Abstract
RNA-binding proteins and microRNAs are potent post-transcriptional regulators of gene expression. Human transformer 2β (Tra2β) is a serine/arginine-rich-like protein splicing factor and is now implicated to have wide-ranging roles in gene expression as an RNA-binding protein. RNA immunoprecipitation (RIP) with an anti-Tra2β antibody and microarray analysis identified a subset of Tra2β-associated mRNAs in HCT116 human colon cancer cells, many of which encoded cell death-related proteins including Bcl-2 (B-cell CLL/lymphoma 2). Tra2β knockdown in HCT116 cells decreased Bcl-2 expression and induced apoptosis. Tra2β knockdown accelerated the decay of BCL2α mRNA that encodes Bcl-2 and full-length 3′ UTR, while it did not affect the stability of BCL2β mRNA having a short, alternatively spliced 3′ UTR different from BCL2α 3′ UTR. RIP assays with anti-Tra2β and anti-Argonaute 2 antibodies, respectively, showed that Tra2β bound to BCL2α 3′ UTR, and that Tra2β knockdown facilitated association of miR-204 with BCL2α 3′ UTR. The consensus sequence (GAA) for Tra2β-binding lies within the miR-204-binding site of BCL2 3′ UTR. Mutation of the consensus sequence canceled the binding of Tra2β to BCL2 3′ UTR without disrupting miR-204-binding to BCL2 3′ UTR. Transfection of an anti-miR-204 or introduction of three-point mutations into the miR-204-binding site increased BCL2 mRNA and Bcl-2 protein levels. Inversely, transfection of precursor miR-204 reduced their levels. Experiments with Tra2β-silenced or overexpressed cells revealed that Tra2β antagonized the effects of miR-204 and upregulated Bcl-2 expression. Furthermore, TRA2β mRNA expression was significantly upregulated in 22 colon cancer tissues compared with paired normal tissues and positively correlated with BCL2 mRNA expression. Tra2β knockdown in human lung adenocarcinoma cells (A549) increased their sensitivity to anticancer drugs. Taken together, our findings suggest that Tra2β regulates apoptosis by modulating Bcl-2 expression through its competition with miR-204. This novel function may have a crucial role in tumor growth.
Transformer 2β (Tra2β) is a prototypical serine/arginine-rich (SR)-like protein splicing factor (SRSF) that is ubiquitously expressed in metazoan genomes.1,2 The human TRA2β gene consists of 10 exons and generates five mRNA isoforms (TRA2β1-TRA2β5) through alternative splicing.3 A functional, full-length Tra2β protein encoded by TRA2β1 mRNA contains two SR domains separated by one RNA recognition motif (RRM).4 Tra2β maintains protein–protein interactions with other SR-containing proteins through the SR domains.5, 6, 7 The SR domains also interact with RNA and support RNA–RNA base pairing.8 RRM is mainly responsible for the specific interaction with RNA. A key priority to understand the biological functions of Tra2β is to identify target RNAs and their associated pathways. Grellscheid et al.9 used high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) and determined the specific G/A-rich motifs as frequent targets in the mouse genome. Cléryl et al.10 demonstrated that Tra2β RRM specifically recognized a 5′-AGAA-3′ motif in the target RNAs. Tra2β is considered to bind directly to the target RNA sequences and to activate splicing inclusion of alternative exons. One well-known target is the survival motor neuron 1 (SMN1) pre-mRNA. Tra2β specifically binds to the splicing enhancer within exon 7 of SMN1 pre-mRNA and facilitates inclusion of exon 7 in the mature SMN1 mRNA in neuroblastoma cell lines.11 Similarly, Tra2β as well as SRSF1 (also known as ASF/SF2) and SRSF9 (SRp30c) promote inclusion of exon 10 of tau pre-mRNA.12,13 Tra2β also facilitates alternative splicing of the CD44 gene via binding to CD44 exons v4 and v5, which is associated with breast cancer progression.14,15 Alternative splicing is regulated in a developmental stage- or tissue-specific manner.3 As Tra2β-deficient mice resulted in early embryonic lethality around E7.5, Tra2β-mediated regulation of alternative splicing was considered to be essential for mouse embryogenesis and spermatogenesis.16 On the other hand, deletion of TRA2β in murine embryonic fibroblasts derived from mice carrying a human SMN2 transgene on murine Smn-null background did not change the splicing pattern of SMN2. This finding suggests that in addition to the regulation of alternative splicing, Tra2β, similar to SRSF family members, has wide-ranging roles in gene expression.
SRSFs regulate transcription and post-splicing processes, including chromatin modification, transcription elongation, mRNA export, translation, and protein modification.17, 18, 19, 20 For example, SRSF1 associates with interphase chromatin, and small interference RNA (siRNA)-mediated knockdown of SRSF1 causes retention of heterochromatin protein 1 on mitotic chromatin.18 SRSF1 promotes translation initiation by suppressing the activity of eukaryotic translation initiation factor 4E-binding protein, a competitive inhibitor of cap-dependent translation.19 SRSF1 directly interacts with the primary miR-7 transcript and promotes the maturation of miR-7 by facilitating cleavage by Drosha in a splicing-independent manner.21 This finding suggests potential coordination between splicing factors and microRNA (miRNA) (miRNA)-mediated gene repression in gene regulatory networks. Similar to the SRSF proteins, Tra2β may regulate both transcription and post-transcriptional processes.
Tra2β is overexpressed in lung, cervical, and ovarian cancers, and is thought to have an important role in their growth.15,22,23 We also reported that Tra2β was expressed preferentially in the proliferative compartment of normal human colonic glands and adenocarcinomas, and that knockdown of Tra2β facilitated apoptosis of human colon cancer cells.24 However, the precise role of Tra2β in tumor growth still remains unclear. To better understand how Tra2β regulates apoptosis of cancer cells, we used mRNA–protein immnoprecipitation and microarray analyses to investigate potential mRNA targets of Tra2β. We identified B-cell CLL/lymphoma 2 (BCL2) mRNA as a potential target responsible for the regulation of apoptosis. Tra2β regulated turnover of BCL2 mRNA by competing with miR-204 for binding to the 3′ UTR. This novel splicing-independent function underscores a potential role of Tra2β in tumor growth.
Results
Identification of Tra2β-associated mRNAs
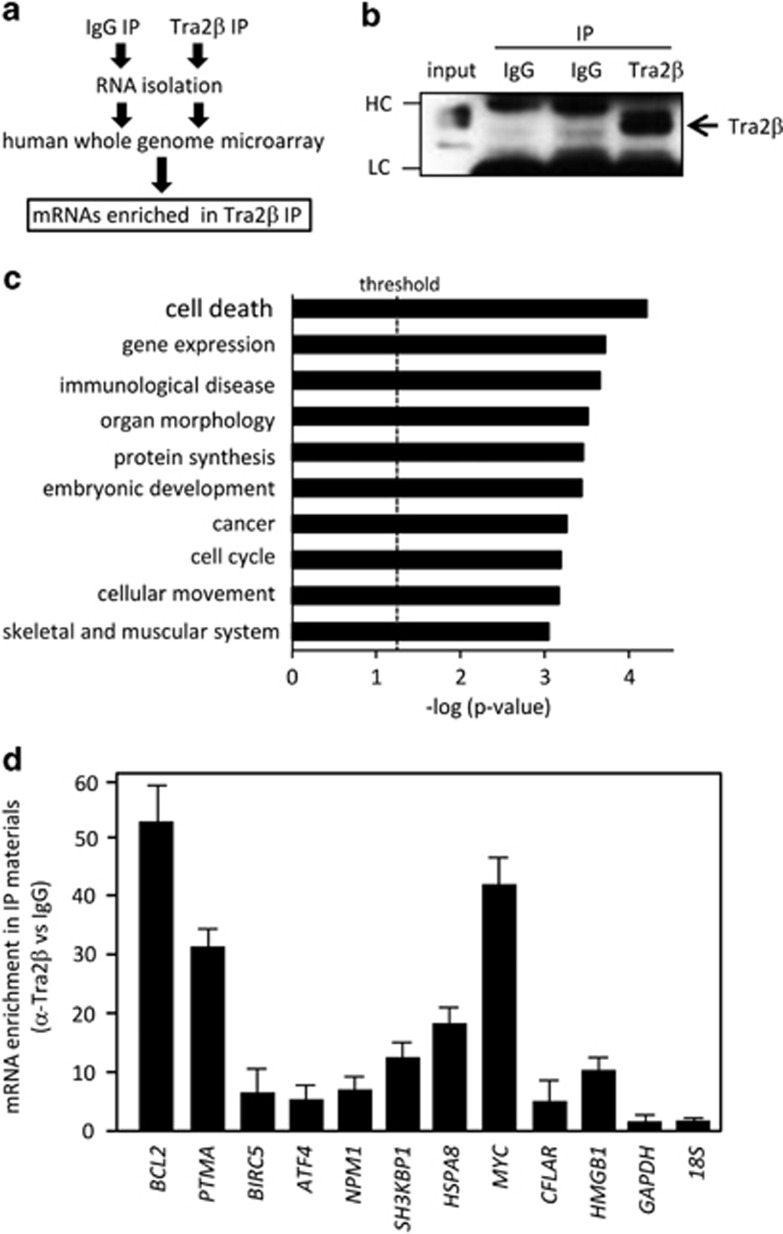
RNA immunoprecipitation (RIP) using an anti-Tra2β antibody (Tra2β immunoprecipitation (IP)) or control IgG (IgG IP) was used to identify Tra2β-associated mRNAs in HCT116 cells (Figure 1a). The antibody effectively immunoprecipitated endogenous Tra2β (Figure 1b). Total RNA was isolated from the Tra2β IP or IgG IP samples and subjected to transcriptome analysis using a human whole-genome microarray. From this analysis, we selected 470 genes in total, whose fluorescence intensities in Tra2β IP samples were >100- and also >5-fold higher than those in IgG IP samples. The raw and normalized values for these samples by microarray analysis were deposited in the Gene Expression Omnibus database (accession number: GSE60904). Biological processes or molecular functions related to the selected mRNAs were analyzed using Ingenuity Pathway Analysis. Consistent with the previous finding that Tra2β knockdown induced apoptosis of HCT116 cells,24 ‘Cell death (P=4.46E−5)' was the top-scored biological function related to the Tra2β-associated mRNAs (Figure 1c). Among the cell death-associated genes extracted, we selected BCL2, BIRC5, PTMA, ATF4, NPM1, SH3KBP1, HSPA8, MYC, CASP8, CFLAR, and HMGB1, and then used quantitative real-time reverse transcription PCR (qPCR) to measure the amounts of these mRNAs in the Tra2β IP and IgG IP samples. The primer sequences used for qPCR are listed in Supplementary Table S1. The results shown in Figure 1d are the fold enrichment of expression in the Tra2β IP samples relative to expression in the IgG IP samples (Figure 1d). The fold enrichment of expression of GAPDH mRNA or 18S RNA between the Tra2β IP and IgG IP samples was used as a negative control. Among the mRNAs that we analyzed, BCL2 mRNA was most abundantly recovered from the Tra2β IP samples, and its levels in Tra2β IP were >50-fold higher in Tra2β IP samples than in the IgG IP samples (Figure 1d).
Figure 1.
Identification of Tra2β target mRNAs by RNA immunoprecipitation and microarray analysis. (a) Lysates were prepared from HCT116 cells and subjected to IP assays with an anti-Tra2β antibody or normal rabbit IgG as described in Materials and Methods section. (b) The relative amount of Tra2β protein the anti-Tra2β IP or IgG IP materials was measured by western blotting. (c) mRNAs in the Tra2β IP materials were subjected to Ingenuity Pathway Analysis. The top 10-scored biological functions are listed. The level of significance was set at a P-value of 0.05 by the Fisher's exact test. (d) The abundance of mRNAs present in the Tra2β IP materials after the RIP assay was validated using qPCR with GAPDH and 18S as background controls. The values shown are the mean±S.D. (n=3). IP, immunoprecipitation; HC, heavy chain; LC, light chain
Binding of Tra2β to BCL2 3′ UTR
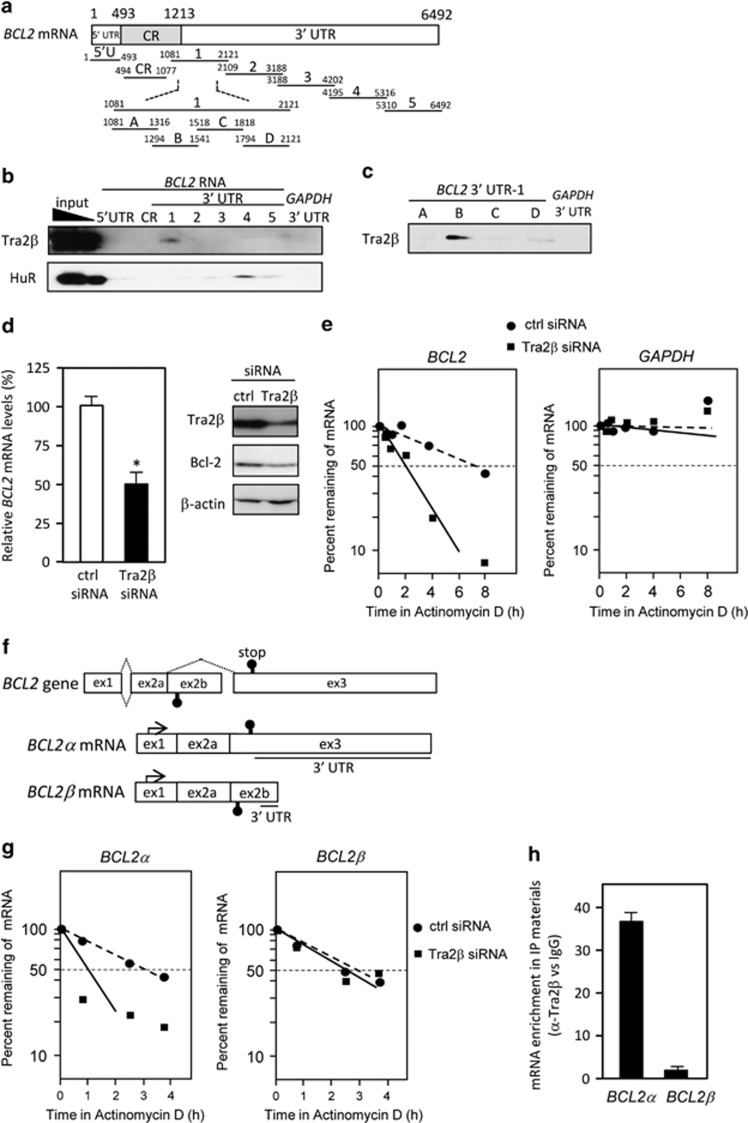
To confirm the association of Tra2β with BCL2 mRNA, we prepared biotinylated transcripts spanning the 3′ UTR, coding region (CR), or 5′ UTR of BCL2 mRNA (Figure 2a). The 3′ UTR was subdivided into five overlapping fragments of ~1000 nucleotides (nt) in length (these fragments are referred to as 3′ UTR -1 to -5). After the biotinylated RNAs were incubated with lysates from HCT116 cells, the RNA–protein complexes were precipitated with streptavidin-coated beads, and the precipitated Tra2β was examined by western blotting. As shown in Figure 2b, Tra2β specifically interacted with 3′ UTR-1, but not with CR or 5′ UTR of BCL2 mRNA or with biotinylated 3′ UTR of GAPDH mRNA (Figure 2b). BCL2 mRNA bears an AU-rich 3′ UTR and was reported to be a target of an RNA-binding protein, ELAV-like protein 1 (HuR).25,26 Consistent with this report, HuR bound to BCL2 3′ UTR-4 (Figure 2b). We further divided 3′ UTR-1 into four overlapping fragments (3′ UTR-A, -B, -C, and -D) and found that Tra2β predominantly bound to the region within 3′ UTR-B (nt 1294–1541; Figure 2c).
Figure 2.
Tra2β binds to BCL2 3′ UTR. (a) Schema of the 5′ UTR, coding region (CR), and 3′ UTR fragments of BCL2 mRNA that were used for in vitro binding assays. (b) A biotin pull-down assay was carried out using lysates prepared from HCT116 cells and biotinylated RNA fragments. RNA–protein complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal), and bound Tra2β or HuR was detected by western blotting. (c) After biotin pull-down using biotinylated fragments of the 3′ UTR of BCL2, bound Tra2β was analyzed by western blotting. (d, left) Forty-eight hours after transfection with 10 nM control (ctrl) or TRA2β siRNA, the amounts of BCL2 and GAPDH mRNA were determined by qPCR. The values shown are the mean±S.D. (n=5). *Significantly decreased compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (d, right) After treatment of HCT116 cells with 10 nM control or TRA2β siRNA for 48 h, levels of Tra2β and Bcl-2 were measured by western blotting. β-actin was used as a loading control. (e) After HCT116 cells were transfected with control or TRA2β siRNA for 48 h, they were treated with actinomycin D (2.5 μg/ml) for the indicated times. BCL2 and GAPDH mRNA levels were measured by qPCR. (f) Schema of two BCL2 mRNA isoforms, BCL2α and BCL2β. The arrow indicates the transcriptional start site. The 3′ UTRs of these isoforms are underlined. (g) After a 48-h transfection of control or TRA2β siRNA, HCT116 cells were treated with actinomycin D (2.5 μg/ml) for the indicated times. BCL2α and BCL2β mRNA levels were measured by qPCR and the percentage of mRNA that remained was plotted. (h) qPCR was used to measure the abundance of BCL2α or BCL2β mRNAs present in the Tra2β-IP materials after the RIP assay was measured by qPCR
Next, we examined whether Tra2β knockdown affected the stability of BCL2 mRNA. Treatment of HCT116 cells with 10 nM Tra2β siRNA for 48 h effectively reduced Tra2β protein levels (Figure 2d, right panel). At the same time, Tra2β knockdown decreased the expression of both BCL2 mRNA (Figure 2d, left panel) and Bcl-2 protein (Figure 2d, right panel). As shown in Figure 2e, the rate of decay for BCL2 mRNA in Tra2β siRNA-treated cells in the presence of actinomycin D (t1/2=2.1±0.3 h) was faster than that of control siRNA-treated cells (t1/2=7.5±0.6 h). To exclude the possibility that Tra2β might change the efficiency of transcription of BCL2, we used a luciferase reporter assay and confirmed that Tra2β knockdown did not affect the activity of the BCL2 promoter (Supplementary Figure S1). The BCL2 gene is alternatively spliced to generate long (BCL2α) and short (BCL2β) mRNA isoforms (Figure 2f). Full-length and functional Bcl-2 protein is translated from BCL2α mRNA. We compared alternative splicing events between Tra2β and control siRNA-treated cells using the GeneChip Human Exon 1.0 ST Array (Affymetrix, Santa Clara, CA, USA). The splicing indices (SIs) of BCL2 were not affected by Tra2β knockdown (SI<1.0 and P>0.05). Treatment with a Tra2β siRNA accelerated the decay of BCL2α mRNA containing full-length 3′ UTR, while it did not modify the stability of BCL2β mRNA having short, alternatively spliced 3′ UTR (Figure 2g). RIP using an anti-Tra2β antibody determined that Tra2β associated with BCL2α more specifically than BCL2β mRNA (Figure 2h). These results suggest that Tra2β may stabilize BCL2 mRNA by binding to BCL2α 3′ UTR.
Tra2β stabilizes BCL2 mRNA
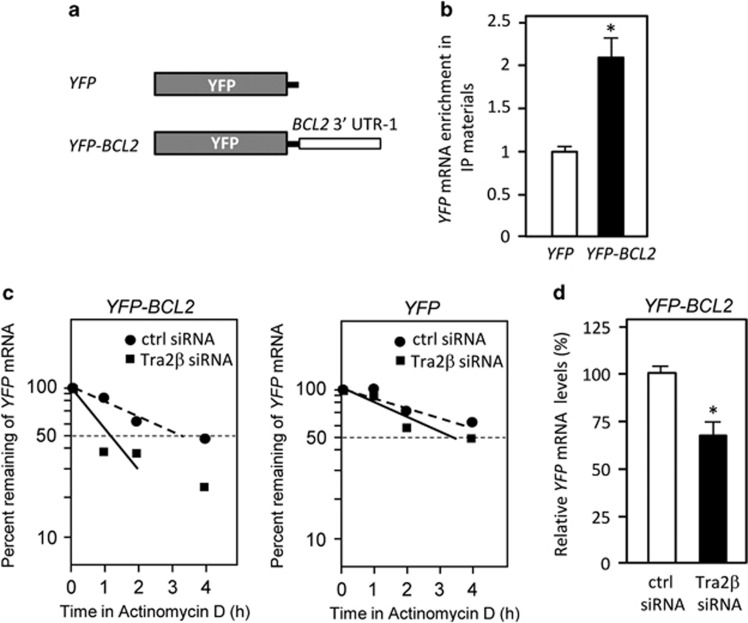
To further confirm that BCL2 mRNA is stabilized by associating with Tra2β (Figure 2b), we prepared an YFP reporter construct of a chimeric RNA that spanned the YFP gene and BCL2 3′ UTR-1 (YFP-BCL2; Figure 3a), and transfected HCT116 cells with this construct. RIP assay demonstrated the association between Tra2β and the chimeric RNA containing BCL2 3′ UTR-1 in the transfected cells (Figure 3b). The rate of YFP mRNA degradation in the presence of actinomycin D was monitored by qPCR. Tra2β silencing significantly shortened the half-life of the chimeric YFP-BCL2 3′ UTR-1 mRNA (Figure 3c, left panel) and significantly decreased YFP mRNA levels (Figure 3d), but it did not change the degradation rate of YFP mRNA lacking BCL2 3′ UTR (Figure 3c, right panel). Together, these findings suggest that Tra2β stabilizes BCL2 mRNA through interacting with BCL2 3′ UTR-1.
Figure 3.
Tra2β regulates Bcl-2 expression through BCL2 3′ UTR. (a) Constructs were prepared to express chimeric RNAs spanning the YFP CR and BCL2 3′ UTR-1 (nt 1213–2121 of BCL2α 3' mRNA) as described in Materials and Methods section. (b) After a 24-h transfection with the YFP or YFP-BCL2 construct, the amounts of Tra2β binding to the chimeric RNAs were analyzed by RIP assay followed by measurement of YFP mRNA levels by qPCR. *Significantly increased compared with cells transfected with the YFP construct (P<0.05 by unpaired Student's t-test). (c) After HCT116 cells were treated with control (ctrl) or TRA2β siRNA for 24 h, the YFP or YFP-BCL2 vector was transfected for 24 h. These cells were then treated with actinomycin D (2.5 μg/ml) for the indicated times. YFP mRNA levels were measured by qPCR and the percentage of YFP mRNA that remained was plotted. (d) After the cells were treated with control or TRA2β siRNA for 24 h, the YFP-BCL2 construct was transfected and then YFP mRNA levels were measured by qPCR. *Significantly decreased compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test)
Binding of miR-204 to BCL2 3′ UTR
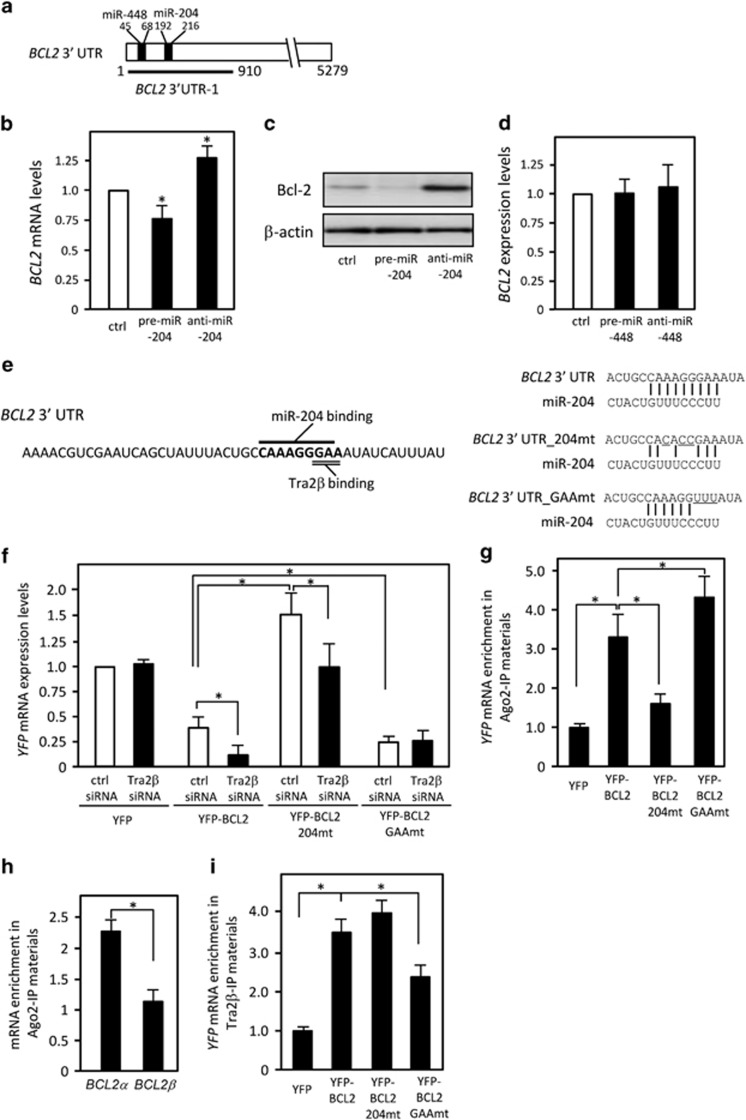
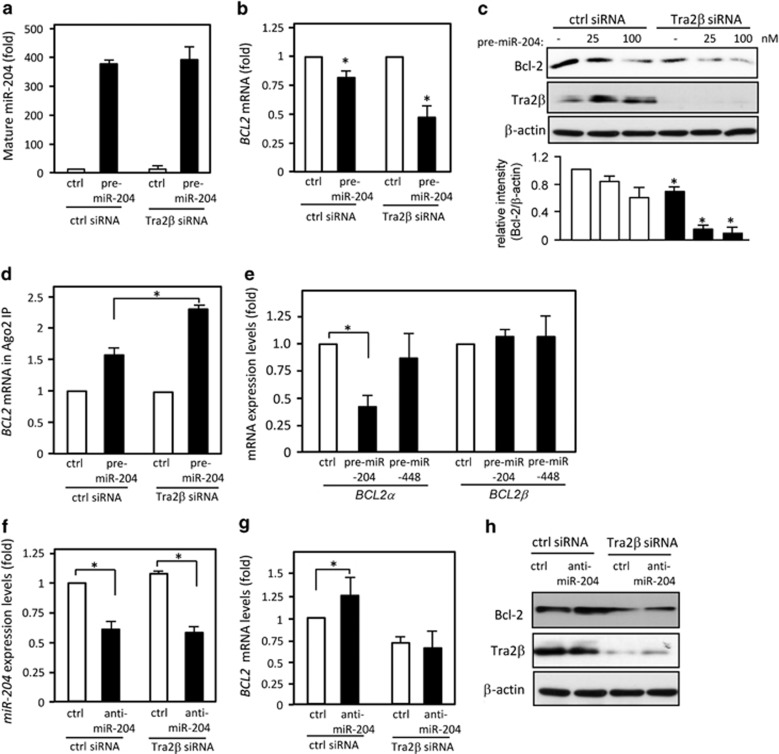
We used the TargetScan (http://www.targetscan.org/) and miRNA org (http://www.microrna.org/microrna/) programs to search for miRNAs that could potentially interact with BCL2 3′ UTR-1. Both programs cited putative binding sites for miR-448 (nt 1258–1281) and miR-204 (nt 1405–1429; Figure 4a). One miR-204-binding site lies within the 3′ UTR-B region (nt 1294–1541). Transfection of precursor miR-204 (pre-miR-204) reduced BCL2 mRNA (Figure 4b) and Bcl-2 protein (Figure 4c) levels. Conversely, transfection of an antisense RNA complementary to miR-204 (anti-miR-204) increased their levels (Figures 4b and c). In contrast, overexpression or reduction of miR-448 did not lead to changes in the levels of BCL2 mRNA (Figure 4d). We also confirmed that neither overexpression nor knockdown of miR-204 affected TRA2β1 mRNA levels (Supplementary Figures S2a and b), and that Tra2β knockdown did not affect miR-204 levels (Supplementary Figure S2c).
Figure 4.
miR-204 targets BCL2 3′ UTR. (a) Schema of miR-204- and miR-448-binding sites on the BCL2 3′ UTR. (b) After a 48-h transfection of 25 nM control (ctrl), precursor (pre)-miR-204, or anti-miR-204, the amounts of BCL2 and GAPDH mRNA were determined by qPCR. The values are mean±S.D. (n=5). *Significantly decreased compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (c) Whole-cell lysates were prepared from HCT116 cells after treatment with control, pre-miR-204, or anti-miR-204 for 48 h. The expression levels of Bcl-2 and β-actin were measured by western blotting. (d) After a 48-h transfection of control, pre-miR-448, or anti-miR-448, the amounts of BCL2 and GAPDH mRNA were determined by qPCR. (e) YFP reporter plasmids with BCL2 3′ UTR-1 containing the intact miR-204-binding site (YFP-BCL2), a mutated miR-204 site (YFP-BCL2_204mt), or a mutated GAA site (YFP-BCL2_GAAmt) were generated. The mutated sequences are underlined. (f) After the cells were treated with control or TRA2β siRNA for 24 h, each construct was transfected and then YFP mRNA levels were measured by qPCR. *Significant changes compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (g) YFP mRNA enrichment in Ago2-IP materials was analyzed by RIP assay with an anti-Ago2 antibody followed by qPCR. *Significant changes compared with the cells transfected with the YFP construct (P<0.05 by unpaired Student's t-test). (h) qPCR was used to measure the abundance of BCL2α or BCL2β mRNAs present in the Ago2-IP materials after the RIP assay. *Significant changes compared with the cells transfected with the YFP construct (P<0.05 by unpaired Student's t-test). (i) The interaction between Tra2β and YFP mRNA was measured by RIP assay followed by qPCR. Values are mean±S.D. (n=3). * Significant changes compared with YFP-transfected cells (P<0.05 by unpaired Student's t-test)
To precisely examine the interaction between Tra2β and miR-204 on BCL2 3′ UTR-1, we prepared reporter plasmids that expressed chimeric RNAs containing the sequence for YFP and one of two mutated BCL2 3′ UTR-1 sequences. One mutant, YFP-BCL2_204mt, harbored three-point mutations within the binding site for miR-204; the other, YFP-BCL2_GAAmt, contained three-point mutations that replaced the consensus Tra2β-binding motif (GAA) with UUU (Figure 4e). In control siRNA-treated cells, the amount of YFP mRNA produced by the YFP-BCL2 3′ UTR-1 construct was significantly less than that of the control YFP construct, probably because of regulation by miR-204 (Figure 4f). Knockdown of Tra2β additionally enhanced the BCL2 3′ UTR-1-dependent reduction of YFP mRNA levels (Figure 4f). In contrast, the reduction was eliminated and levels of YFP mRNA were increased in cells expressing YFP-BCL2_204mt (Figure 4f). Tra2β knockdown also decreased the amount of YFP mRNA produced by the YFP-BCL2_204mt construct (Figure 4f). The mutations in the GAA site (YFP-BCL2_GAAmt) significantly reduced YFP mRNA levels and eliminated the effect of Tra2β siRNA (Figure 4f). RIP assays with an anti-Argonaute 2 (Ago2) antibody showed that miR-204 actually bound to BCL2 3′ UTR-1, and this binding was eliminated by the mutations in YFP-BCL2_204mt that altered of the miR-204-binding site (Figure 4g). The miR-204 seed sequence is partially overlapped with the GAA sequence (Figure 4e). We therefore examined whether the GAA mutation (YFP-BCL2_GAAmt) also disrupted miR-204 binding to BCL2 3′ UTR. Ago2-IP experiments demonstrated that YFP-BCL2_GAAmt rather enhanced its association with miR-204 (Figure 4g). Thus, the GAA mutation did not likely disrupt miR-204 binding to BCL2 3′ UTR. As shown in Figure 4h, miR-204 bound to BCL2α, but not BCL2β, mRNA. We also confirmed that BCL2_GAAmt significantly blocked Tra2β binding to the BCL2 3′ UTR (Figure 4i). These results suggest that the binding sites of Tra2β and miR-204 overlap and these two factors competitively regulate degradation of BCL2α mRNA.
Antagonistic regulation of Bcl-2 expression by Tra2β and miR-204
To further confirm the competitive effect of Tra2β and miR-204 on the expression of Bcl-2, we examined the effects of Tra2β siRNA in HCT116 cells overexpressing miR-204 (Figure 5a). Overexpression of miR-204 significantly decreased BCL2 mRNA levels (Figure 5b). Tra2β knockdown in cells transfected with pre-miR-204 enhanced the overexpressed miR-204-mediated degradation of BCL2 mRNA (Figure 5b) and resulted in decreased levels of Bcl-2 protein (Figure 5c). The miR-204 overexpression facilitated the association between Ago2 and BCL2 mRNA, and this association was further enhanced by silencing of Tra2β (Figure 5d). We also confirmed the specific downregulation of the BCL2α mRNA isoform by overexpression of miR-204, but not miR-448 (Figure 5e).
Figure 5.
Co-regulation of Bcl-2 expression by Tra2β and miR-204. (a and b) After treatment with 10 nM control (ctrl) or TRA2β siRNA for 24 h, HCT116 cells were transfected with 25 nM control or pre-miR-204 was transfected for 24 h. The levels of mature miR-204 and BCL2 mRNA were measured by qPCR. *Significant changes compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (c) After treatment of HCT116 cells with 10 nM control or TRA2β siRNA for 24 h, they were transfected with the indicated concentrations of pre-miR-204 for 24 h. The levels of Tra2β and Bcl-2 were measured by western blotting. β-actin was used as a loading control. Bcl-2 signals were quantified by densitometry. Changes in relative intensities (Bcl-2/β-actin) are shown in the lower panel. Values are mean±S.D. from three independent experiments. *Significant changes compared with cells treated with control siRNA (P<0.05 by unpaired Student's t-test). (d) HCT116 cells were treated as in a and b. RIP assay was performed using an anti-Ago2 antibody or control IgG and lysates from cells transfected with the indicated oligonucleotides, and then BCL2 mRNA levels were measured by qPCR. *Significant changes compared with cells transfected with the YFP construct (P<0.05 by unpaired Student's t-test). (e) HCT116 cells were treated with 10 nM control or TRA2β siRNA for 24 h, and then transfected with 25 nM pre-miR-204 or pre-miR-448. BCL2α and BCL2β mRNA levels were measured by qPCR. *Significant change compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (f and g) After HCT116 cells were treated with 10 nM control or TRA2β siRNA for 24 h, they were transfected with 25 nM control or anti-miR-204 was transfected for 24 h. The levels of mature miR-204 and BCL2 mRNA were measured by qPCR. *Significant changes compared with control siRNA-treated cells (P<0.05 by unpaired Student's t-test). (h) HCT116 cells were treated as in f and g. The levels of Tra2β and Bcl-2 were measured by western blotting. β-actin was used as a loading control
Next, we examined effects of anti-miR-204. Transfection with anti-miR-204 reduced miR-204 levels by 50% in cells treated with control or Tra2β siRNA (Figure 5f). Anti-miR-204 significantly increased levels of BCL2 mRNA (Figure 5g) and Bcl-2 protein (Figure 5h) in control siRNA-treated cells. However, the anti-miR-204-mediated increases in BCL2 mRNA (Figure 5g) and Bcl-2 protein (Figure 5h) levels were canceled when Tra2β was silenced. This discrepancy suggests that the regulatory mechanism for Bcl-2 expression may be more complex than simply competitive binding between Tra2β and miR-204. However, our results suggest that Tra2β may regulate Bcl-2 expression at least in part by acting as a competitive inhibitor of miR-204.
Regulation of Bcl-2 expression by Tra2β
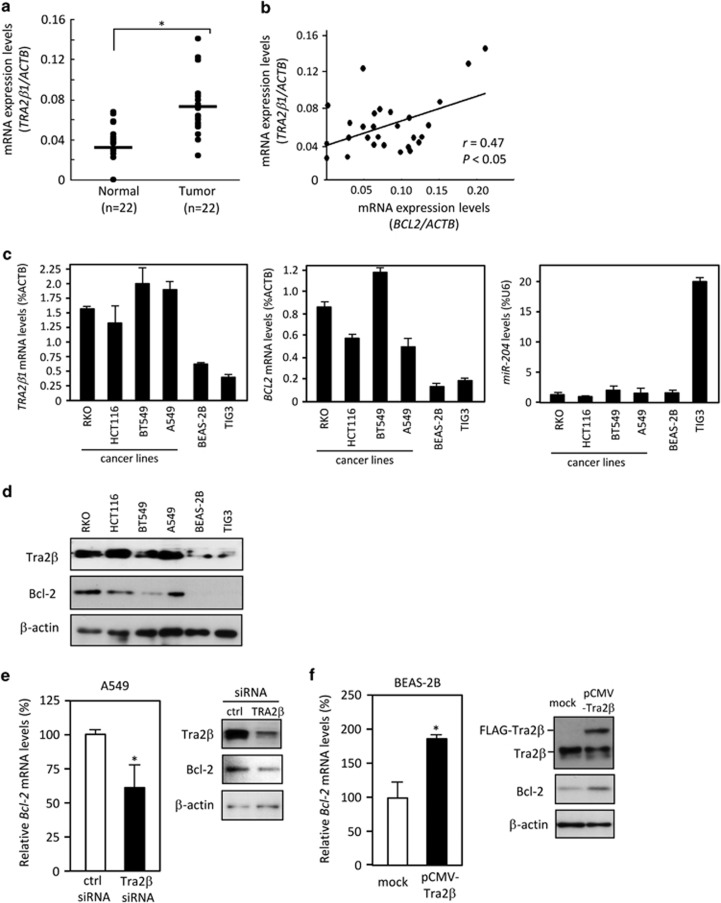
Using qPCR, we measured the expression of TRA2β1 mRNA in cDNA libraries prepared from 22 patients with colon cancer (HCRT103; OriGene, Rockville, MD, USA). As shown in Figure 6a, colon cancer tissues expressed significantly higher levels of TRA2β1 mRNA than did paired normal tissues. The correlation between TRA2β1 and BCL2 expression in qPCR arrays of the colon tissues was analyzed by determining the Pearson product moment correlation coefficients. The levels of TRA2β1 mRNA were positively correlated with those of BCL2 mRNA in human colon tissues and cancers (Figure 6b).
Figure 6.
Tra2β regulates Bcl-2 levels. (a) Using TissueScan Tissue qPCR Arrays (HCRT103), TRA2β1 and ACTB mRNA levels in cDNAs prepared from tumors and paired normal tissues of 22 patients with adenocarcinomas of the colon were measured by qPCR. (b) The correlation between TRA2β1 and BCL2 expression in the array of the colon was analyzed by determining the Pearson product moment correlation. (c) The amounts of TRA2β1 and BCL2 mRNAs in colon cancer cell lines (HCT116 and RKO), breast cancer cells (BT549), lung carcinoma cells (A549), lung epithelial cells (BEAS-2B), and diploid fibroblasts (TIG-3) were measured by qPCR. ACTB mRNA was used as an endogenous quantity control. The values shown are the mean±S.D. from three independent experiments. (d) Whole-cell lysates were prepared from the indicated cell lines, and the amounts of Tra2β and Bcl-2 were measured by western blotting. β-actin was using as a loading control. (e, left) After A549 cells were treated with 10 nM of the indicated siRNA for 48 h, changes in the expression of BCL2 mRNA were measured by qPCR. GAPDH mRNA was used as an endogenous quantity control. The values (mean±S.D., n=3) are expressed as fold changes, compared with those of control siRNA-treated cells. *P<0.05 by unpaired Student's t-test. (e, right) The amounts of Tra2β and Bcl-2 proteins in these cells were measured by western blotting using β-actin a loading control. (f, left) After BEAS-2B cells were transfected with of pCMV (mock) or pCMV-Tra2β for 24 h, changes in the expression of BCL2 mRNA were measured by qPCR. GAPDH mRNA was used as an endogenous quantity control. The values (mean±S.D., n=3) are expressed as fold changes, compared with those of mock-treated cells. *P<0.05 by unpaired Student's t-test. (f, right) The amounts of Tra2β and Bcl-2 proteins in these cells were measured by western blotting. β-actin was used as a loading control
Both mRNA and protein levels of Tra2β and Bcl-2 were also monitored in colon cancer cells (RKO and HCT116), breast cancer cells (BT549), lung cancer cells (A549), lung epithelial cells (BEAS-2B), and diploid fibroblasts (TIG-3). Among these cell lines, cancer cells expressed higher levels of both TRA2β1 and BCL2 mRNAs, whereas the expression of miR-204 was downregulated (Figure 6c). Tra2β and Bcl-2 proteins were overexpressed in all of the cancer cell lines tested (Figure 6d). This limited survey suggests that Tra2β may contribute to Bcl-2 overexpression in cancer cells. In fact, when Tra2β siRNA was transfected into A549 cells, which expressed higher amounts of Tra2β (Figure 6d), it reduced the expression of BCL2 mRNA and Bcl-2 protein as effectively as it reduced that of TRA2β mRNA and Tra2β protein (Figure 6e). In contrast, transient expression of FLAG-tagged Tra2β at a level that was similar to that of endogenous Tra2β in BEAS-2B cells, which expressed limited amounts of Tra2β (Figure 6d), led to doubling in the levels of BCL2 mRNA (Figure 6f). Consequently, BEAS-2B cells increased expression of Bcl-2 protein (Figure 6f).
Tra2β knockdown increases apoptosis in A549 cells
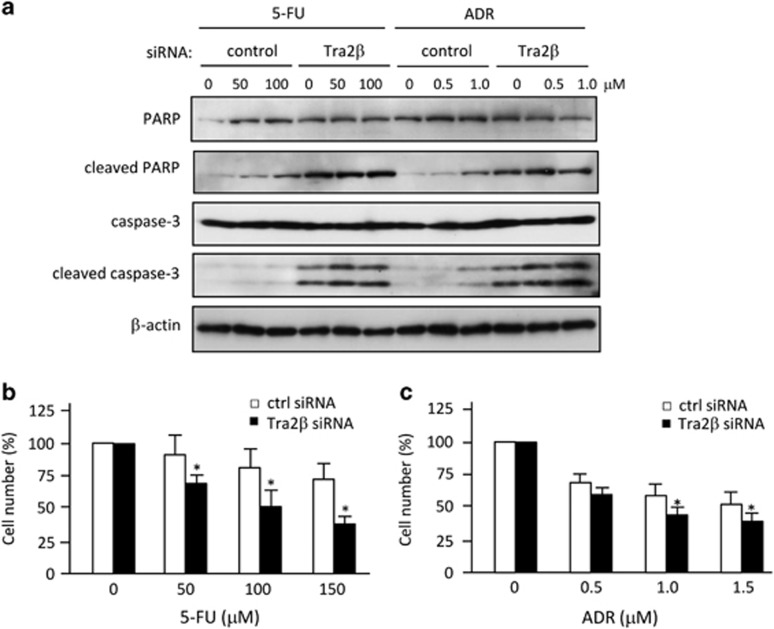
We used A549 lung carcinoma cells to examine whether the decline of Bcl-2 induced by Tra2β knockdown actually increased the susceptibility to apoptotic cell death. Treatment with Tra2β siRNA promoted activation of caspase 3 and cleavage of poly (ADP-ribose) polymerase (PARP; Figure 7a). Moreover, exposure of the Tra2β siRNA-treated cells to 5-fluorouracil (5-FU) or adriamycin (ADR) further enhanced the cleavages of caspase 3 and PARP (Figure 7a). As a result, the numbers of cells that survived after treatment with 5-FU (Figure 7b) or ADR (Figure 7c) were significantly less in the Tra2β siRNA-treated cells than in the control siRNA-treated cells. These results suggest that Tra2β function may be one of the crucial regulators of the anti-apoptotic properties of Bcl-2 in various types of cancer cells.
Figure 7.
Tra2β knockdown increases susceptibility to anticancer drugs. (a) Whole-cell lysates were prepared from A549 cells that had been treated with 10 nM control or Tra2β siRNA for 48 h. The levels of unprocessed or cleaved caspase 3 and PARP were measured by western blotting. β-actin was used as a loading control. (b and c) A549 cells were treated with control (ctrl) or Tra2β siRNA for 36 h, and then these cells were exposed to the indicated concentrations of 5-fluorouracil (FU) or adriamycin (ADR) for 24 h. Subsequently, growing cells were harvested and counted. The values shown are the mean±S.D. (n=3). *Significantly different by ANOVA and the Bonferroni test (P<0.05)
Discussion
The present study suggests that Tra2β functions as an RNA-binding protein to regulate the turnover of distinct target mRNAs. In agreement with the previous finding that Tra2β knockdown induced apoptosis in HCT116 cells,24 microarray analysis of Tra2β-associated mRNAs isolated by Tra2β-RNA IP identified a group of mRNAs encoding cell death-related proteins. Among the potential mRNA targets for Tra2β, BCL2 mRNA may be a critical regulator of the apoptosis induced by Tra2β knockdown. Knockdown of SRSF1 is known to affect the alternative splicing of pre-mRNAs of several BCL2 family members to yield their proapoptotic forms.27 Although Tra2β knockdown did not change the alternative splicing pattern and promoter activity of BCL2, it decreased BCL2 mRNA levels through accelerating the degradation of BCL2α, but not BCL2β mRNA. BCL2α mRNA encodes functional Bcl-2 protein and contains the full-length 3′ UTR, whereas BCL2β mRNA isoform contains a short, alternatively spliced 3′ UTR, whose sequence differs from that of the full-length 3′ UTR. Tra2β bound to the consensus Tra2β-binding motif (GAA) within the miR-204-binding site of BCL2α 3′ UTR and stabilized the transcript.
Post-transcriptional stabilization of mRNA is regulated by RNA-binding proteins and non-coding RNAs. miRNAs associate with the RNA-induced silencing complex (RISC) and recognize target mRNAs containing 3′ UTRs with partially complementary sequence.28 RNA-binding proteins influence mRNA stability and translation by interacting mainly with the 3′ UTR. Nucleolin and heterogeneous nuclear ribonucleoprotein E associate with the 3′ UTR of beta-globin mRNA and extend its half-life.29 Several RNA-binding proteins (HuR, nuclear factor 90, tristetraprolin, butyrate response factor 1, and K homology-type splicing regulatory protein) preferentially interact with U/AU-rich sequences in the 3′ UTR of the target transcripts and regulate their stability, whereas nucleolin preferentially binds to G-rich sequences.30 SR proteins also participate in the regulation of mRNA turnover. For example, SRSF1 selectively enhances the decay of target mRNAs by binding to their 3′ UTRs. 31 SRSF2 (SC35) not only promotes inclusion of tau exon 10 but also stabilizes the tau mRNA.32 However, further evidence is needed to fully understand how SR or SR-like proteins regulate RNA turnover.
Several lines of evidence suggest that RNA-binding proteins post-transcriptionally regulate target mRNAs via the joint influence of miRNAs. For instance, HuR binds to the 3′ UTR of Topoisomerase IIα mRNA and increases its translation by antagonizing the binding of miR-548-3p.33 In contrast, HuR downregulates c-Myc expression by recruiting let-7-loaded RISC to the MYC 3′ UTR.34 Thus, when miRNA-binding sites overlap with or present near binding sites for RNA-binding proteins, RNA-binding proteins could either compete or cooperate with miRNAs via their physical interactions. In a recent high-throughput study of HuR-mRNA interactions using photoactivatable-ribonucleoside-enhanced crosslinking and IP, Lebedeva et al.35 observed that miRNA-binding sites are preferentially located toward the boundaries of 3′ UTR, whereas HuR-binding sites are distributed uniformly along the 3′ UTR, with the exception of the regions surrounding the stop codon and the polyadenylation site.35,36 Bioinformatic analysis has revealed that most of the miRNA-binding sites are found in the immediate vicinity of the HuR-binding sites.35 Direct competition between miRNAs and HuR for the overlapping binding sites is thought to be possible, whereas in the context of non-overlapping sites, competition could occur by steric hindrance or by non-steric hindrance involving changes in the secondary structure of the RNA. Tra2β is likely to use at least one GAA sequence within the miR-204-binding site to bind to the BCL2 3′ UTR and may compete with miR-204 to regulate the stability of BCL2 mRNA.
Tra2β likely has wide-ranging roles in gene expression, and the regulatory mechanism for Bcl-2 expression may be more complex than simply competitive interaction between Tra2β and miR-204. However, the competitive interactions between Tra2β and miR-204 may provide new insight into the regulation of BCL2 expression in colon cancer cells. Importantly, the increased levels of BCL2 mRNA could act in concert with other oncogenes such as TRA2β to inhibit cancer cell apoptosis. In fact, the levels of BCL2 and TRA2β1 mRNAs were significantly elevated in colon cancers and several cancer cell lines, and TRA2β1 levels were positively correlated with those of BCL2 mRNA in cancerous tissues. Knockdown and overexpression experiments demonstrated that BCL2 expression is regulated by Tra2β in various cancer cell lines. In addition, downregulation of miR-204 has been documented in several types of cancers. 37, 38, 39, 40, 41 Decreased expression of miR-204 results in overexpression of its target, myeloid cell leukemia sequence 1 (Mcl-1) mRNA, and induces anti-apoptotic signaling in pancreatic cancers.42 miR-204 also regulates expression of an oncogene, neurotrophic receptor tyrosine kinase B, and promotes metastasis in endometrial carcinoma.43 Imam et al.44 showed that the genomic locus encoding miR-204 is frequently lost in multiple cancers, including breast and ovarian cancers, and pediatric renal tumors. They suggest that miR-204 targets distinct genes involved in tumorigenesis including BDNF. Binding to the target mRNAs is followed by activation of a small GTPase, Rac1, and actin reorganization through the AKT/mTOR signaling pathway, both of which facilitate cancer cell migration and invasion.45 These studies suggest an important role of miR-204 as a potent tumor suppressor.
Here, we show that not only miR-204 but also Tra2β levels affect post-transcriptional regulation of their target, BCL2 mRNA. Increased expression of Tra2β in cancer cells prevents decay of BCL2 mRNA by competitive inhibition of miR-204 binding to BCL2 3′ UTR. Consequently, Tra2β induces resistance to the apoptosis caused by anticancer drugs. Our findings disclose a novel function of the splicing factor Tra2β in mRNA turnover and may provide mechanistic insight into the role of Tra2β in tumor growth.
Materials and Methods
Cell culture
Human colon cancer HCT116 and RKO cells were cultured in McCoy's 5A medium (Gibco, Grand Island, NY, USA) supplemented with 5% (v/v) heat-inactivated fetal calf serum (FCS) and antibiotics. Human lung BEAS-2B and A549 cells were maintained in RPMI-1640 medium (Gibco) supplemented with 10% FCS. Human embryonic kidney 293T (HEK293T) and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS. Human diploid fibroblasts (TIG-3) cells were cultured in minimal essential medium (Gibco) supplemented with 10% FCS. All cells were cultured at 37 °C in 5% CO2.
Reporter constructs
The 3′ UTR of BCL2 mRNA was cloned into the pd2EYFP-N1 reporter vector (Clontech, Palo Alto, CA, USA). In brief, BCL2 3′ UTR-1 (1–968 nt of the 3′ UTR) was amplified by PCR using the following primer set: 5′-GCGGCCGCGATGCCTTTGTGGAACTGTACG-3′ (forward) and 5′-GCGGCCGCATGCGGAAGTCACCGAAATGTTC-3′ (reverse). The amplified fragment was subcloned into pd2EYFP-N1 (YFP) using a Not1 site located downstream of the YFP stop codon (a Not1 sequence is underlined). Three-point mutations in the miR-204-binding site or the GAA site in YFP-BCL2 were introduced by using a site-directed mutagenesis kit (Agilent Technologies, Palo Alto, CA, USA) and the following primers: YFP-BCL_204mt, 5′-GCTATTTACTGCCACACCGAAATATCATTTATTTT-3′ and 5′-AAAATAAATGATATTTCGGTGTGGCAGTAAATAGC-3′ YFP-BCL2_GAAmt, 5′- GCTATTTACTGCCAAAGGTTTATATCATTTATTTTTTAC-3′ and 5′- GTAAAAAATAAATGATATAAACCTTTGGCAGTAAATAGC-3′ (the mutated sites are underlined).
Western blotting
HCT116 cells were treated with 10 nM Tra2β siRNA (Invitrogen, Carlsbad, CA, USA) or control siRNA (AllStars; Invitrogen) for 24 or 48 h. These cells were also transfected with 25 nM pre-miR-204 or -448 (Applied Biosystems, Foster City, CA, USA) or 25 nM anti-miR-204 or -448 (Applied Biosystems) for 24 h. Whole-cell lysates were prepared using RIPA buffer (Thermo Scientific, Rockford, IL, USA) containing a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). The extracted proteins were separated by SDS-polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA). After blocking with 5% nonfat milk (Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature, the membrane was incubated with anti-Tra2β (Abcam, Cambridge, UK), anti-Bcl-2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-FLAG (Sigma-Aldrich, St. Louis, MO, USA), or anti-β-actin (Abcam) antibody overnight at 4 °C. Following incubation with an appropriate secondary antibody for 1 h at room temperature, the bound antibodies were detected with an ECL Western Blotting Detection System (GE Healthcare, Piscataway, NJ, USA). The intensities of the bound antibodies were quantified by using the Image J software.
Quantitative PCR
Total RNAs were extracted from cells using the miRNeasy mini kit (Qiagen, Valencia, CA, USA), and contaminating DNA was removed using RNase-free DNase (Qiagen). One microgram of isolated RNA was reverse-transcribed using the PrimeScript RT reagent kit (Takara, Otsu, Japan). TRA2β1, BCL2, GAPDH, and 18S mRNA levels were measured using a specific primer set and SYBR Green Master Mix (Applied Biosystems) as previously described.24 Total RNA (10 ng) was used as a template to generate specific first-strand cDNA for miRNAs by using a TaqMan-specific miRNA reverse transcription kit (Applied Biosystems). miRNA levels were normalized by using U6 snRNA as an endogenous quantity control.
Biotin pull-down assay
PCR fragments containing the T7 RNA polymerase promoter sequence were used as templates for in vitro transcription. Biotinylated transcripts were prepared using T7 polymerase (Invitrogen) and biotin-CTP (Perkin-Elmer-Cetus, Norwalk, CT, USA), and purified with ssDNA/RNA Clean & Concentrator (Zymo Research, Orange, CA, USA). Biotin pull-down assays were carried out by incubating 40 μg of cell lysates with 1 μg of biotinylated transcripts in TENT buffer (10 mM Tris-HCl buffer, pH 8.0, containing 1 mM EDTA, 250 mM NaCl, and 0.5% Triton X-100) for 1 h at room temperature. The RNA–protein complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway), and the bound proteins in the pull-down materials were analyzed by western blotting with an anti-Tra2β or anti-HuR antibody.
RIP assay
Analysis of mRNA in RIP materials was performed as described previously.34,46,47 In brief, HCT116 cells were lysed with 25 mM Tris-HCl buffer (pH 7.5) containing 150 mM NaCl, 1 mM EDTA, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol, and 100 U/ml RNase inhibitor (Promega, Madison, WI, USA), and then whole-cell extracts were incubated with protein-A Sepharose beads precoated with 3 μg anti-Tra2β antibody or control rabbit IgG for 2 h at 4 °C. After washing with NT2 buffer (50 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl2, 1 mM MgCl2, and 0.05% Nonidet P-40), the beads were incubated with 20 U of RNase-free DNase I (Invitrogen) in NT2 buffer for 15 min at 37 °C and further incubated in NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K for 20 min at 55 °C. RNA in the IP materials was measured by human whole-genome microarray (Agilent Technologies) and qPCR.
Acknowledgments
This research was supported by a research grant (nos. 22790649 and 24659370) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to Y.K) and Takeda Science Foundation.
Glossary
- Tra2β
Transformer 2β
- RIP
RNA immunoprecipitation
- SRSF
serine/arginine-rich splicing factor
- RRM
RNA recognition motif
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by C Borner
Supplementary Material
References
- Mayeda A, Zahler AM, Krainer AR, Roth MB. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci USA. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Nayler O, Cap C, Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics. 1998;53:191–202. doi: 10.1006/geno.1998.5471. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud R, Da Penha Berzaghi M, Siedler F, Hübener M, Stamm S. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur J Neurosci. 1999;11:788–802. doi: 10.1046/j.1460-9568.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci USA. 1996;93:9004–9009. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beil B, Screaton G, Stamm S. Molecular cloning of htra2-beta-1 and htra2-beta-2, two human homologs of tra-2 generated by alternative splicing. DNA Cell Biol. 1997;16:679–690. doi: 10.1089/dna.1997.16.679. [DOI] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Grellscheid S, Dalgliesh C, Storbeck M, Best A, Liu Y, Jakubik M, et al. Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2β in development. PLoS Genet. 2011;7:e1002390. doi: 10.1371/journal.pgen.1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat Struct Mol Biol. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Someya T, Kuwasako K, Takahashi M, He F, Unzai S, et al. Structural basis for the dual RNA-recognition modes of human Tra2-β RRM. Nucleic Acids Res. 2011;39:1538–1553. doi: 10.1093/nar/gkq854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watermann DO, Tang Y, Zur Hausen A, Jäger M, Stamm S, Stickeler E. Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 2006;66:4774–4780. doi: 10.1158/0008-5472.CAN-04-3294. [DOI] [PubMed] [Google Scholar]
- Fischer DC, Noack K, Runnebaum IB, Watermann DO, Kieback DG, Stamm S, et al. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–1090. [PubMed] [Google Scholar]
- Mende Y, Jakubik M, Riessland M, Schoenen F, Rossbach K, Kleinridders A, et al. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum Mol Genet. 2010;19:2154–2167. doi: 10.1093/hmg/ddq094. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Pelisch F, Gerez J, Druker J, Schor IE, Muñoz MJ, Risso G, et al. The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc Natl Acad Sci USA. 2010;107:16119–16124. doi: 10.1073/pnas.1004653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Matsuo N, Ozawa K, Hori O, Higashi T, Nishizaki J, et al. Expression of a novel RNA-splicing factor, RA301/Tra2beta, in vascular lesions and its role in smooth muscle cell proliferation. Am J Pathol. 2001;158:1685–1694. doi: 10.1016/s0002-9440(10)64124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel B, Zur Hausen A, Bouda J, Boudova L, Koprivova M, Hirschfeld M, et al. Significance of nuclear hTra2-beta1 expression in cervical cancer. Acta Obstet Gynecol Scand. 2009;88:216–221. doi: 10.1080/00016340802503021. [DOI] [PubMed] [Google Scholar]
- Kajita K, Kuwano Y, Kitamura N, Satake Y, Nishida K, Kurokawa K, et al. Ets1 and heat shock factor 1 regulate transcription of the Transformer 2β gene in human colon cancer cells. J Gastroenterol. 2013;48:1222–1233. doi: 10.1007/s00535-012-0745-2. [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, et al. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, et al. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xu XS, Russell JE. A nucleolin-binding 3' untranslated region element stabilizes beta-globin mRNA in vivo. Mol Cell Biol. 2006;26:2419–2429. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF) Nat Immunol. 2011;12:853–860. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F. Splicing factor SC35 promotes tau expression through stabilization of its mRNA. FEBS Lett. 2011;585:875–880. doi: 10.1016/j.febslet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, et al. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–1045. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- Lam EK, Wang X, Shin VY, Zhang S, Morrison H, Sun J, et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3:209–218. [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479–2491. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- Sacconi A, Biagioni F, Canu V, Mori F, Di Benedetto A, Lorenzon L, et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3:e423. doi: 10.1038/cddis.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Sangwan V, Banerjee S, Mackenzie T, Dudeja V, Li X, et al. miR-204 mediated loss of Myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol Cancer. 2013;12:105. doi: 10.1186/1476-4598-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, et al. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155. doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7:e52397. doi: 10.1371/journal.pone.0052397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam SZ, Trickler W, Kimura S, Binienda ZK, Paule MG, Slikker W, et al. Neuroprotective efficacy of a new brain-penetrating C-Abl inhibitor in a murine Parkinson's disease model. PLoS One. 2013;8:e65129. doi: 10.1371/journal.pone.0065129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N, et al. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2013;33:1407–1417. doi: 10.1038/onc.2013.86. [DOI] [PubMed] [Google Scholar]
- Masuda K, Kuwano Y, Nishida K, Rokutan K. Application of DNA microarray technology to gerontological studies. Methods Mol Biol. 2013;1048:285–308. doi: 10.1007/978-1-62703-556-9_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.