Abstract
Cellulose accounts for approximately half of photosynthesis-fixed carbon; however, the ecology of its degradation in soil is still relatively poorly understood. The role of actinobacteria in cellulose degradation has not been extensively investigated despite their abundance in soil and known cellulose degradation capability. Here, the diversity and abundance of the actinobacterial glycoside hydrolase family 48 (cellobiohydrolase) gene in soils from three paired pasture-woodland sites were determined by using terminal restriction fragment length polymorphism (T-RFLP) analysis and clone libraries with gene-specific primers. For comparison, the diversity and abundance of general bacteria and fungi were also assessed. Phylogenetic analysis of the nucleotide sequences of 80 clones revealed significant new diversity of actinobacterial GH48 genes, and analysis of translated protein sequences showed that these enzymes are likely to represent functional cellobiohydrolases. The soil C/N ratio was the primary environmental driver of GH48 community compositions across sites and land uses, demonstrating the importance of substrate quality in their ecology. Furthermore, mid-infrared (MIR) spectrometry-predicted humic organic carbon was distinctly more important to GH48 diversity than to total bacterial and fungal diversity. This suggests a link between the actinobacterial GH48 community and soil organic carbon dynamics and highlights the potential importance of actinobacteria in the terrestrial carbon cycle.
INTRODUCTION
Cellulases are responsible for the degradation of cellulose, an insoluble, recalcitrant substrate which comprises approximately half of the biologically fixed CO2 on earth (1). Cellulases are classified as glycosyl hydrolases (GHs) together with other enzymes that target the glycosidic bonds in oligo- and polysaccharides and are grouped into families that reflect their protein folding structure (2). Metagenomic studies have characterized various cellulose-rich environments, such as the bovine rumen (3–6), rabbit cecum (7), ant fungus gardens (8), compost (9, 10), earthworm casts (11), termite gut (12), and forest soil (13, 14). These studies have revealed a rich new GH gene diversity not thus far observed in cultured microorganisms. However, little is known about the role of GH genes in natural environments and the enzymes which they encode.
Glycoside hydrolases are a large and complex group of enzymes, with some GH families showing multiple substrate specificities (15). Horizontal gene transfer has also been documented for many GH families (15–21). The presence of multiple substrate specificities within the same GH family has precluded the design of molecular tools for in-depth investigation of their environmental role. Importantly, all 13 functionally characterized bacterial glycoside hydrolase family 48 (GH48) enzymes have been shown to target cellulose, and in most bacteria that carry the GH48 gene, it is present as a single genomic copy (19). Only in insects have the characterized GH48 enzymes been shown not to target cellulose; in these organisms, GH48 enzymes are chitinases.
In conventional systems for cellulase classification, GH48 enzymes function mostly as cellobiohydrolases, also known as exoglucanases (22). This class of cellulases is known to hydrolyze the ends of the cellulose chain and to act processively, producing glucose or cellobiose as the end product (15). Although their specific activity is low, GH48 cellobiohydrolases are important components of the multienzyme cellulolytic systems that they are part of, acting in synergy with other cellulases in order to achieve efficient depolymerization of cellulose. The deletion of GH48 genes from bacterial genomes has been shown to significantly impair cellulolytic activity (23–25). GH48 cellobiohydrolases are prevalent among Gram-positive cellulose degraders but are also found in anaerobic fungi (order Neocallimastigales), a small number of Gram-negative bacteria, and certain insects (19, 21, 26, 27).
Evolutionary analysis of GH48 sequences obtained from bacterial, fungal, and insect genomes suggests that GH48 genes evolved in the common ancestor of the phyla Actinobacteria, Firmicutes, and Chloroflexi and that their occurrence outside these phyla is due to horizontal gene transfer (19). Sukharnikov et al. (19) also identified a conserved omega loop in all functionally characterized bacterial GH48 proteins, which is absent in insect GH48 proteins, and used the presence of this loop and conserved residues in the catalytic region of the GH48 gene to predict that all known bacterial and fungal GH48 enzymes target cellulose. In addition to cellulose, the GH48 enzymes of some Clostridium species are known to also hydrolyze xylan, mannan, and β-glucan (28–30). Previous studies have investigated the diversity and abundance of GH48 genes from anaerobic Gram-positive bacteria such as Clostridium spp. in thermophilic composts, sulfate-reducing bioreactors, and wastewater sediments (22, 31, 32), and one study investigated actinobacterial GH48 gene diversity in decomposing straw (33).
Members of the phylum Actinobacteria are abundant in soils and are thought to have an important role in organic matter turnover and the breakdown of recalcitrant molecules such as cellulose (34, 35) and polycyclic aromatic hydrocarbons (36). Cultivation-based studies have demonstrated the ability of many actinobacteria to grow on cellulose (37–39), while the properties of actinobacterial cellulolytic enzymes have been characterized for two model cellulose-degrading species, Thermobifida fusca and Cellulomonas fimi (40, 41). Furthermore, analyses of the available actinobacterial genomes have shown the presence of functional GH genes that are similar to characterized cellulase genes in T. fusca and C. fimi (42, 43). Further analysis of their cellulolytic ability indicated that most actinobacteria that contain functional cellulase genes are able to degrade cellulose, whereas the absence of detectable activity in laboratory assays may be a result of the use of modified cellulose substrates and artificial growth media not optimal for cellulase production and activity (43). It is important to note that efficient cellulose degradation can occur only through the concerted action of several enzymes (i.e., endoglucanases, cellobiohydrolases, and β-glucosidases) (15), and therefore, it is not possible to estimate cellulose degradation rates through the detection and quantification of a single cellulose degradation gene. However, the development of tools for the determination of the abundance and diversity of one of the most substrate-specific cellulose degradation genes (the GH48 gene) from actinobacteria will aid in elucidating the ecology and the role of these organisms in terrestrial carbon cycling.
In this study, we investigated the ecology of saprotrophic actinobacteria in soil by developing standard and quantitative PCR (qPCR) primers targeting the catalytic region of the actinobacterial GH48 gene. We aimed to determine whether the GH48 gene community correlates more strongly with soil carbon quantity and quality than do the overall bacterial and fungal communities. Cloning and sequencing were used to determine the phylogeny of actinobacterial GH48 genes amplified from soils. We investigated the presence of the catalytic base residue and residues involved in substrate recognition and cellulose chain accessibility in order to assess whether these residues represented functional GH48 genes likely to be involved in cellulose degradation. Quantitative PCR was used to provide an estimate of the abundance of these genes in the soils analyzed. Using terminal restriction fragment length polymorphism (T-RFLP) analysis, we assessed the diversity of soil actinobacterial GH48 genes from three paired pasture-woodland sites. Specifically, we contrasted actinobacterial GH48 ecology in two systems where plant litter is likely to be chemically and structurally different (44, 45). Furthermore, soil variables were quantified and used to develop multivariate correlation models of actinobacterial GH48, general bacterial 16S rRNA, and fungal internal transcribed spacer (ITS) genes.
MATERIALS AND METHODS
Field sites.
Sampling was conducted at three paired sites, each comprising adjacent 1-ha pasture and woodland plots. The sites were Talmo (34.936976°S, 148.625293°E), Glenrock (34.858413°S, 148.56724°E), and Bogo (34.813746°S, 148.704558°E) and are located on farms near the locality of Bookham, NSW, Australia (46). The woodland sites were dominated by Eucalyptus spp. (Eucalyptus melliodora A. Cunn. ex Schaur [yellow box] and E. albens Benth. [white box]); the Talmo woodland plot had the highest tree densities, with an infrequent population of Acacia dealbata and Acacia implexa and small isolated patches of native Australian grasses such as Themeda triandra Forssk. and Poa sieberiana Spreng. The pasture plots were improved with Trifolium subterraneum (subterranean clover) and showed a mixture of annual and perennial native and introduced grasses, including Phalaris aquatica. The pasture plots were routinely grazed with sheep and received regular inputs of phosphorus fertilizer (∼10 kg P ha−1 year−1), with Talmo and Glenrock having higher overall soil fertility levels and stocking rates (for further site description details, see reference 46 and the supplemental material).
A total of 240 samples were taken across the six plots. Soil cores (0 to 10 cm deep and 5 cm in diameter) were kept cold (4°C) until they were processed. The soils were sieved (5 mm), homogenized, and separated into aliquots that were (i) frozen in liquid nitrogen (for DNA analyses), (ii) kept at 4°C (for quantification of soil nutrients and moisture), or (iii) air dried (for soil pH, mid-infrared [MIR] spectrometry, and C and N analyses). Soil properties (pH; soil moisture; dissolved organic carbon [DOC] content; dissolved organic nitrogen [DON] content; ammonium [NH4+-N] content; nitrate [NO3−-N] content; free amino acid nitrogen [FAA-N] content; microbial biomass C [MBC] content; microbial biomass N [MBN] content; loss on ignition [LOI]; total, inorganic, and organic P contents; total C and N contents; and particulate organic carbon [POC], humus organic carbon [HOC], and resistant organic carbon [ROC] contents) were determined as described previously by de Menezes et al. (46). There is a high correlation between the MIR predictive algorithms used to estimate HOC content and total organic carbon (TOC) content, which indicates that these two organic carbon fractions are predicted based on the same MIR spectral features (47). Therefore, the interpretation of HOC as humic organic carbon should be taken with caution (for more details regarding the organic carbon fraction determination by MIR spectrometry, see the supplemental material).
GH48 primer design. (i) Standard GH48 PCR primers.
All 39 unique available actinobacterial GH48 sequences were obtained from the CAZy Web portal (http://www.cazy.org/) (48) and aligned in MAFFT by using the accuracy-oriented global pair (G-INS-i) method (49). Alignments were visualized in Geneious v. 5.6.6 (Biomatters, New Zealand), and conserved regions that partially covered the GH48 catalytic domain were identified (1,923 bp long in T. bifida [24]). Primers were designed by using the primer design tool in Geneious. Due to the difficulties in designing one primer pair covering all actinobacterial GH48 gene diversity, two primer pairs were developed, primer pair GH48_F1 (5′-RRCATBTACGGBATGCACTGGCT-3′) and GH48_R1 (5′-VCCGCCCCABGMGTARTACC-3′) as well as primer pair GH48_F1_cell (5′-AYGTCGACAACRTSTACGGMTWCG-3′) and GH48_R1_cell (5′ CCGCCCCASGCSWWRTACC-3′), and both primer pairs were used for cloning, sequencing, and T-RFLP analysis. In silico primer specificity was analyzed by using MFEprimer-2.0 (50), which revealed that the combination of the two primer pairs provided good coverage of known actinobacterial GH48 gene diversity (see Results) (for further details of GH48 gene primer design, see Tables S1 and S2 in the supplemental material).
(ii) Quantitative PCR primer design.
GH48 qPCR primers (qPCR_GH48_F8 [5′-GCCADGHTBGGCGACTACCT-3′] and qPCR_GH48_R5 [5′-CGCCCCABGMSWWGTACCA-3′]) were designed as described above, based on sequences obtained from the CAZy database (for further details of GH48 gene primer design, see methods and Tables S1 and S2 in the supplemental material).
DNA extraction.
The MoBio PowerSoil kit (MoBio, Carlsbad, CA) was used to extract DNA from 0.25 g of soil according to the manufacturer's instructions, except for the use of a Qiagen TissueLizer shaker (Qiagen, Venlo, Netherlands) for 2 min at full speed after the introduction of buffer C1. DNA concentrations were normalized across all samples, as described previously by de Menezes et al. (46).
Cloning.
The 40 DNA samples from each of the 6 plots (total of 240 samples from the 3 paired pasture-woodland sites) were pooled and amplified with both GH48 PCR primer pairs (GH48_F1-GH48_R1 and GH48_F1_cell-GH48_R1_cell) to give 12 amplifications. Each of the 12 amplicon fragments were excised and purified from 1% agarose gels by using QIAquick Spin Miniprep kits (Qiagen, Düsseldorf, Germany). Amplicons were then cloned from each of the 12 amplicon mixtures separately by using the Promega pGEM-T Easy vector system (Promega, Madison, WI, USA). The resulting plasmids containing inserts of the correct length were extracted by using Perfectprep plasmid isolation kits (Eppendorf, Hamburg, Germany), and the clones were sequenced by Macrogen (Seoul, South Korea), using M13 primers. Sequences were analyzed with Geneious, and poor-quality sequences were removed from the data set. A total of 80 high-quality GH48 sequences were obtained from the 12 cloning reactions, which were then used for phylogenetic analyses, with a total of 39 sequences from woodlands, 41 sequences from pastures, and 9 to 16 sequences per individual pasture or woodland plot.
Phylogenetic analysis of actinobacterial GH48 sequences.
Available GH48 sequences (excluding duplicates) from cultured actinobacterial strains as well as a selection of sequences from the Firmicutes (Bacillus spp., Paenibacillus spp., Clostridium spp.), Proteobacteria (Hahella chejuensis and Myxobacter sp.), Chloroflexi (Herpetosiphon aurantiacus), anaerobic fungi (Piromyces spp. and Neocallimastix spp.), and Insecta (Leptinotarsa decemlineata) were aligned in MAFFT (49), using the G-INS-i model and default parameters for DNA alignment. The alignment was visualized and manually optimized in Geneious and exported after the removal of the primer regions. After alignment, nucleotide sequences were translated into protein sequences; any sequence that did not translate into a GH48 protein through its entire length was removed from the original nucleotide sequence alignment prior to the construction of phylogenetic trees.
Maximum likelihood phylogenetic trees were produced by exporting the alignment to PhyML (http://atgc.lirmm.fr/phyml/) (51) and RaxML (http://phylobench.vital-it.ch/raxml-bb/) (52). PhyML was run by using the HKY85 substitution model and the Shimodaira-Hasegawa (SH)-like approximate likelihood-ratio test (aLRT) branch support method with 100 bootstraps. RaxML was run with 100 bootstraps and the CAT (category) model of heterogeneity (52). The tree topologies generated with PhyML and RaxML were compared to determine the consistency of tree branching patterns. The resulting RaxML phylogenetic trees were uploaded to iTol (http://itol.embl.de/) (53) for visualization of the phylogeny and metadata (see Fig. S2 in the supplemental material for the PhyML tree).
Analysis of residues of functional significance.
The GH48 sequence region targeted in this study corresponds to the region in the T. fusca gene that contains the catalytic base D225 (aspartic acid), which essential for cellulose hydrolysis (54), as well as 12 other residues conserved in bacterial and fungal GH48 cellobiohydrolases with functional significance in substrate recognition (hydrogen bonding and hydrophobic stacking interactions) and enzyme thermal stability (calcium coordination) (19, 55, 56). In addition, the region targeted also includes two aromatic residues (phenylalanine 195 and tyrosine 213) in the entrance of the T. fusca GH48 active-site tunnel (where the cellulose chain slides in and where hydrolysis takes place), which play a role in facilitating the access of the cellulose chains to the active site and promote enzyme processivity (57). In order to determine if the GH48 sequences obtained here also had these residues, the 80 aligned GH48 sequences produced from our clone libraries were translated into protein sequences and compared with the protein sequence of the model organism T. fusca.
qPCR cycling conditions.
GH48, bacterial 16S rRNA, and fungal ITS gene abundances for each of the 240 individual soil samples were quantified in triplicate reactions using SsoAdvanced SYBR green Supermix (Bio-Rad, Hercules, CA), using the C1000 thermal cycler (Bio-Rad, Hercules, CA), to which 400 nM primers, 2 μl DNA (diluted 1 in 10), and H2O were added to 10 μl. qPCR cycling conditions were as follows: 95°C for 1 min (1 cycle) and 95°C for 5 s and 57°C for 20 s (39 cycles) for the GH48 gene; 95°C for 1 min (1 cycle) and 95°C for 15 s and 56°C for 20 s (39 cycles) for the 16S rRNA gene (primers Eub338 [58] and Eub518 [59]); and 95°C for 1 min (1 cycle) and 95°C for 5 s, 53°C for 20 s, and extension at 72°C (39 cycles) for the ITS1 gene (primers ITS1F [60] and 5.8s [61]). A melt curve from 65°C to 95°C was added at the end of all amplification cycles. Standards were run in triplicates in each qPCR plate with 10-fold dilution series from 108 to 100 copies μl−1. GH48 standards were purified PCR products from a local soil actinobacterial isolate; for bacteria and fungi, the PCR products from the 16S rRNA gene from Pseudomonas fluorescens strain 5.2 or the ITS gene from Fusarium oxysporum f. sp. vasinfectum were used. Amplification efficiencies were >90% and the R2 value was >0.99 for all GH48, bacterial 16S rRNA, and fungal ITS gene calibration curves.
T-RFLP analysis. (i) GH48 gene.
For T-RFLP analysis of the 240 individual soil samples, multiplex PCR was performed by using both primer pairs designed for the actinobacterial GH48 gene (GH48_F1-GH48_R1 and GH48_F1_cell-GH48_R1_cell). Primers GH48_F1 and GH48_F1_cell were labeled with 6-carboxyfluorescein (6-FAM) and 5-hexachlorofluorescein at their 5′ ends, respectively. The PCR mixtures contained 2 ng DNA, forward and reverse primers (200 nM each), MyTaq DNA polymerase (0.25 μl) (Bioline, Alexandria, Australia), deoxynucleoside triphosphates (dNTP) (250 μM each), and 1× supplied buffer. The following touchdown PCR protocol was used for GH48 gene amplification: 95°C for 2 min followed by 2 cycles of 30 s each at 95°C, 65°C, and 72°C; 2 cycles of 30 s each at 95°C, 62°C, and 72°C; 3 cycles of 30 s at 95°C and 59°C and 40 s at 72°C; 4 cycles of 30 s at 95°C and 56°C and 45 s at 72°C; 5 cycles of 30 s at 95°C and 53°C and 50 s at 72°C; 30 cycles of 30 s at 95°C, 45 s at 50°C, and 100 s at 72°C; and, finally, 1 cycle of 10 min at 72°C. The amplified PCR products were cleaned with Agencourt AMPure beads (Beckman Coulter, Lane Cove, Australia) and quantified by using a Quant-iT Picogreen double-stranded DNA (dsDNA) quantification kit (Life Technologies, Mulgrave, Australia), according to the manufacturer's instructions. Twenty-five nanograms of the PCR product was added to a reaction mixture containing water, reaction buffer, and 20 units of the AluI and MboII restriction enzymes (New England BioLabs); double digestion was carried out overnight at 37°C. For the purification of PCR products, digests were precipitated by incubation with 150 μl of cold 75% (vol/vol) isopropanol (Sigma-Aldrich, Sydney, Australia) for 30 min, followed by centrifugation at 4,000 rpm for 45 min. Purified PCR products were added to a reaction mixture containing Hi-Di formamide (9.7 μl) and a GeneScan 600 LIZ size standard (0.3 μl) (Applied Biosystems, Mulgrave, Australia). After denaturation (94°C for 3 min), fragment lengths were determined by electrophoresis using an AB3031xl genetic analyzer (Applied Biosystems, Mulgrave, Australia). GENEMAPPER (Applied Biosystems, Mulgrave, Australia) was used to provide restriction fragment profiles, and these profiles were filtered according to the method of Abdo et al. (62) to remove spurious baseline peaks (minimum height of 20 fluorescence units; peaks smaller than 2 times the standard deviation calculated over all peaks were removed). The resulting GH48 gene sizing data were binned by using Interactive Binner (63) in R (64), with a sliding-window approach and the following parameters: minimum and maximum peak sizes of 40 and 520 bp, respectively; a minimum of 0.099 relative fluorescence units; a window size of 2.5 bp; and a shift size of 0.25 bp.
(ii) Bacterial and fungal DNA.
Briefly, DNA from bacteria was amplified by using primers 27f (58) and 519r (65), and the fungal ITS region was amplified by using primers ITS1f (66) and ITS4 (60). For both fungi and bacteria, the forward primer was 6-FAM labeled at the 5′ end. The amplicon mixtures of both groups were digested with AluI (New England BioLabs), and the resulting digests were processed and analyzed as described previously by de Menezes et al. (46).
Statistical analysis. (i) Data analysis.
Multivariate statistical analyses were carried out with PRIMER 6 and PERMANOVA+ (Primer-E Ltd., Plymouth, United Kingdom) (67). GH48, bacterial 16S rRNA, and fungal ITS gene community ribotype relative abundance data obtained by T-RFLP analysis were square-root transformed, and a Bray-Curtis dissimilarity matrix was calculated. Soil variables were fourth-root transformed (except for pH) and standardized, and a Euclidean distance matrix was calculated. In order to visualize differences in GH48, bacterial 16S rRNA, and fungal ITS gene compositions across sites and land uses, ordination by principal coordinate (PCO) analysis was carried out. The vector overlay function in PRIMER was used to visualize the soil variables that correlated with the first two PCO axes. Only variables that had a vector length of >0.4 were included for visualization in the PCO plot. The vector length is calculated based on the correlations between the soil variable in question and the first two PCO axes and indicates the strength and sign of the relationship between the soil variable and the PCO axes (68).
Permutational multivariate analysis of variance (PERMANOVA) (performed with a nonnested fixed factors design, type III partial sums of squares, and 9,999 permutations under a reduced model) was used to determine differences in GH48 community compositions between pastures and woodlands and between sites and the interaction between site and land use. For PERMANOVA, there were 2 factors: site (3 levels [Talmo, Glenrock, and Bogo]) and land use (2 levels [woodland and pasture]). Multivariate dispersion index (MVDISP) analysis was used to quantify β-diversity (community composition heterogeneity) among samples within each land use as well as among samples in each pasture and woodland plot, while the significance of differences in β-diversity between land uses and sites was determined by using a test of homogeneity of dispersion (PERMDISP), using 9,999 permutations.
SIMPER analysis (analysis of the contribution of variables to similarity) was used to determine the variability of soil properties (67) across land uses and sites and was calculated by using the Euclidean distance matrix of the fourth-root-transformed (except pH) and standardized soil variable data set.
(ii) DistLM.
The relationships between GH48 gene community compositions and soil parameters were determined by using nonparametric multivariate multiple regressions (DistLM) from the PERMANOVA+ add-on in the PRIMER-E package. DistLM was conducted by using the stepwise selection procedure with the adjusted R2 selection criterion and 9,999 permutations. Soil variables that were correlated with each other (r > 0.9) were removed from the data set, except for the predicted organic carbon fractions (HOC, ROC, and POC), as a specific goal was to investigate the importance of carbon fractions of different qualities. Total C was removed, as it was correlated with predicted HOC and ROC, and N was removed, as it was correlated with the predicted POC in the woodlands. Total P was also removed, as it was correlated with organic P.
Nucleotide sequence accession numbers.
The actinobacterial GH48 gene sequences generated in this study were submitted to GenBank (accession numbers KM891594 to KM891673).
RESULTS
In silico PCR and qPCR primer specificity analysis.
A thermodynamic in silico analysis of PCR amplification was performed to determine whether the primers developed here showed good specificity and coverage of known actinobacterial GH48 genes. The two standard GH48 PCR primer pairs developed showed differences in their coverage of actinobacterial GH48 diversity (see Tables S1 and S2 in the supplemental material). Primer pair GH48_F1-GH48_R1 was found to be capable of amplifying GH48 genes from every actinobacterial genus present in the CAZy database except Cellulomonas and Cellvibrio; however, 10 out of 16 GH48 sequences from Streptomyces strains were not predicted to be amplified by primer pair GH48_F1-GH48_R1 (see Table S1 in the supplemental material). Primer pair GH48_F1_cell-GH48_R1_cell was predicted to amplify all available GH48 gene sequences from the genera Cellulomonas, Xylanimonas, and Jonesia as well as Cellvibrio gilvus. In addition, primers GH48_F1_cell and GH48_R1_cell were predicted to amplify seven Streptomyces strains not expected to be amplified by primer pair GH48_F1-GH48_R1. However, GH48_F1_cell and GH48_R1_cell were not predicted to amplify GH48 sequences from several actinobacterial genera, such as Nocardiopsis, Salinispora, Catenulispora, Streptosporangium, and Thermobifida. The overall coverage of actinobacterial GH48 diversity was therefore lower than but complementary to that of primer pair GH48_F1-GH48_R1. Both primer pairs missed three Streptomyces GH48 sequences present in the databases, and in combination, the two primer pairs are predicted to amplify 36 out of 39 actinobacterial GH48 gene sequences (see Table S1 in the supplemental material). Primer pair GH48_F1-GH48_R1 was predicted to amplify Hahella chejuensis, a member of the phylum Gammaproteobacteria that acquired this gene by horizontal gene transfer (19). No non-GH48 genes are predicted to be amplified by primer pair GH48_F1-GH48_R1 or GH48_F1_cell-GH48_R1_cell (see Table S1 in the supplemental material). Therefore, in silico analysis demonstrated that, in combination, the two standard PCR primer pairs developed here provided good specificity and coverage of known actinobacterial GH48 genes.
The GH48 qPCR primers developed here covered a narrower range of actinobacterial GH48 diversity than did the standard GH48 primers and were not predicted to amplify 15 out of 39 GH48 gene sequences from cultured actinobacterial strains (see Table S1 in the supplemental material). As with the standard PCR primers, no non-GH48 genes are predicted to be amplified by primer pair qPCR_GH48_F8-qPCR_GH48_R5 (see Table S1 in the supplemental material). Therefore, the qPCR primer pair developed here underestimates the total abundance of the actinobacterial GH48 gene and was used only to estimate a minimum abundance of actinobacterial GH48 genes in the soils studied.
Phylogenetic analysis of GH48 clones.
Cloning and sequencing of the PCR products obtained with both primer pairs GH48_F1-GH48_R1 and GH48_F1_cell-GH48_R1_cell were conducted for further evaluation of primer specificity and to determine the phylogenetic relationships of the amplified soil GH48 genes with those of cultured actinobacteria. A total of 87 high-quality sequences were obtained, 3 of which were removed due to the presence of stop codons in the sequence. A further four sequences were removed because translation did not result in a GH48 protein sequence for the entire region covered. One sequence was removed because no close database matches were identified by BLASTn analysis. Of the remaining 80 clones, 77 were unique sequences.
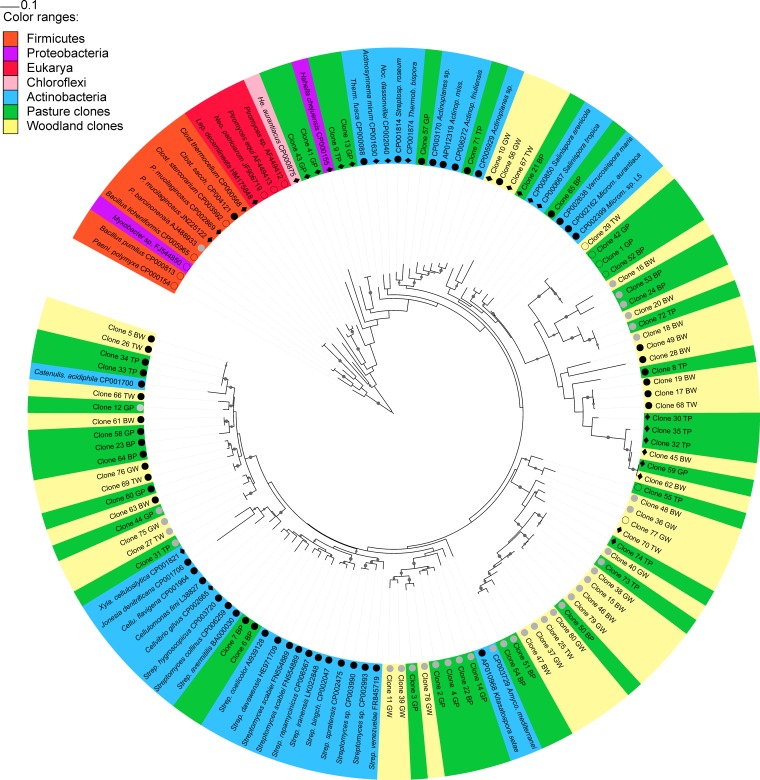
Comparison of phylogenetic trees generated by PhyML and RaxML revealed consistent branching patterns. GH48 sequences from cultured actinobacterial strains formed a cluster separate from those of other cultured bacteria and eukaryotes; however, this cluster had low bootstrap support. All but 2 (PhyML) or 4 (RaxML) of the 80 soil clones from this study clustered with the actinobacteria (Fig. 1; see also Fig. S2 in the supplemental material), and BLASTn analysis of these divergent clones showed that their top hits remained as GH48 genes from cultured actinobacteria. However, the possibility that a small proportion of amplified sequences were not derived from the actinobacteria cannot be ruled out.
FIG 1.
Maximum likelihood tree (RaxML) constructed with GH48 sequences from soil clones and cultured strains from Actinobacteria, Firmicutes, Neocallimastigales (anaerobic fungi), Proteobacteria, Chloroflexi, and Insecta. Nodes in tree branches indicate bootstrap support of >0.8. Sequences from Bacillus spp. and Paenibacillus spp. were used as outgroups. GenBank accession numbers are indicated after the organism name. Colors indicate sequence taxonomy or soil clone provenance; symbols in front of sequence names indicate the presence or absence of aromatic amino acids relevant to cellulolytic action in T. fusca. Filled black circles indicate the presence of both amino acids, filled gray circles indicate the lack of phenylalanine 195, diamonds indicate the lack of tyrosine 213, and open circles indicate the lack of both aromatic amino acids. TW, Talmo woodland; GW, Glenrock woodland; BW, Bogo woodland; TP, Talmo pasture; GP, Glenrock pasture; BP, Bogo pasture; Clost. sacch, Clostridium saccharoperbutylacetonicum N1-4(HMT); Amyco, Amycolatopsis mediterranei S699; Actinop. miss, Actinoplanes missouriensis 431; Strep. bingch., Streptomyces bingchenggensis BCW-1.
Neither PhyML nor RaxML phylogenetic analysis showed any pronounced clustering of sequences arising from either land use category or from any of the sites studied. There was no clustering of sequences arising from the use of either primer pair, and therefore, the diversities recovered by both standard PCR GH48 primer sets designed in this study were similar at the sequencing depth investigated here.
A closer inspection of the phylogenetic tree shown in Fig. 1 and in Fig. S2 in the supplemental material revealed that some of the sequences obtained clustered with known actinobacterial GH48 genes, while the majority of clones formed new clusters without any cultured representative. All streptomycete sequences grouped into one cluster in both the PhyML and RaxML trees, and this cluster, which had high bootstrap support in the RaxML tree (>0.8), included two soil clone sequences. A second cluster with high bootstrap support in the trees generated with both methods included Catenulispora acidiphila and 18 soil clones. RaxML indicated that two clones clustered with high bootstrap support with Actinoplanes spp., and one soil clone clustered with Verrucosispora maris. PhyML also showed the presence of these clusters but with lower bootstrap support. Both the RaxML and PhyML phylogenetic trees revealed that most soil clones were located in clusters that did not contain any GH48 sequences from cultured strains. The largest of these clusters had 22 soil clones, while other clusters containing only soil clones had 16, 7, and 3 clones. There were four soil clones that clustered adjacent to a Salinispora-Micromonospora-Verrucosispora cluster. There were two soil clones that clustered with low bootstrap support with Hahella chejuensis from the phylum Gammaproteobacteria. The phylogenetic tree obtained with RaxML, but not that obtained with PhyML, showed that two other soil clones were located outside the cluster of actinobacteria although clearly separate from the firmicute and eukaryote clusters.
Analysis of residues of functional significance.
In order to evaluate whether the amplified GH48 genes were likely to be functional cellobiohydrolase genes, the presence of conserved residues with known roles in substrate recognition, accessibility, and hydrolysis was determined. All of the clones obtained in this study showed the presence of the catalytic base, a conserved aspartic acid residue in the same position as T. fusca D225 (Fig. 1). Analysis of 12 conserved residues with functional roles in bacterial and fungal GH48 cellobiohydrolases (see Table S3 in the supplemental material) showed that all 8 conserved residues involved in hydrogen bonding were present in >89% of the sequences obtained in this study, and 6 of these residues were present in >98% of the GH48 clones. One residue involved in calcium coordination was present in 71% of the GH48 clones obtained. Two further conserved residues involved in calcium coordination and hydrophobic stacking interactions were present in 47 to 57% of the clones; however, these residues were absent from the GH48 protein sequence of the model cellulose-degrading actinobacterium T. fusca. Figure 1 shows the presence or absence of aromatic residues that have a demonstrated role in providing access to cellulose chains in the T. fusca GH48 enzyme. All but six soil clones showed the presence of either or both aromatic residues of functional significance (phenylalanine 195 and tyrosine 213). Inspection of the GH48 sequences from cultured strains revealed that all actinobacterial sequences had either one or both conserved aromatic residues, while the presence of these residues in the Firmicutes GH48 sequences was more variable, and none of the fungal sequences showed their presence.
GH48 diversity in woodlands and pastures.
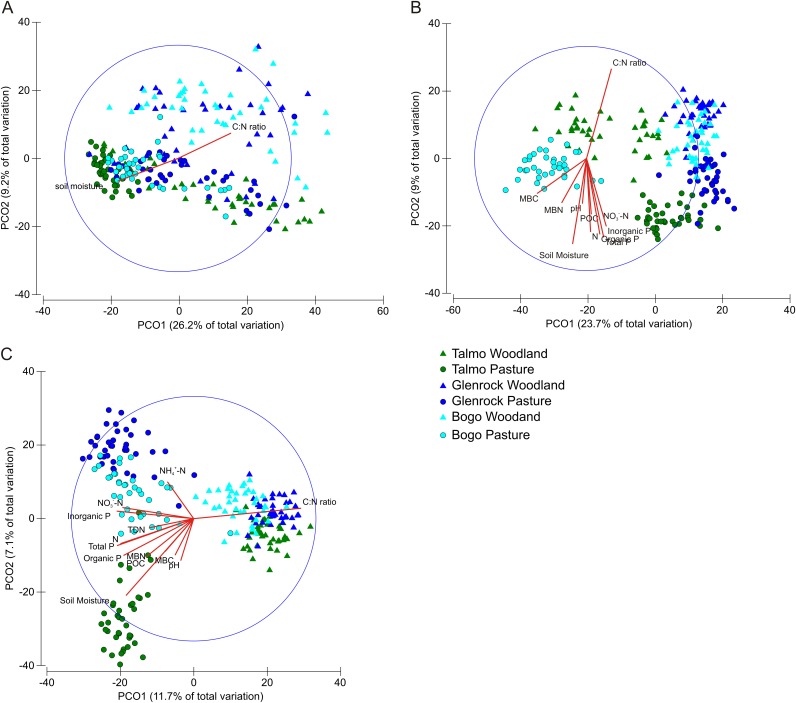
GH48 gene diversity across sites and land uses was determined by T-RFLP analysis, and their community composition was analyzed against measured soil properties to provide an understanding of which environmental variables drive GH48 gene ecology. Principal coordinate analysis of GH48 gene diversity (Fig. 2A) revealed a broad separation of samples by land use, although some overlap of samples from woodlands and pastures was observed, particularly between the Talmo woodland and Glenrock pasture. Separation of samples by site was less clear, and samples from Glenrock and Bogo overlapped substantially within each land use category. PCO analysis also showed a greater spread of woodland samples than of pasture samples. The main PCO axis explained 26.2% of the total variation, and the vector overlay function suggests that the C/N ratio (positive correlation with the main PCO axis) and soil moisture (negative correlation with the main PCO axis) were the main soil variables influencing GH48 community composition in the sites investigated. The soil C/N ratio vector correlated mostly with shifts in woodland GH community composition, while moisture correlated mostly with changes in the GH48 composition of pasture samples, particularly those of the Talmo pasture. The Glenrock and Bogo pasture samples as well as the Talmo woodland samples were spread along the C/N ratio-moisture axis in the PCO plot.
FIG 2.
PCO analysis of GH48 (A), bacterial (B), and fungal (C) community compositions for all sites and land uses generated by T-RFLP. Colors indicate sites, triangles represent woodland samples, and circles represent pasture samples. Vectors included were those that had a length of >0.4. The large circle is a unit circle with a radius of 1.
In order to contrast the ecology of the actinobacterial GH48 gene with that of the overall soil microbial community, the diversity and community composition of bacterial 16S rRNA and fungal ITS genes were also analyzed. PCO analysis of bacterial 16S rRNA gene diversity (Fig. 2B) showed that pasture and woodland samples were broadly separated along the second PCO axis, which explained 9% of the total variation in community composition. The C/N ratio and soil moisture vectors were positively and negatively correlated with the second PCO axis, respectively, and were the two main soil variables explaining shifts in bacterial community composition. However, PCO analysis showed that shifts in pasture bacterial community composition also correlated (>0.4) with the MBC; MBN; pH; NO3−-N; predicted POC; total, inorganic, and organic P; and total N vectors. PCO analysis of fungal community diversity (Fig. 2C) showed that pasture and woodland samples were separated along the main PCO axis, which explained 11.7% of the variation in community composition. The C/N ratio and soil moisture vectors were positively and negatively correlated with the main PCO axis, respectively, and the soil moisture vector also correlated negatively with the second PCO axis. The fungal pasture communities from each site were separated along the second PCO axis. Shifts in fungal community composition in Glenrock pasture samples correlated with the NH4+-N vector, and shifts in fungal community composition in Bogo pasture samples correlated with the NO3−-N, inorganic P, TDN, total N, and total P vectors, while shifts in fungal community composition in Talmo pasture samples correlated with the soil moisture, pH, MBC, MBN, predicted POC, and organic P vectors. Shifts in fungal community composition at all woodland sites correlated with the C/N vector.
PERMANOVA was conducted to determine if the differences in community composition between land uses and sites were statistically significant. Table 1 shows that the GH48 community compositions were significantly different between land uses and between sites (P < 0.001), and the pseudo-F value (see reference 68) determined by PERMANOVA suggests that the GH48 community composition was more different between land uses than among sites; however, an interaction effect between site and land use suggests that this difference was not uniform across sites. Pairwise PERMANOVA comparisons showed that differences in GH48 community compositions between land uses at each site were greater than differences between sites for each land use except when Talmo and Bogo samples were compared.
TABLE 1.
PERMANOVA values for differences in GH48 gene community compositions between land uses and sitesa
| Test | Factor | Pairwise comparison | Pseudo-F | t statistic | P value |
|---|---|---|---|---|---|
| Main test | Land use | 30.886 | <0.001 | ||
| Site | 11.785 | <0.001 | |||
| Site-vs-land use interaction | 10.29 | <0.001 | |||
| Pairwise, woodland | Site | TO vs GK | 2.782 | <0.001 | |
| Pairwise, woodland | Site | TO vs BO | 3.709 | <0.001 | |
| Pairwise, woodland | Site | GK vs BO | 2.210 | <0.001 | |
| Pairwise, pasture | Site | TO vs GK | 4.316 | <0.001 | |
| Pairwise, pasture | Site | TO vs BO | 3.724 | <0.001 | |
| Pairwise, pasture | Site | GK vs BO | 3.313 | <0.001 | |
| Pairwise, TO | Land use | Pasture vs woodland | 5.171 | <0.001 | |
| Pairwise, GK | Land use | Pasture vs woodland | 2.993 | <0.001 | |
| Pairwise, BO | Land use | Pasture vs woodland | 4.352 | <0.001 |
Pairwise comparisons show differences between sites within land uses and between land uses within sites. Pseudo-F values and t statistics are shown for the main test and pairwise comparisons, respectively. For PERMANOVA, there were 2 factors: site (3 levels [Talmo {TO}, Glenrock {GK}, and Bogo {BO}]) and land use (2 levels [woodland and pasture]).
MVDISP analysis was performed to determine whether the GH48 gene showed patterns of community composition heterogeneity across sites and land uses similar to those of the bacterial and fungal communities. In addition, variability in soil properties was also analyzed by using SIMPER. Table 2 shows that the GH48 gene composition was more heterogeneous in the woodlands than in the pastures. PERMDISP analysis indicated that this difference was significant (P < 0.001). The MVDISP and PERMDISP analyses of individual woodland and pasture plots revealed that while the Talmo and Bogo pastures had low heterogeneity levels, the level of heterogeneity of the Glenrock pasture was similar to those of the woodland plots (Table 2). PERMDISP analysis showed that differences in community heterogeneity were not significant between the individual woodland plots and between woodland plots and the Glenrock pasture, but the Talmo and Bogo pastures had significantly lower heterogeneity levels than did all other plots (Table 2).
TABLE 2.
Global multivariate dispersion analysis of GH48, fungal ITS, and bacterial 16S rRNA genes for each land use and SIMPER analysis of soil variablesa
| Test | Factor | MVDISP |
SIMPER avg squared distance for environmental variables | ||
|---|---|---|---|---|---|
| GH48 | Fungi | Bacteria | |||
| Land use | Woodland | 1.186* | 1.323* | 0.999 | 23.76 |
| Pasture | 0.807* | 0.677* | 1.001 | 13.87 | |
| Individual plot | TW | 1.214ab | 1.384abcd | 1.368abcd | 19.45 |
| GW | 1.314f | 1.286ehij | 0.816bf | 22.75 | |
| BW | 1.124ch | 1.561cfikm | 1.345efgh | 31.20 | |
| TP | 0.456acde | 0.514aefg | 0.809ae | 13.68 | |
| GP | 1.232dg | 0.456bhkl | 0.78cg | 15.07 | |
| BP | 0.622befgh | 0.778dgjlm | 0.855dh | 18.00 | |
Shared symbols within each MVDISP column indicate significant differences in the homogeneity of dispersion between land uses (asterisks) or between two specific plots (roman superscript letters). TW, Talmo woodland; GW, Glenrock woodland; BW, Bogo woodland; TP, Talmo pasture; GP, Glenrock pasture; BP, Bogo pasture.
Bacterial heterogeneity levels were similar between the combined woodland and pasture plots, while when each individual plot was analyzed, the Talmo and Bogo woodlands had the highest heterogeneity levels. Fungal communities were more heterogeneous in the woodlands, and higher levels of heterogeneity of the woodland fungal communities were observed both when all woodlands were compared to all pastures and when each individual plot was analyzed (Table 2). The variability of soil parameters (Table 2) followed a pattern similar to that seen for GH48 gene and fungal community heterogeneity and was higher at the woodland plots.
Factors driving GH48, general bacterial, and fungal community composition.
Multivariate correlation models were built by using DistLM to provide a quantitative description of the contributions of each measured soil property to the observed patterns of variability in community composition for the GH48, 16S rRNA, and fungal ITS genes (Table 3). When the combined land use data set was analyzed, a similar set of soil variables made the greatest contribution to the variability of the three groups investigated, in particular the C/N ratio and soil moisture. Notable differences between groups include a greater importance of organic P and MBC in the bacterial model; organic P in particular was the second most important factor in explaining bacterial community composition variability, whereas this variable had little impact on the GH48 functional community and fungal models. Predicted HOC explained a higher proportion of GH48 community composition variability than for the other two groups. Predicted ROC and POC, DON, NH4+-N, MBC, clay, and inorganic P were included in all three models but generally made smaller contributions to explaining community composition variability. A greater proportion of community composition variability was explained in the bacterial model (R2 = 0.35) than for GH48 (R2 = 0.23) or fungal (R2 = 0.25) communities.
TABLE 3.
DistLM models showing variables that best explain variation in GH48, bacterial 16S rRNA, and fungal ITS community compositionsa
| Land use | Microbial community | Factorsb (% contribution) | R2 |
|---|---|---|---|
| Woodland + pasture | GH48 | C/N ratio (9.06), soil moisture (5.51), pH (2.31), clay (2.30), HOC (2.00), POC (1.50), MBC (1.41), DON (0.98), Inorg P (0.83), ROC (0.73), MBN (0.64), NH4+-N (0.57) | 0.23 |
| Bacteria | C/N ratio (7.84), Org P (5.67), MBC (5.13), soil moisture (3.79), pH (2.81), clay (2.50), Inorg P (1.44), ROC (1.27), POC (0.92), DOC (0.88), DON (0.86), HOC (0.69), NH4+-N (0.59), FAA-N (0.57) | 0.35 | |
| Fungi | C/N ratio (9.29), soil moisture (3.85), pH (2.06), Inorg P (1.72), clay (1.38), POC (1.14), HOC (1.02), ROC (0.86), Org P (0.65), FAA-N (0.63), MBC (0.59), DON (0.57), NH4+-N (0.53), NO3−-N (0.47) | 0.25 | |
| Woodland | GH48 | C/N ratio (5.47), pH (5.44), HOC (2.21), soil moisture (2.08), FAA-N (1.49) | 0.16 |
| Bacteria | pH (10.65), C/N ratio (5.86), NH4+-N (3.72), MBC (1.96), DOC (1.96), DON (1.86), clay (1.59), MBN (1.40), LOI (1.36), Inorg P (1.32), FAA-N (1.15), ROC (1.13), NO3−-N (1.02), HOC (0.99) | 0.36 | |
| Fungi | pH (3.51), C/N ratio (2.57), LOI (1.80), MBC (1.26), ROC (1.14), POC (1.05), FAA-N (1.05) | 0.13 | |
| Pasture | GH48 | Soil moisture (11.95), HOC (4.44), clay (2.31), POC (1.71), pH (1.23) | 0.21 |
| Bacteria | Inorg P (16.09), soil moisture (11.80), MBN (5.51), clay (3.40), pH (3.06), ROC (2.12), HOC (2.05), POC (1.19), DON (1.40) | 0.47 | |
| Fungi | Soil moisture (13.81), Inorg P (5.28), clay (1.81), pH (1.65), MBN (1.31), Org P (1.29), HOC (1.17), POC (1.14), ROC (0.97), C/N ratio (0.96) | 0.29 |
Variables are ordered based on decreasing percent contributions to the total explained variability (in parentheses). R2 values indicate the overall amount of variability explained by the model. HOC is shown in boldface type to highlight its importance for the different groups. Org, organic; Inorg, inorganic.
With a P value of <0.05.
In woodland samples alone, pH explained a level of variability similar to (for the GH48 gene and fungal communities) or higher than (for bacteria) that of the C/N ratio compared to the combined land use models. Predicted HOC and soil moisture were relatively important in the GH48 gene model but did not contribute to a significant extent or at all in explaining variability in the bacterial and fungal DistLM models. The bacterial model explained more community composition variability (R2 = 0.36), followed by the GH48 (R2 = 0.16) and fungal (R2 = 0.13) community models.
Soil moisture explained more community composition variability in the pasture than in the woodland DistLM models. While inorganic P was the variable that explained the most variability in the bacterial community and was the second most important variable in the fungal model, it was not part of the GH48 community model. Predicted HOC was the second variable that most explained the variability of the GH48 community model, whereas in the bacterial and fungal models, predicted HOC was comparatively less important. Clay, pH, and predicted POC were included in the models of the three groups. As with the combined land use model, the bacterial model had the largest amount of explained variability (R2 = 0.47), followed by the fungal (R2 = 0.29) and GH48 (R2 = 0.21) models.
Total and relative abundances of GH48, 16S rRNA, and fungal ITS genes.
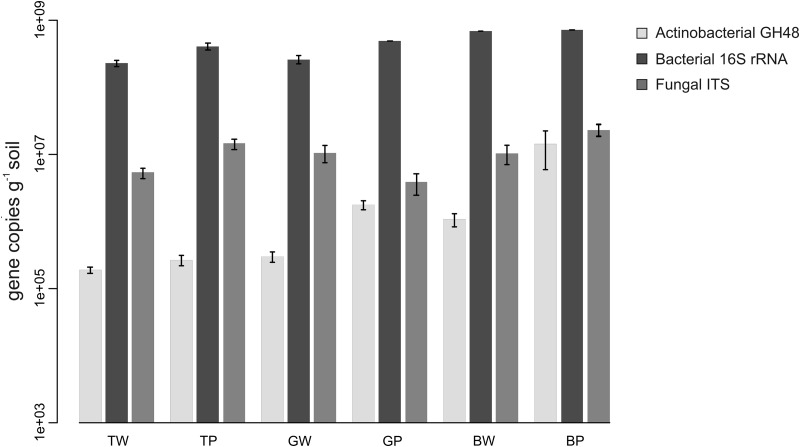
The abundance of the GH48 gene was determined and compared to those of the bacterial 16S rRNA and fungal ITS genes in order to estimate the relative importance of actinobacterial saprotrophs to the overall microbial community (Fig. 3). The results show that while the GH48 abundances at Talmo were similar for both land uses, the abundances in pastures at the other sites were 5- to 13-fold greater than in the woodlands in Glenrock and Bogo, respectively. Bacterial 16S rRNA abundance was greater in the pastures than in the woodlands at Talmo (1.79-fold) and Glenrock (1.9-fold) but not at Bogo (1.04-fold). The fungal ITS total abundance was greater in pastures than in woodlands at Talmo (2.7-fold) and Bogo (2.2-fold) but not at Glenrock (0.34-fold).
FIG 3.
Total abundance per gram of soil of actinobacterial GH48, bacterial 16S rRNA, and fungal ITS genes. Error bars are standard errors. TW, Talmo woodland; GW, Glenrock woodland; BW, Bogo woodland; TP, Talmo pasture; GP, Glenrock pasture; BP, Bogo pasture.
DISCUSSION
GH48 PCR primer specificity.
In silico analysis of standard PCR primers developed here indicated that the two primer sets are broadly complementary in their actinobacterial GH48 diversity coverage and in combination cover almost all the actinobacterial GH48 diversity from cultured strains available in GenBank (see Tables S1 and S2 in the supplemental material). The high level of specificity of the GH48 primers for actinobacterial GH48 genes indicated by in silico analysis is corroborated by the fact that only 1 out of 87 high-quality clone sequences represented a non-GH48 sequence. One nonactinobacterial GH48 sequence (Hahella chejuensis) was a predicted target of primer pair GH48_F1-GH48_R1. This species is a member of the phylum Gammaproteobacteria and acquired the GH48 gene by horizontal gene transfer (19). Phylogenetic analysis of the GH48 clone sequences indeed indicated that 4 clones may be of nonactinobacterial origin. This represents a relatively low level of nonspecificity (<5%), and in any case, the gene amplified was a GH48 gene, indicating that these primers are highly specific to the targeted gene ecological function for which they were designed.
The in silico specificity analysis of the GH48 qPCR primer pair developed here showed that this primer pair was unlikely to amplify any template other than actinobacterial GH48 genes; however, the qPCR primers did not cover all actinobacterial GH48 gene diversity from cultured actinobacterial strains and therefore provided an underestimation of the actual actinobacterial GH48 gene abundances (see Tables S1 and S2 in the supplemental material).
As with any PCR-based approach, the standard and quantitative PCR primers developed here will not amplify GH48 genes that do not share sequence similarity in the PCR primer target sites. However, application of these primers for the determination of diversity and abundance patterns across a large number of samples, in concert with the determination of soil properties, allows a better understanding of the ecological role of saprotrophic actinobacteria in soil ecology and carbon cycling.
Diversity of actinobacterial GH48 genes in soils.
Phylogenetic analysis of soil GH48 sequences revealed significant new diversity not covered in gene databases (Fig. 1; see also Fig. S2 in the supplemental material). The majority of sequences were located in clusters without any cultured representative, although a large number of sequences clustered with high bootstrap support with Catenulispora acidiphila, and a smaller number of soil clones clustered with Streptomyces spp., Actinoplanes spp., and Verrucosispora maris. GH48 gene sequence data are available for only 17 actinobacterial genera, and this low coverage of GH48 gene diversity from cultured actinobacteria precludes a better understanding of the phylogenetic identity of the soil clones obtained here. No obvious clustering of sequences derived from woodlands or pastures, or from a specific site, was observed.
All the soil clones obtained showed the presence of the catalytic base aspartic acid 225, as determined for T. fusca (Fig. 1), which plays the essential role of activating the catalytic water molecule that allows hydrolysis of the β-1,4 glycosidic bonds in cellulose (54). Additionally, conserved residues present in the targeted region that are involved in substrate recognition were conserved in the majority of the clones obtained here (see Table S3 in the supplemental material). The three conserved residues whose presence was more variable in the clones obtained were also absent in the GH48 protein sequence of the model cellulolytic organism T. fusca or were not involved in substrate recognition, which suggests that their presence is not essential for cellulose degradation activity in actinobacteria. Furthermore, all but 6 soil clones showed the presence of either or both aromatic amino acids (F195 and Y213) in the entrance of the GH48 catalytic tunnel (Fig. 1) (57). The absence of the F195 and Y213 residues would not necessarily indicate a lack of cellulolytic function; indeed, the sequences from Xylanimonas cellulolytica, Actinosynnema mirum, and Nocardiopsis dassonvillei, which are known to possess the ability to degrade cellulose (43), lack the Y213 aromatic residue. The presence of the catalytic base, conserved residues involved in substrate recognition, and key aromatic amino acid residues in the majority of the GH48 sequences obtained in this study is an indication that these clones represent functional cellobiohydrolases.
Actinobacterial GH48 gene community diversity across land uses.
PCO analysis showed that C/N ratios and soil moisture were the two main drivers of GH48 community composition across all sites and land uses, opposing most Talmo pasture samples (high soil moisture) to Talmo, Bogo, and Glenrock woodlands (high C/N ratio) (Fig. 2A). The C/N ratio is thought to influence litter C availability, and a high C/N ratio has a negative effect on extracellular hydrolytic enzymes during litter decomposition (69, 70). It is not surprising, therefore, that the C/N ratio was one of the vectors with the strongest correlations to the main PCO axis and was more important than the levels of the organic soil carbon fractions themselves.
As with the GH48 gene PCO analysis, the C/N ratio and soil moisture were also the two main drivers of the overall bacterial and fungal community compositions (Fig. 2B and C); however, other soil properties were comparatively more important in explaining community composition shifts of these broader groups than those of the GH48 gene community composition. As the primers developed here target a more specific microbial functional group (i.e., actinobacterial saprotrophs), the lesser importance of other soil variables may simply reflect the narrower physiological range of the GH48 gene community.
While clustering of samples based on land use and site can be discerned, some overlap between samples from different sites and land uses was observed, particularly between Talmo woodland, Glenrock woodland, and Glenrock pasture samples. PERMANOVA was able to demonstrate that the actinobacterial GH48 community composition was significantly different between all sites and between land uses, while pairwise comparisons confirmed that the GH48 community composition was significantly different between pasture and woodlands at every site and between sites for both land uses (Table 1).
The β-diversity estimated by using multivariate dispersion analysis showed that the actinobacterial GH48 and fungal communities were more heterogeneous in the woodlands, while the β-diversity of the bacterial community was more similar between the two land uses (Table 2). The heterogeneity of the GH48 and fungal communities broadly tracked the variability of soil properties, which was also greater in the woodland samples. These results suggest that the community assembly processes of GH48 and fungal communities are more strongly influenced by the measured soil properties than is the case for the bacterial community. The similarity in heterogeneity patterns between the GH48 and fungal communities is interesting given that a significant proportion of soil fungi have a similar, but not identical, ecological niche in soil (i.e., soil saprotrophs capable of degrading plant cell wall polysaccharides).
In the Glenrock pasture, the GH48 community was as heterogeneous as those in the woodland plots. It is not possible to determine the reason for the high levels of heterogeneity at the Glenrock pasture, but this site had the highest and most variable overall C/N ratios of all pastures (46). Given the importance of C/N ratios in structuring GH48 community composition, it is possible that the ecology of the actinobacterial GH48 community at the Glenrock pasture was more similar to those of the woodlands than to those of the other pastures investigated here.
Influence of soil carbon on GH48, bacterial, and fungal community composition.
Predicted HOC was found to be consistently more important to the variability of the GH48 community than to the general bacterial and fungal communities based on DistLM analysis (Table 3). These results agree with the saprotrophic life-style expected for GH48 gene-carrying actinobacteria (34, 71), which represent a more ecologically defined microbial group than the overall bacterial and fungal communities. While other bacterial and fungal groups are also likely to have a saprotrophic role in these soils, the general fungal and bacterial communities analyzed here represent broad metabolic groups with diverse ecological roles, and their association with soil organic fractions is therefore less obvious. Our data indicate, therefore, that we were successful in targeting actinobacteria with a saprotrophic life-style that are likely involved in organic C dynamics in these soils.
The precise nature of the predicted HOC fraction measured here is currently unknown; further studies are necessary to determine whether cellulose is a component of this fraction and whether saprotrophic actinobacteria are able to scavenge cellulose from a matrix of less bioavailable compounds such as peptidoglycan, lignin, and lipids (72, 73). The association of saprotrophic actinobacteria with soil organic carbon found here agrees with the results of Baldrian et al. (74), who showed increased actinobacterial abundance in the deeper humic horizon of forest soils.
HOC was the most abundant fraction of predicted soil organic carbon in the sites studied, representing ∼50% of the combined carbon from all MIR-resolved C fractions (46), and this is also the case in a range of soils from Australia (47). The data obtained here therefore highlight the potential importance of soil actinobacteria in the turnover of this more recalcitrant C pool and, thus, their impact on the carbon cycle.
Other factors influencing community composition.
Soil moisture in the woodlands was noticeably more important for the GH48 community model than for the general bacterial and fungal models (Table 3). The woodland soils at the time of sampling were relatively dry and considerably drier than the pastures (46), where moisture was one of the most important variables for the three microbial groups investigated. It is possible that the greater importance of moisture in the woodland GH48 than in the bacterial and fungal DistLM models was because their community composition, as tracked by their DNA, more accurately reflected the active community, while a large proportion of the community composition tracked by the general 16S rRNA and ITS genes may have been inactive or senescent, capturing growth that occurred during past periods of higher moisture. This is corroborated by a study showing that actinobacteria were more dominant in the RNA rather than in the DNA fractions in spruce forest soils, particularly in the deeper humic horizon (74). Inorganic P was the first and second most important soil variable explaining variability in the bacterial and fungal models in the pastures, respectively; the impact of P fertilization on soil bacterial and fungal community composition was documented previously (75). In contrast, the best GH48 community pasture DistLM model did not include inorganic P. The soil P levels determined in this study represent an approximation of soluble, plant-available organic and inorganic P (76, 77), and the application of P fertilizer was the major anthropologically driven difference between each of the paired woodland and pasture sites. The low importance of inorganic P in actinobacterial GH48 models suggests that these organisms obtain most of their P requirements directly from decomposing organic matter (which, unlike soluble orthophosphate, is not immediately plant available), even when more easily available inorganic P is present.
DistLM analysis further revealed that for the GH48, bacterial, and fungal communities, pH and particularly the C/N ratio played smaller roles in structuring composition in the pastures than in the woodlands, while moisture was considerably more important in the pastures than in the woodlands. C/N ratios in the pasture soils were lower (52 to 65%) and less variable than those in the woodlands (46), which probably accounts for their smaller impact on community composition in the pastures. Moisture and pH were more correlated with each other in the pastures (r = 0.67) than in the woodlands (r = 0.30); their cocorrelation most likely decreased the additional variability explained by pH, once the variability explained by moisture was included in the DistLM model.
GH48 abundance in relation to bacteria and fungi.
The quantification of GH48 genes showed a higher abundance of GH48 in the Glenrock and Bogo pastures, whereas bacterial and fungal abundances in these pastures were similar to or lower than those in the woodlands (Fig. 3). As the qPCR primers developed here do not cover all of the cultured GH48 gene diversity, it is not possible to determine whether the higher abundances in the Glenrock and Bogo pastures are caused by higher total gene abundances or by differences in the taxonomical composition of the GH48 community between these pastures and the other plots. However, it is interesting that the Bogo pasture had the lowest overall C/N ratio and the highest overall GH48 abundance; likewise, the Bogo woodlands had the lowest C/N ratio and the highest GH48 abundance among woodlands (46). This pattern agrees with data from studies showing that higher C/N ratios are negatively correlated with extracellular hydrolytic enzyme activity during litter decomposition (69).
Conclusions.
This study is to our knowledge the first soil-based, landscape-scale investigation of the diversity of a cellobiohydrolase gene in relation to different land uses and soil properties. Our data revealed significant new diversity of actinobacterial GH48 genes and that the C/N ratio and moisture are primary factors driving GH48 community composition in the soils studied. Given the ubiquity and abundance of actinobacteria in soils, their role in soil carbon cycling clearly merits further attention. Finally, we have laid the groundwork necessary for further studies to investigate the diversity of actinobacterial GH48 genes in soil. Future studies focusing on in situ expression of this cellobiohydrolase gene should provide a better understanding of the ecological role of these organisms in soil C cycling and their interactions with other soil saprotrophs.
Supplementary Material
ACKNOWLEDGMENTS
We are particularly grateful to Tony Armour (Glenrock), Chris Shannon (Talmo), and Malcolm Peake (Bogo) for their enthusiastic support of this research and their willingness to allow us access to their properties. We thank Shamsul Hoque (CPI) for providing excellent technical support in the laboratory and field.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00067-15.
REFERENCES
- 1.Ljungdahl LG, Eriksson KE. 1985. Ecology of microbial cellulose degradation. Adv Microb Ecol 8:237–299. doi: 10.1007/978-1-4615-9412-3_6. [DOI] [Google Scholar]
- 2.Henrissat B. 1998. Glycosidase families. Biochem Soc Trans 26:153–156. [DOI] [PubMed] [Google Scholar]
- 3.Qi M, Wang P, O'Toole N, Barboza PS, Ungerfeld E, Leigh MB, Selinger LB, Butler G, Tsang A, McAllister TA, Forster RJ. 2011. Snapshot of the eukaryotic gene expression in muskoxen rumen—a metatranscriptomic approach. PLoS One 6:e20521. doi: 10.1371/journal.pone.0020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, Feng JX. 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol 107:245–256. doi: 10.1111/j.1365-2672.2009.04202.x. [DOI] [PubMed] [Google Scholar]
- 6.Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Dos Santos V, Strompl C, Elborough K, Jarvis G, Neef A, Yakimov MM, Timmis KN, Golyshin PN. 2005. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol 7:1996–2010. doi: 10.1111/j.1462-2920.2005.00920.x. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Duan CJ, Pang H, Mo XC, Wu CF, Yu Y, Hu YL, Wei J, Tang JL, Feng JX. 2007. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl Microbiol Biotechnol 75:319–328. doi: 10.1007/s00253-006-0820-9. [DOI] [PubMed] [Google Scholar]
- 8.Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomás AA, Foster CE, Pauly M, Weimer PJ, Barry KW, Goodwin LA, Bouffard P, Li L, Osterberger J, Harkins TT, Slater SC, Donohue TJ, Currie CR. 2010. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet 6:e1001129. doi: 10.1371/journal.pgen.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang H, Zhang P, Duan C-J, Mo X-C, Tang J-L, Feng J-X. 2009. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr Microbiol 58:404–408. doi: 10.1007/s00284-008-9346-y. [DOI] [PubMed] [Google Scholar]
- 10.Allgaier M, Reddy A, Park JI, Ivanova N, D'Haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, VanderGheynst JS, Hugenholtz P. 2010. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One 5:e8812. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beloqui A, Nechitaylo TY, López-Cortés N, Ghazi A, Guazzaroni M-E, Polaina J, Strittmatter AW, Reva O, Waliczek A, Yakimov MM, Golyshina OV, Ferrer M, Golyshin PN. 2010. Diversity of glycosyl hydrolases from cellulose-depleting communities enriched from casts of two earthworm species. Appl Environ Microbiol 76:5934–5946. doi: 10.1128/AEM.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang XN, Hernández M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 13.Uroz S, Buee M, Murat C, Frey-Klett P, Martin F. 2010. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288. doi: 10.1111/j.1758-2229.2009.00117.x. [DOI] [PubMed] [Google Scholar]
- 14.Takasaki K, Miura T, Kanno M, Tamaki H, Hanada S, Kamagata Y, Kimura N. 2013. Discovery of glycoside hydrolase enzymes in an avicel adapted forest soil fungal community by a metatranscriptomic approach. PLoS One 8:e55485. doi: 10.1371/journal.pone.0055485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert HJ, Hazlewood GP, Laurie JI, Orpin CG, Xue GP. 1992. Homologous catalytic domains in a rumen fungal xylanase—evidence for gene duplication and prokaryotic origin. Mol Microbiol 6:2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Vallvé S, Palau J, Romeu A. 1999. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol 16:1125–1134. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Vallvé S, Romeu A, Palau J. 2000. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol Biol Evol 17:352–361. doi: 10.1093/oxfordjournals.molbev.a026315. [DOI] [PubMed] [Google Scholar]
- 19.Sukharnikov LO, Alahuhta M, Brunecky R, Upadhyay A, Himmel ME, Lunin VV, Zhulin IB. 2012. Sequence, structure, and evolution of cellulases in glycoside hydrolase family 48. J Biol Chem 287:41068–41077. doi: 10.1074/jbc.M112.405720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomares-Rius JE, Hirooka Y, Tsai IJ, Masuya H, Hino A, Kanzaki N, Jones JT, Kikuchi T. 2014. Distribution and evolution of glycoside hydrolase family 45 cellulases in nematodes and fungi. BMC Evol Biol 14:69. doi: 10.1186/1471-2148-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quillet L, Barray S, Labedan B, Petit F, Guespin-Michel J. 1995. The gene encoding the β-1,4-endoglucanase (celA) from Myxococcus xanthus—evidence for independent acquisition by horizontal transfer of binding and catalytic domains from actinomycetes. Gene 158:23–29. doi: 10.1016/0378-1119(95)00091-J. [DOI] [PubMed] [Google Scholar]
- 22.Izquierdo JA, Sizova MV, Lynd LR. 2010. Diversity of bacteria and glycosyl hydrolase family 48 genes in cellulolytic consortia enriched from thermophilic biocompost. Appl Environ Microbiol 76:3545–3553. doi: 10.1128/AEM.02689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson DG, Tripathi SA, Giannone RJ, Lo J, Caiazza NC, Hogsett DA, Hettich RL, Guss AM, Dubrovsky G, Lynd LR. 2010. Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc Natl Acad Sci U S A 107:17727–17732. doi: 10.1073/pnas.1003584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin DC, Zhang S, Wilson DB. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur J Biochem 267:4988–4997. doi: 10.1046/j.1432-1327.2000.01546.x. [DOI] [PubMed] [Google Scholar]
- 25.Devillard E, Goodheart DB, Karnati SKR, Bayer EA, Lamed R, Miron J, Nelson KE, Morrison M. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J Bacteriol 186:136–145. doi: 10.1128/JB.186.1.136-145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steenbakkers PJM, Freelove A, van Cranenbroek B, Sweegers BMC, Harhangi HR, Vogels GD, Hazlewood GP, Gilbert HJ, Op den Camp HJ. 2002. The major component of the cellulosomes of anaerobic fungi from the genus Piromyces is a family 48 glycoside hydrolase. DNA Seq 13:313–320. doi: 10.1080/1042517021000024191. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez-Ramírez N, Romero-García ER, Calderón VC, Avitia CI, Téllez-Valencia A, Pedraza-Reyes M. 2008. Expression, characterization and synergistic interactions of Myxobacter sp AL-1 Cel9 and Cel48 glycosyl hydrolases. Int J Mol Sci 9:247–257. doi: 10.3390/ijms9030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen H, Gilkes NR, Kilburn DG, Miller RC, Warren RAJ. 1995. Cellobiohydrolase B, a second exo-cellobiohydrolase from the cellulolytic bacterium Cellulomonas fimi. Biochem J 311:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CC, Doi RH. 1998. Properties of exgS, a gene for a major subunit of the Clostridiun cellulovorans cellulosome. Gene 211:39–47. doi: 10.1016/S0378-1119(98)00081-X. [DOI] [PubMed] [Google Scholar]
- 30.Berger E, Zhang D, Zverlov VV, Schwarz WH. 2007. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol Lett 268:194–201. doi: 10.1111/j.1574-6968.2006.00583.x. [DOI] [PubMed] [Google Scholar]
- 31.Pereyra LP, Hiibel SR, Riquelme MVP, Reardon KF, Pruden A. 2010. Detection and quantification of functional genes of cellulose-degrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl Environ Microbiol 76:2192–2202. doi: 10.1128/AEM.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CY, Wang W, Du MF, Li CF, Ma CQ, Xu P. 2013. Pulp mill wastewater sediment reveals novel methanogenic and cellulolytic populations. Water Res 47:683–692. doi: 10.1016/j.watres.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zhong Y, Yang S, Zhang W, Xu M, Ma A, Zhuang G, Chen G, Liu W. 2014. Diversity and dynamics of the microbial community on decomposing wheat straw during mushroom compost production. Bioresour Technol 170:183–195. doi: 10.1016/j.biortech.2014.07.093. [DOI] [PubMed] [Google Scholar]
- 34.Goodfellow M, Williams ST. 1983. Ecology of Actinomycetes. Annu Rev Microbiol 37:189–216. doi: 10.1146/annurev.mi.37.100183.001201. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy AJ. 1987. Lignocellulose-degrading actinomycetes. FEMS Microbiol Rev 46:145–163. doi: 10.1111/j.1574-6968.1987.tb02456.x. [DOI] [Google Scholar]
- 36.de Menezes A, Clipson N, Doyle E. 2012. Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil. Environ Microbiol 14:2577–2588. doi: 10.1111/j.1462-2920.2012.02781.x. [DOI] [PubMed] [Google Scholar]
- 37.Phelan MB, Crawford DL, Pometto AL. 1979. Isolation of lignocellulose-decomposing actinomycetes and degradation of specifically labelled 14C-labelled lignocelluloses by six selected Streptomyces strains. Can J Microbiol 25:1270–1276. doi: 10.1139/m79-200. [DOI] [PubMed] [Google Scholar]
- 38.Ulrich A, Klimke G, Wirth S. 2008. Diversity and activity of cellulose-decomposing bacteria, isolated from a sandy and a loamy soil after long-term manure application. Microb Ecol 55:512–522. doi: 10.1007/s00248-007-9296-0. [DOI] [PubMed] [Google Scholar]
- 39.de Menezes AB, Lockhart RJ, Cox MJ, Allison HE, McCarthy AJ. 2008. Cellulose degradation by micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA gyrase B gene sequencing. Appl Environ Microbiol 74:7080–7084. doi: 10.1128/AEM.01092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabinovich ML, Melnik MS, Boloboba AV. 2002. Microbial cellulases (review). Appl Biochem Microbiol 38:305–322. doi: 10.1023/A:1016264219885. [DOI] [Google Scholar]
- 41.Teeri TT. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol 15:160–167. [Google Scholar]
- 42.Koeck DE, Pechtl A, Zverlov VV, Schwarz WH. 2014. Genomics of cellulolytic bacteria. Curr Opin Biotechnol 29:171–183. doi: 10.1016/j.copbio.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Anderson I, Abt B, Lykidis A, Klenk H-P, Kyrpides N, Ivanova N. 2012. Genomics of aerobic cellulose utilization systems in Actinobacteria. PLoS One 7:e39331. doi: 10.1371/journal.pone.0039331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kögel-Knabner I. 2002. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol Biochem 34:139–162. doi: 10.1016/S0038-0717(01)00158-4. [DOI] [Google Scholar]
- 45.Austin AT, Ballaré CL. 2010. Dual role of lignin in plant litter decomposition in terrestrial ecosystems. Proc Natl Acad Sci U S A 107:4618–4622. doi: 10.1073/pnas.0909396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH. 11 July 2014. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol doi: 10.1111/1462-2920.12559. [DOI] [PubMed] [Google Scholar]
- 47.Baldock JA, Hawke B, Sanderman J, Macdonald LM. 2013. Predicting contents of carbon and its component fractions in Australian soils from diffuse reflectance mid-infrared spectra. Soil Res 51:577–583. doi: 10.1071/SR13077. [DOI] [Google Scholar]
- 48.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu W, Zhou Y, Zhang Y, Lu Y, Wang X, Zhao D, Yang Y, Zhang C. 2012. MFEprimer-2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res 40:W205–W208. doi: 10.1093/nar/gks552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 52.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 53.Letunic I, Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 54.Kostylev M, Wilson DB. 2011. Determination of the catalytic base in family 48 glycosyl hydrolases. Appl Environ Microbiol 77:6274–6276. doi: 10.1128/AEM.05532-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsiegla G, Reverbel C, Tardif C, Driguez H, Haser R. 2008. Structures of mutants of cellulase Ce148F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action. J Mol Biol 375:499–510. doi: 10.1016/j.jmb.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 56.Guimarães BG, Souchon H, Lytle BL, Wu JHD, Alzari PM. 2002. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J Mol Biol 320:587–596. doi: 10.1016/S0022-2836(02)00497-7. [DOI] [PubMed] [Google Scholar]
- 57.Kostylev M, Alahuhta M, Chen M, Brunecky R, Himmel ME, Lunin VV, Brady J, Wilson DB. 2014. Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues. Biotechnol Bioeng 111:664–673. doi: 10.1002/bit.25139. [DOI] [PubMed] [Google Scholar]
- 58.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 59.Muyzer G, Dewaal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White TJ, Burns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis M, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols. Academic Press, San Diego, CA. [Google Scholar]
- 61.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdo Z, Schüette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol 8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- 63.Ramette A. 2009. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl Environ Microbiol 75:2495–2505. doi: 10.1128/AEM.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Development Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 65.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 67.Clarke K, Gorley R. 2006. PRIMER v6: user manual/tutorial. Primer-E Ltd, Plymouth, United Kingdom. [Google Scholar]
- 68.Anderson M, Gorley R, Clarke K. 2008. PERMANOVA+ for PRIMER, user manual. Primer-E Ltd, Plymouth, United Kingdom. [Google Scholar]
- 69.Leitner S, Wanek W, Wild B, Haemmerle I, Kohl L, Keiblinger KM, Zechmeister-Boltenstern S, Richter A. 2012. Influence of litter chemistry and stoichiometry on glucan depolymerization during decomposition of beech (Fagus sylvatica L.) litter. Soil Biol Biochem 50:174–187. doi: 10.1016/j.soilbio.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geisseler D, Horwath WR. 2009. Relationship between carbon and nitrogen availability and extracellular enzyme activities in soil. Pedobiologia 53:87–98. doi: 10.1016/j.pedobi.2009.06.002. [DOI] [Google Scholar]
- 71.Goodfellow M. 2012. Phylum XXVI. Actinobacteria phyl. nov. In Goodfellow M, Kampfer P, Busse H, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology, vol 5 The Actinobacteria, part A. Springer, New York, NY. [Google Scholar]
- 72.Simpson AJ, Song G, Smith E, Lam B, Novotny EH, Hayes MHB. 2007. Unraveling the structural components of soil humin by use of solution-state nuclear magnetic resonance spectroscopy. Environ Sci Technol 41:876–883. doi: 10.1021/es061576c. [DOI] [PubMed] [Google Scholar]
- 73.Spence A, Simpson AJ, McNally DJ, Moran BW, McCaul MV, Hart K, Paull B, Kelleher BP. 2011. The degradation characteristics of microbial biomass in soil. Geochim Cosmochim Acta 75:2571–2581. doi: 10.1016/j.gca.2011.03.012. [DOI] [Google Scholar]
- 74.Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Vv̆trovský T, Zifčáková L, Šnajdr J, Rídl J, Vlček C, Voříšková J. 2012. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rooney DC, Clipson NJW. 2009. Phosphate addition and plant species alters microbial community structure in acidic upland grassland soil. Microb Ecol 57:4–13. doi: 10.1007/s00248-008-9399-2. [DOI] [PubMed] [Google Scholar]
- 76.Irving GCJ, McLaughlin MJ. 1990. A rapid and simple field test for phosphorus in Olsen and Bray no. 1 extracts of soil. Commun Soil Sci Plant Anal 21:2245–2255. doi: 10.1080/00103629009368377. [DOI] [Google Scholar]
- 77.Olsen SR, Sommers LE. 1982. Phosphorus, p 403–430. In Page AL. (ed), Methods of soil analysis, part 2. Chemical and microbiological properties. American Society of Agronomy, Madison, WI. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.