Abstract
Extracellular non-flagellar appendages, called pili or fimbriae, are widespread in gram-negative bacteria. They are involved in many different functions, including motility, adhesion, biofilm formation, and uptake of DNA. Sequencing data for a large number of cyanobacterial genomes revealed that most of them contain genes for pili synthesis. However, only for a very few cyanobacteria structure and function of these appendages have been analyzed. Here, we review the structure and function of type IV pili in Synechocystis sp. PCC 6803 and analyze the distribution of type IV pili associated genes in other cyanobacteria. Further, we discuss the role of the RNA-chaperone Hfq in pilus function and the presence of genes for the chaperone-usher pathway of pilus assembly in cyanobacteria.
Keywords: type IV pili, motility, surface structure
1. Introduction
Surface appendages are widespread among gram-negative bacteria, but have been found also on the surface of gram-positive bacteria. These structures can be observed by transmission electron microscopy coupled with negative staining and also by scanning electron microscopy. From the early days of bacterial genetics conjugative pili have attracted a lot of attention. Though, the most prominent extension beyond the surface of a bacterial cell is most probably the flagellum. Many bacteria use flagella for movement, but they are also important factors for virulence and adhesion [1,2]. On the other hand several bacteria are able to move across surfaces using non-flagellar motility machineries, such as type IV pili. In addition, most gram-negative bacteria have short, thin appendages, which are not involved in motility, but rather facilitate adherence to surfaces or other cells. These structures are not mutually exclusive and some bacteria synthesize a range of different surface appendages [3].
The gram-negative cyanobacteria do not contain genes encoding flagella components. Examination of cyanobacteria by electron microscopy revealed the existence of surface appendages in several unicellular cyanobacteria [4]. The most advanced studies on the function of these structures have been published for the model cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803). Molecular approaches support the role of type IV pili in motility and natural competence [5,6,7]. As most cyanobacteria harbor genes encoding proteins homologous to those of the type IV pilus apparatus, we are convinced that type IV pili or alternative structures encoded by these genes have important functions in many other cyanobacteria. Based on a brief overview of the structure, function and regulation of type IV pili biogenesis in Synechocystis 6803, we discuss the potential presence of similar structures in other cyanobacteria and hint at specific differences in comparison to other gram-negative bacteria.
2. Appendages of Bacteria: Classification and Nomenclature
According to Fronzes et al. [8], non-flagellar appendages of gram-negative bacteria are classified into five major groups: Conjugative pili (homologous to the type IV secretion system), type IV pili, type III secretion needle/pili, curli and chaperone-usher (CU) pili (also known as type I pili). Type IV secretion pili are well known from bacteria that are able to transfer genetic material by conjugation. They establish a close contact from the host bacterium to other cells, even to eukaryotic cells. Some pathogenic bacteria use these appendages to transfer virulence factors, including DNA (Agrobacterium tumefaciens) or proteins (Helicobacter pylori). The size of type IV secretion pili varies from <1–20 μm depending on the specific system. Electron cryo-microscopy studies showed that the F-pilus has a central lumen of 3 nm in diameter that is theoretically large enough to allow for the passage of single-stranded DNA [9]. Transfer of the genetic material through the pilus lumen has been proposed, but never demonstrated [10,11].
Type IV pili are 6–8 nm in diameter and several μm in length and can form bundles. They are essential for many different processes like biofilm formation, aggregation, twitching motility and virulence [12,13]. Genes that encode core components of the type IV pilus show high sequence similarity, even if they originate from evolutionary divergent bacterial groups [14]. Type IV pili are assembled by a complex machinery at the inner membrane [15]. The main structural component of the pilus (PilA) is produced as a precursor. PilD removes the N-terminal leader sequence and methylates the protein. The assembly of the pilus requires a large ATP driven complex. This machinery is also essential for the reversibility of assembly and retraction of the pilus, a major feature of type IV pili. The PilB ATPase is required for pilus assembly, whereas PilT energizes depolymerisation of the pilus. Both ATPases are located at the base of the pilus and most probably interact with PilC, which is embedded in the inner membrane. PilQ forms the pore for the pilus to cross the outer membrane (Figure 1a). According to differences in their subunits, assembly system, and gene organization, two type IV pili subfamilies can be distinguished. Type IVa pili are widely distributed and highly conserved among many bacterial phyla and typically associated with twitching motility. The more heterogeneous type IVb pili are only found in a small subset of genera and often lack an ATPase for pilus retraction [14].
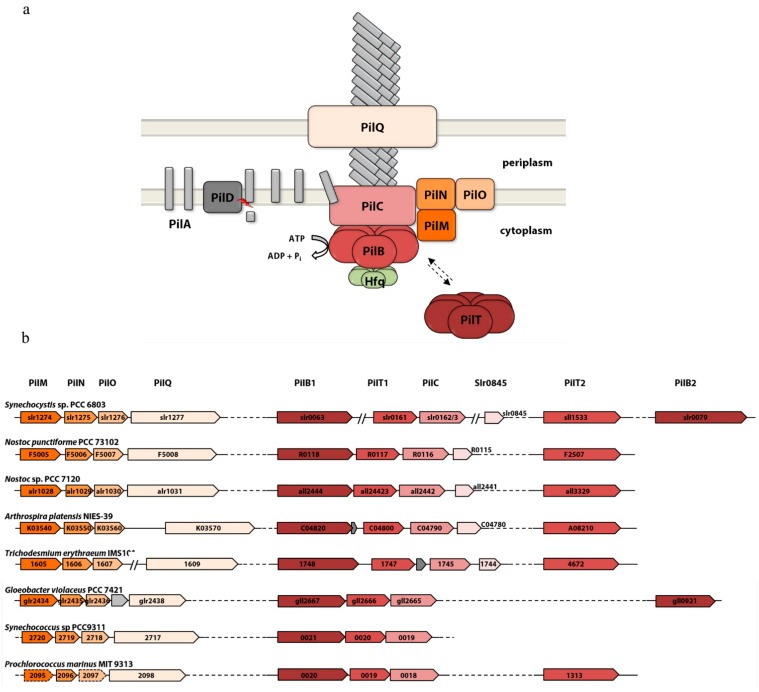
Figure 1.
(a) Model of type IV pili and their assembly machineries in cyanobacteria. Most cyanobacteria harbor more than one PilA homolog. Their specific functions are unknown so far. (b) Synteny of Synechocystis 6803 gene clusters encoding type IV pili proteins in comparison with similar genes clusters of diverse cyanobacteria. Color-coding of open reading frames refers to the schematic view in (a).
Type III secretion needles are mostly known from pathogenic bacteria like Salmonella typhimurium. They use this, so called injectisome, to secrete effector proteins into the eukaryotic cell thereby assisting colonization of the host cell. This short, rigid structure is approximately 60 nm long and has an inner diameter of 2–3 nm. Assembly of the type III secretion system shares similarities to flagella assembly [16].
Curli have been described as abundant amyloid extracellular fibers of 6–10 nm produced by many Enterobacteria (reviewed in [17]). These protein structures are the major component of E. coli biofilms and contribute to the adhesion of pathogenic bacteria to cells.
CU pili are found in many bacteria and it seems that they form the most abundant group of bacterial surface appendages [8]. The surface structures assembled by the chaperon-usher pathway can differ in their morphology and function and may even form non-fimbrial structures. Based on operon structure, phylogeny and morphology, CU pili are classified into α-, β-, γ-, κ-, π- and σ-fimbriae [18]. CU operons contain at least one major structural subunit, a periplasmic chaperon and a membrane protein for translocation of the subunits (the so called usher protein) [18]. The Sec translocase is involved in transport of the subunits across the inner membrane, which are then folded and stabilized with the help of the chaperone in the periplasm. After binding to the usher protein, which forms a pore in the outer membrane, the chaperone is displaced and the pilin is integrated into the filament on the surface. Further details are discussed in Fronzes et al. [8].
Appendages of Cyanobacteria
Vaara [4] demonstrated the presence of pili in 11 out of 22 analyzed unicellular cyanobacterial strains. The author showed that these surface structures are not specific for cyanobacteria and resemble those of other bacteria. However, in contrast to well-characterized appendages of model Proteobacteria, these structures are largely genetically and biochemically uncharacterized. Moreover, appendages are referred to in different studies as pilus-like structures [19], fibers [20] or spinae [21] resulting in terminological confusion. Without detailed analysis of these components, it is not possible to evaluate whether we deal with different structures described in these studies.
In Synechocystis 6803, two different types of pili have been defined using negative staining [6,7]. One morphotype is defined by an average diameter of 3–4 nm and a length of approximately 1 μm. These thin pili cover the whole surface of the cell. The second morphotype is specified as thick and long pili with a diameter of 6–8 nm and more than 2 μm in length. The thick pili often appear as tuffs and have been identified as type IVa pili by genetic methods [5,6,7].
There is also evidence that hormogonia developed by the symbiotic cyanobacterium Nostoc punctiforme possess pilus-like structures all over the surface, whereas the vegetative cells of the parent filaments lack pili [22,23]. However, by using antibodies against the putative major pilin PilA Risser et al. [24] demonstrated that this subunit accumulated exclusively in rings at the junction between cells. The authors suggest that this putative pilin is part of the previously described junctional pore complex involved in motility [25].
Transmission electron microscope studies showed pilus-like structures on the surface of Microcystis aeruginosa PCC 7806 cells [26]. Appendages from cells grown in liquid culture were comparable to the thick pili shown by Bhaya et al. [6]. In contrast, when cells from agar plates were analyzed, they exhibited thicker pili with diameters of 20 to 35 nm, which may consist of bundles of thinner filaments [26]. Thus, growth conditions may influence composition and morphology of appendages. Homologs to type IV pili genes have been identified in this organism.
3. Type IV Pili of Cyanobacteria
The type IV pili apparatus is a complex, multi-protein machinery conserved in a wide variety of bacteria. Phylogenetic analyses suggest that several of its evolutionary ancient protein constituents are homologous to those of the type II secretion system and the archaellum [27,28]. The function of most cyanobacterial type IV pili proteins was not investigated on a biochemical and structural level in detail. Nevertheless, their role in pilus assembly can be inferred from mutant phenotypes observed in Synechocystis 6803 and Nostoc punctiforme and their similarity to the widely studied homologs from gram-negative model organisms like Neisseria, Pseudomonas or Myxococcus xanthus (reviewed in [15,29]). The type IV pili apparatus is composed of four distinct subcomplexes that together span the inner and outer membranes: The pilus rod, the outer membrane complex, the cytoplasmic pilus platform and the secretion ATPases (see Figure 1a). The genes encoding components of the cyanobacterial type IV pili apparatus were originally identified in Synechocystis 6803. Figure 1b shows gene organization of operons and orphan genes in the genome of Synechocystis 6803 in comparison to other cyanobacteria.
3.1. The Membrane Complexes
Not much is known about the membrane components of the cyanobacterial type IV pili apparatus. PilQ is a secretin family protein that forms a homo-multimeric pore complex, which facilitates the transport of the pilus subunits across the outer membrane. PilMNO proteins are thought to be important for aligning the pore complex with the pilus platform [30,31,32]. Genes encoding these membrane proteins are conserved within a putative operon structure in most cyanobacteria (Figure 1b). In Synechocystis 6803 disruption of these genes leads to loss of type IV pili, motility and natural competence, while cells retain (bundles) of thin pili [7]. Remarkably, we were not able to identify PilP homologs in cyanobacteria, although this protein is encoded in the highly conserved pilMNOPQ gene cluster in essentially all other gram-negative bacteria harboring type IVa pili genes. The lipoprotein PilP is thought to connect the PilQ secretin pore with the inner membrane proteins PilN and PilO, thereby bridging the periplasm [33]. Despite their overall gram-negative structure, the peptidoglycan layer of cyanobacteria is substantially thicker than that of most gram-negative bacteria [34]. Hence, we speculate that the periplasmic structure of the alignment complex in cyanobacteria may differ considerably from that of other gram-negative bacteria, and PilP may have been replaced by other unidentified proteins. Probably for the same reason, pilotins like Pseudomonas PilF or Neisseria PilW and other accessory proteins, which are involved in the formation of the secretin pore in many gram-negative bacteria [35] have not been identified in cyanobacteria. Disruption of the pilus platform protein PilC abolished motility and natural competence [6]. Moreover both pili morphotypes (thin and thick pili) were absent from the cell surface of this mutant.
3.2. The Secretion ATPases
The two ATPases PilB and PilT are required for pilus growth and its depolymerization, respectively. They belong to the type II/IV secretion ATPases [36] which are involved in protein transport across the outer membrane. The ATPases consist of an N-terminal and a C-terminal domain and assemble into hexameric ring structures [37,38,39]. Their function as molecular motors requires binding and hydrolysis of ATP. In case of PilB ATP hydrolysis leads to a conformational change of the protein, which is suggested to trigger the integration of a pilin subunit from the inner membrane into the growing pilus rod [39].
All cyanobacteria encoding pil-like genes harbor a homolog of each of the secretion ATPases (PilB1/PilT1), which are typically encoded upstream of the pilC gene in a conserved locus (Figure 1b). Furthermore, most cyanobacteria encode at least one copy of a second PilT homolog (PilT2), which is characterized by a proline rich N-terminal extension. Albeit not as frequent and with a far greater sequence variability than PilT2, additional PilB homologs (designated as PilB2 in Synechocystis 6803) can also be found in a variety of different cyanobacteria. Both Synechocystis 6803 and Nostoc punctiforme encode two copies of pilT and pilB-like genes, respectively. A Synechocystis 6803 pilB1 mutant lost type IV pili, competency and motility, whereas inactivation of pilB2 did not affect pilus biogenesis or motility [7]. In Synechocystis 6803 and Nostoc punctiforme pilT1 mutants are non-motile but hyperpiliated. This phenotype is consistent with the supposed function of PilT in pilus retraction [6,23]. Moreover PilT1 from Synechocystis 6803 and Microcystis aeruginosa show ATPase activity in vitro and the Microcystis pilT1 complements the pilT mutant of Pseudomonas aeruginosa (but interestingly not that of Synechocystis 6803) [40,41]. This suggests that PilB1 and PilT1 constitute the essential motor proteins. PilT2 mutants show negative phototaxis in Synechocystis 6803 [6] and a slightly reduced number of pili and infection frequency in Nostoc punctiforme [23]. Therefore, PilT2 is thought to be involved in the regulation of pilus function. Whether it exerts its function as a homomer or a heteromultimer composed of PilT1 and PilT2 subunits is not known. The function of the additional PilB homologs in cyanobacteria has not been elucidated.
We hypothesize that additional proteins are associated with the secretion ATPases in many cyanobacteria and regulate their activity (Figure 1a). Cyanobacterial PilB1 homologs (with the exception of some Prochlorococcus species) are characterized by a highly conserved C-terminal domain with an invariant tetra cysteine motif resembling a zinc finger. This domain is lacking in PilB proteins from other bacteria and in cyanobacterial PilB2 homologs. In Synechocystis 6803, this specific C-terminal domain of PilB1 interacts with Hfq and is responsible for the correct localization and function of the RNA-chaperone which in turn is essential for biogenesis of both thick and thin pili [42,43]. Considering the co-occurrence of hfq and pilB1 genes in cyanobacterial genomes we speculate that this interaction may be conserved among different species. Another candidate is the Synechocystis 6803 slr0845 gene and its homologs, which are specific for clade B cyanobacteria [44]. They show a high degree of synteny and are predominantly located directly downstream of the pilB/T/C gene locus. In Synechocystis 6803 we could co-purify Slr0845 together with Hfq and PilB1 (unpublished) and a protein interaction study demonstrated an interaction between Slr0845 and PilL (TaxAY3) a histidine kinase involved in the regulation of motility [45,46].
3.3. The Pilus Rod
The pilus is composed predominantly of a major structural subunit, the so-called major pilin, and contains additional minor pilin subunits that may be required for assembly or specific pilus functions. Pilin proteins as well as the homologous pseudopilins of type II secretion systems are synthesized as prepilin precursors, which are characterized (and easily identified) by a distinct N-terminal signal peptide. During maturation the prepilin peptidase PilD removes the leader peptide and methylates the N-terminal amino acid of the mature protein [47]. Both pili morphotypes are absent on the cell surface of a Synechocystis 6803 pilD mutant which consequently is non-motile and lost transformation competency [6,7]. Interestingly, Linhartová et al. [47] showed that a pilD mutant in the non-motile Synechocystis 6803 wild-type background (the so called glucose-tolerant strain [48]) did not grow photoautotrophically because of a defect in the synthesis of photosystem II subunits. Most probably the unprocessed pilins interfere with translocation of photosystem II membrane subunits. Inactivation of pilD in Nostoc punctiforme led to non-motile mutant cells. However, pilD mutants were still able to assemble shortened pili. In addition, infection efficiency and symbiotic growth were severely attenuated in this mutant. [23]. From the gene products of eleven putative prepilin genes (pilA1-pilA11) identified in Synechocystis 6803, PilA1 constitutes the major pilin and is critical for type IV pili biogenesis, motility and transformation [5]. While the pilins PilA2–PilA8 seem to be dispensable for pilus biogenesis and motility [6,7], pilA9–pilA11 mutants show a non-motile phenotype but retain normal type IV pili on the cell surface [49,50]. The PilA3 protein can be disregarded as a pilin since it shows no clear conservation of the signal peptide processing site and was recently characterized as a TatA homolog [51]. There are no conclusive data about the composition of pili in other cyanobacteria. Mutagenesis studies on prepilin genes have been further performed only for Nostoc punctiforme, which contains five (pseudo-)pilin-like genes. Duggan et al. [23] inactivated three of these genes, but only one mutant (in the locus NpF0069) was successfully segregated. This pilA mutant showed normal pilus biogenesis in hormogonia, although its infection efficiency was attenuated [23]. In Synechocystis 6803 post-translational modification of pilin subunits is thought to be critical for the biogenesis of functional pili. This includes O-linked glycosylation of pilin subunits and trimethylation of pilin at the C-terminal lysine [52,53,54].
3.4. Distribution of Pili Genes in Cyanobacterial Genomes
A systematic search using NCBI PSI-BLAST with default parameters revealed that type IV pilus biogenesis genes are conserved in almost all cyanobacterial species from all the major clades, including the phylogenetic distant Gloeobacter lineage (Figure 1b). While the number and organization of pilA genes coding for prepilins is variable, the organization of other core genes is conserved among cyanobacteria. The pilMNOQ genes encoding the outer membrane pore and the alignment complex as well as genes encoding the inner platform protein PilC together with the essential secretion ATPase genes pilB1 and pilT1 are generally clustered in the same order in two distinct loci. This implies that pili genes in cyanobacteria are phylogenetically ancient and were already present in a common ancestor. Remarkably, a complete set of these core genes is conserved in many species for which type IV pili (or other cell appendages) have never been described. Moreover, these species are not known to exhibit twitching motility or natural competence. Hence, it should be considered that at least in some cyanobacteria the type IV pilus apparatus has a function unrelated to motility or DNA uptake. Regarding the homology to type II secretion systems, a role in macromolecule secretion via a pseudopilus seems likely as discussed in the next paragraph.
4. Function of Type IV Pili
Already more than 30 years have passed since the motility of certain filamentous cyanobacteria was analyzed in detail [55], however the mechanism of gliding motility of these cyanobacteria on a solid surface was not well defined. It was proposed that motility in Phormidium unicinatum and possibly in several other filamentous cyanobacteria is based on a junctional pore complex for directional extrusion of slime [25]. In addition, there is also evidence suggesting that arrays of contractile fibrils function in generating thrust for gliding motility [56]. However, several studies demonstrate the involvement of type IV pili in movement of filamentous cyanobacteria. Hormogonia, which are shorter filaments involved in dispersal and symbiotic associations with plants and fungi, are peritrichously piliated [23], suggesting that type IV pili are involved at least in hormogonial gliding. The core genes required for biogenesis of type IV pili are encoded in the genome of Nostoc punctiforme and pilT mutants are non-motile [23]. Risser et al. [24] used immunofluorescent staining to detect PilA on the surface of hormogonia. They demonstrated a fluorescence signal localized to a ring at the junction between individual cells showing a bias to one cell pole that is consistent among the cells of a particular hormogonium. Non-motile strains did not show a directional bias of PilA localization to one side of the septum, in contrast to motile strains. The authors hypothesize that the putative PilA protein they have analyzed might be part of the junctional pore complex. Thus, it is possible that type IV pili in hormogonia are mainly involved in secretion of polysaccharides. Although there is considerable evidence that polysaccharide export is important for motility in Nostoc punctiforme [57] this is no clear evidence for polysaccharide “jet propulsion”: The requirement may simply be to provide a suitable surface for gliding, as appears to be the case in Synechocystis 6803 or Myxococcus xanthus [58,59].
Apart from their importance in motility type IV pili are essential for natural competence of Synechocystis 6803. Inactivation of genes encoding components of the pilus apparatus inhibit the uptake of exogenous DNA [7] suggesting that type IV pili are part of the DNA-uptake machinery in Synechocystis 6803. Most DNA-uptake complexes of gram-negative naturally competent bacteria include a type IV-like pilus as a core component. Based on data obtained with Vibrio cholera Seitz and Blokesch [60] suggest a two-step process for uptake of exogenous DNA, where the type IV pili are responsible for the translocation of incoming DNA across the outer membrane. Several competency proteins in a pilus-independent process then facilitate further transport into the cytoplasm.
In addition, there is evidence that pili function as nanowires facilitating electron transfer to extracellular electron acceptors [61]. Lamb et al. [62] published data on Synechocystis 6803 showing that a pilA1 mutant grows slower than the wild type on oxidized iron minerals. They speculate that the PilA1 protein may be important for transport of electrons to these iron oxides. However, in this study a non-motile strain harboring a mutation in pilC was used. The importance of an intact type IV pili structure and function for the transport of electrons has still to be analyzed.
A new function of type IV pili has been demonstrated in our lab [43]. The RNA chaperone Hfq facilitates the action of small non-coding regulatory RNA molecules (sRNA) in many bacteria [63]. Synechocystis 6803 encodes several sRNAs but involvement of Hfq in the function of these RNA regulators has not been shown so far, though the expression of some mRNAs and sRNAs was altered in an hfq mutant. Inactivation of hfq in Synechocystis 6803 leads to a phenotype: Mutant cells have lost all detectable appendages on their surface and are non-motile and non-transformable [42]. Surprisingly, Hfq binds to the pilus base via the C-terminal part of the PilB1 protein, which is unique in cyanobacterial PilB proteins. Whereas Hfq seems to be important for type IV pili assembly, pili are also important for Hfq function. Expression of Hfq-dependent transcripts is altered when pilB1 or pilC are inactivated and when Hfq is not correctly localized to the pilus base due to different single amino acid changes in the PilB1 binding sites of Hfq. Consequently, all Synechocystis 6803 strains with mutations in pili genes or which are non-motile due to mutations in regulatory components might also be defective in Hfq function.
The composition of the type IV pili apparatus shares important similarities to type II secretion systems [64]. Secretion of proteins, which are not related to pilins was shown to be type-IV-pili-dependent in a range of bacteria (e.g., Kirn et al. [65]). Most cyanobacteria harbor genes that are similar to pilB and other genes for components of the type IV pili apparatus, but do not assemble pili on their surface [66,67]. The role of these gene products is unclear, however a function in protein secretion might be possible. Schatz et al. [68] showed that biofilm formation of Synechococcus elongatus PCC 7942 cells is regulated by an extracellular factor. This factor is most probably not secreted anymore to the medium in pilB and pilC mutants. Assuming that type IV pili are also involved in Synechocystis 6803 protein secretion, deletion of hfq might affect protein secretion thereby having secondary effects on putative extracellular signals.
5. Pili Assembled by the Chaperone-Usher Pathway (CU pili)
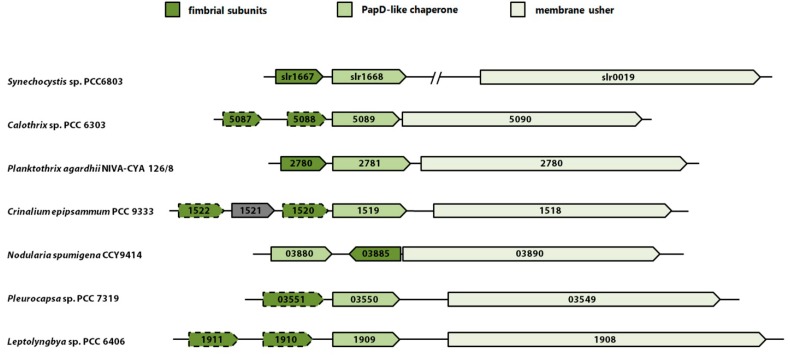
Yoshimura et al. [69] suggested that two proteins encoded by the genes slr1667 and slr1668 (named construction of cell surface components—cccS and cccP, respectively) are involved in biogenesis of the thick pili in Synechocystis 6803. Immunocytochemical analysis supported the idea that CccS was localized at the cell surface region and CccP in the cell periplasm. Both cccS and cccP mutants are non-motile and have no or less thick pili on their surface as shown by electron microscopy. Interestingly, cccP and cccS transcripts are the most differentially accumulated mRNAs in sycrp1 as well as hfq mutants, which are also non-motile. In their paper Yoshimura et al. [69] proposed a model, where CccS and/or CccP support targeting or stable assembly of the PilQ channel, thereby affecting assembly of thick pili. In addition, they state, that thin pili are not affected, suggesting that CccP and CccS are not involved in assembly of these structures. However, a different hypothesis can be drawn from the analysis of the amino acid sequences of both proteins. CccS exhibits a spore coat protein U-domain (SCPU, PFAM05229), whereas CccP contains a PapD-N domain (PFAM00345) [69]. These domains are typically found in proteins involved in biogenesis of CU pili in gram-negative bacteria via the chaperon/usher pathway. Usually, the respective genes are organized in operons containing at least one subunit of the fimbriae, a chaperone and a membrane protein for translocation of the subunits, the so called usher [18]. During assembly of the CU pili the signal peptide of a fimbriae subunit is cleaved off during its translocation across the cytoplasmic membrane. Further, the chaperone protein assists the correct folding and stabilization of the subunit. After binding to the usher protein, which forms a pore in the outer membrane, the chaperone is displaced and the fimbriae subunit is integrated into a filament on the cell surface [70]. In accordance with the domain structure, CccS might represent a subunit of the CU pilus and CccP the corresponding chaperone. It was already shown that CccS has a signal peptide, which is cleaved off, and that CccS is transported across the outer membrane in a CccP-dependent way [69]. These authors did not link CccSP with assembly of CU pili-like structures on the surface of Synechocystis 6803, most probably because there is no usher homolog encoded in this operon. However, in a phylogenetic analysis Nuccio and Bäumler [18] identified an usher homolog (slr0019, PFAM00577) in Synechocystis 6803. A search in the JGI IMG database (http://img.jgi.doe.gov) for proteins containing usher domains revealed the existence of further slr0019 homologs in different cyanobacteria. Remarkably, in most cases these genes are located directly downstream of a putative cccSP operon and thus represent the classical chaperone/usher operon structure (Figure 2). Thus, it seems plausible that CccSP together with Slr0019 could form CU-like pilus structures defined as the thin pili in Synechocystis 6803 by electron microscopy analysis. Further, their incorrect assembly may affect biogenesis of thick pili, indirectly. This hypothesis is contradictory to the data shown by Yoshimura et al. [69], which suggest that formation of thin pili was not affected in cccS and cccP mutants. However, in this paper it was also mentioned that, whereas bundles of thin-pili-like structures were clearly visualized by negative straining, this was not possible for thin pili. As the subunit composition of these different structures is not known so far, it is hardly possible to conclude from electron microscopy data about the nature of the various pili structures. Clearly, a comprehensive inspection of surface structures, including analysis of an slr0019 mutant, is needed to link putative CU pili related gene products to the respective surface appendages.
Figure 2.
Synteny of gene clusters encoding all the major components of putative CU pili in different cyanobacteria compared to Synechocystis 6803 where the usher homolog is located elsewhere in the genome. All putative CU pili related cyanobacterial gene clusters exhibiting a conserved domain structure (putative subunits PFAM05229; chaperone PFAM00345; usher PFAM00577) are shown. Weak homology is indicated by broken lines.
6. Conclusions
Many cyanobacteria harbor genes for the assembly of pili, but their function is largely unknown apart from one model strain. Functionally, non-flagellar appendages should have important roles in biofilm formation, aggregation, adhesion, natural competence or secretion, also in non-motile cyanobacteria. In addition, cyanobacterial appendages might have specialized roles and specific implications for these phototrophic organisms, which are different from other non-phototrophic bacteria. A different mechanism of Hfq function as well as effects of mutations in pili genes on photosynthetic functions might be only the beginning of the understanding how appendages are involved in cyanobacterial growth. In this respect it is important to note that various cyanobacterial wild-type strains are used in different laboratories all over the world. At least for Synechocystis 6803, the most important model strain for the study of motility and the function of pili, many laboratories work with the non-motile GT-strain, which was sequenced in 1996 [71]. This strain harbors several mutations, including a frame shift mutation in pilC and a mutation in the serine threonine kinase spkA, which is also involved in motility [72]. Over the few last years, motile sub-strains of this species have been re-sequenced [73,74] revealing even more differences between the strains. For this reason, physiological studies or microarray analyses in laboratories using different strains of Synechocystis 6803 may give conflicting results. Because there are so many genes involved in pilus assembly and regulation of motility mutations occur very often. There is also a bias for picking non-motile strains, especially because these colonies appear to be nicely shaped. It should be stressed here that known differences in genome sequences have to be considered when planning certain experiments and when comparing mutant strains from different wild-type backgrounds even in the same laboratory.
Acknowledgments
This work was partly supported by a grant from the German Research Foundation (DFG) to Annegret Wilde. We thank Brian Barnhart for critical reading of the manuscript.
Author Contributions
Nils Schuergers performed bioinformatic analyses, prepared the figures on these data and wrote the manuscript. Annegret Wilde wrote the manuscript. Both authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Terashima H., Kojima S., Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 2.Duan Q., Zhou M., Zhu L., Zhu G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013;53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 3.Conrad J.C., Gibiansky M.L., Jin F., Gordon V.D., Motto D.A., Mathewson M.A., Stopka W.G., Zelasko D.C., Shrout J.D., Wong G.C.L. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys. J. 2011;100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaara T. The outermost surface structures in chroococcacean cyanobacteria. Can. J. Microbiol. 1982;28:929–941. doi: 10.1139/m82-140. [DOI] [Google Scholar]
- 5.Bhaya D., Watanabe N., Ogawa T., Grossman A.R. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya D., Bianco N.R., Bryant D., Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara S., Geng X., Okamoto S., Yura K., Murata T., Go M., Ohmori M., Ikeuchi M. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2001;42:63–73. doi: 10.1093/pcp/pce007. [DOI] [PubMed] [Google Scholar]
- 8.Fronzes R., Remaut H., Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008;27:2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y.A., Yu X., Silverman P.M., Harris R.L., Egelman E.H. The structure of F-pili. J. Mol. Biol. 2009;385:22–29. doi: 10.1016/j.jmb.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zechner E.L., Lang S., Schildbach J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babic A., Lindner A.B., Vulic M., Stewart E.J., Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 12.Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 13.Craig L., Volkmann N., Arvai A.S., Pique M.E., Yeager M., Egelman E.H., Tainer J.A. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Pelicic V. Type IV pili: E pluribus unum? Mol. Microbiol. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 15.Craig L., Li J. Type IV pili: Paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis G.R. The type III secretion injectisome. Nat. Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 17.Evans M.L., Chapman M.R. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuccio S.-P., Bäumler A.J. Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lounatmaa K., Vaara T., Osterlund K., Vaara M. Ultrastructure of the cell wall of a Synechocystis strain. Can. J. Microbiol. 1980;26:204–208. doi: 10.1139/m80-031. [DOI] [PubMed] [Google Scholar]
- 20.Jensen T.E., Sicko L.M. The fine structure of the cell wall of Gloeocapsa alpicola, a blue-green alga. Cytobiologie. 1972;6:439–446. [Google Scholar]
- 21.Perkins F.O., Haas L.W., Phillips D.E., Webb K.L. Ultrastructure of a marine Synechococcus possessing spinae. Can. J. Microbiol. 1981;27:318–329. doi: 10.1139/m81-049. [DOI] [PubMed] [Google Scholar]
- 22.Dick H., Stewart W.D.P. The occurrence of fimbriae on a N2-fixing cyanobacterium which occurs in lichen symbiosis. Arch. Microbiol. 1980;124:107–109. doi: 10.1007/BF00407037. [DOI] [Google Scholar]
- 23.Duggan P.S., Gottardello P., Adams D.G. Molecular analysis of genes in Nostoc punctiforme involved in pilus biogenesis and plant infection. J. Bacteriol. 2007;189:4547–4551. doi: 10.1128/JB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risser D.D., Chew W.G., Meeks J.C. Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme. Mol. Microbiol. 2014;92:222–233. doi: 10.1111/mmi.12552. [DOI] [PubMed] [Google Scholar]
- 25.Hoiczyk E., Baumeister W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 1998;8:1161–1168. doi: 10.1016/S0960-9822(07)00487-3. [DOI] [PubMed] [Google Scholar]
- 26.Nakasugi K., Neilan B.A. Identification of pilus-like structures and genes in Microcystis aeruginosa PCC7806. Appl. Environ. Microbiol. 2005;71:7621–7625. doi: 10.1128/AEM.71.11.7621-7625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peabody C.R., Chung Y.J., Yen M.-R., Vidal-Ingigliardi D., Pugsley A.P., Saier M.H. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 28.Hobbs M., Mattick J.S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: A general system for the formation of surface-associated protein complexes. Mol. Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 29.Burrows L.L. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 30.Li C., Wallace R.A., Black W.P., Li Y., Yang Z. Type IV pilus proteins form an integrated structure extending from the cytoplasm to the outer membrane. PLoS One. 2013;8:e70144. doi: 10.1371/journal.pone.0070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgiadou M., Castagnini M., Karimova G., Ladant D., Pelicic V. Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: Characterization of a subcomplex involved in pilus assembly. Mol. Microbiol. 2012;84:857–873. doi: 10.1111/j.1365-2958.2012.08062.x. [DOI] [PubMed] [Google Scholar]
- 32.Ayers M., Sampaleanu L.M., Tammam S., Koo J., Harvey H., Howell P.L., Burrows L.L. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 2009;394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 33.Tammam S., Sampaleanu L.M., Koo J., Manoharan K., Daubaras M., Burrows L.L., Howell P.L. PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol. 2013;195:2126–2135. doi: 10.1128/JB.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoiczyk E., Hansel A. Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J. Bacteriol. 2000;182:1191–1199. doi: 10.1128/JB.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo J., Burrows L.L., Lynne Howell P. Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 2012;328:1–12. doi: 10.1111/j.1574-6968.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 36.Planet P.J., Kachlany S.C., DeSalle R., Figurski D.H. Phylogeny of genes for secretion NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeo H.J., Savvides S.N., Herr A.B., Lanka E., Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/S1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 38.Robien M.A., Krumm B.E., Sandkvist M., Hol W.G. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Yamagata A., Tainer J.A. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakasugi K., Alexova R., Svenson C.J., Neilan B.A. Functional analysis of PilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J. Bacteriol. 2007;189:1689–1697. doi: 10.1128/JB.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto S., Ohmori M. The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol. 2002;43:1127–1136. doi: 10.1093/pcp/pcf128. [DOI] [PubMed] [Google Scholar]
- 42.Dienst D., Dühring U., Mollenkopf H.J., Vogel J., Golecki J., Hess W.R., Wilde A. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology. 2008;154:3134–3143. doi: 10.1099/mic.0.2008/020222-0. [DOI] [PubMed] [Google Scholar]
- 43.Schuergers N., Ruppert U., Watanabe S., Nürnberg D.J., Lochnit G., Dienst D., Mullineaux C.W., Wilde A. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014;92:840–852. doi: 10.1111/mmi.12595. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R.S., Mathews D.W. Signature proteins for the major clades of Cyanobacteria. BMC Evol. Biol. 2010;10:24. doi: 10.1186/1471-2148-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato S., Shimoda Y., Muraki A., Kohara M., Nakamura Y., Tabata S. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshihara S., Geng X., Ikeuchi M. pilG Gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2002;43:513–521. doi: 10.1093/pcp/pcf061. [DOI] [PubMed] [Google Scholar]
- 47.Linhartová M., Bučinská L., Halada P., Ječmen T., Šetlík J., Komenda J., Sobotka R. Accumulation of the Type IV prepilin triggers degradation of SecY and YidC and inhibits synthesis of Photosystem II proteins in the cyanobacterium Synechocystis PCC 6803. Mol. Microbiol. 2014;93:1207–1223. doi: 10.1111/mmi.12730. [DOI] [PubMed] [Google Scholar]
- 48.Williams J.G.K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 49.Bhaya D., Takahashi A., Shahi P., Grossman A.R. Novel motility mutants of Synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 2001;183:6140–6143. doi: 10.1128/JB.183.20.6140-6143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshihara S., Ikeuchi M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 2004;3:512–518. doi: 10.1039/b402320j. [DOI] [PubMed] [Google Scholar]
- 51.Aldridge C., Spence E., Kirkilionis M.A., Frigerio L., Robinson C. Tat-dependent targeting of Rieske iron-sulphur proteins to both the plasma and thylakoid membranes in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 2008;70:140–150. doi: 10.1111/j.1365-2958.2008.06401.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y.H., Park K.H., Kim S.-Y., Ji E.S., Kim J.Y., Lee S.K., Yoo J.S., Kim H.S., Park Y.M. Identification of trimethylation at C-terminal lysine of pilin in the cyanobacterium Synechocystis PCC 6803. Biochem. Biophys. Res. Commun. 2011;404:587–592. doi: 10.1016/j.bbrc.2010.11.133. [DOI] [PubMed] [Google Scholar]
- 53.Kim Y.H., Kim J.Y., Kim S.-Y., Lee J.H., Lee J.S., Chung Y.-H., Yoo J.S., Park Y.M. Alteration in the glycan pattern of pilin in a nonmotile mutant of Synechocystis sp. PCC 6803. Proteomics. 2009;9:1075–1086. doi: 10.1002/pmic.200800372. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y.H., Park Y.M., Kim S.-J., Park Y.-I., Choi J.-S., Chung Y.-H. The role of Slr1443 in pilus biogenesis in Synechocystis sp. PCC 6803: Involvement in post-translational modification of pilins. Biochem. Biophys. Res. Commun. 2004;315:179–186. doi: 10.1016/j.bbrc.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 55.Häder D.P. Photosensory behavior in procaryotes. Microbiol. Rev. 1987;51:1–21. doi: 10.1128/mr.51.1.1-21.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Read N., Connell S., Adams D.G. Nanoscale visualization of a fibrillar array in the cell wall of filamentous cyanobacteria and its implications for gliding motility. J. Bacteriol. 2007;189:7361–7366. doi: 10.1128/JB.00706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Risser D.D., Meeks J.C. Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol. Microbiol. 2013;87:884–893. doi: 10.1111/mmi.12138. [DOI] [PubMed] [Google Scholar]
- 58.Li Y., Sun H., Ma X., Lu A., Lux R., Zusman D., Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burriesci M., Bhaya D. Tracking phototactic responses and modeling motility of Synechocystis sp. strain PCC6803. J. Photochem. Photobiol. B. 2008;91:77–86. doi: 10.1016/j.jphotobiol.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Seitz P., Blokesch M. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2013;110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorby Y.A., Yanina S., McLean J.S., Rosso K.M., Moyles D., Dohnalkova A., Beveridge T.J., Chang I.S., Kim B.H., Kim K.S., et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamb J.J., Hill R.E., Eaton-Rye J.J., Hohmann-Marriott M.F. Functional role of PilA in iron acquisition in the cyanobacterium Synechocystis sp. PCC 6803. PLoS One. 2014;9:e105761. doi: 10.1371/journal.pone.0105761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korotkov K.V., Sandkvist M., Hol W.G.J. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirn T.J., Bose N., Taylor R.K. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 2003;49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- 66.Samuel A.D., Petersen J.D., Reese T.S. Envelope structure of Synechococcus sp. WH8113, a nonflagellated swimming cyanobacterium. BMC Microbiol. 2001;1 doi: 10.1186/1471-2180-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palenik B., Brahamsha B., Larimer F.W., Land M., Hauser L., Chain P., Lamerdin J., Regala W., Allen E.E., McCarren J., et al. The genome of a motile marine Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 68.Schatz D., Nagar E., Sendersky E., Parnasa R., Zilberman S., Carmeli S., Mastai Y., Shimoni E., Klein E., Yeger O., et al. Self-suppression of biofilm formation in the cyanobacterium Synechococcus elongatus. Environ. Microbiol. 2013;15:1786–1794. doi: 10.1111/1462-2920.12070. [DOI] [PubMed] [Google Scholar]
- 69.Yoshimura H., Kaneko Y., Ehira S., Yoshihara S., Ikeuchi M., Ohmori M. CccS and CccP are involved in construction of cell surface components in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2010;51:1163–1172. doi: 10.1093/pcp/pcq081. [DOI] [PubMed] [Google Scholar]
- 70.Waksman G., Hultgren S.J. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 72.Kamei A., Yuasa T., Orikawa K., Geng X.X., Ikeuchi M. A eukaryotic-type protein kinase, SpkA, is required for normal motility of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2001;183:1505–1510. doi: 10.1128/JB.183.5.1505-1510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanesaki Y., Shiwa Y., Tajima N., Suzuki M., Watanabe S., Sato N., Ikeuchi M., Yoshikawa H. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 2012;19:67–79. doi: 10.1093/dnares/dsr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trautmann D., Voss B., Wilde A., Al-Babili S., Hess W.R. Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012;19:435–448. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]