Abstract
Hydrocarbon seepage is widespread and patchy in the Gulf of Mexico, and six species of symbiont containing bathymodiolin mussels are found on active seeps over wide and overlapping depth and geographic ranges. We use mitochondrial genes to discriminate among the previously known and a newly discovered species and to assess the connectivity among populations of the same species in the northern Gulf of Mexico (GoM). Our results generally validate the morphologically based distribution of the three previously known GoM species of Bathymodiolus, although we found that approximately 10% of the morphologically based identifications were incorrect and this resulted in some inaccuracies with respect to their previously assigned depth and geographical distribution patterns. These data allowed us to confirm that sympatry of two species of Bathymodiolus within a single patch of mussels is common. A new species of bathymodiolin, Bathymodiolus sp. nov., closely related to B. heckerae was also discovered. The two species live at the same depths but have not been found in sympatry and both have small effective population sizes. We found evidence for genetic structure within populations of the three species of Bathymodiolinae for which we had samples from multiple sites and suggest limited connectivity for populations at some sites. Despite relatively small sample sizes, genetic diversity indices suggest the largest population sizes for B. childressi and Tamu fisheri and the smallest for B. heckerae and B. sp. nov. among the GoM bathymodiolins. Moreover, we detected an excess of rare variants indicating recent demographic changes and population expansions for the four species of bathymodiolins from the Gulf of Mexico.
Introduction
Individual species occur in distinct geographic ranges where the members of its populations live, feed, reproduce and die. The current distribution of a species is determined by the suitability of habitats and the colonization and extinction processes over time. Distribution patterns are not permanent and can change as a result of changes in abiotic factors as well as through biotic interactions. Gause’s theory of competitive exclusion [1] suggests that two species with the same ecological niches cannot stably coexist.
When a geographic barrier divides a population, the two separated populations have the potential to evolve into two independent species. New species resulting from allopatric speciation are known as vicariant species. They are often quite similar genetically and occupy very similar niches, but live in geographically separated areas. Secondary contact between such relatively new species is a field of interest for evolutionary ecologists [2]–[4]. Indeed, these closely related species are often phenotypically different where the species occur together (‘sympatry’) but are often indistinguishable where each species occurs alone (‘allopatry’) (reviewed in [5]). When sympatric species are phenotypically similar, postzygotic reproductive isolation can keep co-occurring species distinct [6]. In the case of secondary contact between sibling species, we can distinguish a continuum of contrasting scenarios based on reproductive isolation and/or ecological differentiation. If the species are completely reproductively isolated and sufficiently ecologically differentiated, their interspecific competition will be weak and it is likely that they will coexist in sympatry [3]. On the other hand, if the species are completely reproductively isolated but ecologically identical, interspecific competition will be intense. Principles of competitive exclusion and limiting similarity predict one species will go extinct [7] or diverge through ecological character displacement [8]–[10]. The characters involved in character displacement may be morphological, behavioral, or physiological. Natural selection will favor, in each population, individuals able to use resources or habitats not used by the other species, reducing resource competition and permitting coexistence.
The deep-sea chemosynthetic habitats in the Gulf of Mexico (GoM) provide a natural laboratory to study secondary contact and sympatric habits of species with different levels of differentiation. There are three described species of Bathymodiolus in the Gulf of Mexico (B. heckerae, B. brooksi, and B. childressi) [11]. Based primarily on morphology, B. childressi were reported from 400m to 2200m depth, B. brooksi from approximately 1080m to 3300m, and B. heckerae from 2200m to 3300m [12]. Previous phylogenetic analysis of worldwide collections of bathymodiolin mussels revealed that B. brooksi and B. heckerae species are closely-related species and belong within the same clade, whereas B. childressi is in another more divergent clade with other Bathymodiolus spp. [13]–[15]. Another deep-sea bathymodiolin, Tamu fisheri, is reported from 540m to 700m in the Gulf of Mexico [11], [16] and while most often collected from among vestimentiferan tubeworms is occasionally collected with Bathymodiolus childressi (CRF pers. Obs.). This species was included in our study because of the risk of misidentification of juveniles, young or broken individuals. Only one species in known in the genus Tamu and the phylogenetic position of this unique and divergent lineage remains uncertain [14]–[15].
Tamu fisheri harbors thiotrophic symbionts (Species III in [16]), B. childressi harbor only methanotrophic symbionts, while Bathymodiolus brooksi harbor both a methanotroph- and thiotroph-related symbiont and B. heckerae harbor four co-occurring symbionts (a methanotroph, two phylogenetically distinct thiotrophs and a methylotroph-related phylotype) [17]. B. childressi are found in a range of physico-chemical environments on the Upper Louisiana Slope, including brine-dominated and petroleum-dominated seep sites, and are exposed to a range of concentrations of methane, oil and hydrogen sulfide [18]. They are also found on the lower Louisiana slope and despite these well differentiated habitats and the large range of depth, there has previously no evidence of significant B. childressi population subdivision in the GoM [19]. B. brooksi and B. heckerae have been collected from a range of environments on the lower Louisiana slope [12], [17], however there is little published data on potential correlations between symbiont complements and environmental conditions.
In this study, we present first an analysis of morphological and genetic diversity for the Bathymodiolin mussels from the Gulf of Mexico. We compare these identifications with a barcoding approach, using two mitochondrial genes. We then use additional genetic analyses to test for population structure within species. Finally, we examine the demographic stability of the populations in the Gulf of Mexico and the evolution and interbreeding potential of Bathymodiolus species in a larger Atlantic context.
Material and Methods
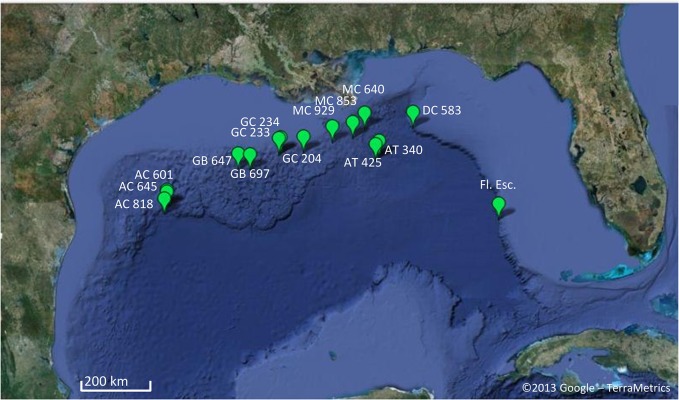
1. Sample collection
Mussels were collected from the deep-sea localities on the Florida Escarpment (Fl.Esc.) and other sites named for the Bureau of Ocean Energy Management Lease Block in which they occur. The lease blocks are designated by a two letter abbreviation for the region (Alaminos canyon, AC; Garden Banks, GB; Green Canyon, GC; Mississippi Canyon, MC; Atwater Valley, AT; Desota Canyon, DC) followed by a three digit number referring to a 3 x 3 nm block. Collections were made using the remotely operated vehicle (ROV) Jason II and the Alvin and Johnson Sea Link II manned submersibles. We extracted DNA and analyzed mussels from 15 discrete sites, ranging from 527 to 3288m and from 84°55’ W to 94°34’ W across the Gulf of Mexico (Fig. 1, Table 1). A total of 238 mussels, collected during 20 dives between October 2002 and August 2009 in the GoM, were included in the analyses. Mussels were collected with either mussel pot collection devices or nets as described in [20]. Upon recovery of the ROV or submersible, the mussels were transferred to chilled seawater, preliminarily identified using morphological criteria, and dissected on board ship. Pieces of tissue (mantle and gills) were frozen (−80°C) or kept in 70% ethanol until nucleic acid extraction using the CTAB+PVP method [21] at The Pennsylvania State University. This work was carried out under a contract from the US Bureau of Ocean Energy Management in the Department of the Interior, the cognizant federal agency charged with overseeing and protecting these communities. All samples were collected in US territorial waters. No permits are required for sampling of these species for scientific purposes in US or international waters.
Fig 1. Map of collection sites in the Gulf of Mexico.
Sampling effort, depth and coordinates are indicated in Table 1.
Table 1. Information on mussel samples sorted by depth.
| Site | Latitude | Longitude | Depth (m) | Dive# | Date of collection | B. childressi | B. brooksi | B. heckerae | B. nov. sp GoM | Tamu fisheri | Accession number | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Morpho. ID | Genetic ID | Morpho. ID | Genetic ID | Morpho. ID | Genetic ID | Morpho. ID | Genetic ID | Morpho. ID | Genetic ID | ||||||||||||||
| CO1 | ND4 | CO1 | ND4 | CO1 | ND4 | CO1 | ND4 | CO1 | ND4 | CO1 | ND4 | ||||||||||||
| Upper slope | |||||||||||||||||||||||
| GC234 | 27°44.1'N | 91°13.5'W | 527 | JSL 4717 | 17 July. 04 | 10 | 10 | 10 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024170—KM024179 | (Bc) KM044648—KM044657 | |
| JSL 4590 | 05 Sept. 03 | - | - | - | - | - | - | - | - | - | - | - | - | 5 | 5 | 5 | (Tf) KM024269—KM024273 | (Tf) KM044791—KM044795 | |||||
| MC929 | 28°01.1'N | 89°43.1'W | 636 | JSL 3340 | 08 Oct. 02 | 15 | 15 | 15 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024180—KM024194 | (Bc) KM044658—KM044672 | |
| GC233 | 27°43.4'N | 91°16.8'W | 651 | JSL 4711 | 09 July 04 | 10 | 10 | 10 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024195—KM024204 | (Bc) KM044673—KM044682 | |
| GC204 | 27°46.0’N | 90°32.7’W | 870 | JSL 3354 | 17 Oct. 02 | 15 | 15 | 9 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024205—KM024219 | (Bc) KM044683—KM044691 | |
| GB647 | 27°19.8'N | 92°25.8'W | 1007 | J2–280 | 26 June 07 | 6 | 6 | 0 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024220—KM024225 | - | |
| GB697 | 27°19.2'N | 92°06.7'W | 1015 | J2–274 | 17 June 07 | 4 | 4 | 4 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024226—KM024229 | (Bc) KM044692—KM044692 | |
| MC853 | 28°07.6'N | 89°08.5'W | 1075 | AD 4178 | 14 May 06 | 3 | 8 | 7 | 9 | 4 | 4 | - | - | - | - | - | - | - | - | - | (Bc) KM024230—KM024237; (Bb) KM024103—KM024106 | (Bc) KM044698—KM044704; (Bb) KM044583—KM044586 | |
| Deeper slope | |||||||||||||||||||||||
| MC640 | 28°21.4'N | 88°47.7'W | 1414 | AD 4182 | 18 May 06 | 0 | 3 | 2 | 4 | 1 | 1 | - | - | - | - | - | - | - | - | - | (Bc) KM024238—KM024240; (Bb) KM024107 | (Bc) KM044703—KM044704; (Bb) KM044587 | |
| AT425 | 27°34.2’N | 88°29.6’W | 1869 | AD 3918 | 14 Oct. 03 | - | - | - | 0 | 3 | 3 | 3 | 0 | 0 | - | - | - | - | - | - | (Bb) KM024108—KM024110 | (Bb) KM044588—KM044590 | |
| AC645 | 26°21.3'N | 94°30.1'W | 2195 | AD 4197 | 01 June 06 | - | - | - | 0 | 3 | 3 | 3 | 0 | 0 | - | - | - | - | - | - | (Bb) KM024134—KM024136 | (Bb) KM044607—KM044609 | |
| 2197 | J2–281 | 29 June 07 | - | - | - | 0 | 3 | 3 | 3 | 0 | 0 | - | - | - | - | - | - | (Bb) KM024137—KM024139 | (Bb) KM044610—KM044612 | ||||
| 2200 | AD 3923 | 18 Oct. 03 | - | - | - | 0 | 8 | 7 | 8 | 0 | 0 | - | - | - | - | - | - | (Bb) KM024126—KM024133 | (Bb) KM044613—KM044619 | ||||
| 2222 | AD 3924 | 19 Oct. 03 | 27 | 25 | 17 | 15 | 17 | 16 | - | - | - | - | - | - | - | - | - | (Bc) KM024241—KM024265; (Bb) KM024111—KM024125, KM024140—KM024141 | (Bc) KM044705—KM044721; (Bb) KM044591—KM044606 | ||||
| AT340 | 27°38.7'N | 88°21.9'W | 2190 | J2–270 | 10 June 07 | - | - | - | 4 | 4 | 4 | - | - | - | - | - | - | - | - | - | (Bb) KM024142—KM024145 | (Bb) KM044620 - ‘KM044623 | |
| 2216 | AD 4180 | 16 May 06 | - | - | - | 0 | 1 | 1 | 20 | 19 | 19 | - | - | - | - | - | - | (Bb) KM024146; (Bh) KM024274—KM024292 | (Bb) KM044624; (Bh) KM044724—KM044742 | ||||
| AC601 | 26°23.6'N | 94°30.9'W | 2335 | J2–283 | 07 July 07 | 3 | 3 | 2 | - | - | - | - | - | - | - | - | - | - | - | - | (Bc) KM024266—KM024268 | (Bc) KM044722—KM044723 | |
| DC583 | 28°23.1'N | 87°23.3'W | 2445 | J2–454 | 22 Aug. 09 | - | - | - | 19 | 0 | 0 | 13 | 0 | 0 | 0 | 32 | 32 | - | - | - | (BNovSP) KM024071—KM024102 | (BNovSp) KM044759—KM044790 | |
| AC818 | 26°11.1'N | 94°34.4'W | 2744 | J2–282 | 01 July 07 | - | - | - | 0 | 6 | 6 | 6 | 0 | 0 | - | - | - | - | - | - | (Bb) KM024163—KM024168 | (Bb) KM044641—KM044646 | |
| 2785 | AD 4192 | 27 May 06 | - | - | - | 16 | 16 | 16 | 2 | 2 | 2 | - | - | - | - | - | - | (Bb) KM024147—KM024162; (Bh) KM024293—KM024294 | (Bb) KM044625—KM044640; (Bh) KM044743—KM044744 | ||||
| Fl.Esc. | 26°01.8'N | 84°55.1'W | 3288 | AD 3915 | 11 Oct. 03 | - | - | - | 0 | 1 | 1 | 15 | 14 | 14 | - | - | - | - | - | - | (Bb) KM024169; (Bh) KM024295—KM024208 | (Bb) KM044647; (Bh) KM044745—KM044758 | |
| Total morph. (N = 238) | 93 | - | - | 67 | - | - | 73 | - | - | 0 | - | - | 5 | - | - | - | - | ||||||
| Total CO1 (N = 238) | - | 99 | - | - | 67 | - | - | 35 | - | - | 32 | - | - | 5 | - | - | - | ||||||
| Total ND4 (N = 213) | - | - | 76 | - | - | 65 | - | - | 35 | - | - | 32 | - | - | 5 | - | - | ||||||
Morph, Morphological Sample
JSL, Johnson Sea Link dive number
J2, Jason 2 dive number; AD, Alvin dive number.
(Bc): Bathymodiolus childressi
(Bb) B. brooksi
(Bh) B. heckerae
(BNovSp) B. nov. sp. GoM
(Tf) Tamu fisheri.
2. PCR amplification and sequencing
DNA sequences were obtained for 2 mitochondrial genes: cytochrome c oxidase 1 (CO1) and NADH dehydrogenase subunit 4 (ND4). Fragments of the mitochondrial gene CO1 were amplified using the primers CO1 Bathymodiolus sense/antisense [22], and HCO/LCO [23] with the amplification conditions described in [24]. The ND4 segments were amplified with the sense primers ND46S or ArgBL with the anti-sense primers NAP2H using the conditions described [13].
The PCR products were run on a 1% agarose gel stained with ethidium bromide to check the quantity and the quality of the products and then purified with the ExoSap-It protocol (USB, Affimetrix). Both strands of the purified PCR products for the two mitochondrial genes CO1 and ND4 were directly sequenced (Sanger DNA sequencing, Applied Biosystems 3730XL sequencer at the Huck Institutes, PennState: http://www.huck.psu.edu/facilities).
3. Data analyses
Quality of the sequence reads from the two DNA strands were checked from the chromatogram with the Chromas 2.22 computer program (Technelysium Pty. Ltd., Helensvale, Australia). Sequences were aligned with Clustal W [25] in the BioEdit program [26], with manual adjustments to insure that indels were scored consistently among individuals. The distribution of pairwise Kimura’s two-parameter (K2P) distance [27] was plotted for each species. Neighbor-joining trees were constructed using MEGA v4.0 [28] using a K2P distance matrix to infer gene genealogies and 1,000 bootstrap replicates to assess the significance of observed clades [29]. We also included NCBI sequences for B. thermophilus (CO1: FJ766893 [30], ND4: AY649808 [31]), B. azoricus (CO1: FJ766849 [30], ND4: AF128534 [32]), B. puteoserpentis (CO1: FJ766949 [30], ND4: AF128533 [32]) in our phylogenetic trees to further refine relations among taxa. We added CO1 sequences for B. boomerang Barbados (DQ513444 [22]), B. boomerang Zaire (DQ513442 [22]), B. nov. sp. 5°S from the Mid Atlantic Ridge (JQ844817 [33], JQ844789 [33]), and B. nov. sp. 9°S MAR (JQ844819 [33], JQ844822 [33]) but ND4 sequences for these species were not available at NCBI.
The number of haplotypes, nucleotide diversity, divergence (average number of nucleotides differences per site between two sequences [34]) and the Tajima’s [35] D statistics were estimated using the DnaSP version 4.20.2 software package [36]. Pairwise distances between individuals were used to determine genetic differences among populations based on mtDNA sequences (Φst statistic) with Arlequin [37] and 1,000 permutations of the data were used to evaluate the statistical significance of the differentiation values.
Mitochondrial (CO1) divergence time was estimated as T = D/2r, where D is the pairwise sequence divergence and T is the time of divergence, multiplied by 2 to account for the age of each lineage [38]. To estimate the time of divergence between species, a genus specific CO1 molecular clock was calibrated using the divergence time between Bathymodiolus thermophilus from between 13°N and 21°S on the East Pacific Rise and its sister species from 37°S on the East Pacific Rise as in [30]. This particular pair likely split about 5.9 MYA and its CO1 pairwise divergence is 4.62%. Consequently, we estimated a mitochondrial mutation rate of 0.39% per MY. We used this mutation rate (r) to estimate the time of divergence (T) between Atlantic species from their pairwise divergence (D).
For the five GoM species of mussels, intraspecific relationships between haplotypes were estimated with the software Network (Network 4.5.1.6, Fluxus Technology Ltd). As our sampling represents only a subset of the natural haplotype diversity, we used the median-joining method [39] to provide the best estimates of the genealogy when internal node haplotype are not sampled [40].
Results and Discussion
1. Morphological and genetic identification
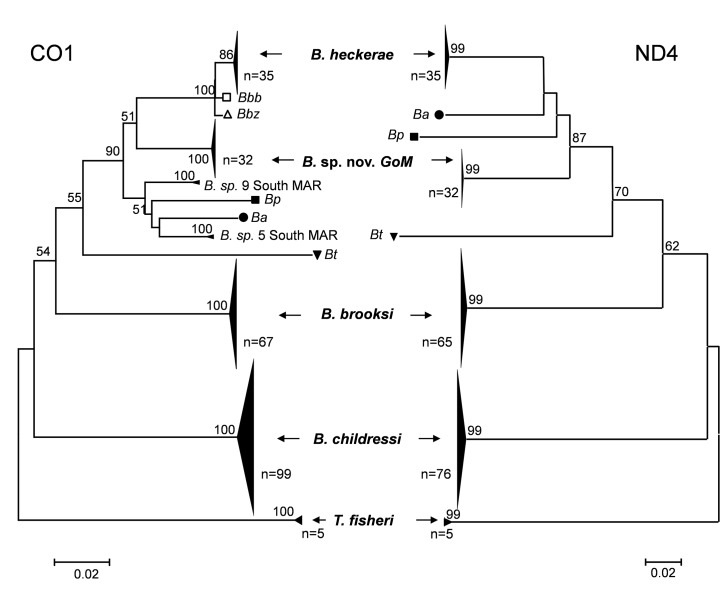
The rapid identification of closely related mussels, such as those in the subfamily Bathymodiolinae, at sea is not straight forward, especially for small or incomplete individuals. We used the widely utilized mitochondrial markers CO1 and ND4 to check the species identification of our collections. These markers provide a robust separation of the three previously known GoM Bathymodiolus species, B. childressi, B. brooksi, and B. heckerae, as well as numerous other closely related bathymodiolins [13], and the results were 100% consistent between the two markers when both were successfully amplified (we were unable to amplify the ND4 loci for 25 of the 238 individuals). Amplification success and morphological and genetic identifications are presented in Table 1 and the phylogenetic trees are shown in Fig. 2.
Fig 2. Neighbor-joining trees for mitochondrial loci (CO1 and ND4) using Kimura’s two-parameter method [27].
Bootstrap support above 50% are shown next to the branches (1000 replicates). CO1: n = 247 (238 individuals from the Gulf of Mexico, Bathymodiolus azoricus from the Mid Atlantic Ridge (Ba: FJ766849), B. puteoserpentis from the Mid Atlantic Ridge (Bp: FJ766949), B. boomerang from Barbados (Bbb: DQ513444), B. aff. boomerang from Zaire (Bbz: DQ513442), B. thermophilus from the East Pacific Rise (Bt: FJ766893), B. nov.sp 5°S from the Mid Atlantic Ridge (B.sp 5 South MAR: JQ844817, JQ844789), and B. nov. sp 9°S MAR (B. sp 9 South MAR: JQ844819, JQ844822). ND4: n = 216 (213 individuals from the GoM, Bathymodiolus azoricus from the Mid Atlantic Ridge (Ba: AF128534), B. puteoserpentis from the Mid Atlantic Ridge (Bp: AF128533), and B. thermophilus from the East Pacific Rise (Bt: AY649808)).
Table 2 summarizes the reassignment of the shipboard morphological identifications resulting from the molecular analyses. The discovery of a new monophyletic group of mussels, which we propose is a new species, led to a significant number of misidentifications of B. brooksi and B. heckerae in that collection. In addition, B. brooksi were incorrectly identified on board ship as B. childressi 12% of the time and B. heckerae were incorrectly identified on board ship as B. brooksi 33% of the time. B. childressi identified on board ship based on morphological criteria were confirmed with molecular analysis 98% of the time, although as noted above, 12% of the mussels originally identified as B. brooksi were later genetically identified as B. childressi. T. fisheri were correctly identified based on morphological criteria 100% of the time. Although the original shipboard misidentifications were significant overall, when those associated with Bathymodiolus sp. nov. are excluded, about 95% of those remaining were either noted as troublesome in notebooks at sea and/or were associated with small individuals or broken shells.
Table 2. Assignment of species after genetic identification (as percentage of total).
| Genetic ID | |||||
|---|---|---|---|---|---|
| Morphological ID | B. childressi | B. brooksi | B. heckerae | B. sp. nov. GoM | T. fisheri |
| From Bathymodiolus childressi to… | 98 (98) | 2 (2) | 0 | 0 | 0 |
| From B. brooksi to… | 12 (17) | 60 (83) | 0 | 28 | 0 |
| From B. heckerae to… | 1 (2) | 33 (40) | 48 (58) | 18 | 0 |
| From T. fisheri to… | 0 | 0 | 0 | 0 | 100 |
Percentage of assignments after removing individuals of the new species from site DC583 from the calculation are shown in parentheses.
Although the molecular reanalysis of the morphological species identifications changed numerous details with respect to the confirmed sites and relative abundance for the three described species of Bathymodiolus, the re-identifications did not result in large range extensions for any species from that previously published [12]. However, the molecular analyses only confirmed B. heckerae in three of the original nine collections where the preliminary morphology-based identification indicated they occurred [12]. Our molecular identifications are consistent with previous reports of B. brooksi and B. childressi, but not B. heckerae at the relatively well visited Alaminos Canyon (AC) 645 site [11], [41]-[42] and also confirmed the presence of B. heckerae at the nearby but deeper AC818 site. Our analyses support the wide geographic range of this species from east to west across the northern GoM but suggest its upper depth limit is below 2,200m. Our analyses also confirm the widespread occurrence and large depth and geographic range of B. brooksi from 1075 to 3288m, and from Alaminos Canyon to the Florida Escarpment. Our analyses slightly extend the already large depth range of the dominant upper slope mussel B. childressi from a maximum depth of 2220m in AC645 [42] to 2335m in AC601. Although B. brooksi may occur sympatrically with either B. childressi or B. heckerae, the latter two species were never found in the same sites. Tamu fisheri was only found in sympatry with B. childressi (GC234).
A completely unexpected result from our molecular analyses was the discovery of a new species of Bathymodiolus (n = 32) at a site in DC583. These mussels are clearly divergent from the 3 other species of Bathymodiolus in the GoM, as well as all other Bathymodiolus species with published CO1 or ND4 sequences, including those on the Blake Ridge, the North and South Mid-Atlantic Ridge, and the west African Seeps [22], [33], [43–45] (Fig. 2). Two other symbiont-containing species of Bathymodiolinae have also been described in the Gulf of Mexico, Idas macdonaldi and Tamu fisheri [11], and the new species is genetically distinct from these mussels as well. Phylogenetic trees including closely related mussels confirm the new species belongs in the Bathymodiolus thermophilus group as defined by [45] (Fig. 2). Discovery of this new species of Bathymodiolus at 2,445m is surprising because this is within the depth and longitudinal range of two other GoM Bathymodiolus species (B. brooksi: 1075 to 3288m, and B. heckerae: 2216 to 3288m depth).
The limited distribution of this new species within the relatively well-sampled GoM and three other widespread species of Bathymodiolus suggests that this site is in some way oceanographically isolated from the other sites we have visited. The large size range of animals in our collection (96 to 151 cm in shell length), and the presence of very small mussels on the video record of the site suggests that this population is not the result of a single recent settlement event. Bathymodiolus species have a planktonic larval phase and a long larval life period that has the potential for long distance dispersal [46]. Such extensive dispersal potential has been suggested in the amphi-Atlantic distribution of closely related congeners [22], [47]. There is no evidence that closely related species differ dramatically in their dispersal abilities as all the Bathymodiolus species studied show evidence of high-rate gene flow and long-range dispersal [48]. Therefore, it is unlikely that this Bathymodiolus sp. nov. has limited dispersal ability. We know of no unique current patterns in this area of the gulf that would physically prevent dispersal of Bathymodiolus sp. nov. It is possible that Bathymodiolus sp. nov. is only able to survive in a very specific habitat that is unsuitable for other bathymodiolins, however we have no data to support this hypothesis and the metabolic versatility of the symbionts of the other GoM Bathymodiolus spp. suggest this is unlikely [17]. The occurrence of this species and no other bathymodiolins in this location remains an enigma.
Deep-sea mussels from the GoM are often found in sympatry: B. childressi with T. fisheri (GC234), B. childressi with B. brooksi (MC853, MC640, AC645) or B. brooksi with B. heckerae (AT340, AC818, Fl. Esc.). In sympatry, the principle of competitive exclusion and limiting similarity predict one species will go extinct [7] or diverge through ecological character displacement [8]–[10]. GoM bathymodiolins are clearly in the second situation where sympatric and phenotypically close species diverge through their symbiotic composition [17].Without observation of sympatry between B. sp. nov and other Bathymodiolus species, we cannot exclude a case of interspecific competition leading to extinction of one species on sites where two species previously occurred. Clearly, additional exploration and oceanographic, evolutionary, and physiological studies are needed to understand the origin and the distribution of this new species of Bathymodiolus, and the sympatric occurrence of the others.
2. Population structure within mussel species
The power to detect differences with these types of analyses is strongly correlated with sample sizes. Because (as expected) no genetic differentiation was detected between the different sampling dates within a site (at AC645, AT340 and AC818), we combined different collections from the same site for the population structure analysis. Populations with less than three individuals sampled were excluded from these analyses.
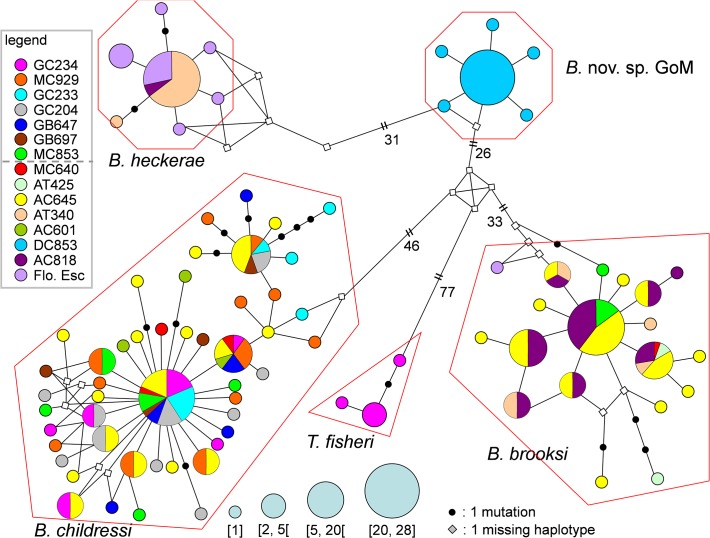
Haplotype networks provide a graphical representation of the genetic structure and relationship among haplotypes within species. The number of haplotypes detected in each species is indicated in Table 3 and the CO1 network of haplotypes for 5 species from the GoM is presented in Fig. 3. In order to maximize the number of individuals for this analysis we used only CO1 (n = 238) (Table 1). Because of the large data set used for this analysis, we simplified the graphical representation of the haplotype network and present only four diameters of haplotype circles (rather than 15 which would otherwise be required): (1) unique haplotypes (n = 1), (2) rare haplotypes (n = 2–5), (3) common haplotypes (n = 5–20), and major haplotypes (n = 20–28).
Table 3. Summary statistics for mitochondrial loci polymorphism.
| Species | Locus | N | h | Hd | S | π | Θw | Tajima's D |
|---|---|---|---|---|---|---|---|---|
| B. childressi | CO1 | 99 | 48 | 0.909 | 45 | 0.00501 | 0.01598 | −2.24** |
| ND4 | 76 | 38 | 0.947 | 48 | 0.00465 | 0.01451 | −2.27** | |
| (CO1+ND4) | 76 | 60 | 0.988 | 84 | 0.00489 | 0.01488 | −2.26** | |
| B. brooksi | CO1 | 67 | 19 | 0.829 | 21 | 0.00290 | 0.00807 | −1.97* |
| ND4 | 65 | 25 | 0.694 | 30 | 0.00172 | 0.00937 | −2.62*** | |
| (CO1+ND4) | 65 | 38 | 0.936 | 51 | 0.00226 | 0.00881 | −2.48** | |
| B. heckerae | CO1 | 35 | 7 | 0.363 | 8 | 0.00094 | 0.00356 | −2.16* |
| ND4 | 35 | 10 | 0.494 | 13 | 0.00125 | 0.00466 | −2.34** | |
| (CO1+ND4) | 35 | 13 | 0.610 | 21 | 0.00111 | 0.00417 | −2.49*** | |
| B. sp. nov. | CO1 | 32 | 6 | 0.292 | 5 | 0.00057 | 0.00228 | −2.01* |
| ND4 | 32 | 5 | 0.238 | 4 | 0.00037 | 0.00146 | −1.89* | |
| (CO1+ND4) | 32 | 10 | 0.490 | 9 | 0.00046 | 0.00183 | −2.29** |
Locus: CO1 (545bp)
ND4 (678bp)
concatenate C01+ND4: 1123bp)
N: number of sequences
h: number of haplotypes
Hd: haplotype diversity
S: Number of segregating sites
π: nucleotide diversity
Θw: Watterson’s Theta per site from S
Tajima’s D: Tajima’s statistic [35]
Statistical significance
*,P < 0.05
**, P < 0.01
***, P < 0.001
Fig 3. Median-joining network showing genetic structure of the five species of bathymodiolin mussels from the GoM (Bathymodiolus heckerae, B. childressi, B. brooksi, B. nov. sp. GoM and Tamu fisheri) (CO1 data only).
Size of the haplotype circle are proportional to their frequencies (Unique (n = 1); rare (n = 2 to 4), common (n = 5 to 20) and abundant (n = 20 to 28). Black points represent mutations. Diamonds represent missing haplotype. Colours represent locations were the haplotype was found. In the legend, the dashed line represents the virtual limit between “shallow population (less than or about 1000m deep) and deep populations (more than 1400m deep). The number of sequences used for each species and population is indicated on Table 1.
As predicted, every species forms a distinct cluster in the network with no shared haplotypes between species (Fig. 3). The most closely related species (B. heckerae and B. nov. sp.) are only separated by 33 mutations while the most divergent species (B. childressi and Tamu fisheri) are separated by at least 126 mutations. B. heckerae and B. nov. sp. are typical examples of star-like haplotype networks with a single major haplotype (found at 71% and 84% respectively) and a small number of derived haplotypes (n = 7 haplotypes for B. heckerae, and n = 6 for B. nov. sp.) that are directly derived from the major haplotype.
The B. childressi network is more genetically structured than the other species. A total of 48 haplotypes are present with the rare and unique haplotypes articulated around one dominant haplotype (shared by 27 individuals) and two common haplotypes (shared by 9 and 10 indiv.). The majority (81%) of the haplotypes are unique (39 out of a total of 48 haplotypes) and all the unique and rare haplotypes are derived with less than 3 mutations from one of the 3 more common haplotypes. The presence of three major haplotypes may indicate past or intermittent isolation of three sub-populations during its evolutionary history. The haplotypes found at MC853 (in green) are only from (or derived from) the major haplotype. This major haplotype, however, is never found in the well-sampled population MC929 (n = 15, in orange). We detected a significant genetic differentiation between these two populations in the CO1 locus (φst = 0.106, p < 0.05; data not shown in table) suggesting reduced connectivity between these two sites.
Although originally suggested to occur at nine of the study sites [12], B. heckerae was confirmed in only three of the study sites (AT340, AC818 and on the Florida Escarpment) and we only collected sufficient sample sizes of individuals from AT340 and the Florida Escarpment for robust population analyses. We detected genetic differentiation in the CO1 locus between AT340 and the Florida Escarpment populations (φst = 0.04, p<0.01; data not shown). Because of this, we examined the genetic diversity within the populations (Table 4). For both loci, the highest diversity is found for the Florida Escarpment population (for example Fl.Esc. nucleotide diversity (π = 0.0018) is almost 5 times higher than AT340 diversity (π = 0.0004)). Although variation levels are higher than found in the other GoM populations, this diversity is very low compared to that of other species of Bathymodiolus (Atlantic: π = 0.0006 to 0.0058 [29], Pacific: up to π = 0.003 [56]). B. heckerae from AT340 are almost genetically monomorphic for the CO1 locus (only two segregation sites and two haplotypes for 19 individuals, π = 0.00039). This diversity is 4.5 times lower than the diversity detected in samples from the Florida Escarpment. A similar trend is observed with the locus ND4 (π = 0.00147 at the Florida Escarpment and π = 0.00109 at AT340).
Table 4. B. heckerae site-specific genetic diversity.
| Species | Locus | Location | N | h | Hd | S | π |
|---|---|---|---|---|---|---|---|
| B. heckerae | CO1 | Fl.Esc. | 14 | 6 | 0.681 | 6 | 0.00179 |
| (545bp) | AT340 | 19 | 2 | 0.105 | 2 | 0.00039 | |
| AC818 | 2 | 1 | 0 | 0 | 0 | ||
| ND4 | Fl.Esc. | 14 | 5 | 0.505 | 7 | 0.00147 | |
| (678bp) | AT340 | 19 | 6 | 0.468 | 7 | 0.00109 | |
| AC818 | 2 | 2 | 1 | 1 | 0.00147 |
N: number of individuals
h: number of haplotypes
Hd: Haplotype diversity
S: number of segregating site
π: Average number of nucleotide differences per site between two sequences [34]
The two genes, CO1 and ND4 are in linkage disequilibrium because they are linked on the circular mitochondrial genome. For this reason, we concatenated the two genes and then treated them as a single longer mitochondrial gene for each individual. For this analysis, we removed the individuals for which only one gene was amplified, leaving 213 individuals with concatenated sequences for the population structure analyses (Table 1).
We estimated population differentiation (φst) for B. childressi (Table 5), B. brooksi (Table 6) and B. heckerae (Table 7). Unlike with CO1 alone, no genetic differentiation was detected among the seven B. childressi populations nor between the two B. heckerae populations in this analysis. This may reflect to lower power due to the reduced sample sizes in key populations. However, two significant differences were detected among B. brooksi populations using the concatenated data set: between AT340 and MC853 and between AC818 and MC853. We note that this is consistent with the observation of significant differentiation in the CO1 gene in B. childressi from MC853 and suggests limited connectivity between this site and the others sampled in this study. Moreover, and despite several attempts, we were unable to amplify the ND4 locus for any B. childressi from the GB647 site. This may result from a mutation on the PCR primer regions and suggest the possibility of reduced connectivity for this population as well.
Table 5. B. childressi φst population differentiation, tested by 1023 permutations with the Arlequin software for the concatenated CO1 and ND4 genes.
| Bc_GC234 | Bc_MC929 | Bc_GC233 | Bc_GC204 | Bc_GB697 | Bc_MC853 | |
|---|---|---|---|---|---|---|
| (n = 10) | (n = 15) | (n = 10) | (n = 9) | (n = 4) | (n = 7) | |
| Bc_MC929 (n = 15) | 0.01 | |||||
| Bc_GC233 (n = 10) | −0.03 | 0.02 | ||||
| Bc_GC204 (n = 9) | −0.01 | 0.01 | −0.01 | |||
| Bc_GB697 (n = 4) | 0.01 | 0.01 | 0.01 | 0.01 | ||
| Bc_MC853 (n = 7) | 0.03 | 0.03 | 0.03 | 0.02 | 0.03 | |
| Bc_AC645 (n = 17) | 0.01 | −0.01 | 0.01 | 0.01 | 0.01 | 0.03 |
(*: p<0.05)
Table 6. B. brooksi ɸst population differentiation, tested by 1023 permutations with the Arlequin software for the concatenated CO1 and ND4 genes.
| Bb_MC853 | Bb_AC645 | Bb_AT340 | |
|---|---|---|---|
| (n = 4) | (n = 29) | (n = 5) | |
| Bb_AC645 (n = 29) | 0.07 | ||
| Bb_AT340 (n = 5) | 0.23* | 0.01 | |
| Bb_AC818 (n = 19) | 0.15* | 0.01 | 0.01 |
(*: p<0.05)
Table 7. B. heckerae ɸst population differentiation, tested by 1023 permutations with the Arlequin software for the concatenated CO1 and ND4 genes.
| Bh_AT340 | |
|---|---|
| (n = 19) | |
| Bh_Fl.Esc (n = 14) | 0.03 |
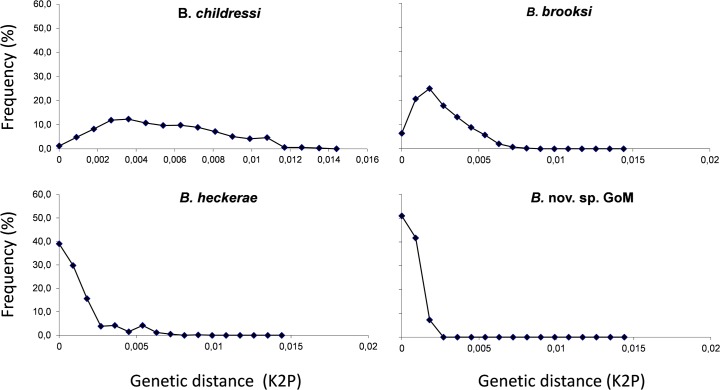
Bimodal distributions of pairwise genetic distances can reveal hidden population structure. The distributions of intraspecific distances for the four GoM species are shown in Fig. 4. The highest maximum intraspecific pairwise distance is observed for B. childressi (K2P distance = 0.0135) and the smallest maximum distance is observed of B. nov. sp. (K2P distance = 0.0018). We never observed a bimodal distribution of the K2P distances which would have been observed if divergent species were analyzed together. Under the assumption of no selection, we reject the hypothesis that any of the bathymodiolin species represent a mixture of deep and shallow water species [49]. The pairwise difference distributions suggest that the populations of B. childressi and B. brooksi are expanding and that B. heckerae and B. nov. sp. appear to be at equilibrium [50].
Fig 4. Distribution of pairwise K2P distance-frequency histogram from CO1 and ND4 concatenated data.
Our preliminary analysis suggests detectable population differentiation in GoM mussel populations against a generally high level of connectivity in the Bathymodiolus spp. populations throughout the northern GoM. The data from MC853 and GB647 suggest these populations have limited connectivity with other populations in the GoM and the discovery of a new species limited to a singe site suggests that currently unknown oceanographic or ecological isolating mechanisms may be important in the diversification of these species. Additional studies using larger sample sizes and additional molecular tools such as microsatellites and nuclear markers are needed to better constrain levels of isolation between populations and understand the population biology of the GoM bathymodiolins.
3. Low diversity and population expansion in the GoM
As suspected from the haplotype network, levels of genetic diversity are variable between the species. Bathymodiolus childressi and B. brooksi show a high level of genetic diversity (for example CO1 nucleotide diversity π = 0.0050 and 0.0029 respectively) when B. heckerae and B. sp. nov GoM nucleotide diversity is almost 3 to 5 times smaller (0.0009 and 0.0006 respectively) (Table 3). This trend (B. childressi > B. brooksi > B. heckerae > B. sp. nov. GoM) is present in both CO1 and ND4.
We used Tajima’s D statistic to test the equilibrium conditions in the genetic history of the population. Tajima’s D is a difference statistic between two estimates of the neutral mutation parameter (4N eμ) on a per gene basis, k and Θw, where k is based on the number of pairwise differences [51] and Θw is based on the number of segregating sites [52]. A significant difference between the two estimates may indicate the action of selection [35] or demographic changes in the population [53]. The mitochondrial dataset (CO1 and ND4) are considered to be of one locus and were thus analyzed together here. Surprisingly, both concatenated and independent loci have a significantly negative Tajima’s D for each species (Table 3), indicating an excess of rare variants. Although an excess of rare mutations is a commonly observed pattern for mtDNA and may be due to segregating slightly deleterious mutations [54], star-like network of haplotypes (Fig. 3), low genetic diversity, and significant negative Tajima’s D (Table 3) are strongly suggestive of a non-equilibrium dynamic, which could be interpreted as recent demographic or selective events.
There is no evidence that a selective pressure is acting on the mitochondrial genome. Demographic or geographic population expansion can also result in negative Tajima’s D. One can discriminate between these two hypotheses by examining multiple independent loci. Selection is likely to affect just a single locus, but demography is expected to affect all loci in the genome [50]–[56]. This second hypothesis seems more probable and suggests that, despite the relative stability of the cold seep habitats over time (in comparison to the hydrothermal vent habitats of many bathymodiolins), populations are not demographically stable. It could either indicate recent extinction and recolonization of populations (and genetic bottlenecks) or a recent colonization of the GoM by this group. Because of the independent evolution of the four species from the GoM (evidenced by their very dissimilar haplotype networks), a recent or simultaneous colonization seems unlikely. We can only suggest that species and populations in the GoM are not at equilibrium and are subject to a constant flux in population size. This result is similar to those found for animals from deep sea hydrothermal vent habitats [30], [57]. In vent environments, sudden stoppage of hydrothermal activity, plate tectonics and transform faults are all known to promote bottlenecks in populations, recurrence of extinction/recolonization within populations, and allopatric speciation. Changes in seepage appear to also affect cold seep mussel populations, albeit on longer time scale of decades to centuries [58] rather than years to decades as for hydrothermal vent populations [59]–[61]. In both cases the life spans of the mussel species are of the same order as the life span of the habitat [58], [62]. In addition, natural selection may favor some characters for colonization of new habitats (through resistance to greater depths, to certain chemicals, or acquisition of new symbionts). Additional nuclear genetic loci may clarify whether selection or demography are responsible for the population structure observed in the Bathymodiolus species in the GoM.
4. Evolution and divergence between GoM and Atlantic Bathymodiolinae
In a recent paper, Van Der Heijden et al [33] suggest that the GoM and mid-Atlantic Ridge were not colonized by species coming from the Indian Ridge based on a lack of close relationships between the south mid-Atlantic Ridge and Indian Ocean taxa. Our data document the species richness of Bathymodiolus spp. living in GoM (B. childressi, B. brooksi, B. heckerae and B. nov. sp. GoM) and also that the GoM and Atlantic species are sister taxa with close phylogenetic relationships (Fig. 2). The phylogenetic relationships need further investigation (with more genes and more species, as, for example in [14] and [15]) to understand the evolutionary scenarios which promoted GoM and Atlantic Bathymodiolus diversity and widespread distribution. Indeed, cold-seep Bathymodiolinae are widespread and are present on both sides of the Atlantic equatorial belt [47]. For example, the B. childressi complex is found from the Gulf of Mexico across to the Nigerian Margin and the B. boomerang complex is found from Florida escarpment across to the Congo margin. Moreover, Van Der Heijden et al [33] suggest there was no evolutionary dispersal barrier for seeps species across the Atlantic Equatorial Belt. Hydrothermal vent Bathymodiolus species are also widely distributed: the complex of the closely related and hybridizing species B. azoricus and B. puteoserpentis extends over 3000km on the Mid-Atlantic Ridge, whereas B. thermophilus complex is found over 4600km on the East Pacific Rise (13°N to 38°S) [63]. So far, B. brooksi has only been collected in the GoM and, other than the new GoM species detected in this study, is the Bathymodiolinae taxon with the most limited distribution. However, as the Bathymodiolinae often disperse over long distances and new seep sites are still being discovered, we cannot exclude the possibility that other sites, thousands kilometers distant from the documented occurrences of the GoM species, will also harbor B. brooksi and/or the new GoM B. sp.
We used the mutation rate r = 0.39 [30] to estimate the time of divergence (T) between Atlantic species from their pairwise divergence (D) as (T = D / 2r) (Table 8, Table 9) and compared this with the divergence time previously estimated between B. azoricus and B. puteoserpentis [30]. The results from these two methods are of the same order of magnitude (8.4MYA in this study against 0.37 to 8.6MYA in the literature). Using the same methods we estimate that, B. heckerae and B. nov. sp. diverged approximately 8 MYA. During such a long divergence time it is unlikely that the new species distribution was restricted to very limited areas in the GoM, suggesting that Bathymodiolus sp. nov. will be found elsewhere in the Gulf of Mexico and possibly outside the GoM.
Table 8. CO1 divergence between GoM and Atlantic Bathymodiolus species.
Divergence is estimated as the average number of nucleotidic differences per site between populations.
| CO1 divergence | |||||||
|---|---|---|---|---|---|---|---|
| B. brooksi | B. heckerae | B. nov. sp. GoM | B. azoricus | B. puteoserpentis | B. boomerang | B. sp South MAR | |
| B childressi | 0.143 | 0.144 | 0.136 | 0.142 | 0.145 | 0.139 | 0.131 |
| B. brooksi | 0.121 | 0.112 | 0.119 | 0.125 | 0.113 | 0.113 | |
| B. heckerae | 0.064 | 0.078 | 0.080 | 0.011 | 0.072 | ||
| B. nov. sp. GoM | 0.078 | 0.062 | 0.058 | 0.059 | |||
| B. azoricus | 0.066 | 0.078 | 0.056 | ||||
| B. puteoserpentis | 0.074 | 0.053 | |||||
| B. boomerang | 0.064 | ||||||
Table 9. Estimated time of divergence between GoM and Atlantic Bathymodiolus species (in MY).
Time of divergence is estimated using the CO1 mutation rate of 0.39% / MY and divergence values from Table 8.
| CO1 divergence | |||||||
|---|---|---|---|---|---|---|---|
| B. brooksi | B. heckerae | B. nov. sp. GoM | B. azoricus | B. puteoserpentis | B. boomerang | B. sp South MAR | |
| B childressi | 18.23 | 18.36 | 17.34 | 18.10 | 18.49 | 17.72 | 16.70 |
| B. brooksi | 15.43 | 14.28 | 15.17 | 15.94 | 14.41 | 14.41 | |
| B. heckerae | 8.16 | 9.94 | 10.20 | 1.40 | 9.18 | ||
| B. nov. sp. GoM | 9.94 | 7.90 | 7.39 | 7.52 | |||
| B. azoricus | 8.41 | 9.94 | 7.14 | ||||
| B. puteoserpentis | 9.43 | 6.76 | |||||
| B. boomerang | 8.16 | ||||||
Conclusion
In a well-studied geographic area such as the GoM, Bathymodiolus spp. morphological identifications are generally reliable. They are fast and particularly useful for onboard identifications but present a risk of confusion as soon as new sites are explored and sampled. Genetic barcoding markers work well for the Bathymodiolus spp and are appropriate to confirm morphological identifications, especially in cases of juveniles and incomplete individuals and newly discovered sites.
When geographic distance is smaller than the effective dispersal distance, all individuals are potential mating partners. Panmictic populations are generally assumed for species with long-range dispersal capabilities such as Bathymodiolus spp. in the GoM [46]. However, although our results are generally in agreement with long range dispersal and considerable connectivity among widely dispersed bathymodilinae populations over a wide depth range, we also find evidence of genetic differentiation within each Bathymodiolus species in the GoM. Even more importantly, a site harboring a completely new species was discovered. Those results are not consistent with complete panmixia of Bathymodiolus spp. in the Gulf of Mexico and the as yet unconstrained extrinsic (oceanographic or local chemistry) or intrinsic (selection) factors that drive these patterns need to be investigated.
Acknowledgments
We would like to thank Liz Podowski, Stephane Hourdez, Erik Cordes, Stephanie Lessard-Pilon, Erin Becker and Susan. L. Carney for their contribution during the sampling, Iliana Baums for her collaboration with lab work, and Maria Pia Miglietta and Didier Jollivet for helpful discussions. Thanks to the captains and crews of the research vessels Seward Johnson, and Atlantis and the NOAA Ship Ronald H. Brown, and the pilots and crews of the Johnson Sea-Link, the Alvin submersible and the ROV Jason.
Funding Statement
This work was supported by a Lavoisier grant from the French Ministry of Foreign Affairs and by subcontracts to CRF from BOEM contracts 1435-01-05-39187 and M08PC20038 to TDI Brooks, with vessel and submergence facilities support provided by National Oceanic and Atmospheric Administration’s Office of Ocean Exploration and Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gause GF (1932) Experimental studies on the struggle for the existence: I mixed populations of two species of yeast. Journal of Experimental Biology 9: 389–402. [Google Scholar]
- 2. Dobzhansky T (1937) Genetics and the origin of species New York: Columbia University Press. [Google Scholar]
- 3. Mayr E (1942) Systematics and the Origin of Species. New York: Columbia University Press. [Google Scholar]
- 4. Gavrilets S (2003) Perspective: Models of speciation: What have we learned in 40 years? Evolution 57 (10): 2197–2215. [DOI] [PubMed] [Google Scholar]
- 5. Rice AM, Pfennig DW (2007) Character displacement: in situ evolution of novel phenotypes or sorting of pre-existing variation? Journal of Evolutionary Biology 20 (2): 448–459. [DOI] [PubMed] [Google Scholar]
- 6. Noor MA, Grams KL, Bertucci LA, Reiland J (2001) Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences of the United States of America 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Macarthur R, Levins R (1967) The limiting similarity, convergence, and divergence of coexisting species. The American Naturalist 101 (921): 377–385. [Google Scholar]
- 8. Brown WL Jr, Wilson EO (1956) Character displacement. Systematic zoology 5 (2): 49–64. [Google Scholar]
- 9. Slatkin M (1980). Ecological character displacement. Ecology 61 (1): 163–177. [Google Scholar]
- 10. Losos JB (2000) Ecological character displacement and the study of adaptation. Proceedings of the National Academy of Sciences of the United States of America 97 (11): 5693–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gustafson RG, Turner RD, Lutz RA, Vrijenhoek RC (1998) A new genus and five new species of mussels (Bivalvia, Mytilidae) from deep-sea sulfide/hydrocarbon seeps in the Gulf of Mexico. Malacologia 40 (1–2): 63–112. [Google Scholar]
- 12. Cordes EE, Bergquist DC, Fisher CR (2009) Macro-ecology of Gulf of Mexico cold seeps. Annual Review of Marine Science 1: 143–168. [DOI] [PubMed] [Google Scholar]
- 13. Iwasaki H, Kyuno A, Shintaku M, Fujita Y, Fujiwara Y, et al. (2006) Evolutionary relationships of deep-sea mussels inferred by mitochondrial DNA sequences. Marine Biology 149 (5): 1111–1122. [Google Scholar]
- 14. Lorion J, Kiel S, Faure B, Kawato M, Ho SYW, et al. (2013) Adaptive radiation of chemosymbiotic deep-sea mussels. Preceedings of the Royal Society B 280: 20131243 10.1098/rspb.2013.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thubaut J, Puillandre N, Faure B, Cruaud C, Samadi S (2013) The contrasted evolutionary fates of deep-sea chemosynthetic mussels (Bivalvia, Bathymodiolinae). Ecology and Evolution 3(14): 4748–4766. 10.1002/ece3.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fisher CR (1993) Oxidation of methane by deep sea mytilids in the Gulf of Mexico In: Oremland RS, editor. Biogeochemistry of Global Change: Radiatively Active Trace Gases. New York: Chapman and Hall Inc. pp. 606–618. [Google Scholar]
- 17. Duperron S, Sibuet M, MacGregor BJ, Kuypers MMM, Fisher CR, et al. (2007) Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico. Environmental Microbiology 9 (6): 1423–1438. [DOI] [PubMed] [Google Scholar]
- 18. Bergquist DC, Fleckenstein C, Szalai EB, Knisel J, Fisher CR (2004) Environment drives physiological variability in the cold seep mussel Bathymodiolus childressi. Limnology and Oceanography 49 (3): 706–715. [Google Scholar]
- 19. Carney SL, Formica MI, Divatia H, Nelson K, Fisher CR, et al. (2006) Population structure of the mussel "Bathymodiolus" childressi from Gulf of Mexico hydrocarbon seeps. Deep-Sea Research Part I-Oceanographic Research Papers 53 (6): 1061–1072. [Google Scholar]
- 20. Becker EL, Lee RW, Macko SA, Faure BM, Fisher CR (2010) Stable carbon and nitrogen isotope compositions of hydrocarbon-seep bivalves on the Gulf of Mexico lower continental slope. Deep-Sea Research Part II-Topical Studies in Oceanography 57 (21–23): 1957–1964. 21264038 [Google Scholar]
- 21. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bullettin 19: 11–15. [Google Scholar]
- 22. Olu-Le Roy K, Von Cosel R, Hourdez S, Carney SL, Jollivet D (2007) Amphi-Atlantic cold-seep Bathymodiolus species complexes across the equatorial belt. Deep-Sea Research I 54: 1890–1911. [Google Scholar]
- 23. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3 (5): 294–299. [PubMed] [Google Scholar]
- 24. Faure B, Bierne N, Tanguy A, Bonhomme F, Jollivet D (2007) Evidence for a slightly deleterious effect of intron polymorphisms at the EF1 alpha gene in the deep-sea hydrothermal vent bivalve Bathymodiolus. Gene 406 (1–2): 99–107. [DOI] [PubMed] [Google Scholar]
- 25. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. [Google Scholar]
- 27. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 28. Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics 5(2): 150–163. [DOI] [PubMed] [Google Scholar]
- 29. Felsenstein J (1985) Phylogenies from Gene-Frequencies—a Statistical Problem. Systematic zoology 34 (3): 300–311. [Google Scholar]
- 30. Faure B, Jollivet D, Tanguy A, Bonhomme F, Bierne N (2009) Speciation in the deep-sea: multi-locus analysis of divergence and gene flow between two hybridizing species of hydrothermal vent mussels. PLoSOne 4 (8): e6485 10.1371/journal.pone.0006485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones WJ, Won Y-J, Maas PAY, Smith PJ, Lutz RA, et al. (2006) Evolution of habitat use by deep-sea mussels. Marine Biology 148: 841–851. [Google Scholar]
- 32. Maas PAY, O’Mullan GD, Lutz RA, Vrijenhoek RC (1999) Genetic and morphometric characterization of mussels (Bivalvia: Mytilidae) from Mid-Atlantic hydrothermal vents. Biological Bulletin 196: 265–272. [DOI] [PubMed] [Google Scholar]
- 33. Van Der Heijden K, Petersen JM, Dubilier N, Borowski C (2012) Genetic connectivity between north and south mid-atlantic ridge chemosynthetic bivalves and their symbionts. PLOS ONE 7(7): e39994 10.1371/journal.pone.0039994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nei M (1987) Molecular Evolutionary Genetics. New York: Columbia Univ. Press. [Google Scholar]
- 35. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rozas J, Sànchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 37. Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online (1): 47–50. [PMC free article] [PubMed] [Google Scholar]
- 38. Li W-H, Graur D (1991) Fundamentals of molecular evolution Sunderland, MA: Sinauer Associates. [Google Scholar]
- 39. Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16 (1): 37–48. [DOI] [PubMed] [Google Scholar]
- 40. Cassens I, Mardulyn P, Milinkovitch MC (2005) Evaluating intraspecific "Network" construction methods using simulated sequence data: Do existing algorithms outperform the global maximum parsimony approach? Systematic Biology 54 (3): 363–372. [DOI] [PubMed] [Google Scholar]
- 41. Fisher CR, Brooks JM, Vodenichar JS, Zande JM, Childress JJ, et al. (1993) The Cooccurrence of Methanotrophic and Chemoautotrophic Sulfur-Oxidizing Bacterial Symbionts in a Deep-Sea Mussel. Marine Ecology-Pubblicazioni Della Stazione Zoologica Di Napoli I 14 (4): 277–289. [Google Scholar]
- 42. Craddock C, Hoeh WR, Gustafson RG, Lutz RA, Hashimoto J, et al. (1995) Evolutionary relationships among deep-sea Mytilids (Bivalvia, Mytilidae) from hydrothermal vents and cold-water methane sulfide seeps. Marine Biology 121 (3): 477–485. [Google Scholar]
- 43. Won YJ, Maas PAY, Van Dover CL, Vrijenhoek RC (2002) Habitat reversal in vent and seep mussels: seep species, Bathymodiolus heckerae, derived from vent ancestors. Cahiers De Biologie Marine 43 (3–4): 387–390. [Google Scholar]
- 44. Genio L, Johnson SB, Vrijenhoek RC, Cunha MR, Tyler PA, et al. (2008) New record of "Bathymodiolus" mauritanicus Cosel 2002 from the Gulf of Cadiz (NE Atlantic) mud volcanoes. Journal of Shellfish Research 27 (1): 53–61. [Google Scholar]
- 45. Won YJ, Jones WJ, Vrijenhoek RC (2008) Absence of cospeciation between deep-sea Mytilids and their thiotrophic endosymbionts. Journal of Shellfish Research 27 (1): 129–138. [Google Scholar]
- 46. Arellano SM, Young CM (2009) Spawning, Development, and the Duration of Larval Life in a Deep-Sea Cold-Seep Mussel. Biological Bulletin 216 (2): 149–162. [DOI] [PubMed] [Google Scholar]
- 47. Olu K, Cordes EE, Fisher CR, Brooks JM, Sibuet M, et al. (2010) Biogeography and Potential Exchanges Among the Atlantic Equatorial Belt Cold-Seep Faunas. PLoS ONE 5 (8): e11967, 11961–11911. 10.1371/journal.pone.0011967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vrijenhoek RC (2010) Genetic diversity and connectivity of deep-sea hydrothermal vent metapopulations. Molecular Ecology 19 (20): 4391–4411. 10.1111/j.1365-294X.2010.04789.x [DOI] [PubMed] [Google Scholar]
- 49. Meyer CP, Paulay G (2005) DNA barcoding: Error rates based on comprehensive sampling. Plos Biology 3 (12): 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 51. Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theoretical Population Biology 7: 256–276. [DOI] [PubMed] [Google Scholar]
- 53. Schaeffer SW (2002) Molecular population genetics of sequence length diversity in the Adh region of Drosophila pseudoobscura . Genetics Research. 80: 163–175. [DOI] [PubMed] [Google Scholar]
- 54. Rand DM, Kann LM (1996) Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, Mice, and Humans. Mol. Biol. Evol. 13(6): 735–748. [DOI] [PubMed] [Google Scholar]
- 55. Hudson RR, Kreitman M, Aguadé M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Galtier N, Depaulis F, Barton NH (2000) Detecting bottlenecks and selective sweeps from DNA sequence polymorphysm. Genetics 155: 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Plouviez S, Shank TM, Faure B, Daguin-Thiebaut C, Viard F, et al. (2009) Comparative phylogeography among hydrothermal vent species along the East Pacific Rise reveals vicariant processes and population expansion in the South. Molecular Ecology 18 (18): 3903–3917. 10.1111/j.1365-294X.2009.04325.x [DOI] [PubMed] [Google Scholar]
- 58. Nix E, Fisher CR, Scott KM, Vodenichar J (1995) Physiological ecology of a mussel with methanotrophic symbionts at three hydrocarbon seep sites in the Gulf of Mexico. Marine Biology 122: 605–617 [Google Scholar]
- 59. Bergquist DC, Andras J, McNelis T, Howlett S, van Horn MJ, et al. (2003) Succession in upper Louisiana slope cold seep vestimentiferan aggregations: the importance of spatial variability. Marine Ecology Progress Series 24: 31–44. [Google Scholar]
- 60. Cordes EE, Hourdez S, Predmore BL, Redding ML, Fisher CR (2005) Succession of hydrocarbon seep communities associated with the long-lived foundation species Lamellibrachia luymesi . Marine Ecology Progress Series 305: 17–29. [Google Scholar]
- 61. Fisher CR, Roberts H, Cordes E, Bernard B (2007) Cold seeps and associated communities of the Gulf of Mexico. Oceanography 20–4: 68–79. [Google Scholar]
- 62. Smith EB, Scott KM, Nix ER, Korte C, Fisher CR (2000) Growth and condition of seep mussels (Bathymodiolus childressi) at a Gulf of Mexico Brine Pool. Ecology 81: 2392–2403. [Google Scholar]
- 63. Plouviez S, Faure B, Le Guen D, Lallier FH, Bierne N, et al. (2013) A new barrier to dispersal trapped old genetic clines that escaped the easter microplate tension zone of the Pacific vent mussels. PLoS ONE 8(12): e81555 10.1371/journal.pone.0081555 [DOI] [PMC free article] [PubMed] [Google Scholar]