in Central Metabolism
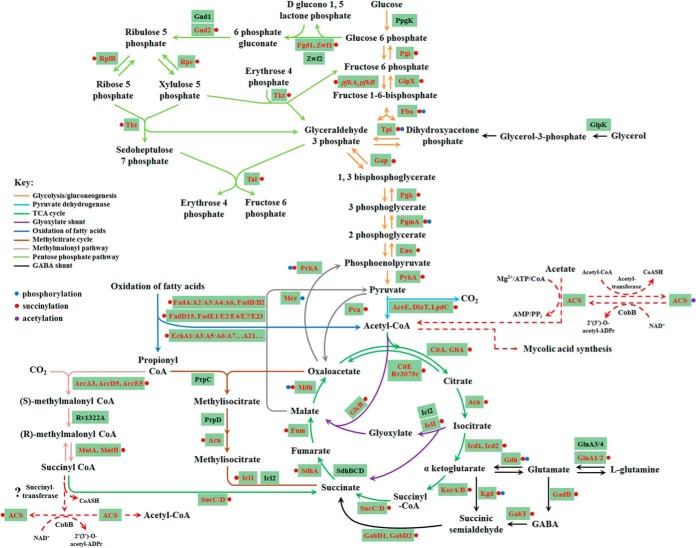
Increasing evidences show that lysine succinylation may play a regulatory role in metabolic pathways, similar with lysine acetylation (4, 67). We investigated the succinylation of metabolic enzymes in Mtb by mapping succinylated proteins to KEGG metabolic pathways (supplemental Table S8). As shown in Fig. 6, a large proportion of metabolic enzymes were succinylated. The fact that nearly all enzymes in major metabolic pathways are subjected to succinylation implies that this PTM may regulate cellular metabolic process at multiple levels in Mtb. For example, a large proportion of metabolic enzymes were mapped to two essential pathways for the growth and survival of Mtb, i.e. mycolic acid biosynthesis (supplemental Fig. S9A) and purine nucleotides de novo biosynthesis pathways (supplemental Fig. S9B), were identified to be succinylated.
Fig. 6.
Central carbon metabolism network in Mtb H37Rv. The identified succinylated proteins were highlighted in red.
Confirmation of Succinylated Proteins by Immunoprecipitation and Western blotting
Based on how well their biological functions and importance are understood, ten newly identified succinylated proteins, including AtpA, AtpF, KasA, DesA1, FadD31, Fgd1, GlcB, HspX, MtrA, and Pks2, were selected for verification. These proteins were subjected to IP and Western analysis (supplemental Fig. S10). The immunoprecipitated proteins were first confirmed by their specific antibodies. Anti-succinyl lysine antibody detected the respective proteins, confirming that they contain succinylated lysine residues. In comparison, the unmodified BSA (negative control protein) was not detected by anti-succinyl lysine antibodies, whereas the immuno-reactive signal was strongly observed when the succinylated BSA was used as positive control (supplemental Fig. S10). These data provided additional biochemical evidence for these identified succinylated proteins.
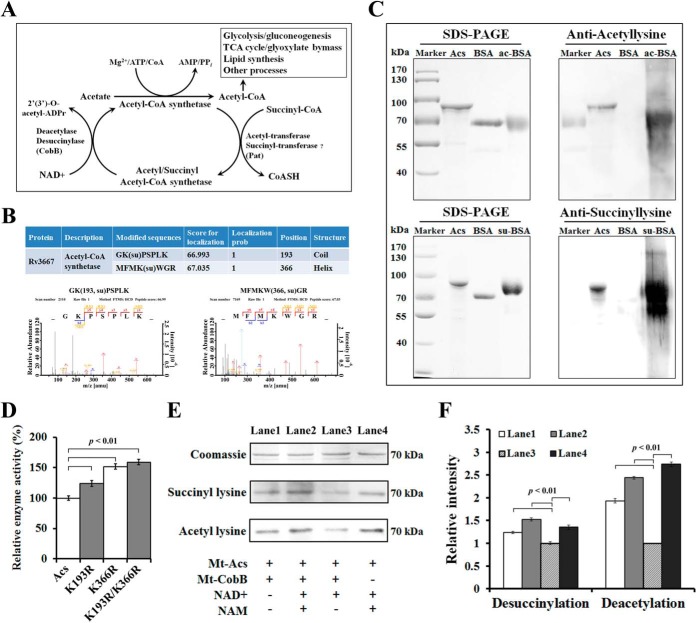
Effect of Succinylation on Acs Activity
Acs plays a crucial role in central metabolism and is important for maintaining adequate levels of Ac-CoA, a metabolite of many anabolic and catabolic processes. It is reported that acetylation of Acs completely abolishes the conversion of acetate to Ac-CoA (57, 68, 69) (Fig. 7A). In this study, we identified two reliable succinylation sites on Acs, Lys193 and Lys366 (Fig. 7B). We speculate that the Acs activity may be regulated by reversible succinylation. To test our hypothesis, the acs gene of Mtb was cloned and was overexpressed as a His-tagged fusion protein, which was purified by Ni2+ chromatography. As shown in Fig. 7C, the recombinant Acs were both acetylated and succinylated. Based on the conservation analysis, lysine at 193 (in coil) and 366 (in α helix) positions of Acs were conserved in the Mtb orthologs, suggesting that these residues could be important for an evolutionarily conserved function (supplemental Fig. S11A). To determine the functional consequences of succinylation on Acs acitivity, we converted the modified residues at the two positions (Lys193 and Lys366) to arginine (R), which essentially locks Acs into a constitutively desuccinylated state. All mutations were verified by both DNA sequencing (supplemental Fig. S11B) and MS analysis (supplemental Fig. S11C–S11F). Then, we measured the enzymatic activity of Acs and its mutants. The results showed that mutation of Lys193 and Lys366 to R (K193R, K366R, K193R/K366R), which conserved the positive charge and structure with unmodified lysine (15), led to a significant increase of the enzymatic activity (p < 0.01) (Fig. 7D). These data suggest that succinylation on Lys193 and Lys366 was a negative regulatory modification on Acs activity.
Fig. 7.
Succinylation profiling of Acs of Mtb. A, Schematic illustration of regulation of Acs activity by reversible acetylation and succinylation. B, Identification of the modification sites of Acs. The representation of the corresponding MS/MS spectra of succinylated peptides from Acs annotated with a comprehensive series of b and y fragment ions. Details of succinylation sites were listed in the supplemental Table S1. C, The acetylation/succinylation levels of purified recombinant Acs were determined by immunoblotting using specific anti-acetyl/succinyl lysine antibodies. D, Succinylation of Lys193 and Lys366 affect the enzyme activity of Acs. The Acs and its mutants (Acs-K193R, Acs-K366R, and Acs-K193R/K366R) were expressed in E. coli BL21 (DE3) and the specific activity of Acs was determined. Data are means ± S.D. from three independent assays. E, NAD+-dependent deacetylation/desuccinylation of Acs by CobB protein (Rv1151c). Acs was incubated with CobB in the presence of NAD+ (0.5 mm) for 10h at 25 °C. Nicotinamide (NAM, 2 mm) was added to the reaction as CobB inhibitor. F, Quantification of acetylated/succinylated Acs in the reactions from E. Each sample was standardized by comparing to the Coomassie blue stained gel. Error bars indicate S.D. of three measurements.
NAD+ Dependent Desuccinylase Activity of Mtb
The sirtuin proteins, which are one conserved family of deacetylases, have a unique ability to remove acetyl and other acyl groups from protein lysine side chains in a nicotinamide adenine dinucleotide (NAD+)-dependent manner (70). Recently, the human sirtuin protein (Sirt5) was showed to be an efficient NAD+-dependent protein lysine desuccinylase and demalonylase (8, 71). In E. coli, a sirtuin-like protein CobB has deacetylase/desuccinylase activity (14). In Mtb H37Rv, the sirtuin-like protein, CobB (Rv1151c) is able to deacetylate Acs (57, 69). Thus, CobB may have desuccinylase activity in Mtb H37Rv. To test this possibility, we expressed and purified CobB to measure the protein desuccinylase activity in the presence of NAD+. As shown in Fig. 7E and 7F, CobB can desuccinylate Acs, while no reduction of succinylation was observed when CobB or NAD+ was withdrawn from the reaction. Consistent with previous reports, CobB can also deacetylate Acs with similar efficiency (69, 72). Therefore, our data suggest that CobB is a bifunctional enzyme with both lysine desuccinylation and deacetylation activities in Mtb.
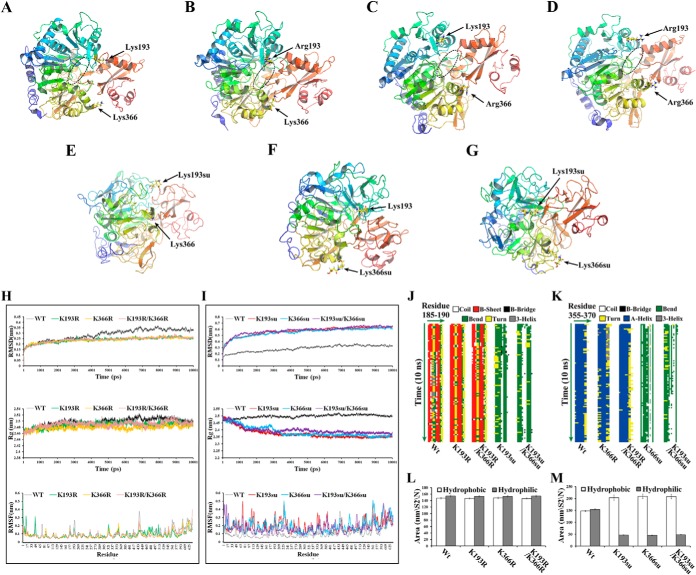
Molecular Dynamics Simulations of Acs
To gain insight into potential mechanisms by which succinylation might affect Acs enzymatic activity, we used molecular dynamics (MD) simulations to determine how succinylation of K193 and K366 influences the structure of Acs. It is reported that Acs uses the CoA as substrate to form the Ac-CoA complex and the residues in Acs that interact with the CoA may affect the binding of Acs with CoA by changing the conformation (73). In this study, we found the CoA-binding region marked with circular section in the unmodified Acs of Mtb, suggesting that the Acs could interact with the substrate CoA (Fig. 8A and supplemental Movie S1). When the lysine residues at 193 and 366 positions were mutated to R, we found that the CoA binding region in the mutants and the protein structures were almost accordance with the unmodified Acs structure (Fig. 8B–8D and supplemental Movies S2–S4). Therefore, the mutation of lysine residue to R can lock Acs into a constitutively desuccinylated state and maintain the protein structures to bind with CoA. The MD simulations of the succinylated Acs (K193su, K366su, K193su/K366su) showed that succinylation of K193 and K366 resulted in the change of charge at each modified position and led to the conformation change at this region, which may prevent the binding of Acs with CoA (Fig. 8E–8G and supplemental Movies S5–S7). Therefore, succinylation at K193 and K366 could change the residue charge status from +1 to −1 and consequently disrupt the favorable interaction with the near helix region, leading to partially or completely perturbation of the neighboring regions and destabilization of the protein conformation. As shown in Fig. 8H and 8I, the structures of succinylated (K193su, K366su, and K193su/K366su) and unmodified Acs were significantly different, whereas the R mutants (K193R, K366R, and K193R/K366R) showed nearly identical structures with the unmodified Acs during the entire course of simulations. Furthermore, K193 and K366, which were positively charged before succinylation, were located in coil and α helix region, respectively. The simulations showed that the mutants and unmodified Acs shared the similar secondary structures and underwent little change from the starting to their final states. However, the secondary structures of succinylated Acs were significantly different from that of unmodified Acs (Fig. 8J and 8K). Owing to the perturbation of the secondary structures and the conformational fluctuations, the structures of succinylated Acs have a strong tendency to form the hydrophobic solvent-accessible surface area (Fig. 8L and 8M). Together, these observations suggest that the R mutants mimic the structure of unmodified Acs at the molecular level and the succinylation could influence the conformation of Acs.
Fig. 8.
Structural modeling of Acs. Final structures of the simulations of Acs were shown. The circular section marks the CoA-binding region A–D. A, Nonsuccinylated (Wt) Acs. Site-directed mutants of B, Acs-K193R; C, Acs-K366R; and D, Acs-K193R/K366R. Acs succinylated at E, K193; F, K366; and G, K193 and K366. H–I, Analysis of the root mean square deviations (RMSDs), root mean square fluctuations (RMSFs) and radius of gyrations (Rgs) for the different Acs systems. (Top) Trajectories of the overall RMSDs of the different Acs systems. (Middle) Radius of gyrations profiles of the different Acs systems. (Bottom) Residue-specific RMSF profiles of the different Acs systems. J–K, Secondary structure evolution of Acs and its mutants. The evolution in secondary structure at each frame was monitored using the dictionary of protein secondary structure (DSSP) algorithm. In the stripes each pixel represents the secondary structure (color-coded) of a residue (185–190 or 355–370, x-dimension) at a given time in simulation (y-dimension). L–M, Time evolution of the solvent accessible surface area (SAS) calculated for the different systems that were simulated.
DISCUSSION
In this study, we report the first extensive data on lysine succinylome in the virulent Mtb strain H37Rv. A total of 1561 unique modified peptides encompassing 1545 succinylation sites (class I) were identified from 626 H37Rv proteins. Because lysine succinylation has emerged as an important regulatory PTM and may impact diverse metabolic pathways, our data set provides a rich resource of putative regulatory modification sites in this life-threatening pathogen. We show that proteins in major metabolic pathways are targeted by succinylation. These pathways include glycolysis, fatty acid oxidation, TCA cycle, and mycolic acid biosynthesis, which are crucial components of Mtb's energy metabolic networks.
Among all metabolic pathways, mycolic acid biosynthesis pathway is unique to mycobacteria and is crucial for their success as pathogens (74). Our data showed that a large proportion of enzymes in mycolic acids biosynthesis pathway were succinylated (supplemental Fig. S9A). For example, the single fatty acid synthase (FAS) I type protein FAS, which generates CoA esters from Ac-CoA primers, creating precursors for elongation by all other fatty acid and polyketide systems, was found to be succinylated. In FAS-II system of mycolic acid biosynthesis, sets of enzymes, which can be inhibited by many drugs used in treatment of TB (75), were also succinylated. Therefore, the reversible succinylation, although the mechanism remains unknown, could be an important mechanism for regulating the interaction of these drugs with FAS-II-related enzymes, and could also be targets for novel drug therapies. The recent observation showed that Mtb is metabolically flexible and can even alter the host cellular metabolism, such as the pathogen-induced dysregulation of host lipid synthesis, to create a niche that is ideally suited to its persistent lifestyle (76). Mtb can utilize the lipid bodies accumulated in the infected host as a nutrient source via β-oxidation pathway and this seems to sustain intracellular Mtb in a physiological state. Thus, aside from fatty acid biosynthesis pathway, the β-oxidation of fatty acids, which serves as carbon and energy source, is also required for survival of Mtb in the infected host cell (77). Several enzymes involved in this pathway were found to be succinylated in this study (Fig. 6), indicating that lysine succinylation may play a crucial role in regulating the fatty acid metabolism of Mtb during infection.
It is now widely accepted that metabolic adaptation to the host environment is a defining feature of the pathogenicity of Mtb (38, 78). However, we lack biochemical knowledge of its metabolic networks and how they are regulated. The succinyl-CoA is a succinyl donor to lysine and the concentration of succinyl-CoA may regulate succinylation levels globally. Our understanding of the role of succinyl-CoA and lysine succinylation on the control of Mtb metabolism remains rudimentary. However, their importance in the control of metabolism in bacteria, yeast and human, and the conservation of the enzymes involved in these pathways in Mtb, suggest that lysine succinylation may work together with many other PTMs to regulate the metabolic pathways of Mtb. Mtb may have unique mechanisms involved in controlling metabolic flux via succinyl-CoA and protein succinylation, because it resides in humans as its only known natural host and a large part of the energy status in Mtb occurs through the interaction with the human cells (35). Reversible succinylation is a strong candidate for dynamically controlling metabolic pathways and enabling Mtb to adapt to the changing host environment. To better understand the role of succinyl-CoA and protein succinylation in Mtb metabolism, it is essential to acquire an in-depth description of the succinylome in Mtb. Furthermore, it is important to understand how the succinylation of metabolic enzymes regulates metabolic pathways in Mtb. More studies need to be done to accomplish this. Firstly, examine how environmental conditions as well as the carbon and energy balance of Mtb cells can influence the level of lysine succinylation. Secondly, determine the effects of succinylation on enzymatic activity. Thirdly, identify lysine succinyltransferase and desuccinylase involved in controlling the succinylation status of proteins in Mtb. In mammalian cell, the histone H3 propionylation might be generated by the histone acetyltransferase p300 and that selection of donor molecules (propionyl-CoA versus acetyl-CoA) may determine the difference of modifications (79). In this study, we identified the first desuccinylase, CobB, in Mtb, however, it remains unknown whether succinyltransferase and more desuccinylases exist in Mtb. Acetylation of Acs by the acetyltransferase (Pat) inactivated the enzyme in S. enterica, whereas deacetylation by the CobB reactivated it (80). We found the Pat homolog in Mtb (Rv0998) and further studies are thus necessary to identify the succinyltransferase in Mtb.
Being only one of many PTMs, lysine succinylation competes with other modifications, such as acetylation, for the same lysine residues. Accumulating evidence indicated that lysine acetylation is a conserved PTM that links Ac-CoA metabolism and cellular signaling (81, 82). Acs is the enzyme catalyzes the conversion of acetate into Ac-CoA and the level of Ac-CoA can fluctuate according to the changing metabolic conditions (83). In this study, we demonstrated that the enzymatic activity of Acs is affected by the lysine succinylation. It is thus expected that crosstalk exists between protein succinylation and acetylation may modulate protein activity and metabolic pathways in both Mtb and human cells. Jungblut and Schlüter proposed a systematic nomenclature for the comprehensive description of protein species and suggested that protein species sequence including PTMs should replace the basic unit protein sequence (84, 85). Therefore, our succinylome data may help to provide the accurate and detailed description of protein species in Mtb and to elucidate potential biological roles of each protein species.
In summary, we report the first large-scale, high resolution MS-based survey of lysine succinylation in Mtb. The provided dataset may serve as important starting point for assessing the functional relevance of succinylation sites in this life-threatening pathogen. Given that the high abundance of lysine succinylation and its induced conformation fluctuation, it is likely that reversible succinylation may be an important regulatory mechanism in metabolism. It would be interesting and important to investigate the biological functions of reversible succinylation in metabolic regulation in Mtb.
Supplementary Material
Footnotes
Author contributions: F.G. designed research; M.Y., Y.W., Y.C., C.C., and R.M. performed research; J.G., J.D., and X.W. contributed new reagents or analytic tools; M.Y., Y.W., Y.C., Z.C., L.B., and F.G. analyzed data; M.Y., Z.C., and F.G. wrote the paper.
* This work was supported by the National Basic Research Program of China (973 Program, 2012CB518700), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB14030202), the Key Program of the Chinese Academy of Sciences (KJZD-EW-L02), and National Natural Science Foundation of China to C.Z. (31271362).
 This article contains supplemental Figs. S1 to S11, Tables S1 to S7, and Movies S1 to S8.
This article contains supplemental Figs. S1 to S11, Tables S1 to S7, and Movies S1 to S8.
1 The abbreviations used are:
- TB
- Tuberculosis
- Mtb
- Mycobacterium tuberculosis
- PTM
- post-translational modification
- Acs
- acetyl-CoA synthetase
- FDR
- false discovery rate
- GO
- Gene Ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- SPC
- simple point charge
- LINCS
- linear constraint solver
- PME
- particle mesh Ewald
- MD
- molecular dynamics
- Ac-CoA
- acetyl-CoA
- CoA
- Coenzyme A
- NAD+
- nicotinamide adenine dinucleotide
- NAM
- Nicotinamide
- Sirt5
- sirtuin
- RMSD
- root mean square deviation
- Rgs
- gyrations
- RMSF
- residue-specific root mean square fluctuation.
REFERENCES
- 1. Olsen J. V., Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao Y., Jensen O. N. (2009) Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miao J., Lawrence M., Jeffers V., Zhao F., Parker D., Ge Y., Sullivan W. J., Jr., Cui L. (2013) Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol. Microbiol. 89, 660–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y., Zhao W. H., Yang J. S., Cheng Z. Y., Luo H., Lu Z. K., Tan M. J., Gu W., Zhao Y. M. (2012) Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol. Cell. Proteomics 11, 1048–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J. D., Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B. M., Tishkoff D., Ho L., Lombard D., He T. C., Dai J., Verdin E., Ye Y., Zhao Y. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111 012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Montellier E., Rousseaux S., Zhao Y., Khochbin S. (2012) Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays 34, 187–193 [DOI] [PubMed] [Google Scholar]
- 10. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang K., Chen Y., Zhang Z., Zhao Y. (2009) Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J. Proteome Res. 8, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S. S., Gu W., Zhao Y. (2009) Molecular characterization of propionyllysines in nonhistone proteins. Mol. Cell. Proteomics 8, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colak G., Xie Z., Zhu A. Y., Dai L., Lu Z., Zhang Y., Wan X., Chen Y., Cha Y. H., Lin H., Zhao Y., Tan M. (2013) Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics 12, 3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinert B. T., Schölz C., Wagner S. A., Iesmantavicius V., Su D., Daniel J. A., Choudhary C. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 [DOI] [PubMed] [Google Scholar]
- 17. Lin H., Su X., He B. (2012) Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olsen C. A. (2012) Expansion of the lysine acylation landscape. Angew. Chem. Int. Ed. Engl. 51, 3755–3756 [DOI] [PubMed] [Google Scholar]
- 19. Park J., Chen Y., Tishkoff D. X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B. M., Skinner M. E., Lombard D. B., Zhao Y. (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rardin M. J., He W., Nishida Y., Newman J. C., Carrico C., Danielson S. R., Guo A., Gut P., Sahu A. K., Li B., Uppala R., Fitch M., Riiff T., Zhu L., Zhou J., Mulhern D., Stevens R. D., Ilkayeva O. R., Newgard C. B., Jacobson M. P., Hellerstein M., Goetzman E. S., Gibson B. W., Verdin E. (2013) SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newman J. C., He W., Verdin E. (2012) Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J. Biol. Chem. 287, 42436–42443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He W., Newman J. C., Wang M. Z., Ho L., Verdin E. (2012) Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol. Metab. 23, 467–476 [DOI] [PubMed] [Google Scholar]
- 23. Ehrt S., Rhee K. (2013) Mycobacterium tuberculosis metabolism and host interaction: mysteries and paradoxes. Curr. Top. Microbiol. Immunol. 374, 163–188 [DOI] [PubMed] [Google Scholar]
- 24. Team E. E. (2013) WHO publishes Global tuberculosis report 2013. Euro. Surveill. 18, 20615. [PubMed] [Google Scholar]
- 25. Corbett E. L., Watt C. J., Walker N., Maher D., Williams B. G., Raviglione M. C., Dye C. (2003) The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163, 1009–1021 [DOI] [PubMed] [Google Scholar]
- 26. Sun G., Luo T., Yang C., Dong X., Li J., Zhu Y., Zheng H., Tian W., Wang S., Barry C. E., 3rd, Mei J., Gao Q. (2012) Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 206, 1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Li D., Zhao L., Fleming J., Lin N., Wang T., Liu Z., Li C., Galwey N., Deng J., Zhou Y., Zhu Y., Gao Y., Wang T., Wang S., Huang Y., Wang M., Zhong Q., Zhou L., Chen T., Zhou J., Yang R., Zhu G., Hang H., Zhang J., Li F., Wan K., Wang J., Zhang X. E., Bi L. (2013) Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45, 1255–1260 [DOI] [PubMed] [Google Scholar]
- 28. Srinivasan L., Ahlbrand S., Briken V. (2014) Interaction of Mycobacterium tuberculosis with host cell death pathways. Cold Spring Harb. Perspect. Med. 4, a022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gengenbacher M., Kaufmann S. H. (2012) Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boon C., Dick T. (2012) How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol. 7, 513–518 [DOI] [PubMed] [Google Scholar]
- 31. Wirth T., Hildebrand F., Allix-Beguec C., Wolbeling F., Kubica T., Kremer K., van Soolingen D., Rusch-Gerdes S., Locht C., Brisse S., Meyer A., Supply P., Niemann S. (2008) Origin, spread, and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4, e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Carvalho L. P., Fischer S. M., Marrero J., Nathan C., Ehrt S., Rhee K. Y. (2010) Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131 [DOI] [PubMed] [Google Scholar]
- 33. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010) Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi S., Ehrt S. (2006) Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect. Immun. 74, 56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boshoff H. I., Barry C. E., 3rd (2005) Tuberculosis – metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3, 70–80 [DOI] [PubMed] [Google Scholar]
- 36. Mehrotra P., Jamwal S. V., Saquib N., Sinha N., Siddiqui Z., Manivel V., Chatterjee S., Rao K. V. (2014) Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PLoS Pathog. 10, e1004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chopra T., Hamelin R., Armand F., Chiappe D., Moniatte M., McKinney J. D. (2014) Quantitative Mass Spectrometry reveals plasticity of metabolic networks in Mycobacterium smegmatis. Mol. Cell. Proteomics 13, 3014–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010) Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412 [DOI] [PubMed] [Google Scholar]
- 39. Arora N., Banerjee A. K. (2012) Targeting tuberculosis: a glimpse of promising drug targets. Mini Rev. Med. Chem. 12, 187–201 [DOI] [PubMed] [Google Scholar]
- 40. Camus J. C., Pryor M. J., Medigue C., Cole S. T. (2002) Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148, 2967–2973 [DOI] [PubMed] [Google Scholar]
- 41. Meena L. S., Rajni (2010) Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. FEBS J. 277, 2416–2427 [DOI] [PubMed] [Google Scholar]
- 42. Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., Ge F., Zhao J. D. (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. Strain PCC 7002. J. Proteome Res. 12, 1909–1923 [DOI] [PubMed] [Google Scholar]
- 43. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 44. Chen Y., Kwon S. W., Kim S. C., Zhao Y. M. (2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 45. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 46. Conesa A., Gotz S. (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 50. Hou J., Cui Z., Xie Z., Xue P., Wu P., Chen X., Li J., Cai T., Yang F. (2010) Phosphoproteome analysis of rat L6 myotubes using reversed-phase C18 prefractionation and titanium dioxide enrichment. J. Proteome Res. 9, 777–788 [DOI] [PubMed] [Google Scholar]
- 51. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 52. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y., Cui T., Zhang C., Yang M., Huang Y. X., Li W. H., Zhang L., Gao C. H., He Y., Li Y. Q., Huang F., Zeng J. M., Huang C., Yang Q. O., Tian Y. X., Zhao C. C., Chen H. C., Zhang H., He Z. G. (2010) Global protein–protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome Res. 9, 6665–6677 [DOI] [PubMed] [Google Scholar]
- 54. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 55. Petersen B., Petersen T. N., Andersen P., Nielsen M., Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rose I. A., Grunberg-Manago M., Korey S. R., Ochoa S. (1954) Enzymatic phosphorylation of acetate. J. Biol. Chem. 211, 737–756 [PubMed] [Google Scholar]
- 57. Li R., Gu J., Chen P., Zhang Z. P., Deng J. Y., Zhang X. E. (2011) Purification and characterization of the acetyl-CoA synthetase from Mycobacterium tuberculosis. Acta Biochim. Biophys. Sin. 43, 891–899 [DOI] [PubMed] [Google Scholar]
- 58. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 59. Kaminski G. A., Friesner R. A., Tirado-Rives J., Jorgensen W. L. (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 105, 6474–6487 [Google Scholar]
- 60. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Hermans J. (1981) Interaction models for water in relation to protein hydration. Intermolecular Forces, pp. 331–342, Springer; Netherlands [Google Scholar]
- 61. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126:014101 [DOI] [PubMed] [Google Scholar]
- 62. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M. (1997) LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 63. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 64. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 65. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 66. Weinert B. T., Iesmantavicius V., Wagner S. A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272 [DOI] [PubMed] [Google Scholar]
- 67. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Starai V. J., Escalante-Semerena J. C. (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 69. Xu H., Hegde S. S., Blanchard J. S. (2011) Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50, 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 71. Du J. T., Zhou Y. Y., Su X. Y., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., Lin H. N. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gu J., Deng J. Y., Li R., Wei H., Zhang Z., Zhou Y., Zhang Y., Zhang X. E. (2009) Cloning and characterization of NAD-dependent protein deacetylase (Rv1151c) from Mycobacterium tuberculosis. Biochemistry 74, 743–748 [DOI] [PubMed] [Google Scholar]
- 73. Reger A. S., Carney J. M., Gulick A. M. (2007) Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry 46, 6536–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffe M. (2008) Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Grzegorzewicz A. E., Kordulakova J., Jones V., Born S. E., Belardinelli J. M., Vaquie A., Gundi V. A., Madacki J., Slama N., Laval F., Vaubourgeix J., Crew R. M., Gicquel B., Daffe M., Morbidoni H. R., Brennan P. J., Quemard A., McNeil M. R., Jackson M. (2012) A common mechanism of inhibition of the Mycobacterium tuberculosis mycolic acid biosynthetic pathway by isoxyl and thiacetazone. J. Biol. Chem. 287, 38434–38441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Russell D. G., Cardona P. J., Kim M. J., Allain S., Altare F. (2009) Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shi L., Sohaskey C. D., Pfeiffer C., Datta P., Parks M., McFadden J., North R. J., Gennaro M. L. (2010) Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol. Microbiol. 78, 1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Munoz-Elias E. J., McKinney J. D. (2006) Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22 [DOI] [PubMed] [Google Scholar]
- 79. Liu B., Lin Y. H., Darwanto A., Song X. H., Xu G. L., Zhang K. L. (2009) Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 284, 32288–32295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 81. Crosby H. A., Heiniger E. K., Harwood C. S., Escalante-Semerena J. C. (2010) Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol. 76, 874–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wellen K. E., Thompson C. B. (2012) A two-way street: reciprocal regulation of metabolism and signaling. Nat. Rev. Mol. Cell Bio. 13, 270–276 [DOI] [PubMed] [Google Scholar]
- 84. Schluter H., Apweiler R., Holzhutter H. G., Jungblut P. R. (2009) Finding one's way in proteomics: a protein species nomenclature. Chem. Cent. J. 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jungblut P. R., Holzhutter H. G., Apweiler R., Schluter H. (2008) The speciation of the proteome. Chem. Cent. J. 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.