Abstract
In this study, we investigated the transcriptomic response of Streptococcus pneumoniae D39 to sialic acid (N-acetylneuraminic acid [Neu5Ac]). Transcriptome comparison of wild-type D39 grown in M17 medium with and without sialic acid revealed the elevated expression of various genes and operons, including the nan gene cluster (nan operon I and nanA gene). Our microarray analysis and promoter-lacZ fusion studies showed that the transcriptional regulator NanR acts as a transcriptional activator of nan operon I and the nanA gene in the presence of sialic acid. The putative regulatory site of NanR in the promoter region of nan operon I is predicted and confirmed by promoter truncation experiments. Furthermore, the role of CcpA in the regulation of the nan gene cluster is demonstrated through microarray analysis and promoter-lacZ fusion studies, suggesting that in the presence of sialic acid and glucose, CcpA represses the expression of nan operon I while the expression of the nanA gene is CcpA independent.
INTRODUCTION
The low-GC Gram-positive bacterium Streptococcus pneumoniae is a major human pathogen and the causal agent of many diseases, including pneumonia, sepsis, meningitis, otitis media, and conjunctivitis, which result in over a million deaths each year worldwide (1, 2). S. pneumoniae colonizes the human nasopharynx during the first few months of life (3). For survival in the host, bacteria rely not only on the different virulence factors they possess but also on the appropriate use of nutrients available in their habitat (4, 5). The ability of S. pneumoniae to utilize a variety of carbohydrate sources is one of the crucial factors in successful colonization and in causing pneumococcal infections (6). Metabolic selection enables a bacterium to choose a preferred source of carbon over a nonpreferred one through a mechanism called carbon catabolite repression (CCR) (5). CcpA (carbon catabolite protein A) is a transcription factor that mediates CCR in the presence of a preferred sugar source, e.g., glucose, by binding to catabolite repression elements (cre boxes) found in the promoter regions of CcpA-targeted genes (5, 7–9). Despite the importance of carbohydrates in the pathogenesis of S. pneumoniae, research concerning metabolic pathways of S. pneumoniae still demands more attention.
Sialic acid is one of the most important carbohydrates for S. pneumoniae, since it plays a vital role as a carbon/energy source, a receptor for adhesion and invasion, and a molecular signal for the promotion of biofilm formation, nasopharyngeal carriage, and invasion of the lungs (10). It has been shown that S. pneumoniae can utilize sialic acid as a carbon source, which results in improved pneumococcal biofilm formation in vitro, at concentrations comparable to those of free sialic acid in human saliva (11, 12). Of the 43 known naturally occurring derivatives of the nine-carbon sugar neuraminic acid, N-acetylneuraminic acid (Neu5Ac) is the only one found in humans (13). Another form of sialic acid is N-glycolylneuraminic acid (Neu5Gc), which is not synthesized by humans. Some factors, including a number of extracellular glycosidases and carbohydrate transporters, affect the interaction of the bacterium with the host (10). At least 10 surface-associated glycosidases (NanA, NanB, NanC, BgaA, EndoD, Eng, Hyl, SpuA, BgaC, and StrH) are expressed in S. pneumoniae, and these glycosidases scavenge and modify the host glycans and sialic acid residues (14) to use them as a carbon source for growth, making host clearance multifunctional molecules, helping the pneumococcus to fight against other bacteria for a niche, assisting it in driving through the mucin film, and stimulating its attachment to epithelial cells (15–20). Three neuraminidases are encoded by the genome of S. pneumoniae (NanA, NanB, and NanC), and their importance in pathogenicity has made pneumococcal neuraminidases the most-studied surface-located glycosyl-hydrolases (10). The regulatory mechanisms of neuraminidase expression are important, since release of sialic acid from O-glycans has been linked to pathogenesis. Moreover, sialic acid has been proposed as a molecular signal in stimulating in vitro biofilm production and in vivo nasopharyngeal carriage and lung invasion by S. pneumoniae (12, 21). NanA is the best-understood neuraminidase and aids the progression of otitis media in a chinchilla animal model (22) and of respiratory tract infection and sepsis in mice (23). Less is known about NanB, but NanB has also been suggested to play a significant role during pneumococcal infection of the respiratory tract and sepsis (23). NanA and NanB stimulate the pneumococcal colonization of the upper respiratory tract by cleaving sialic acid from cell surface glycans and mucin and by presenting host cell surface receptors for pneumococcal adherence (23, 24). Mouse model studies have also revealed that NanA and NanB are essential for S. pneumoniae infection (23).
The present study was aimed at elucidating the effect of sialic acid (N-acetylneuraminic acid [Neu5Ac]) on the global gene expression of S. pneumoniae. We characterized the nan regulon (nan operon I and the nanA gene) in S. pneumoniae D39 and demonstrated that the transcriptional regulator NanR acts as a transcriptional activator for nan operon I and the nanA gene in the presence of sialic acid. We also demonstrated that regulation of nan operon I is CcpA dependent, whereas the regulation of the nanA gene is CcpA independent in the presence of glucose and sialic acid. The putative regulatory site (5′-TCTGAAASTACTTTCARA-3′) of NanR in the promoter region of nanE is also predicted and confirmed by promoter truncation experiments and was found to be highly conserved in other pneumococcal strains and streptococci.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA isolation and modification.
Bacterial strains and plasmids used in this study are listed in Table 1. The S. pneumoniae strain D39 was grown as described previously (25). For β-galactosidase assays, derivatives of the S. pneumoniae D39 strain were grown in M17 medium supplemented with different sugars (arabinose, cellobiose, dextrose, fructose, fucose, glucose, galactose, lactose, maltose, mannitol, mannose, melibiose, sorbitol, trehalose, and xylose) at concentrations given in Results. For selection on antibiotics, the medium was supplemented with 150 μg/ml spectinomycin and 2.5 μg/ml tetracycline for S. pneumoniae and with 100 μg/ml ampicillin for Escherichia coli. All bacterial strains used in this study were stored in 10% (vol/vol) glycerol at −80°C. All DNA manipulations in this study were performed as described before (26). For PCR amplification, chromosomal DNA of S. pneumoniae D39 (27) was used as a template. Primers used in this study are based on the sequence of the D39 genome and are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. pneumoniae strains | ||
| D39 | Serotype 2 strain, cps-2 | Laboratory of P. Hermans |
| ΔccpA strain | D39 ΔccpA Specr | 8 |
| MA200 | D39 ΔnanR Specr | This study |
| MA201 | D39 ΔbgaA::PnanA-lacZ Tetr | This study |
| MA202 | D39 ΔbgaA::Pspd_1503-lacZ Tetr | This study |
| MA203 | D39 ΔbgaA::PnanE-lacZ Tetr | This study |
| MA204 | MA200 ΔbgaA::PnanA-lacZ Tetr | This study |
| MA205 | MA200 ΔbgaA::Pspd_1503-lacZ Tetr | This study |
| MA206 | MA200 ΔbgaA::PnanE-lacZ Tetr | This study |
| MA207 | ΔccpA ΔbgaA::PnanA-lacZ Tetr | This study |
| MA208 | ΔccpA ΔbgaA::Pspd_1503-lacZ Tetr | This study |
| MA209 | ΔccpA ΔbgaA::PnanE-lacZ Tetr | This study |
| MA210 | D39 ΔbgaA::PnanE-F-lacZ Tetr | This study |
| MA211 | D39 ΔbgaA::PnanE-H-lacZ Tetr | This study |
| MA212 | D39 ΔbgaA::PnanE-N-lacZ Tetr | This study |
| E. coli EC1000 | Kmr; MC1000 derivative carrying a single copy of the pWV1 repA gene in glgB | Laboratory collection |
| Plasmids | ||
| pPP2 | Ampr Tetr; promoterless lacZ; for replacement of bgaA with promoter lacZ fusion; derivative of pPP1 | 30 |
| pORI38 | Specr ori+; repA deletion; derivative of pWV01 | 47 |
| pMA201 | pPP2 PnanA-lacZ | This study |
| pMA202 | pPP2 Pspd_1503-lacZ | This study |
| pMA203 | pPP2 PnanE-lacZ | This study |
| pMA204 | pPP2 PnanE-F-lacZ | This study |
| pMA205 | pPP2 PnanE-H-lacZ | This study |
| pMA206 | pPP2 PnanE-N-lacZ | This study |
TABLE 2.
Primers used in this study
| Primer | Nucleotide sequence (5′→3′) | Restriction site |
|---|---|---|
| nanA-R | CATGGGATCCATATTATTCCCCTTTTCTAAGC | BamHI |
| nanA-F | CATGGAATTCGCTGACTTCGTCAGTTCTATCC | EcoRI |
| spd-1503-R | CATGGGATCCCATCTCTTCTAACCTCCTTTCTC | BamHI |
| spd-1503-F | CATGGAATTCGTCCACTCGAAACAAGTATTGTAAG | EcoRI |
| nanE-Rv | CATGGGATCCCTAATCTGTGGCATCCTCTTTCC | BamHI |
| nanE-Fr | CATGGAATTCCAGAGATGGATGGAGCAATTGC | EcoRI |
| nanE-N | CATGGAATTCGCTGTTTGACTTGGCTAGTTTTTG | EcoRI |
| nanE-H | CATGGAATTCACTTTTAGAGGAGCTGTTTGACTTGG | EcoRI |
| nanE-F | CATGGAATTCCGACTTGCTCCTCTGAAAGTAC | EcoRI |
| nanR-1 | GCCGTCATTTTATTGCTACGCG | |
| nanR-2 | GCATAGGCGCGCCCGTTTGGTCAGTGGACTGTGC | AscI |
| nanR-3 | CGATTGCGGCCGCCCTATACAGAGACTGTTCTTGTAGC | NotI |
| nanR-4 | GGGTATATGCATATGCAGGATGG | |
| Spec-R | GCTAAGCGGCCGCACTAAACGAAATAAACGC | NotI |
| Spec-F | GCTATGGCGCGCCCTAATCAAAATAGTGAGGAGG | AscI |
| RT-PCR primers | ||
| IR-I-F | GGATTGAGCAGGAAGTATGG | |
| IR-I-R | GGAGACGTTCCAAATACCACTGCTCC | |
| IR-II-F | GTCCACTCGAAACAAGTATTGTAAG | |
| IR-II-R | CATCTCTTCTAACCTCCTTTCTC | |
| IR-III-F | GGTTCATTTGAGAGCTGGTG | |
| IR-III-R | GCCGCTAGCGCAATTCCAGC | |
| IR-IV-F | GAGGCTGTTCAGTAGATGTA | |
| IR-IV-R | CCTACGATAATCATAGCTGGC | |
| IR-V-F | GCTATCATCAGTAAGTGGG | |
| IR-V-R | ACTGCAACGATGGTCGCACC | |
| IR-VI-F | GCCAAAGAGTCTATTAGTGCTAG | |
| IR-VI-R | GGCTAATTTTTGTAATAATCATC | |
| IR-VII-F | CGTGGTCGAGAAATGAATTGC | |
| IR-VII-R | GCGAGCCAATTCAGCTCC | |
| IR-VIII-F | CAGAGATGGATGGAGCAATTGC | |
| IR-VIII-R | CTAATCTGTGGCATCCTCTTTCC | |
| IR-IX-F | GGTGGCGCCATTACTAGACC | |
| IR-IX-R | GCAGGAAGTATAGCTATAGG | |
| IR-X-F | GGTGTAGATGATTAAGTACTTACTG | |
| IR-X-R | CCAGAATTGCCACAAGCAGC | |
| IR-XI-F | GCTATGAAACAATAGTCCTTAGTTATTC | |
| IR-XI-R | TGAAGCCCATCACCATCGGAGC | |
| IR-XII-F | GCAGTCATGATTGCTATCG | |
| IR-XII-R | GGAAAGATGAACAGCACAGTC | |
| IR-XIII-F | GGGTATTACTATGGGAGCGG | |
| IR-XIII-R | GAGGTAGTCGATAGCCTTGTC | |
| IR-XIV-F | CCCAGGTGAGCCACATCAG | |
| IR-XIV-R | CTAGCTCTTGCAGCATAC | |
| IR-XV-F | GCTTTACTGACAAAAAAGAAAACAGTTTACAAA | |
| IR-XV-R | CTCAAACATGGTCTCGTAGAGGC | |
| IR-XVI-F | GCAATTATTTCTGAAACTACTTTCAAAGGC | |
| IR-XVI-R | CCTTTGGCTACCGCCATGACTTC | |
| IR-XVII-F | CTGTTCGTTCACCATTGACACC | |
| IR-XVII-R | TGCAACGTAGTATGTCATAAATAC |
Construction of a nanR mutant.
The nanR deletion mutant was made by allelic replacement with a spectinomycin resistance marker as described before (28). Briefly, primers nanR-1/nanR-2 and nanR-3/nanR-4 were used to generate PCR fragments of the left and right flanking regions of nanR. PCR products of the left and right flanking regions of nanR contain AscI and NotI restriction enzyme sites, respectively. The spectinomycin resistance marker was amplified with primers Spec-F/Spec-R from plasmid pORI38 (29). The spectinomycin resistance marker also contains AscI and NotI restriction enzyme sites. Then, by restriction and ligation, the left and right flanking regions of nanR were fused to the spectinomycin resistance cassette. The resulting ligation product was transformed into wild-type S. pneumoniae D39. For transformation, cells were grown at 37°C without shaking until an optical density at 600 nm (OD600) of ∼0.1 was reached. Bovine serum albumin (BSA; 0.2%) and 1 mM CaCl2 were added to the grown cells. A 1-ml portion of the grown culture was transferred to a 1.5-ml Eppendorf tube, and 100 ng/μl of competence-stimulating peptide 1 (CSP1) was added to the culture. Cells were incubated at 37°C for 10 to 12 min. Then the ligation mixture was added to the incubated cells, and the cells were allowed to grow for 90 to 120 min at 37°C. After growth, the culture was spun for 1 min at 7,000 rpm, and most of the supernatant was discarded. The cell pellet was dissolved in the remaining medium (50 to 100 μl) and plated on GM17 (0.5% glucose plus M17) plus 1% sheep blood agar. Selection of the nanR mutant strain was done on the appropriate concentration of spectinomycin. The nanR mutant was further confirmed by PCR and DNA sequencing.
Construction of promoter-lacZ fusions and β-galactosidase assays.
Chromosomal transcriptional lacZ fusions to the nanA, spd_1503, and nanE promoters were constructed in the integration plasmid pPP2 (30) via double crossover in the bgaA locus with primer pairs listed in Table 2, resulting in pMA201 to pMA203, respectively. These constructs were further introduced into wild-type D39, resulting in strains MA201 to MA203, respectively. pMA201, pMA202, and pMA203 were also transformed into the ΔnanR strain, resulting in strains MA204, MA205, and MA206, respectively. Similarly, pMA201, pMA202, and pMA203 were transformed into a ΔccpA strain (8), resulting in strains MA207, MA208, and MA209. The following subclones of PnanE were made in pPP2 (30) using the primer pairs mentioned in Table 2: PnanE-F (truncated just a few bases upstream of the NanR regulatory site), PnanE-H (deletion of half of the NanR regulatory site), and PnanE-N (deletion of the full NanR regulatory site), resulting in plasmids pMA204 to pMA206. These constructs were introduced into the wild type, resulting in strains MA210, MA211, and MA212. All plasmid constructs were further checked for the presence of insert by PCR and DNA sequencing.
β-Galactosidase assays were performed as described before (30) using cells that were harvested in the mid-exponential growth phase and grown in M17 medium with appropriate sugars, as described in Results.
Microarray analysis.
For DNA microarray analysis of S. pneumoniae in the presence of sialic acid, the transcriptome of wild-type S. pneumoniae D39, grown in replicates in SM17 (0.5% sialic acid plus M17) medium, was compared to that grown in M17 medium (no added sugar). To analyze the effect of nanR deletion on the transcriptome of S. pneumoniae in the presence of sialic acid, wild-type D39 and its isogenic mutant nanR, were grown in replicates in SM17 medium and harvested at the mid-exponential growth phase. For the identification of differentially expressed genes, a Bayesian P value of <0.001 and a cutoff of a >2-fold change were applied. nanR is downregulated less than 2-fold but is listed in the table to show the deletion of nanR. Similarly, to analyze the effect of a ccpA deletion on the transcriptome of S. pneumoniae in the presence of sialic acid and glucose, wild-type D39 and its isogenic ΔccpA mutant were grown in replicates in SM17 and GM17 medium and harvested at the mid-exponential growth phase.
For RNA isolation, the following procedure was performed. The pellet of the harvested cells was resuspended in 400 μl Tris-EDTA (TE) containing diethyl pyrocarbonate (DEPC), and the resuspended cells were added to RNA-free screw-cap tubes containing 0.5 g glass beads, 50 μl 10% SDS, 500 μl phenol-chloroform–isoamyl alcohol (IAA), and a Macaloïd layer (Bentone MA; Rheox Inc.) (150 to 175 μl; the value is not exact, as the layer is very viscous). To break the cells, the screw-cap tubes were placed in a bead beater and two 60-s pulses were applied with a 60-s interval on ice. The samples were centrifuged for 10 min at 10,000 rpm (4°C). Chloroform-IAA (500 μl; 24:1) was added to tubes containing the upper phase from the centrifuged tubes, and the samples were again centrifuged for 5 min at 10,000 rpm (4°C). A 500-μl portion of the upper phase was transferred to the fresh tubes, and 2 volumes (1 ml) of lysis/binding buffer was added and mixed by pipetting up and down. Total RNA was isolated using the RNA isolation kit, and to remove contaminating DNA from total RNA, 100 μl DNase I mix (90 μl DNase buffer and 10 μl DNase I) was added and the samples were incubated for 20 to 30 min at 15 to 25°C. RNA was wash-cleaned using the RNase kit, and 50 μl of eluted volume was obtained. To check the quality of the RNA isolated, the RNA sample was diluted with DEPC water to get a concentration of 20 to 200 ng/μl. A 1-μl portion of the diluted RNA sample was used to check quality on the Bioanalyzer according to the manufacturer's instructions, and a ratio of 23S to 16S of around 2.0 was found, which is considered good. Microarray analysis was performed as described before (26, 31, 32). Homemade slides were used for sample hybridization, and in-house-made software packages were used for data analysis (33, 34).
All other procedures regarding the DNA microarray experiment and data analysis were performed as previously described (35–37).
Reverse transcription-PCR.
To confirm that nan operons I and II are transcribed into single transcriptional units, wild-type D39 was grown in SM17, and total RNA was isolated as described above. Reverse transcription-PCR (RT-PCR) was performed as described before (25) on all possible intergenic regions of nan operons I and II with primer pairs mentioned in Table 2. For a fair comparison of the PCR products, 100 ng of RNA and 20 ng of DNA were used.
Microarray data accession number.
Microarray data have been submitted to the Gene Expression Omnibus (GEO) under accession number GSE66561.
RESULTS
Genetic organization of the nan gene cluster in S. pneumoniae D39.
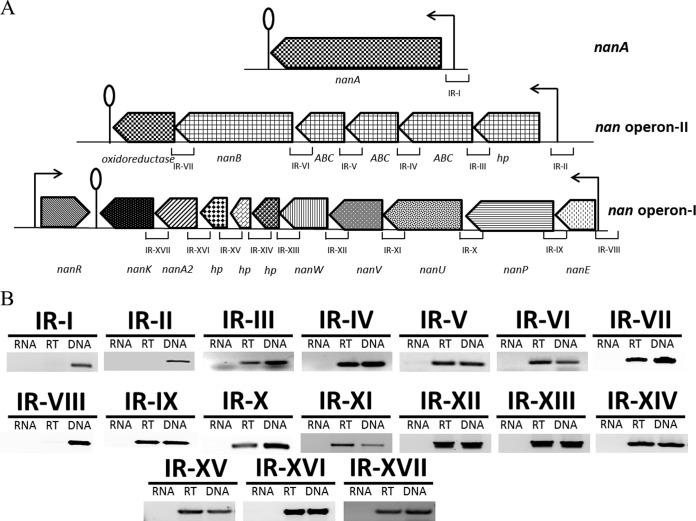
Based on the annotation of the D39 genome, the nan gene cluster consists of nan operon I (spd_1497 to spd_1488), the nanA gene (spd_1504), and nan operon II (spd_1503 to spd_1498) (Fig. 1A). nanA encodes a sialidase that has been shown to cleave sialic acid and plays a role in pneumococcal pathogenesis (21). nan operon I consists of 10 genes (spd_1488 to spd_1497), which code for an N-acetylmannosamine-6-phosphate 2-epimerase (NanE) (spd_1497), a phosphotranferase system (PTS) transporter (NanP) (spd_1496), three ABC transporters (NanUVW) (spd_1493 to spd_1495), three hypothetical proteins (spd_1490 to spd_1492), an N-acetylneuraminate lyase (spd_1489), and a ROK family protein (NanK) (spd_1488). The putative nan operon II (spd_1498 to spd_1503) is composed of six genes, which code for a hypothetical protein (spd_1503), three ABC transporters (spd_1500 to spd_1502), a neuraminidase nanB (spd_1499), and an oxidoreductase (spd_1498). The gene encoding a phosphosugar-binding transcriptional regulator, NanR, belonging to the RpiR family is present downstream of the nan gene cluster (Fig. 1A). The presence of nanR next to the nan gene cluster may indicate its putative function in the regulation of the nan gene cluster.
FIG 1.
(A) Organization of the nan gene cluster in S. pneumoniae D39. A lollipop structure represents a putative transcriptional terminator, while black arrows indicate the promoter regions. See the text for further details. (B) RT-PCR analysis to confirm the polycistronic nature of S. pneumoniae nan operons I and II. RT-PCR was performed on total RNA isolated from wild-type D39 grown in SM17 (0.5% sialic acid plus M17) medium with (RT) and without (RNA) reverse transcriptase treatment using the intergenic region primer pairs. DNA was used as a positive control. The sizes of RT-PCR products range from 100 to 300 bp.
To confirm whether nan operons I and II are transcribed into single transcriptional units and whether nanA has its own promoter, we performed RT-PCR on all possible intergenic regions present in nan operon I, in nan operon II, and on the upstream and downstream regions of nanA with the primer pairs listed in Table 2. RT-PCR data revealed that nan operons I and II are transcribed separately as single transcriptional units and that nanA has its own promoter (Fig. 1B).
Sialic acid-dependent gene expression in S. pneumoniae.
To elucidate the transcriptomic response of S. pneumoniae to sialic acid, transcriptome comparison of wild-type D39 grown in SM17 medium (0.5% sialic acid plus M17) to that grown in M17 (no added sugar) medium was performed. Table 3 summarizes the transcriptome changes induced in S. pneumoniae in the presence of sialic acid. After the criteria of a >2.0-fold difference and a P value of <0.001 were applied, 34 genes were upregulated in the presence of sialic acid, whereas 13 genes were downregulated under the tested conditions, including nan operon I and the nanA gene. Upregulation of nan operon I and the nanA gene in the presence of sialic acid indicates that the nan gene cluster is functional in S. pneumoniae D39 and responds to sialic acid. No change in the expression of nan operon II was observed. This indicates that nan operon II does not respond to sialic acid under our tested conditions. Some other genes and operons were found to be up- and downregulated in our transcriptome experiment (Table 3). The glutamine regulon, consisting of genes involved in nitrogen metabolism and known to contribute to the colonization of the nasopharynx by S. pneumoniae (26), was downregulated in the presence of sialic acid. The expression of this regulon is repressed in the presence of a nitrogen source (which in this case is sialic acid). There were other amino acid utilization and transport genes (spd_0334 and spd_0901) that were also downregulated in our microarray analysis (Table 3). Two bacteriocin secretion accessory proteins (encoded by spd_0115 and spd_0116) and some hypothetical proteins (encoded by spd_0091, spd_0373, spd_1159, spd_1265, spd_1505, spd_1515, spd_1516, and spd_1800) were also upregulated in the presence of sialic acid in addition to some ABC transporters (encoded by spd_1263, spd_1264, spd_1267, spd_1330, and spd_1514) (Table 3). spd_1265 codes for an integral membrane protein, whereas spd_1800 codes for an ABC-type multidrug transport system permease. An alpha-glycerophosphate oxidase gene and a glycerol kinase gene were also among the genes upregulated in our microarray experiment.
TABLE 3.
Summary of transcriptome comparison of wild-type S. pneumoniae D39 grown in SM17 (0.5% sialic acid plus M17) to that grown in M17
| Category and D39 locus tag | Functiona | Ratiob |
|---|---|---|
| Upregulated genes | ||
| spd_0091 | Hypothetical protein | 2.4 |
| spd_0115 | Hypothetical protein | 2.5 |
| spd_0116 | Hypothetical protein | 3.0 |
| spd_0373 | Hypothetical protein | 2.2 |
| spd_1263 | ABC transporter, ATP-binding/permease protein | 2.2 |
| spd_1264 | ABC transporter, ATP-binding protein | 2.7 |
| spd_1265 | Hypothetical protein | 2.9 |
| spd_1267 | ABC transporter, ATP-binding protein | 3.1 |
| spd_1330 | Amino acid ABC transporter, permease protein | 5.6 |
| spd_1488 | ROK family protein, NanK | 4.8 |
| spd_1489 | N-Acetylneuraminate lyase, putative | 3.8 |
| spd_1490 | Hypothetical protein | 2.6 |
| spd_1491 | Hypothetical protein | 2.8 |
| spd_1492 | Hypothetical protein | 2.6 |
| spd_1493 | Sugar ABC transporter, permease protein, NanW | 2.7 |
| spd_1494 | Sugar ABC transporter, permease protein, NanV | 2.9 |
| spd_1495 | Sugar ABC transporter, sugar-binding protein, NanU | 4.4 |
| spd_1496 | PTS, IIBC components, NanP | 4.2 |
| spd_1497 | N-Acetylmannosamine-6-phosphate 2-epimerase 2, NanE | 2.8 |
| spd_1504 | Sialidase A precursor, NanA | 6.8 |
| spd_1505 | Hypothetical protein | 2.0 |
| spd_1800 | Hypothetical protein | 2.1 |
| spd_2012 | Alpha-glycerophosphate oxidase | 3.2 |
| spd_2013 | Glycerol kinase | 3.3 |
| Downregulated genes | ||
| spd_0334 | Oligopeptide ABC transporter, oligopeptide-binding protein AliA | −2.4 |
| spd_0447 | Transcriptional regulator, GlnR | −5.5 |
| spd_0448 | Glutamine synthetase, GlnA | −6.0 |
| spd_0901 | Dihydrodipicolinate synthase | −2.0 |
| spd_1098 | Amino acid ABC transporter, GlnP | −4.0 |
| spd_1099 | Amino acid ABC transporter, GlnQ | −4.0 |
| spd_1158 | NADP-specific glutamate dehydrogenase | −3.5 |
| spd_1514 | ABC transporter, ATP-binding protein | −3.3 |
| spd_1515 | Hypothetical protein | −3.9 |
| spd_1516 | Hypothetical protein | −3.6 |
| spd_1956 | Dihydroxy-acid dehydratase | −2.0 |
D39 annotation or TIGR4 annotation (27). PTS, phosphotransferase system.
Fold increase or decrease in the expression of genes in SM17 compared to M17. Errors in the ratios never exceeded 10% of the given values.
Sialic acid induces, while glucose represses, the expression of nan operon I and the nanA gene.
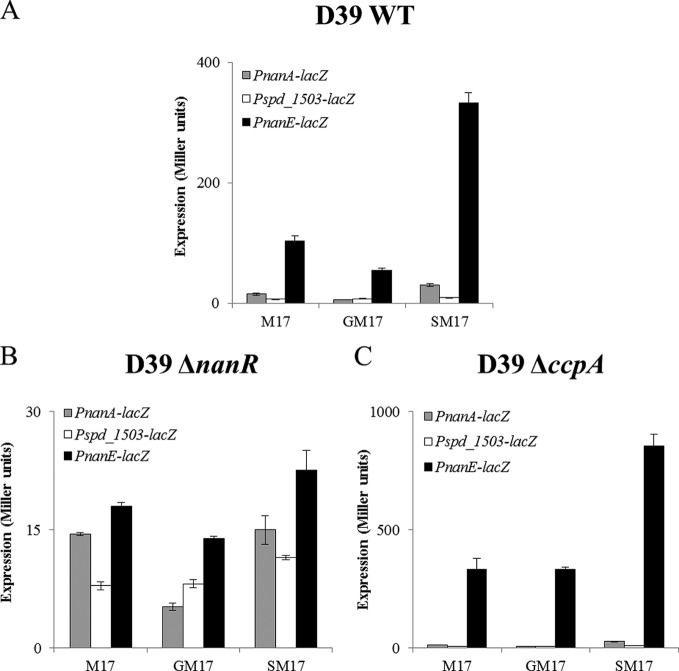
To further confirm sialic acid-dependent transcriptome results, we made transcriptional lacZ fusions of PnanA, Pspd_1503, and PnanE and transformed these lacZ fusions into the D39 wild type. β-Galactosidase assays were performed with the strains containing these transcriptional lacZ fusions grown in M17 (no added sugar), GM17 (0.5% glucose plus M17), and SM17 medium. β-Galactosidase assays showed that the expression of PnanA-lacZ and PnanE-lacZ was strikingly higher in the presence of sialic acid in the medium than in the presence of glucose (Fig. 2A). However, no significant change in the expression of Pspd_1503-lacZ was observed in the presence of sialic acid compared to glucose. This may suggest that nan operon II does not respond to sialic acid under our tested conditions. These results further suggest that the expression of nan operon I and the nanA gene is activated in the presence of sialic acid. Moreover, these results are in accordance with our transcriptome data mentioned above.
FIG 2.
Expression levels (in Miller units) of PnanA-lacZ, Pspd_1503-lacZ, and PnanE-lacZ in wild-type D39 (A), D39 ΔnanR (B), and D39 ΔccpA (C) grown in M17 (without sugar), GM17 (0.5% glucose plus M17), and SM17 (0.5% sialic acid plus M17). Standard deviations from three independent experiments are indicated by error bars.
To investigate the impact of other sugars (arabinose, cellobiose, dextrose, fructose, fucose, glucose, galactose, lactose, maltose, mannitol, mannose, melibiose, sorbitol, trehalose, and xylose) on the expression of the nan gene cluster, we checked the activity of PnanE-lacZ in the presence of various sugars (Table 4). β-Galactosidase assay data revealed that the activity of PnanE-lacZ was highest in the presence of sialic acid and lowest in the presence of glucose. These data suggest that the nan gene cluster specifically responds to sialic acid.
TABLE 4.
Expression levels of the PnanE-lacZ transcriptional fusion in wild-type S. pneumoniae D39 grown in the presence of various sugars in M17 medium
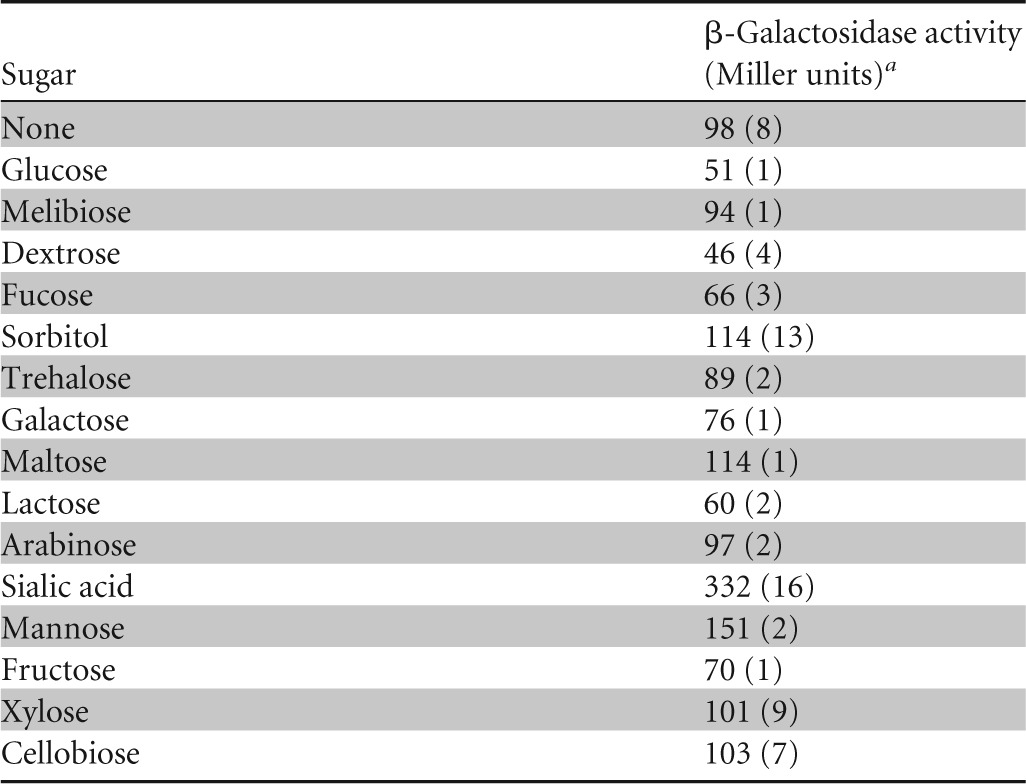
Standard deviations of three independent experiments are in parentheses.
NanR acts as a transcriptional activator of nan operon I and the nanA gene.
NanR, a phosphosugar-binding transcriptional regulator belonging to the RpiR family of transcriptional regulators, is present downstream of the nan gene cluster. To investigate the role of NanR in the regulation of the nan gene cluster, we constructed a nanR deletion mutant in S. pneumoniae D39 by replacing nanR with a spectinomycin resistance marker and transformed PnanA-lacZ, Pspd_1503-lacZ, and PnanE-lacZ transcriptional fusions into the ΔnanR strain. β-Galactosidase assays were performed with the strains containing these transcriptional lacZ fusions grown in M17 (no added sugar), GM17, and SM17 medium. The assays showed that the deletion of nanR led to inactivation of PnanA-lacZ and PnanE-lacZ even in the presence of sialic acid, suggesting the role of nanR as a transcriptional activator of nan operon I and the nanA gene in the presence of sialic acid (Fig. 2B). However, no effect of the nanR deletion was observed on the activity of Pspd_1503, suggesting that NanR plays no role in the regulation of nan operon II under our tested conditions.
DNA microarray analysis with the ΔnanR strain.
To elucidate the effect of the nanR deletion on the transcriptome of S. pneumoniae, DNA microarray analyses were performed with wild-type D39 against its isogenic ΔnanR mutant grown in SM17 medium. SM17 medium was used, as our β-galactosidase assays showed that NanR activates its targets in the presence of sialic acid (data shown above). Table 5 lists the results of transcriptome changes induced in S. pneumoniae due to the deletion of nanR. nan operon I and the nanA gene were downregulated significantly in the ΔnanR strain, confirming the role of NanR as a transcriptional activator of nan operon I and the nanA gene in the presence of sialic acid. We did not observe any significant differences in the expression of nan operon II in our microarray study, confirming that NanR has no role in the regulation of nan operon II. These data are also in accordance with the β-galactosidase assay data mentioned above.
TABLE 5.
Summary of transcriptome comparison of S. pneumoniae D39 ΔnanR and wild-type D39 grown in SM17 (0.5% sialic acid plus M17)
| D39 locus tag | Functiona | Ratiob |
|---|---|---|
| spd_1487 | Phosphosugar-binding transcriptional regulator, NanR | −1.4 |
| spd_1488 | ROK family protein, NanK | −8.0 |
| spd_1489 | N-Acetylneuraminate lyase, NanA2 | −6.1 |
| spd_1490 | Hypothetical protein | −3.4 |
| spd_1491 | Hypothetical protein | −2.1 |
| spd_1492 | Hypothetical protein | −2.2 |
| spd_1493 | Sugar ABC transporter, permease protein, NanW | −4.1 |
| spd_1494 | Sugar ABC transporter, permease protein, NanV | −5.2 |
| spd_1495 | Sugar ABC transporter, sugar-binding protein, NanU | −4.1 |
| spd_1496 | PTS, IIBC components, NanP | −5.0 |
| spd_1497 | N-Acetylmannosamine-6-phosphate 2-epimerase 2, NanE | −4.6 |
| spd_1504 | Sialidase A, NanA | −5.9 |
D39 annotation or TIGR4 annotation (27). PTS, phosphotransferase system.
Fold decrease in the expression of genes in the ΔnanR mutant compared to the wild type. nanR is downregulated less than 2-fold but is listed in the table to confirm the deletion of nanR. Errors in the ratio numbers never exceeded 10% of the given values.
Role of CcpA in the regulation of the nan gene cluster.
CcpA is the global transcriptional regulator in S. pneumoniae that represses the expression of genes involved in the utilization of nonpreferred sugars in the presence of a preferred one (8). To study the role of CcpA in the regulation of the nan gene cluster, we transformed PnanA-lacZ, Pspd_1503-lacZ, and PnanE-lacZ into the ΔccpA strain and performed β-galactosidase assays. The β-galactosidase assay data showed that ccpA deletion led to the increased expression of PnanE-lacZ in the presence of sialic acid and glucose (Fig. 2C), whereas no change in the expression of PnanA-lacZ and Pspd_1503-lacZ was observed in the presence of sialic acid and glucose (Fig. 2C). These results suggest that CcpA represses the expression of nan operon I in the presence of glucose and sialic acid but that regulation of the nanA gene and nan operon II is CcpA independent. Furthermore, we analyzed the promoter regions of nanA, spd_1503, and nanE to check for the presence of a putative cre box. Interestingly, a cre box (5′-TTGAAAGCGTTTTAAT-3′) is present in the nanE promoter region, supporting the putative role of CcpA in the regulation of nan operon I. However, no cre box was found in PnanA and Pspd_1503, which suggests the CcpA-independent regulation of nanA by the transcriptional activator NanR and nan operon II by another, unknown transcriptional regulator.
DNA microarray analysis of the D39 ΔccpA strain.
To further verify our β-galactosidase results and to find more targets of CcpA in the presence of sialic acid, we performed microarray analysis with ΔccpA against the wild-type D39 strain in the presence of sialic acid and glucose. Table 6 summarizes the transcriptome changes induced in S. pneumoniae due to the deletion of ccpA in the presence of sialic acid and glucose. nan operon I was significantly upregulated in ΔccpA in the presence of sialic acid and glucose (Table 6), which also confirms our β-galactosidase assay results mentioned above. These results confirm the role of CcpA in the regulation of nan operon I. We could not observe any changes in the expression of the nanA gene and nan operon II in our microarray analysis, which also confirms our β-galactosidase assay results mentioned above. These findings are also consistent with the findings of Carvalho et al. (8), who performed transcriptome analysis with a ccpA mutant in the presence of glucose and galactose (in minimal medium). Various other genes and operons were also regulated in our microarray experiment (see Table S1 in the supplemental material). These genes are grouped in COG functional categories according to their putative function (see Table S1 in the supplemental material). Most of these genes have a cre box in their promoter regions, suggesting that CcpA regulates these genes in the presence of sialic acid. Most of the genes affected in the microarray experiment were carbohydrate transport and metabolism genes. Amino acid transport and utilization genes were also among the ones that were affected significantly in the transcriptome analysis.
TABLE 6.
Summary of transcriptome comparison of S. pneumoniae D39 ΔccpA and wild-type D39 grown in SM17 (0.5% sialic acid plus M17) and GM17 (0.5% glucose plus M17)
| D39 locus tag | Functiona | Ratiob in: |
|
|---|---|---|---|
| SM17 | GM17 | ||
| spd_1488 | ROK family protein, NanK | 1.6 | 4.2 |
| spd_1489 | N-Acetylneuraminate lyase, NanA2 | 4.5 | |
| spd_1490 | Hypothetical protein | 1.6 | 3.7 |
| spd_1491 | Hypothetical protein | 2.0 | 4.7 |
| spd_1492 | Hypothetical protein | 2.0 | 4.4 |
| spd_1493 | Sugar ABC transporter, permease protein, NanW | 2.5 | 6.2 |
| spd_1494 | Sugar ABC transporter, permease protein, NanV | 2.2 | 6.4 |
| spd_1495 | Sugar ABC transporter, sugar-binding protein, NanU | 5.4 | |
| spd_1496 | PTS, IIBC components, NanP | 4.7 | 8.8 |
| spd_1497 | N-Acetylmannosamine-6-phosphate 2-epimerase 2, NanE | 1.6 | 4.6 |
D39 annotation or TIGR4 annotation (27). PTS, phosphotransferase system.
Fold increase in the expression of genes in the ΔccpA mutant compared to the wild type. Errors in the ratio numbers never exceeded 10% of the given values.
Prediction and verification of a NanR regulatory site in PnanE.
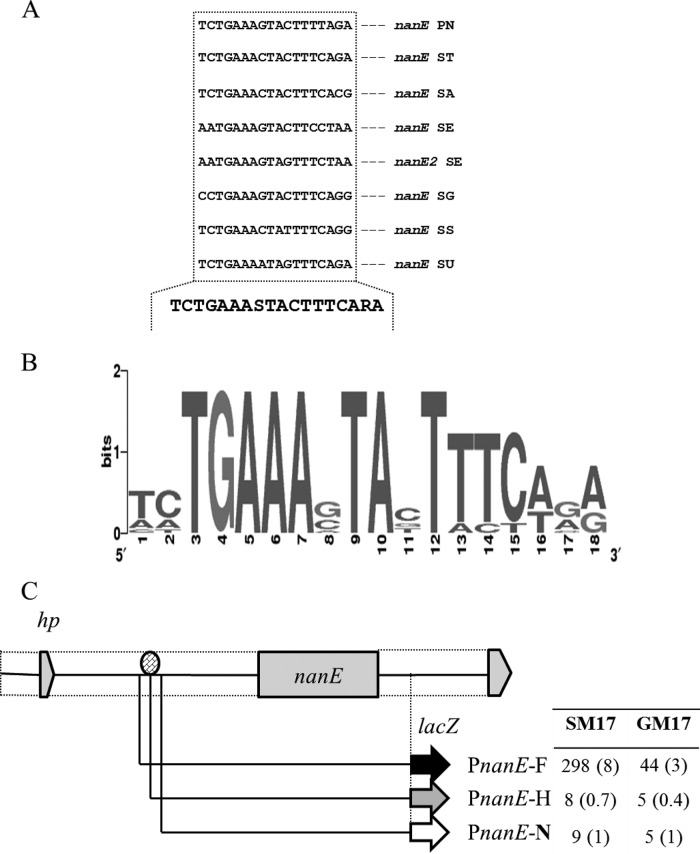
By using Genome2D software (34) and a MEME motif sampler search (38), an 18-bp palindromic sequence was found in the promoter regions of nanA and nanE in S. pneumoniae D39 (Fig. 3A). A weight matrix of these putative NanR regulatory sites (5′-TCTGAAASTACTTTCARA-3′) was constructed using these DNA regions. This DNA sequence may serve as the NanR regulatory site in S. pneumoniae. We also analyzed the spd_1503 promoter region for the presence of a putative NanR regulatory site. We could not find any DNA stretch matching the NanR regulatory site in the promoter region of spd_1503, which further confirms the NanR-independent regulation of nan operon II. PnanA and PnanE of other streptococcal species were studied to check if the NanR regulatory site is also conserved in those streptococci. From this study, we conclude that the NanR regulatory sequence is highly conserved in these streptococci as well (Fig. 3B).
FIG 3.
Identification of the NanR regulatory site in PnanE. (A) Position of the NanR regulatory site in the PnanE of different streptococci. PN, S. pneumoniae; ST, S. mitis; SA, S. agalactiae; SD, S. dysgalactiae; SE, S. equi; SM, S. mutans; SP, S. pyogenes; SG, S. sanguinis; SS, S. suis; SU, S. uberis. (B) Weight matrix of the identified NanR regulatory site in PnanE of different streptococci. (C) Verification of NanR regulatory site in the PnanE. A schematic illustration of PnanE truncations is shown. The oval indicates the putative NanR regulatory site. The table gives the β-galactosidase activity (Miller units) of the truncated promoters fused with lacZ in wild-type S. pneumoniae D39 grown in GM17 (0.5% glucose plus M17) and SM17 (0.5% sialic acid plus M17) medium. Standard deviations from three independent experiments are given in parentheses.
We further conducted a genome-wide search with the putative NanR regulatory site. The NanR regulatory site was also found in the upstream region of the nanA2 gene. We had already shown by RT-PCR that nanA2 is part of nan operon I (Fig. 1B). To confirm this further, we constructed a transcriptional lacZ fusion with the upstream region of the nanA2 and performed a β-galactosidase assay with strain D39 containing PnanA2-lacZ in the presence of glucose and sialic acid. We could not see any changes in the expression of PnanA2-lacZ, even in the presence of sialic acid (data not shown). Our β-galactosidase assay with PnanA2-lacZ also suggests that the NanR regulatory site present upstream of spd_1489 is most likely not functional.
To verify the NanR regulatory site present in PnanE, transcriptional lacZ fusions to 5′ truncations of PnanE were constructed (Fig. 3C). β-Galactosidase assays revealed that deletion of half or full of the predicted NanR regulatory site in PnanE abolished activity in M17, SM17, or GM17 medium. However, we observed expression similar to that of the full-length promoter when the promoter (PnanE) was truncated only a few bases upstream of the predicted regulatory site (Fig. 3C). These data confirm that the NanR regulatory site present in PnanE is functional and acts as a NanR regulatory site in S. pneumoniae.
DISCUSSION
In this study, we explored the regulatory mechanism of the nan gene cluster and found that the nan gene cluster consisting of nan operon I and the nanA gene is functional and responds to sialic acid in S. pneumoniae D39. The results presented in this study will increase our understanding regarding sialic acid-dependent regulation of the nan gene cluster in S. pneumoniae significantly. The nan gene cluster, putatively responsible for the utilization of sialic acid in S. pneumoniae, shows many variations among various pneumococcal strains. The provided annotation of S. pneumoniae TIGR4 genome suggests that there are two putative systems for sialic acid utilization, which are most likely regulated by two different transcriptional regulators (NanR1 and NanR2) (39). However, in S. pneumoniae D39, only one system for sialic acid utilization is present that is regulated by a transcriptional regulator NanR (this study). Based on the number of sialic acid systems, pneumococcal strains available in the RegPrecise database (http://regprecise.lbl.gov/RegPrecise/) can be divided into two groups. The first group consists of S. pneumoniae CDC0288-04, CGSP14, G54, Hungary19A-6, JJA, MLV-016, SP11-BS70, and SP9-BS68. Like the TIGR4 strain, these strains have two nan systems, whereas the second group possesses only one nan system, like D39, and includes R6, SP195, SP23-BS72, SP3-BS71, SP6-BS73, SP14-BS69, and P1031. These variations in the nan gene cluster among pneumococcal strains suggest the importance of the nan gene cluster in the physiology and lifestyle of S. pneumoniae.
Sialic acid may enter the cell through either the ABC transporters (NanUVW), a secondary transporter, or tripartite ATP-independent periplasmic (TRAP) transporters (40). Once internalized, NanK and NanE lead the metabolism of sialic acid. NanK converts ManNAc into ManNAc-6P by phosphorylating it at C-6. NanE then converts ManNAc-6P to GlcNAc-6P. NagA (N-acetylglucosamine-6-phosphate deacetylase) and NagB (glucosamine-6-phosphate deaminase), which perform the final two reactions of the sialic acid utilization pathway, are the two enzymes not encoded by the nan gene cluster (40). The S. pneumoniae D39 nan gene cluster codes for NanA, which may start the catabolism of sialic acid, and NanK and NanE, which may lead the further metabolism of ManNAc. Interestingly, the genome of S. pneumoniae D39 also encodes NagA and NagB. We could not see any change in expression of nagA and nagB in our transcriptome study in the presence of sialic acid. Further studies focusing on the regulation of nagA and nagB are required.
NanA and NanB are the pneumococcal neuraminidases that cleave sialic acid, although the localization and cleavage specificity of NanA differ from those of NanB, which suggests distinct roles of these two neuraminidases in S. pneumoniae (41). NanA is a hydrolytic enzyme that is attached to the cell surface and cleaves α2-3- and α2-6-linked sialic acid, whereas NanB is a secreted neuraminidase that shows strict specificity for α2-3-linked sialic acid (41, 42). NanA is important for pathogenesis in the host, as it helps in unmasking carbohydrate receptors for attachment, providing a carbon source for the bacteria, changing the surface of other bacteria in the same niche, and disturbing the function of host defense molecules (41). NanA was highly expressed under our tested conditions. Moreover, we show that NanR activates the nanA gene and nan operon I in the presence of sialic acid. These observations suggest that NanA is a suitable target for drugs and vaccines against pneumococci. The gene coding for NanB is one of the six genes present in nan operon II. However, nan operon II is not regulated under our tested conditions, and NanR and CcpA also have no role in its regulation of nan operon II under our tested conditions. Future investigations focusing on the conditions and mechanisms under which nan operon II is regulated are required.
CcpA is the master transcriptional regulator of the sugar utilization systems in S. pneumoniae and regulates multiple sugar systems in a pleiotropic manner (8, 9, 43). However, many systems dedicated to the utilization of nonpreferred sugars are regulated independently of CcpA in S. pneumoniae, like the cel and lac systems (25, 31). In this study, we show that CcpA plays an important role in the regulation of nan operon I in S. pneumoniae D39. Our results also show that the regulation of the nanA gene and nan operon II is CcpA independent. Similarly, recent transcriptome analysis of a ccpA mutant in the presence of glucose and galactose revealed that CcpA represses the expression of nan operon I in the presence of glucose and galactose (8). However, no effect of ccpA deletion on the expression of nan operon II and the nanA gene was observed in the presence of glucose and galactose (8). Therefore, data presented in our study are consistent with the observations presented in a previous study on CcpA (8).
In this study, we have shown that NanR (an RpiR-type transcriptional regulator), located downstream of the nan gene cluster, acts as a transcriptional activator of nan operon I and the nanA gene in the presence of sialic acid. NanR has a DNA-binding helix-turn-helix (HTH) domain and a sugar isomerase (SIS) domain and shares >90% sequence identity to its counterparts in S. oralis, S. tigurinus, S. mitis, and S. pseudopneumoniae. The SIS domain is a phosphosugar-binding domain found in many phosphosugar isomerases and phosphosugar binding proteins that regulate the expression of genes involved in the synthesis of phosphosugars possibly by binding to the end product of the pathway (44). RpiR-type transcriptional regulators have been shown to regulate various sugar systems in different bacteria. These regulators mostly control sugar and sugar phosphate catabolic pathways, including maltose, glucose, and ribose metabolism, the pentose phosphate pathway, inositol catabolism, and N-acetylmuramic acid catabolism (39). For example, HpxU is an RpiR-type transcriptional regulator in Klebsiella pneumoniae that has been shown to act as a transcriptional repressor of allantoate catabolism (45). In E. coli, NanR has also been shown to act as a transcriptional activator of the sialic acid gene cluster (46).
Supplementary Material
ACKNOWLEDGMENT
M.A. is supported by the G C University, Faisalabad, Pakistan, under the faculty development program of HEC Pakistan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00499-15.
REFERENCES
- 1.Ispahani P, Slack RCB, Donald FE, Weston VC, Rutter N. 2004. Twenty year surveillance of invasive pneumococcal disease in Nottingham: serogroups responsible and implications for immunisation. Arch Dis Child 89:757–762. doi: 10.1136/adc.2003.036921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team . 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 3.Gray BM, Turner ME, Dillon HC Jr. 1982. Epidemiologic studies of Streptococcus pneumoniae in infants. The effects of season and age on pneumococcal acquisition and carriage in the first 24 months of life. Am J Epidemiol 116:692–703. [DOI] [PubMed] [Google Scholar]
- 4.Phillips NJ, John CM, Reinders LG, Gibson BW, Apicella MA, Griffiss JM. 1990. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed Environ Mass Spectrom 19:731–745. doi: 10.1002/bms.1200191112. [DOI] [PubMed] [Google Scholar]
- 5.Titgemeyer F, Hillen W. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59–71. doi: 10.1023/A:1020628909429. [DOI] [PubMed] [Google Scholar]
- 6.Linke CM, Woodiga SA, Meyers DJ, Buckwalter CM, Salhi HE, King SJ. 2013. The ABC transporter encoded at the pneumococcal fructooligosaccharide utilization locus determines the ability to utilize long- and short-chain fructooligosaccharides. J Bacteriol 195:1031–1041. doi: 10.1128/JB.01560-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckwalter CM, King SJ. 2012. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol 20:517–522. doi: 10.1016/j.tim.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. 2011. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae D39. PLoS One 6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:1366–1381. doi: 10.1128/JB.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gualdi L, Hayre JK, Gerlini A, Bidossi A, Colomba L, Trappetti C, Pozzi G, Docquier J-D, Andrew P, Ricci S, Oggioni MR. 2012. Regulation of neuraminidase expression in Streptococcus pneumoniae. BMC Microbiol 12:200. doi: 10.1186/1471-2180-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. 2009. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis 199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- 13.Traving C, Schauer R. 1998. Structure, function and metabolism of sialic acids. Cell Mol Life Sci 54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King SJ. 2010. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol 25:15–24. doi: 10.1111/j.2041-1014.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- 15.Burnaugh AM, Frantz LJ, King SJ. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol 190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalia AB, Standish AJ, Weiser JN. 2010. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun 78:2108–2116. doi: 10.1128/IAI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King SJ, Hippe KR, Weiser JN. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol 59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 18.Marion C, Burnaugh AM, Woodiga SA, King SJ. 2011. Sialic acid transport contributes to pneumococcal colonization. Infect Immun 79:1262–1269. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakhnovich EA, King SJ, Weiser JN. 2002. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect Immun 70:7161–7164. doi: 10.1128/IAI.70.12.7161-7164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett 278:231–235. doi: 10.1111/j.1574-6968.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- 21.Parker D, Soong G, Planet P, Brower J, Ratner AJ, Prince A. 2009. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun 77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong HH, Blue LE, James MA, DeMaria TF. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun 68:921–924. doi: 10.1128/IAI.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun 74:4014–4020. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu Rev Microbiol 47:89–115. doi: 10.1146/annurev.mi.47.100193.000513. [DOI] [PubMed] [Google Scholar]
- 25.Afzal M, Shafeeq S, Kuipers OP. 2014. LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae. Appl Environ Microbiol 80:5349–5358. doi: 10.1128/AEM.01370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloosterman TG, Hendriksen WT, Bijlsma JJ, Bootsma HJ, van Hijum SA, Kok J, Hermans PW, Kuipers OP. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J Biol Chem 281:25097–25109. doi: 10.1074/jbc.M601661200. [DOI] [PubMed] [Google Scholar]
- 27.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafeeq S, Kloosterman TG, Rajendran V, Kuipers OP. 2012. Characterization of the ROK-family transcriptional regulator RokA of Streptococcus pneumoniae D39. Microbiology 158:2917–2926. doi: 10.1099/mic.0.062919-0. [DOI] [PubMed] [Google Scholar]
- 29.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- 30.Halfmann A, Hakenbeck R, Bruckner R. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol Lett 268:217–224. doi: 10.1111/j.1574-6968.2006.00584.x. [DOI] [PubMed] [Google Scholar]
- 31.Shafeeq S, Kloosterman TG, Kuipers OP. 2011. CelR-mediated activation of the cellobiose-utilization gene cluster in Streptococcus pneumoniae. Microbiology 157:2854–2861. doi: 10.1099/mic.0.051359-0. [DOI] [PubMed] [Google Scholar]
- 32.Shafeeq S, Kuipers OP, Kloosterman TG. 2013. Cellobiose-mediated gene expression in Streptococcus pneumoniae: a repressor function of the novel GntR-type regulator BguR. PLoS One 8:e57586. doi: 10.1371/journal.pone.0057586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hijum SAFT, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2:241–244. [PubMed] [Google Scholar]
- 34.Baerends RJS, Smits WK, de Jong A, Hamoen LW, Kok J, Kuipers OP. 2004. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol 5:R37. doi: 10.1186/gb-2004-5-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afzal M, Shafeeq S, Henriques-Normark B, Kuipers OP. 2015. UlaR activates expression of the ula operon in Streptococcus pneumoniae in the presence of ascorbic acid. Microbiology 161:41–49. doi: 10.1099/mic.0.083899-0. [DOI] [PubMed] [Google Scholar]
- 36.Shafeeq S, Kloosterman TG, Kuipers OP. 2011. Transcriptional response of Streptococcus pneumoniae to Zn(2+) limitation and the repressor/activator function of AdcR. Metallomics 3:609–618. doi: 10.1039/c1mt00030f. [DOI] [PubMed] [Google Scholar]
- 37.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 38.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 39.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, Rodionov DA. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res 38:D111–D118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gut H, King SJ, Walsh MA. 2008. Structural and functional studies of Streptococcus pneumoniae neuraminidase B: an intramolecular trans-sialidase. FEBS Lett 582:3348–3352. doi: 10.1016/j.febslet.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Li X, Andrew PW, Taylor GL. 2008. Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:772–775. doi: 10.1107/S1744309108024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lulko AT, Buist G, Kok J, Kuipers OP. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J Mol Microbiol Biotechnol 12:82–95. doi: 10.1159/000096463. [DOI] [PubMed] [Google Scholar]
- 44.Bateman A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem Sci 24:94–95. doi: 10.1016/S0968-0004(99)01357-2. [DOI] [PubMed] [Google Scholar]
- 45.Guzmán K, Campos E, Aguilera L, Toloza L, Giménez R, Aguilar J, Baldoma L, Badia J. 2013. Characterization of the gene cluster involved in allantoate catabolism and its transcriptional regulation by the RpiR-type repressor HpxU in Klebsiella pneumoniae. Int Microbiol 16:165–176. [DOI] [PubMed] [Google Scholar]
- 46.Jaeger T, Mayer C. 2008. The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli. J Bacteriol 190:6598–6608. doi: 10.1128/JB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leenhouts K, Venema G, Kok J. 1998. A lactococcal pWV01 based integration toolbox for bacteria. Methods Cell Sci 20:35–50. doi: 10.1023/A:1009862119114. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.