Abstract
The signaling component of the mammalian Fibroblast Growth Factor (FGF) family is comprised of eighteen secreted proteins that interact with four signaling tyrosine kinase FGF receptors (FGFRs). Interaction of FGF ligands with their signaling receptors is regulated by protein or proteoglycan cofactors and by extracellular binding proteins. Activated FGFRs phosphorylate specific tyrosine residues that mediate interaction with cytosolic adaptor proteins and the RAS-MAPK, PI3K-AKT, PLCγ, and STAT intracellular signaling pathways. Four structurally related intracellular non-signaling FGFs interact with and regulate the family of voltage gated sodium channels. Members of the FGF family function in the earliest stages of embryonic development and during organogenesis to maintain progenitor cells and mediate their growth, differentiation, survival, and patterning. FGFs also have roles in adult tissues where they mediate metabolic functions, tissue repair, and regeneration, often by reactivating developmental signaling pathways. Consistent with the presence of FGFs in almost all tissues and organs, aberrant activity of the pathway is associated with developmental defects that disrupt organogenesis, impair the response to injury, and result in metabolic disorders, and cancer. © 2015 Wiley Periodicals, Inc.
Introduction
The Fibroblast Growth Factor (FGF) family is comprised of secreted signaling proteins (secreted FGFs) that signal to receptor tyrosine kinases and intracellular non-signaling proteins (intracellular FGFs (iFGFs)) that serve as cofactors for voltage gated sodium channels and other molecules (Table1(a) and Figure 1(a)). Additionally, secreted FGFs and iFGFs may have direct functions in the nucleus and functional interactions with other cellular proteins. Members of both branches of the FGF family are related by core sequence conservation and structure and are found in vertebrates and invertebrates.1,2 Secreted FGFs are expressed in nearly all tissues and they serve essential roles in the earliest stages of embryonic development, during organogenesis, and in the adult, where they function as homeostatic factors that are important for tissue maintenance, repair, regeneration, and metabolism (Table2(a)). In general, secreted FGFs function as autocrine or paracrine factors (canonical FGFs; also called paracrine FGFs), however, three members of the secreted FGFs have evolved to function as endocrine factors (endocrine FGFs) with essential roles in the adult where they regulate phosphate, bile acid, carbohydrate and lipid metabolism in addition to the canonical FGF functions that control cell proliferation, differentiation and survival.75–77,98,149–163
Table 1.
Nomenclature of the Mammalian Fgf and Fgfr family
| HUGO/MGI Symbol | Name | Alternative Symbol | Name, Comments |
|---|---|---|---|
| (a) Fgf | |||
| FGF1/Fgf1 | Fibroblast Growth Factor 1 | aFgf | Acidic Fgf |
| Hbgf1 | Heparin-binding growth factor 1 | ||
| Ecgr | Endothelial cell growth factor | ||
| FGF2/Fgf2 | Fibroblast Growth Factor 2 | bFgf | Basic Fgf |
| Hbgf2 | Heparin-binding growth factor 2 | ||
| FGF3/Fgf3 | Fibroblast Growth Factor 3 | Int-2 | Int-2 oncogene |
| V-Int-2 | MMTV integration site 2 | ||
| FGF4/Fgf4 | Fibroblast Growth Factor 4 | Hst1 | Human stomach tumor oncogene |
| Hstf1 | Heparin secretory transforming protein 1 | ||
| K-Fgf, Kfgf | Kaposi sarcoma Fgf | ||
| FGF5/Fgf5 | Fibroblast Growth Factor 5 | ||
| FGF6/Fgf6 | Fibroblast Growth Factor 6 | Hst2 | Hst2 oncogene |
| FGF7/Fgf7 | Fibroblast Growth Factor 7 | Kgf | Keratinocyte growth factor |
| FGF8/Fgf8 | Fibroblast Growth Factor 8 | Aigf | Androgen induced growth factor |
| Kal6 | |||
| FGF9/Fgf9 | Fibroblast Growth Factor 9 | Gaf | Glia activating factor |
| Eks | Elbow knee synostosis | ||
| FGF10/Fgf10 | Fibroblast Growth Factor 10 | Kgf-2 | Keratinocyte growth factor 2 |
| FGF11/Fgf11 | Fibroblast Growth Factor 11 | Fhf3 | Fibroblast Growth Factor homologous factor 3 |
| FGF12/Fgf12 | Fibroblast Growth Factor 12 | Fhf1 | Fibroblast Growth Factor homologous factor 1 |
| FGF13/Fgf13 | Fibroblast Growth Factor 13 | Fhf2 | Fibroblast Growth Factor homologous factor 2 |
| FGF14/Fgf14 | Fibroblast Growth Factor 14 | Fhf4 | Fibroblast Growth Factor homologous factor 4 spinocerebellar ataxia 27 |
| Sca27 | |||
| Fgf15 | Fibroblast Growth Factor 15 | Rodent ortholog of vertebrate Fgf19 | |
| FGF16/Fgf16 | Fibroblast Growth Factor 16 | ||
| FGF17/Fgf17 | Fibroblast Growth Factor 17 | Called FGF-13 in some older literature | |
| FGF18/Fgf18 | Fibroblast Growth Factor 18 | ||
| FGF19 | Fibroblast Growth Factor 19 | Human ortholog of rodent Fgf15 | |
| FGF20/Fgf20 | Fibroblast Growth Factor 20 | ||
| FGF21/Fgf21 | Fibroblast Growth Factor 21 | ||
| FGF22/Fgf22 | Fibroblast Growth Factor 22 | ||
| FGF23/Fgf23 | Fibroblast Growth Factor 23 | ||
| (b) Fgfr | |||
| FGFR1/Fgfr1 | Fgf receptor 1 | Flg | Fms-like gene |
| Flt2 | Fms-like tyrosine kinase 2 | ||
| Cek | Chicken embryo kinase 1 | ||
| KAL2 | Kallman syndrome 2 | ||
| K-sam | KATO-III cell-derived stomach cancer amplified gene | ||
| FGFR2/Fgfr2 | Fgf Receptor 2 | Bek | Bacterial expressed kinase |
| Cek3 | Chicken embryo kinase 3 | ||
| Kgfr | KGF receptor | ||
| FGFR3/Fgfr3 | Fgf Receptor 3 | Cek2 | Chicken embryo kinases 2 |
| Ach | Achondroplasia | ||
| FGFR4/Fgfr4 | Fgf Receptor 4 | Tkf | Tyrosine kinase related to Fibroblast Growth Factor receptor |
| FGFRL1/Fgfrl1 | Fgf receptor like 1 | Fgfr5 | Fgf receptor 5 |
Figure 1.
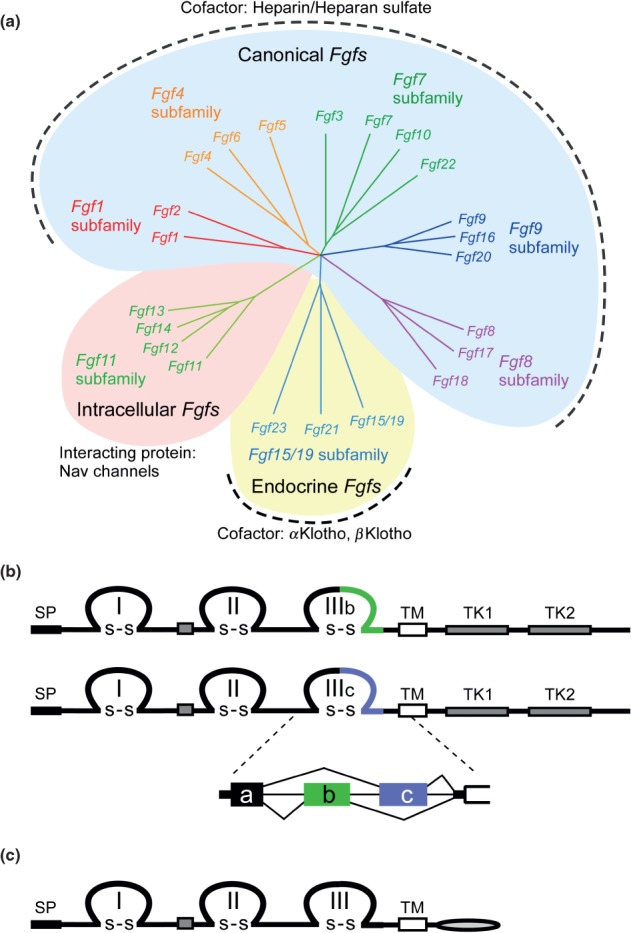
FGF and FGFR families. (a) Phylogenetic analysis suggests that 22 Fgf genes can be arranged into seven subfamilies containing two to four members each. Branch lengths are proportional to the evolutionary distance between each gene. The Fgf1, Fgf4, Fgf7, Fgf8, and Fgf9 subfamily genes encode secreted canonical FGFs, which bind to and activate FGFRs with heparin/HS as a cofactor. The Fgf15/19 subfamily members encode endocrine FGFs, which bind to and activate FGFRs with the Klotho family protein as a cofactor. The Fgf11 subfamily genes encode intracellular FGFs, which are non-signaling proteins serving as cofactors for voltage gated sodium channels and other molecules. (b) Schematic representations of FGFR protein structures are shown. FGFR is a receptor tyrosine kinase of ∼800 amino acids with several domains including three extracellular immunoglobulin-like domains (I, II, and III), a transmembrane domain (TM), and two intracellular tyrosine kinase domains (TK1 and TK2). SP indicates a cleavable secreted signal sequence. The Fgfr gene family is comprised of four members, Fgfr1-Fgfr4. Among them, Fgfr1–Fgfr3 generate two major splice variants of immunoglobulin-like domain III, referred to as IIIb and IIIc, which are essential determinants of ligand-binding specificity. (c) The schematic representation of FGFRL1/FGFR5 protein structure is shown. FGFRL1, with structural similarity to FGFRs, is a membrane protein of ∼500 amino acids with three extracellular immunoglobulin-like domains (I, II, and III), a transmembrane domain (TM), and a short intracellular tail with no tyrosine kinase domain. SP indicates a cleavable secreted signal sequence.
Table 2.
Phenotypes of Null and Tissue-Specific Fgf Mutations
| Gene Name | Viability /Age at Death of Null Mutant | Null Phenotype (Organ, Structure, or Cell Type Affected) | Tissue-Specific (Conditional) Phenotypes, Redundant Phenotypes, Phenotypes Induced by Physiological Challenge | Selected References |
|---|---|---|---|---|
| (a) Phenotypes of germline and conditional loss-of-function Fgf mutations in mice | ||||
| Fgf1 | Viable | No apparent phenotype | An aggressive diabetic phenotype with white adipocyte remodeling on high-fat diet | 3,4 |
| Fgf2 | Viable | Cortical neuron, vascular smooth muscle, blood pressure, skeletal development, and wound healing | Decreased cardiac hypertrophy induced by ischemic injury and delayed wound healing; Increased bone mineralization in high molecular weight isoform knockout | 5–11 |
| Fgf3 | Viable | Inner ear and skeletal development | Heart development (redundant with Fgf10) | 12,13 |
| Fgf4 | E4-5 | Blastocyst inner cell mass | Limb bud development (redundant with Fgf8) | 14–17 |
| Fgf5 | Viable | Hair follicle development | 18 | |
| Fgf6 | Viable | Muscle development | Muscle regeneration | 19–21 |
| Fgf7 | Viable | Hair follicle and ureteric bud development and synaptogenesis | Thymus regeneration (radiation injury) and wound healing | 22–26 |
| Fgf8 | E7 | Gastrulation | Heart field, limb, somitogenesis, kidney, CNS, inner ear development, spermatogenesis | 27–39 |
| Fgf9 | P0 | Lung, heart, skeletal, gonad, inner ear, and intestine development | Migration of cerebellar granule neurons and kidney agenesis (redundant with Fgf20) | 40–53 |
| Fgf10 | P0 | Limb bud, lung bud, trachea, thymus, pancreas, pituitary, palate, tongue epithelium, cecum, kidney, submandibular, salivary, lacrimal, and mammary gland, heart, stomach, and white adipose tissue | Lung branching morphogenesis and inner ear development (redundant with Fgf3) | 54–68 |
| Fgf11 | Viable | No identified phenotype | (unpublished) | |
| Fgf12 | Viable | No identified phenotype | Severe ataxia and motor weakness (redundant with Fgf14) | 69 |
| Fgf13 | Viable | Neuronal migration, learning and memory deficits, and microtubule binding | 69–73 | |
| Fgf14 | Viable | Ataxia, motor weakness, learning and memory deficits, and impaired neuronal excitability | Severe ataxia and motor weakness (redundant with Fgf12) | 69,74 |
| Fgf15 | E13.5-P7 | Cardiac outflow tract development, neurogenesis, and bile acid metabolism | Liver regeneration | 75–80 |
| Fgf16 | Viable | Heart development | Promotes cardiac remodeling induced by angiotensin II | 81–83 |
| Fgf17 | Viable | Cerebellum and frontal cortex development | 31,84 | |
| Fgf18 | P0 | CNS, skeletal, palate, and lung development | 40,85–89 | |
| Fgf20 | Viable | Guards hair, teeth, cochlea, and kidney development | Kidney agenesis (redundant with Fgf9) | 40,90–92 |
| Fgf21 | Viable | Energy/lipid metabolism | 75,93,94 | |
| Fgf22 | Viable | Synaptogenesis | Decreased skin papillomas formation following carcinogenesis challenge | 22,95–97 |
| Fgf23 | PW4-13 | Phosphate and vitamin D homeostasis, deafness, middle ear development | 98–102 | |
| (b) Phenotypes of germline and conditional loss-of-function Fgfr mutations in mice | ||||
| Fgfr1 | E7.5-9.5 | Gastrulation, Blastocyst inner cell mass | Hematopoietic cell engraftment | 39,103–114 |
| Osteoblast maturation | ||||
| Limb bud development | ||||
| Hippocampal progenitor cell proliferation | ||||
| Inner ear sensory epithelium | ||||
| Deletion of Ig domain 1 (defect in node regression) | ||||
| Adipocyte metabolism | ||||
| Endothelial Tgfβ expression and endothelial-mesenchymal transition; Endothelial regulation of CXCR4 in liver regeneration and fibrosis | ||||
| Spermatogenesis | ||||
| Fgfr2 | E10-11 | Placenta, no limb buds | Skeletal, lung, limb bud, CNS, GI tract, skin, and adrenal cortex development in Fgfr2b null mice | 115–119 |
| Fgfr1/2 | Myelin sheath thickness in oligodendrocyte | 120–126 | ||
| Kidney, metanephric mesenchyme, ureteric bud, ocular gland development | ||||
| Angiogenesis, vascular integrity | ||||
| Hepato-cytoprotective through regulation of cytochrome P450 enzymes | ||||
| Fgfr3 | Viable | Skeletal overgrowth, inner ear, brain, articular cartilage, oligodendrocyte differentiation, pancreatic growth, intestinal crypt cell growth arrest | Alveolar septation and elastogenesis (redundant with Fgfr4) | 10,127–139 |
| Fgfr4 | Viable | Cholesterol metabolism and bile acid synthesis | Increased liver injury and fibrosis induced by carbon tetrachloride | 131,139–146 |
| Alveolar septation and elastogenesis (redundant with Fgfr3) | ||||
| Vitamin D homeostasis (redundant with Fgfr3) | ||||
| Phosphate homeostasis (redundant with Fgfr1) | ||||
| Fgfrl1 | P0 | Kidney, diaphragm, skeleton | 147,148 | |
At the cellular level, secreted FGFs regulate fundamental cellular processes that include positive and negative regulation of proliferation, survival, migration, differentiation, and metabolism. During early development, FGFs regulate differentiation of the inner cell mass into epiblast and primitive endoderm lineages.164–167 Later in development, FGFs have key roles in organogenesis, for example in the regulation of the anterior and secondary heart fields,168,169 induction of limb buds54,55,170 and lung buds,54,55 ventral liver and pancreas,171,172 kidney development,27,40,120,121,147, inner ear development,12,28,41,56,90,103,104,127,173 and brain development.174,175
In the adult, FGFs have important roles in response to injury and tissue repair.176 FGF signaling is cardioprotective following ischemic injury to the heart,177–179 and is important for epithelial repair in the lung and in wound healing.180–182 FGF signaling, however, may also increase or decrease tissue fibrosis.81,183–185 Endocrine FGFs mediate mineral, metabolic, energy, and bile acid homeostasis.75,98,186,187 FGF receptor (Fgfr) mutation, amplification, and gene fusions can drive abnormal morphogenesis, the progression of several types of cancer, and provide escape pathways for drugs that target other oncogenic tyrosine kinase receptors.152,188–196
Given the ubiquitous roles for FGF signals in development, homeostasis, and disease, tight regulation of the pathway is essential. Canonical FGFs are tightly bound to heparin/heparan sulfate (HS) proteoglycans (HSPGs), which function to limit diffusion through the extracellular matrix (ECM) and serve as cofactors that regulate specificity and affinity for signaling FGFRs.153,197–201 The endocrine FGFs, evolved with reduced affinity for heparin/HS and the requirement for a protein cofactor, αKlotho, βKlotho, or KLPH for receptor binding.75,202 Additional regulation is provided by a fifth non-tyrosine kinase FGFR (FGFRL1) which can bind FGF ligands and possibly function as a decoy receptor, dimerization-induced inhibitor of tyrosine kinase FGFRs, or modulator of receptor turnover or signaling.203 Downstream of the signaling tyrosine kinase FGFRs, intracellular signaling cascades are also tightly regulated by specialized adaptor proteins such as FGFR substrate 2α (FRS2α) and regulators of the RAS-MAPK and PI3K-AKT pathways such as Sprouty (SPRY) proteins151,204–207 (Figure 3(a)).
Figure 3.
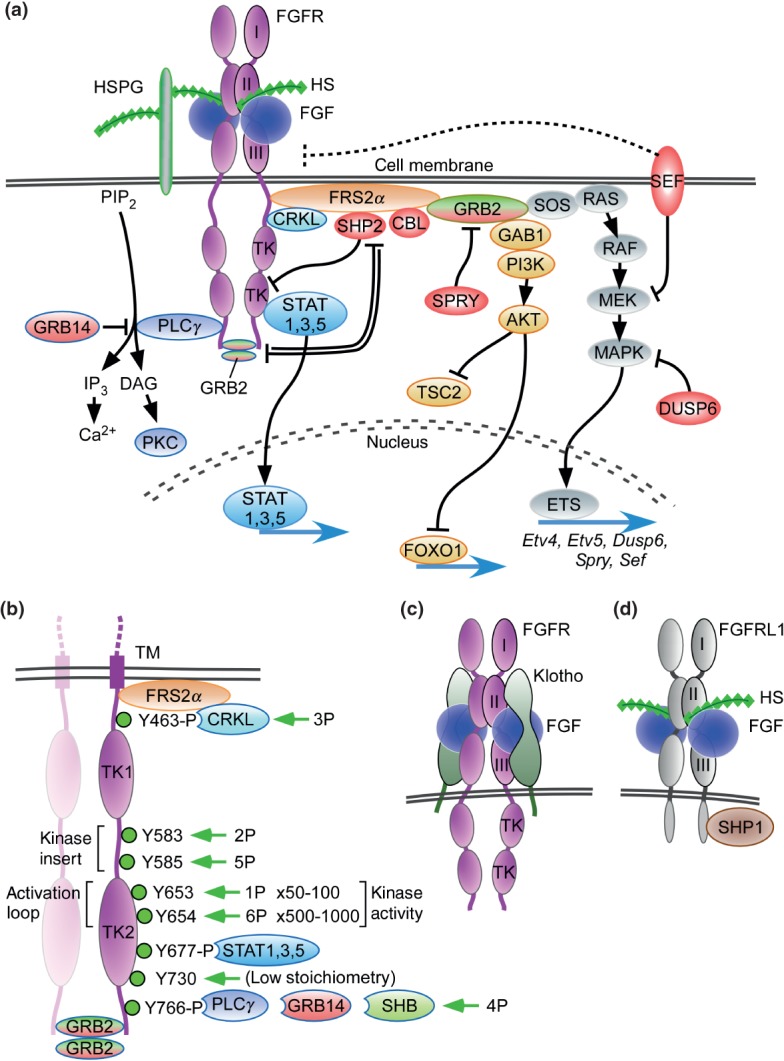
FGF signaling pathways. (a) Binding of canonical FGFs to FGFR with HS (or HSPG) as a cofactor induces the formation of ternary FGF-FGFR-HS complex, which activates the FGFR intracellular tyrosine kinase domain by phosphorylation of specific tyrosine residues. The activated receptor is coupled to intracellular signaling pathways including the RAS-MAPK, PI3K-AKT, PLCγ, and STAT pathways. The RAS-MAPK pathway: The major FGFR kinase substrate, FRS2α, which is constitutively associated with the juxtamembrane region of FGFR (peptide: MAVHKLAKSIPLRRQVTVSADS), interacts with CRKL bound to pY463 and is phosphorylated by the activated FGFR kinase. Phosphorylated FRS2α recruits the adaptor protein GRB2, which then recruits the guanine nucleotide exchange factor SOS. The recruited SOS activates the RAS GTPase, which then activates the MAPK pathway. MAPK activates members of the Ets transcription factor family such as Etv4 (Pea3) and Etv5 (Erm) and negative regulators of the FGF signaling pathways such as SHP2, CBL, SPRY, SEF, and DUSP6. The PI3-AKT pathway: The recruited GRB2 also recruits the adaptor protein GAB1, which then activates the enzyme PI3K, which then phosphorylates the enzyme AKT. AKT has multiple activities including activation of the mTOR complex 1 through inhibition of TSC2 and phosphorylation of the FOXO1 transcription factor causing it to exit the nucleus. The PLCγ pathway: Activated FGFR kinase recruits and activates the enzyme PLCγ, which produces IP3 and DAG by the hydrolysis of PIP2. IP3 induces calcium ion release from intracellular stores and the activation of downstream signaling pathways. DAG activates the enzyme PKC and its downstream signaling pathways. GRB14 inhibits activation of PLCγ. The STAT pathway: FGFR kinase also activates STAT1, 3, and 5. STAT3 interacts with phosphorylated tyrosine 677 (pYxxQ motif). These activated signaling pathways mostly regulate gene expression in the nucleus. SPRY interacts with GRB2 to inhibit the RAS-MAPK pathway and to regulate the PI3K-AKT pathway. GRB2 dimers are docked at the c-terminus of FGFR2 where they inhibit SHP2, allowing low-level receptor kinase activity. Molecules shaded red generally function to inhibit FGFR signaling. (b) Dimerization of the FGFR1 kinase domain leads to sequential phosphorylation of tyrosine residues (1P–6P) leading to increasing activity of the FGFR kinase and phosphorylation of tyrosine substrates for CRKL, STAT, GRB14, and PLCγ binding. In the first phase of activation, Y653 (1P), in the activation loop, is phosphorylated, resulting in a 50- to 100-fold increase in kinase activity. In the third phase of activation, Y654 (6P), in the activation loop, is phosphorylated, resulting in an overall 500–1000 fold increase in kinase activity. Y730 is weakly phosphorylated. Phosphorylation of Y677 allows docking of STAT3 and phosphorylation of Y766 allows docking of either GRB14 or PLCγ. Ligand-induced receptor activation phosphorylates GRB2, leading to its dissociation from the receptor. Tyrosine residues correspond to human FGFR1 (accession NP_075598). (c) Binding of endocrine FGF to FGFR with Klotho as a cofactor induces the formation of ternary FGF-FGFR-Klotho complex, which leads to activation of the FGFR tyrosine kinase. (d) FGFRL1 is a protein containing three extracellular immunoglobulin-like domains with similarity to FGFRs. FGFRL1 has a single transmembrane domain, and a short intracellular tail with no tyrosine kinase domain. The short cytoplasmic domain contains an SH2 binding motif that interacts with SHP1. FGFRL1 is not simply a decoy receptor, but rather a non-tyrosine kinase signaling molecule.
iFGFs (also known as FGF homologous factors (FHFs)) are essential regulators of neuronal and myocardial excitability. However, whether iFGFs are required during normal embryonic development is currently not known. Several proteins are known to directly interact with iFGFs. These include members of the voltage gated sodium channel family,154 IB2 (MAPK8IP2, Mitogen-activated protein kinase 8-interacting protein 2),208 β-tubulin,70 and NEMO209 (NF-κB essential modulator). Analysis of evolutionary relationships in the FGF family suggests that iFGFs may be the first members of the family to evolve, followed by the acquisition of a signal peptide for secretion, and affinity for heparin/HS to limit diffusion and regulate receptor binding.210 The most recent evolutionary event led to the endocrine branch of the FGF family, which has reduced affinity for heparin/HS and a requirement for Klotho family cofactors for receptor binding.
In this review we will focus on the roles and regulation of FGF signaling pathways that function during vertebrate organogenesis and on how gain and loss-of-function mutations in the FGF pathway result in developmental or metabolic disease and cancer.
Pathway Components
Fibroblast Growth Factors
Historical Perspective
Embryo extracts and brain extracts were shown to promote the growth of chicken periosteal fibroblast as early as 1939.211 A proteinaceous ‘Fibroblast Growth Factor’ activity was first identified in an extract from bovine pituitary in 1973.212 This activity was shown to be protease sensitive and thermolabile and could stimulate the proliferation of 3 T3 fibroblasts at low (ng/ml) concentrations. This activity was partially purified in 1975213 and purified to homogeneity in 1983214 and would later be referred to as basic FGF (bFGF or FGF2) due the overall basic composition of amino acids and high isoelectric point. Purification of a factor with similar mitogenic activity from bovine brain that was free of myelin basic protein fragments identified a second Fibroblast Growth Factor-like activity with a low isoelectric point that was eventually referred to as acidic FGF215–220 (aFGF or FGF1). This factor was also found to be identical to an activity called endothelial cell growth factor221 (ECGF) and related to FGF2.217 In addition to stimulation of 3 T3 cell proliferation, these growth factors were found to promote proliferation of a wide variety of mesoderm-derived cells such as endothelial cells.217,220–222 cDNA clones for FGF1 were first isolated from a human brain cDNA library in 1986.223 cDNA clones for Fgf1 and Fgf2 were also isolated from bovine pituitary cDNA libraries in 1986.224 Additional members of the FGF family were identified as growth factors for cultured cells, as oncogenes tagged by retroviral insertions, as genes responsible for hereditary diseases, or by homology-based PCR or homology-based searches of DNA databases.152,153,99
The mammalian Fgf family contains 22 genes, 18 of which encode molecules known to signal through FGF tyrosine kinase receptors (Table1(a)). The secreted signaling FGFs can be grouped into subfamilies based on biochemical function, sequence similarities, and evolutionary relationships. The current consensus is that there are 5 subfamilies of paracrine FGFs, one subfamily of endocrine FGFs, and one subfamily of intracellular FGFs150,153,157,158,210,225,226 (Figure 1(a)). Fgf15 and Fgf19 are likely to be orthologs in vertebrates. The orthologs were named Fgf15 in rodents and Fgf19 in other vertebrates. In this review, we refer to these as Fgf15/19.
Canonical (Secreted) FGFs
FGF1 Subfamily
The FGF1 subfamily is comprised of FGF1 and FGF2 (Figure 1(a)). These FGFs lack classical secretory signal peptides but are nevertheless readily exported from cells by direct translocation across the cell membrane.227 The mechanism of translocation is thought to involve a chaperone complex that includes synaptotagmin-1 and the calcium binding protein S100A13.228,229 FGF1 and FGF2 have also been found in the nucleus of some cells. The mechanisms by which FGFs transit through the cell are poorly understood, but are thought to require binding to and activating cell surface tyrosine kinase FGFRs with heparin/HS as a cofactor and interaction with HSP90.230,231 Several studies have shown that extracellular FGF1 passes through the plasma membrane, moves through the cytosol, and enters the nucleus.232,233 Potential functions of nuclear FGF1 include regulation of the cell cycle, cell differentiation, survival, and apoptosis.234,235 FGF1 is the only FGF that can activate all FGFR splice variants (Figure 2; see below).
Figure 2.
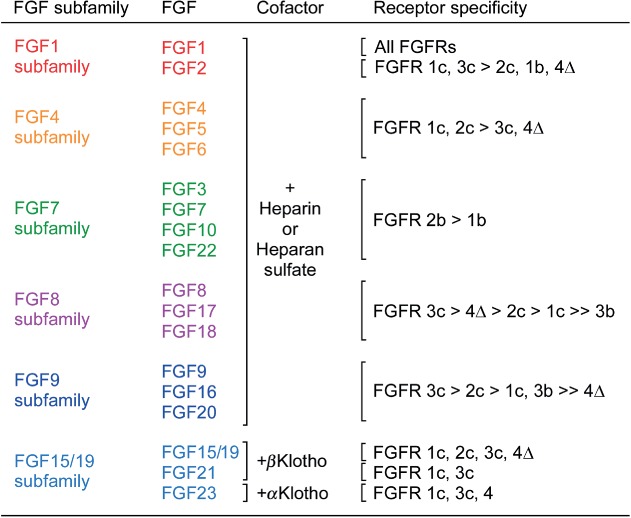
Receptor specificity of canonical and endocrine FGFs. The six subfamilies of signaling FGFs use either heparin-like molecules or Klotho molecules as cofactors for receptor binding. Data is derived from receptor activation assays using BaF3 cells, L6 myoblasts, or HEK293 cells transfected with individual splice variants of FGFRs or by direct binding studies.140,141,162,237,238,246–254 FGFR4Δ is a two immunoglobulin-like domain form of FGFR4.
FGF4 Subfamily
Phylogenetic analysis suggests that the FGF4 family is comprised of FGF4, FGF5 and FGF6236 (Figure 1(a)). However, there is some controversy as to whether FGF5 should be included in this subfamily, because synteny relationships could be used to place FGF5 in the FGF1 subfamily.210 All members of this subfamily are secreted proteins with cleavable N-terminal signal peptides that mediate biological responses as extracellular proteins by binding to and activating FGFRs.158 These FGFs activate IIIc splice variants of FGFRs 1–3 and FGFR4237,238 (Figure 2; see below).
FGF7 Subfamily
Phylogenetic analysis suggests that the FGF7 family is comprised of FGF3, FGF7, FGF10 and FGF22236 (Figure 1(a)). However, there is some controversy as to whether FGF3 should be included in this subfamily. Sequence homology and biochemical properties support inclusion in the FGF7 subfamily, while chromosomal localization supports inclusion with FGF4 and FGF6.210 One recent study proposed an eighth subfamily composed of only FGF3.226 FGFs, 3, 7, 10, and 22 preferentially activate the IIIb splice variant of FGFR2 and FGF3 and FGF10 also activate the IIIb splice variant of FGFR1237,238 (Figure 2; see below).
FGF8 Subfamily
The FGF8 subfamily is comprised of FGF8, FGF17, and FGF18236 (Figure 1(a)). Members of this subfamily contain an N-terminal cleaved signal peptide. These FGFs activate IIIc splice variants of FGFRs 1–3 and FGFR4237,238 (Figure 2; see below).
FGF9 Subfamily
The FGF9 subfamily is comprised of FGF9, FGF16, and FGF20 (Figure 1(a)). This subfamily does not have a classical N-terminal signal peptide but does have an internal hydrophobic sequence that functions as a non-cleaved signal for transport into the endoplasmic reticulum and secretion from cells.239–241 This subfamily has the unique properties of activation of the IIIb splice variant of FGFR3 in addition to FGFR4 and the IIIc splice variants of FGFRs 1, 2 and 3237,238 (Figure 2; see below).
FGF15/19 Subfamily (Endocrine FGFs)
The FGF15/19 subfamily is comprised of FGF15/19, FGF21, and FGF2375,242 (Figure 1(a)). These FGFs are unique in that they primarily function as endocrine factors and are referred to as endocrine FGFs. In contrast to canonical FGFs, endocrine FGFs bind to heparin/HS with very low affinity.243 The reduced heparin-binding affinity facilitates release from ECM and allows these FGFs to function as endocrine factors. However, endocrine FGFs still mediate their biological responses in an FGFR-dependent manner, but instead of heparin/HS as cofactors for receptor binding and activation, endocrine FGFs require members of the Klotho family, αKlotho (Klotho), βKlotho, and Klotho-LPH related protein (KLPH), which has also been called Lactase-like Klotho (Lctl) or γKlotho. αKlotho and βKlotho are structurally related single-pass transmembrane proteins of ∼1000 amino acids with a short cytoplasmic domain. FGF15/19 and FGF21 signaling requires βKlotho1,75,156,244–246 (see below). In vitro assays for receptor activation using BaF3 cells or L6 myoblasts that co-express FGFR splice variants and βKlotho shows that FGF19 can activate FGFR1c, FGFR2c, FGFR3c, and FGFR4, while FGF21 only activates FGFR1c and FGFR3c162,246 (Figure 2). In vivo studies show that FGF21 directly regulates hepatocyte and adipocyte metabolism through interactions with FGFR1 and βKlotho.162,245,255 By contrast, FGF19, but not FGF21, activates FGFR4, which functions in hepatocytes as a proliferative signal and as a regulator of bile acid synthesis, and has been implicated in the etiology or progression of hepatocellular carcinoma.143,162,256,257 KLPH has been shown to enhance signaling of FGF19 in HEK293 cells,160 however, the in vivo function of KLPH is not known. FGF23 signaling is mediated through the activation of FGFR1c, FGFR3c, and FGFR4, together with the cofactor, αKlotho140,141,247,248,258 (Figure 2; see below).
Intracellular FGFs
FGF11 Subfamily
The FGF11 subfamily (FGF11, FGF12, FGF13, FGF14) is also known as iFGFs259 (Figure 1(a)). iFGFs are not secreted and have no identified interaction with signaling FGFRs.260 iFGFs interact with the cytosolic carboxy terminal tail of voltage gated sodium (Nav) channels. This interaction may help to regulate the subcellular localization of Nav channels at the axon initial segment during development and the ion-gating properties of the channel in mature neurons and other excitable cells such as cardiomyocytes.69,261–264 Additional interacting proteins have been identified for some iFGFs. For example, FGF12 (FHF1) was shown to interact with the MAP kinase scaffolding protein, IB2 (MAPK8IP2),265 and FGF13 (FHF2) was shown to interact with microtubules.70
Fibroblast Growth Factor Receptors
Historical Perspective
Tyrosine kinase activity was first associated with signaling by brain-derived growth factor, an activity with similar properties to FGF1.266 Subsequently, purified FGF1 and FGF2 were shown to cause phosphorylation of a 90 kDa protein in Swiss 3 T3 cells.267 Crosslinking of 125I-FGF2 was used to tag and purify a receptor protein from chicken embryo membrane fractions. Sequence of tryptic peptides from the chicken FGF receptor, were found to match a partial human cDNA clone called FLG (Fms-like gene),268 now referred to as FGF receptor 1 (FGFR1) (Table1(b)). This information was used to clone a full-length cDNA from a chicken library. The cDNA encoded a 91.7 kDa protein with an N-terminal hydrophobic signal sequence, three extracellular immunoglobulin-like domains, and an intracellular tyrosine kinase domain (Figures 1(b) and 3). The chicken cDNA showed high homology to a cDNA isolated from a human library (90–100% in the tyrosine kinase domain) and the partial FLG cDNA clone, and 84% sequence identity to a mouse partial cDNA called Bek (bacterial expressed kinase).269 Bek is now referred to as FGF receptor 2 (FGFR2) (Table1(b)). Homology based cloning was used to identify Fgfr3 and Fgfr4.270–273 A receptor for FGF7/KGF was isolated by functional cloning in NIH3T3 cells that expressed FGF7.274 Sequencing revealed a two immunoglobulin-like domain variant with identity to BEK in the tyrosine kinase domain.
Determinants of Ligand Binding Affinity and Specificity of FGFRs
Immunoglobulin-like domains II and III, and the linker region between these domains regulates the ligand binding specificity of the four FGFR proteins.275–277 Immunoglobulin-like domain I and the acidic amino acid sequence (acidic box) located between immunoglobulin-like domains I and II are thought to inhibit ligand binding.278 Consistent with this, an alternative splicing event that results in receptor variants lacking immunoglobulin-like domain I and the I-II linker have increased affinity for FGF ligands.279,280 Fgfr1–Fgfr3 generate two additional major splice variants of immunoglobulin-like domain III, referred to as IIIb and IIIc281–283 (Figure 1(b)). The FGFRb and FGFRc splice variants are essential determinants of ligand-binding specificity (Figure 2).237,238,275,281,282 Alternative splicing of Fgfrs is critical to pathway function as evidenced by the highly conserved intronic control elements in species ranging from sea urchin to mammals.284 Immunoglobulin-like domain III of Fgfr4 is not alternatively spliced.271 Among the other three Fgfrs, alternative splicing of Fgfr2 is functionally the most important. Fgfr1 splicing and ligand binding properties parallels that of Fgfr2, and these two receptors often show functional redundancy during development. Other splice variants of Fgfrs have also been identified. For example, an Fgfr1 cDNA encoding immunoglobulin-like domains II and III generates a secreted FGFR binding domain that can functionally inhibit FGFR signaling.285 An Fgfr3 splice variant in which exons 8–10, which encode the transmembrane domain, are skipped has been identified in both normal epithelial cells and some cancer cell lines. This splice variant produces a secreted protein that can bind FGF ligands and functionally inhibit FGFR signaling.286,287
Ligand binding specificity of the 18 secreted FGFs have been compared using various mitogenic assays and by directly measuring affinity for FGFRs. The BaF3 cell line and L6 myoblasts have been particularly useful, as they have little or no endogenous Fgfr expression. Studies in BaF3 cells identified strong mitogenic response to FGFR1 and FGFR2 and weak responses to FGFR3 and FGFR4, suggesting that the strength or the specific downstream signaling pathways activated by FGFRs may be unique.238,249,288 Using BaF3 cells or L6 myoblasts that express unique extracellular splice variants of Fgfrs (Fgfr1b, 1c, 2b, 2c, 3b, 3c, 4) fused to either the FGFR1 or FGFR2 cytoplasmic domain, the mitogenic activity of all secreted FGFs were compared in the presence of heparin.162,237,238,249–254 Additionally, the mitogenic activity of FGF15/19 and FGF21 were assayed on BaF3 cells or L6 myoblasts that also co-expressed βKlotho and FGF23 was assayed on HEK293 cells that co-expressed αKlotho.162,246,248 This analysis showed that FGF1 was the only ligand that could activate all receptor splice variants (Figure 2). This analysis also showed that members of FGF subfamilies have very similar receptor specificities. Direct binding, using iodinated FGFs and using surface plasmon resonance has also been used to evaluate FGF binding specificity.281,282,289,290 The binding studies are qualitatively in agreement with mitogenic assays.
Expression of alternative splice variants of Fgfr1 and Fgfr2 are regulated in a tissue-specific manner. Mesenchymal tissue expresses IIIc splice variants of Fgfr1 and Fgfr2 and often are activated by FGF ligands that are expressed in epithelial cells, such as members of the Fgf4 and Fgf8 subfamilies.54,55,291,292 By contrast, epithelial tissues express IIIb splice variants of Fgfr1 and Fgfr2 and bind ligands that are normally expressed in mesenchymal tissues, such as members of the Fgf7 subfamily.293,294 This epithelial/mesenchymal expression of alternative splice variants of FGFRs and reciprocal expression of interacting FGF ligands is essential for the development of many organs, particularly those that undergo branching morphogenesis such as the lung or salivary gland, and structures such as the limb bud, and skin (Figure 4).
Figure 4.
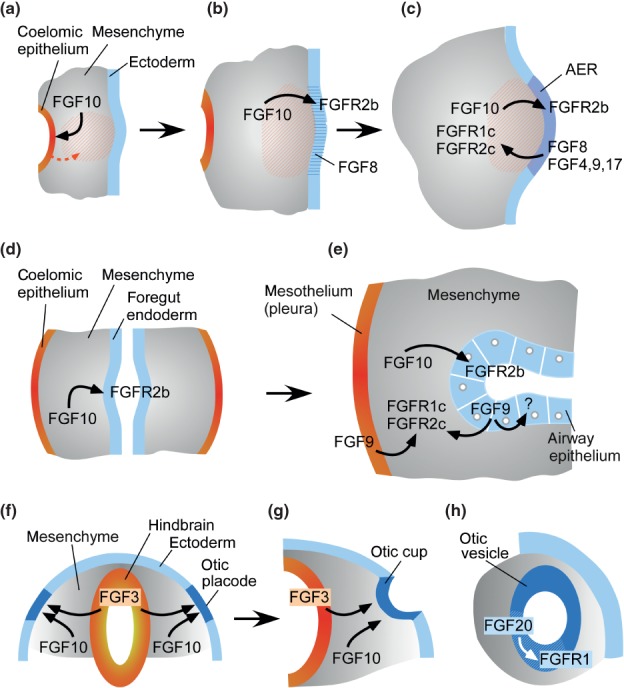
Mechanisms of FGF signaling during organogenesis. (a–c) Limb bud development uses a classical reciprocal epithelial-mesenchymal FGF signal. The earliest identified event in limb bud development involves an FGF10 signal to coelomic epithelium (a). This induces an epithelial to mesenchymal transition (orange arrow) that increases the amount of mesenchyme (orange hash) at the forming limb bud, resulting in a bulge. As development progresses (b), FGF10 signals to ectoderm to induce the formation of the apical ectodermal ridge (AER). Initially FGF8 (blue hash) is expressed throughout its length of the AER (b) and later FGF4, FGF9, and FGF17 are also expressed in the posterior half of the AER. AER FGFs signal to FGFR1 and FGFR2 in distal mesenchyme. (d, e) Lung development uses a modified reciprocal mesothelial/epithelial-mesenchymal FGF signal. The lung bud is initiated with an FGF10 signal from foregut mesenchyme to FGFR2b in foregut epithelium. Continued FGF10 expression is required for epithelial branching. Reciprocal signals from mesothelial FGF9 regulates mesenchymal proliferation through FGFR1 and FGFR2, while epithelial FGF9 functions as an autocrine factor to regulate epithelial branching through an as yet unidentified receptor. (f–h) Induction of the otic placode and differentiation of the otic vesicle. (f, g) FGF3, derived from the hindbrain and FGF10 derived from head mesenchyme, together, induce formation of the otic placode and its progression to the otic cup and otic vesicle. (h) After formation of the otic vesicle, FGF20 signals to FGFR1 within the prosensory epithelium (white hash) as a permissive autocrine factor required for differentiation of outer hair cells and outer supporting cells in the organ of Corti.
Although this pattern of reciprocal signaling is essential for the development of some organs, it is not universal. For example, the tissue-specific regulation of alternative splicing is less stringent for Fgfr3, where both splice variants have been found in epithelial cell types.295,296 The FGF9 subfamily, though primarily expressed in epithelial cells, has the unique ability to activate FGFR3b in addition to IIIc splice variants of FGFRs 1–3237,253 (Figure 2). Fgf10 expression can be found in some epithelial cell types, such as the developing inner ear where it likely signals in an autocrine manner to epithelial cells.237 During somitogenesis, Fgf4 and Fgf8 are expressed and signal within presomitic mesenchyme and nascent somites where they suppress differentiation.29
Pathway Regulation
Extracellular FGF Associated Cofactors and Binding Proteins
Heparan Sulfate Proteoglycans
HS is now recognized to function as a potent cofactor for canonical FGF signaling as well as a wide range of other signaling pathways including bone morphogenetic proteins (BMPs), WNTs, and Hedgehogs197,297–299 (Figure 3(a)). Heparin was found to potentiate the biological activity of FGF1 in 1985220–222 and was first shown to directly enhance FGFR binding and activity in 1991.200,201,254 Using a cell line (BaF3) that lacks cell surface HS, or through inhibition of HS sulfation (with chlorate), the signaling ability of all canonical FGFs were shown to require heparin/HS200,237,238,254,300 (Figure 2).
HS is a long linear carbohydrate chain of repeating sulfated disaccharides, glucuronic acid linked to N-acetylglucosamine. The HS chains are covalently linked to specific core proteins such as syndecan, perlecan, glypican, and agrin. These HS proteoglycans (HSPGs) are cell surface transmembrane type proteins (e.g. syndecans), cell surface glycerophosphatidylinositide-anchored type proteins (e.g. glypicans), or diffusible proteins localized in the ECM198,298,301–304 (e.g. perlecan and agrin). HS independently can interact with both FGFs and FGFRs and is proposed to cooperatively increase the affinity of a 1:1 FGF-FGFR dimer by binding to a cleft formed between the HS binding sites on FGFs and the N-terminal region of immunoglobulin-like domain 2301,305 (Figure 3(a)). This 1:1:1 FGF-HS-FGFR complex leads to conformational changes that stabilize a symmetric 2:2:2 dimer. FGFR dimerization then directs the juxtaposition and activation of the intracellular tyrosine kinase domains, followed by the activation of intracellular signaling pathways.198,301 As a component of the ECM, HS also functions to sequester FGFs and modulate their diffusion through tissue to effectively regulate the shape of a gradient. For example, differences in binding affinity of FGF7 and FGF10 for HS, underlie differences in epithelial branching patterns during glandular organogenesis.306
The structure of HS is complex and heterogeneous; with variations in chain length, and patterns of sulfation and acetylation along the length of the glycosaminoglycan (GAG) chain.307–309 Synthesis of the HS chain is catalyzed by the glycosyltransferases, EXT1 and EXT2.310 The HS molecule consists of repeating disaccharide units of N-acetylglucosamine and glucuronic acid. The HS chain matures in the Golgi where N-acetylglucosamine residues are partially N-deacetylated and N-sulfated by a family of four N-deacetylase/N-sulfotransferase enzymes311 (NDST1-4). Subsequently, 2-O-sulfotransferases, 6-O-sulfotransferases, and 3-O-sulfotransferases add O linked sulfate groups.309 The pattern and density of deactylation and sulfation varies along the length of the GAG chain. In the extracellular environment, the 6-O-endosulfatases, SULF1 and SULF2, can also selectively desulfate HS.
The sulfation pattern and length of HS chains regulate FGF signaling.198,312 In the embryo, specific HS chains can regulate the cell-specific patterns of FGF and FGFR binding to the extracellular matrix, the direct interactions between FGFs and FGFRs, and activation of FGFR signaling.313–315 In general, higher levels of sulfation of HS chains facilitate FGF signaling and the formation of ternary complexes with FGFs and FGFRs.316,317 Furthermore, oligosaccharides with eight or more sugar residues are most active, but shorter HS chains can also facilitate the formation of ternary complexes with FGFs and FGFRs.254,316,318,319 Cleavage of the HSPG core protein also modulates FGF signaling.198 The cleavage by serine proteinases possibly facilities FGF signaling by releasing FGFs that were sequestered at the cell-surface.315 In addition, the cleavage by endoglycosidases such as heparanase possibly modulates FGF signaling.198 For example, FGF10 in the basement membrane, that is released by heparanase, promotes FGF signaling in branching morphogenesis.320
Klotho Family Proteins
The αKlotho gene was originally identified as a candidate gene responsible for a premature aging syndrome.321 Based on the phenotypic similarity of αKlotho and Fgf23 knockout mice, αKlotho was identified as a cofactor for FGF23 signaling through FGFR1c, FGFR3c and FGFR4140,141,247,248,322 (Figure 2). The αKlotho gene is highly expressed in the distal convoluted tubules in the kidney and choroid plexus in the brain.321 A major function of FGF23-αKlotho-FGFR signaling in the kidney is to regulate phosphate and calcium homeostasis. Mice lacking Fgf23 or αKlotho develop hyperphosphatemia and hypercalcemia by two weeks of age.258,323
The Klotho family is comprised of three members including αKlotho, βKlotho, and KLPH.98,324 αKlotho contains ∼1000 amino acid, a single transmembrane domain, and a short cytoplasmic domain (Figure 3(c)). There are no known functions of the cytoplasmic domains of Klotho proteins. The large extracellular part of the Klotho molecule has two repeated internal domains, KL1 and KL2, which are structurally similar to β-glucosidases. However, there is no evidence for glucosidase activity of αKlotho. βKlotho is also a single-pass transmembrane protein similar to αKlotho. The βKlotho gene is predominantly expressed in the liver and white adipose tissue.325 βKlotho is a cofactor for FGF15/19 and FGF21 signaling through FGFR4 and FGFR1c, respectively (Figure 3(b)).322 KLPH is also a single-pass transmembrane protein similar to αKlotho.326 The KLPH gene is expressed in the eye and brown adipose tissue. KLPH efficiently interacts with FGFR1b, FGFR1c, FGFR2c, and FGFR4. In KLPH-transfected HEK293 cells, FGF19, but not FGF21 and FGF23, causes ERK phosphorylation.160 However, the physiological function of KLPH remains unclear. Although Klotho proteins act as cofactors for the endocrine FGFs through formation of an FGF-FGFR-Klotho ternary complex, they also directly compete with a receptor docking site for canonical FGF8 family ligands, and thus may actively suppress these canonical FGFs while activating endocrine FGFs.327
FGF Binding Proteins
FGFBP1 (FGF Binding Protein 1)
FGFBP1 (HBP17) was originally isolated as a heparin-binding protein that co-eluted with FGF2 from a heparin affinity column.328 The Fgfbp1 cDNA encodes a secreted 234 amino acid polypeptide (Mr 17,000) that binds both heparin and FGF1 and FGF2.328 In these initial studies, FGFBP1 was shown to inhibit the biological activity of these FGFs by inhibiting receptor binding. However, in later studies, FGFBP1 was shown to mobilize FGF from HS binding sites in the extracellular matrix and function to present FGF to the FGFR.329
FGFBP1 is expressed in several human tumors, including breast and colon cancer, and FGFBP1 can be rate-limiting for tumor growth, but pro-angiogenic, thus acting to facilitate tumor invasion.330 In mice, Fgfbp1 is abundantly expressed in the colon, stomach, ileum, and eye.160 FGFBP1 also binds to and activates FGF7, FGF10 and FGF22, and functions to enhance wound healing.331,332
FGFRL1/FGFR5
FGFRL1 was identified as a protein structurally similar to FGFRs203,333 (Figure 1(c)). The Fgfrl1 cDNA, originally cloned from human cartilage, encodes a ∼500 amino acid protein containing three extracellular immunoglobulin-like domains with similarity to FGFRs, a single transmembrane domain, and a short intracellular tail with no tyrosine kinase domain.333,334 Fgfrl1 (termed Fgfr5) was also cloned from human and mouse cDNA libraries.335,336 A soluble form of FGFRL1 binds to heparin and to FGF2, 3, 4, 8, 10, 22, and ectopic expression antagonized FGF signaling during Xenopus development and inhibited cell proliferation in vitro.334,337 Interestingly, the short cytoplasmic domain of FGFRL1 contains an SH2 binding motif that interacts with the tyrosine phosphatase SHP1338 (Figure 3(d)). Overexpression of Fgfrl1 results in increased ERK1/2 signaling.338 This result suggests that FGFRL1 is not a decoy receptor, but rather a non-tyrosine kinase signaling molecule.
Fgfrl1 knockout mice die immediately after birth from respiratory failure due to a hypoplastic diaphragm.148 Analysis of these mice reveals agenesis of slow muscle fibers.339 These mice also show kidney agenesis due to a reduction in mesenchymal nephron progenitors (cap mesenchyme), arrested branching of the urogenic epithelium, failure to form functional nephrons, and a hypoplastic collecting duct system147 (Table2(b)). Interestingly, mice that lack the intracellular domain of FGFRL1 are viable, fertile, and phenotypically normal, suggesting that the extracellular domain of FGFRL1 mediates most of its activity (Box 1).340
BOX 1 Inhibitory Mechanisms that Regulate FGFR Signaling
Inhibition of FGFR signaling is important for the precise control of cellular functions. Several mechanisms have evolved to regulate FGF signaling. These range from internalization and degradation of the receptor to modulation of receptor kinase activity by phosphatases and regulation of accessibility to downstream signaling pathways.
In recent studies,389,749 a dimeric form of GRB2, the adaptor protein that couples FRS2 to the RAS-MAPK and PI3K-AKT pathways (Figure 3(a)), was found to interact directly with the FGFR2 C-terminal 10 amino acid residues, where it stabilized a FGFR dimer which could autophosphorylate a limited number of tyrosine residues including Y653 and Y654 in the activation loop (Figure 3(b)). However, additional C-terminal phosphorylation and recruitment of signaling proteins was sterically hindered by the bound GRB2 dimer. Following ligand mediated receptor activation, phosphorylation of GRB2 caused GRB2 to dissociate from the FGFR C-terminus permitting full receptor activation.749 Additionally, high levels of GRB2 inhibited phosphorylation-independent binding of PLCγ (through its SH3 domain) to the very C-terminus of the FGFR. Lower levels of GRB2 allowed PLCγ binding and increased phospholipase activity, resulting in increased cell motility, an activity that can promote metastatic behavior of melanoma cells.750
The RAS-MAPK pathway can also exert direct negative feedback inhibition of FGFRs. ERK1 and ERK2, which are activated by FGFR and other receptor tyrosine kinases, can phosphorylate the C-terminus of FGFR2 at Ser777 to functionally inhibit FGFR2 tyrosine kinase activity.751 This provides a negative feedback pathway for FGF signaling and a means for other receptor tyrosine kinases that use the RAS-MAPK pathways to communicate with FGFRs.
Intracellular Signal Transduction
Cytosolic Signaling Pathways
FGF binding activates the FGFR tyrosine kinase by inducing receptor dimerization and trans-autophosphorylation of the kinase domain1 (Figure 3(a)). For FGFR1, six tyrosine residues are sequentially phosphorylated to fully activate the kinase domain341,342 (Figure 3(b)). In the first phase of activation, Y653 is phosphorylated, resulting in a 50- to 100-fold increase in tyrosine kinase activity. In the second phase of activation, Y583, and then Y463, Y766, and Y585 are phosphorylated. In the third phase of activation, Y654 is phosphorylated, resulting in a further 10 fold (overall 500- to 1000-fold) increase in tyrosine kinase activity. Phosphorylation of two additional tyrosine residues, 677 and 766, is required, respectively, for STAT3 and phospholipase Cγ (PLCγ) binding.343–345 The adaptor protein, FGFR substrate 2α (FRS2α) is constitutively docked to its binding site in the juxtamembrane region of FGFRs and anchored to the cell membrane through myristoylation207,346,347 (Figure 3(b)).
The activated FGFR phosphorylates adaptor proteins for four major intracellular signaling pathways, RAS-MAPK, PI3K-AKT, PLCγ, and signal transducer and activator of transcription (STAT)1,151–153,344 (Figure 3(a) and (b)). Activation of the RAS-MAPK and PI3K-AKT pathway is initiated by phosphorylation of FRS2α. FRS2α phosphorylation and ERK1/2 activation is partially dependent on phosphorylation of Y463 and the presence of CRKL.348,349 pY463 directly interacts with the adapter protein CRKL and with much lower affinity to the related protein, CRK.348–350 Downstream of RAS and PI3K, FGFR signaling has been shown to regulate several distinct MAP kinases including ERK1/2, JNK and p38.178,179,351–353
Activated (phosphorylated) FRS2α binds the membrane anchored adaptor protein, growth factor receptor-bound 2 (GRB2) and the tyrosine phosphatase SHP2.207,354 GRB2 further activates the RAS-MAPK pathway through recruitment of SOS, and the PI3K-AKT pathway through recruitment of GAB1 to the signaling complex (Figure 3(a)).207,355 The RAS-MAPK pathway regulates the expression of diverse target genes through activation of E26 transformation-specific (ETS) transcription factors. Etv4 (Pea3) and Etv5 (Erm) are ETS transcription factors that are often transcriptionally induced by FGF signaling.356–359 Phosphorylation of ETS transcription factors by activated MAPK allows interaction with DNA and regulation of target gene expression.352
In contrast to the RAS-MAPK pathway, the PI3K-AKT pathway functions to inhibit the activity of target molecules such as the forkhead box class transcription factor, FOXO1, and the cytosolic tuberous sclerosis complex 2, TSC2. FOXO1, a pro-apoptotic effector, is inactivated by AKT phosphorylation, causing it to exit the nucleus and promote cell survival.360 AKT also activates the mTOR complex 1 through phosphorylation and inhibition of TSC2, ultimately stimulating cell growth and proliferation.360 Phosphorylation of PLCγ by the activated FGFR tyrosine kinase leads to the hydrolysis of phosphatidylinositol 4,5-bisphosphate to produce inositol triphosphate (IP3) and diacylglycerol (DAG) (Figure 3(a)). IP3 increases intracellular calcium ion levels and DAG activates protein kinase C (PKC). The adaptor protein, GRB14, also interacts with the activated FGFR1 at multiple sites, including pY766361 (and possibly pY776). Binding of GRB14 to pY766 inhibits tyrosine phosphorylation and activation of PLCγ362 (Figure 3(a) and (b)). Additionally, the SRC homology-2 protein, SHB, interacts with pY766 and acts to enhance phosphorylation of FRS2α and the mitogenic response to FGFs in an immortalized brain endothelial cell line.363
The activated FGFR also phosphorylates and activates STAT1, STAT3, and STAT5, to regulate STAT pathway target gene expression343,364–367 (Figure 3(a) and (b)). STAT1 was activated in chondrocytes derived from Thanatophoric dysplasia patients with a constitutively active mutant of FGFR3,364 and STAT1 activation in response to FGF1 in primary growth plate chondrocytes was necessary to suppress proliferation.368 However, using a rat chondrosarcoma cell line that stops growing in response to FGF1, it has been controversial as to whether STAT1 or MAPK signaling mediates the observed growth arrest.368,369 In cancer cells, under conditions of gene amplification or overexpression of FGFR3, STAT3 was phosphorylated resulting in activation of downstream target genes.343 In brain microvascular endothelial cells, FGF signaling was found to activate STAT5, which was necessary for migration, invasion, and tube formation.370
To the best of our knowledge, quantitative similarities and differences in the signaling output of the four FGFR kinase domains have not been assessed. However, activation of downstream signaling pathways are thought to be qualitatively similar for Fgfr1 and Fgfr2 and different from Fgfr3 and Fgfr4.238,249,288 Similarities and differences in signaling of the four FGFRs could be mediated by differential rates of endocytosis,371 by differential subcellular trafficking after ligand activation,372,373 or by differences in the affinity or specificity for adaptor proteins that couple to downstream signaling cascades.374
Inhibitors of FGFR Signaling
Sprouty (SPRY) is an intracellular negative regulator of receptor tyrosine kinases including FGFR, vascular-endothelial growth factor receptor, platelet-derived growth factor receptor, and nerve growth factor receptor.375,376 The human/mouse SPRY family is composed of four members, SPRY1-SPRY4. Most Spry genes are ubiquitously expressed in both embryos and adult tissues. In FGF signaling, SPRY interacts with GRB2 to inhibit the RAS-MAPK pathway and to regulate the PI3K-AKT pathway206,377 (Figure 3(a)). The phenotypes of Spry knockout mice indicate that SPRYs are essential for development and growth. The deregulation of SPRY function often results in human cancers and autoimmune diseases.375,376
SEF (similar expression to Fgf) is a transmembrane protein that functions as an antagonist of FGF signaling through the Ras-MAPK pathway378,379 (Figure 3(a)). SEF functions by binding to activated MEK to inhibit dissociation of the MEK–MAPK (ERK1/2) complex, thus blocking nuclear translocation of activated MAPK.377,380 The extracellular domain of SEF may also interact directly with the FGFR to inhibit receptor phosphorylation.381
Dusp6 (Dual-specificity phosphatase 6) encodes an ERK-specific MAPK phosphatase (MKP3)382 Dusp6 expression is transcriptionally upregulated by FGFR signaling and Dusp6 expression patterns closely resemble those of Fgfs.383–386 DUSP6 serves in vivo as a negative feedback regulator of FGFR signaling by directly dephosphorylating MAPK (ERK1 and ERK2) on phosphotyrosine and phosphothreonine residues382 (Figure 3(a)).
CBL, an E3 ubiquitin ligase, forms a ternary complex with phosphorylated FRS2α and GRB2, resulting in the ubiquitination and degradation of FGFR and FRS2 in response to FGF stimulation387 (Figure 3(a)). FGFR2 activation can also increase CBL-PI3K interactions, leading to PI3K degradation and attenuated signaling.388
SHP2 binds to phosphorylated FRS2 following ligand activation of the FGFR.354 SHP2 functions to dephosphorylate FGFR2 and GRB2 (Figure 3(a)). However, activation of SHP2 (by phosphorylation) and access to the FGFR are also inhibited by receptor-bound GRB2.389,390
Regulation of the Cellular Response to FGFR Activation
The cellular response to FGFR signaling is regulated by differences in the intrinsic signaling properties of FGFRs and by the dynamics of subcellular FGFR trafficking in response to ligand binding. Cytosolic signaling pathways can be differentially activated by cell surface FGFRs and internalized FGFRs. Furthermore, regulating synthesis and degradation of FGFRs can modulate the strength of the FGFR signal. Differential cellular response can also result from differences in signal output from multiple FGFRs. For example, FGF1 stimulates lung epithelial cells to form buds resulting in branching, while FGF7 stimulates lung epithelial cells to form cyst-like structures.391,393 This could be due to activation of FGFR2 and FGFR4 by FGF1 and only activation of FGFR2b in response to FGF7. Two FGFs that are even more similar, FGF7 and FGF10, still can elicit different cellular responses. FGF10 specifically induced the formation of a Y734-phosphorylated FGFR2b-PI3K-SH3BP4 complex that targets FGFR2b to recycling endosomes and controls cell migration and epithelial branching, whereas FGF7 leads to cell proliferation and degradation of FGFR2b.373,392,393
Function of FGFs and FGFRs in the Nucleus
Both FGF ligands and receptors can localize to the cell nucleus where they carry out signaling functions that can be independent of receptor tyrosine kinase activity.394,395 FGF1 localization in the nucleus was found to stimulate DNA synthesis independent of FGFRs, and FGF2 nuclear localization was associated with glioma cell proliferation.396,397 It is not clear whether FGFs have direct transcriptional functions or exert their activity in the nucleus through interactions with other molecules.
Following ligand-mediated internalization, FGFR1 can be transported to the nucleus by interactions with importin β. Nuclear FGFR1 is required for neuronal differentiation and functions by activating transcription in cooperation with cyclic AMP response element-binding protein (CREB).398,399 Nuclear translocation of FGFR1, along with its ligand, FGF2, promoted pancreatic stellate cell proliferation and changes in the elaborated ECM, making it more permissive for pancreatic cancer cell invasion.400 In breast cancer cells, activation of FGFR1b by FGF10 activated granzyme B cleavage of FGFR1. Transport of the resulting C-terminal fragment of FGFR1 to the nucleus was required for cell migration.401
MicroRNA Regulation of FGF and FGFR Expression and Signaling
MicroRNAs (miRNAs) are small (approximately 21–24 nucleotides) non-coding RNAs, which are post-transcriptional regulators of gene expression.402 miRNAs participate in diverse biological processes including development, differentiation, cell proliferation, metabolism, as well as in human diseases including metabolic disorders and cancers.403,404 FGF pathway activity during development or regeneration can be regulated by miRNAs and loss of miRNA regulation of FGF signaling can result in disease progression or cancer.
During development, miRNAs can affect cell differentiation by directly regulating Fgf or Fgfr expression. For example, in the osteoblast, miR-338 was found to directly regulate the 3′ untranslated region (UTR) of Fgfr2 to suppress Fgfr2 expression. Decreased miR-338 increased Fgfr2 expression resulting in enhanced osteoblast differentiation.405 miRNAs can also affect FGF signaling during development by regulating downstream effectors of the pathway. For example, the miR-17 family directly targets Stat3 and Mapk14 in lung epithelium to modulate the response to FGF10-FGFR2b signaling.406
In disease pathogenesis, such as in pulmonary arterial hypertension (PAH), hyperproliferation of pulmonary artery endothelial and smooth-muscle cells leads to destruction of the pulmonary vascular plexus. miR-424 and miR-503 directly regulate (suppress) Fgf2 and Fgfr1 expression in pulmonary artery endothelial cells. Decreased expression of miR-424 and miR-503 in PAH leads to increased FGF2 and FGFR1 and consequent vascular hyperproliferation.407 In a model for tissue repair, inhibition of miR-710, a direct regulator of Fgf15 expression in myofibroblasts, increased FGF15 in conditioned media and enhanced in vitro intestinal epithelial wound repair.408
The metabolic functions of endocrine FGFs can be regulated by miRNAs. miR-34a is highly elevated in adipose tissue in obese mice and in liver in patients with steatosis. Elevated miR-34a in obesity attenuates hepatic FGF19 signaling and adipose FGF21 signaling by directly targeting the 3′ UTR of β-Klotho and Fgfr1.409,410 Downregulation of miR-34a increases the levels of the FGF21 receptor components, FGFR1 and βKlotho (and also SIRT1), resulting in FGF21/SIRT1-dependent induction of genes that favor brown fat and improved hepatic FGF21 signaling and lipid oxidation.410
In several cancers, decreased expression of miRNAs that normally suppress FGF expression have been identified as a potential mechanism for promoting cancer progression. For example, in non-small-cell lung cancer (NSCLC) miR-152 and miR-198 are downregulated, and FGF2, a direct target of miR-152, and Fgfr1, a direct target of miR-198, are overexpressed, leading to decreased apoptosis and increased proliferation and invasion.411,412 In a breast cancer cell line, miR-503 expression is suppressed by HBXIP (hepatitis B X-interacting protein). Reduced expression of miR-503, which directly targets the 3′ UTR of FGF8, results in increased FGF8 and consequent increased angiogenesis and proliferation of the breast cancer cells.413 In gastric cancer and hepatocellular carcinoma, miR-26a and miR-140-5p, respectively, are strongly downregulated, and FGF9, a direct target of both of these miRNAs is increased.414,415 Interestingly, decreased miR-140-5p and miR-99b expression has also been observed in NSCLC tissue.416,417 High FGF9 expression observed in 10% of human NSCLC specimens,418 suggests an additional pathogenic relationship between miR-140-5p and FGF9 in lung cancer. Increased expression of FGFR3, a direct target of miR-99b, was observed in human NSCLC tissue.417 Of relevance to this mechanism, FGFR3 is the obligate FGFR mediating FGF9 induced adenocarcinoma in a mouse model for lung cancer.419
Developmental, Genetic, and Pathological Functions
FGF Signaling during Peri-implantation Development
The earliest requirement for FGF signaling is in the preimplantation embryo, where Fgf4 is first expressed in the morula and later in the epiblast cells of the inner cell mass (ICM).420 Fgf4 gene inactivation in mice shows that FGF4 is required for ICM proliferation and for formation of the primitive ectoderm.167,14 The receptor for FGF4 in the ICM is more controversial. Campbell et al. detected Fgfr1 (Flg) but not Fgfr2 (Bek) transcripts in mouse blastocysts,421 Orr-Urtreger et al. concluded that both Fgfr1 and Fgfr2 are expressed in the ICM and Fgfr2 is expressed in the embryonic ectoderm,422 while Guo et al. concluded that Fgfr2 is not expressed in the epiblast lineage but is highly expressed in embryonic ectoderm.423 Fgfr knockout studies are also controversial (Table2(b)). Arman et al. generated a mutant allele of Fgfr2 and found defects in the outgrowth, differentiation, and maintenance of the inner cell mass424; however, it is possible that this allele functions as a dominant negative that partially interferes with Fgfr1 signaling, as mice homozygous for two other engineered null alleles of Fgfr2 survived until embryonic day 10–11115,116 (Table2(b)). Inactivation of Fgfr1 or Fgf8, which are also expressed in the blastocyst, indicates a function slightly later in development, with phenotypes affecting axis formation and mesoderm specification105,425,426 (Table2(a) and (b)). We are not aware of studies in which both Fgfr1 and Fgfr2 have been conditionally inactivated in the ICM.
FGF Signaling in Organogenesis
FGF signaling is involved almost ubiquitously throughout organogenesis.161 A key function of FGF signaling is to regulate interactions between epithelial (and mesothelial) cells and mesenchyme. A general principle that applies to branched organs (lung, salivary gland, lacrimal gland), intestine, liver, and limb bud development involves mesenchymal expressed FGFs, such as FGF10 signaling to the epithelial IIIb splice variant of FGFR1 and FGFR2.171,117,427 Reciprocal signaling, from epithelium to mesenchyme is mediated by FGFs expressed in epithelia, such as FGF8 and FGF9, which signal to mesenchymal IIIc splice variants of FGFR1 and FGFR2.428,429 However, this general principle does not apply to all tissues. For example, in the developing central nervous system, FGF8 signals as an autocrine/paracrine factor in the anterior neural primordium430 and during development of the inner ear, autocrine/paracrine FGF signaling regulates differentiation of the cochlear sensory epithelium.90,103,104
Epithelial-Mesenchymal Signaling in Limb, Lung, and Neurogenic Placode Development
FGF signaling is essential for initiation and proximal-distal growth of the limb bud (Figure 4(a)–(c)). Fgf10 is expressed diffusely in the lateral plate mesoderm.54 FGF10 was recently shown to signal to coelomic epithelium where it induces an epithelial-mesenchymal transition to generate mesenchyme in the presumptive limb fields.431 Later, FGF10 signals to overlying ectoderm to initiate formation of the apical ectodermal ridge (AER), a specialized thickening of epithelium at the tip of the growing limb that is required for proximal-distal limb growth. Inactivation of FGFR2 in the AER at different times during development results in blunt truncations of the limb117,427 (Table2(b)). FGF10 signaling to the AER activates expression of Wnt3a and expression of the downstream transcription factors SP6 and SP8, which are required for Fgf8 expression.432–434 Fgf8 is first expressed as the lateral ectoderm begins to swell and then throughout the AER. Fgf4, Fgf9, and Fgf17 are subsequently expressed in the posterior AER.15,30,435,436 AER FGFs signal to distal limb mesenchyme through FGFR1 and FGFR2 to activate ETV1 and EWSR1, which are required to maintain Fgf10 expression117,437 (Table2(b)).
FGF signaling in lung development follows similar principles to that in limb development (Figure 4(d) and (e)). Fgf10 expression in mesenchyme adjacent to the sites of lung bud formation is regulated by the transcription factor Tbx4.293,438,439 FGF10 signals to FGFR2 in foregut endoderm to induce expression of Nkx2.1, a transcription factor that demarcates the lung field in the foregut.118,393,439 In the absence of FGF10, primary lung buds fail to form.54,55,440 Conditional inactivation of FGF10 or FGFR2, after initial lung bud formation, results in reduced epithelial branching.57,441 FGF10 signaling in lung epithelium is inhibited by Spry1, Spry2, and Spry4, which are expressed in the distal ductal epithelium proximal to sites of Fgf10 expression in mesenchyme.442,443 Inactivation of Spry1 and Spry2 results in increased epithelial proliferation, branching, and differentiation toward distal airway cell-types.444,445 Inactivation of Spry2 and Spry4 results in epithelial dilation and reduced branching.446 Interestingly, Fgf10 appears to be expressed in a lung mesenchymal progenitor that can give rise to parabronchial cells, vascular smooth muscle cells and lipofibroblasts.447
FGF9 has a complementary role to that of FGF10. Fgf9 is expressed in the mesothelium and epithelium.448,449 Mice lacking Fgf9 have severely hypoplastic lung development, characterized by reduced distal mesenchyme and decreased epithelial branching.42,43 The primary target of FGF9 in lung mesenchyme is FGFR1 and FGFR2.43 Most of the mesenchymal proliferation can be accounted for by FGF9 derived from the mesothelium, whereas epithelial-derived FGF9 is important for branching.44 In lung mesenchyme, an interaction with FGFR and canonical WNT signaling is essential for development. FGFR activation is required for the expression of Wnt2a and WNT/β-catenin signaling is required to maintain mesenchymal Fgfr1 and Fgfr2 expression.450 Thus WNT/β-catenin signaling functions to modulate the tissue responsiveness to FGF signals.
FGF signaling is required for the induction of neurogenic placodes.451 For example, the otic placode, which gives rise to the entire inner ear including sensory hair cells, specialized supporting cells and the innervating sensory neurons, requires direct signaling from FGF3 and FGF10 (Figure 4(f) and (g)). FGF3 is derived from the hindbrain and FGF10 is expressed in head mesenchyme. Both of these FGFs signal to pre-otic ectoderm to induce the otic placode.452,453 The size of the otic placode is initially regulated by FGF-induced proliferation and expression of the FGF pathway inhibitors, Spry1 and Spry2.454,455 FGF8 is also necessary for otic placode development; however, FGF8 functions indirectly, signaling from cranial endoderm to regulate Fgf10 expression in adjacent head mesenchyme.28
Canonical FGF Signaling within the Nervous System
Canonical FGF signaling within an epithelial or mesenchymal compartment is used in an autocrine, paracrine, or juxtacrine manner during the development of some neuronal tissues. For example, in the developing central nervous system, Fgf8 is expressed in localized organizing centers such as the anterior neural primordium (neuroepithelium) where it signals as a paracrine factor to regulate anterior–posterior patterning of the telencephalon430 and maintain the survival of telencephalic progenitors.456 Similarly, FGF signaling is important for patterning around the midbrain-hindbrain junction and around rhombomere 4.31,457–464
During development of the cochlear duct in the inner ear, Fgf20 is expressed in the prosensory epithelium and signals as an autocrine/paracrine factor to FGFR1 to regulate differentiation of the cochlear sensory epithelium103,90 (Figure 4(h)). FGF signaling is also required for neuronal migration in the cortical ventricular zone and for the translocation of astroglial cells from the ventricular zone to the cortical surface.465,466 In myelinating nerves, FGFs expressed in neurons, signal to FGFR1 and FGFR2 in oligodendrocytes to regulate myelination122 and in synaptogenesis, FGF7 and FGF22, expressed in specific neuronal populations, are required for the induction of inhibitory and excitatory synapses, respectively, in the neurons that they innervate.22,467
Canonical FGF Diffusion Controlled by ECM Interactions Regulates Development
Interactions of FGF ligands and the ECM affect receptor affinity and their diffusion through tissue.197,468 Receptor binding and diffusion through tissue can have synergistic or antagonistic effects on overall FGF signaling. An example of this is the elbow knee synostosis (EKS) mutation and multiple synostosis syndrome, both of which result from missense mutations in Fgf9469,470 (discussed under Heritable disease mutations below). The Fgf9EKS mutation reduces its affinity for heparan sulfate proteoglycan and increases diffusion of FGF9 through developing joint tissue. This increases FGF9 signaling distally in the presumptive joint space and results in failure to form a joint cavity. In lacrimal gland development, Fgf10 is expressed in perioccular mesenchyme. Lacrimal gland development was impaired in mice in which the mesenchymal biosynthetic enzyme for glycosaminoglycans, UDP-glucose dehydrogenase, or enzymes required for heparan sulfation, NDST1 and NDST2, were inactivated.471 Phenotypic analysis indicated that these mutations resulted in increased FGF10 diffusion, decreased local concentrations, and defective epithelial branching into the FGF10-deficient mesenchyme.
Loss-of-Function Fgf and Fgfr Mutations in Mice
Fgf1 Subfamily
FGF1 and FGF2 appear to have relatively minor roles in embryonic development but are important regulators of the injury response.3,5,6,181,472–475 Fgf1 expression in adipose tissue is induced in response to a high fat diet and mice lacking Fgf1 develop a diabetes phenotype when placed on a high fat diet4 (Table2(a)). Mice lacking Fgf2 also develop normally, but show reduced vascular tone, impaired cardiac hypertrophy, reduced cortical neuron density, and defects in response to cutaneous, pulmonary, or cardiac injury3,5,7,181,473,476–478 (Table2(a)).
Fgf4 Subfamily
Fgf4 knockout mice die at early embryonic stages due to impaired proliferation of the blastocyst inner cell mass14 (Table2(a)). Conditional inactivation of Fgf4 in limb bud apical ectodermal ridge cells identified redundancy with Fgf8 for survival of cells located distal to the apical ectodermal ridge.292 Similarly, Fgf4 and Fgf8 show redundancy in somitogenesis and conditional loss of both genes results in loss of presomitic mesoderm and its premature differentiation.29 Genetic analysis in domestic dog breeds identified a retrovirus-mediated duplication of Fgf4 associated with a short-legged phenotype resembling chondrodysplasia.16 Fgf5 and Fgf6 knockout mice are viable. Inactivation of the Fgf5 gene results in a long hair phenotype in angora mice and in engineered knockouts18 (Table2(a)). Fgf6 knockout mice have defects in muscle regeneration19 and the combined loss of Fgf2, Fgf6 and the Mdx gene leads to severe dystrophic changes with reduced formation of new myotubes in regenerating muscle20 (Table2(a)).
Fgf7 Subfamily
Fgf3 knockout mice are viable, but have phenotypes that include inner ear agenesis and dysgenesis, microtia, and microdontia12,56,479 (Table2(a)). Fgf7 knockout mice, which are also viable, have impaired hair and kidney development23,24 and defects in the formation of neuronal synapses22 (Table2(a)). Fgf10 knockout mice die shortly after birth. Fgf10 is critical for epithelial-mesenchymal interactions necessary for the development of epithelial components of multiple organs including the limb, lung, salivary glands kidney, and white adipose tissue54,55,58,480 (Table2(a)). Fgf22 knockout mice are viable, but like Fgf7 have defects in synaptogenesis.467 Interestingly, Fgf22 knockout mice have defects in the formation of excitatory synapses, while Fgf7 knockout mice have defects in inhibitory synapses. Consistent with this, Fgf7 and Fgf22 knockout mice are either resistant to or prone to epileptic seizures, respectively22,95 (Table2(a)).
Fgf8 Subfamily
Fgf8 knockout mice lack all embryonic mesoderm and endoderm-derived structures and die by embryonic day 9.5.426 Subsequent analysis revealed that FGF8 is required for Fgf4 expression in the primitive streak resulting in impaired migration away from the primitive streak.32 Conditional inactivation of Fgf8 identified additional roles in limb bud development and organogenesis15 (Table2(a)). Fgf17 knockout mice are viable, but show impaired hindbrain development and a selective reduction in the size of the dorsal frontal cortex31,84 (Table2(a)). Fgf18 knockout mice die shortly after birth. Fgf18 has essential roles in the development of mesenchymal components of multiple organs including the skeleton, lung, and brain.85–88,481 Late in embryonic development FGF18 is involved in lung alveolar development88 (Table2(a)).
Fgf9 Subfamily
Mice lacking Fgf9 have hypoplastic lungs, sex reversal and impaired survival of male germ cells, impaired skeletal growth, impaired cardiomyocyte growth, impaired growth of the small intestine and cecum, and defects in inner ear development41,42,45–49,482 (Table2(a)). Mice lacking Fgf16 are viable but have decreased proliferation of cardiomyocytes in embryos and neonatal mice82,83 and enhanced cardiac hypertrophy and fibrosis in response to angiotensin II as adults81 (Table2(a)). Mice lacking Fgf20 are viable but lack guard hairs, have impaired differentiation of sensory cells in the cochlea, small kidneys, and defects in tooth development.40,90–92 Fgf9 and Fgf20 show redundancy in their requirement for kidney development, where they function to maintain the stemness of cap mesenchyme progenitor cells40 (Table2(a)).
Fgf15/19 Subfamily
Mice lacking Fgf15 develop normally until E10.5, but then gradually die due to variably penetrant defects in morphogenesis of the cardiac outflow tract.76,483 At postnatal stages, intestinal FGF15/19 functions to regulate hepatic bile acid synthesis78 (Table2(a)). Following partial hepatectomy, mice lacking Fgf15 have severe defects in regeneration; showing reduced or delayed expression of early response genes and transcription factors that regulate the cell cycle.79,484 Mice lacking Fgf21 are phenotypically normal under homeostatic conditions. However, when fasted, Fgf21 expression is rapidly upregulated in the liver,485–487 and in response to fasting, mice lacking Fgf21 showed increased lipolysis93 and an impaired adaptation to a ketogenic diet.488 Subsequent studies showed that FGF21 is an upstream effector of adiponectin in white adipocytes and that adiponectin mediates many of the systemic effects of FGF21 on energy metabolism and insulin sensitivity in liver and skeletal muscle489,490 (Table2(a)). Fgf23 knockout mice survive until birth, but then gradually die.100 Fgf23 knockout mice and mice in which FGF23 is inhibited with antibodies show hyperphosphatemia and increased levels of the active form of vitamin D, 1,25-dihydroxyvitamin D (1,25(OH)2D). Fgf23, which is expressed in osteocytes, signals to the kidney where it induces the vitamin D activating enzyme Cyp27b1 and inhibits Cyp24, which inactivates vitamin D. Injection of recombinant FGF23 rapidly reduces circulating parathyroid hormone (PTH) and levels of the sodium-dependent phosphate co-transporters, NPT2a and NPT2c, in the kidney, resulting in phosphaturia.491,492 FGF23 has also been shown to signal directly to cardiomyocytes to induce hypertrophy,493 and increase myocyte Ca2+ levels and cardiac contractility494 (Table2(a)).
Fgf11 Subfamily
Mice lacking Fgf13, though viable, have defects in neuronal migration and deficits in learning and memory70 (Table2(a)). Mice lacking Fgf14 have paroxysmal dyskinesia, movement disorders, and impaired spatial learning71,72,74 (Table2(a)). FGF14 and other members of the iFGF family interact with the cytoplasmic carboxy terminal tail of voltage gated sodium channel (Nav) α subunits69,263,264,495–498. FGF13 was also found to interact directly with and stabilize microtubules70 and bind junctophilin-2, a protein that regulates L-type Ca2+ channels.499
Mice lacking Fgf14 have defective neuronal firing due to altered Nav channel physiology69,72,73,500 (Table2(a)). Inactivation of Fgf14 in adult mouse Purkinje neurons results in loss of spontaneous firing and deficits in coordination,501 suggesting that FGF14 functions as a physiological regulator of Nav channels in vivo. Interestingly, FGF14 interactions with Nav channels may be regulated downstream of glycogen synthase kinase 3 providing a pathway that could link intercellular signaling and neuronal excitability.498,502–504 Consistent with phenotypes seen in Fgf14 deficient mice, mutations in Fgf14 in humans result in a progressive spinocerebellar ataxia syndrome (SCA27)505,506 (see below).
Fgfr Family
Most embryos lacking both alleles of Fgfr1 do not survive past embryonic day 8.5. Analysis of earlier stages of development shows that Fgfr1-null embryos are smaller, but do initiate gastrulation (mesoderm formation), have impaired mesoderm migration, but fail to initiate somitogenesis105,425 (Table2(b)). Mice lacking Fgfr2 survive until embryonic day 10–11. These embryos fail to form a functional placenta and do not form limb buds115,116 (Table2(b)). As discussed above, another presumed null allele of Fgfr2 that dies earlier in development may have dominant negative effects on other Fgfrs, uncovering potential redundancies and resulting in earlier and more severe phenotypes.424 At later stages of development, several studies have demonstrated redundant function of Fgfr1 and Fgfr2 in organogenesis.43,49,121,507
Mice lacking Fgfr3 are viable. In the absence of Fgfr3, the most prominent phenotype is skeletal overgrowth127,128 (Table2(b)). However, close examination of Fgfr3 null mice revealed defects in inner ear development resulting in sensorineural hearing loss,127,129,508 decreased growth of the cerebral cortex and telencephalon,509 reduced numbers of differentiated oligodendrocytes,130 and fewer intestinal crypts with impaired paneth cell differentiation.510
Mice lacking Fgfr4 are viable and overtly healthy.131 Although, they have normal liver histology and regenerative response to partial hepatectomy, mice lacking Fgfr4 exhibit depleted gallbladders, elevated bile acid reserves, elevated bile acid excretion, increased mass of white adipose tissue, hyperlipidemia, glucose intolerance, insulin resistance, and hypercholesterolemia143,511 (Table2(b)). The role of FGFR4 in tumorigenesis is controversial. In one study, mice lacking Fgfr4 have increased susceptibility to chemically induced hepatocellular carcinoma, indicating that FGFR4 may function as a tumor suppressor in the liver.512 However, in a second study, FGFR4 was found to be required for FGF15/19 induced hepatocellular carcinoma and mice lacking Fgf15 are resistant to chemically induced hepatocellular carcinogenesis.257,513 FGFR3 and FGFR4 show redundant function in the regulation of vitamin D levels and in regulating alveolar septation131,140,142 (Table2(b)). FGFR1 and FGFR4 show redundant function in phosphate homeostasis141 (Table2(b)).
Heritable Disease Mutations in FGFs and FGFRs in Humans and Other Mammals
FGF4 Subfamily
Chondrodysplasia is a short-legged phenotype that defines at least 19 dog breeds. The expression of a recently acquired expressed retrogene encoding Fgf4 is strongly associated with the chondrodysplasia phenotype16 (Table3(a)). Genome-wide association studies (GWAS) in dogs identified a mutation in Fgf5 that is associated with hair length518 (Table3(a)). A missense mutation in Fgf5 was also found in longhaired cats and the Angora mouse mutant18,519 (Table3(a)).
Table 3.
Heritable Mutations in FGFs Associated with Disease in Humans (and Mice)
| Gene Name | Mutation | Associated Disease | Selected References |
|---|---|---|---|
| (a) Heritable mutations in FGFs associated with disease in humans (and other mammals) | |||
| FGF1 | |||
| FGF2 | |||
| FGF3 | Haploinsufficiency | Oto-dental syndrome | 479,514–517 |
| Missense/frameshift mutation | Michel aplasia (inner ear agenesis, microtia, and microdontia), LAMM syndrome (labyrinthine aplasia, microtia, and microdontia) | ||
| FGF4 | Retroviral overexpression | Chondrodysplasia (dogs) | 16 |
| FGF5 | Deletion mutation | Angora mutation (mice) | 18,518–521 |
| Missense/splice-site mutation | Coat variability (pure bred dogs) | ||
| Missense/insertion/deletion mutation | Long-hair (cats) | ||
| FGF6 | |||
| FGF7 | Polymorphism | Chronic obstructive pulmonary disease risk | 522 |
| FGF8 | Nonsense mutation | Hypogonadotropic hypogonadism | 523–528 |
| Missense mutation | Cleft lip and palate, Holoprosencephaly, craniofacial defects, Hypothalamo-pituitary dysfunction, Kallman syndrome type 6 | ||
| Hypomorphic allele | Lack of hypothalamic GnRH neurons | ||
| FGF9 | Missense mutation | Multiple synostoses syndrome, Elbow knee synostosis (mice) | 469,470,529 |
| Promoter polymorphism | Sertoli cell-only syndrome | ||
| FGF10 | Nonsense mutation | Aplasia of lacrimal and salivary glands, LADD syndrome | 530–534 |
| Polymorphism | Extreme myopia | ||
| FGF11 | |||
| FGF12 | Missense mutation | Brugada syndrome (candidate gene) | 535 |
| FGF13 | Nonsense mutation | Börjeson-Forssman-Lehmann syndrome (BFLS) (candidate gene) | 536,537 |
| Position effect | X-linked congenital generalized hypertrichosis | ||
| FGF14 | Missense mutation/translocation/deletion | Spinocerebellar ataxia 27 (SCA27) | 505,538,539 |
| FGF15/19 | |||
| FGF16 | Nonsense mutation | Metacarpal 4–5 fusion | 540,541 |
| FGF17 | Missense mutation | Hypogonadotropic hypogonadism | 542 |
| FGF18 | Polymorphism | Nonsyndromic cleft lip and palate | 524 |
| FGF20 | Polymorphism | Parkinson disease risk | 40,543–545 |
| Missense mutation | Kidney agenesis (human) | ||
| FGF21 | Polymorphism | Macronutrient intake, obesity, and type-2 diabetes risk | 546–548 |
| FGF22 | |||
| FGF23 | Missense mutation | Autosomal dominant hypophosphataemic rickets, Familial hyperphosphatemic tumoral calcinosis | 242,549–555 |
| Polymorphism | Cardiac abnormality risk in Kawasaki syndrome (increased serum FGF23) | ||
| (b) Heritable mutations in FGFRs associated with disease in humans (and other mammals) | |||
| FGFR1 | Missense mutation | Pfeifer syndrome, Kallman syndrome 2, Normosmic idiopathic hypogonadotropic hypogonadism, Split hand/foot malformation, Osteoglophonic dyplasia, Harstfield syndrome | 556–566 |
| Missense or frameshift mutation | Jackson-Weiss syndrome | ||
| FGFR2 | Missense mutation | Apert syndrome, Crouzon syndrome, Jackson-Weiss syndrome, Pfeifer syndrome, Non syndromic craniosynostosis, Bent bone dysplasia | 567–579 |
| Deletion | Saethre-Chotzen-syndrome | ||
| FGFR3 | Missense mutation | Hypochondroplasia, Achondroplasia, Thanatophoric dysplasia, Coronal craniosynostosis, Crouzon syndrome with acanthosis nigricans, Platyspondylic lethal skeletal dysplasia, Achondroplasia with developmental delay, and acanthosis nigricans (SADDAN), Muenke syndrome, Saethre-Chotzen-syndrome, CATSHL syndrome, Mouse models for aberrant osteogenesis, Achondroplasia, Muenke syndrome | 288,574,580–611 |
| FGFR4 | Overexpression | Facioscapulohumeral muscular dystrophy | 612–614 |
| Missense mutation | Gallstone disease | ||
| Polymorphism | Bronchopulmonary dysplasia, Neonatal respiratory distress syndrome | ||
| FGFRL1 | Frameshift mutation | Craniosynostosis, Antley–Bixler-like syndrome | 615–617 |
| Deletion | Wolf-Hirshchhorn syndrome | ||
FGF7 Subfamily
Michel aplasia is a unique autosomal recessive syndrome characterized by type I microtia, microdontia, and profound congenital deafness associated with a complete absence of inner ear structures. Michel aplasia is caused by mutations in FGF3514 (Table3(a)). Human chronic obstructive pulmonary disease (COPD) is a type of obstructive lung disease characterized by chronically poor airflow. Genome wide association studies identified single nucleotide polymorphisms in FGF7 significantly associated with COPD522 (Table3(a)). Aplasia of the lacrimal and salivary glands (ALSG) is an autosomal dominant congenital anomaly characterized by aplasia, atresia, or hypoplasia of the lacrimal and salivary systems. Lacrimo-auriculo-dento-digital (LADD) syndrome is an autosomal-dominant multiple congenital anomaly disorder characterized by aplasia, atresia, or hypoplasia of the lacrimal and salivary systems, cup-shaped ears, hearing loss, and dental and digital anomalies. Both ALSG and LADD syndromes are caused by FGF10 mutations530,531 (Table3(a)). Severe myopia (nearsightedness) is associated with a single nucleotide polymorphism in FGF10532 (Table3(a)). In strong support of an FGF10-FGFR2b signal, loss-of-function mutations in FGFR2 are also a cause of LADD syndrome (see below).
FGF8 Subfamily
Nonsense mutations in FGF8 and destabilizing missense mutations in FGF17 were found in familial hypogonadotropic hypogonadism with variable degrees of gonadotropin-releasing hormone deficiency and olfactory phenotypes523,542 (Table3(a)). Cleft lip and/or palate (CLP) appear when the two halves of the palatal shelves fail to fuse completely. A missense mutation in FGF8 which is predicted to cause loss-of-function by destabilizing the N-terminal structure of the protein (important for FGFR binding affinity and specificity) was found in a patient with CLP524 (Table3(a)).
FGF9 Subfamily
An autosomal dominant missense mutation in FGF9 was found in patients with multiple synostosis syndrome (SYNS). The mutation leads to significantly impaired FGF9 receptor binding, reduced chondrocyte proliferation, increased osteoblast differentiation, and matrix mineralization resulting in joint fusions (synostosis)470 (Table3(a)). An autosomal dominant missense mutation in Fgf9 is also responsible for the mouse mutant, elbow knee synostosis (EKS), showing elbow and knee joint synostosis, and premature fusion of cranial sutures. The mutation prevents homodimerization of the FGF9 protein, resulting in reduced affinity for heparin. Even though receptor binding affinity is decreased by this mutation, the EKS phenotype resembles that of a gain-of-function mutation. The reduced affinity for heparan sulfate results in increased diffusion of FGF9 through tissue, leading to ectopic FGF9 signaling and repression of joint and suture development469 (Table3(a)). Overexpression of an activated form of FGFR1 in developing chondrocytes results in a similar joint fusion phenotype.618
Sertoli cell–only syndrome (SCOS) patients commonly have atrophic testes, azoospermia, and hypogonadism. FGF9 is expressed in the Leydig cells of the testis and FGF9 expression is significantly decreased in patients with SCOS. A promoter mutation in FGF9 results in weak promoter activity and the resulting low expression of testicular FGF9529 (Table3(a)). Metacarpal 4–5 fusion is a rare congenital malformation of the hand characterized by the partial or complete fusion of the fourth and fifth metacarpals in humans. Nonsense mutations in FGF16 are associated with X-linked recessive metacarpal 4–5 fusion, indicating the involvement of FGF16 in the fine tuning of skeletal development540,619 (Table3(a)). Parkinson disease is a common neurodegenerative disorder resulting in the inability to control movement. The disease has been attributed to the severe loss of dopaminergic neurons within the substantia nigra. The significant correlation of Parkinson disease with single nucleotide polymorphisms in FGF20 indicates that the genetic variability of FGF20 may be a risk factor for Parkinson disease543–545 (Table3(a)). A frameshift mutation in FGF20 also results in bilateral renal agenesis in humans, indicating that FGF20 is essential for metanephric kidney development40 (Table3(a)).
FGF15/19 Subfamily
Dietary intake of macronutrients has been associated with risk of obesity and type 2 diabetes. Polymorphisms in FGF21 are potentially associated with macronutrient consumption and risk of obesity and type 2 diabetes in humans546–548 (Table3(a)). Mutations resulting in either gain- or loss-of-function of FGF23 result in human disease620 (Table3(a)). Autosomal dominant hypophosphatemic rickets (AHDR) is caused by gain-of-function mutations in FGF23.242 Tumors that over-produce FGF23 also cause tumor-induced osteomalacia (TIO), which is a paraneoplastic disease characterized by renal phosphate wasting and resulting hypophosphatemia.621 Reduced FGF23 signaling also causes familial tumoral calcinosis (FTC); a disease characterized by ectopic calcification and hyperphosphatemia549,550 (Table3(a)). Kawasaki syndrome (KS) is a childhood vascular inflammatory disease with an increased risk of developing subsequent cardiac abnormalities. Thirty three percent of patients examined were found to have a polymorphism in FGF23 and elevated serum levels of FGF23.551,552 FGF23 polymorphisms were significantly associated with cardiac abnormalities (Table3(a)).
FGF11 Subfamily
Brugada syndrome (BrS) is a potentially life-threatening inherited cardiac arrhythmia. FGF12 (FHF1) is the major intracellular FGF expressed in the human ventricle. A single missense mutation in FGF12 in Brugada syndrome patients reduces binding to the voltage gated sodium channel (NaV1.5) C-terminus, resulting in reduced Na+ channel current density and availability without affecting Ca2+ channel function535 (Table3(a)). Börjeson–Forssman–Lehmann syndrome (BFLS) is an X-linked mental retardation syndrome. A duplication breakpoint identified in a patient with BFLS maps near the FGF13 (FHF2) gene at Xq26.3. This disease association and the high expression of FGF13 in brain and skeletal muscle makes it a good candidate gene for BFLS536 (Table3(a)). X-linked congenital generalized hypertrichosis is an extremely rare condition of hair overgrowth on different body sites. This disease maps to Xq24-27 and a large interchromosomal insertion at Xq27.1 co-segregates with the disease. In patients with this disease, FGF13 expression is significantly decreased throughout the outer root sheath of affected hair follicles, suggesting a role for FGF13 in hair follicle growth and in the hair cycle537 (Table3(a)). Spinocerebellar ataxias (SCAs) are neurodegenerative disorders with multiple genetic etiologies. SCA27 is characterized by early onset tremor, dyskinesia, and slowly progressive cerebellar ataxia. SCA27 is caused by missense, translocation, or deletion mutations in FGF14506,622,623 (Table3(a)). Loss of binding of the mutant FGF14 protein to Nav channel α subunits and instability of the mutant protein are thought to be the primary factors leading to this disease505,624 (Table3(a)).
FGFR1
Gain-of-function missense mutations in FGFR1 are found in several craniosynostosis syndromes including Pfeiffer syndrome, Jackson-Weiss syndrome, Muenke syndrome, and osteoglophonic dysplasia556,557,567,625,626 (Table3(a)). These are autosomal dominant syndromes that affect cranial suture closure and have various additional skeletal and soft tissue phenotypes. Interestingly, Pfeiffer syndrome, Jackson-Weiss syndrome, and Muenke syndrome phenotypes also can be caused by activating mutations in FGFR2 (Pfeiffer) or FGFR3 (Pfeiffer, Muenke), suggesting possible redundant or parallel function of these FGFRs in skeletal development.568,626
Loss-of-function missense mutations have also been identified in FGFR1 as a cause of Kallmann syndrome 2 (hypogonadotropic hypogonadism 2) with or without anosmia558,559 (Table3(a)). Dominant or recessive mutations in FGFR1 that are likely loss-of-function are found in Harstfield syndrome (holoprosencephaly and ectrodactyly, with or without cleft lip and palate)560 (Table3(a)).
FGFR2
Autosomal dominant gain-of-function missense mutations, deletions, and insertions in FGFR2 result in Apert syndrome, Crouzon syndrome, non syndromic craniosynostosis syndrome, Saethe-Chotzen syndrome, Pfeiffer syndrome, and Jackson-Weiss syndrome567–574,627,628 (Table3(b)). Pfeiffer and Jackson-Weiss syndromes also result from mutations in FGFR1, as described above. All of these syndromes have in common synostosis of at least one cranial suture; many of these syndromes also affect the appendicular skeleton and other organs. For example, the Crouzon syndrome mutation, FGFR2C342Y, affects the shape of the brain, but not its overall volume.629
The biochemical consequences of the classic Apert syndrome mutations (FGFR2S252W and FGFR2P253R) and a relatively rare Alu element insertion, or deletion of an intronic splicing element in the intron between exon 8 (IIIb) and exon 9 (IIIc) of FGFR2 is to change the ligand binding affinity to an extent that mesenchymal ligands such as FGF10 are able to activate mesenchymal splice variants of FGFR2575,630 (Figure 5(b)). Importantly, the Apert mutations all remain ligand dependent. The Alu element insertion acts by disrupting splicing to exon 9, encoding the IIIc splice variant (Figure 1(b)), leading to alternative mesenchymal misexpression of exon 8, encoding the IIIb splice variant. The missense mutations directly affect ligand affinity for the mutant receptor.575,576,631–638
Figure 5.
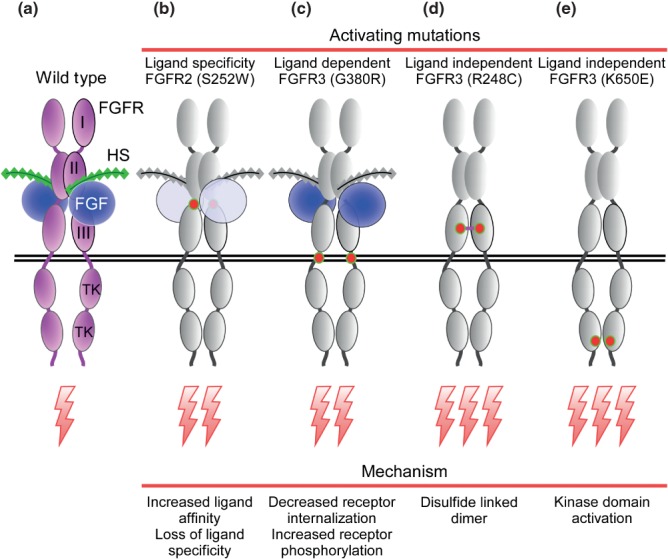
Activating mutations in FGFRs in heritable and acquired disease. (a) Wild type FGFR-FGF-HS complex. (b) Missense mutations in the linker between immunoglobulin-like domain II and III affect the affinity and specificity of the receptor. The Apert syndrome mutation, S252W, allows FGF10 to activate the IIIc splice variant of FGFR2. (c) Missense mutations in the transmembrane domain, as seen in the G380R Achondroplasia mutation, weakly activates the receptor in a ligand dependent manner by impeding receptor internalization. (d) The strongly activating ligand independent mutation, R248C, in Thanatophoric dysplasia, type I, causes constitutively active disulfide linked receptor dimers. (e) Mutations in the tyrosine kinase domain, as seen in the K640E Thanatophoric dysplasia, type II mutation, are often ligand independent and result in receptor autophosphorylation and signaling from intracellular sites such as the endoplasmic reticulum.
Bent bone dysplasia, which is a perinatal lethal skeletal dysplasia characterized by osteopenia, craniofacial dysmorphology and bent bones, results from mutations in FGFR2 that decrease plasma membrane signaling without affecting nuclear localization of the mutant receptor577 (Table3(b)). The consequence of this mutation is enhanced nucleolar occupancy of the receptor at the ribosomal DNA promoter where it activates rDNA transcription.639
Loss-of-function mutations in FGFR2 are seen in lacrimo-auriculo-dento-digital (LADD) syndrome, which is characterized by lacrimal-duct aplasia, dysplastic ears, hearing loss, small teeth, and digital malformations531 (Table3(b)). Mutations in FGFR2 disrupt the catalytic pocket of the tyrosine kinase domain resulting in reduced substrate binding and reduced tyrosine kinase activity.640,641 Other individuals with LADD syndrome have inactivating mutations in FGF10 (see above), a ligand for FGFR2b,533 or a missense mutation in FGFR3531 (Box 2).
BOX 2 Developing a Pharmacological Treatment for Achondroplasia
Achondroplasia is caused by a ligand dependent autosomal dominant mutation in FGFR3. Because the disease phenotypes form during the prepubertal years when bones are actively growing, it was anticipated that direct or indirect inhibition of the FGFR3 signaling pathway could form the basis of a therapy for Achondroplasia.738 The direct inhibition of the FGFR3 kinase has thus far not succeeded in vivo, possibly because of difficulties in achieving therapeutic levels of FGFR3 kinase inhibitors in the avascular growth plate. However, over the past 20 years, other therapies have been aimed at indirectly augmenting skeletal growth or indirectly suppressing FGFR3 signaling. One of the first therapies to be evaluated was the use of human growth hormone; however, no long-term benefit was observed.581,739 More recently, it was discovered that C-type natriuretic peptide (CNP) signaling through its receptor, natriuretic peptide receptor 2 (guanylate cyclase B) in chondrocytes, inhibits the MAPK signaling pathway at the level of RAF1, to regulate skeletal growth. Overexpression of CNP in mice or humans results in skeletal overgrowth through attenuation of FGFR3 signaling.740,741 BMN-111, a CNP agonist with an extended half-life, was found to normalize skeletal growth in a mouse model for Achondroplasia604,742,743 and this drug is currently being evaluated in a clinical trial for the treatment of Achondroplasia.
Other indirect strategies involve the use of a soluble FGFR3 extracellular domain (sFGFR3) to interfere with endogenous FGFR3 signaling by binding FGF ligands (FGF9 and FGF18) that normally are required to activate the receptor during postnatal skeletal development.47,85–87,744 In a mouse model for Achondroplasia,132 subcutaneous injections of recombinant sFGFR3 throughout the growth period normalized skeletal growth and decreased mortality without having any apparent toxic side effects. Several inhibitory antibodies have also been developed to target the FGFR3 extracellular domain for potential cancer therapeutics, but these have not yet been evaluated for treatment of Achondroplasia.745–747
Statins (drugs that inhibit cholesterol biosynthesis) were recently identified through a screen for drugs that could improve chondrogenic differentiation of induce pluripotent stem cells (iPSCs) derived from patients with Thanatophoric dysplasia.748 Treatment of a mouse model for Achondroplasia132 with Rosuvastatin, which is one of the statin drugs, increased anteroposterior skull length and the lengths of the ulnas, femurs and tibiae.748 Although the mechanism is poorly defined, statin treatment was found to increase degradation of the mutant FGFR3.
FGFR3
Hypochondroplasia, Achondroplasia, Thanatophoric dysplasia, and Platyspondylic lethal skeletal dysplasia are autosomal dominant disorders characterized by short-limbed dwarfism.580,581 These syndromes are caused by gain-of-function missense mutations in FGFR3. Among the mutations, the G380R mutation in the transmembrane domain of FGFR3 activates the receptor in a ligand dependent manner resulting in Achondroplasia, the most common form of skeletal dwarfism in humans (Figure 5(c)). By contrast, in the lethal skeletal dysplasia syndrome, Thanatophoric dysplasia, type I or type II, the R248C mutation in the extracellular domain or the K650E mutation in the intracellular domain activates FGFR3 in a ligand independent manner288,582–595,642,643 (Figure 5(d) and (e)). Muenke syndrome (Muenke nonsyndromic coronal craniosynostosis) is an autosomal dominant disorder characterized by synostosis, macrocephaly, midfacial hypoplasia, and hearing loss caused by gain-of-function missense mutations in FGFR3596–602,644 (Table3(b)). Mouse models for aberrant osteogenesis, Achondroplasia, and Muenke syndrome have been developed603–606 (Table3(b)). Two craniosynostosis syndromes, Crouzon syndrome and Saethe-Chotzen syndrome, can result from mutations in FGFR2 or FGFR3, suggesting overlapping or redundant functions of these FGFRs.592,607,608
Loss-of-function missense mutations, that likely function through a dominant negative mechanism, have been identified in FGFR3 as the cause of CATSHL syndrome (autosomal dominant syndrome characterized by camptodactyly, tall stature, and hearing loss)609 (Table3(b)). A recessive loss-of-function mutations in FGFR3 has also been identified in two siblings with tall stature, severe skeletal abnormalities, camptodactyly, arachnodactyly, scoliosis and hearing impairment.610 A similar disease, spider-lamp syndrome in sheep, is characterized by abnormally long limbs, kyphoscoliosis, malformed ribs and sternebrae, hooked or ‘Roman’ nose, lack of body fat, and muscular atrophy.645,646 This disease is associated with a missense mutation in the FGFR3 tyrosine kinase domain coupled with poorly described interactions with other genetic and environmental factors.
A mutation in FGFR3 has also been associated with LADD syndrome (Table3(b)). Although the function of the mutation, localized to the conserved proximal tyrosine kinase domain (TK1, Figure 1(b)), is not known, the phenotypes of affected individuals are distinct from both known gain-of-function mutations causing chondrodysplasia syndromes and loss-of-function mutations resulting in skeletal overgrowth and hearing loss.531,581,609
FGFR4
Facioscapulohumeral muscular dystrophy is an autosomal dominant disorder, ranging from mild dysfunction to severe respiratory failure. Overexpression of FGFR4 in muscle and surrounding connective tissue and overexpression of FGF1 and FGF2 on the sarcolemma may be associated with this disease.612 Bronchopulmonary dysplasia, characterized by impaired alveolar development and inflammation is the most common chronic lung disease resulting from premature birth. Neonatal respiratory distress syndrome is a pulmonary disease affecting preterm neonates. A single nucleotide polymorphism (I > V) in exon 10 of FGFR4 is a potential risk factor for these diseases613 (Table3(b)). The common allelic variant (G388R) in FGFR4 is associated with breast cancer progression and increased insulin secretion and risk of diabetes.647
FGFRL1/FGFR5
Antley–Bixler syndrome is a disorder characterized by craniosynostosis, radio-ulnar synostosis, and genital abnormalities. A C-terminal frameshift mutation in FGFRL1 was found in one patient with this disease. The mutation results in preferential localization of the mutant protein to the plasma membrane, compared to the localization of wild-type FGFRL1 to vesicular structures and the Golgi complex615 (Table3(b)). Wolf-Hirschhorn syndrome (WHS) is a disease resulting in growth delay, craniofacial dysgenesis, developmental delay, and epilepsy. Micro deletions containing FGFRL1, but not the WHSC1 gene have craniofacial features resembling that seen in WHS patients, suggesting that FGFRL1 could be a possible candidate gene616 (Table3(b)). Analysis of a new null allele for Fgfrl1 in mice revealed skeletal and other defects that resemble WHS.617
FGFs and FGFRs: Mutations and Expression in Cancer
Deregulation of FGF signaling pathways have been implicated in many types of human and animal cancers. Deregulation can occur at the level of gene/protein expression of ligands or receptors, which can result from changes in transcriptional activity or gene amplification. Deregulation can also result from mutations in FGF ligands, receptors, or downstream signaling pathways. A more detailed discussion of FGF signaling in cancer can be found in a review by Turner and Grose.152
FGF Family
Mechanisms of FGF ligand activation involve aberrant expression, gene amplification leading to overexpression, or mutations that increase diffusion through tissue or increase affinity for FGFRs (Table4(a)). Aberrant expression and mutations in FGFs have been observed in many human cancers.163,418,649–653,656–662,664,665,667–669,671,672 Gene amplification of FGFs has also been observed.648,654 Overexpression and gene amplification leads to excessive FGF signaling, which can result in cancer initiation or progression. In contrast to the oncogenic properties of many FGF ligands, in some human colon and endometrial cancers that lack β-catenin activation, homozygosity for loss-of-function somatic mutations in FGF9 have been observed. Additionally, mice lacking Fgf22 have normal skin, but show increased papilloma formation in a DMBA/TPA induced tumorigenesis model96 (Table4(a)). These examples show that in at least some cases, FGF signaling can also function to suppress tumorigenesis, possibly by promoting cell differentiation.664 Single nucleotide polymorphisms in FGF23 have been associated with an increased risk of prostate cancer, although it remains unclear whether polymorphisms result in gain- or loss-of-function.673
Table 4.
Acquired and Heritable Mutations in FGFs and FGFRs in Malignancy
| Gene Name | Mutation | Type of Cancer | Selected References |
|---|---|---|---|
| (a) Contributions of FGFs to malignancy (in vivo) | |||
| FGF1 | Amplification | Ovarian cancer | 648 |
| FGF2 | Over expression | Bladder cancer, Prostate cancer, Small cell lung carcinoma, Melanoma, Hepatocellular carcinoma | 649–653 |
| FGF3 | Amplification | Breast cancer | 654 |
| FGF4 | Amplification | Breast cancer | 655 |
| FGF5 | Over expression | Glioblastoma | 656 |
| FGF6 | Over expression | Prostate cancer | 657 |
| FGF7 | Over expression | Lung adenocarcinoma | 658 |
| FGF8 | Over expression | Breast cancer, Prostate cancer, Hepatocellular carcinoma, Colorectal cancer | 659–663 |
| Fgf9 | Frameshift/missense/nonsense mutation | Colorectral and endometrial carcinomas | 418,664–666 |
| Over expression | Non small cell lung cancer | ||
| FGF10 | Over expression | Breast carcinomas, Prostate cancer | 667,668 |
| FGF15/19 | Over expression | Prostate cancer, Hepatocellular carcinoma | 162,163,513,669,670 |
| FGF16 | Over expression | Ovarian cancer | 671 |
| FGF17 | Over expression | Prostate cancer, Hepatocellular carcinoma | 660,672 |
| FGF18 | Over expression | Hepatocellular carcinoma | 660 |
| FGF22 | Knockout | Suppresses skin papilloma (in mice) | 96 |
| FGF23 | Polymorphism | Increased risk of prostate cancer | 673 |
| (b) Contributions of FGFRs to malignancy (in vivo) | |||
| FGFR1 | Amplification | Small cell lung cancer, Squamous cell lung cancer, Breast cancer, Ovarian cancer, Pancreatic ductal adenocarcinoma, Tongue squamous cell carcinoma | 191,192,674–682 |
| Missense mutation | Melanoma, Pilocytic astrocytoma | 683 | |
| Translocation | Leukemia, Lymphoma, Alveolar rhabdomyosarcoma, Glioblastoma, Myeloproliferative syndrome (fusion with CUX1, FGFROP2, FIM, RANBP2/NUP358, SQSTM1, TRP, ZNF198) | 681,684–692 | |
| Over expression | Glioblastoma | 656 | |
| FGFR2 | Amplification | Gastric cancer, Breast cancer | 693–697 |
| Missense mutation | Endometrial carcinoma, Gastric cancer | 693,698 | |
| Translocation | Cholangiocarcinoma | 699–702 | |
| FGFR3 | Missense mutation | Gastric cancer, Colorectal cancer, Breast cancer, Endometrial carcinoma, Urothelial carcinoma, Bladder tumor, Skin tumor, Myeloma | 608,693,703–706 |
| Mis-localization | Brest cancer | 707 | |
| Translocation | Myeloma, Squamous cell lung cancer, Bladder cancer, Glioblastoma, Lymphoma | 708–712 | |
| Over expression | Breast cancer, Colon cancer (FGFR3c) | 713,714 | |
| FGFR4 | Missense mutation | Rhabdomyosarcoma, Adenoid cystic carcinoma, Breast Cancer (resistance to adjuvant therapy) | 715–717 |
| Over expression | Ovarian cancer, hepatocellular carcinoma | 256,718,719 | |
FGFR Family
FGFRs can be activated by gene amplification leading to receptor overexpression, by activating mutations (Figure 5), or by translocations resulting in activating gene fusions.720,721 FGFR1 gene amplification has been identified in 20% of lobular breast cancer, in 3% of lung adenocarcinomas and 21% of squamous cell lung cancer.192,674,722,723 FGFR1 or FGFR2 was amplified in 47% of hormone resistant prostate cancers.724 FGFR3 was amplified in 3% of bladder cancers.725 FGFR4 overexpression (65% of cases) and amplification (30% of adult tumors) were observed in adrenocortical tumors and amplification was associated with worse prognosis.726 FGFR4 amplification was also found in 10% of primary breast tumors.727 Thus, FGFR gene amplification may be pathogenic in a large fraction of some of the major cancer subtypes (Table4(b)).
Oncogenic gene fusions that activate the FGFR tyrosine kinase domain is a relatively common occurrence in myeloproliferative syndrome, glioblastoma, bladder, lung, breast, thyroid, oral, and prostate cancers.193,684 In the 8p11 myeloproliferative syndrome (myeloid and lymphoid neoplasms with FGFR1 abnormalities), FGFR1 translocations result in at least 10 fusion proteins with N-terminal dimer forming partners fused to the C-terminal FGFR1 tyrosine kinase domain684 (Table4(b)). FGFR1–FGFR3 are also closely linked to the transforming, acidic coiled-coil containing protein 1–3 genes (TACC1–TACC3).236 FGFR1 and TACC1 or FGFR3 and TACC3 gene fusions have been identified in glioblastoma, non–small cell lung cancers (NSCLC), bladder cancer, multiple myeloma, and lung squamous cell carcinoma685,708,728–730 (Table4(b)). These gene fusions can generate constitutively active FGFR kinase domains that are localized to the mitotic spindle. FGFR2 translocations resulting in gene fusions with AHCYL1, BICC1, MGEA5, AFF3, and TACC3 have been identified in subtypes of cholangiocarcinoma.699–701 Gene fusions can also result in 3′ UTR deletion, allowing escape from regulation by microRNAs, as seen in an FGFR3-TACC3 fusion in multiple myeloma729 (Table4(b)).
Activation of FGFR3 in multiple myeloma can occur through several mechanisms and is thought to contribute to the neoplastic transformation. A common translocation between the immunoglobulin heavy chain locus on chromosome 14q32 and the FGFR3 and MMSET (multiple myeloma set domain) region of chromosome 4 is found in 15–20% of multiple myeloma cases and many of these result in increased expression of FGFR3731,732 (Table4(b)). However, although this translocation is associated with poor survival, survival does not correlate with FGFR3 expression.733,734
Activation of FGFRs by somatic acquisition of missense mutations is another common tumorigenic mechanism. Missense mutations in FGFR2 have been found in gastric and endometrial cancer693,698,735 (Table4(b)). Missense mutations in FGFR3 have been observed in 25% of cervical carcinomas and 35% of bladder carcinomas736 (Table4(b)). Interestingly, these mutations are identical to the activating mutations that cause Thanatophoric dysplasia. Tyrosine kinase domain mutations were found in 7.5% of rhabdomyosarcomas.715 In gastric cancer, at least one allele of the common G388R variant of FGFR4 was present in 57% of patients, and expression of this allele was associated with worse prognosis737 (Table4(b)).
Conclusion
Since the purification of the first FGF over thirty years ago, an amazing amount of research has uncovered biochemical and biological functions of FGFs, FGFRs, and other interacting molecules that are essential for almost all aspects of life through the regulation of developmental, physiological, and pathological processes, from the earliest stages of embryonic development, to organogenesis, tissue maturation, homeostasis, response to injury, and cancer. Biochemical studies have identified mechanisms that regulate the expression of FGFs, their bioavailability, and their ability to activate cellular responses through interaction with cell surface receptors. Within the cell, signal transduction mechanisms have been identified that reveal interactions with multiple cellular signaling pathways, complex feedback mechanisms, and regulatory molecules that control FGF signaling, both extracellularly and intracellularly. Developmental studies have uncovered redundant functions of FGFs and FGFRs, and interactions with most of the other major signaling pathways, including BMP, WNT, Notch and Hedgehog. The discovery of endocrine FGFs has uncovered new mechanisms that regulate metabolism, lipid, and mineral homeostasis, and has provided potential therapeutic targets for a variety of common diseases, including type 2 diabetes, chronic kidney disease, and obesity. Understanding pathogenic mechanisms resulting from mutations, gene fusions, and gene amplifications in FGFs and FGFRs has led to therapeutic approaches for chondrodysplasia and craniosynostosis syndromes, as well as a variety of cancers. Future directions will be aimed at acquiring a deeper mechanistic understanding of the roles of FGF signaling in development and in adult tissues with a goal of understanding how these pathways become reactivated during injury response and cancer. The development of highly selective pharmacological agonists and antagonists that function at all levels of FGF signaling should provide new tools to protect tissues from injury, enhance cell and tissue repair, treat a variety of metabolic diseases, and inhibit cancer.
Acknowledgments
This work was supported by NIH grants HL105732, HL111190, and HD049808, March of Dimes (FY14-329), Action on Hearing Loss, and a Grant-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (25460065).
Further Reading
General Reviews
Belov AA, Mohammadi M. Grb2, a double-edged sword of receptor tyrosine kinase signaling. Sci Signal 2012, 5:pe49.
Coleman SJ, Bruce C, Chioni AM, Kocher HM, Grose RP. The ins and outs of Fibroblast Growth Factor receptor signalling. Clin Sci (Lond) 2014, 127:217–231.
Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol 2013, 14:166–180.
Itoh N, Ornitz DM. Fibroblast Growth Factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 2011,149:121–130.
Oulion S, Bertrand S, Escriva H. Evolution of the FGF gene family. Int J Evol Biol 2012, 2012:298147.
FGFs and Development
Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. J Cell Physiol 2012, 227:3731–3743.
El Agha E, Bellusci S. Walking along the Fibroblast Growth Factor 10 route: a key pathway to understand the control and regulation of epithelial and mesenchymal cell-lineage formation during lung development and repair after injury. Scientifica (Cairo) 2014, 2014:538379.
Freter S, Muta Y, Mak SS, Rinkwitz S, Ladher RK. Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Development 2008, 135:3415–3424.
Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 2014, 15:123–138.
Miraoui H, Marie PJ. Fibroblast Growth Factor receptor signaling crosstalk in skeletogenesis. Sci Signal 2010, 3:re9.
Ornitz DM, Yin Y. Signaling networks regulating development of the lower respiratory tract. Cold Spring Harb Perspect Biol 2012, 4:1–19.
Pownall ME, Isaacs HV. FGF Signalling in Vertebrate Development. San Rafael CA: Morgan & Claypool Life Sciences; 2010.
Towers M, Wolpert L, Tickle C. Gradients of signalling in the developing limb. Curr Opin Cell Biol 2012, 24:181–187.
FGFs and Genetic Diseases
Laederich MB, Horton WA. FGFR3 targeting strategies for achondroplasia. Expert Rev Mol Med 2012, 14:e11.
Melville H, Wang Y, Taub PJ, Jabs EW. Genetic basis of potential therapeutic strategies for craniosynostosis. Am J Med Genet A 2010, 152A:3007–3015.
Valdes-Socin H, Rubio Almanza M, Tome Fernandez-Ladreda M, Debray FG, Bours V, et al. Reproduction, smell, and neurodevelopmental disorders: genetic defects in different hypogonadotropic hypogonadal syndromes. Front Endocrinol (Lausanne) 2014, 5:109.
FGFs and Cancer
Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast Growth Factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov 2013, 3:264–279.
Kelleher FC, O'Sullivan H, Smyth E,McDermott R, Viterbo A. Fibroblast Growth Factor receptors, developmental corruption and malignant disease. Carcinogenesis 2013, 34:2198–2205.
Turner N, Grose R. Fibroblast Growth Factor signalling: from development to cancer. Nat Rev Cancer 2010, 10:116–129.
Endocrine FGFs
Hu MC, Shiizaki K, Kuro-o M, Moe OW. Fibroblast Growth Factor 23 and Klotho: physiology and pathophysiology of an endocrine network of mineral metabolism. Annu Rev Physiol 2013, 75:503–533.
Itoh N. FGF21 as a hepatokine, adipokine, and myokine in metabolism and diseases. Front Endocrinol (Lausanne) 2014, 5:107.
Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine Fibroblast Growth Factors 15/19 and 21: from feast to famine. Genes Dev 2012, 26:312–324.
Ray K. Liver: Fgf15 maintains bile acid homeostasis and is a key mediator of liver regeneration in mice. Nat Rev Gastroenterol Hepatol 2013, 10:65.
Sapir-Koren R, Livshits G. Bone mineralization is regulated by signaling cross talk between molecular factors of local and systemic origin: the role of Fibroblast Growth Factor 23. Biofactors 2014, 40:555–568.
Smith ER, McMahon LP, Holt SG. Fibroblast Growth Factor 23. Ann Clin Biochem 2014, 51:203–227.
References
- Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Dover K, Laezza F, Shtraizent N, Huang X, Tchetchik D, Eliseenkova AV, Xu CF, Neubert TA, Ornitz DM. Crystal structure of a Fibroblast Growth Factor homologous factor (FHF) defines a conserved surface on FHFS for binding and modulation of voltage-gated sodium channels. J Biol Chem. 2009;284:17883–17896. doi: 10.1074/jbc.M109.001842. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Ortega S, Bashayan O, Basch R, Basilico C. Compensation by Fibroblast Growth Factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol Cell Biol. 2000;20:2260–2268. doi: 10.1128/mcb.20.6.2260-2268.2000. Mol Cell Biol [published erratum appears in 2000, 20:3752] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker JW, Suh JM, Atkins AR, Ahmadian M, Li P, Whyte J, He M, Juguilon H, Yin YQ, Phillips CT. A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485:391–394. doi: 10.1038/nature10998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Sutliff RL, Paul RJ, Lorenz JN, Hoying JB, Haudenschild CC, Yin M, Coffin JD, Kong L, Kranias EG. Fibroblast Growth Factor 2 control of vascular tone. Nat Med. 1998;4:201–207. doi: 10.1038/nm0298-201. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JE, Witt SA, Nieman ML, Reiser PJ, Engle SJ, Zhou M, Pawlowski SA, Lorenz JN, Kimball TR, Doetschman T. Fibroblast Growth Factor-2 mediates pressure-induced hypertrophic response. J Clin Invest. 1999;104:709–719. doi: 10.1172/JCI7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Ittmann M, Tsang SH, Ehrlich M, Basilico C. Neuronal defects and delayed wound healing in mice lacking Fibroblast Growth Factor 2. Proc Natl Acad Sci USA. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic Fibroblast Growth Factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the Fibroblast Growth Factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer M, Cesnulevicius K, Winkler C, Kolb J, Lipokatic-Takacs E, Jungnickel J, Grothe C. Fibroblast Growth Factor (FGF)-2 and FGF receptor 3 are required for the development of the substantia nigra, and FGF-2 plays a crucial role for the rescue of dopaminergic neurons after 6-hydroxydopamine lesion. J Neurosci. 2007;27:459–471. doi: 10.1523/JNEUROSCI.4493-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer-Bouthiette C, Doetschman T, Xiao L, Hurley MM. Knockout of nuclear high molecular weight FGF2 isoforms in mice modulates bone and phosphate homeostasis. J Biol Chem. 2014;289:36303–36314. doi: 10.1074/jbc.M114.619569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Urness LD, Bleyl SB, Wright TJ, Moon AM, Mansour SL. Redundant and dosage sensitive requirements for Fgf3 and Fgf10 in cardiovascular development. Dev Biol. 2011;356:383–397. doi: 10.1016/j.ydbio.2011.05.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Sun X, Lewandoski M, Meyers EN, Liu YH, Maxson RE, Jr, Martin GR. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Boulet AM, Capecchi MR. Normal limb development in conditional mutants of Fgf4. Development. 2000;127:989–996. doi: 10.1242/dev.127.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, Rosenquist T, Götz J, Martin GR. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Floss T, Arnold HH, Braun T. A role for Fgf-6 in skeletal muscle regeneration. Genes Dev. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus P, Oustanina S, Loch T, Kruger M, Bober E, Dono R, Zeller R, Braun T. Reduced mobility of Fibroblast Growth Factor (FGF)-deficient myoblasts might contribute to dystrophic changes in the musculature of FGF2/FGF6/mdx triple-mutant mice. Mol Cell Biol. 2003;23:6037–6048. doi: 10.1128/MCB.23.17.6037-6048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand AS, Pariset C, Laziz I, Launay T, Fiore F, Della Gaspera B, Birnbaum D, Charbonnier F, Chanoine C. FGF6 regulates muscle differentiation through a calcineurin-dependent pathway in regenerating soleus of adult mice. J Cell Physiol. 2005;204:297–308. doi: 10.1002/jcp.20302. [DOI] [PubMed] [Google Scholar]
- Terauchi A, Johnson-Venkatesh EM, Toth AB, Javed D, Sutton MA, Umemori H. Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature. 2010;465:783–787. doi: 10.1038/nature09041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Degenstein L, Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- Qiao J, Uzzo R, Obara-Ishihara T, Degenstein L, Fuchs E, Herzlinger D. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- Alpdogan O, Hubbard VM, Smith OM, Patel N, Lu S, Goldberg GL, Gray DH, Feinman J, Kochman AA, Eng JM. Keratinocyte growth factor (KGF) is required for postnatal thymic regeneration. Blood. 2006;107:2453–2460. doi: 10.1182/blood-2005-07-2831. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, He Q, Luo C. Lack of keratinocyte growth factor retards angiogenesis in cutaneous wounds. J Int Med Res. 2011;39:416–423. doi: 10.1177/147323001103900209. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche LA, Holder N, Lewandoski M. FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc Natl Acad Sci USA. 2011;108:4018–4023. doi: 10.1073/pnas.1007417108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CB, Wenning JM, Lu MM, Epstein DJ, Meyers EN, Epstein JA. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004;267:190–192. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–391. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Saga Y. FGF8-FGFR1 signaling acts as a niche factor for maintaining undifferentiated spermatogonia in the mouse. Biol Reprod. 2014;91:145. doi: 10.1095/biolreprod.114.121012. [DOI] [PubMed] [Google Scholar]
- Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschke P, Salomon R, Antignac C. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev Biol. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White A, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang F, Ornitz DM. Mesothelial and epithelial derived FGF9 have distinct functions in the regulation of lung development. Development. 2011;138:3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking Fibroblast Growth Factor 9. Cell. 2001b;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- Dinapoli L, Batchvarov J, Capel B. FGF9 promotes survival of germ cells in the fetal testis. Development. 2006;133:1519–1527. doi: 10.1242/dev.02303. [DOI] [PubMed] [Google Scholar]
- Hung IH, Yu K, Lavine KJ, Ornitz DM. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev Biol. 2007;307:300–313. doi: 10.1016/j.ydbio.2007.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske MJ, Zhang X, Patel KK, Ornitz DM, Stappenbeck TS. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Kim Y, Colvin JS, Ornitz DM, Capel B. Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development. 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- Lin Y, Chen L, Lin C, Luo Y, Tsai RY, Wang F. Neuron-derived FGF9 is essential for scaffold formation of Bergmann radial fibers and migration of granule neurons in the cerebellum. Dev Biol. 2009;329:44–54. doi: 10.1016/j.ydbio.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerie A, Bajolle F, Zaffran S, Brown NA, Dickson C, Buckingham ME, Kelly RG. Congenital heart defects in Fgfr2-IIIb and Fgf10 mutant mice. Cardiovasc Res. 2006;71:50–60. doi: 10.1016/j.cardiores.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Nyeng P, Norgaard GA, Kobberup S, Jensen J. FGF10 signaling controls stomach morphogenesis. Dev Biol. 2006;303:295–310. doi: 10.1016/j.ydbio.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. et al. [DOI] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. et al. [DOI] [PubMed] [Google Scholar]
- Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn. 2009;238:1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskoll T, Abichaker G, Witcher D, Sala FG, Bellusci S, Hajihosseini MK, Melnick M. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev Biol. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, Rice R, Spencer-Dene B, Mailleux AA, Rice DP. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006;133:2325–2335. doi: 10.1242/dev.02394. et al. [DOI] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Carbe C, Tao C, Powers A, Lawrence R, van Kuppevelt TH, Cardoso WV, Grobe K, Esko JD, Zhang X. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 2011;286:14435–14444. doi: 10.1074/jbc.M111.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala FG, Del Moral PM, Tiozzo C, Alam DA, Warburton D, Grikscheit T, Veltmaat JM, Bellusci S. FGF10 controls the patterning of the tracheal cartilage rings via Shh. Development. 2011;138:273–282. doi: 10.1242/dev.051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn WJ, Jung HI, Choi MA, Han JH, Gwon GJ, Yamamoto H, Lee S, Ryoo ZY, Park EK, Shin HI. Reciprocal interactions of Fgf10/Fgfr2b modulate the mouse tongue epithelial differentiation. Cell Tissue Res. 2011;345:265–273. doi: 10.1007/s00441-011-1204-8. et al. [DOI] [PubMed] [Google Scholar]
- Al Alam D, Sala FG, Baptista S, Galzote R, Danopoulos S, Tiozzo C, Gage P, Grikscheit T, Warburton D, Frey MR. FGF9-Pitx2-FGF10 signaling controls cecal formation in mice. Dev Biol. 2012;369:340–342. doi: 10.1016/j.ydbio.2012.07.008. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb M, Schoorlemmer J, Williams A, Diwakar S, Wang Q, Huang X, Giza J, Tchetchik D, Kelley K, Vega A. Fibroblast Growth Factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron. 2007;55:449–463. doi: 10.1016/j.neuron.2007.07.006. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QF, Yang L, Li S, Wang Q, Yuan XB, Gao X, Bao L, Zhang X. Fibroblast Growth Factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149:1549–1564. doi: 10.1016/j.cell.2012.04.046. [DOI] [PubMed] [Google Scholar]
- Wozniak DF, Xiao M, Xu L, Yamada KA, Ornitz DM. Impaired spatial learning and defective theta burst induced LTP in mice lacking Fibroblast Growth Factor 14. Neurobiol Dis. 2007;26:14–26. doi: 10.1016/j.nbd.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Xu L, Laezza F, Yamada K, Feng S, Ornitz DM. Impaired hippocampal synaptic transmission and plasticity in mice lacking Fibroblast Growth Factor 14. Mol Cell Neurosci. 2007;34:366–377. doi: 10.1016/j.mcn.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Shakkottai VG, Xiao M, Xu L, Wong M, Nerbonne JM, Ornitz DM, Yamada KA. FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol Dis. 2009;33:81–88. doi: 10.1016/j.nbd.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Bardgett ME, Wong M, Wozniak DF, Lou J, McNeil BD, Chen C, Nardi A, Reid DC, Yamada K. Ataxia and paroxysmal dyskinesia in mice lacking axonally transported FGF14. Neuron. 2002;35:25–38. doi: 10.1016/s0896-6273(02)00744-4. et al. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine Fibroblast Growth Factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, McWhirter JR, Murre C, Baldini A, Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- Borello U, Cobos I, Long JE, McWhirter JR, Murre C, Rubenstein JL. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Dev. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA. Fibroblast Growth Factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. et al. [DOI] [PubMed] [Google Scholar]
- Kong B, Huang J, Zhu Y, Li G, Williams J, Shen S, Aleksunes LM, Richardson JR, Apte U, Rudnick DA. Fibroblast Growth Factor 15 deficiency impairs liver regeneration in mice. Am J Physiol Gastrointest Liver Physiol. 2014;306:G893–G902. doi: 10.1152/ajpgi.00337.2013. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Moschetta A, Bookout AL, Peng L, Umetani M, Holmstrom SR, Suino-Powell K, Xu HE, Richardson JA, Gerard RD. Identification of a hormonal basis for gallbladder filling. Nat Med. 2006;12:1253–1255. doi: 10.1038/nm1501. et al. [DOI] [PubMed] [Google Scholar]
- Matsumoto E, Sasaki S, Kinoshita H, Kito T, Ohta H, Konishi M, Kuwahara K, Nakao K, Itoh N. Angiotensin II-induced cardiac hypertrophy and fibrosis are promoted in mice lacking Fgf16. Genes Cells. 2013;18:544–553. doi: 10.1111/gtc.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Sasaki S, Konishi M, Kinoshita H, Kuwahara K, Nakao K, Itoh N. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev Dyn. 2008;237:2947–2954. doi: 10.1002/dvdy.21726. [DOI] [PubMed] [Google Scholar]
- Lu SY, Sheikh F, Sheppard PC, Fresnoza A, Duckworth ML, Detillieux KA, Cattini PA. FGF-16 is required for embryonic heart development. Biochem Biophys Res Commun. 2008;373:270–274. doi: 10.1016/j.bbrc.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci USA. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by Fibroblast Growth Factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H, Shibayama M, Ohbayashi N, Konishi M, Takada S, Itoh N. Fgf18 is required for embryonic lung alveolar development. Biochem Biophys Res Commun. 2004;322:887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Clark JC, Picard L, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT. Fibroblast Growth Factor 18 influences proximal programming during lung morphogenesis. J Biol Chem. 2002;277:22743–22749. doi: 10.1074/jbc.M202253200. [DOI] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haara O, Harjunmaa E, Lindfors PH, Huh SH, Fliniaux I, Aberg T, Jernvall J, Ornitz DM, Mikkola ML, Thesleff I. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development. 2012;139:3189–3199. doi: 10.1242/dev.079558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Närhi K, Lindfors PH, Häärä O, Yang L, Ornitz DM, Mikkola ML. Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes Dev. 2013;27:450–458. doi: 10.1101/gad.198945.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Nakamura H, Konishi M, Murata Y, Takagi H, Matsumura S, Inoue K, Fushiki T, Itoh N. Fibroblast Growth Factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150:4625–4633. doi: 10.1210/en.2009-0119. [DOI] [PubMed] [Google Scholar]
- Murata Y, Nishio K, Mochiyama T, Konishi M, Shimada M, Ohta H, Itoh N. Fgf21 impairs adipocyte insulin sensitivity in mice fed a low-carbohydrate, high-fat ketogenic diet. PLoS One. 2013;8:e69330. doi: 10.1371/journal.pone.0069330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Umemori H. Suppression of epileptogenesis-associated changes in response to seizures in FGF22-deficient mice. Front Cell Neurosci. 2013;7:43. doi: 10.3389/fncel.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jarosz M, Robbez-Masson L, Chioni AM, Cross B, Rosewell I, Grose R. Fibroblast Growth Factor 22 is not essential for skin development and repair but plays a role in tumorigenesis. PLoS One. 2012;7:e39436. doi: 10.1371/journal.pone.0039436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Su J, Brooks J, Terauchi A, Umemori H, Fox MA. Fibroblast Growth Factor 22 contributes to the development of retinal nerve terminals in the dorsal lateral geniculate nucleus. Front Mol Neurosci. 2012;4:61. doi: 10.3389/fnmol.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Shiizaki K, Kuro-o M, Moe OW. Fibroblast Growth Factor 23 and Klotho: physiology and pathophysiology of an endocrine network of mineral metabolism. Annu Rev Physiol. 2013;75:503–533. doi: 10.1146/annurev-physiol-030212-183727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, White KE. FGF23 and disorders of phosphate homeostasis. Cytokine Growth Factor Rev. 2005;16:221–232. doi: 10.1016/j.cytogfr.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, Fukumoto S, Tomizuka K, Yamashita T. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J Clin Invest. 2004;113:561–568. doi: 10.1172/JCI19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, Juppner H, Lanske B. Homozygous ablation of Fibroblast Growth Factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol. 2004;23:421–432. doi: 10.1016/j.matbio.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysaght AC, Yuan Q, Fan Y, Kalwani N, Caruso P, Cunnane M, Lanske B, Stankovic KM. FGF23 deficiency leads to mixed hearing loss and middle ear malformation in mice. PLoS One. 2014;9:e107681. doi: 10.1371/journal.pone.0107681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Ono K, Kita T, Sato S, O'Neill P, Mak SS, Paschaki M, Ito M, Gotoh N, Kawakami K, Sasai Y. FGFR1-Frs2/3 signalling maintains sensory progenitors during inner ear hair cell formation. PLoS Genet. 2014;10:e1004118. doi: 10.1371/journal.pgen.1004118. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. FGFR-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- Zhao M, Ross JT, Itkin T, Perry JM, Venkatraman A, Haug JS, Hembree MJ, Deng CX, Lapidot T, He XC. FGF signaling facilitates postinjury recovery of mouse hematopoietic system. Blood. 2012;120:1831–1842. doi: 10.1182/blood-2011-11-393991. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast Growth Factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyden JM, Lewandoski M, Deng C, Harfe BD, Sun X. Conditional inactivation of Fgfr1 in mouse defines its role in limb bud establishment, outgrowth and digit patterning. Development. 2005;132:4235–4245. doi: 10.1242/dev.02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu X, Nelson DK, Williams T, Kuehn MR, Deng CX. FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development. 2005;132:4755–4764. doi: 10.1242/dev.02065. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Uchida AO, Shin D, Partanen J, Vaccarino FM. Fibroblast Growth Factor receptor 1 is required for the proliferation of hippocampal progenitor cells and for hippocampal growth in mouse. J Neurosci. 2004;24:6057–6069. doi: 10.1523/JNEUROSCI.1140-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Li CL, Takahashi K, Slavkin HC, Shum L, Deng CX. Murine Fibroblast Growth Factor receptor 1 alpha isoforms mediate node regression and are essential for posterior mesoderm development. Dev Biol. 1999;208:293–306. doi: 10.1006/dbio.1999.9227. [DOI] [PubMed] [Google Scholar]
- Adams AC, Yang C, Coskun T, Cheng CC, Gimeno RE, Luo Y, Kharitonenkov A. The breadth of FGF21's metabolic actions are governed by FGFR1 in adipose tissue. Mol Metab. 2012;2:31–37. doi: 10.1016/j.molmet.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Tellides G, Simons M. Fibroblast Growth Factor receptor 1 is a key inhibitor of TGFbeta signaling in the endothelium. Sci Signal. 2014;7:ra90. doi: 10.1126/scisignal.2005504. [DOI] [PubMed] [Google Scholar]
- Ding BS, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, Penfold ME, Shido K, Rabbany SY, Rafii S. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast Growth Factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Yu K, Ornitz DM. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development. 2008;135:483–491. doi: 10.1242/dev.013268. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of Fibroblast Growth Factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Guasti L, Candy Sze WC, McKay T, Grose R, King PJ. FGF signalling through Fgfr2 isoform IIIb regulates adrenal cortex development. Mol Cell Endocrinol. 2013;37:182–188. doi: 10.1016/j.mce.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kegg H, Grady S, Truong HT, Robinson ML, Baum M, Bates CM. Role of Fibroblast Growth Factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poladia DP, Kish K, Kutay B, Hains D, Kegg H, Zhao H, Bates CM. Role of Fibroblast Growth Factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291:325–339. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Furusho M, Dupree JL, Nave KA, Bansal R. Fibroblast Growth Factor receptor signaling in oligodendrocytes regulates myelin sheath thickness. J Neurosci. 2012;32:6631–6641. doi: 10.1523/JNEUROSCI.6005-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladipupo SS, Smith C, Santeford A, Park C, Sene A, Wiley LA, Osei-Owusu P, Hsu J, Zapata N, Liu F. Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc Natl Acad Sci USA. 2014;111:13379–13384. doi: 10.1073/pnas.1324235111. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Nguyen LT, Zhuang ZW, Moodie KL, Carmeliet P, Stan RV, Simons M. The FGF system has a key role in regulating vascular integrity. J Clin Invest. 2008;118:3355–3366. doi: 10.1172/JCI35298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Huang J, Liu Y, Dattilo LK, Huh SH, Ornitz D, Beebe DC. FGF signaling activates a Sox9-Sox10 pathway for the formation and branching morphogenesis of mouse ocular glands. Development. 2014;141:2691–2701. doi: 10.1242/dev.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm F, Speicher T, Hellerbrand C, Dickson C, Partanen J, Ornitz D, Werner S. FGF receptors 1 and 2 control chemically-induced injury and compound detoxification in regenerating livers of mice. Gastroenterology. 2010;139:1385–1396. doi: 10.1053/j.gastro.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking Fibroblast Growth Factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast Growth Factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, Fritzsch B, Kelley MW. Disruption of Fibroblast Growth Factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh LY, Denninger A, Colvin JS, Vyas A, Tole S, Ornitz DM, Bansal R. Fibroblast Growth Factor receptor 3 signaling regulates the onset of oligodendrocyte terminal differentiation. J Neurosci. 2003;23:883–894. doi: 10.1523/JNEUROSCI.23-03-00883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by Fibroblast Growth Factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Szarama KB, Stepanyan R, Petralia RS, Gavara N, Frolenkov GI, Kelley MW, Chadwick RS. Fibroblast Growth Factor receptor 3 regulates microtubule formation and cell surface mechanical properties in the developing organ of Corti. Bioarchitecture. 2012;2:214–219. doi: 10.4161/bioa.22332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Schlessinger J. Skeletal overgrowth is mediated by deficiency in a specific isoform of Fibroblast Growth Factor receptor 3. Proc Natl Acad Sci USA. 2007;104:3937–3942. doi: 10.1073/pnas.0700012104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Franco G, Binette JS, Li W, Wang H, Chai S, Laflamme F, Tran-Khanh N, Quenneville E, Meijers T, Poole AR. Defects in articular cartilage metabolism and early arthritis in Fibroblast Growth Factor receptor 3 deficient mice. Hum Mol Genet. 2006;15:1783–1792. doi: 10.1093/hmg/ddl100. et al. [DOI] [PubMed] [Google Scholar]
- Valverde-Franco G, Liu H, Davidson D, Chai S, Valderrama-Carvajal H, Goltzman D, Ornitz DM, Henderson JE. Defective bone mineralization and osteopenia in young adult FGFR3−/− mice. Hum Mol Genet. 2004;13:271–284. doi: 10.1093/hmg/ddh034. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Jacques BE, Kelley MW. Fibroblast Growth Factor signaling regulates pillar cell development in the organ of corti. J Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Xu X, Li C, He Q, Zhao L, Chen S, Luo F, Yi L, Du X, Huang H. Generation of Fgfr3 conditional knockout mice. Int J Biol Sci. 2010;6:327–332. doi: 10.7150/ijbs.6.327. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, Mariani TJ. Fibroblast Growth Factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. Am J Respir Crit Care Med. 2010;181:838–850. doi: 10.1164/rccm.200904-0544OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattineni J, Twombley K, Goetz R, Mohammadi M, Baum M. Regulation of serum 1,25(OH)2 vitamin D3 levels by Fibroblast Growth Factor 23 is mediated by FGF receptors 3 and 4. Am J Physiol Renal Physiol. 2011;301:F371–F377. doi: 10.1152/ajprenal.00740.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattineni J, Alphonse P, Zhang Q, Mathews N, Bates CM, Baum M. Regulation of renal phosphate transport by FGF23 is mediated by FGFR1 and FGFR4. Am J Physiol Renal Physiol. 2014;306:F351–F358. doi: 10.1152/ajprenal.00232.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Martin A, David V, Quarles LD. Compound deletion of Fgfr3 and Fgfr4 partially rescues the Hyp mouse phenotype. Am J Physiol Endocrinol Metab. 2011;300:E508–E517. doi: 10.1152/ajpendo.00499.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- Yu C, Wang F, Jin C, Wu X, Chan WK, McKeehan WL. Increased carbon tetrachloride-induced liver injury and fibrosis in FGFR4-deficient mice. American Journal of Pathology. 2002;161:2003–2010. doi: 10.1016/S0002-9440(10)64478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud-Dabernat S, Kritzik M, Kayali AG, Zhang YQ, Liu G, Ungles C, Sarvetnick N. FGFR3 is a negative regulator of the expansion of pancreatic epithelial cells. Diabetes. 2007;56:96–106. doi: 10.2337/db05-1073. [DOI] [PubMed] [Google Scholar]
- Arnaud-Dabernat S, Yadav D, Sarvetnick N. FGFR3 contributes to intestinal crypt cell growth arrest. J Cell Physiol. 2008;216:261–268. doi: 10.1002/jcp.21401. [DOI] [PubMed] [Google Scholar]
- Gerber SD, Steinberg F, Beyeler M, Villiger PM, Trueb B. The murine Fgfrl1 receptor is essential for the development of the metanephric kidney. Dev Biol. 2009;335:106–119. doi: 10.1016/j.ydbio.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274:6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast Growth Factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast Growth Factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by Fibroblast Growth Factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast Growth Factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb M. Fibroblast Growth Factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A. FGFs and metabolism. Curr Opin Pharmacol. 2009;9:805–810. doi: 10.1016/j.coph.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Angelin B, Larsson TE, Rudling M. Circulating Fibroblast Growth Factors as metabolic regulators--a critical appraisal. Cell Metab. 2012;16:693–705. doi: 10.1016/j.cmet.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Itoh N. Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010;342:1–11. doi: 10.1007/s00441-010-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Fibroblast Growth Factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, Goetz R, Mohammadi M, Kuro-o M, Mangelsdorf DJ. Research resource: comprehensive expression atlas of the Fibroblast Growth Factor system in adult mouse. Mol Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall ME, Isaacs HV. FGF Signalling in Vertebrate Development. San Rafael CA: Morgan & Claypool Life Sciences; 2010. [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Vonderfecht S, Weiszmann J, Hecht R, Gupte J, Hager T, Wang Z, Lindberg R. FGF19-induced hepatocyte proliferation is mediated through FGFR4 activation. J Biol Chem. 2010;285:5165–5170. doi: 10.1074/jbc.M109.068783. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Dakhova O, Creighton CJ, Ittmann M. Endocrine Fibroblast Growth Factor FGF19 promotes prostate cancer progression. Cancer Res. 2013;73:2551–2562. doi: 10.1158/0008-5472.CAN-12-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk D, Honma-Yamanaka N, Anani S, Yamanaka Y. FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Dev Biol. 2013;384:65–71. doi: 10.1016/j.ydbio.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Kelly RG. Molecular inroads into the anterior heart field. Trends Cardiovasc Med. 2005;15:51–56. doi: 10.1016/j.tcm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG. New developments in the second heart field. Differentiation. 2012;84:17–24. doi: 10.1016/j.diff.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. et al. [DOI] [PubMed] [Google Scholar]
- Wang J, Rhee S, Palaria A, Tremblay KD. FGF signaling is required for anterior but not posterior specification of the murine liver bud. Dev Dyn. 2015;244:431–443. doi: 10.1002/dvdy.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naye F, Voz ML, Detry N, Hammerschmidt M, Peers B, Manfroid I. Essential roles of zebrafish bmp2a, fgf10, and fgf24 in the specification of the ventral pancreas. Mol Biol Cell. 2012;23:945–954. doi: 10.1091/mbc.E11-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R, Jukkola T, Saarimaki J, Peltopuro P, Naserke T, Weisenhorn DM, Trokovic N, Wurst W, Partanen J. Fgfr1-dependent boundary cells between developing mid- and hindbrain. Dev Biol. 2005;278:428–439. doi: 10.1016/j.ydbio.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Blak AA, Naserke T, Saarimaki-Vire J, Peltopuro P, Giraldo-Velasquez M, Vogt Weisenhorn DM, Prakash N, Sendtner M, Partanen J, Wurst W. Fgfr2 and Fgfr3 are not required for patterning and maintenance of the midbrain and anterior hindbrain. Dev Biol. 2007;303:231–243. doi: 10.1016/j.ydbio.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Muller AK, Meyer M, Werner S. The roles of receptor tyrosine kinases and their ligands in the wound repair process. Semin Cell Dev Biol. 2012;23:963–970. doi: 10.1016/j.semcdb.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Kardami E, Detillieux K, Ma X, Jiang Z, Santiago JJ, Jimenez SK, Cattini PA. Fibroblast Growth Factor-2 and cardioprotection. Heart Fail Rev. 2007;12:267–277. doi: 10.1007/s10741-007-9027-0. [DOI] [PubMed] [Google Scholar]
- House SL, Branch K, Newman G, Doetschman T, Schultz Jel J. Cardioprotection induced by cardiac-specific overexpression of Fibroblast Growth Factor-2 is mediated by the MAPK cascade. Am J Physiol Heart Circ Physiol. 2005;289:H2167–H2175. doi: 10.1152/ajpheart.00392.2005. [DOI] [PubMed] [Google Scholar]
- Liao S, Porter D, Scott A, Newman G, Doetschman T, Schultz Jel J. The cardioprotective effect of the low molecular weight isoform of Fibroblast Growth Factor-2: the role of JNK signaling. J Mol Cell Cardiol. 2007;42:106–120. doi: 10.1016/j.yjmcc.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, auf dem Keller U, Steiling H, Werner S. Fibroblast Growth Factors in epithelial repair and cytoprotection. Philos Trans R Soc Lond B Biol Sci. 2004;359:753–757. doi: 10.1098/rstb.2004.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Stoilov I, Elton TJ, Mecham RP, Ornitz DM. FGF2 is required for epithelial recovery, but not for pulmonary fibrosis, in response to bleomycin. Am J Respir Cell Mol Biol. 2015;52:116–128. doi: 10.1165/rcmb.2014-0184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Muller AK, Yang J, Moik D, Ponzio G, Ornitz DM, Grose R, Werner S. FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. J Cell Sci. 2012;125:5690–5701. doi: 10.1242/jcs.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Du Y, Shen Y, He Y, Zhao H, Li Z. TGF-beta 1 induced fibroblast proliferation is mediated by the FGF-2/ERK pathway. Front Biosci (Landmark Ed) 2012;17:2667–2674. doi: 10.2741/4077. [DOI] [PubMed] [Google Scholar]
- Warburton D. Developmental responses to lung injury: repair or fibrosis. Fibrogenesis Tissue Repair. 2012;5(Suppl 1):S2. doi: 10.1186/1755-1536-5-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte VV, Ramasamy SK, Reddy R, Lee J, Weinreb PH, Violette SM, Guenther A, Warburton D, Driscoll B, Minoo P. Overexpression of Fibroblast Growth Factor-10 during both inflammatory and fibrotic phases attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Respir Crit Care Med. 2009;180:424–436. doi: 10.1164/rccm.200811-1794OC. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YC, Kharitonenkov A. Hormone-like Fibroblast Growth Factors and metabolic regulation. Biochim Biophys Acta. 1812;2011:791–795. doi: 10.1016/j.bbadis.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Itoh N. FGF21 as a hepatokine, adipokine, and myokine in metabolism and diseases. Front Endocrinol (Lausanne) 2014;5:107. doi: 10.3389/fendo.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Kilgour E, Smith PD. Molecular pathways: Fibroblast Growth Factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. J Cell Physiol. 2012;227:3731–3743. doi: 10.1002/jcp.24083. [DOI] [PubMed] [Google Scholar]
- Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, Edwards MG, Bohm D, Perner S, Helfrich BA. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299–3309. doi: 10.1158/1078-0432.CCR-13-3060. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, Wolf J, Buttner R, Schildhaus HU. Fibroblast Growth Factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol. 2014;27:214–221. doi: 10.1038/modpathol.2013.141. [DOI] [PubMed] [Google Scholar]
- Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, Fernandez-Cuesta L, Heuckmann JM, Gautschi O, Diebold J. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014;4:246–257. doi: 10.1158/2159-8290.CD-13-0323. et al. [DOI] [PubMed] [Google Scholar]
- Parker BC, Engels M, Annala M, Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol. 2014;232:4–15. doi: 10.1002/path.4297. [DOI] [PubMed] [Google Scholar]
- Yadav V, Zhang X, Liu J, Estrem S, Li S, Gong XQ, Buchanan S, Henry JR, Starling JJ, Peng SB. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 2012;287:28087–28098. doi: 10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, Cummings CT, Graham DK, Astling D, Tan AC. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai H, Soejima K, Yasuda H, Nakayama S, Hamamoto J, Arai D, Ishioka K, Ohgino K, Ikemura S, Sato T. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res. 2013;11:759–767. doi: 10.1158/1541-7786.MCR-12-0652. et al. [DOI] [PubMed] [Google Scholar]
- Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kimura-Yoshida C. Extracellular modulation of Fibroblast Growth Factor signaling through heparan sulfate proteoglycans in mammalian development. Curr Opin Genet Dev. 2013;23:399–407. doi: 10.1016/j.gde.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:1–33. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic Fibroblast Growth Factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- Smith ER, McMahon LP, Holt SG. Fibroblast Growth Factor 23. Ann Clin Biochem. 2014;51:203–227. doi: 10.1177/0004563213510708. [DOI] [PubMed] [Google Scholar]
- Trueb B. Biology of FGFRL1, the fifth Fibroblast Growth Factor receptor. Cell Mol Life Sci. 2011;68:951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Yaguchi Y, Echevarria D, Martinez S, Basson MA. Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development. 2011;138:2957–2968. doi: 10.1242/dev.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Gong Y, Tang Y, Li H, He Q, Gower L, Liaw L, Friesel RE. Spry1 and Spry4 differentially regulate human aortic smooth muscle cell phenotype via Akt/FoxO/myocardin signaling. PLoS One. 2013;8:e58746. doi: 10.1371/journal.pone.0058746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, Goldfarb M. Fibroblast Growth Factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J Biol Chem. 2002;277:49111–49119. doi: 10.1074/jbc.M205520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig HG, Fenner BJ, Byrne JC, Schwamborn RF, Bernas T, Jefferies CA, Prehn JH. Fibroblast Growth Factor homologous factor 1 interacts with NEMO to regulate NF-kappaB signaling in neurons. J Cell Sci. 2012;125:6058–6070. doi: 10.1242/jcs.111880. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- Trowell OA, Chir B, Willmer EN. Studies on the growth of tissues in vitro. J Exp Biol. 1939;16:60–70. [Google Scholar]
- Armelin HA. Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proc Natl Acad Sci USA. 1973;70:2702–2706. doi: 10.1073/pnas.70.9.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz D. Purification of a Fibroblast Growth Factor from bovine pituitary. J Biol Chem. 1975;250:2515–2520. [PubMed] [Google Scholar]
- Lemmon SK, Bradshaw RA. Purification and partial characterization of bovine pituitary Fibroblast Growth Factor. J Cell Biochem. 1983;21:195–208. doi: 10.1002/jcb.240210302. [DOI] [PubMed] [Google Scholar]
- Thomas KA, Rios-Candelore M, Fitzpatrick S. Purification and characterization of acidic Fibroblast Growth Factor from bovine brain. Proc Natl Acad Sci USA. 1984;81:357–361. doi: 10.1073/pnas.81.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KA, Riley MC, Lemmon SK, Baglan NC, Bradshaw RA. Brain Fibroblast Growth Factor: nonidentity with myelin basic protein fragments. J Biol Chem. 1980;255:5517–5520. [PubMed] [Google Scholar]
- Lemmon SK, Riley MC, Thomas KA, Hoover GA, Maciag T, Bradshaw RA. Bovine Fibroblast Growth Factor: comparison of brain and pituitary preparations. J Cell Biol. 1982;95:162–169. doi: 10.1083/jcb.95.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Gallego G, Conn G, Hatcher VB, Thomas KA. The complete amino acid sequence of human brain-derived acidic Fibroblast Growth Factor. Biochem Biophys Res Commun. 1986;138:611–617. doi: 10.1016/s0006-291x(86)80540-x. [DOI] [PubMed] [Google Scholar]
- Gimenez-Gallego G, Rodkey J, Bennett C, Rios-Candelore M, DiSalvo J, Thomas K. Brain-derived acidic Fibroblast Growth Factor: complete amino acid sequence and homologies. Science. 1985;230:1385–1388. doi: 10.1126/science.4071057. [DOI] [PubMed] [Google Scholar]
- Thomas KA, Rios-Candelore M, Gimenez-Gallego G, DiSalvo J, Bennett C, Rodkey J, Fitzpatrick S. Pure brain-derived acidic Fibroblast Growth Factor is a potent angiogenic vascular endothelial cell mitogen with sequence homology to interleukin 1. Proc Natl Acad Sci USA. 1985;82:6409–6413. doi: 10.1073/pnas.82.19.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber AB, Kenney J, Kowalski WJ, Friesel R, Mehlman T, Maciag T. Interaction of endothelial cell growth factor with heparin: characterization by receptor and antibody recognition. Proc Natl Acad Sci USA. 1985;82:6138–6142. doi: 10.1073/pnas.82.18.6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber AB, Kenney J, Kowalski J, Thomas KA, Gimenez-Gallego G, Rios-Candelore M, Di Salvo J, Barritault D, Courty J, Courtois Y. A unique family of endothelial cell polypeptide mitogens: the antigenic and receptor cross-reactivity of bovine endothelial cell growth factor, brain-derived acidic Fibroblast Growth Factor, and eye-derived growth factor-II. J Cell Biol. 1985;101:1623–1626. doi: 10.1083/jcb.101.4.1623. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaye M, Howk R, Burgess W, Ricca GA, Chiu I-M, Ravera MW, O'Brien SJ, Modi WS, Maciag T, Drohan WN. Human endothelial cell growth factor: cloning, nucleotide sequence, and chromosome localization. Science. 1986;233:541–545. doi: 10.1126/science.3523756. [DOI] [PubMed] [Google Scholar]
- Abraham JA, Mergia A, Whang JL, Tumolo A, Friedman J, Hjerrild KA, Gospodarowicz D, Fiddes JC. Nucleotide sequence of a bovine clone encoding the angiogenic protein, basic Fibroblast Growth Factor. Science. 1986;233:545–548. doi: 10.1126/science.2425435. [DOI] [PubMed] [Google Scholar]
- Krejci P, Prochazkova J, Bryja V, Kozubik A, Wilcox WR. Molecular pathology of the Fibroblast Growth Factor family. Hum Mutat. 2009;30:1245–1255. doi: 10.1002/humu.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulion S, Bertrand S, Escriva H. Evolution of the FGF gene family. Int J Evol Biol. 2012;2012:298147. doi: 10.1155/2012/298147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudovsky I, Kumar TK, Sterling S, Neivandt D. Protein-phospholipid interactions in nonclassical protein secretion: problem and methods of study. Int J Mol Sci. 2013;14:3734–3772. doi: 10.3390/ijms14023734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudovsky I, Mandinova A, Soldi R, Bagala C, Graziani I, Landriscina M, Tarantini F, Duarte M, Bellum S, Doherty H. The non-classical export routes: FGF1 and IL-1alpha point the way. J Cell Sci. 2003;116:4871–4881. doi: 10.1242/jcs.00872. et al. [DOI] [PubMed] [Google Scholar]
- Landriscina M, Soldi R, Bagala C, Micucci I, Bellum S, Tarantini F, Prudovsky I, Maciag T. S100A13 participates in the release of Fibroblast Growth Factor 1 in response to heat shock in vitro. J Biol Chem. 2001;276:22544–22552. doi: 10.1074/jbc.M100546200. [DOI] [PubMed] [Google Scholar]
- Wesche J, Malecki J, Wiedlocha A, Skjerpen CS, Claus P, Olsnes S. FGF-1 and FGF-2 require the cytosolic chaperone Hsp90 for translocation into the cytosol and the cell nucleus. J Biol Chem. 2006;281:11405–11412. doi: 10.1074/jbc.M600477200. [DOI] [PubMed] [Google Scholar]
- Sorensen V, Wiedlocha A, Haugsten EM, Khnykin D, Wesche J, Olsnes S. Different abilities of the four FGFRs to mediate FGF-1 translocation are linked to differences in the receptor C-terminal tail. J Cell Sci. 2006;119:4332–4341. doi: 10.1242/jcs.03209. [DOI] [PubMed] [Google Scholar]
- Olsnes S, Klingenberg O, Wiedlocha A. Transport of exogenous growth factors and cytokines to the cytosol and to the nucleus. Physiol Rev. 2003;83:163–182. doi: 10.1152/physrev.00021.2002. [DOI] [PubMed] [Google Scholar]
- Planque N. Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun Signal. 2006;4:7. doi: 10.1186/1478-811X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouleau S, Grimal H, Rincheval V, Godefroy N, Mignotte B, Vayssiere JL, Renaud F. FGF1 inhibits p53-dependent apoptosis and cell cycle arrest via an intracrine pathway. Oncogene. 2005;24:7839–7849. doi: 10.1038/sj.onc.1208932. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enfedaque A, Bouleau S, Laurent M, Courtois Y, Mignotte B, Vayssiere JL, Renaud F. FGF1 nuclear translocation is required for both its neurotrophic activity and its p53-dependent apoptosis protection. Biochim Biophys Acta. 2009;1793:1719–1727. doi: 10.1016/j.bbamcr.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the Fibroblast Growth Factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the Fibroblast Growth Factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Miyakawa K, Hatsuzawa K, Kurokawa T, Asada M, Kuroiwa T, Imamura T. A hydrophobic region locating at the center of Fibroblast Growth Factor-9 is crucial for its secretion. J Biol Chem. 1999;274:29352–29357. doi: 10.1074/jbc.274.41.29352. [DOI] [PubMed] [Google Scholar]
- Revest JM, DeMoerlooze L, Dickson C. Fibroblast Growth Factor 9 secretion is mediated by a non-cleaved amino-terminal signal sequence. J Biol Chem. 2000;275:8083–8090. doi: 10.1074/jbc.275.11.8083. [DOI] [PubMed] [Google Scholar]
- Miyakawa K, Imamura T. Secretion of FGF-16 requires an uncleaved bipartite signal sequence. J Biol Chem. 2003;278:35718–35724. doi: 10.1074/jbc.M300690200. [DOI] [PubMed] [Google Scholar]
- Consortium A. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ. Molecular insights into the klotho-dependent, endocrine mode of action of Fibroblast Growth Factor 19 subfamily members. Mol Cell Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Mangelsdorf DJ. Fibroblast Growth Factor 21: from pharmacology to physiology. Am J Clin Nutr. 2010;91:254s–257s. doi: 10.3945/ajcn.2009.28449B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Boney-Montoya J, Owen BM, Bookout AL, Coate KC, Mangelsdorf DJ, Kliewer SA. betaKlotho is required for Fibroblast Growth Factor 21 effects on growth and metabolism. Cell Metab. 2012;16:387–393. doi: 10.1016/j.cmet.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Uehara Y, Motomura-Matsuzaka K, Oki J, Koyama Y, Kimura M, Asada M, Komi-Kuramochi A, Oka S, Imamura T. betaKlotho is required for Fibroblast Growth Factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol. 2008;22:1006–1014. doi: 10.1210/me.2007-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW. Regulation of Fibroblast Growth Factor-23 signaling by klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Leder P. Ligand specificity and heparin dependence of Fibroblast Growth Factor receptors 1 and 3. J Biol Chem. 1992;267:16305–16311. [PubMed] [Google Scholar]
- Wang JK, Gao GX, Goldfarb M. Fibroblast growth-factor receptors have different signaling and mitogenic potentials. Mol Cell Biol. 1994;14:181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Chatelain E, Ornitz D, Bresnick J, Mason I, Kiefer P, Dickson C. Receptor binding and mitogenic properties of mouse Fibroblast Growth Factor 3. Modulation of response by heparin. J Biol Chem. 1995;270:24197–24203. doi: 10.1074/jbc.270.41.24197. [DOI] [PubMed] [Google Scholar]
- Blunt AG, Lawshe A, Cunningham ML, Seto ML, Ornitz DM, Macarthur CA. Overlapping expression and redundant activation of mesenchymal Fibroblast Growth Factor (FGF) receptors by alternatively spliced FGF-8 ligands. J Biol Chem. 1997;272:3733–3738. doi: 10.1074/jbc.272.6.3733. [DOI] [PubMed] [Google Scholar]
- Santos-Ocampo S, Colvin JS, Chellaiah AT, Ornitz DM. Expression and biological activity of mouse Fibroblast Growth Factor-9. J Biol Chem. 1996;271:1726–1731. doi: 10.1074/jbc.271.3.1726. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Heparin is required for cell-free binding of basic Fibroblast Growth Factor to a soluble receptor and for mitogenesis in whole cells. Mol Cell Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AL, Kolumam G, Stawicki S, Chen Y, Li J, Zavala-Solorio J, Phamluong K, Feng B, Li L, Marsters S. Amelioration of type 2 diabetes by antibody-mediated activation of Fibroblast Growth Factor receptor 1. Sci Transl Med. 2011;3:113ra126. doi: 10.1126/scitranslmed.3002669. et al. [DOI] [PubMed] [Google Scholar]
- Ho HK, Pok S, Streit S, Ruhe JE, Hart S, Lim KS, Loo HL, Aung MO, Lim SG, Ullrich A. Fibroblast Growth Factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J Hepatol. 2009;50:118–127. doi: 10.1016/j.jhep.2008.08.015. [DOI] [PubMed] [Google Scholar]
- French DM, Lin BC, Wang M, Adams C, Shek T, Hotzel K, Bolon B, Ferrando R, Blackmore C, Schroeder K. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One. 2012;7:e36713. doi: 10.1371/journal.pone.0036713. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y. The discovery of alpha-Klotho and FGF23 unveiled new insight into calcium and phosphate homeostasis. Cell Mol Life Sci. 2008;65:3218–3230. doi: 10.1007/s00018-008-8177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Fibroblast Growth Factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast Growth Factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- Hsu WC, Nilsson CL, Laezza F. Role of the axonal initial segment in psychiatric disorders: function, dysfunction, and intervention. Front Psychiatry. 2014;5:109. doi: 10.3389/fpsyt.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Bosch MK, Nerbonne JM, Ornitz DM. FGF14 localization and organization of the axon initial segment. Mol Cell Neurosci. 2013;56:393–403. doi: 10.1016/j.mcn.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou JY, Laezza F, Gerber BR, Xiao M, Yamada KA, Hartmann H, Craig AM, Nerbonne JM, Ornitz DM. Fibroblast Growth Factor 14 is an intracellular modulator of voltage-gated sodium channels. J Physiol (Lond) 2005;569:179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Hennessey JA, Kirkton RD, Graham V, Puranam RS, Rosenberg PB, Bursac N, Pitt GS. Fibroblast Growth Factor homologous factor 13 regulates Na+ channels and conduction velocity in murine hearts. Circ Res. 2011;109:775–782. doi: 10.1161/CIRCRESAHA.111.247957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, Goldfarb M. Fibroblast Growth Factor homologous factors are intracellular signaling proteins. Curr Biol. 2001;11:793–797. doi: 10.1016/s0960-9822(01)00232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Huang JS. Association of bovine brain-derived growth factor receptor with protein tyrosine kinase activity. J Biol Chem. 1986;261:9568–9571. [PubMed] [Google Scholar]
- Coughlin SR, Barr PJ, Cousens LS, Fretto LJ, Williams LT. Acidic and basic Fibroblast Growth Factors stimulate tyrosine kinase activity in vivo. J Biol Chem. 1988;263:988–993. [PubMed] [Google Scholar]
- Lee PL, Johnson DE, Cousens LS, Fried VA, Williams LT. Purification and complementary DNA cloning of a receptor for basic Fibroblast Growth Factor. Science. 1989;245:57–60. doi: 10.1126/science.2544996. [DOI] [PubMed] [Google Scholar]
- Kornbluth S, Paulson KE, Hanafusa H. Novel tyrosine kinase identified by phosphotyrosine antibody screening of cDNA libraries. Mol Cell Biol. 1988;8:5541–5544. doi: 10.1128/mcb.8.12.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan K, Johnson DE, Williams LT, Hayman MJ. Isolation of an additional member of the Fibroblast Growth Factor receptor family, FGFR-3. Proc Natl Acad Sci USA. 1991;88:1095–1099. doi: 10.1073/pnas.88.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen J, Makela TP, Eerola E, Korhonen J, Hirvonen H, Claesson-Welsh L, Alitalo K. FGFR-4, a novel acidic Fibroblast Growth Factor receptor with a distinct expression pattern. EMBO J. 1991;10:1347–1354. doi: 10.1002/j.1460-2075.1991.tb07654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, McMahon JA, McMahon AP. FGFR-4, a new member of the Fibroblast Growth Factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development. 1991;113:641–651. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. A distinctive family of embryonic protein-tyrosine kinase receptors. Proc Natl Acad Sci USA. 1990;87:5812–5816. doi: 10.1073/pnas.87.15.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Fleming TP, Bottaro DP, Rubin JS, Ron D, Aaronson SA. Expression cDNA cloning of the KGF receptor by creation of a transforming autocrine loop. Science. 1991;251:72–75. doi: 10.1126/science.1846048. [DOI] [PubMed] [Google Scholar]
- Yeh BK, Igarashi M, Eliseenkova AV, Plotnikov AN, Sher I, Ron D, Aaronson SA, Mohammadi M. Structural basis by which alternative splicing confers specificity in Fibroblast Growth Factor receptors. Proc Natl Acad Sci USA. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- Kalinina J, Dutta K, Ilghari D, Beenken A, Goetz R, Eliseenkova AV, Cowburn D, Mohammadi M. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure. 2012;20:77–88. doi: 10.1016/j.str.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kan M, Yan G, Xu J, McKeehan WL. Alternately spliced NH2-terminal immunoglobulin-like loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J Biol Chem. 1995;270:10231–10235. doi: 10.1074/jbc.270.17.10231. [DOI] [PubMed] [Google Scholar]
- Roghani M, Moscatelli D. Prostate cells express two isoforms of Fibroblast Growth Factor receptor 1 with different affinities for Fibroblast Growth Factor-2. Prostate. 2007;67:115–124. doi: 10.1002/pros.20448. [DOI] [PubMed] [Google Scholar]
- Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM, Aaronson SA. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM. Fibroblast Growth Factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–11627. [PubMed] [Google Scholar]
- Werner S, Duan DS, de Vries C, Peters KG, Johnson DE, Williams LT. Differential splicing in the extracellular region of Fibroblast Growth Factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol Cell Biol. 1992;12:82–88. doi: 10.1128/mcb.12.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry N, Harrington W, Lasda E, Wagner EJ, Garcia-Blanco MA. Of urchins and men: evolution of an alternative splicing unit in Fibroblast Growth Factor receptor genes. RNA. 2003;9:209–217. doi: 10.1261/rna.2470903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan DS, Werner S, Williams LT. A naturally occurring secreted form of Fibroblast Growth Factor (FGF) receptor 1 binds basic FGF in preference over acidic FGF. J Biol Chem. 1992;267:16076–16080. [PubMed] [Google Scholar]
- Tomlinson DC, L'Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of Fibroblast Growth Factor receptor 3 produces a secreted isoform that inhibits Fibroblast Growth Factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–10449. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- Terada M, Shimizu A, Sato N, Miyakaze SI, Katayama H, Kurokawa-Seo M. Fibroblast Growth Factor receptor 3 lacking the Ig IIIb and transmembrane domains secreted from human squamous cell carcinoma DJM-1 binds to FGFs. Mol Cell Biol Res Commun. 2001;4:365–373. doi: 10.1006/mcbr.2001.0306. [DOI] [PubMed] [Google Scholar]
- Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of Fibroblast Growth Factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Ibrahimi OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M. Insights into the molecular basis for Fibroblast Growth Factor receptor autoinhibition and ligand-binding promiscuity. Proc Natl Acad Sci USA. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, Mohammadi M. Plasticity in interactions of Fibroblast Growth Factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J Biol Chem. 2012;287:3067–3078. doi: 10.1074/jbc.M111.275891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur CA, Lawshé A, Xu J, Santos-Ocampo S, Heikinheimo M, Chellaiah AT, Ornitz DM. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development. 1995;121:3603–3613. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast Growth Factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Werner S, Dickson C, Grose R. Fibroblast Growth Factor 22 and its potential role during skin development and repair. Exp Cell Res. 2003;287:228–236. doi: 10.1016/s0014-4827(03)00139-3. [DOI] [PubMed] [Google Scholar]
- Scotet E, Houssaint E. The choice between alternative IIIb and IIIc exons of the FGFR-3 gene is not strictly tissue-specific. Biochim Biophys Acta. 1995;1264:238–242. doi: 10.1016/0167-4781(95)00156-b. [DOI] [PubMed] [Google Scholar]
- Murgue B, Tsunekawa S, Rosenberg I, deBeaumont M, Podolsky DK. Identification of a novel variant form of Fibroblast Growth Factor receptor 3 (FGFR3 IIIb) in human colonic epithelium. Cancer Res. 1994;54:5206–5211. [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Kuo WJ, Digman MA, Lander AD. Heparan sulfate acts as a bone morphogenetic protein coreceptor by facilitating ligand-induced receptor hetero-oligomerization. Mol Biol Cell. 2010;21:4028–4041. doi: 10.1091/mbc.E10-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olwin B, Rapraeger A. Repression of myogenic differentiation by aFGF, bFGF, and K-FGF is dependent on cellular heparan sulfate. J Cell Biol. 1992;118:631–639. doi: 10.1083/jcb.118.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov AA, Mohammadi M. Molecular mechanisms of Fibroblast Growth Factor signaling in physiology and pathology. Cold Spring Harb Perspect Biol. 2013;5:1–24. doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Cotman SL, Halfter W, Cole GJ. The heparan sulfate proteoglycan agrin modulates neurite outgrowth mediated by FGF-2. J Neurobiol. 2003;55:261–277. doi: 10.1002/neu.10213. [DOI] [PubMed] [Google Scholar]
- Cotman SL, Halfter W, Cole GJ. Identification of extracellular matrix ligands for the heparan sulfate proteoglycan agrin. Exp Cell Res. 1999;249:54–64. doi: 10.1006/excr.1999.4463. [DOI] [PubMed] [Google Scholar]
- Aviezer D, Hecht D, Safran M, Eisinger M, David G, Yayon A. Perlecan, basal lamina proteoglycan, promotes basic Fibroblast Growth Factor-receptor binding, mitogenesis, and angiogenesis. Cell. 1994;79:1005–1013. doi: 10.1016/0092-8674(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J Biol Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, Patel VN, Lang RA, Mohammadi M. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, Kjellen L. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim Biophys Acta. 2002;1573:209–215. doi: 10.1016/s0304-4165(02)00386-0. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 2007;282:32802–32810. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]
- Kjellen L. Glucosaminyl N-deacetylase/N-sulphotransferases in heparan sulphate biosynthesis and biology. Biochem Soc Trans. 2003;31:340–342. doi: 10.1042/bst0310340. [DOI] [PubMed] [Google Scholar]
- Cai Z, Grobe K, Zhang X. Role of heparan sulfate proteoglycans in optic disc and stalk morphogenesis. Dev Dyn. 2014;243:1310–1316. doi: 10.1002/dvdy.24142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Meyer K, Rapraeger AC, Friedl A. Differential ability of heparan sulfate proteoglycans to assemble the Fibroblast Growth Factor receptor complex in situ. FASEB J. 2000;14:137–144. doi: 10.1096/fasebj.14.1.137. [DOI] [PubMed] [Google Scholar]
- Allen BL, Rapraeger AC. Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J Cell Biol. 2003;163:637–648. doi: 10.1083/jcb.200307053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa K, Kimura-Yoshida C, Nagai N, Mukai K, Matsubara K, Watanabe H, Matsuda Y, Mochida K, Matsuo I. Cell surface heparan sulfate chains regulate local reception of FGF signaling in the mouse embryo. Dev Cell. 2011;21:257–272. doi: 10.1016/j.devcel.2011.06.027. [DOI] [PubMed] [Google Scholar]
- Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, Yates EA, Turnbull JE, Ron D, Hoffman MP. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. J Biol Chem. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, van Kuppevelt TH, Zcharia E, Vlodavsky I, Lindahl U. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 2007;3:773–778. doi: 10.1038/nchembio.2007.41. et al. [DOI] [PubMed] [Google Scholar]
- Ostrovsky O, Berman B, Gallagher J, Mulloy B, Fernig DG, Delehedde M, Ron D. Differential effects of heparin saccharides on the formation of specific Fibroblast Growth Factor (FGF) and FGF receptor complexes. J Biol Chem. 2002;277:2444–2453. doi: 10.1074/jbc.M108540200. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Herr AB, Nilsson M, Westman J, Svahn CM, Waksman G. FGF binding and FGF receptor activation by synthetic heparan-derived di- and trisaccharides. Science. 1995;268:432–436. doi: 10.1126/science.7536345. [DOI] [PubMed] [Google Scholar]
- Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, Whitelock JM, Elkin M, Vlodavsky I, Hoffman MP. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development. 2007;134:4177–4186. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. et al. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Endocrine FGFs and Klothos: emerging concepts. Trends Endocrinol Metab. 2008;19:239–245. doi: 10.1016/j.tem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Kuro OM. The Klotho gene family as a regulator of endocrine Fibroblast Growth Factors. Mol Cell Endocrinol. 2009;299:72–78. doi: 10.1016/j.mce.2008.10.052. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Klotho in health and disease. Curr Opin Nephrol Hypertens. 2012;21:362–368. doi: 10.1097/MNH.0b013e32835422ad. [DOI] [PubMed] [Google Scholar]
- Ito S, Kinoshita S, Shiraishi N, Nakagawa S, Sekine S, Fujimori T, Nabeshima YI. Molecular cloning and expression analyses of mouse betaklotho, which encodes a novel Klotho family protein. Mech Dev. 2000;98:115–119. doi: 10.1016/s0925-4773(00)00439-1. [DOI] [PubMed] [Google Scholar]
- Ito S, Fujimori T, Hayashizaki Y, Nabeshima Y. Identification of a novel mouse membrane-bound family 1 glycosidase-like protein, which carries an atypical active site structure. Biochim Biophys Acta. 2002;1576:341–345. doi: 10.1016/s0167-4781(02)00281-6. [DOI] [PubMed] [Google Scholar]
- Goetz R, Ohnishi M, Ding X, Kurosu H, Wang L, Akiyoshi J, Ma J, Gai W, Sidis Y, Pitteloud N. Klotho coreceptors inhibit signaling by paracrine Fibroblast Growth Factor 8 subfamily ligands. Mol Cell Biol. 2012;32:1944–1954. doi: 10.1128/MCB.06603-11. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DQ, Kan MK, Sato GH, Okamoto T, Sato JD. Characterization and molecular cloning of a putative binding protein for heparin-binding growth factors. J Biol Chem. 1991;266:16778–16785. [PubMed] [Google Scholar]
- Czubayko F, Liaudet-Coopman ED, Aigner A, Tuveson AT, Berchem GJ, Wellstein A. A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nat Med. 1997;3:1137–1140. doi: 10.1038/nm1097-1137. [DOI] [PubMed] [Google Scholar]
- Abuharbeid S, Czubayko F, Aigner A. The Fibroblast Growth Factor-binding protein FGF-BP. Int J Biochem Cell Biol. 2006;38:1463–1468. doi: 10.1016/j.biocel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Beer HD, Bittner M, Niklaus G, Munding C, Max N, Goppelt A, Werner S. The Fibroblast Growth Factor binding protein is a novel interaction partner of FGF-7, FGF-10 and FGF-22 and regulates FGF activity: implications for epithelial repair. Oncogene. 2005;24:5269–5277. doi: 10.1038/sj.onc.1208560. [DOI] [PubMed] [Google Scholar]
- Tassi E, McDonnell K, Gibby KA, Tilan JU, Kim SE, Kodack DP, Schmidt MO, Sharif GM, Wilcox CS, Welch WJ. Impact of Fibroblast Growth Factor-binding protein-1 expression on angiogenesis and wound healing. Am J Pathol. 2011;179:2220–2232. doi: 10.1016/j.ajpath.2011.07.043. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- Trueb B, Zhuang L, Taeschler S, Wiedemann M. Characterization of FGFRL1, a novel Fibroblast Growth Factor (FGF) receptor preferentially expressed in skeletal tissues. J Biol Chem. 2003;278:33857–33865. doi: 10.1074/jbc.M300281200. [DOI] [PubMed] [Google Scholar]
- Kim I, Moon S, Yu K, Kim U, Koh GY. A novel Fibroblast Growth Factor receptor-5 preferentially expressed in the pancreas(1) Biochim Biophys Acta. 2001;1518:152–156. doi: 10.1016/s0167-4781(00)00282-7. [DOI] [PubMed] [Google Scholar]
- Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, Murison JG. Identification of a new Fibroblast Growth Factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- Steinberg F, Zhuang L, Beyeler M, Kalin RE, Mullis PE, Brandli AW, Trueb B. The FGFRL1 receptor is shed from cell membranes, binds FGFs and antagonizes FGF signaling in Xenopus embryos. J Biol Chem. 2010;285:2193–2202. doi: 10.1074/jbc.M109.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PN, Altamentova SM, Kilkenny DM, Rocheleau JV. Fibroblast Growth Factor receptor like-1 (FGFRL1) interacts with SHP-1 phosphatase at insulin secretory granules and induces beta-cell ERK1/2 protein activation. J Biol Chem. 2013;288:17859–17870. doi: 10.1074/jbc.M112.440677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Wyder S, Slavotinek AM, Trueb B. The FgfrL1 receptor is required for development of slow muscle fibers. Dev Biol. 2014;394:228–241. doi: 10.1016/j.ydbio.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Bluteau G, Zhuang L, Amann R, Trueb B. Targeted disruption of the intracellular domain of receptor FgfrL1 in mice. PLoS One. 2014;9:e105210. doi: 10.1371/journal.pone.0105210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the Fibroblast Growth Factor receptor is activated by receptor amplification. Cancer Res. 2010;70:3391–3401. doi: 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KG, Marie J, Wilson E, Ives HE, Escobedo J, Del Rosario M, Mirda D, Williams LT. Point mutation of an FGF receptor abolishes phosphatidylinositol turnover and Ca2+ flux but not mitogenesis. Nature. 1992;358:678–681. doi: 10.1038/358678a0. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Honneger AM, Rotin D, Fisher R, Bellot F, Li W, Dionne CA, Jaye M, Rubinstein M, Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the Fibroblast Growth Factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-g1. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Lee KW, Goldfarb M. Novel recognition motif on Fibroblast Growth Factor receptor mediates direct association and activation of SNT adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]
- Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J, Lax I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on Fibroblast Growth Factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Guris DL, Seo JH, Li L, Hammond J, Talbot A, Imamoto A. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10:71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JH, Suenaga A, Hatakeyama M, Taiji M, Imamoto A. Structural and functional basis of a role for CRKL in a Fibroblast Growth Factor 8-induced feed-forward loop. Mol Cell Biol. 2009;29:3076–3087. doi: 10.1128/MCB.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson H, Klint P, Landgren E, Claesson-Welsh L. Fibroblast Growth Factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J Biol Chem. 1999;274:25726–25734. doi: 10.1074/jbc.274.36.25726. [DOI] [PubMed] [Google Scholar]
- Kanazawa S, Fujiwara T, Matsuzaki S, Shingaki K, Taniguchi M, Miyata S, Tohyama M, Sakai Y, Yano K, Hosokawa K. bFGF regulates PI3-kinase-Rac1-JNK pathway and promotes fibroblast migration in wound healing. PLoS One. 2010;5:e12228. doi: 10.1371/journal.pone.0012228. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb MJ. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for Fibroblast Growth Factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamothe B, Yamada M, Schaeper U, Birchmeier W, Lax I, Schlessinger J. The docking protein Gab1 is an essential component of an indirect mechanism for Fibroblast Growth Factor stimulation of the phosphatidylinositol 3-kinase/Akt antiapoptotic pathway. Mol Cell Biol. 2004;24:5657–5666. doi: 10.1128/MCB.24.13.5657-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnberg N, Neubuser A. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol. 2002;247:237–250. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–507. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131:3885–3896. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly JF, Mickey G, Maher PA. Association of Fibroblast Growth Factor receptor 1 with the adaptor protein Grb14. Characterization of a new receptor binding partner. J Biol Chem. 2000;275:7771–7778. doi: 10.1074/jbc.275.11.7771. [DOI] [PubMed] [Google Scholar]
- Browaeys-Poly E, Blanquart C, Perdereau D, Antoine AF, Goenaga D, Luzy JP, Chen H, Garbay C, Issad T, Cailliau K. Grb14 inhibits FGF receptor signaling through the regulation of PLCgamma recruitment and activation. FEBS Lett. 2010;584:4383–4388. doi: 10.1016/j.febslet.2010.09.048. et al. [DOI] [PubMed] [Google Scholar]
- Cross MJ, Lu L, Magnusson P, Nyqvist D, Holmqvist K, Welsh M, Claesson-Welsh L. The Shb adaptor protein binds to tyrosine 766 in the FGFR-1 and regulates the Ras/MEK/MAPK pathway via FRS2 phosphorylation in endothelial cells. Mol Biol Cell. 2002;13:2881–2893. doi: 10.1091/mbc.E02-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WC, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- Yang X, Qiao D, Meyer K, Pier T, Keles S, Friedl A. Angiogenesis induced by signal transducer and activator of transcription 5A (STAT5A) is dependent on autocrine activity of proliferin. J Biol Chem. 2012;287:6490–6502. doi: 10.1074/jbc.M111.254631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19:3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- Heath C, Cross NC. Critical role of STAT5 activation in transformation mediated by ZNF198-FGFR1. J Biol Chem. 2004;279:6666–6673. doi: 10.1074/jbc.M308743200. [DOI] [PubMed] [Google Scholar]
- Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci P, Salazar L, Goodridge HS, Kashiwada TA, Schibler MJ, Jelinkova P, Thompson LM, Wilcox WR. STAT1 and STAT3 do not participate in FGF-mediated growth arrest in chondrocytes. J Cell Sci. 2008;121:272–281. doi: 10.1242/jcs.017160. [DOI] [PubMed] [Google Scholar]
- Yang X, Qiao D, Meyer K, Friedl A. Signal transducers and activators of transcription mediate Fibroblast Growth Factor-induced vascular endothelial morphogenesis. Cancer Res. 2009;69:1668–1677. doi: 10.1158/0008-5472.CAN-07-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugsten EM, Zakrzewska M, Brech A, Pust S, Olsnes S, Sandvig K, Wesche J. Clathrin- and dynamin-independent endocytosis of FGFR3–implications for signalling. PLoS One. 2011;6:e21708. doi: 10.1371/journal.pone.0021708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auciello G, Cunningham DL, Tatar T, Heath JK, Rappoport JZ. Regulation of Fibroblast Growth Factor receptor signalling and trafficking by Src and Eps8. J Cell Sci. 2013;126:613–624. doi: 10.1242/jcs.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla C, Rigbolt KT, Emdal KB, Carraro G, Vernet E, Bekker-Jensen DB, Streicher W, Wikstrom M, Sundstrom M, Bellusci S. Functional proteomics defines the molecular switch underlying FGF receptor trafficking and cellular outputs. Mol Cell. 2013;51:707–722. doi: 10.1016/j.molcel.2013.08.002. et al. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Dikic I, Giordano S. Negative receptor signalling. Curr Opin Cell Biol. 2003;15:128–135. doi: 10.1016/s0955-0674(03)00004-8. [DOI] [PubMed] [Google Scholar]
- Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol. 2009;203:191–202. doi: 10.1677/JOE-09-0110. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of Fibroblast Growth Factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Kovalenko D, Yang X, Chen PY, Nadeau RJ, Zubanova O, Pigeon K, Friesel R. A role for extracellular and transmembrane domains of Sef in Sef-mediated inhibition of FGF signaling. Cell Signal. 2006;18:1958–1966. doi: 10.1016/j.cellsig.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Camps M, Nichols A, Gillieron C, Antonsson B, Muda M, Chabert C, Boschert U, Arkinstall S. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science. 1998;280:1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekerot M, Stavridis MP, Delavaine L, Mitchell MP, Staples C, Owens DM, Keenan ID, Dickinson RJ, Storey KG, Keyse SM. Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem J. 2008;412:287–298. doi: 10.1042/BJ20071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RJ, Eblaghie MC, Keyse SM, Morriss-Kay GM. Expression of the ERK-specific MAP kinase phosphatase PYST1/MKP3 in mouse embryos during morphogenesis and early organogenesis. Mech Dev. 2002;113:193–196. doi: 10.1016/s0925-4773(02)00024-2. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Lunn JS, Dickinson RJ, Munsterberg AE, Sanz-Ezquerro JJ, Farrell ER, Mathers J, Keyse SM, Storey K, Tickle C. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr Biol. 2003;13:1009–1018. doi: 10.1016/s0960-9822(03)00381-6. [DOI] [PubMed] [Google Scholar]
- Wong A, Lamothe B, Lee A, Schlessinger J, Lax I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc Natl Acad Sci USA. 2002;99:6684–6689. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour C, Guenou H, Kaabeche K, Bouvard D, Sanjay A, Marie PJ. FGFR2-Cbl interaction in lipid rafts triggers attenuation of PI3K/Akt signaling and osteoblast survival. Bone. 2008;42:1032–1039. doi: 10.1016/j.bone.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, George R, Lin CC, Suen KM, Levitt JA, Suhling K, Ladbury JE. Direct binding of Grb2 SH3 domain to FGFR2 regulates SHP2 function. Cell Signal. 2010;22:23–33. doi: 10.1016/j.cellsig.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Lin CC, Suen KM, Melo FA, Levitt JA, Suhling K, Ladbury JE. Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity. J Cell Biol. 2013;200:493–504. doi: 10.1083/jcb.201204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso WV, Itoh A, Nogawa H, Mason I, Brody JS. FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev Dyn. 1997;208:398–405. doi: 10.1002/(SICI)1097-0177(199703)208:3<398::AID-AJA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Belleudi F, Leone L, Nobili V, Raffa S, Francescangeli F, Maggio M, Morrone S, Marchese C, Torrisi MR. Keratinocyte growth factor receptor ligands target the receptor to different intracellular pathways. Traffic. 2007;8:1854–1872. doi: 10.1111/j.1600-0854.2007.00651.x. [DOI] [PubMed] [Google Scholar]
- Volckaert T, De Langhe SP. Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev Dyn. 2015;244:342–366. doi: 10.1002/dvdy.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Stow JL. Nuclear translocation of cell-surface receptors: lessons from Fibroblast Growth Factor. Traffic. 2005;6:947–954. doi: 10.1111/j.1600-0854.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Coleman SJ, Bruce C, Chioni AM, Kocher HM, Grose RP. The ins and outs of Fibroblast Growth Factor receptor signalling. Clin Sci (Lond) 2014;127:217–231. doi: 10.1042/CS20140100. [DOI] [PubMed] [Google Scholar]
- Wiedlocha A, Falnes PO, Madshus IH, Sandvig K, Olsnes S. Dual mode of signal transduction by externally added acidic Fibroblast Growth Factor. Cell. 1994;76:1039–1051. doi: 10.1016/0092-8674(94)90381-6. [DOI] [PubMed] [Google Scholar]
- Joy A, Moffett J, Neary K, Mordechai E, Stachowiak EK, Coons S, Rankin-Shapiro J, Florkiewicz RZ, Stachowiak MK. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14:171–183. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Stachowiak EK. Integrative nuclear signaling in cell development--a role for FGF receptor-1. DNA Cell Biol. 2007;26:811–826. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- Fang X, Stachowiak EK, Dunham-Ems SM, Klejbor I, Stachowiak MK. Control of CREB-binding protein signaling by nuclear Fibroblast Growth Factor receptor-1: a novel mechanism of gene regulation. J Biol Chem. 2005;280:28451–28462. doi: 10.1074/jbc.M504400200. [DOI] [PubMed] [Google Scholar]
- Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, Grose RP. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6:467–481. doi: 10.1002/emmm.201302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197:801–817. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–r776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH. microRNAs in early vertebrate development. Cell Cycle. 2009;8:3513–3520. doi: 10.4161/cc.8.21.9847. [DOI] [PubMed] [Google Scholar]
- Liu H, Sun Q, Wan C, Li L, Zhang L, Chen Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229:1494–1502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA, McLean DL, Park H, Comhair SA, Greif DM. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med. 2013;19:74–82. doi: 10.1038/nm.3040. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Naito Y, Takagi T, Mizushima K, Hayashi N, Harusato A, Hirata I, Omatsu T, Handa O, Ishikawa T. Carbon monoxide enhance colonic epithelial restitution via FGF15 derived from colonic myofibroblasts. Biochem Biophys Res Commun. 2010;391:1122–1126. doi: 10.1016/j.bbrc.2009.12.035. et al. [DOI] [PubMed] [Google Scholar]
- Fu T, Choi SE, Kim DH, Seok S, Suino-Powell KM, Xu HE, Kemper JK. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor beta-Klotho. Proc Natl Acad Sci USA. 2012;109:16137–16142. doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T, Seok S, Choi S, Huang Z, Suino-Powell K, Xu HE, Kemper B, Kemper JK. MiR-34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte FGF21 signaling and SIRT1 function. Mol Cell Biol. 2014;34:4130–4142. doi: 10.1128/MCB.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ma R, Tan W, Zhang L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. 2014;46:e112. doi: 10.1038/emm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao H, Xin Y, Fan L. MicroRNA-198 inhibits proliferation and induces apoptosis of lung cancer cells via targeting FGFR1. J Cell Biochem. 2014;115:987–995. doi: 10.1002/jcb.24742. [DOI] [PubMed] [Google Scholar]
- Liu F, You X, Wang Y, Liu Q, Liu Y, Zhang S, Chen L, Zhang X, Ye L. The oncoprotein HBXIP enhances angiogenesis and growth of breast cancer through modulating FGF8 and VEGF. Carcinogenesis. 2014;35:1144–1153. doi: 10.1093/carcin/bgu021. [DOI] [PubMed] [Google Scholar]
- Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu YX, Liu JF, He ZM. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8:e72662. doi: 10.1371/journal.pone.0072662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and Fibroblast Growth Factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shen Y, Xue L, Fan H. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8:e73604. doi: 10.1371/journal.pone.0073604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lee SY, Kim YJ, Park JY, Kwon SJ, Na MJ, Lee EJ, Jeon HS, Son JW. microRNA-99b acts as a tumor suppressor in non-small cell lung cancer by directly targeting Fibroblast Growth Factor receptor 3. Exp Ther Med. 2012;3:149–153. doi: 10.3892/etm.2011.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgino K, Soejima K, Yasuda H, Hayashi Y, Hamamoto J, Naoki K, Arai D, Ishioka K, Sato T, Terai H. Expression of Fibroblast Growth Factor 9 is associated with poor prognosis in patients with resected non-small cell lung cancer. Lung Cancer. 2014;83:90–96. doi: 10.1016/j.lungcan.2013.10.016. et al. [DOI] [PubMed] [Google Scholar]
- Yin Y, Betsuyaku T, Garbow JR, Miao J, Govindan R, Ornitz DM. Rapid induction of lung adenocarcinoma by Fibroblast Growth Factor 9 signaling through FGF receptor 3. Cancer Res. 2013;73:5730–5741. doi: 10.1158/0008-5472.CAN-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Campbell WJ, Miller KA, Anderson TM, Shull JD, Rizzino A. Expression of Fibroblast Growth Factor receptors by embryonal carcinoma cells and early mouse embryos. In Vitro Cell Dev Biol. 1992;>28a:61–66. doi: 10.1007/BF02631080. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Developmental expression of two murine Fibroblast Growth Factor receptors, flg and bek. Development. 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of Fibroblast Growth Factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Lu P, Yu Y, Perdue Y, Werb Z. The apical ectodermal ridge is a timer for generating distal limb progenitors. Development. 2008;135:1395–1405. doi: 10.1242/dev.018945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Yin Y. Signaling networks regulating development of the lower respiratory tract. Cold Spring Harb Perspect Biol. 2012;4:1–19. doi: 10.1101/cshperspect.a008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazet JD, Zeller R. Vertebrate limb development: moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb Perspect Biol. 2009;1:a001339. doi: 10.1101/cshperspect.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Gros J, Tabin CJ. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science. 2014;343:1253–1256. doi: 10.1126/science.1248228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danopoulos S, Parsa S, Al Alam D, Tabatabai R, Baptista S, Tiozzo C, Carraro G, Wheeler M, Barreto G, Braun T. Transient Inhibition of FGFR2b-ligands signaling leads to irreversible loss of cellular beta-catenin organization and signaling in AER during mouse limb development. PLoS One. 2013;8:e76248. doi: 10.1371/journal.pone.0076248. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Esteban CR, Matsui T, Rodriguez-Leon J, Kato S, Izpisua Belmonte JC. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development. 2004;131:4763–4774. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- Haro E, Delgado I, Junco M, Yamada Y, Mansouri A, Oberg KC, Ros MA. Sp6 and Sp8 transcription factors control AER formation and dorsal-ventral patterning in limb development. PLoS Genet. 2014;10:e1004468. doi: 10.1371/journal.pgen.1004468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Yamamoto-Shiraishi YI, Higuchi H, Yamamoto S, Hirano M, Kuroiwa A. Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge-derived Fibroblast Growth Factor signal. Dev Biol. 2014;394:181–190. doi: 10.1016/j.ydbio.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Sakiyama J, Yamagishi A, Kuroiwa A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development. 2003;130:1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Bellusci S. Walking along the Fibroblast Growth Factor 10 route: a key pathway to understand the control and regulation of epithelial and mesenchymal cell-lineage formation during lung development and repair after injury. Scientifica (Cairo) 2014;2014:538379. doi: 10.1155/2014/538379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK. Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol. 2007;307:237–247. doi: 10.1016/j.ydbio.2007.04.033. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Nakano H, Singh G, Katyal S. Expression of Spred and Sprouty in developing rat lung. Gene Expr Patterns. 2002;2:347–353. doi: 10.1016/s1567-133x(02)00053-4. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lin Y, Itaranta P, Yagi A, Vainio S. Expression of Sprouty genes 1, 2 and 4 during mouse organogenesis. Mech Dev. 2001;109:367–370. doi: 10.1016/s0925-4773(01)00526-3. [DOI] [PubMed] [Google Scholar]
- Tefft JD, Lee M, Smith S, Leinwand M, Zhao J, Bringas P, Jr, Crowe DL, Warburton D. Conserved function of mSpry-2, a murine homolog of Drosophila sprouty, which negatively modulates respiratory organogenesis. Curr Biol. 1999;9:219–222. doi: 10.1016/s0960-9822(99)80094-3. [DOI] [PubMed] [Google Scholar]
- Tang N, Marshall WF, McMahon M, Metzger RJ, Martin GR. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–345. doi: 10.1126/science.1204831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Ayada T, Ichiyama K, Kohno R, Yonemitsu Y, Minami Y, Kikuchi A, Maehara Y, Yoshimura A. Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and regulation of FGF signaling. Biochem Biophys Res Commun. 2007;352:896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, Moiseenko A, Chao CM, Minoo P, Seeger W. Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development. 2014;141:296–306. doi: 10.1242/dev.099747. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse Fibroblast Growth Factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D, Wang K, Warburton D, Bellusci S. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassiter RN, Stark MR, Zhao T, Zhou CJ. Signaling mechanisms controlling cranial placode neurogenesis and delamination. Dev Biol. 2014;389:39–49. doi: 10.1016/j.ydbio.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulding K, Padanad MS, Dong J, Riley BB. Mesodermal Fgf10b cooperates with other Fibroblast Growth Factors during induction of otic and epibranchial placodes in zebrafish. Dev Dyn. 2014;243:1275–1285. doi: 10.1002/dvdy.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wright KD, Mahoney Rogers AA, Barrett MM, Shim K. Compensatory regulation of the size of the inner ear in response to excess induction of otic progenitors by Fibroblast Growth Factor signaling. Dev Dyn. 2014;243:1317–1327. doi: 10.1002/dvdy.24148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney Rogers AA, Zhang J, Shim K. Sprouty1 and Sprouty2 limit both the size of the otic placode and hindbrain Wnt8a by antagonizing FGF signaling. Dev Biol. 2011;353:94–104. doi: 10.1016/j.ydbio.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek H, Gutin G, Hebert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136:2457–2465. doi: 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Picker A, Brand M. Global and local mechanisms of forebrain and midbrain patterning. Curr Opin Neurobiol. 2006;16:5–12. doi: 10.1016/j.conb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Liu A, Li JY, Bromleigh C, Lao Z, Niswander LA, Joyner AL. FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development. 2003;130:6175–6185. doi: 10.1242/dev.00845. [DOI] [PubMed] [Google Scholar]
- Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- Partanen J. FGF signalling pathways in development of the midbrain and anterior hindbrain. J Neurochem. 2007;101:1185–1193. doi: 10.1111/j.1471-4159.2007.04463.x. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- Saarimaki-Vire J, Peltopuro P, Lahti L, Naserke T, Blak AA, Vogt Weisenhorn DM, Yu K, Ornitz DM, Wurst W, Partanen J. Fibroblast Growth Factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci. 2007;27:8581–8592. doi: 10.1523/JNEUROSCI.0192-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/s0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Zimmer C. From cradle to grave: the multiple roles of Fibroblast Growth Factors in neural development. Neuron. 2011;71:574–588. doi: 10.1016/j.neuron.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Smith KM, Ohkubo Y, Maragnoli ME, Rasin MR, Schwartz ML, Sestan N, Vaccarino FM. Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat Neurosci. 2006;9:787–797. doi: 10.1038/nn1705. [DOI] [PubMed] [Google Scholar]
- Umemori H, Linhoff MW, Ornitz DM, Sanes JR. FGF22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cell. 2004;118:257–270. doi: 10.1016/j.cell.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Flaumenhaft R, Moscatelli D, Rifkin DB. Heparin and heparan sulfate increase the radius of diffusion and action of basic Fibroblast Growth Factor. J Cell Biol. 1990;111:1651–1659. doi: 10.1083/jcb.111.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y, Futatsugi N, Mizutani-Koseki Y. FGF9 monomer/dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat Genet. 2009;41:289–298. doi: 10.1038/ng.316. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Gu MM, Huang L, Liu XS, Zhang HX, Ding XY, Xu JQ, Cui B, Wang L, Lu SY. Multiple synostoses syndrome is due to a missense mutation in exon 2 of FGF9 gene. Am J Hum Genet. 2009;85:53–63. doi: 10.1016/j.ajhg.2009.06.007. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Pan Y, Carbe C, Powers A, Grobe K, Zhang X. Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development. 2012;139:2730–2739. doi: 10.1242/dev.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann K, Faulhaber J, Campean V, Balajew V, Dono R, Mall G, Ehmke H. Impaired myocardial capillarogenesis and increased adaptive capillary growth in FGF2-deficient mice. Lab Invest. 2006;86:45–53. doi: 10.1038/labinvest.3700359. [DOI] [PubMed] [Google Scholar]
- Virag JA, Rolle ML, Reece J, Hardouin S, Feigl EO, Murry CE. Fibroblast Growth Factor-2 regulates myocardial infarct repair: effects on cell proliferation, scar contraction, and ventricular function. Am J Pathol. 2007;171:1431–1440. doi: 10.2353/ajpath.2007.070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusayr E, Doetschman T. Cardiac development and physiology are modulated by FGF2 in an isoform- and sex-specific manner. Physiol Rep. 2013;1:e00087. doi: 10.1002/phy2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusayr E, Sadideen DT, Doetschman T. FGF2 modulates cardiac remodeling in an isoform- and sex-specific manner. Physiol Rep. 2013;1:e00088. doi: 10.1002/phy2.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House SL, House BE, Glascock B, Kimball T, Nusayr E, Schultz JE, Doetschman T. Fibroblast Growth Factor 2 mediates isoproterenol-induced cardiac hypertrophy through activation of the extracellular regulated kinase. Mol Cell Pharmacol. 2010;2:143–154. doi: 10.4255/mcpharmacol.10.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dono R, Faulhaber J, Galli A, Zuniga A, Volk T, Texido G, Zeller R, Ehmke H. FGF2 signaling is required for the development of neuronal circuits regulating blood pressure. Circ Res. 2002;90:E5–E10. [PubMed] [Google Scholar]
- Grose R, Werner S. Wound healing studies in transgenic and knockout mice: a review. Methods Mol Med. 2003;78:191–216. doi: 10.1385/1-59259-332-1:191. [DOI] [PubMed] [Google Scholar]
- Tekin M, Hismi BO, Fitoz S, Ozdag H, Cengiz FB, Sirmaci A, Aslan I, Inceoglu B, Yuksel-Konuk EB, Yilmaz ST. Homozygous mutations in Fibroblast Growth Factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am J Hum Genet. 2007;80:338–344. doi: 10.1086/510920. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue H, Konishi M, Ogawa W, Asaki T, Mori T, Yamasaki M, Takata M, Ueno H, Kato S, Kasuga M. Requirement of Fibroblast Growth Factor 10 in development of white adipose tissue. Genes Dev. 2002;16:908–912. doi: 10.1101/gad.983202. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Ashigaki S, Takamatsu M, Suzuki-Migishima R, Ohbayashi N, Itoh N, Takada S, Tanabe Y. Laminar patterning in the developing neocortex by temporally coordinated Fibroblast Growth Factor signaling. J Neurosci. 2004;24:8711–8719. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, Ornitz DM. Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development. 2006;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TJ, Ladher R, McWhirter J, Murre C, Schoenwolf GC, Mansour SL. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–275. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HC, Carotti S, Vespasiani-Gentilucci U, Morini S, Vicente E, Concepcion AR. Identification of Fibroblast Growth Factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. et al. [DOI] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic Fibroblast Growth Factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V. Endocrine regulation of the fasting response by PPARalpha-mediated induction of Fibroblast Growth Factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. et al. [DOI] [PubMed] [Google Scholar]
- Lundasen T, Hunt MC, Nilsson LM, Sanyal S, Angelin B, Alexson SE, Rudling M. PPARalpha is a key regulator of hepatic FGF21. Biochem Biophys Res Commun. 2007;360:437–440. doi: 10.1016/j.bbrc.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Badman MK, Koester A, Flier JS, Kharitonenkov A, Maratos-Flier E. Fibroblast Growth Factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology. 2009;150:4931–4940. doi: 10.1210/en.2009-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Tian H, Lam KS, Lin S, Hoo RC, Konishi M, Itoh N, Wang Y, Bornstein SR, Xu A. Adiponectin mediates the metabolic effects of FGF21 on glucose homeostasis and insulin sensitivity in mice. Cell Metab. 2013;17:779–789. doi: 10.1016/j.cmet.2013.04.005. et al. [DOI] [PubMed] [Google Scholar]
- Holland WL, Adams AC, Brozinick JT, Bui HH, Miyauchi Y, Kusminski CM, Bauer SM, Wade M, Singhal E, Cheng CC. An FGF21-adiponectin-ceramide axis controls energy expenditure and insulin action in mice. Cell Metab. 2013;17:790–797. doi: 10.1016/j.cmet.2013.03.019. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fukumoto S. FGF23 as a novel therapeutic target. Adv Exp Med Biol. 2012;728:158–170. doi: 10.1007/978-1-4614-0887-1_10. [DOI] [PubMed] [Google Scholar]
- Tang WJ, Wang LF, Xu XY, Zhou Y, Jin WF, Wang HF, Gao J. Autocrine/paracrine action of vitamin D on FGF23 expression in cultured rat osteoblasts. Calcif Tissue Int. 2010;86:404–410. doi: 10.1007/s00223-010-9355-2. [DOI] [PubMed] [Google Scholar]
- Faul C, Amaral AP, Oskouei B, Hu MC, Sloan A, Isakova T, Gutierrez OM, Aguillon-Prada R, Lincoln J, Hare JM. FGF23 induces left ventricular hypertrophy. J Clin Invest. 2011;121:4393–4408. doi: 10.1172/JCI46122. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchberry CD, Green TM, Tchikrizov V, Mannix JE, Mao TF, Carney BW, Girgis M, Vincent RJ, Wetmore LA, Dawn B. FGF23 is a novel regulator of intracellular calcium and cardiac contractility in addition to cardiac hypertrophy. Am J Physiol Endocrinol Metab. 2013;304:E863–E873. doi: 10.1152/ajpendo.00596.2012. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Dib-Hajj SD, Waxman SG. Fibroblast Growth Factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNav1.9a (NaN) J Biol Chem. 2001;276:18925–18933. doi: 10.1074/jbc.M101606200. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Dib-Hajj SD, Renganathan M, Cummins TR, Waxman SG. Modulation of the cardiac sodium channel Na(v)1.5 by Fibroblast Growth Factor homologous factor 1B. J Biol Chem. 2003;278:1029–1036. doi: 10.1074/jbc.M207074200. [DOI] [PubMed] [Google Scholar]
- Laezza F, Lampert A, Kozel MA, Gerber BR, Rush AM, Nerbonne JM, Waxman SG, Dib-Hajj SD, Ornitz DM. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol Cell Neurosci. 2009;42:90–101. doi: 10.1016/j.mcn.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavkunov A, Panova N, Prasai A, Veselenak R, Bourne N, Stoilova-McPhie S, Laezza F. Bioluminescence methodology for the detection of protein-protein interactions within the voltage-gated sodium channel macromolecular complex. Assay Drug Dev Technol. 2012;10:148–160. doi: 10.1089/adt.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey JA, Wei EQ, Pitt GS. Fibroblast Growth Factor homologous factors modulate cardiac calcium channels. Circ Res. 2013;113:381–388. doi: 10.1161/CIRCRESAHA.113.301215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover K, Solinas S, D'Angelo E, Goldfarb M. Long-term inactivation particle for voltage-gated sodium channels. J Physiol. 2010;588:3695–3711. doi: 10.1113/jphysiol.2010.192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch MK, Carrasquillo Y, Ransdell JL, Kanakamedala A, Ornitz DM, Nerbonne JM. Intracellular FGF14 (iFGF14) is required for spontaneous and evoked firing in cerebellar Purkinje neurons and for motor coordination and balance. J Neurosci doi: 10.1523/JNEUROSCI.2663-14.2015. . In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavkunov AS, Wildburger NC, Nenov MN, James TF, Buzhdygan TP, Panova-Elektronova NI, Green TA, Veselenak RL, Bourne N, Laezza F. The Fibroblast Growth Factor 14.voltage-gated sodium channel complex is a new target of glycogen synthase kinase 3 (GSK3) J Biol Chem. 2013;288:19370–19385. doi: 10.1074/jbc.M112.445924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildburger NC, Laezza F. Control of neuronal ion channel function by glycogen synthase kinase-3: new prospective for an old kinase. Front Mol Neurosci. 2012;5:80. doi: 10.3389/fnmol.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Shavkunov A, Panova N, Stoilova-McPhie S, Laezza F. Modulation of the FGF14:FGF14 homodimer interaction through short peptide fragments. CNS Neurol Disord Drug Targets. 2014;13:1559–1570. doi: 10.2174/1871527313666141126103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Swieten JC, Brusse E, De Graaf BM, Krieger E, Van De Graaf R, De Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA. A mutation in the Fibroblast Growth Factor 14 gene is associated with autosomal dominant cerebellar ataxia. Am J Hum Genet. 2003;72:191–199. doi: 10.1086/345488. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusse E, de Koning I, Maat-Kievit A, Oostra BA, Heutink P, van Swieten JC. Spinocerebellar ataxia associated with a mutation in the Fibroblast Growth Factor 14 gene (SCA27): a new phenotype. Mov Disord. 2006;21:396–401. doi: 10.1002/mds.20708. [DOI] [PubMed] [Google Scholar]
- Yang J, Meyer M, Muller AK, Bohm F, Grose R, Dauwalder T, Verrey F, Kopf M, Partanen J, Bloch W. Fibroblast Growth Factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J Cell Biol. 2010;188:935–952. doi: 10.1083/jcb.200910126. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of corti. Dev Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- Moldrich RX, Mezzera C, Holmes WM, Goda S, Brookfield SJ, Rankin AJ, Barr E, Kurniawan N, Dewar D, Richards LJ. Fgfr3 regulates development of the caudal telencephalon. Dev Dyn. 2011;240:1586–1599. doi: 10.1002/dvdy.22636. et al. [DOI] [PubMed] [Google Scholar]
- Vidrich A, Buzan JM, Brodrick B, Ilo C, Bradley L, Fendig KS, Sturgill T, Cohn SM. Fibroblast Growth Factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. Am J Physiol Gastrointest Liver Physiol. 2009;297:G168–G178. doi: 10.1152/ajpgi.90589.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang C, Luo Y, Jin C, Wang F, McKeehan WL. FGFR4 prevents hyperlipidemia and insulin resistance but underlies high-fat diet induced fatty liver. Diabetes. 2007;56:2501–2510. doi: 10.2337/db07-0648. [DOI] [PubMed] [Google Scholar]
- Huang X, Yang C, Jin C, Luo Y, Wang F, McKeehan WL. Resident hepatocyte Fibroblast Growth Factor receptor 4 limits hepatocarcinogenesis. Mol Carcinog. 2009;48:553–562. doi: 10.1002/mc.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriarte I, Latasa MU, Carotti S, Fernandez-Barrena MG, Garcia-Irigoyen O, Elizalde M, Urtasun R, Vespasiani-Gentilucci U, Morini S, de Mingo A. Ileal FGF15 contributes to fibrosis-associated hepatocellular carcinoma development. Int J Cancer. 2014 doi: 10.1002/ijc.29287. et al.. In press. doi: 10.1002/ijc.29287. [DOI] [PubMed] [Google Scholar]
- Tekin M, Ozturkmen Akay H, Fitoz S, Birnbaum S, Cengiz FB, Sennaroglu L, Incesulu A, Yuksel Konuk EB, Hasanefendioglu Bayrak A, Senturk S. Homozygous FGF3 mutations result in congenital deafness with inner ear agenesis, microtia, and microdontia. Clin Genet. 2008;73:554–565. doi: 10.1111/j.1399-0004.2008.01004.x. et al. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans CY, Moosajee M, Hodges MD, Mackay DS, Game L, Vargesson N, Bloch-Zupan A, Ruschendorf F, Santos-Pinto L, Wackens G. SNP genome scanning localizes oto-dental syndrome to chromosome 11q13 and microdeletions at this locus implicate FGF3 in dental and inner-ear disease and FADD in ocular coloboma. Hum Mol Genet. 2007;16:2482–2493. doi: 10.1093/hmg/ddm204. et al. [DOI] [PubMed] [Google Scholar]
- Alsmadi O, Meyer BF, Alkuraya F, Wakil S, Alkayal F, Al-Saud H, Ramzan K, Al-Sayed M. Syndromic congenital sensorineural deafness, microtia and microdontia resulting from a novel homoallelic mutation in Fibroblast Growth Factor 3 (FGF3) Eur J Hum Genet. 2009;17:14–21. doi: 10.1038/ejhg.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensi A, Ceruti S, Trevisi P, Gualandi F, Busi M, Donati I, Neri M, Ferlini A, Martini A. LAMM syndrome with middle ear dysplasia associated with compound heterozygosity for FGF3 mutations. Am J Med Genet A. 2011;>155a:1096–1101. doi: 10.1002/ajmg.a.33962. [DOI] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C, Rufenacht S, Wichert B, Leeb T. Mutations within the FGF5 gene are associated with hair length in cats. Anim Genet. 2007;38:218–221. doi: 10.1111/j.1365-2052.2007.01590.x. [DOI] [PubMed] [Google Scholar]
- Kehler JS, David VA, Schaffer AA, Bajema K, Eizirik E, Ryugo DK, Hannah SS, O'Brien SJ, Menotti-Raymond M. Four independent mutations in the feline Fibroblast Growth Factor 5 gene determine the long-haired phenotype in domestic cats. J Hered. 2007;98:555–566. doi: 10.1093/jhered/esm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks C, Momke S, Philipp U, Distl O. Allelic heterogeneity of FGF5 mutations causes the long-hair phenotype in dogs. Anim Genet. 2013;44:425–431. doi: 10.1111/age.12010. [DOI] [PubMed] [Google Scholar]
- Brehm JM, Hagiwara K, Tesfaigzi Y, Bruse S, Mariani TJ, Bhattacharya S, Boutaoui N, Ziniti JP, Soto-Quiros ME, Avila L. Identification of FGF7 as a novel susceptibility locus for chronic obstructive pulmonary disease. Thorax. 2011;66:1085–1090. doi: 10.1136/thoraxjnl-2011-200017. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trarbach EB, Abreu AP, Silveira LF, Garmes HM, Baptista MT, Teles MG, Costa EM, Mohammadi M, Pitteloud N, Mendonca BB. Nonsense mutations in FGF8 gene causing different degrees of human gonadotropin-releasing deficiency. J Clin Endocrinol Metab. 2010;95:3491–3496. doi: 10.1210/jc.2010-0176. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arauz RF, Solomon BD, Pineda-Alvarez DE, Gropman AL, Parsons JA, Roessler E, Muenke M. A hypomorphic allele in the FGF8 gene contributes to holoprosencephaly and is allelic to gonadotropin-releasing hormone deficiency in humans. Mol Syndromol. 2010;1:59–66. doi: 10.1159/000302285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MJ, Gaston-Massuet C, Tziaferi V, Gregory LC, Alatzoglou KS, Signore M, Puelles E, Gerrelli D, Farooqi IS, Raza J. Novel FGF8 mutations associated with recessive holoprosencephaly, craniofacial defects, and hypothalamo-pituitary dysfunction. J Clin Endocrinol Metab. 2011;96:E1709–E1718. doi: 10.1210/jc.2011-0454. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Socin H, Rubio Almanza M, Tome Fernandez-Ladreda M, Debray FG, Bours V, Beckers A. Reproduction, smell, and neurodevelopmental disorders: genetic defects in different hypogonadotropic hypogonadal syndromes. Front Endocrinol (Lausanne) 2014;5:109. doi: 10.3389/fendo.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CL, Lu CW, Cheng YS, Lin CY, Sun HS, Lin YM. Association of aberrant expression of sex-determining gene Fibroblast Growth Factor 9 with Sertoli cell-only syndrome. Fertil Steril. 2013;100:1547–1554.e1541-1544. doi: 10.1016/j.fertnstert.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Entesarian M, Dahlqvist J, Shashi V, Stanley CS, Falahat B, Reardon W, Dahl N. FGF10 missense mutations in aplasia of lacrimal and salivary glands (ALSG) Eur J Hum Genet. 2007;15:379–382. doi: 10.1038/sj.ejhg.5201762. [DOI] [PubMed] [Google Scholar]
- Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nurnberg G, Lew ED, Dobbie A, Eswarakumar VP, Uzumcu A, Ulubil-Emeroglu M. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–417. doi: 10.1038/ng1757. et al. [DOI] [PubMed] [Google Scholar]
- Hsi E, Chen KC, Chang WS, Yu ML, Liang CL, Juo SH. A functional polymorphism at the FGF10 gene is associated with extreme myopia. Invest Ophthalmol Vis Sci. 2013;54:3265–3271. doi: 10.1167/iovs.13-11814. [DOI] [PubMed] [Google Scholar]
- Milunsky JM, Zhao G, Maher TA, Colby R, Everman DB. LADD syndrome is caused by FGF10 mutations. Clin Genet. 2006;69:349–354. doi: 10.1111/j.1399-0004.2006.00597.x. [DOI] [PubMed] [Google Scholar]
- Entesarian M, Matsson H, Klar J, Bergendal B, Olson L, Arakaki R, Hayashi Y, Ohuchi H, Falahat B, Bolstad AI. Mutations in the gene encoding Fibroblast Growth Factor 10 are associated with aplasia of lacrimal and salivary glands. Nat Genet. 2005;37:125–127. doi: 10.1038/ng1507. et al. [DOI] [PubMed] [Google Scholar]
- Hennessey JA, Marcou CA, Wang C, Wei EQ, Wang C, Tester DJ, Torchio M, Dagradi F, Crotti L, Schwartz PJ. FGF12 is a candidate Brugada syndrome locus. Heart Rhythm. 2013;10:1886–1894. doi: 10.1016/j.hrthm.2013.09.064. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Baker E, Donnelly A, Ming JE, McDonald-McGinn DM, Spinner NB, Zackai EH, Sutherland GR, Mulley JC. Fibroblast Growth Factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum Genet. 1999;104:56–63. doi: 10.1007/s004390050910. [DOI] [PubMed] [Google Scholar]
- DeStefano GM, Fantauzzo KA, Petukhova L, Kurban M, Tadin-Strapps M, Levy B, Warburton D, Cirulli ET, Han Y, Sun X. Position effect on FGF13 associated with X-linked congenital generalized hypertrichosis. Proc Natl Acad Sci USA. 2013;110:7790–7795. doi: 10.1073/pnas.1216412110. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalski A, Atici J, Kreuz FR, Hellenbroich Y, Schwinger E, Zuhlke C. Mutation analysis in the Fibroblast Growth Factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. Eur J Hum Genet. 2005;13:118–120. doi: 10.1038/sj.ejhg.5201286. [DOI] [PubMed] [Google Scholar]
- Coebergh JA. van de Putte DE, Snoeck IN, Ruivenkamp C, van Haeringen A, Smit LM. A new variable phenotype in spinocerebellar ataxia 27 (SCA 27) caused by a deletion in the FGF14 gene. Eur J Paediatr Neurol. 2014;18:413–415. doi: 10.1016/j.ejpn.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Jamsheer A, Zemojtel T, Kolanczyk M, Stricker S, Hecht J, Krawitz P, Doelken SC, Glazar R, Socha M, Mundlos S. Whole exome sequencing identifies FGF16 nonsense mutations as the cause of X-linked recessive metacarpal 4/5 fusion. J Med Genet. 2013;50:579–584. doi: 10.1136/jmedgenet-2013-101659. [DOI] [PubMed] [Google Scholar]
- Laurell T, Nilsson D, Hofmeister W, Lindstrand A, Ahituv N, Vandermeer J, Amilon A, Anneren G, Arner M, Pettersson M. Identification of three novel FGF16 mutations in X-linked recessive fusion of the fourth and fifth metacarpals and possible correlation with heart disease. Mol Genet Genomic Med. 2014;2:402–411. doi: 10.1002/mgg3.81. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraoui H, Dwyer AA, Sykiotis GP, Plummer L, Chung W, Feng B, Beenken A, Clarke J, Pers TH, Dworzynski P. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92:725–743. doi: 10.1016/j.ajhg.2013.04.008. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt JM, Noureddine MA, Kittappa R, Hauser MA, Scott WK, McKay R, Zhang F, Stajich JM, Fujiwara K, Scott BL. Fibroblast Growth Factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am J Hum Genet. 2004;74:1121–1127. doi: 10.1086/421052. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ohta H. Roles of FGF20 in dopaminergic neurons and Parkinson's disease. Front Mol Neurosci. 2013;6:15. doi: 10.3389/fnmol.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre H, Mattay VS, Sambataro F, Verchinski B, Straub RE, Callicott JH, Kittappa R, Hyde TM, Lipska BK, Kleinman JE. Genetic variation in FGF20 modulates hippocampal biology. J Neurosci. 2010;30:5992–5997. doi: 10.1523/JNEUROSCI.5773-09.2010. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu AY, Workalemahu T, Paynter NP, Rose LM, Giulianini F, Tanaka T, Ngwa JS, Qi Q, Curhan GC, Rimm EB. Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet. 2013;22:1895–1902. doi: 10.1093/hmg/ddt032. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, Houston DK, Kanoni S, Lemaitre RN, Luan J. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97:1395–1402. doi: 10.3945/ajcn.112.052183. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zeng L, Wang YJ, An ZM, Ying BW. Associations of Fibroblast Growth Factor 21 gene 3' untranslated region single-nucleotide polymorphisms with metabolic syndrome, obesity, and diabetes in a Han Chinese population. DNA Cell Biol. 2012;31:547–552. doi: 10.1089/dna.2011.1302. [DOI] [PubMed] [Google Scholar]
- Benet-Pages A, Orlik P, Strom TM, Lorenz-Depiereux B. An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum Mol Genet. 2005;14:385–390. doi: 10.1093/hmg/ddi034. [DOI] [PubMed] [Google Scholar]
- Masi L, Gozzini A, Franchi A, Campanacci D, Amedei A, Falchetti A, Franceschelli F, Marcucci G, Tanini A, Capanna R. A novel recessive mutation of Fibroblast Growth Factor-23 in tumoral calcinosis. J Bone Joint Surg Am. 2009;91:1190–1198. doi: 10.2106/JBJS.H.00783. et al. [DOI] [PubMed] [Google Scholar]
- Masi L, Franceschelli F, Leoncini G, Gozzini A, Rigante D, La Torre F, Matucci-Cerinic M, Brandi ML, Falcini F. Can Fibroblast Growth Factor (FGF)-23 circulating levels suggest coronary artery abnormalities in children with Kawasaki disease? Clin Exp Rheumatol. 2013;31:149–153. [PubMed] [Google Scholar]
- Falcini F, Rigante D, Masi L, Covino M, Franceschelli F, Leoncini G, Tarantino G, Matucci Cerinic M, Brandi ML. Fibroblast Growth Factor 23 (FGF23) gene polymorphism in children with Kawasaki syndrome (KS) and susceptibility to cardiac abnormalities. Ital J Pediatr. 2013;39:69. doi: 10.1186/1824-7288-39-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefetz I, Heller R, Galli-Tsinopoulou A, Richard G, Wollnik B, Indelman M, Koerber F, Topaz O, Bergman R, Sprecher E. A novel homozygous missense mutation in FGF23 causes familial tumoral calcinosis associated with disseminated visceral calcification. Hum Genet. 2005;118:261–266. doi: 10.1007/s00439-005-0026-8. et al. [DOI] [PubMed] [Google Scholar]
- Araya K, Fukumoto S, Backenroth R, Takeuchi Y, Nakayama K, Ito N, Yoshii N, Yamazaki Y, Yamashita T, Silver J. A novel mutation in Fibroblast Growth Factor (FGF)23 gene as a cause of tumoral calcinosis. J Clin Endocrinol Metab. 2005;90:5523–5527. doi: 10.1210/jc.2005-0301. et al. [DOI] [PubMed] [Google Scholar]
- Garringer HJ, Malekpour M, Esteghamat F, Mortazavi SM, Davis SI, Farrow EG, Yu X, Arking DE, Dietz HC, White KE. Molecular genetic and biochemical analyses of FGF23 mutations in familial tumoral calcinosis. Am J Physiol Endocrinol Metab. 2008;295:E929–E937. doi: 10.1152/ajpendo.90456.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S. A common mutation in the Fibroblast Growth Factor receptor 1 gene in Pfeiffer syndrome. Nat Genet. 1994;8:269–274. doi: 10.1038/ng1194-269. et al. [DOI] [PubMed] [Google Scholar]
- Roscioli T, Flanagan S, Kumar P, Masel J, Gattas M, Hyland VJ, Glass IA. Clinical findings in a patient with FGFR1 P252R mutation and comparison with the literature. Am J Med Genet. 2000;93:22–28. doi: 10.1002/1096-8628(20000703)93:1<22::aid-ajmg5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. et al. [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Acierno JS, Jr, Meysing AU, Dwyer AA, Hayes FJ, Crowley WF., Jr Reversible kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the Fibroblast Growth Factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- Simonis N, Migeotte I, Lambert N, Perazzolo C, de Silva DC, Dimitrov B, Heinrichs C, Janssens S, Kerr B, Mortier G. FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. J Med Genet. 2013;50:585–592. doi: 10.1136/jmedgenet-2013-101603. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koika V, Varnavas P, Valavani H, Sidis Y, Plummer L, Dwyer A, Quinton R, Kanaka-Gantenbein C, Pitteloud N, Sertedaki A. Comparative functional analysis of two Fibroblast Growth Factor receptor 1 (FGFR1) mutations affecting the same residue (R254W and R254Q) in isolated hypogonadotropic hypogonadism (IHH) Gene. 2012;516:146–151. doi: 10.1016/j.gene.2012.12.041. et al. [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YX, Xu X, Chen L, Li C, Brodie SG, Deng CX. A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum Mol Genet. 2000;9:2001–2008. doi: 10.1093/hmg/9.13.2001. [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Acierno JS, Jr, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA. Mutations in Fibroblast Growth Factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva C, Jacobson-Dickman E, Xu C, Manouvrier S, Dwyer AA, Sykiotis GP, Beenken A, Liu Y, Tommiska J, Hu Y. Congenital hypogonadotropic hypogonadism with split hand/foot malformation: a clinical entity with a high frequency of FGFR1 mutations. Genet Med. 2014 doi: 10.1038/gim.2014.166. et al.. In press. doi: 10.1038/gim.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Wang DD, Gu JQ, Zhang N, Shan ZY. A female patient with normosmic idiopathic hypogonadotropic hypogonadism carrying a novel mutation in FGFR1. Genet Mol Res. 2014;13:9472–9476. doi: 10.4238/2014.November.11.12. [DOI] [PubMed] [Google Scholar]
- Rutland P, Pulleyn LJ, Reardon W, Baraitser M, Hayward R, Jones B, Malcolm S, Winter RM, Oldridge M, Slaney SF. Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat Genet. 1995;9:173–176. doi: 10.1038/ng0295-173. et al. [DOI] [PubMed] [Google Scholar]
- Schell U, Hehr A, Feldman GJ, Robin NH, Zackai EH, de Die-Smulders C, Viskochil DH, Stewart JM, Wolff G, Ohashi H. Mutations in FGFR1 and FGFR2 cause familial and sporadic Pfeiffer syndrome. Hum Mol Genet. 1995;4:323–328. doi: 10.1093/hmg/4.3.323. et al. [DOI] [PubMed] [Google Scholar]
- Jabs EW, Li X, Scott AF, Meyers G, Chen W, Eccles M, Mao J, Charnas LR, Jackson CE, Jaye M. Jackson-Weiss and Crouzon syndromes are allelic with mutations in Fibroblast Growth Factor receptor 2. Nat Genet. 1994;8:275–279. doi: 10.1038/ng1194-275. [DOI] [PubMed] [Google Scholar]
- Park WJ, Meyers GA, Li X, Theda C, Day D, Orlow SJ, Jones MC, Jabs EW. Novel FGFR2 mutations in Crouzon and Jackson-Weiss syndromes show allelic heterogeneity and phenotypic variability. Hum Mol Genet. 1995;4:1229–1233. doi: 10.1093/hmg/4.7.1229. [DOI] [PubMed] [Google Scholar]
- Reardon W, Winter RM, Rutland P, Pulleyn LJ, Jones BM, Malcolm S. Mutations in the Fibroblast Growth Factor receptor 2 gene cause Crouzon syndrome. Nat Genet. 1994;8:98–103. doi: 10.1038/ng0994-98. [DOI] [PubMed] [Google Scholar]
- Oldridge M, Wilkie AO, Slaney SF, Poole MD, Pulleyn LJ, Rutland P, Hockley AD, Wake MJ, Goldin JH, Winter RM. Mutations in the third immunoglobulin domain of the Fibroblast Growth Factor receptor-2 gene in Crouzon syndrome. Hum Mol Genet. 1995;4:1077–1082. doi: 10.1093/hmg/4.6.1077. et al. [DOI] [PubMed] [Google Scholar]
- Steinberger D, Mulliken JB, Muller U. Crouzon syndrome: previously unrecognized deletion, duplication, and point mutation within FGFR2 gene. Hum Mutat. 1996;8:386–390. doi: 10.1002/(SICI)1098-1004(1996)8:4<386::AID-HUMU18>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Paznekas WA, Cunningham ML, Howard TD, Korf BR, Lipson MH, Grix AW, Feingold M, Goldberg R, Borochowitz Z, Aleck K. Genetic heterogeneity of Saethre-Chotzen-Syndrome, due to twist and Fgfr mutations. Am J Hum Genet. 1998;62:1370–1380. doi: 10.1086/301855. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, Johnson D, Wall SA, Jiang W, Theda C. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am J Hum Genet. 1999;64:446–461. doi: 10.1086/302245. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AOM, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, Hayward RD, David DJ, Pulleyn LJ, Rutland P. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9:165–172. doi: 10.1038/ng0295-165. et al. [DOI] [PubMed] [Google Scholar]
- Merrill AE, Sarukhanov A, Krejci P, Idoni B, Camacho N, Estrada KD, Lyons KM, Deixler H, Robinson H, Chitayat D. Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am J Hum Genet. 2012;90:550–557. doi: 10.1016/j.ajhg.2012.02.005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Wall SA, Mann S, Wilkie AO. A novel mutation, Ala315Ser, in FGFR2: a gene-environment interaction leading to craniosynostosis? Eur J Hum Genet. 2000;8:571–577. doi: 10.1038/sj.ejhg.5200499. [DOI] [PubMed] [Google Scholar]
- Park WJ, Theda C, Maestri NE, Meyers GA, Fryburg JS, Dufresne C, Cohen MM, Jr, Jabs EW. Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. Am J Hum Genet. 1995;57:321–328. [PMC free article] [PubMed] [Google Scholar]
- Foldynova-Trantirkova S, Wilcox WR, Krejci P. Sixteen years and counting: the current understanding of Fibroblast Growth Factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum Mutat. 2012;33:29–41. doi: 10.1002/humu.21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton WA, Hall JG, Hecht JT. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- Chen L, Li C, Qiao W, Xu X, Deng C. A Ser(365)–>Cys mutation of Fibroblast Growth Factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet. 2001;10:457–465. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- Winterpacht A, Hilbert K, Stelzer C, Schweikardt T, Decker H, Segerer H, Spranger J, Zabel B. A novel mutation in FGFR-3 disrupts a putative N-glycosylation site and results in hypochondroplasia. Physiol Genomics. 2000;2:9–12. doi: 10.1152/physiolgenomics.2000.2.1.9. [DOI] [PubMed] [Google Scholar]
- Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in Fibroblast Growth Factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie SG, Kitoh H, Lachman RS, Nolasco LM, Mekikian PB, Wilcox WR. Platyspondylic lethal skeletal dysplasia, San Diego type, is caused by FGFR3 mutations. Am J Med Genet. 1999;84:476–480. [PubMed] [Google Scholar]
- Bellus GA, Bamshad MJ, Przylepa KA, Dorst J, Lee RR, Hurko O, Jabs EW, Curry CJ, Wilcox WR, Lachman RS. Severe achondroplasia with developmental delay and acanthosis nigricans (SADDAN): phenotypic analysis of a new skeletal dysplasia caused by a Lys650Met mutation in Fibroblast Growth Factor receptor 3. Am J Med Genet. 1999;85:53–65. et al. [PubMed] [Google Scholar]
- Matsui Y, Yasui N, Kimura T, Tsumaki N, Kawabata H, Ochi T. Genotype phenotype correlation in achondroplasia and hypochondroplasia. J Bone Joint Surg Br. 1998;80:1052–1056. doi: 10.1302/0301-620x.80b6.9277. [DOI] [PubMed] [Google Scholar]
- Webster MK, D'Avis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of Fibroblast Growth Factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MK, Donoghue DJ. Constitutive activation of Fibroblast Growth Factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Shiang R, Thompson LM, Zhu Y, Wilkin DJ, Lachman RS, Wilcox WR, Rimoin DL, Cohn DH, Wasmuth JJ. Thanatophoric dysplasia (types I and II) caused by distinct mutations in Fibroblast Growth Factor receptor 3. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW. Fibroblast Growth Factor receptor 3 (FGFR3) transmembrane mutation in Crouzon syndrome with acanthosis nigricans. Nat Genet. 1995;11:462–464. doi: 10.1038/ng1295-462. [DOI] [PubMed] [Google Scholar]
- Bellus GA, McIntosh I, Smith EA, Aylesworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of Fibroblast Growth Factor receptor 3 causes hypochondroplasia. Nat Genet. 1995;10:357–359. doi: 10.1038/ng0795-357. [DOI] [PubMed] [Google Scholar]
- Shiang R, Thompson LM, Zhu Y-Z, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev Cell. 2002;3:439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- Muenke M, Gripp KW, McDonald-McGinn DM, Gaudenz K, Whitaker LA, Bartlett SP, Markowitz RI, Robin NH, Nwokoro N, Mulvihill JJ. A unique point mutation in the Fibroblast Growth Factor receptor 3 gene (FGFR3) defines a new craniosynostosis syndrome. Am J Hum Genet. 1997;60:555–564. et al. [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, McDonald-McGinn DM, Gaudenz K, Whitaker LA, Bartlett SP, Glat PM, Cassileth LB, Mayro R, Zackai EH, Muenke M. Identification of a genetic cause for isolated unilateral coronal synostosis: a unique mutation in the Fibroblast Growth Factor receptor 3. J Pediatr. 1998;132:714–716. doi: 10.1016/s0022-3476(98)70366-x. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, El Ghouzzi V, Le Merrer M, Munnich A, Bonaventure J, Renier D. Sex related expressivity of the phenotype in coronal craniosynostosis caused by the recurrent P250R FGFR3 mutation. J Med Genet. 1999;36:9–13. [PMC free article] [PubMed] [Google Scholar]
- Graham JM, Jr, Braddock SR, Mortier GR, Lachman R, Van Dop C, Jabs EW. Syndrome of coronal craniosynostosis with brachydactyly and carpal/tarsal coalition due to Pro250Arg mutation in FGFR3 gene. Am J Med Genet. 1998;77:322–329. doi: 10.1002/(sici)1096-8628(19980526)77:4<322::aid-ajmg14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Moloney DM, Wall SA, Ashworth GJ, Oldridge M, Glass IA, Francomano CA, Muenke M, Wilkie AO. Prevalence of Pro250Arg mutation of Fibroblast Growth Factor receptor 3 in coronal craniosynostosis. Lancet. 1997;349:1059–1062. doi: 10.1016/s0140-6736(96)09082-4. [DOI] [PubMed] [Google Scholar]
- Golla A, Lichmer P, von Gernet S, Winterpacht A, Fairley J, Murken J, Schuffenhauer S. Phenotypic expression of the Fibroblast Growth Factor receptor 3 (FGFR3) mutation P250R in a large craniosynostosis family. J Med Genet. 1997;34:683–684. doi: 10.1136/jmg.34.8.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler S, Friedrich M, Wagener H, Lorenz B, Preising MN. Heterozygous P250L mutation of Fibroblast Growth Factor receptor 3 in a case of isolated craniosynostosis. J Med Genet. 2002;39:764–766. doi: 10.1136/jmg.39.10.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Sun Q, Li C, Lu X, Qi H, Chen S, Yang J, Du X, Zhao L, He Q. Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum Mol Genet. 2010;19:1199–1210. doi: 10.1093/hmg/ddp590. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier S, Couloigner V, Messaddeq N, Elmaleh-Berges M, Munnich A, Romand R, Legeai-Mallet L. Activating Fgfr3 Y367C mutation causes hearing loss and inner ear defect in a mouse model of chondrodysplasia. Biochim Biophys Acta. 2009;1792:140–147. doi: 10.1016/j.bbadis.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Twigg SR, Freeland RM, Wall SA, Li C, Wilkie AO. Hearing loss in a mouse model of Muenke syndrome. Hum Mol Genet. 2009;18:43–50. doi: 10.1093/hmg/ddn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Li C, Urness LD. Genetic rescue of Muenke syndrome model hearing loss reveals prolonged FGF-dependent plasticity in cochlear supporting cell fates. Genes Dev. 2013;27:2320–2331. doi: 10.1101/gad.228957.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes D, Rutland P, Pulleyn LJ, Reardon W, Moss C, Ellis JP, Winter RM, Malcolm S. A recurrent mutation, ala391glu, in the transmembrane region of FGFR3 causes Crouzon syndrome and acanthosis nigricans. J Med Genet. 1996;33:744–748. doi: 10.1136/jmg.33.9.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin P, Tarnow P, Martinsson T, Stenman G. Germline mutation in the FGFR3 gene in a TWIST1-negative family with saethre-chotzen syndrome and breast cancer. Genes Chromosomes Cancer. 2009;48:285–288. doi: 10.1002/gcc.20637. [DOI] [PubMed] [Google Scholar]
- Toydemir RM, Brassington AE, Bayrak-Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, Longo N, Viskochil DH, Carey JC, Bamshad MJ. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am J Hum Genet. 2006;79:935–941. doi: 10.1086/508433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrythanasis P, Temtamy S, Aglan MS, Otaify GA, Hamamy H, Antonarakis SE. A novel homozygous mutation in FGFR3 causes tall stature, severe lateral tibial deviation, scoliosis, hearing impairment, camptodactyly and arachnodactyly. Hum Mutat. 2014;35:959–963. doi: 10.1002/humu.22597. [DOI] [PubMed] [Google Scholar]
- Iwata T, Chen L, Li C, Ovchinnikov DA, Behringer RR, Francomano CA, Deng CX. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum Mol Genet. 2000;9:1603–1613. doi: 10.1093/hmg/9.11.1603. [DOI] [PubMed] [Google Scholar]
- Saito A, Higuchi I, Nakagawa M, Saito M, Uchida Y, Inose M, Kasai T, Niiyama T, Fukunaga H, Arimura K. An overexpression of Fibroblast Growth Factor (FGF) and FGF receptor 4 in a severe clinical phenotype of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2000;23:490–497. doi: 10.1002/(sici)1097-4598(200004)23:4<490::aid-mus6>3.0.co;2-k. et al. [DOI] [PubMed] [Google Scholar]
- Rezvani M, Wilde J, Vitt P, Mailaparambil B, Grychtol R, Krueger M, Heinzmann A. Association of a FGFR-4 gene polymorphism with bronchopulmonary dysplasia and neonatal respiratory distress. Dis Markers. 2013;35:633–640. doi: 10.1155/2013/932356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Li WJ, Wan YY, Yu CD, Li WG. Fibroblast Growth Factor receptor 4 Gly388Arg polymorphism associated with severity of gallstone disease in a Chinese population. Genet Mol Res. 2012;11:548–555. doi: 10.4238/2012.March.8.3. [DOI] [PubMed] [Google Scholar]
- Rieckmann T, Zhuang L, Fluck CE, Trueb B. Characterization of the first FGFRL1 mutation identified in a craniosynostosis patient. Biochim Biophys Acta. 2009;1792:112–121. doi: 10.1016/j.bbadis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Engbers H, van der Smagt JJ, van‘t Slot R, Vermeesch JR, Hochstenbach R, Poot M. Wolf-Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet. 2009;17:129–132. doi: 10.1038/ejhg.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Green RP, Zhao G, Ornitz DM. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development. 2001;128:3867–3876. doi: 10.1242/dev.128.19.3867. [DOI] [PubMed] [Google Scholar]
- Jones B, Byers H, Watson JS, Newman WG. Identification of a novel familial FGF16 mutation in metacarpal 4–5 fusion. Clin Dysmorphol. 2014;23:95–97. doi: 10.1097/MCD.0000000000000043. [DOI] [PubMed] [Google Scholar]
- Razzaque MS. The FGF23-Klotho axis: endocrine regulation of phosphate homeostasis. Nat Rev Endocrinol. 2009;5:611–619. doi: 10.1038/nrendo.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Mizutani S, Muto T, Yoneya T, Hino R, Takeda S, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc Natl Acad Sci USA. 2001;98:6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D, Fannemel M, Baroy T, Roberto R, Tvedt B, Jaeger T, Bryn V, Stromme P, Frengen E. SCA27 caused by a chromosome translocation: further delineation of the phenotype. Neurogenetics. 2009;10:371–374. doi: 10.1007/s10048-009-0197-x. [DOI] [PubMed] [Google Scholar]
- Shimojima K, Okumura A, Natsume J, Aiba K, Kurahashi H, Kubota T, Yokochi K, Yamamoto T. Spinocerebellar ataxias type 27 derived from a disruption of the Fibroblast Growth Factor 14 gene with mimicking phenotype of paroxysmal non-kinesigenic dyskinesia. Brain Dev. 2012;34:230–233. doi: 10.1016/j.braindev.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Laezza F, Gerber BR, Lou JY, Kozel MA, Hartman H, Craig AM, Ornitz DM, Nerbonne JM. The FGF14(F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. J Neurosci. 2007;27:12033–12044. doi: 10.1523/JNEUROSCI.2282-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KE, Cabral JM, Davis SI, Fishburn T, Evans WE, Ichikawa S, Fields J, Yu X, Shaw NJ, McLellan NJ. Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am J Hum Genet. 2005;76:361–367. doi: 10.1086/427956. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, Linhardt RJ, Mohammadi M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum Mol Genet. 2004;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Ma HW, Bonaventure J, Munnich A, LeMerrer M. FGFR2 mutations in Pfeiffer syndrome. Nat Genet. 1995;9:108. doi: 10.1038/ng0295-108. [DOI] [PubMed] [Google Scholar]
- Meyers GA, Day D, Goldberg R, Daentl DL, Przylepa KA, Abrams LJ, Graham JM, Jr, Feingold M, Moeschler JB, Rawnsley E. FGFR2 exon IIIa and IIIc mutations in Crouzon, Jackson-Weiss, and Pfeiffer syndromes: evidence for missense changes, insertions, and a deletion due to alternative RNA splicing. Am J Hum Genet. 1996;58:491–498. et al. [PMC free article] [PubMed] [Google Scholar]
- Martinez-Abadias N, Motch SM, Pankratz TL, Wang Y, Aldridge K, Jabs EW, Richtsmeier JT. Tissue-specific responses to aberrant FGF signaling in complex head phenotypes. Dev Dyn. 2013;242:C1. doi: 10.1002/dvdy.23903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick AL, Bowdin SC, Klatt RE, Wilkie AO. A deletion of FGFR2 creating a chimeric IIIb/IIIc exon in a child with Apert syndrome. BMC Med Genet. 2011;12:122. doi: 10.1186/1471-2350-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Burns HD, Enriquez-Harris P, Wilkie AOM, Heath JK. Apert syndrome mutations in Fibroblast Growth Factor receptor 2 exhibit increased affinity for FGF ligand. Hum Mol Genet. 1998;7:1475–1483. doi: 10.1093/hmg/7.9.1475. [DOI] [PubMed] [Google Scholar]
- Yu K, Herr AB, Waksman G, Ornitz DM. Loss of Fibroblast Growth Factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci USA. 2000;97:14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Wilson S, De Moerlooze L, Dickson C. A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc Natl Acad Sci USA. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, Mohammadi M. Structural basis for Fibroblast Growth Factor receptor 2 activation in Apert syndrome. Proc Natl Acad Sci USA. 2001;5:5. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Ornitz DM. Uncoupling Fibroblast Growth Factor receptor 2 ligand binding specificity leads to Apert syndrome-like phenotypes. Proc Natl Acad Sci USA. 2001;98:3641–3643. doi: 10.1073/pnas.081082498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Duarte R, Pegrum J, Donjacour A, Lana-Elola E, Rice DP, Sharpe J, Dickson C. Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev Dyn. 2009;238:376–385. doi: 10.1002/dvdy.21648. [DOI] [PubMed] [Google Scholar]
- Holmes G, Basilico C. Mesodermal expression of Fgfr2(S252W) is necessary and sufficient to induce craniosynostosis in a mouse model of Apert syndrome. Dev Biol. 2012;368:283–293. doi: 10.1016/j.ydbio.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita J, Nakamura M, Kobayashi Y, Deng CX, Funato N, Moriyama K. Soluble form of FGFR2 with S252W partially prevents craniosynostosis of the apert mouse model. Dev Dyn. 2014;243:560–567. doi: 10.1002/dvdy.24099. [DOI] [PubMed] [Google Scholar]
- Neben CL, Idoni B, Salva JE, Tuzon CT, Rice JC, Krakow D, Merrill AE. Hum Mol Genet. 2014;23:5659–5671. doi: 10.1093/hmg/ddu282. . Bent bone dysplasia syndrome reveals nucleolar activity for FGFR2 in ribosomal DNA transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew ED, Bae JH, Rohmann E, Wollnik B, Schlessinger J. Structural basis for reduced FGFR2 activity in LADD syndrome: implications for FGFR autoinhibition and activation. Proc Natl Acad Sci USA. 2007;104:19802–19807. doi: 10.1073/pnas.0709905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams I, Rohmann E, Eswarakumar VP, Lew ED, Yuzawa S, Wollnik B, Schlessinger J, Lax I. Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the Fibroblast Growth Factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol Cell Biol. 2007;27:6903–6912. doi: 10.1128/MCB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding Fibroblast Growth Factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- Bonaventure J, Rousseau F, Legeai-Mallet L, Le Merrer M, Munnich A, Maroteaux P. Common mutations in the Fibroblast Growth Factor receptor 3 (FGFR 3) gene account for achondroplasia, hypochondroplasia, and thanatophoric dwarfism. Am J Med Genet. 1996;63:148–154. doi: 10.1002/(SICI)1096-8628(19960503)63:1<148::AID-AJMG26>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Agochukwu NB, Solomon BD, Muenke M. Hearing loss in syndromic craniosynostoses: introduction and consideration of mechanisms. Am J Audiol. 2014;23:135–141. doi: 10.1044/2014_AJA-13-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beever JE, Smit MA, Meyers SN, Hadfield TS, Bottema C, Albretsen J, Cockett NE. A single-base change in the tyrosine kinase II domain of ovine FGFR3 causes hereditary chondrodysplasia in sheep. Anim Genet. 2006;37:66–71. doi: 10.1111/j.1365-2052.2005.01398.x. [DOI] [PubMed] [Google Scholar]
- Smith LB, Dally MR, Sainz RD, Rodrigue KL, Oberbauer AM. Enhanced skeletal growth of sheep heterozygous for an inactivated Fibroblast Growth Factor receptor 3. J Anim Sci. 2006;84:2942–2949. doi: 10.2527/jas.2006-255. [DOI] [PubMed] [Google Scholar]
- Ezzat S, Zheng L, Florez JC, Stefan N, Mayr T, Hliang MM, Jablonski K, Harden M, Stancakova A, Laakso M. The cancer-associated FGFR4-G388R polymorphism enhances pancreatic insulin secretion and modifies the risk of diabetes. Cell Metab. 2013;17:929–940. doi: 10.1016/j.cmet.2013.05.002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, Welch WR, Berkowitz RS, Mok SC. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified Fibroblast Growth Factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- Shariat SF, Youssef RF, Gupta A, Chade DC, Karakiewicz PI, Isbarn H, Jeldres C, Sagalowsky AI, Ashfaq R, Lotan Y. Association of angiogenesis related markers with bladder cancer outcomes and other molecular markers. J Urol. 2010;183:1744–1750. doi: 10.1016/j.juro.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Cuevas R, Korzeniewski N, Tolstov Y, Hohenfellner M, Duensing S. FGF-2 disrupts mitotic stability in prostate cancer through the intracellular trafficking protein CEP57. Cancer Res. 2013;73:1400–1410. doi: 10.1158/0008-5472.CAN-12-1857. [DOI] [PubMed] [Google Scholar]
- Reed JA, McNutt NS, Albino AP. Differential expression of basic Fibroblast Growth Factor (bFGF) in melanocytic lesions demonstrated by in situ hybridization. Implications for tumor progression. Am J Pathol. 1994;144:329–336. [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Inoue Y, Kawaguchi T, Hosoe S, Kawahara M. Increased serum levels of basic Fibroblast Growth Factor in lung cancer patients: relevance to response of therapy and prognosis. Lung Cancer. 2001;31:213–219. doi: 10.1016/s0169-5002(00)00187-2. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Higashi T, Nouso K, Kariyama K, Nakamura S, Suzuki M, Nakatsukasa H, Kobayashi Y, Hanafusa T, Tsuji T. Altered expression of vascular endothelial growth factor, Fibroblast Growth Factor-2 and endostatin in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20:583–588. doi: 10.1111/j.1440-1746.2005.03726.x. et al. [DOI] [PubMed] [Google Scholar]
- Gruel N, Lucchesi C, Raynal V, Rodrigues MJ, Pierron G, Goudefroye R, Cottu P, Reyal F, Sastre-Garau X, Fourquet A. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–2407. doi: 10.1016/j.ejca.2010.05.013. et al. [DOI] [PubMed] [Google Scholar]
- Theillet C, LeRoy X, deLapeyriere O, Grosgeorges J, Adnane J, Raynaud SD, Simony-Lafontaine J, Goldfarb M, Escot C, Birnbaum D. Amplification of FGF-related genes in human tumors: possible involvement of HST in breast carcinomas. Oncogene. 1989;4:915–922. [PubMed] [Google Scholar]
- Allerstorfer S, Sonvilla G, Fischer H, Spiegl-Kreinecker S, Gauglhofer C, Setinek U, Czech T, Marosi C, Buchroithner J, Pichler J. FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene. 2008;27:4180–4190. doi: 10.1038/onc.2008.61. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropiquet F, Giri D, Kwabi-Addo B, Mansukhani A, Ittmann M. Increased expression of Fibroblast Growth Factor 6 in human prostatic intraepithelial neoplasia and prostate cancer. Cancer Res. 2000;60:4245–4250. [PubMed] [Google Scholar]
- Yamayoshi T, Nagayasu T, Matsumoto K, Abo T, Hishikawa Y, Koji T. Expression of keratinocyte growth factor/Fibroblast Growth Factor-7 and its receptor in human lung cancer: correlation with tumour proliferative activity and patient prognosis. J Pathol. 2004;204:110–118. doi: 10.1002/path.1617. [DOI] [PubMed] [Google Scholar]
- Zammit C, Coope R, Gomm JJ, Shousha S, Johnston CL, Coombes RC. Fibroblast Growth Factor 8 is expressed at higher levels in lactating human breast and in breast cancer. Br J Cancer. 2002;86:1097–1103. doi: 10.1038/sj.bjc.6600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauglhofer C, Sagmeister S, Schrottmaier W, Fischer C, Rodgarkia-Dara C, Mohr T, Stattner S, Bichler C, Kandioler D, Wrba F. Up-regulation of the Fibroblast Growth Factor 8 subfamily in human hepatocellular carcinoma for cell survival and neoangiogenesis. Hepatology. 2011;53:854–864. doi: 10.1002/hep.24099. et al. [DOI] [PubMed] [Google Scholar]
- Gnanapragasam VJ, Robinson MC, Marsh C, Robson CN, Hamdy FC, Leung HY. FGF8 isoform b expression in human prostate cancer. Br J Cancer. 2003;88:1432–1438. doi: 10.1038/sj.bjc.6600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SK, Bansal GS, Zammit C, Barnard R, Coope R, Roberts-Clarke D, Gomm JJ, Coombes RC, Johnston CL. Increased expression of Fibroblast Growth Factor 8 in human breast cancer. Oncogene. 1999;18:1053–1060. doi: 10.1038/sj.onc.1202392. [DOI] [PubMed] [Google Scholar]
- Liu R, Huang S, Lei Y, Zhang T, Wang K, Liu B, Nice EC, Xiang R, Xie K, Li J. FGF8 promotes colorectal cancer growth and metastasis by activating YAP1. Oncotarget. 2014;6:935–952. doi: 10.18632/oncotarget.2822. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, Eliseenkova AV, Butzow R, Mohammadi M, Peltomaki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous beta-catenin. Hum Mutat. 2008;29:390–397. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- Chen TM, Shih YH, Tseng JT, Lai MC, Wu CH, Li YH, Tsai SJ, Sun HS. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 2014;42:2932–2944. doi: 10.1093/nar/gkt1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai D, Hegab AE, Soejima K, Kuroda A, Ishioka K, Yasuda H, Naoki K, Shizuko K, Hamamoto J, Yin Y. Characterization of the cell of origin and propagation potential of the Fibroblast Growth Factor 9-induced mouse model of lung adenocarcinoma. J Pathol. 2015;235:593–605. doi: 10.1002/path.4486. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Boer M, Weigelt B, Jonkers J, van der Valk M, Hilkens J. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene. 2004;23:6047–6055. doi: 10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
- Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, Witte ON. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Mitsuhashi N, Shimizu H, Kimura F, Yoshidome H, Otsuka M, Kato A, Shida T, Okamura D, Miyazaki M. Fibroblast Growth Factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer. 2012;12:56. doi: 10.1186/1471-2407-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Dakhova O, Creighton CJ, Ittmann MM. The endocrine Fibroblast Growth Factor FGF19 promotes prostate cancer progression. Cancer Res. 2013;73:2551–2562. doi: 10.1158/0008-5472.CAN-12-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M, Mukhopadhyay S, Chatterjee U, Roy SS. FGF16 promotes invasive behavior of SKOV-3 ovarian cancer cells through activation of mitogen-activated protein kinase (MAPK) signaling pathway. J Biol Chem. 2014;289:1415–1428. doi: 10.1074/jbc.M113.535427. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Polnaszek N, Kwabi-Addo B, Wang J, Ittmann M. FGF17 is an autocrine prostatic epithelial growth factor and is upregulated in benign prostatic hyperplasia. Prostate. 2004;60:18–24. doi: 10.1002/pros.20026. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim KH, Lee J, Oh JJ, Cheong HS, Wong EL, Yang BS, Byun SS, Myung SC. Single nucleotide polymorphisms in Fibroblast Growth Factor 23 gene, FGF23, are associated with prostate cancer risk. BJU Int. 2013;114:303–310. doi: 10.1111/bju.12396. [DOI] [PubMed] [Google Scholar]
- Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SM, Kim HR, Shim HS, Soo RA, Cho BC. Role of FGF receptors as an emerging therapeutic target in lung squamous cell carcinoma. Future Oncol. 2013;9:377–386. doi: 10.2217/fon.12.190. [DOI] [PubMed] [Google Scholar]
- Lehnen NC, von Massenhausen A, Kalthoff H, Zhou H, Glowka T, Schutte U, Holler T, Riesner K, Boehm D, Merkelbach-Bruse S. Fibroblast Growth Factor receptor 1 gene amplification in pancreatic ductal adenocarcinoma. Histopathology. 2013;63:157–166. doi: 10.1111/his.12115. et al. [DOI] [PubMed] [Google Scholar]
- Yang F, Gao Y, Geng J, Qu D, Han Q, Qi J, Chen G. Elevated expression of SOX2 and FGFR1 in correlation with poor prognosis in patients with small cell lung cancer. Int J Clin Exp Pathol. 2013;6:2846–2854. [PMC free article] [PubMed] [Google Scholar]
- Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand EK, Grand FH, Chase AJ, Ross FM, Corcoran MM, Oscier DG, Cross NC. Identification of a novel gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer. 2004;40:78–83. doi: 10.1002/gcc.20023. [DOI] [PubMed] [Google Scholar]
- Kornmann M, Ishiwata T, Matsuda K, Lopez ME, Fukahi K, Asano G, Beger HG, Korc M. IIIc isoform of Fibroblast Growth Factor receptor 1 is overexpressed in human pancreatic cancer and enhances tumorigenicity of hamster ductal cells. Gastroenterology. 2002;123:301–313. doi: 10.1053/gast.2002.34174. [DOI] [PubMed] [Google Scholar]
- Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- Young RJ, Lim AM, Angel C, Collins M, Deb S, Corry J, Wiesenfeld D, Kleid S, Sigston E, Lyons B. Frequency of Fibroblast Growth Factor receptor 1 gene amplification in oral tongue squamous cell carcinomas and associations with clinical features and patient outcome. Oral Oncol. 2013;49:576–581. doi: 10.1016/j.oraloncology.2013.01.006. et al. [DOI] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol. 2010;41:461–476. doi: 10.1016/j.humpath.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Guzman MA, Pezanowski D, Patel D, Hauptman J, Keisling M, Hou SJ, Papenhausen PR, Pascasio JM, Punnett HH. FOXO1-FGFR1 fusion and amplification in a solid variant of alveolar rhabdomyosarcoma. Mod Pathol. 2011;24:1327–1335. doi: 10.1038/modpathol.2011.98. et al. [DOI] [PubMed] [Google Scholar]
- Popovici C, Zhang B, Gregoire MJ, Jonveaux P, Lafage-Pochitaloff M, Birnbaum D, Pebusque MJ. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to Fibroblast Growth Factor receptor 1. Blood. 1999;93:1381–1389. [PubMed] [Google Scholar]
- Gervais C, Dano L, Perrusson N, Helias C, Jeandidier E, Galoisy AC, Ittel A, Herbrecht R, Bilger K, Mauvieux L. A translocation t(2;8)(q12;p11) fuses FGFR1 to a novel partner gene, RANBP2/NUP358, in a myeloproliferative/myelodysplastic neoplasm. Leukemia. 2013;27:1186–1188. doi: 10.1038/leu.2012.286. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito Y, Wakimoto N, Kakegawa E, Uchida Y, Bessho M. A novel fusion of SQSTM1 and FGFR1 in a patient with acute myelomonocytic leukemia with t(5;8)(q35;p11) translocation. Blood Cancer J. 2014;4:e265. doi: 10.1038/bcj.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhai YP, Tang YM, Wang LP, Wan PJ. Identification of a novel partner gene, TPR, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer. 2012;51:890–897. doi: 10.1002/gcc.21973. [DOI] [PubMed] [Google Scholar]
- Wasag B, Lierman E, Meeus P, Cools J, Vandenberghe P. The kinase inhibitor TKI258 is active against the novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic leukemia/lymphoma and t(7;8)(q22;p11) Haematologica. 2011;96:922–926. doi: 10.3324/haematol.2010.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter A, Sohal J, Kulkarni S, Chase A, Macdonald DH, Aguiar RC, Goncalves C, Hernandez JM, Jennings BA, Goldman JM. Consistent fusion of ZNF198 to the Fibroblast Growth Factor receptor-1 in the t(8;13)(p11;q12) myeloproliferative syndrome. Blood. 1998;92:1735–1742. et al. [PubMed] [Google Scholar]
- Jang JH, Shin KH, Park JG. Mutations in Fibroblast Growth Factor receptor 2 and Fibroblast Growth Factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541–3543. [PubMed] [Google Scholar]
- Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Di Bacco A, Elbi C, Lutterbach B. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- Su X, Zhan P, Gavine PR, Morgan S, Womack C, Ni X, Shen D, Bang YJ, Im SA, Ho Kim W. FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br J Cancer. 2014;110:967–975. doi: 10.1038/bjc.2013.802. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Arao T, Hamaguchi T, Shimada Y, Kato K, Oda I, Taniguchi H, Koizumi F, Yanagihara K, Sasaki H. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer. 2012;106:727–732. doi: 10.1038/bjc.2011.603. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, Ojima H, Furuta K, Shimada K, Okusaka T. Fibroblast Growth Factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–1434. doi: 10.1002/hep.26890. et al. [DOI] [PubMed] [Google Scholar]
- Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, McCullough AE, Barrett MT, Hunt K, Patel MD. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, Murphy SJ, McWilliams RR, Hart SN, Halling KC. Fibroblast Growth Factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45:1630–1638. doi: 10.1016/j.humpath.2014.03.014. et al. [DOI] [PubMed] [Google Scholar]
- Al-Ahmadie HA, Iyer G, Janakiraman M, Lin O, Heguy A, Tickoo SK, Fine SW, Gopalan A, Chen YB, Balar A. Somatic mutation of Fibroblast Growth Factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. J Pathol. 2011;224:270–279. doi: 10.1002/path.2892. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez S, Toll A, Baselga E, Ribe A, Azua-Romeo J, Pujol RM, Real FX. Fibroblast Growth Factor receptor 3 mutations in epidermal nevi and associated low grade bladder tumors. J Invest Dermatol. 2007;127:1664–1666. doi: 10.1038/sj.jid.5700705. [DOI] [PubMed] [Google Scholar]
- Logie A, Dunois-Larde C, Rosty C, Levrel O, Blanche M, Ribeiro A, Gasc JM, Jorcano J, Werner S, Sastre-Garau X. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005;14:1153–1160. doi: 10.1093/hmg/ddi127. et al. [DOI] [PubMed] [Google Scholar]
- Chesi M, Brents LA, Fly SA, Bais C, Robbiani DF, Mesri E, Kuehl WM, Bergsagel PL. Activated Fibroblast Growth Factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- Zammit C, Barnard R, Gomm J, Coope R, Shousha S, Coombes C, Johnston C. Altered intracellular localization of Fibroblast Growth Factor receptor 3 in human breast cancer. J Pathol. 2001;194:27–34. doi: 10.1002/path.846. [DOI] [PubMed] [Google Scholar]
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Mittempergher L, Davidson NM, Bosma A, Willems SM, Horlings HM, de Rink I, Greger L, Hooijer GK, Peters D. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230:270–276. doi: 10.1002/path.4209. et al. [DOI] [PubMed] [Google Scholar]
- Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, Rocchi M, Otsuki T, Lombardi L, Maiolo AT. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90:4062–4070. et al. [PubMed] [Google Scholar]
- Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of Fibroblast Growth Factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagasaki F, Wakao D, Yokoyama Y, Uchida Y, Murohashi I, Kayano H, Taniwaki M, Matsuda A, Bessho M. Fusion of ETV6 to Fibroblast Growth Factor receptor 3 in peripheral T-cell lymphoma with a t(4;12)(p16;p13) chromosomal translocation. Cancer Res. 2001;61:8371–8374. [PubMed] [Google Scholar]
- Tomlinson DC, Knowles MA, Speirs V. Mechanisms of FGFR3 actions in endocrine resistant breast cancer. Int J Cancer. 2012;130:2857–2866. doi: 10.1002/ijc.26304. [DOI] [PubMed] [Google Scholar]
- Sonvilla G, Allerstorfer S, Heinzle C, Stattner S, Karner J, Klimpfinger M, Wrba F, Fischer H, Gauglhofer C, Spiegl-Kreinecker S. Fibroblast Growth Factor receptor 3-IIIc mediates colorectal cancer growth and migration. Br J Cancer. 2010;102:1145–1156. doi: 10.1038/sj.bjc.6605596. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JGT, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen QR, Shah K, Youngblood V. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–798. doi: 10.1038/ng.2643. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–3755. doi: 10.1200/JCO.2005.04.8587. et al. [DOI] [PubMed] [Google Scholar]
- Zaid TM, Yeung TL, Thompson MS, Leung CS, Harding T, Co NN, Schmandt RS, Kwan SY, Rodriguez-Aguay C, Lopez-Berestein G. Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin Cancer Res. 2013;19:809–820. doi: 10.1158/1078-0432.CCR-12-2736. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauglhofer C, Paur J, Schrottmaier WC, Wingelhofer B, Huber D, Naegelen I, Pirker C, Mohr T, Heinzle C, Holzmann K. Fibroblast Growth Factor receptor 4: a putative key driver for the aggressive phenotype of hepatocellular carcinoma. Carcinogenesis. 2014;35:2331–2338. doi: 10.1093/carcin/bgu151. et al. [DOI] [PubMed] [Google Scholar]
- Acevedo VD, Ittmann M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009;8:580–588. doi: 10.4161/cc.8.4.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- Brunello E, Brunelli M, Bogina G, Calio A, Manfrin E, Nottegar A, Vergine M, Molino A, Bria E, Massari F. FGFR-1 amplification in metastatic lymph-nodal and haematogenous lobular breast carcinoma. J Exp Clin Cancer Res. 2012;31:103. doi: 10.1186/1756-9966-31-103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Krishna NS, Witton CJ, Bartlett JM. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res. 2003;9:5271–5281. [PubMed] [Google Scholar]
- Fischbach A, Rogler A, Erber R, Stoehr R, Poulsom R, Heidenreich A, Schneevoigt BS, Hauke S, Hartmann A, Knuechel R. Fibroblast Growth Factor Receptor (FGFR) amplifications are rare events in bladder cancer. Histopathology. 2014 doi: 10.1111/his.12473. et al.. doi: 10.1111/his.12473. [DOI] [PubMed] [Google Scholar]
- Brito LP, Ribeiro TC, Almeida MQ, Jorge AA, Soares IC, Latronico AC, Mendonca BB, Fragoso MC, Lerario AM. The role of Fibroblast Growth Factor receptor 4 overexpression and gene amplification as prognostic markers in pediatric and adult adrenocortical tumors. Endocr Relat Cancer. 2012;19:L11–L13. doi: 10.1530/ERC-11-0231. [DOI] [PubMed] [Google Scholar]
- Jaakkola S, Salmikangas P, Nylund S, Partanen J, Armstrong E, Pyrhonen S, Lehtovirta P, Nevanlinna H. Amplification of fgfr4 gene in human breast and gynecological cancers. Int J Cancer. 1993;54:378–382. doi: 10.1002/ijc.2910540305. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang L, Li Y, Hu H, Shen L, Shen X, Pan Y, Ye T, Zhang Y, Luo X. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20:4107–4114. doi: 10.1158/1078-0432.CCR-14-0284. et al. [DOI] [PubMed] [Google Scholar]
- Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58:165–170. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- Kalff A, Spencer A. The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer J. 2012;2:e89. doi: 10.1038/bcj.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, Belch AR, Pilarski LM. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, Mohammadi M, Futreal PA, Stratton MR, Trent JM. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shi Y, Zhou Y, Du C, Wang C, Zhan H, Zheng B, Cao X, Sun MH, Fu H. The Fibroblast Growth Factor receptor-4 Arg388 allele is associated with gastric cancer progression. Ann Surg Oncol. 2010;17:3354–3361. doi: 10.1245/s10434-010-1323-6. [DOI] [PubMed] [Google Scholar]
- Laederich MB, Horton WA. FGFR3 targeting strategies for achondroplasia. Expert Rev Mol Med. 2012;14:e11. doi: 10.1017/erm.2012.4. [DOI] [PubMed] [Google Scholar]
- Kanazawa H, Tanaka H, Inoue M, Yamanaka Y, Namba N, Seino Y. Efficacy of growth hormone therapy for patients with skeletal dysplasia. J Bone Miner Metab. 2003;21:307–310. doi: 10.1007/s00774-003-0425-7. [DOI] [PubMed] [Google Scholar]
- Bocciardi R, Giorda R, Buttgereit J, Gimelli S, Divizia MT, Beri S, Garofalo S, Tavella S, Lerone M, Zuffardi O. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum Mutat. 2007;28:724–731. doi: 10.1002/humu.20511. et al. [DOI] [PubMed] [Google Scholar]
- Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10:80–86. doi: 10.1038/nm971. et al. [DOI] [PubMed] [Google Scholar]
- Lorget F, Kaci N, Peng J, Benoist-Lasselin C, Mugniery E, Oppeneer T, Wendt DJ, Bell SM, Bullens S, Bunting S. Evaluation of the therapeutic potential of a CNP analog in a Fgfr3 mouse model recapitulating achondroplasia. Am J Hum Genet. 2012;91:1108–1114. doi: 10.1016/j.ajhg.2012.10.014. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasoda A, Kitamura H, Fujii T, Kondo E, Murao N, Miura M, Kanamoto N, Komatsu Y, Arai H, Nakao K. Systemic administration of C-type natriuretic peptide as a novel therapeutic strategy for skeletal dysplasias. Endocrinology. 2009;150:3138–3144. doi: 10.1210/en.2008-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Dirat B, Tognacci T, Rochet N, Mouska X, Bonnafous S, Patouraux S, Tran A, Gual P, Le Marchand-Brustel Y. Postnatal soluble FGFR3 therapy rescues achondroplasia symptoms and restores bone growth in mice. Sci Transl Med. 2013;5:203ra124. doi: 10.1126/scitranslmed.3006247. et al. [DOI] [PubMed] [Google Scholar]
- Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of Fibroblast Growth Factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11:6280–6290. doi: 10.1158/1078-0432.CCR-05-0282. [DOI] [PubMed] [Google Scholar]
- Qing J, Du X, Chen Y, Chan P, Li H, Wu P, Marsters S, Stawicki S, Tien J, Totpal K. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216–1229. doi: 10.1172/JCI38017. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadari Y, Schlessinger J. FGFR3-targeted mAb therapy for bladder cancer and multiple myeloma. J Clin Invest. 2009;119:1077–1079. doi: 10.1172/JCI38948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Morioka M, Kishi H, Kimura T, Yahara Y, Okada M, Fujita K, Sawai H, Ikegawa S, Tsumaki N. Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature. 2014;513:507–511. doi: 10.1038/nature13775. [DOI] [PubMed] [Google Scholar]
- Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, Arold ST, Ahmed Z, Ladbury JE. Inhibition of basal FGF receptor signaling by Dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Timsah Z, Ahmed Z, Lin CC, Melo FA, Stagg LJ, Leonard PG, Jeyabal P, Berrout J, O'Neil RG, Bogdanov M. Competition between Grb2 and Plcgamma1 for FGFR2 regulates basal phospholipase activity and invasion. Nat Struct Mol Biol. 2014;21:180–188. doi: 10.1038/nsmb.2752. et al. [DOI] [PubMed] [Google Scholar]
- Zakrzewska M, Haugsten EM, Nadratowska-Wesolowska B, Oppelt A, Hausott B, Jin Y, Otlewski J, Wesche J, Wiedlocha A. ERK-Mediated phosphorylation of Fibroblast Growth Factor receptor 1 on Ser777 inhibits signaling. Sci Signal. 2013;6:ra11. doi: 10.1126/scisignal.2003087. [DOI] [PubMed] [Google Scholar]


