Abstract
The introduction of next generation sequencing (NGS) has led to an exponential increase of elucidated genetic causes in both extremely rare diseases and common but heterogeneous disorders. It can be applied to the whole or to selected parts of the genome (genome or exome sequencing, gene panels). NGS is not only useful in large extended families with linkage information, but may also be applied to detect de novo mutations or mosaicism in sporadic patients without a prior hypothesis about the mutated gene. Currently, NGS is applied in both research and clinical settings, and there is a rapid transition of research findings to diagnostic applications. These developments may greatly help to minimize the “diagnostic odyssey” for patients as whole-genome analysis can be performed in a few days at reasonable costs compared with gene-by-gene analysis based on Sanger sequencing following diverse clinical tests. Despite the enthusiasm about NGS, one has to keep in mind its limitations, such as a coverage and accuracy of < 100 %, resulting in missing variants and false positive findings. In addition, variant interpretation is challenging as there is usually more than one candidate variant found. Therefore, there is an urgent need to define standards for NGS with respect to run quality and variant interpretation, as well as mechanisms of quality control. Further, there are ethical challenges including incidental findings and how to guide unaffected probands seeking direct-to-customer testing. However, taken together, the application of NGS in research and diagnostics provides a tremendous opportunity to better serve our patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-014-0288-8) contains supplementary material, which is available to authorized users.
Keywords: Exome sequencing, Massively parallel sequencing, Incidental findings, Diagnostic yield, Gene panel
Introduction
Next generation sequencing (NGS) represents an entirely new principle of sequencing technology following Sanger (first generation) sequencing, which was first described in 1977 [1]. Technical improvements of this sequencing technology such as the introduction of fluorescent dyes (to replace radiolabelling) and capillary array electrophoresis (to replace gel-based polyacrylamide gel electrophoresis), enabled automation of this technique, thereby increasing the sequencing capacity from a few hundred base pairs to several thousands of them within a single analysis [2]. Even more strikingly, with the advent of NGS, throughput exploded to up to > 1 tera bases (=1012 or, in other words, 1000 billion bases). This enormous improvement has been achieved by massive parallel sequencing. While a Sanger sequence reaction produces DNA chains arbitrarily terminated at each of the different positions by introducing a dideoxynucleotide and subsequently separating the pool of these chains according to size by electrophoresis, NGS is based on the principle of “sequencing-by-synthesis” [3]. This means that the complementary integration of a nucleotide during chain prolongation (i.e., the sequencing reaction) is directly monitored by the sequencing machine. Remarkably, the increasing sequencing capacity is paralleled by dramatically decreasing costs to sequence a human genome and will probably soon meet the magic threshold of the “$1000 genome”. For comparison, the costs for a whole genome will then be in the range of those of a magnetic resonance imaging scan.
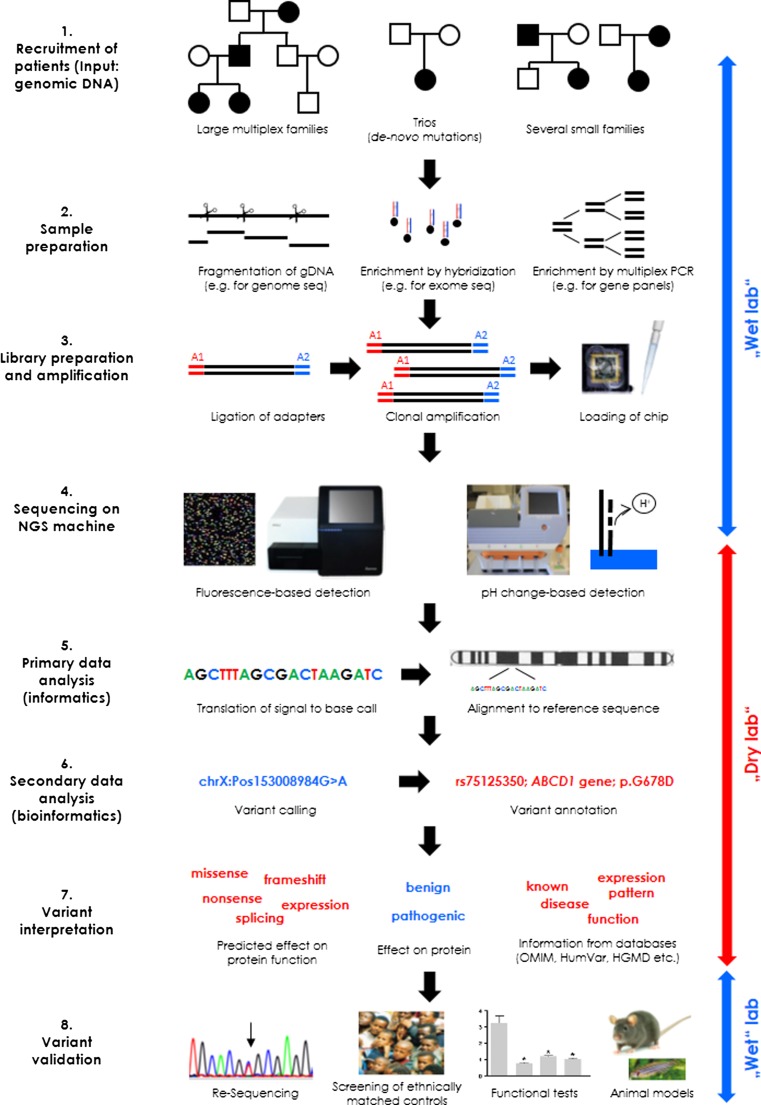
NGS became available to the community in 2008–09 when the first NGS machines entered the market. The process of applying NGS in a research or diagnostic setting comprises a wet laboratory workflow, including library preparation and the actual sequencing of the library. This is followed by a dry laboratory workflow involving informatic (translation of light signals or pH changes into sequence information) and bioinformatic analyses (sequence alignment, variant calling), as well as variant filtering and interpretation (annotation, mapping against variant databases) (Fig. 1).
Fig. 1.
Workflow of a next generation sequencing (NGS) analysis. The figure provides a simplified overview of the main phases in NGS and demonstrates some of the different options at each step. For the sequencing analysis, DNA is required that can be derived from large families with many affected, sporadic patients, and their healthy parents (trios), or several small families with the same disease (for gene discovery) or from individual patients (for diagnostic purposes). In the second step, DNA needs to be prepared for the sequencing by fragmentation if the whole genome is the target for NGS. Alternatively, specific target sequences need to be enriched (for instance, by hybridization or polymerase chain reaction). Third, the equally sized fragments have to be ligated to universal adapters, clonally amplified, and loaded onto a chip. Next, the chip is placed into an NGS machine and the sequencing reaction (integration of a nucleotide) is monitored by a light signal (fluorescence) or by the release of a proton resulting in a pH change. While all of these first steps are carried out at the bench in a genetic laboratory (“wet lab”), the next 3 steps are performed at a desk using a computer and several software packages and databases (“dry lab”). Specifically, the recorded signal from the NGS machine needs to be translated into a sequence, which is aligned to the reference genome. Next, mismatches with the reference sequence are retrieved and annotated with respect to the coding part of the genome. This is followed by the critical step of variant interpretation involving the separation of likely benign from possibly pathogenic variants. For this, information is collected and evaluated for categorization of the effect on an encoded protein, for in silico prediction of the consequences on protein function, and for previously reported knowledge on the gene and the specific mutation in question from databases. Finally, when one or a few candidate variants are selected as potential disease cause, comprehensive validation is needed and takes place in the “wet lab”. Validation must include resequencing to separate false positive from true positive variants, screening of ethnically matched controls, tests for the effect of the mutation on protein function, or studies of the mutated protein in animal models
NGS is a powerful tool to detect variants within the genome of any given individual. With genome sequencing, about 4 million variants per individual can be detected [4], whereas exome sequencing (covering mainly the 1 % protein-coding part of the genome) results in about 20,000 variants [5]. Finding a disease-causing variant among these many alterations mirrors the proverbial search for the needle in the haystack. Nevertheless, to date, NGS is the best available tool to elucidate disease-causing mutations. It is not only useful in large extended families, where linkage information provides information about the disease locus, but may also be applied to detect disease-causing de novo mutations in sporadic patients, a research and diagnostic question impossible to address by conventional Sanger sequencing without having a candidate gene [6].
In this review, we will summarize different applications of NGS, highlight major achievements in unraveling the cause of neurological diseases, and comment on the utility and challenges of using this technology in a diagnostic setting.
Targets of NGS: Selected Genes, Whole Exome, or Whole Genome
NGS can be applied to any species (bacteria, plants, animals, humans, etc.) and source of DNA, including genomic DNA (such as in genome sequencing), complementary DNA (RNA-Seq), methylated DNA (for epigenome sequencing), or specifically enriched DNA, such as binding sites for transcription factors (ChIP-Seq). In this review, we will focus on the different applications in which unmodified genomic (or part of the genomic) DNA is the target for NGS.
In particular, whole-genome sequencing allows for a hypothesis-free approach to genetic testing and screening. All available NGS machines have a certain output per run/analysis varying between 1 Gb and > 1 Tb. This capacity is divided up across the different target sequences. If, for instance, the target sequence comprises 50 Mb (such as the human exome), one will have a mean sequencing depth of 20 reads if the capacity is 1 Gb or a depth of 200 reads if the capacity is 10 Gb. This means that, on average, each base pair will be read 20 or 200 times. Currently, a mean sequencing depth of about 30–40 is aimed for to cover most of the target sequences with sufficient power. This is important for 2 reasons. First, coverage is not equally distributed across the target sequence. Depending on the composition of the sequence, some sequences are read more often than others. Therefore, a mean sequencing depth of 30 implies that the vast majority of sequences will be covered and read at least 10 times. Second, coverage is a stochastic process. Therefore, either the wild-type or the mutant allele will be sequenced by chance for heterozygous variants in a given read. If there are only 5 reads, the likelihood that all of these reads are derived from the wild-type sequence will be 1:25 = 3 %, which will result in quite a large number of false negatives given the enormous number of variants in the target sequence.
Although the sequencing capacity is immense, it still remains somewhat limited. To counteract this restriction, the target sequence had to be narrowed at least in the early days of NGS. Initially, researchers focused on target enrichment by which, for instance, NGS was limited to a linked region in a given family. This can be achieved by using custom-specific probe sets or microarrays, as well as by microdissection of chromosomal regions [7]. However, there are only a few success stories using this approach, such as the identification of the genetic cause of intellectual disability in a family with autosomal recessive inheritance [8].
Currently, NGS can easily be applied to the whole exome, which represents about 1 % of the human genome. For this, nearly all exonic sequences are enriched mainly by in-solution enrichment kits. These kits are commercially available and, owing to the high demand, have been extensively validated, are being constantly improved, and sold at a reasonable price. The number of newly identified disease genes has grown exponentially through the application of exome sequencing within the last years in all fields of medicine [9], including neurological diseases [10], such as epilepsy [11], intellectual disability [12], Parkinson disease [13, 14], and other forms of neurodegeneration [15]; dystonia [16]; paroxysmal dyskinesia [17]; or neuropathies [18].
Currently, genome sequencing can also be carried out at reasonable costs but is still about 5 times more expensive than exome sequencing. As the sequence-specific enrichment step is omitted, the major advantage of genome sequencing compared with exome sequencing comprises the unbiased analysis of the genome. Thus, potential protein-coding regions that have not yet been annotated as genes, as well as regulatory regions, such as noncoding RNAs or transcription factor binding sites, are included in the sequence analysis. In this context, it is worth noting that according to data from the ENCODE project, a regulatory role might be assigned to about 80 % of the human genome [19]. Genome sequencing has been successfully applied to identify genes for amyotrophic lateral sclerosis [20], dystonia [21], retinitis pigmentosa [22], autism [23], and other diseases.
As genome and exome sequencing are usually overpowered for a diagnostic setting, the analysis often focuses (at least in a first step before exome sequencing) on known disease-related genes. For this, the exonic region only of these genes is enriched by respective probes and subsequently sequenced as part of so-called gene panels. Another level of optimization for the ratio of sequencing depth and output (with respect to the costs) includes the parallel analysis of several patients in one run. This is possible through the integration of “molecular barcodes” during library preparation. These barcodes tag all sequences derived from a single patient and thus allow for pooled analyses of up to 400 samples.
Advantages of NGS Compared With Other Mutational Screening Methods: Detection of Mosaicism, de novo Mutations, Digenic Disease Causes, and Reverse Phenotyping
In addition to the immense throughput of the NGS technology, the possibility of sequencing an entire exome or genome within days at reasonable costs and the resulting opportunity to gain knowledge about all exonic/genomic variants within an individual, NGS even enables the detection of otherwise missed genetic disease causes.
Based on the separate sequence analysis of single nucleotide strands, it is possible to detect accurately mosaicism if the sequencing depth is high and the variant calling tools are sensitive enough [24, 25]. Mosaicism frequently contributes to severe, early-onset diseases, such as in patients with Cornelia de Lange syndrome [26]. A related frequent cause of severe, early-onset diseases are de novo mutations, which can also be identified by NGS without prior knowledge about disturbed pathways or candidate genes. For the detection of de novo mutations, a trio analysis is necessary, including the affected child and both healthy parents. This approach has recently been successfully applied to a number of diseases [6], and led to the discovery of de novo mutations in genes for neurodegeneration with brain iron accumulation (WDR45 [15]), leukoencephalopathy (TUBB4A [27]), epilepsy (CHD2 [28]), and even for complex disorders such as autism [29], intellectual disability [12], and many others.
The power of NGS and elucidating the whole spectrum of variants in a given individual will also stimulate the discovery of digenic or polygenic disease causes as, after identifying a first seemingly disease-causing mutation, additional analyses can be carried out because the data are already available. A remarkable example includes the detection of heterozygous mutations in the 2 functionally related genes GUCY1A3 and CCT7 in an extended family with myocardial infarction [30]. Digenic inheritance is a phenomenon probably also seen in neurological diseases such as Parkinson disease with, for example, mutations in PINK1 and DJ-1 [31]. However, it remains to be proven that the combination of the two mutations is, indeed, the cause of the disease versus simple co-occurrence of 2 mutations by chance [32].
With NGS, it may even be possible to find the disease-causing mutation despite incomplete phenotypic information. For example, if a pathogenic mutation is found in a known disease-causing gene by the combination of NGS and segregation analysis, in some cases the gene will have previously been linked to a broader or even to a different phenotype. In such cases, a retrospective clinical investigation of the patient and family members may reveal additional, previously unrecognized features in a process called “reverse phenotyping” [9]. A recent example includes a patient with dystonia and other movement disorders who was shown to carry a mutation in OPA3, a known gene for optic atrophy. This prompted an ophthalmologic examination in the family and did, indeed, reveal optic atrophy [33].
Applications of NGS: From Gene Discovery to Diagnostics
Patients with rare diseases often undergo a “diagnostic odyssey” until they receive the correct diagnosis, if at all. This is owing to clinical and genetic heterogeneity of hereditary conditions, unusual presentations, and lack of specific clinical–genetic knowledge of the attending physicians (“one sees only what one knows”). There are many awe-inspiring examples in the scientific literature illustrating the diagnostic odyssey that patients have undergone, some reports even written by the involved parties themselves. They point out 2 major technical improvements that were instrumental in elucidating the cause of their diseases: NGS and information exchange through the internet and social media [34, 35]. Identification of the disease-related mutation is of great direct importance to patients for a number of different reasons: knowing the cause of the condition relieves uncertainty, facilitates communication with healthcare professionals, employers, and, in case of children, with teachers and counselors, and enables informed family and career planning. In some cases, it also leads to a more specific prognosis and may even impact on therapy.
We are currently witnessing a remarkably rapid transition of research findings to diagnostic applications, that is newly detected genes are quickly fed into the pipelines of high-quality diagnostic testing companies, where it can take fewer than 3 months from the publication of a new gene to its incorporation into commercial diagnostic gene panels. In contrast, access to expert genetic NGS testing and counseling is still, overall, limited. However, primary care physicians should be increasingly alerted to the new diagnostic options for patients with rare, unclassified conditions who may benefit from NGS-based genetic testing, and refer such patients to a center of rare diseases or similar tertiary care facility. As an additional caveat, genetic testing reports can be difficult to read and interpret. However, reports of commercial testing companies differ widely in their clarity, specificity, and interpretability. Ideally, a genetic testing company should be a certified testing laboratory, have a turnaround time for genetic testing reports of 4–8 weeks, and provide a meaningful testing report. For this, the minimum requirements are a literature search on the specific variant/mutation, a report on (in-house and database) frequencies of this finding in controls, conservation across species, and the predicted result of the change on the protein using at least 3 different in silico prediction programs. This information should be followed by an interpretable summary of these findings, acknowledging, however, that a clear-cut distinction between pathogenic and benign sequence changes is not always possible. Employing medical advisors assisting with the interpretation of variants in the respective clinical setting is a further added value that is provided by some genetic testing companies.
One should keep in mind that the application of NGS is suitable for both rare disorders as well as for phenotypically and genetically heterogeneous, common diseases such as epilepsy or intellectual disability. For example, we recently reported a patient with epilepsy, intellectual disability, and other features who had undergone more than 20 different genetic tests before the disease-causing mutation was found in the SCN2A gene by exome sequencing of the patient and her unaffected parents [36]. When compared with the multitude of clinical and laboratory tests commonly applied to patients with unknown diagnoses over many years, NGS may actually represent a relatively inexpensive alternative to the above-mentioned “diagnostic odyssey” of many of such patients. In this regard, studies comparing the utility and cost-effectiveness of NGS diagnostic testing are warranted and—given that patients and indications are carefully chosen—will hopefully lead to wider access to and broader coverage of genetic testing by insurance companies.
Limitations of NGS
Despite all the enthusiasm about NGS and its possibilities, one also has to consider the still existing limitations that are outlined below. While we expect that these limitations will be addressed and the method further improved over the next few years, in our personal view, NGS, at least with its present technological basis, will never achieve 100 % coverage and accuracy. Likewise, correct interpretation of detected variants will remain challenging and thus it may remain impossible to identify all genetic variants contributing to a given disease or trait with complete certainty.
The Problem of Having too Many Variants
After careful filtering for rare and protein-changing variants, a single exome harbors about 100–200 potential disease-causing changes. Along the same lines, each individual carries about 100 deleterious mutations that cause loss of function within protein-coding regions of genes [37], including clearly pathogenic variants not related to the disease in question (incidental findings) [38]. Thus, in the era of genomic medicine, interpretation of variants rather than detection of variants represents one of the bottlenecks for translating sequence information into clinical practice.
For rare diseases, when there is only a single patient with a possible mutation in a gene that has not yet been linked to the given disease, it is difficult to know whether the mutation is causal or not. In this context, weak standards for declaring genetic diagnoses using NGS data can actually cause harm by alienating patients and doctors alike. To further elucidate the pathogenic role of a possible mutation, a number of in silico prediction programs are available. However, all of these software tools have limitations and, as the name implies, they only predict possible pathogenicity [39]. Therefore, functional tests are needed, which may include, but are not limited to, quantitative measurements of the respective mRNA or protein levels, or enzymatic activity. However, it is important to note that conclusive functional tests are not available for the majority of proteins encoded by the human genome. Usually, basic research projects first need to be carried out to identify measurable readouts of the encoded (dysfunctional) proteins, a process that can take several years.
The Problem of Limited Coverage, Sequencing Depth, and Accuracy
Currently, owing to technical limitations, no single test platform provides complete coverage of the whole exome or genome. However, even conventional polymerase chain reaction applications cannot cover the entire genome [4]. Further, a low sequencing depth (<10×) can result in the random detection of only the wild-type allele for actually heterozygous variants (see above). Even though NGS has an accuracy of > 99.9 %, owing to the immense number of variants, there are still several thousands of false positive variant calls. Finally, erroneous results can even be caused by misinterpretation of “true positive” variants that can be confirmed by Sanger sequencing, and segregate with the disease but do not represent the pathogenic mutation. This scenario may account for much more than 1 % of the published findings [40]. In the field of dystonia, for example, this may apply to variants in CIZ1 (DYT23) and ANO3 (DYT24) [41–43], which currently await independent confirmation. Both of these genes harbor a large number of variants, making it particularly difficult to disentangle nonpathogenic from putative pathogenic variants.
The Problem of Quality Control
If NGS is applied in a diagnostic setting, the result of the test should be independent of the laboratory at which it was carried out. For conventional mutational tests such as Sanger sequencing, analyte-specific testing has been established. Here, the so called “proficiency testing” serves as an external measure of laboratory quality. In the USA, laboratories are certified under the Clinical Laboratory Improvement Amendments, and accredited by professional organizations. Also for NGS, quality control of the analysis is required and initial pilot surveys have been started in the USA and Europe for external quality control using methods-based proficiency testing [44]. In these studies, laboratories will be evaluated for their proficiency to correctly call variants in a test genome that has been extensively sequenced by the committee in charge of the quality control testing [44]. However, these tests only help to control the wet laboratory procedure and the informatic and bioinformatic algorithms. Clear-cut guidelines for minimum requirements still have to be developed regarding run quality (with respect to coverage and sequencing depth) and variant interpretation [41].
Ethical Aspects
The blessing of exome or genome sequencing to detect (theoretically) all variants within a given individual is a curse at the same time, as not only variants related to the disease in question, but also to other disorders will be detected (as incidental or secondary findings). This is also known from other genome-wide screening tools, such as microarray analysis, or from brain-wide neuroimaging, such as magnetic resonance imaging scans. Interestingly, in a recent study that evaluated 1000 exomes for “actionable” pathogenic single-nucleotide variants, that is those that cause treatable or even preventable conditions, 23 participants (~2 %) carried such substitutions [38]. The American College of Medical Genetics and Genomics recently published a policy statement on clinical molecular analysis emphasizing the importance of alerting the patient/family to the possibility of such results in pretest counseling discussions, as well as reporting of results [45]. Furthermore, the American College of Medical Genetics and Genomics issued guidelines for clinical testing laboratories that list 56 genes in which incidentally found known pathogenic or expected pathogenic mutations should be reported to the patients (http://www.acmg.net) [45]. The selection of these 56 genes is based on pathogenicity and the possibility of the genetic result leading to a specific therapeutic option (“actionable” findings). However, there is an ongoing discussion how to best proceed with incidental findings [46, 47].
While in the research setting, these incidental findings usually do not attract much attention, it is notable that there is growing interest in receiving information about incidental findings on the patient side [48]. This is also reflected by the increasing availability and popularity of direct-to-consumer genetic testing (DTCGT). DTCGT enables individuals to purchase genetic tests and receive results without the intervention of a health professional. Shortly after DTCGT became available, the American Society of Human Genetics provided a statement on DTCGT including the following recommendation: “Companies offering DTCGT should disclose the sensitivity, specificity, and predictive value of the test, and the populations for which this information is known, in a readily understandable and accessible fashion” [49]. However, even if provided in a transparent fashion, it is difficult for most individuals and even for many doctors to adequately interpret the test results and risk assessment. Importantly, most of these test results pertain to low-risk variants for common diseases or traits, and the important difference between a causal mutation and a gene variant that only mildly increases the lifetime risk for a certain condition is often unclear to both patients and their attending physicians. For example, even if several risk variants for Parkinson disease happen to coincide, the risk of developing the disorder will only be increased 2.5-fold [50]. While at first sight this may look considerable, given the low prevalence of 0.14 % of Parkinson disease in the general population, the corresponding lifetime risk for the disease will still be as low as 0.35 % [51]. Thus, the results of DTCGT can cause considerable uncertainty and anxiety, requiring extensive post-test counseling. According to a recent systematic review, the authors of position statements, policies, and recommendations described more potential harms than benefits. But, although some stated that DTCGT should be actively discouraged, others supported consumer rights to make autonomous choices [52]. Notably, large companies providing DTCGT have currently suspended their health-related genetic tests to comply with the US Food and Drug Administration’s directive to discontinue new consumer access until they can provide satisfactory evidence that the results are reliable and will not jeopardize consumers’ health.
Current Status
Research Setting
NGS has become the method of choice in research laboratories. This is true even for small laboratories that cannot afford their own NGS platform and that use NGS services offered by a number of different companies worldwide at reasonable prices. In fact, owing to the rapidly evolving technology, the technical prerequisites, and the high throughput, NGS platforms have currently mostly been established in core facilities and sequencing centers.
Diagnostic Setting
Gene panels have become the most attractive diagnostic application of NGS with the ability to provide multigene sequence data at similar costs as single-gene Sanger sequencing and to reduce the time for establishing a clinical management plan compared with successive rounds of single-locus testing [53]. Another advantage of gene panels is that less DNA is required (depending on the system, as little as 20 ng may be enough) compared with exon-by-exon Sanger sequencing (10 ng/exon). This is particularly important when the amount of DNA is limited such as for biopsy material, buccal swaps, or in newborns. A third advantage of gene panels is largely avoiding the detection of incidental findings, which have been discussed above. Gene panels have been developed for, and are being applied to, many neurological disorders, including epilepsy [11], ataxia [54], and dementia [55].
The diagnostic yield of exome sequencing seems to be much higher than that of screening (many) candidate genes by Sanger sequencing, especially for highly heterogeneous disorders such as deafness, blindness, mitochondrial diseases, and movement disorders, and may reach 25–52 % [56, 57]. In a recent study from the exome sequencing center at the St Radboud University Medical Center in Nijmegen, the Netherlands, the diagnostic yield using exome sequencing was at least 50 % higher than Sanger sequencing [57]. Although the time required for analysis of NGS sequence data is longer than for single-gene Sanger sequencing, it is possible to run the complete workflow of whole genome sequencing in about 50 h [58].
Even though diagnostic tests based on NGS have already entered the clinic and seem to be of advantage, surprisingly, no formal evaluation of the overall clinical utility has yet been reported. In this regard, initiatives such as the Evaluation of Genomic Applications in Practice and Prevention have been formed to address this important issue [59].
It should also be noted that the clinical utility of a detected disease-causing variant may be low owing to the phenomenon of reduced penetrance that is observed in many, mostly dominantly inherited, disorders. For example, while the movement disorder DYT1 dystonia is caused by a 3-base pair deletion in the TOR1A gene, the penetrance of this mutation is reduced to 30 %, thereby considerably limiting the diagnostic value in a presymptomatic individual. As outlined above, the situation becomes even more problematic for genetic risk variants, that is relatively common variants (polymorphisms) that occur in both patients and controls but are significantly more frequently found among patients leading to the interpretation that they contribute to (but are not sufficient to cause) a disease.
Future Perspectives
Through the introduction of NGS in both research and diagnostic settings, our knowledge of how genetic variants affect human health has been rapidly increasing. Together with declining costs for whole genome sequencing, the overall development is driven in the direction of individualized genetic medicine. This will primarily include genetic diagnoses in affected and presymptomatic individuals, as well as pharmacogenetic aspects.
To evaluate the power of genomic medicine, several multicenter large-scale projects have been initiated. One of these projects is the SickKids Genome Clinic Project in which the utility of pediatric genomic medicine will be addressed in a multidisciplinary clinical/research setting by comparing the outcomes of conventional genetic diagnostics and whole-genome sequencing in > 100 children/year over 5 years [60]. This study will not only focus on the evaluation of potential disease-causing mutations (and incidental findings) but also on pharmacogenetic variants [60].
A future challenge—especially in the field of rare and extremely rare disorders—will be to further extend collaborations between laboratories spread all over the world and an even closer relationship between patients (or the parents of children) and the attending physicians [34]. One helpful tool to better collect information about rare variants and rare phenotypes may be the establishment of phenotype–genotype databases, such as the Leiden Open Variation Database (www.lovd.nl). This platform is also used to establish country-specific subdatabases such as the Finnish Disease Heritage Database (http://findis.org) [61]. The main idea is that in such databases case descriptions and findings from NGS are being collected and thus become searchable with certain search terms, so that subsequent cases can be checked and compared for similarities. The idea is based on DECIPHER (Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources), an interactive web-based database that incorporates a suite of tools designed to aid the interpretation of submicroscopic chromosomal imbalances. However, all of these databases depend on reliable and large-scale data input.
To ensure that information, at least about genetic variants derived from NGS studies, is systematically collected in databases, some of the genetic journals already require the submission of detected variants to public databases by the authors. For instance, the European Journal of Human Genetics requires authors to submit all variants described in the paper to a public database, for example the relevant gene variant database (or the Locus Specific database), prior to acceptance (http://www.nature.com/ejhg).
An important future aim should be to standardize the analysis and develop guidelines for variant interpretation. To reduce the number of false positive findings in the literature, this should include minimal requirements before novel disease-causing genes are published. Given the limitations and challenges of NGS, perhaps not surprisingly, there is evidence for an increasing number of putative disease genes that have been reported to be linked to a certain disease but cannot be confirmed independently. While NGS findings are, overall, easily publishable and quickly entrenched in the literature, erroneous findings usually remain unreported, are included in gene panels and gene reviews, and may prompt additional research, such as screening of multiethnic cohorts, functional studies, and even development of animal models, thereby potentially drawing on already scarce research resources.
In conclusion, it is no exaggeration to say that NGS has truly revolutionized molecular genetic research and diagnostics, thereby providing a wealth of previously unimaginable opportunities. That being said, a careful appreciation of the limitations of NGS and a strong focus on developing standards of clinical evaluation, systematic data collection, and ethical considerations will make NGS an even more useful and powerful tool in the future.
Electronic supplementary material
(PDF 1225 kb)
Acknowledgments
KL receives funding from the German Research Foundation (LO 1555/3-2, LO 1555/8-1) and the Dystonia Coalition (NS065701). CK is supported by a career development award from the Hermann and Lilly Schilling Foundation, and by the German Research Foundation (SFB936 and SFB TR134).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 3.Lin B, Wang J, Cheng Y. Recent patents and advances in the next-generation sequencing technologies. Recent Pat Biomed Eng. 2008;2008:60–67. doi: 10.2174/1874764710801010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam HY, Clark MJ, Chen R, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 7.Weise A, Timmermann B, Grabherr M, et al. High-throughput sequencing of microdissected chromosomal regions. Eur J Hum Genet. 2010;18:457–462. doi: 10.1038/ejhg.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selmer KK, Gilfillan GD, Stromme P, et al. A mild form of Mucopolysaccharidosis IIIB diagnosed with targeted next-generation sequencing of linked genomic regions. Eur J Hum Genet. 2012;20:58–63. doi: 10.1038/ejhg.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar KR, Lohmann K, Klein C. Genetics of Parkinson disease and other movement disorders. Curr Opin Neurol. 2012;25:466–474. doi: 10.1097/WCO.0b013e3283547627. [DOI] [PubMed] [Google Scholar]
- 11.Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–1398. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 12.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 13.Zimprich A, Benet-Pages A, Struhal W, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilarino-Guell C, Wider C, Ross OA, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haack TB, Hogarth P, Kruer MC, et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet. 2012;91:1144–1149. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs T, Saunders-Pullman R, Masuho I, et al. Mutations in GNAL cause primary torsion dystonia. Nat Genet. 2012;45:88–92. doi: 10.1038/ng.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WJ, Lin Y, Xiong ZQ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet. 2011;43:1252–1255. doi: 10.1038/ng.1008. [DOI] [PubMed] [Google Scholar]
- 18.Soong BW, Huang YH, Tsai PC, et al. Exome sequencing identifies GNB4 mutations as a cause of dominant intermediate Charcot-Marie-Tooth disease. Am J Hum Genet. 2013;92:422–430. doi: 10.1016/j.ajhg.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herdewyn S, Zhao H, Moisse M, et al. Whole-genome sequencing reveals a coding non-pathogenic variant tagging a non-coding pathogenic hexanucleotide repeat expansion in C9orf72 as cause of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2412–2419. doi: 10.1093/hmg/dds055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann K, Wilcox RA, Winkler S, et al. Whispering dysphonia (DYT4 dystonia) is caused by a mutation in the TUBB4 gene. Ann Neurol. 2013;73:537–545. doi: 10.1002/ana.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiguchi KM, Tearle RG, Liu YP, et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc Natl Acad Sci U S A. 2013;110:16139–16144. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagnamenta AT, Lise S, Harrison V, et al. Exome sequencing can detect pathogenic mosaic mutations present at low allele frequencies. J Hum Genet. 2012;57:70–72. doi: 10.1038/jhg.2011.128. [DOI] [PubMed] [Google Scholar]
- 25.Tapper WJ, Foulds N, Cross NC, et al. Megalencephaly syndromes: exome pipeline strategies for detecting low-level mosaic mutations. PLoS One. 2014;9:e86940. doi: 10.1371/journal.pone.0086940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huisman SA, Redeker EJ, Maas SM, Mannens MM, Hennekam RC. High rate of mosaicism in individuals with Cornelia de Lange syndrome. J Med Genet. 2013;50:339–344. doi: 10.1136/jmedgenet-2012-101477. [DOI] [PubMed] [Google Scholar]
- 27.Simons C, Wolf NI, McNeil N, et al. A de novo mutation in the beta-tubulin gene TUBB4A results in the leukoencephalopathy hypomyelination with atrophy of the basal ganglia and cerebellum. Am J Hum Genet. 2013;92:767–773. doi: 10.1016/j.ajhg.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suls A, Jaehn JA, Kecskes A, et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelson JJ, Shi Y, Gujral M, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdmann J, Stark K, Esslinger UB, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432–436. doi: 10.1038/nature12722. [DOI] [PubMed] [Google Scholar]
- 31.Tang B, Xiong H, Sun P, et al. Association of PINK1 and DJ-1 confers digenic inheritance of early-onset Parkinson's disease. Hum Mol Genet. 2006;15:1816–1825. doi: 10.1093/hmg/ddl104. [DOI] [PubMed] [Google Scholar]
- 32.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arif B, Kumar KR, Seibler P, et al. A novel OPA3 mutation revealed by exome sequencing: an example of reverse phenotyping. JAMA Neurol. 2013;70:783–787. doi: 10.1001/jamaneurol.2013.1174. [DOI] [PubMed] [Google Scholar]
- 34.Might M, Wilsey M. The shifting model in clinical diagnostics: how next-generation sequencing and families are altering the way rare diseases are discovered, studied, and treated. Genet Med 2014 Mar 20. [DOI] [PubMed]
- 35.Erdmann J, Schunkert H. Forty-five years to diagnosis. Neuromuscul Disord. 2013;23:503–505. doi: 10.1016/j.nmd.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Baasch AL, Huning I, Gilissen C, et al. Exome sequencing identifies a de novo SCN2A mutation in a patient with intractable seizures, severe intellectual disability, optic atrophy, muscular hypotonia, and brain abnormalities. Epilepsia 2014;55:e25-29. [DOI] [PubMed]
- 37.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 40.Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat. 2013;34:1216–1220. doi: 10.1002/humu.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein C, Konig IR, Lohmann K. Exome sequencing for gene discovery: time to set standard criteria. Ann Neurol. 2012;72:627–628. doi: 10.1002/ana.23658. [DOI] [PubMed] [Google Scholar]
- 42.Ma L, Chen R, Wang L, Yang Y, Wan X. No mutations in CIZ1 in twelve adult-onset primary cervical dystonia families. Mov Disord. 2013;28:1899–1901. doi: 10.1002/mds.25542. [DOI] [PubMed] [Google Scholar]
- 43.Zech M, Gross N, Jochim A, et al. Rare sequence variants in ANO3 and GNAL in a primary torsion dystonia series and controls. Mov Disord. 2014;29:143–147. doi: 10.1002/mds.25715. [DOI] [PubMed] [Google Scholar]
- 44.Schrijver I, Aziz N, Jennings LJ, et al. Methods-based proficiency testing in molecular genetic pathology. J Mol Diagn. 2014;16:283–287. doi: 10.1016/j.jmoldx.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burke W, Matheny Antommaria AH, Bennett R, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15:854–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holtzman NA. ACMG recommendations on incidental findings are flawed scientifically and ethically. Genet Med. 2013;15:750–751. doi: 10.1038/gim.2013.96. [DOI] [PubMed] [Google Scholar]
- 48.Shahmirzadi L, Chao EC, Palmaer E, et al. Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med. 2014;16:395–399. doi: 10.1038/gim.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson K, Javitt G, Burke W, Byers P. ASHG Statement on direct-to-consumer genetic testing in the United States. Obstet Gynecol. 2007;110:1392–1395. doi: 10.1097/01.AOG.0000292086.98514.8b. [DOI] [PubMed] [Google Scholar]
- 50.Nalls MA, Plagnol V, Hernandez DG, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein C, Ziegler A. From GWAS to clinical utility in Parkinson's disease. Lancet. 2011;377:613–614. doi: 10.1016/S0140-6736(11)60062-7. [DOI] [PubMed] [Google Scholar]
- 52.Skirton H, Goldsmith L, Jackson L, O'Connor A. Direct to consumer genetic testing: a systematic review of position statements, policies and recommendations. Clin Genet. 2012;82:210–218. doi: 10.1111/j.1399-0004.2012.01863.x. [DOI] [PubMed] [Google Scholar]
- 53.Kassahn KS, Scott HS, Caramins MC. Integrating massively parallel sequencing into diagnostic workflows and managing the annotation and clinical interpretation challenge. Hum Mutat. 2014;35:413–423. doi: 10.1002/humu.22525. [DOI] [PubMed] [Google Scholar]
- 54.Nemeth AH, Kwasniewska AC, Lise S, et al. Next generation sequencing for molecular diagnosis of neurological disorders using ataxias as a model. Brain. 2013;136:3106–3118. doi: 10.1093/brain/awt236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beck J, Pittman A, Adamson G, et al. Validation of next-generation sequencing technologies in genetic diagnosis of dementia. Neurobiol Aging. 2014;35:261–265. doi: 10.1016/j.neurobiolaging.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neveling K, Feenstra I, Gilissen C, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- 58.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veenstra DL, Piper M, Haddow JE, et al. Improving the efficiency and relevance of evidence-based recommendations in the era of whole-genome sequencing: an EGAPP methods update. Genet Med. 2013;15:14–24. doi: 10.1038/gim.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowdin S, Ray P, Cohn RD, Meyn MS. The genome clinic: A multidisciplinary approach to assessing the opportunities and challenges of integrating genomic analysis into clinical care. Hum Mutat. 2014;35:513–519. doi: 10.1002/humu.22536. [DOI] [PubMed] [Google Scholar]
- 61.Polvi A, Linturi H, Varilo T, et al. The Finnish disease heritage database (FinDis) update-a database for the genes mutated in the Finnish disease heritage brought to the next-generation sequencing era. Hum Mutat. 2013;34:1458–1466. doi: 10.1002/humu.22389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1225 kb)