Abstract
Nickel is an essential component of all H2-uptake hydrogenases. A fragment of DNA that complements a H2-uptake-deficient but nickel-cured mutant strain (JHK7) of Bradyrhizobium japonicum was isolated and sequenced. This 4.5-kb DNA fragment contains four open reading frames designated as ORF1, hupN, hupO, and hupP, which encode polypeptides with predicted masses of 17, 40, 19, and 63.5 kDa, respectively. The last three open reading frames (hupNOP) are most likely organized as an operon with a putative sigma 54-type promoter. Based on its hydropathy profile, HupN is predicted to be a transmembrane protein. It has 56% identity to the previously described HoxN (high-affinity nickel transport protein) of Alcaligenes eutrophus. A subclone (pJF23) containing the hupNOP genes excluding ORF1 completely complemented (in trans) strain JHK7 for hydrogenase activity in low nickel conditions. pJF26 containing only a functional hupN complemented the hydrogenase activity of mutant strain JHK7 to 30-55% of the wild-type level. Mutant strain JHK70, with a chromosomal deletion in hupP but with an intact hupNO, showed greater activities than pJF26-complemented JHK7 but still had lower activities than the wild type at all nickel levels tested. pJF25, containing the entire hupO and hupP, but without hupN (a portion of hupN was deleted), did not complement hydrogenase activity of mutant strain JHK7. The results suggest that the products of the hupNOP operon are all involved in nickel incorporation/metabolism into the hydrogenase apoprotein. Based on (previous) nickel transport studies of strain JHK7, the hupNOP genes appear not to be involved in nickel transport by whole cells. The deleterious effects on hydrogenase expression are most pronounced by lack of the HupN product.
Full text
PDF
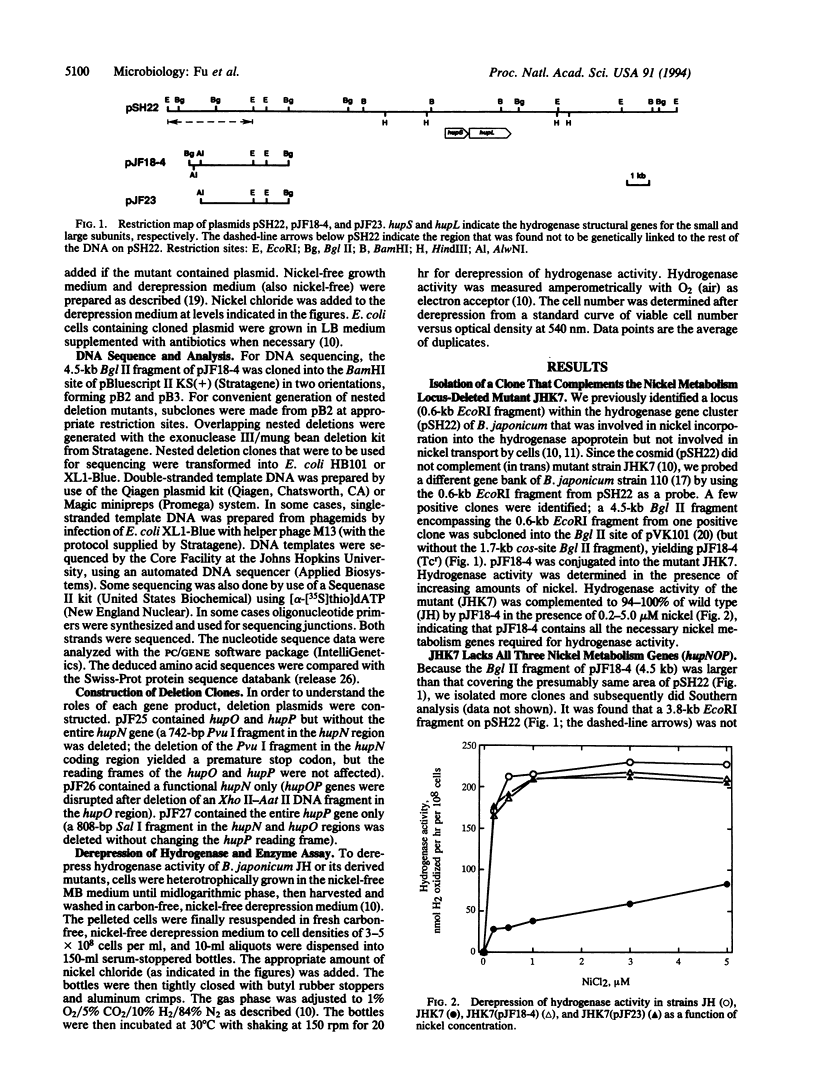

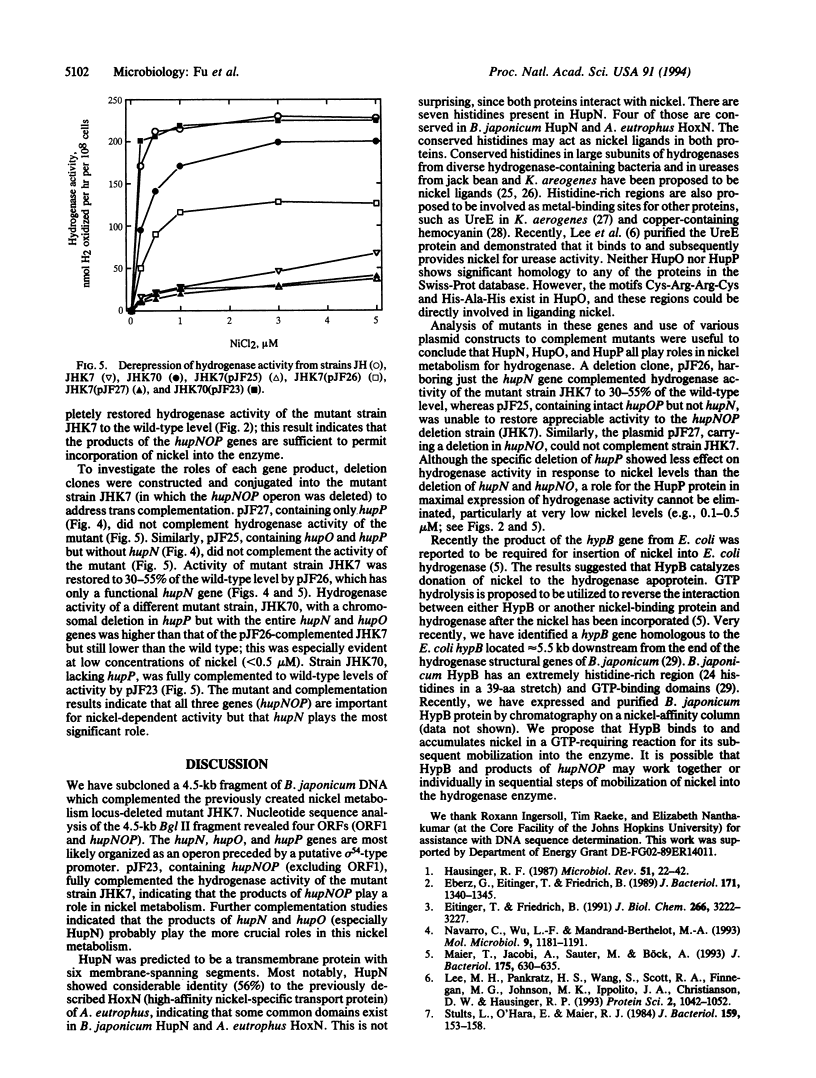

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bishop P. E., Guevara J. G., Engelke J. A., Evans H. J. Relation between Glutamine Synthetase and Nitrogenase Activities in the Symbiotic Association between Rhizobium japonicum and Glycine max. Plant Physiol. 1976 Apr;57(4):542–546. doi: 10.1104/pp.57.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberz G., Eitinger T., Friedrich B. Genetic determinants of a nickel-specific transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J Bacteriol. 1989 Mar;171(3):1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T., Friedrich B. Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem. 1991 Feb 15;266(5):3222–3227. [PubMed] [Google Scholar]
- Eskew D. L., Welch R. M., Cary E. E. A simple plant nutrient solution purification method for effective removal of trace metals using controlled pore glass-8-hydroxyquinoline chelation column chromatography. Plant Physiol. 1984 Sep;76(1):103–105. doi: 10.1104/pp.76.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. L., Maier R. J. Competitive inhibition of an energy-dependent nickel transport system by divalent cations in Bradyrhizobium japonicum JH. Appl Environ Microbiol. 1991 Dec;57(12):3511–3516. doi: 10.1128/aem.57.12.3511-3516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. L., Maier R. J. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 1991 Dec;57(12):3502–3510. doi: 10.1128/aem.57.12.3502-3510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Nickel-dependent reconstitution of hydrogenase apoprotein in Bradyrhizobium japonicum Hupc mutants and direct evidence for a nickel metabolism locus involved in nickel incorporation into the enzyme. Arch Microbiol. 1992;157(6):493–498. doi: 10.1007/BF00276768. [DOI] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Nucleotide sequences of two hydrogenase-related genes (hypA and hypB) from Bradyrhizobium japonicum, one of which (hypB) encodes an extremely histidine-rich region and guanine nucleotide-binding domains. Biochim Biophys Acta. 1994 Feb 8;1184(1):135–138. doi: 10.1016/0005-2728(94)90163-5. [DOI] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Rapid and efficient selection of recombinant site-directed mutants of Bradyrhizobium japonicum by colony hybridization. FEMS Microbiol Lett. 1993 May 1;109(1):33–38. doi: 10.1111/j.1574-6968.1993.tb06139.x. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P. Nickel utilization by microorganisms. Microbiol Rev. 1987 Mar;51(1):22–42. doi: 10.1128/mr.51.1.22-42.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom S. S., Graham L. A., Maier R. J. Isolation of genes (nif/hup cosmids) involved in hydrogenase and nitrogenase activities in Rhizobium japonicum. J Bacteriol. 1985 Mar;161(3):882–887. doi: 10.1128/jb.161.3.882-887.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kim H., Maier R. J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990 Nov 5;265(31):18729–18732. [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993 Jun;2(6):1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990 Oct;172(10):5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Wu L. F., Mandrand-Berthelot M. A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993 Sep;9(6):1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- O'brian M. R., Maier R. J. Isolation of a cytochrome aa(3) gene from Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1987 May;84(10):3219–3223. doi: 10.1073/pnas.84.10.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla A. E., Robbins J., Menon N., Peck H. D., Jr Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992 Feb;8(2):109–135. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- Russell P., Schell M. G., Nelson K. K., Halverson L. J., Sirotkin K. M., Stacey G. Isolation and characterization of the DNA region encoding nodulation functions in Bradyrhizobium japonicum. J Bacteriol. 1985 Dec;164(3):1301–1308. doi: 10.1128/jb.164.3.1301-1308.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults L. W., O'Hara E. B., Maier R. J. Nickel is a component of hydrogenase in Rhizobium japonicum. J Bacteriol. 1984 Jul;159(1):153–158. doi: 10.1128/jb.159.1.153-158.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Hennecke H. The -24/-12 promoter comes of age. FEMS Microbiol Rev. 1989 Dec;5(4):341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Wu L. F. Putative nickel-binding sites of microbial proteins. Res Microbiol. 1992 Mar-Apr;143(3):347–351. doi: 10.1016/0923-2508(92)90027-l. [DOI] [PubMed] [Google Scholar]


