Significance
The levels of aggregated specific proteins in the CNS in neurodegenerative diseases are minute, hampering analysis of structure and growth kinetics. Here we describe a generally applicable method based on binary epitope mapping. It was applied to analysis of superoxide dismutase aggregation in amyotrophic lateral sclerosis model mice. Two different strains of aggregates with different structures, physical stabilities, and growth kinetics were readily distinguished. Moreover, they were different from superoxide dismutase aggregates formed in vitro under a variety of conditions, revealing a key role of the CNS in shaping the aggregation process.
Keywords: protein aggregation, neurodegeneration, strain, amyotrophic lateral sclerosis, transgenic mice
Abstract
Despite considerable progress in uncovering the molecular details of protein aggregation in vitro, the cause and mechanism of protein-aggregation disease remain poorly understood. One reason is that the amount of pathological aggregates in neural tissue is exceedingly low, precluding examination by conventional approaches. We present here a method for determination of the structure and quantity of aggregates in small tissue samples, circumventing the above problem. The method is based on binary epitope mapping using anti-peptide antibodies. We assessed the usefulness and versatility of the method in mice modeling the neurodegenerative disease amyotrophic lateral sclerosis, which accumulate intracellular aggregates of superoxide dismutase-1. Two strains of aggregates were identified with different structural architectures, molecular properties, and growth kinetics. Both were different from superoxide dismutase-1 aggregates generated in vitro under a variety of conditions. The strains, which seem kinetically under fragmentation control, are associated with different disease progressions, complying with and adding detail to the growing evidence that seeding, infectivity, and strain dependence are unifying principles of neurodegenerative disease.
The mechanistic intricacies of protein aggregation in vitro and in neurodegenerative disease are now being puzzled together using new generations of structural approaches and careful quantitative treatment of kinetic data (1–5). Adding to this development, we use here an antibody-based method that allows direct structural and quantitative analysis of the minute amounts of aggregates present in neural tissue. The strength and versatility of the method is exemplified for a transgenic-mouse model of the neurodegenerative disease amyotrophic lateral sclerosis (ALS), which is associated with intracellular aggregation of the radical scavenger superoxide dismutase-1 (SOD1) (Fig. 1). ALS is characterized by adult-onset degeneration of motor neurons, which begins focally and then spreads contiguously through the neural tissue (6). A conspicuous cause of ALS is a large number of structurally diverse mutations in SOD1 (7) (alsod.iop.kcl.ac.uk/), whose common denominator seems to be decreased structural stability and reduced repulsive charge (8). However, pathological aggregation of SOD1 need not always be triggered by mutation; it occurs also in ALS patients lacking SOD1 mutations (9–11). Consistently, it is also found that mice that are bred to express wild-type human SOD1 (hSOD1wt) at a high rate develop a fatal ALS-like disease, with accumulation of large amounts of hSOD1 aggregates in remaining motor neurons (12). This suggests that mutant and wild-type hSOD1 might confer disease by the same mechanism: Human SOD1 seems to be an oversaturated protein with an intrinsic propensity to aggregate (13), and this tendency can be critically augmented by mutation. Consistently, mutant and wild-type hSOD1 show indistinguishable fibrillation behavior in vitro with kinetics determined by structural stability and net charge (14, 15). The in vitro fibrillation occurs moreover by the recruitment of globally unfolded hSOD1 monomers, following exponential time courses controlled by fibril fragmentation (15).
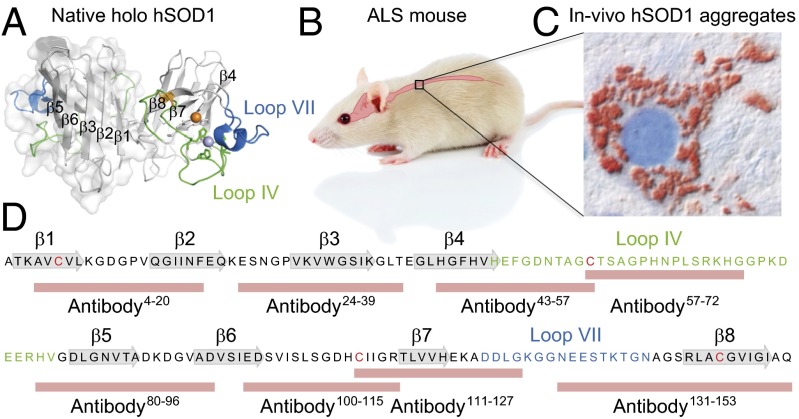
Fig. 1.
Human SOD1 and its involvement in the motor-neuron disease ALS. (A) Structure of the native hSOD1 homodimer showing the positions of β-strands 1–8 and loops IV and VII of the active site (PDB ID code 1HL5). (B) The pathology of transgenic mice expressing hSOD1 variants is mainly confined to the spinal cord, giving disease characteristics similar to those of human ALS. (C) Immunohistochemistry of spinal ventral horn in hSOD1G85R mouse using the 57–72 antibody showing typical accumulation of hSOD1 deposits in a motor neuron. (D) The amino acid sequence of the wild-type hSOD1 subunit, showing the positions of native secondary structure and the peptides used for generation of the antibodies used in this study.
An outstanding feature of hSOD1 aggregation, however, is a pronounced malleability, leading to different aggregate structures upon point mutation and altered experimental conditions (14, 16). Like proteins in general, hSOD1 contains several “sticky” sequence regions, allowing a large number of competing aggregation pathways (17, 18). The pressing question is, then, what role these generic features of the protein-aggregation process play in disease at the organism level. To find out, we use here the antibody assay for systematic analysis of hSOD1 aggregation and neurodegeneration in ALS model mice. The results reveal two strains of coexisting and structurally distinct hSOD1 aggregates, whose relative abundance correlates with disease progression. Moreover, we find that the strain associated with the most rapid disease progression is also the most fragile, and that the aggregation kinetics on the whole comply with fragmentation-controlled growth as observed in vitro. Despite this basic resemblance to in vitro fibrillation behavior, the hSOD1 aggregates emerging in mice and in vitro are structurally dissimilar, revealing a key role of the CNS in shaping—or selecting for—the pathological aggregate structures at play in disease at the organism level.
Results
Fingerprinting of Aggregate Structures by Their Exposed Disordered Sequence Epitopes.
Atomic-level characterization of protein aggregates normally requires much more material than found in the matchstick-sized spinal cord of a mouse. To overcome this problem, we opted here for an alternative, sequenced-based strategy with the binary readout “disordered” or “structured.” The principle is simple and uses antibodies raised to short peptides covering the whole sequence of the monomeric aggregate precursor. In this study, we generated eight polyclonal rabbit antibodies covering 90% of the SOD1 sequence (Fig. 1D). Because the configurational space of each peptide is very large (i.e., >> n3), where n is the number of amino acids, their “randomly” induced antibody-binding epitopes are unlikely to match any part of the uniquely defined native hSOD1 structure (19). As proof of principle, none of the antibodies generated captured native hSOD1 in solution, irrespective of concealment or exposure of the target segment in the folded structure (Fig. 2F and Fig. S1). Native hSOD1 is very rigid and cannot adapt to the antigen-binding sites of the peptide antibodies (20). In sharp contrast, all of the antibodies recognized the flexible globally unfolded hSOD1 (Fig. 2E).
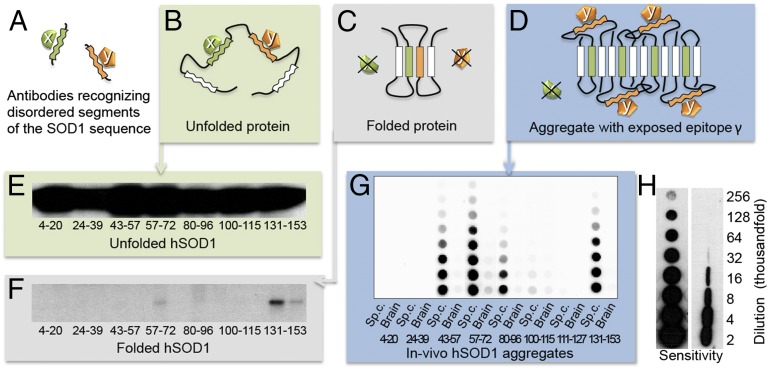
Fig. 2.
Discrimination between “disordered” and “structured” sequence segments by the binary epitope-mapping assay. (A) Antibodies (x and y) were raised against short, consecutive segments of the hSOD1 sequence (Fig. 1). (B) In the globally unfolded state of hSOD1, the individual sequence epitopes are flexible and can adapt to the antigen-binding sites of the anti-peptide antibodies. (C) In the rigid folded hSOD1, the sequence epitopes adopt a fixed structure incompatible with antibody binding. (D) In partially ordered hSOD1 aggregates, antibody x cannot bind to its sequence epitope because this is ordered/hidden in the aggregate core, whereas antibody y can, because the epitope protrudes freely from the aggregate surface. (E) Western blot of hSOD1 captured by immobilized anti-peptide antibodies incubated with unfolded/denatured hSOD1 in solution. All of the antibodies bind, showing full exposure of flexible sequence segments in Fig. 1D. (F) Corresponding data for natively folded hSOD1, giving no binding because the sequence epitopes are now structured/hidden. Two sequential incubations were carried out, the first to capture any traces of disordered hSOD1 present in the preparation. E and F reproduced from ref. 9. (G) Aggregates in spinal cord and brain from a terminally ill hSOD1G93A mouse captured on a filter in a dot-blot apparatus and stained with the anti-peptide antibodies, showing the binary fingerprint of disordered and structured/hidden sequence regions of the constituent hSOD1 monomers. (H) The benchmarking 57–72 antibody detects hSOD1 in filter-captured aggregates with 10-fold higher sensitivity than hSOD1 restricted on a Western immunoblot membrane (SI Materials and Methods). The figures indicate the degree of dilution of the spinal-cord tissue.
Now, upon exposure of the antibodies to hSOD1 aggregates from spinal cord of a terminally ill mouse expressing G93A mutant hSOD1 (hSOD1G93A), captured on a filter in a dot-blot apparatus, a distinct pattern emerges: the binary fingerprint of the pathological aggregate structure (Fig. 2G). “Recognition” shows the hSOD1G93A sequence regions not involved in ordered aggregate structure (i.e., flexible segments that are free to adapt to the antigen-binding sites), whereas “no signal” shows the sequence regions that are either directly engaged in ordered aggregate structure or otherwise hidden (Fig. 2 A–D).
To ensure that soluble disordered hSOD1 species present in vivo react with the antibodies, a 25,000 × g supernatant of a hSOD1G93A spinal-cord homogenate, predicted to contain high levels of globally unfolded monomers, was incubated with antibodies immobilized on Sepharose beads (21, 22). In contrast to our findings with hSOD1G93A aggregates, all of the antibodies reacted with this material (Fig. S2A). They also all reacted with hSOD1 when such a supernatant was dot-blotted onto a cellulose nitrate membrane (Fig. S2B). On this basis, we conclude that the simplistic epitope assay is a versatile tool for “course-grained” structural analysis of low-level proteinaceous aggregates present in vivo.
The sensitivity of the assay is very high. The benchmarking 57–72 antibody gives 10-fold higher sensitivity when hSOD1 is analyzed in filter-captured aggregates compared with hSOD1 restricted on a Western blot membrane (Fig. 2H).
Malleability of the hSOD1 Aggregation Process.
Systematic analysis of hSOD1 aggregate structures was done in four well-characterized ALS mouse lines expressing hSOD1 variants with widely different molecular properties: hSOD1G93A (23), hSOD1G85R (24), hSOD1D90A (25), and hSOD1wt (12). The transgenes are, moreover, expressed at different rates, so the levels of disordered hSOD1 species in spinal cords vary considerably. They are highest in the short-lived hSOD1G93A model and lowest in the hSOD1D90A and hSOD1wt mice (21, 22, 26).
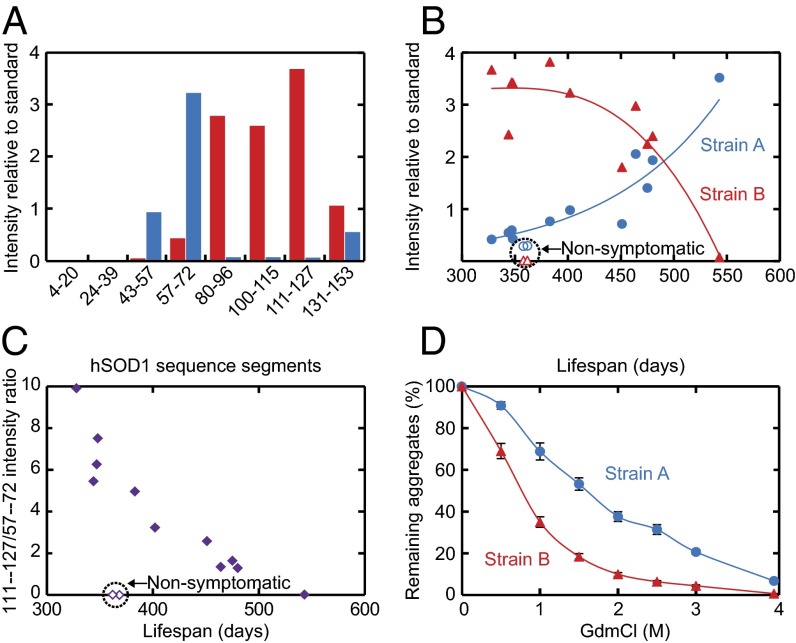
To facilitate data comparison, a spinal-cord homogenate from a terminal hSOD1G93A mouse was frozen in multiple aliquots and designated as standard. It was present in a dilution series on all filters and stained with the 57–72 antibody. In what follows, the staining intensities of all eight antibodies of the individual mouse samples have been normalized against this hSOD1G93A standard. The results show that the aggregates of terminally ill hSOD1G93A, hSOD1G85R, and hSOD1wt mice are all very similar, despite pronounced differences in expression levels and structural properties of the causative protein variants (Fig. 3A and Table S1). We denote this aggregate structure strain A. In contrast, aggregates in hSOD1D90A mice are usually distinctly different and denoted strain B. The sequence regions comprising β-strands 5–7, which are well hidden from the antibodies in the strain-A structure, are now exposed. By strain, we refer here to any distinct topological arrangement of the fibrillar structure that remains robust to point mutation.
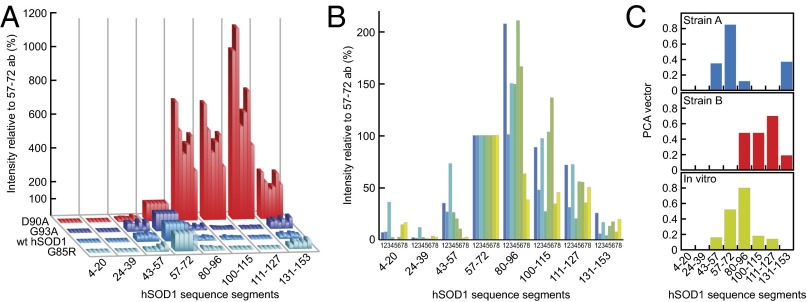
Fig. 3.
Human SOD1 aggregation is malleable and forms two structural strains (A and B) in spinal cord, which both differ from hSOD1 fibrils produced in vitro. The data are presented normalized against the staining with the 57–72 antibody (100%) to enhance appreciation of patterns. (A) Epitope-mapping patterns of hSOD1 aggregates from terminally ill mice of the four different transgenic ALS models. Aggregate results for all hSOD1G85R, hSOD1wt, and hSOD1G93A mice presented in Table S1 are shown. The hSOD1D90A aggregates are from the six youngest mice shown in Fig. 4. (B) Epitope-mapping patterns of hSOD1 aggregates generated in vitro under eight different conditions (SI Materials and Methods). (C) Model-free PCA analysis of antibody data revealing two strains of in vivo aggregates (A and B) that differ from the hSOD1 fibrils produced in vitro (SI Materials and Methods).
To test for the possibility that the patterns resulted from binding competition with other proteins, spinal cord homogenates containing strains A and B were before filter capture harshly treated with sonication in the presence of guanidinium chloride (GdmCl) or urea to dislodge any protein adhering to their surface. No effects were seen on the antibody-binding characteristics of the aggregate strains (Fig. S3A).
In most assays of terminal mice, brain was analyzed in parallel with spinal cord (Fig. 2G). Aggregates were found in all cases, and the strain patterns resembled those in spinal cord. Even so, the amounts were smaller in brain: on average 4, 4, and 40% of those found in hSOD1G93A, hSOD1G85R, and hSOD1wt spinal cords, respectively. For hSOD1D90A mice, see Fig. 5C and Fig. S4 A and B. No aggregates were found in liver, kidney, heart, and skeletal muscle. No (co)aggregation of murine SOD1 was detected in spinal cords (Fig. S5A).
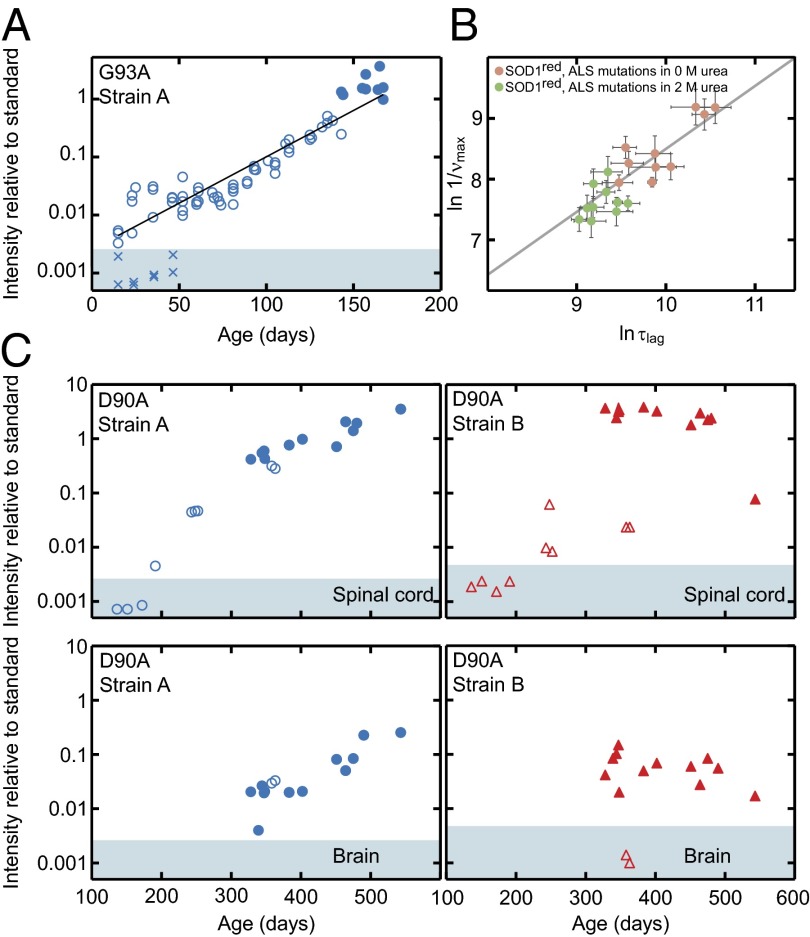
Fig. 5.
In vivo aggregation kinetics of hSOD1 in spinal cord and brain. (A) Strain-A aggregation in spinal cord of hSOD1G93A mice shows essentially simplistic exponential growth (open and filled circles). The filled circles represent end-stage mice. X indicates brains from some of the young hSOD1G93A mice, showing absence of detectable aggregates. Brains contain as much mutant hSOD1 as spinal cords in hSOD1G93A mice (26). (B) In vitro aggregation data of monomeric reduced apo-hSOD1 variants, destabilized by single point mutations in buffer (pink circles) and 2 M urea (green circles), show a strong correlation (slope δlogτlag/δlogνmax = 1.03 ± 0.18, r = 0.88) between the apparent lag time, logτlag, and the aggregate growth rate, δlogνmax, indicating exponential kinetics analogous to the in vivo data in A. (C) Time courses of the simultaneous strain-A and -B aggregation in hSOD1D90A mice (open symbols, nonsymptomatic; closed, end-stage mice). The top panels show data from spinal cord, and the bottom panels show the matching samples from brain. The shaded areas in A and C indicate staining intensities below means + 2 SD of the results for nontransgenic controls (Table S1).
The strain A and B patterns arising in vivo seem to contrast with the somewhat variable aggregate structures formed by the hSOD1 variants in vitro (14, 16). One explanation would be that the CNS modulates the hSOD1 aggregation process by funneling toward uniform aggregate structures distinct from those that spontaneously form under simplified conditions in vitro. As an independent control of such malleability, we produced hSOD1 fibrils under a variety of conditions in vitro and mapped out their structures with the epitope assay (Fig. 3B). To mimic the conditions of the CNS homogenate analyses, the preparations were added to brain homogenates from SOD1 knockout mice. The addition to the brain homogenate, however, had only a negligible effect on the staining characteristics (Fig. S3B). The results show that the in vitro fibrils, despite some variability, are generally more similar to each other than to the aggregates formed in vivo (Fig. 3A). Even so, a shared feature of the in vivo aggregates and in vitro fibrils is the structural engagement of the N-terminal β-strands 1–3 (Fig. 1D), which is fully consistent with earlier peptide work and computational predictions of high aggregation propensities for these sequence regions (17, 18).
Finally, we confirmed the type A and B epitopes by model-free principal component analysis (PCA) (SI Materials and Methods). Antibody-binding data from all mice and also the in vitro-generated fibrils were used to identify the principal vectors corresponding to the most abundant aggregate epitopes. This analysis identified that more than 96% of the data can be fully explained by two strains of in vivo aggregates (A and B), which in turn differed from the in vitro fibrils. The structural variability within each in vivo subpopulation is small; in contrast, the aggregates generated in vitro show a higher degree of structural variation. Despite this variability, the observed orthogonality of the PCA vectors shows that the in vitro aggregates are not mixed populations of strain A and B structure, but represent structurally distinct species (Fig. 3C, SI Materials and Methods, and Table S2).
Coupling Between Aggregate Structure and Disease Progression in D90A Mice.
Closer examination of hSOD1 aggregation in hSOD1D90A mice revealed the most remarkable manifestation of aggregation malleability. The model shows a broad distribution of lifespans (Fig. 4B and SI Materials and Methods) (25). We analyzed mice that became terminally ill throughout that interval, ranging from 328 to 543 d. The epitope-mapping patterns of most mice seemed to be of strain-B type (Fig. 4A and Fig. S4 A and B). The most persistent 543-d-old mouse, however, showed typical strain-A structure (Fig. 4A), following the pattern of the other mouse models (Fig. 3A). The expression of hSOD1D90A in this seemingly deviant mouse was found not to differ from that in younger mice developing strain-B aggregates (Fig. S5B).
Fig. 4.
Correlation between disease progression, strain levels, and their structural properties. Material from spinal-cord samples of hSOD1D90A mice. (A) Epitope-mapping patterns of hSOD1 aggregates from the youngest (328 d, red bars) and oldest (543 d, blue bars) terminally ill hSOD1D90A mice that were analyzed. (B) Coexisting strain A and B plotted against lifespans of mice. (C) Ratios of the strain-B (intensity with 111–127 antibody) and -A (intensity with 57–72 antibody) levels in B, showing clear correlation with survival time. (D) Chemical–mechanical titration of hSOD1 aggregates by sonication of spinal cord homogenates from four terminal hSOD1D90A mice in indicated concentrations of GdmCl followed by analysis in the epitope-mapping assay. Strain-B aggregates are more fragile than their strain-A counterparts.
The identification of this mouse prompted a more systematic analysis of the epitope-mapping patterns in hSOD1D90A mice. As tentative measures of strain-A and -B levels, we used the staining intensities of the 57–72 and 111–127 antibodies (cf. Figs. 3C and 4A), respectively, and plotted these against the lifespans of the individual terminal mice. The plot indicates a strict correlation between aggregate structure and survival time (Fig. 4 B and C and Fig. S4 A and B): Mice with intermediate lifespans (400–500 d) show mixed aggregate structures, which progressively adjust toward cleaner strain-A and -B structures at either end of the lifespan distribution. It is thus apparent that the two strains coexist in hSOD1D90A mice—and the shorter the lifespan, the more the strain B aggregates predominate. Notably, in mice still not symptomatic within the age span of terminal disease, the strain A aggregate levels follow the trajectory of strain-A aggregation in terminally ill mice, whereas the levels of strain B are very low.
This takes us to the question of onset versus progression. To explore this, we plotted the disease duration (i.e., the time between onset of symptoms and death) against lifespan for hSOD1D90A mice. Again, a distinct trend emerges, indicating an acceleration of end-stage disease progression for the early-onset mice. No such trend was seen in the other transgenic model examined (Fig. S6). On this basis, we conclude that the anomalous strain-B aggregates are not only linked to earlier onset but also to faster progression from first symptoms.
Aggregate Fragility Suggests a Link to Simplistic in Vitro Fibrillation Behavior.
In several other protein-disease models, there is evidence for a positive correlation between aggregate “fragility” (i.e., how easily they break apart under mechanical stress and neuroinvasiveness/toxicity) (27–30). A corresponding relationship between aggregate fragility and growth has also been predicted theoretically (4):
| [1] |
Here, is the maximum growth rate, [protein] is the concentration of monomers, e is Euler´s number, and k+ and k− are the rate constants for aggregate elongation and fragmentation, respectively. In this limit of fragmentation control, the aggregation kinetics shows exponential growth with a concentration dependence of δlog (4). Fibrillation of hSOD1 in vitro is found to obey precisely this simplistic behavior (15), as do several other precursors of protein-aggregation disease (4). In further agreement with Eq. 1, we have observed that the lifespan of ALS mice is correlated with the hSOD1 expression levels (26).
Now, we take this analysis one step further by examining the coupling to aggregate fragility. Following standard protocols from the prion field (27), we added spinal-cord homogenates from terminally ill hSOD1D90A mice to PBS containing various concentrations of the ionic chaotrope GdmCl and induced mechanical shear by sonication. After dilution, the fragmentation of the strain-A and -B aggregates was quantified by filter capture, essentially measuring the progressive reduction in aggregate size. The result shows, strikingly, that the more “aggressive” strain B breaks apart more rapidly than strain A (Fig. 4D). Assuming that the midpoints for these fragmentation transitions are proportional to k−, Eq. 1 predicts a ratio of , which is in noticeable accord with the ratio of the hSOD1D90A lifespans at either extreme of their broad distribution, that is, (Fig. 4B and SI Materials and Methods). This overall consistency between in vivo data and in vitro behavior indicates that aggregation of hSOD1 in ALS mice, despite the complexity of the live tissue, is at some level tractable by basic analytical tools.
In Vivo Aggregation Kinetics.
The high sensitivity of the assay allows wide dynamic range recording of in vivo aggregation kinetics for direct comparison with in vitro data and theory. Using the 57–72 antibody, aggregates could be detected in hSOD1G93A spinal cords from 14 d of age. This is far earlier than histopathological evidence of injury, which appears at around 40 d (31). These early aggregates then underwent an essentially exponential buildup with a doubling time of around 14 d (Fig. 5A). Notably, there is no indication of any lag period in the early phase or surge in aggregation in the terminal phase. This simplistic in vivo kinetics closely resembles the exponential, fragmentation-assisted, fibrillar growth observed for apo-hSOD1 monomers in vitro (Fig. 5B), where the apparent lag time directly correlates with the growth rate, clearly indicating a single exponential mechanism for the complete aggregation event (4, 15).
We extended the kinetic analysis to the hSOD1D90A model, including terminal mice and also nonsymptomatic and younger mice. As expected, the picture is here more complex with different kinetics for strains A and B. Detectable amounts of strain-A aggregates first appear around 200 d and grow evenly and exponentially, but much more slowly than in hSOD1G93A mice, to finally slow down in the terminal regime at over 300 d (Fig. 5C). In contrast, strain-B aggregates first appear at around 250 d and are highly variable between different nonsymptomatic mice (Fig. 5C). Furthermore, there is a precipitous jump to the levels seen in terminally ill mice. The data suggest that strain-B aggregation initiates “stochastically” as a side reaction to the steadier buildup of strain A. The strain-B aggregates then proliferate explosively, which is consistent with their higher degree of fragility. If strain-B initiation does not occur, the mice enjoy a long lifespan and eventually die bearing mainly strain-A aggregates (Fig. 4 B and C and Fig. S4 A and B).
Comparison of Spinal Cord and Brain.
The remarkable correlation between aggregate structures in spinal cord of hSOD1D90A mice and age at terminal disease prompted closer examination also of the brain (Figs. 4 and 5C and Fig. S4 A and B). The antibody-binding patterns were found to be overall similar in the individual mice. The patterns turn from overall B dominance to more A as the mice reach the terminal stage at a higher age. Note also the essentially pure strain-A patterns in both spinal cord and brain of the oldest terminal stage mouse, and in the two mice that were still without symptoms at around 360 d. The amount of hSOD1 aggregates in terminal brain is overall lower than in spinal cord—only around 4% (Fig. S4A). Still, the growth of strain-A and -B aggregates in the hSOD1D90A brain closely follows the kinetics and correlation with lifespan observed in spinal cord (Fig. 5C and Fig. S4 A and B). The only difference may be that the relative levels of the more slowly developing strain-A aggregates lag behind somewhat in the brain tissue (Fig. 5C). A speculative interpretation of the findings is that the aggregation is not cell-autonomous but instead spreads within the CNS.
Despite there being similar levels of transgenically expressed hSOD1 variants in spinal cord and brain (12, 26), the levels of aggregates are generally 20-fold lower in brain of hSOD1D90A mice and the other mouse models than in spinal cord. One explanation could be that unfolded disulfide-reduced hSOD1 monomers, which are the likely precursors of aggregates, are overall present at higher levels in the spinal cord than in brain (21, 22). Another feature is the uneven distribution of pathology in the brain, as suggested by immunohistochemistry (Fig. S7). In certain areas of the brain, the hSOD1 aggregation pathology may approach that found in spinal cords.
Discussion
The generally low levels and elusive structures of pathogenic aggregates in neural tissue pose a great challenge for establishing the cause and mechanism of neurodegenerative disease. In spinal cord of terminally ill ALS mice, the level of hSOD1 aggregates is typically less than 100 µg/g wet weight (Fig. S5A), masked by a 1,000-fold background of other cellular proteins. Tailored to work under such circumstances, the antibody assay used here allows quantification and structural fingerprinting of protein aggregates at levels down to 0.1 µg/g wet weight (Fig. 5 A and C). In contrast to previous antibody-based approaches (32), this assay does not rely on the specific 3D structure of the target aggregates but is based on recognizing their local disorder (i.e., the sequence regions protruding from the surface of the aggregate) (Fig. 2 A–G). Accordingly, a single antibody assay can fingerprint at the binary level all combinatorial variants of aggregate structures arising from a given protein precursor (Fig. 3 A–C), and the only input needed to generate this antibody assay is the amino acid sequence of the protein precursor itself (Fig. 1). The usefulness of the antibody assay was tested on transgenic mice expressing a series of ALS-provoking hSOD1 variants, each of which with distinct disease progression. In doing this, we focused on four experimental readouts, chosen to capture the general mechanistic aspects of the disease mechanism and allowing comparison with previous data in the literature: (i) aggregation malleability and structure of aggregates (Figs. 3 and 4), (ii) coupling of structure to disease progression (Fig. 4 A–D), (iii) aggregate growth (Fig. 5 A–C), and (iv) distribution in the CNS (Fig. 5C and Fig. S4 A and B).
Despite stemming from neurons with apparently amorphous inclusions (Fig. 1C), hSOD1 aggregates show distinct order and seem to conform with the generic outline of fibrils: a central spine of repetitively stacked β-strands, with unrecruited sequence material looping into the surrounding solvent (2, 3). Consistent with multiple aggregation sites in the hSOD1 sequence (17, 18), the antibodies recognize two aggregate structures in mice: strain A, hiding ∼ 60% of the primary sequence, and the seemingly less complex strain B, hiding just ∼ 30% (Fig. 3A). Similar strain plasticity has been observed in other neurodegenerative diseases (1, 27, 28, 33–36). Considering the quite different and more malleable appearance of the hSOD1 fibrils in vitro (Fig. 3B), the channeling into merely two well-defined species in vivo is remarkable and supports the idea that the in vivo environment shapes the aggregation process. As to mechanisms, we can only speculate at this point. The major binding partner of disordered hSOD1 in the spinal cord is HSC-70 (37–39), and such chaperone binding would be expected to influence exposure of aggregation-competent sequence elements. Additional modulation of in vivo aggregation would be expected from crowding by surrounding macromolecules, and from interactions with intracellular membranes (40). Furthermore, resistance to recognition and degradation, as well as efficiency in cell-to-cell transmission, would be expected to select for certain fibril structures among multiple types formed.
The stable strain A was detectable in the spinal cord of all transgenic mouse lines examined in this study (Figs. 3A and 4 A and B and Table S1). The less robust strain B, however, emerges only in hSOD1D90A mice and then in coexistence with strain A (Fig. 4 A and B). This coexistence distinguishes the hSOD1D90A mice from previous disease models where the variation of strains is typically observed between individuals (1, 34, 35, 41). Even so, the hSOD1D90A mice highlight the notion that strain variation influences pathology, as previously observed for prions (27, 28), and in patients and models for synucleopathies (33), Alzheimer’s disease (1, 35, 36), and tauopathies (34).
Despite the complexity of hSOD1D90A aggregation kinetics, it is evident that the terminal stage is reached at comparable hSOD1 aggregate loads in all mouse models of the current study (Fig. 4 and Table S1). The consistent “toxic” effect of aggregates of diverse hSOD1 variants, and perhaps also different aggregate strains, lends further fuel to the discussion of whether neural damage in protein disease really needs the assumption of “toxic oligomers” or can simply be explained by aggregate load.
The ability of the antibody assay to follow, both structurally and quantitatively, these strain variations in vivo opens new analytical possibilities. Of immediate interest is not only examination of the origin of strain divergence, but also determination of whether strain heterogeneity as observed in hSOD1D90A mice is an underlying cause of the poorly predictable progression and clinical symptoms of human ALS patients.
Finally, there is currently an intense exploration of immunotherapy directed against aggregating proteins in neurodegenerative diseases. The concept and results of the epitope-mapping assay should provide valuable guidance in selection of epitopes/sequence segments against which to direct passive and active immunotherapy.
Materials and Methods
Additional materials and methods are provided in SI Materials and Methods.
Epitope-Mapping Assay for hSOD1 Aggregate Structure.
Mice were killed by i.p. injection of pentobarbital. The whole spinal cord and brain, dissected free from midbrain, pons, medulla, and cerebellum, were mostly examined. The dissected tissues were homogenized with an Ultraturrax apparatus (IKA) for 30 s and by sonication for 1 min in 25 volumes of ice-cold PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4, pH 7.0) supplemented with 1.8 mM EDTA, 1 mM DTT, and the antiproteolytic mixture Complete without EDTA (Roche Diagnostics). The tissue homogenates were added to 20 volumes of PBS with DTT and EDTA containing 1% of the detergent Nonidet P-40, sonicated for 30 s, and then centrifuged at 200 × g for 10 min. The supernatants were diluted stepwise 1 + 1 in the PBS, and 100 μL was captured on 0.2-μm cellulose acetate filters in a 96-well dot-blot apparatus similar to a previous description (42) (Whatman GmbH). The wells were then washed three times with 300 μL of the PBS without Nonidet P-40. Following blocking in TBS with 50 g/L dry milk and 0.1% (vol/vol) Tween 20 for 1 h, the filters were cut in slices and incubated with the eight anti-hSOD1 peptide antibodies overnight at 4 °C. We aimed at using antibody concentrations that would bind equally well to soluble misfolded hSOD1 species in spinal-cord extracts. When equal amounts of the anti-peptide antibodies immobilized on Sepharose were incubated with a 25,000 × g supernatant of a hSOD1G93A spinal-cord supernatant, most were found to bind equal amounts of misfolded hSOD1 (Fig. S2A). They were used at 0.01 μg/mL in blocking buffer. The 4–20 and 100–115 antibodies bound about half as much and were used at 0.02 μg/mL. After washing, the blots were thereafter developed with HRP-substituted goat anti-rabbit Ig antibodies (Dako 1/42000) and ECL Advance (GE Healthcare), recorded with a Chemidoc apparatus, and evaluated with Quantity One software (BioRad).
To allow comparison and quantification, one homogenate of a spinal cord from a terminal hSOD1G93A mouse was designated as a standard (set to 1) and kept in multiple aliquots at −80 °C. Dilutions series (1 + 1) of this standard were run in one or two lanes of all filters and were stained with the 57–72 antibody. All blots of all homogenates with all antibodies were quantified against this standard. To facilitate comparison of staining patterns, in some cases the staining intensities of the eight antibodies of individual homogenates were normalized against the staining of the homogenate with the 57–72 antibody (taken as 100%).
All animal use and procedures were approved by the Umeå Regional Ethics Committee for Animal Research.
Supplementary Material
Acknowledgments
We thank Eva Bern, Karin Hjertkvist, Åsa Larefalk, Ulla-Stina Spetz, and Agneta Öberg for skillful technical assistance. This work was supported by the Swedish Research Council, The Knut and Alice Wallenberg Foundation, the Bertil Hållsten Foundation, the Torsten and Ragnar Söderberg Foundation, the Swedish Brain Fund, the Västerbotten County Council, and the Kempe Foundations.
Footnotes
Conflict of interest statement: S.L.M., P.M.A., T.B., and M.O. have filed a patent partially related to concepts presented in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419228112/-/DCSupplemental.
References
- 1.Lu JX, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154(6):1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43(1):8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Knowles TP, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326(5959):1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson N, et al. General structural motifs of amyloid protofilaments. Proc Natl Acad Sci USA. 2006;103(44):16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravits JM, La Spada AR. ALS motor phenotype heterogeneity, focality, and spread: Deconstructing motor neuron degeneration. Neurology. 2009;73(10):805–811. doi: 10.1212/WNL.0b013e3181b6bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wroe R, Wai-Ling Butler A, Andersen PM, Powell JF, Al-Chalabi A. ALSOD: The Amyotrophic Lateral Sclerosis Online Database. Amyotroph Lateral Scler. 2008;9(4):249–250. doi: 10.1080/17482960802146106. [DOI] [PubMed] [Google Scholar]
- 8.Sandelin E, Nordlund A, Andersen PM, Marklund SS, Oliveberg M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J Biol Chem. 2007;282(29):21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- 9.Forsberg K, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE. 2010;5(7):e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsberg K, Andersen PM, Marklund SL, Brännström T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121(5):623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13(11):1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graffmo KS, et al. Expression of wild-type human superoxide dismutase-1 in mice causes amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22(1):51–60. doi: 10.1093/hmg/dds399. [DOI] [PubMed] [Google Scholar]
- 13.Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Reports. 2013;5(3):781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan PK, et al. Structural similarity of wild-type and ALS-mutant superoxide dismutase-1 fibrils using limited proteolysis and atomic force microscopy. Proc Natl Acad Sci USA. 2013;110(27):10934–10939. doi: 10.1073/pnas.1309613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang L, Kurnik M, Danielsson J, Oliveberg M. Fibrillation precursor of superoxide dismutase 1 revealed by gradual tuning of the protein-folding equilibrium. Proc Natl Acad Sci USA. 2012;109(44):17868–17873. doi: 10.1073/pnas.1201795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa Y, Kaneko K, Yamanaka K, Nukina N. Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2010;285(29):22221–22231. doi: 10.1074/jbc.M110.113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova MI, et al. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc Natl Acad Sci USA. 2014;111(1):197–201. doi: 10.1073/pnas.1320786110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousseau F, Schymkowitz J, Oliveberg M. ALS precursor finally shaken into fibrils. Proc Natl Acad Sci USA. 2008;105(48):18649–18650. doi: 10.1073/pnas.0810568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38(3):245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 20.Tainer JA, et al. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984;312(5990):127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- 21.Zetterström P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104(35):14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zetterström P, Graffmo KS, Andersen PM, Brännström T, Marklund SL. Composition of soluble misfolded superoxide dismutase-1 in murine models of amyotrophic lateral sclerosis. Neuromolecular Med. 2013;15(1):147–158. doi: 10.1007/s12017-012-8204-z. [DOI] [PubMed] [Google Scholar]
- 23.Gurney ME. Transgenic-mouse model of amyotrophic lateral sclerosis. N Engl J Med. 1994;331(25):1721–1722. doi: 10.1056/NEJM199412223312516. [DOI] [PubMed] [Google Scholar]
- 24.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18(2):327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 25.Jonsson PA, et al. Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J Neuropathol Exp Neurol. 2006;65(12):1126–1136. doi: 10.1097/01.jnen.0000248545.36046.3c. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson PA, et al. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129(Pt 2):451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 27.Bett C, et al. Biochemical properties of highly neuroinvasive prion strains. PLoS Pathog. 2012;8(2):e1002522. doi: 10.1371/journal.ppat.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3(1):a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue WF, Hellewell AL, Hewitt EW, Radford SE. Fibril fragmentation in amyloid assembly and cytotoxicity: When size matters. Prion. 2010;4(1):20–25. doi: 10.4161/pri.4.1.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442(7102):585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 31.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 32.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 33.Bousset L, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clavaguera F, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci USA. 2013;110(23):9535–9540. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heilbronner G, et al. Seeded strain-like transmission of β-amyloid morphotypes in APP transgenic mice. EMBO Rep. 2013;14(11):1017–1022. doi: 10.1038/embor.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watts JC, et al. Serial propagation of distinct strains of Aβ prions from Alzheimer’s disease patients. Proc Natl Acad Sci USA. 2014;111(28):10323–10328. doi: 10.1073/pnas.1408900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA. 2009;106(5):1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe Y, et al. Adherent monomer-misfolded SOD1. PLoS ONE. 2008;3(10):e3497. doi: 10.1371/journal.pone.0003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zetterström P, Graffmo KS, Andersen PM, Brännström T, Marklund SL. Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic amyotrophic lateral sclerosis (ALS) model mice. J Biol Chem. 2011;286(23):20130–20136. doi: 10.1074/jbc.M111.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aisenbrey C, et al. How is protein aggregation in amyloidogenic diseases modulated by biological membranes? Eur Biophys J. 2008;37(3):247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 41.Watts JC, et al. Transmission of multiple system atrophy prions to transgenic mice. Proc Natl Acad Sci USA. 2013;110(48):19555–19560. doi: 10.1073/pnas.1318268110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Xu G, Borchelt DR. High molecular weight complexes of mutant superoxide dismutase 1: Age-dependent and tissue-specific accumulation. Neurobiol Dis. 2002;9(2):139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.