Abstract
‘Will my study answer my research question?’ is the most fundamental question a researcher can ask when designing a study, yet when phrased in statistical terms – ‘What is the power of my study?’ or ‘How precise will my parameter estimate be?’ – few researchers in ecology and evolution (EE) try to answer it, despite the detrimental consequences of performing under- or over-powered research. We suggest that this reluctance is due in large part to the unsuitability of simple methods of power analysis (broadly defined as any attempt to quantify prospectively the ‘informativeness’ of a study) for the complex models commonly used in EE research. With the aim of encouraging the use of power analysis, we present simulation from generalized linear mixed models (GLMMs) as a flexible and accessible approach to power analysis that can account for random effects, overdispersion and diverse response distributions.
We illustrate the benefits of simulation-based power analysis in two research scenarios: estimating the precision of a survey to estimate tick burdens on grouse chicks and estimating the power of a trial to compare the efficacy of insecticide-treated nets in malaria mosquito control. We provide a freely available R function, sim.glmm, for simulating from GLMMs.
Analysis of simulated data revealed that the effects of accounting for realistic levels of random effects and overdispersion on power and precision estimates were substantial, with correspondingly severe implications for study design in the form of up to fivefold increases in sampling effort. We also show the utility of simulations for identifying scenarios where GLMM-fitting methods can perform poorly.
These results illustrate the inadequacy of standard analytical power analysis methods and the flexibility of simulation-based power analysis for GLMMs. The wider use of these methods should contribute to improving the quality of study design in EE.
Keywords: experimental design, sample size, precision, generalized linear mixed model, random effects, simulation, overdispersion, long-lasting insecticidal net
Introduction
‘Will my study answer my research question?’ is the most fundamental question a researcher can ask when designing a study, yet when phrased in statistical terms – ‘What is the power of my study?’ or ‘How precise will my parameter estimate be?’ – few researchers in ecology and evolution (EE) try to answer it (e.g. Taborsky 2010). Consequently many, possibly most, studies are underpowered (Jennions & Møller 2003; Smith, Hardy & Gammell 2011) and likely to be uninformative or misleading (Ioannidis 2005). Failure to consider power can also result in overpowered studies. Both under- and overpowering waste resources and can raise ethical concerns (e.g. in animal studies, by potentially causing needless suffering; and in disease control by causing potentially promising control methods to be prematurely dismissed). Hence, researchers should take all reasonable steps to ensure sufficient, but not wastefully excessive, power.
Power is defined as the probability of rejecting the null hypothesis when it is false and is equal to one minus the type II (false negative) error rate or 1-β. In other words, it is the probability of detecting an effect, given that it exists. It depends on the sample size, the effect size, the amount of variability in the response variable and the significance level. Generally, the aim of a power analysis is to predict the power of a particular experimental design, or the sample size required to achieve an acceptable level of power [80% power is conventionally deemed adequate, although often without justification (Di Stefano 2003)]. Power analysis therefore exists within the framework of null hypothesis significance testing (NHST). In this article, we define power analysis more broadly as any attempt to quantify prospectively the ‘informativeness’ of a study (Bolker 2008; Cumming 2013). This definition of power analysis covers, for example, predicting the precision of an estimate, and could be applied within alternative inference frameworks such as Bayesian or information theoretic.
A major obstacle to power analysis is that standard methods are suitable for only the simplest statistical analyses, such as comparing means using t-tests or anova, or proportions using χ2 tests, and are inadequate when confronted with the more complex analyses generally required to analyse ecological data. Such analyses commonly accommodate multiple sources of random variation (e.g. within and between study sites), where random effects models (also known as mixed effects models) are recommended. In addition, response measures such as counts that are common in EE are not readily shoehorned into t-tests, anova or χ2 tests, and consequently, the associated power analysis methods are inappropriate. Neither random effects nor count responses are handled in relatively sophisticated power analysis software such as g*power (Faul et al. 2007).
The generalized linear mixed model (GLMM) is an analysis framework widely used in EE that can accommodate these complexities. GLMMs allow modelling of diverse response distributions and multiple sources of random variation termed random effects, both of which are common in EE (Bolker et al. 2009; Zuur, Hilbe & Leno 2013). Although analytical formulae for estimating power are available for the simplest Gaussian GLMMs (Snijders & Bosker 1993), more general formulae are not available. A more flexible approach is to use Monte Carlo simulation (Thomas & Juanes 1996). Simulation-based power analysis has many additional advantages over analytical power analysis, beyond its greater flexibility. It is more accurate, conceptually simpler and easy to extend beyond hypothesis testing (Bolker 2008). Simulation-based power analysis methods are available for Gaussian GLMMs (Martin et al. 2011), but there is a lack of guidance and software facilitating power analysis for scenarios with non-Gaussian responses and more complex random effect structures.
The first aim of this study is to illustrate the value of power analysis in the broad sense of predicting the informativeness of a study. The second is to present simulation from GLMMs as a flexible and accessible power analysis method, with examples taken from two ecological systems where standard power analysis methods are inadequate: estimating tick density on game birds and assessment of insecticide-treated nets for malaria mosquito control.
Materials and methods
Power Analysis using Simulation
Estimating the power of a test of a null hypothesis by simulation requires the following steps (Bolker 2008):
Simulate many data sets assuming that the alternative hypothesis is true, that is, the effect of interest is not zero. ‘Many’ means enough to give an adequately precise power estimate. As a guide, with 100 simulations and 80% power, the power estimate will fall within 72–88% with 95% probability, while using 1000 simulations will reduce this range to 77·5–82·4%.
Using each simulated data set, perform a statistical test of the null hypothesis that the effect size is zero.
Calculate the proportion of simulated data sets in which the null hypothesis was rejected. This proportion is the power estimate.
The effect of different designs and assumptions (e.g. sample size, effect size, random effect variances) on power can be explored by repeating steps 1–3 across a range of realistic scenarios.
This scheme can be easily adapted to quantify the informativeness of a study in the broader sense of power analysis defined above. The precision of an effect estimate could be predicted by averaging CI width over the simulated data sets, or, in an information theoretic framework (Burnham & Anderson 2001), the expected difference in the Akaike information criterion (AIC) between models could be estimated.
An Overview of GLMMs
We focus on counts and proportions because these are common types of response data in EE. First, we introduce generalized linear models (GLMs). Like a standard linear regression model (LM), a GLM models, the relationship between the response of the ith observation, yi, and a set of p predictor variables or covariates, x1i, …, xpi via p regression coefficients, β1,…, βp. Unlike a LM, the response can follow distributions other than normal (Gaussian), including binomial, Poisson and negative binomial. As in standard linear regression, the predictors, weighted by the regression coefficients, are summed to form the linear predictor,
where β0 is the intercept. The expected value of yi and the linear predictor, ηi, are related through the link function. For example, in a Poisson GLM, where 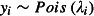 and λi is the expected value of yi, the link function is ηi = log(λi) (where ‘log’ means the natural logarithm, loge, throughout the text). If the responses were binomially distributed with ni Bernoulli trials and pi probability of success in each trial, that is
and λi is the expected value of yi, the link function is ηi = log(λi) (where ‘log’ means the natural logarithm, loge, throughout the text). If the responses were binomially distributed with ni Bernoulli trials and pi probability of success in each trial, that is 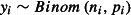 , then we would model the responses using a binomial GLM, usually with a logit (log of the odds) link function,
, then we would model the responses using a binomial GLM, usually with a logit (log of the odds) link function,
There is often a need to account for additional sources of random variation, for example where observations are clustered within study sites, or where multiple observations are taken over time on each study subject. In such data sets, the assumption of the GLM that the yi values are conditionally independent (i.e. independent after adjusting for the effects of covariates) is violated, because clustered observations are correlated. To account for these correlations, a random effect can be added to the linear predictor, allowing each cluster (e.g. site or individual) to have its own mean value. The resulting model is a GLMM. In the example of intersite variation, we are now modelling the response of the ith observation in the jth site, yij. The only change to the model is the addition of a single random effect, γj, to the linear predictor, representing the ‘effect’ of the jth site. Now
where 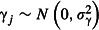 . This is a random intercepts GLMM, so-called because the random effect allows the intercept to vary randomly among sites. Multiple random effects can be included, and random effects can take more complex forms that allow greater flexibility in modelling correlations between observations (e.g. due to familial relationships or proximity in space or time). GLMMs in which regression coefficients, or slopes, are allowed to vary randomly between clusters are termed random intercepts-and-slopes GLMMs. Random intercepts-and-slopes models have been applied to modelling inter-individual variation in regression coefficients both where this variation is the focus of enquiry, as in random regression models (Nussey, Wilson & Brommer 2007), and where it is a nuisance variable that must nevertheless be modelled to guard against overconfident inference (Schielzeth & Forstmeier 2009; Barr et al. 2013). Power analysis for random intercepts-and-slopes GLMMs is beyond the scope of this article, although simulation-based power analysis methods have been developed for Gaussian random regression models and implemented in the pamm package for the R statistical environment (Martin et al. 2011).
. This is a random intercepts GLMM, so-called because the random effect allows the intercept to vary randomly among sites. Multiple random effects can be included, and random effects can take more complex forms that allow greater flexibility in modelling correlations between observations (e.g. due to familial relationships or proximity in space or time). GLMMs in which regression coefficients, or slopes, are allowed to vary randomly between clusters are termed random intercepts-and-slopes GLMMs. Random intercepts-and-slopes models have been applied to modelling inter-individual variation in regression coefficients both where this variation is the focus of enquiry, as in random regression models (Nussey, Wilson & Brommer 2007), and where it is a nuisance variable that must nevertheless be modelled to guard against overconfident inference (Schielzeth & Forstmeier 2009; Barr et al. 2013). Power analysis for random intercepts-and-slopes GLMMs is beyond the scope of this article, although simulation-based power analysis methods have been developed for Gaussian random regression models and implemented in the pamm package for the R statistical environment (Martin et al. 2011).
Simulating from a GLMM
The first step of simulation-based power analysis is to simulate a large number of data sets. When simulating responses from a GLMM, we must assume values for the intercept and the regression coefficients (the fixed effects) and the variances and covariances of the random effects. Estimates of these parameters, or a range of plausible values, will sometimes be available from previous studies; otherwise, a pilot study should be conducted. If no data are available, a plausible range of parameter values could, in some cases and with careful justification, be assumed based on knowledge of the study system.
While designing a study, we will often suspect which predictor variables are most strongly related to the response variable. To simulate responses from a GLMM, we must make assumptions about these relationships, which amounts to assuming values for each of the βm. The interpretation of the βm depends on the type of GLMM. In a Poisson GLMM, they are log relative abundances, or rate ratios, and in a binomial GLMM with a logit link they are log odds ratios. Generally, we will have little knowledge of the size of the effect that is the focus of the study, but this is not an obstacle to power analysis because the study should be powered to detect the smallest biologically meaningful effect, which is purely a question of scientific judgement (see Discussion).
In addition to the βm, we must assume a value for the intercept, β0, which is the expected value of ηij where all the xmij are zero. If setting the xmij to zero is meaningful – for example, if the xmij represent time from the start of the study, or all but one of the levels of a categorical variable – then the intercept is the value of the linear predictor at baseline or in the reference category, respectively. Thus, in a Poisson GLMM, β0 is the log expected count in the baseline or reference category, and in a binomial GLMM with a logit link, β0 is the logit expected proportion or prevalence in the baseline or reference category.
Next, we must make assumptions about the random effects. In the example of a single between-site random effect, only a single variance needs to be assumed. In more complex models, such as random intercepts-and-slopes, multiple random effects and their covariances might need to be considered, but here, we consider only uncorrelated random effects and so assume zero covariance. The value assumed for a random effect variance should be based on an estimate from previous studies or pilot data, where available. Uncertainty around variance estimates can be considerable, and the sensitivity of the power analysis to this uncertainty can be assessed by repeating the power analysis across a range of plausible variance values in place of the point estimate.
An additional source of variation that needs to be considered is overdispersion. Overdispersion is variation exceeding what would be expected from a given distribution and can be thought of as unexplained variation. For example, a Poisson distribution with mean λ also has variance λ. If the variance in a set of counts is greater than the mean then they will not fit a Poisson distribution and are overdispersed. Similarly, a set of binomial responses is overdispersed if the variance exceeds np (1 – p), where n is the number of trials, n > 1, and p is the probability of success in each trial. Overdispersion in a GLMM fit can be artefactual or real (Zuur, Hilbe & Leno 2013). Artefactual overdispersion arises from model misspecification and has several potential causes including missing or poorly modelled covariates, interactions or random effects; wrong choice of distribution or link function; outliers; and zero-inflation. Once these potential causes have been investigated and remedied, any overdispersion that remains is ‘real’ and should be modelled. Fitting a Poisson or binomial GLMM that does not allow for overdispersion is equivalent to assuming that all of the variation that does not arise from the Poisson or binomial distribution is explained by the fixed and random effects. Biological data rarely justify this assumption, so overdispersion should be considered as a matter of course (but note that overdispersion does not apply to the normal distribution because the variance is independent of the mean). While EE researchers have long been alert to overdispersion in count data (Bliss & Fisher 1953; Eberhardt 1978; O'Hara & Kotze 2010), awareness of overdispersion in binomial data (Crowder 1978; Warton & Hui 2011) is comparatively limited. Various methods of accounting for overdispersion are available, including negative binomial and quasi-Poisson for counts, and beta-binomial and quasi-binomial for binomial data (Bolker et al. 2009; O'Hara & Kotze 2010; Warton & Hui 2011; Zuur, Hilbe & Leno 2013). Here, we model overdispersion in both Poisson and binomial GLMMs by adding a normally distributed random intercept,  ij, to the linear predictor of each observation, giving
ij, to the linear predictor of each observation, giving
where 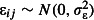 (Elston et al. 2001; Warton & Hui 2011). By absorbing excess (i.e. unexplained) variation, this random effect performs the same role as the residual error term in a linear regression model. Like the random effects variances, the value assumed for the overdispersion variance in the simulation model should be estimated from pilot data.
(Elston et al. 2001; Warton & Hui 2011). By absorbing excess (i.e. unexplained) variation, this random effect performs the same role as the residual error term in a linear regression model. Like the random effects variances, the value assumed for the overdispersion variance in the simulation model should be estimated from pilot data.
Examples
We illustrate power analysis for GLMMs with two contrasting examples. In the first, the responses are counts and the random effects are nested. This example is a power analysis in the broad sense because the aim is to predict not power but the precision of an estimate in terms of confidence interval (CI) width. The second example, in which the responses are binomial and the random effects are crossed, is a narrow sense power analysis where the aim is to estimate the power of a hypothesis test.
Count response example: estimating tick burden on grouse chicks
This example is based on an analysis of tick burdens recorded on grouse chicks on an Aberdeenshire moor from 1995 to 1997 (Elston et al. 2001). Our sampling scheme is a simplified version of Elston et al. (2001), with chicks nested within broods and broods within geographical locations. The aim of the study is to estimate the mean tick burden on grouse chicks in a single year with an expected margin of error of ± 25%. We define margin of error as the average distance from the 95% confidence limits to the estimate (it is necessary to average because a CI for a Poisson mean estimated from a GLM or GLMM is asymmetrical, due to having been back-transformed from a symmetrical 95% CI on log scale). For example, a 95% CI of 8 to 13 around an estimate of 10 ticks per chick has confidence limits that are on average (2+3)/2 = 2·5 units from the estimate, giving an adequately precise margin of error of 2·5/10 = 25%. The aim of the power analysis is to determine the sampling effort required to give adequate precision. Because we are aiming for 25% expected margin of error, and the margin of error is subject to sampling error, we are implicitly prepared to accept a 50% risk of a higher margin of error. If estimating tick burden with poorer precision were expected to have undesirable consequences, the specification could be changed to give greater confidence (e.g. 80% or 90%) of adequate precision. It would be straightforward to adapt the power analysis to estimate the additional sampling effort required.
Predicting the precision to which we can measure tick burden requires the following assumptions (Table1):
Table 1.
Study design choices and effect parameter assumptions for the two example studies
| Example study | Study variable | Simulated values |
|---|---|---|
| Tick burden survey | No of locations | 10, 20, 50, 100, 200 |
| No of broods per location | 2 | |
| No of chicks per brood | 3 | |
Mean tick burden per chick, 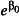
|
1, 5, 10 | |
Location-level variance, 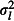
|
0, 1 | |
Brood-level variance, 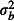
|
0, 0·7 | |
Chick-level variance, 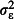
|
0, 0·3 | |
| LLIN trial | No of rotations of the Latin square | 1, 2, 3, 4, 5 |
| No of huts, No of nets, No of weeks per rotation | 6 | |
| No of A. gambiae entering each hut each night, nijk | 5, 25 | |
Mortality using the control net, 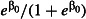
|
70% | |
Minimum acceptable mortality using LLIN type E1, 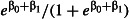 (odds ratio, (odds ratio, 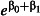 ) ) |
80% (1·7) | |
| Mortality assumed using the four secondary LLINs (odds ratio) | 80% (1·7) | |
Between-hut and -week variances, 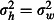
|
0, 0·5 | |
Observation-level (overdispersion) variance, 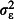
|
0, 0·5, 1 |
The mean tick burden per chick. Mean tick burden is highly variable, ranging from 1·2 to 11·1 over the three years. To account for this uncertainty, we simulated mean tick burdens of 1, 5 and 10 ticks per chick.
The effect sizes of factors affecting tick burden. Tick burden varies substantially between locations, between broods within locations and between chicks within broods (Elston et al. 2001). We modelled variation between locations, broods and chicks with a random effect at each hierarchical level. These random effects are nested, because each brood belongs to only one location, and each chick to only one brood. As each chick provided only one tick count, the chick-level random effect models overdispersion. We simulated naïve (zero) and pessimistic (high) variances for each hierarchical level (Table1). Pessimistic random effect variances were selected by choosing values towards the upper ends of the 95% CIs in Table1 of Elston et al. (2001). In the interests of simplicity, we did not include any fixed effects in the simulation model. Fixed effects are introduced in the second example.
The number of chicks sampled. We assumed that every brood consisted of three chicks and that two broods were sampled at every location. We varied sampling effort by increasing the number of locations sampled from 10 to 200.
The tick burden on the ith chick from the jth brood in the kth location, yijk, was modelled as Poisson distributed, that is 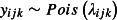 . The expected log tick burden, log(λijk), is
. The expected log tick burden, log(λijk), is
where β0 is the global mean log tick burden, and lk, bjk and ɛijk are the location, brood and chick random effects, which are normally distributed with zero means and variances 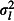 ,
, 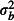 and
and 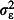 , respectively.
, respectively.
Binomial response example: comparing mortality of malaria mosquitoes exposed to long-lasting insecticidal nets
Long-lasting insecticidal nets (LLINs) are made of fabric with incorporated insecticide which maintains residual activity for several years. LLINs are a key intervention against malaria (Roll Back Malaria Partnership 2008; World Health Organization 2013b). They provide personal protection from malaria mosquitoes by constituting a physical barrier and community protection by killing potentially infectious mosquitoes upon contact (Lengeler 2004). To evaluate their insecticidal activity, LLINs are tested in standardized experimental huts (World Health Organization 2013a) against free-flying, wild mosquitoes. The mosquitoes can enter but not leave the huts, allowing assessment of LLIN efficacy under controlled conditions. Despite their importance in large-scale malaria prevention programs, power analysis has not generally been performed for LLIN hut trials. Power analysis for LLIN hut trials is hindered by the difficulty of accounting for the effects of multiple sources of variation in mosquito mortality, including variation between huts and over time. We show how simulation can be used to account for these complexities.
The aim is to optimize the design of an experimental hut trial comparing mortality in the malaria vector Anopheles gambiae among different types of LLIN. This is achieved by estimating power across a realistic range of assumptions and designs, identifying those that give adequate (≥80%) power. In this example, six types of LLIN are to be tested in six experimental huts, in accordance with the World Health Organization Pesticide Evaluation Scheme (WHOPES) guidelines (World Health Organization 2013a). To prevent confounding of hut and net effects, the LLIN types are rotated through the huts weekly, so that after six weeks, one Latin square rotation has been completed, with each LLIN type having passed one week in each hut (Table2). Simple rotation, where net type E2 always follows E1, etc, would lead to confounding between LLIN type and any carry-over effects, so Table2 presents a design balanced against such effects, where each net type follows each other net type only once (Williams 1949). Data are collected on six nights per week. Nets are replaced after each night so that six replicate nets of each type are used per week. The outcome of interest is mosquito mortality, calculated as the proportion of mosquitoes entering the hut at night that are found dead in the morning or in the following 24 hours. Any mosquito found alive inside the hut is transferred to an insectary and mortality recorded after a holding period of 24 hours. We aim to estimate the number of six-week Latin square rotations that will be required to give adequate power.
Table 2.
Latin square design for trialling one control (C) and five experimental (E1 to E5) types of long-lasting insecticidal net, rotated through six huts over six weeks according to a design balanced against carry-over effects (Williams 1949)
| Week | Hut 1 | Hut 2 | Hut 3 | Hut 4 | Hut 5 | Hut 6 |
|---|---|---|---|---|---|---|
| 1 | E1 | E2 | E3 | E4 | E5 | C |
| 2 | E2 | E3 | E4 | E5 | C | E1 |
| 3 | C | E1 | E2 | E3 | E4 | E5 |
| 4 | E3 | E4 | E5 | C | E1 | E2 |
| 5 | E5 | C | E1 | E2 | E3 | E4 |
| 6 | E4 | E5 | C | E1 | E2 | E3 |
Performing the power analysis requires the following information:
The primary aim of the experiment and the consequent primary analysis. This example follows the standard WHOPES design in which multiple net types are tested simultaneously, allowing multiple secondary research questions to be asked. This power analysis, however, is concerned only with the primary aim of the study, which is to evaluate the efficacy, in terms of mosquito mortality, of an experimental LLIN (labelled E1 in Table2) compared with a positive control LLIN (labelled C in Table2), which would typically be a WHOPES-recommended LLIN (World Health Organization 2013a). The primary analysis will be a two-tailed test of the null hypothesis that the control and E1 LLIN types cause the same mortality. The other four experimental LLIN types (labelled E2 to E5 in Table2) do not feature in the primary analysis, but are included in the simulation because they contribute to the estimation of the hut effect.
The number of mosquitoes in each treatment group. The total number of A. gambiae expected to be sampled in each of the six treatment groups, n, is the product of the number of nights per week when mosquitoes are collected (six), the number of weeks in one rotation (six), the number of mosquitoes caught per night in each hut (nm) and the duration of the study in number of six-week rotations (nr), so that n = 36nmnr The number of mosquitoes entering a hut is difficult to predict. We investigated low (nm = 5) and high (nm = 25) A. gambiae abundance, which is a realistic range (Malima et al. 2008; Winkler et al. 2012; Ngufor et al. 2014). The study was run for nr = 1,… 5 complete six-week rotations of the nets through the huts.
The mortality in huts using the positive control LLIN type. We assumed a realistic mortality rate of 70% (Malima et al. 2008; Winkler et al. 2012; Ngufor et al. 2014).
The size of the smallest treatment effect worth detecting (the sensitivity of the study). We considered mortality of at least 80% using LLIN type E1 to represent a worthwhile improvement in efficacy relative to the control LLIN. This treatment effect can be represented as an odds ratio of
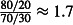 . For simplicity, we also assumed 80% mortality with the four secondary LLINs.
. For simplicity, we also assumed 80% mortality with the four secondary LLINs.
The impact of non-treatment factors on mortality. Variation in mortality among huts and over time is expected, which we simulated as simple random effects between huts and between weeks. The justification for modelling week-to-week variation as random rather than fixed is that we cannot predict the form of the relationship between mortality and time, so any choice of fixed effect form would be arbitrary. A random effect is a simple and conservative way of modelling week-to-week variation without foreknowledge of its nature. The hut and week random effects are completely crossed, because data are collected from each hut during each week (Table2). Between-hut and between-week random effect variances of up to 0·5 are plausible based on analysis data from Winkler et al. (2012). In order to limit the number of parameter combinations, we set the variances between huts and between weeks to be equal. We simulated two hut and week random effect scenarios:
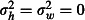 , representing a naïve assumption that mortality does not vary between huts or over weeks and a more realistic assumption of
, representing a naïve assumption that mortality does not vary between huts or over weeks and a more realistic assumption of 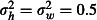 . Another potential source of variation in mortality is the six sleepers who rotate nightly through the huts, in accordance with the WHOPES guidelines (World Health Organization 2013a). We ignored this aspect here for the sake of simplicity and assumed that sleepers have no influence on mosquito mortality.
. Another potential source of variation in mortality is the six sleepers who rotate nightly through the huts, in accordance with the WHOPES guidelines (World Health Organization 2013a). We ignored this aspect here for the sake of simplicity and assumed that sleepers have no influence on mosquito mortality.
The amount of unexplained variation in mortality (overdispersion). We explored three scenarios: a naïve assumption of no overdispersion (
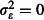 ); realistic overdispersion (
); realistic overdispersion (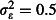 , similar to the estimated value of 0·4 from analysis of Winkler et al. (2012)); and a pessimistic assumption of strong overdispersion (
, similar to the estimated value of 0·4 from analysis of Winkler et al. (2012)); and a pessimistic assumption of strong overdispersion (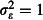 ).
).
These assumptions are summarized in Table1. The GLMM that fits the design described above models the number of dead mosquitoes recorded on the ith night from the jth hut in the kth week among the nijk mosquitoes entering the hut, yijk, as binomially distributed, that is 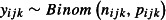 . The log odds of mortality,
. The log odds of mortality, 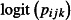 , can be modelled as
, can be modelled as
where the intercept, β0, is the predicted log odds of mortality when using the positive control net (type C) and βm is the log odds ratios representing the difference in the log odds of mortality between the mth of the five experimental nets (types E1-E5) and the control net. The covariate xmjk is an indicator, or dummy variable, that takes the value 1 when the mth of the five experimental nets is in use and 0 otherwise. For example, when the first experimental net, type E1, is in use, x1jk = 1 and x2jk = x3jk = x4jk = x5jk = 0, so the log odds of mortality on the ith night in the jth hut in the kth week is ηijk = β0+β1+hj+wk+ɛijk. The hut random effect, hj, the week random effect, wk, and the observation-level random effect, ɛijk, are normally distributed with zero mean and variances 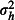 ,
, 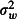 and
and 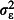 , respectively. For simplicity, we simulated nijk as a constant, although in reality this quantity is random. Simulations of nijk as random indicated that power was not sensitive to this simplification (data not shown).
, respectively. For simplicity, we simulated nijk as a constant, although in reality this quantity is random. Simulations of nijk as random indicated that power was not sensitive to this simplification (data not shown).
Simulation Methods
All combinations of the parameter values in Table1 were simulated, giving a total of 120 scenarios for the tick burden survey and 60 for the LLIN trial. For each example study, 1000 data sets were simulated from each scenario. The responses were simulated using a function, sim.glmm, for the statistical environment R (R Core Team 2014) which is freely available at https://github.com/pcdjohnson/sim.glmm. The function simulates from Gaussian, Poisson, binomial and negative binomial GLMMs; a tutorial is provided as Appendix S1. The simulate.merMod function included in recent versions (≥1·0) of the lme4 R package has similar functionality (Bates et al. 2014).
The next step is to analyse the simulated data set. Ideally, this should be done using the same methods that would be used for the real data, but this is problematic for non-Gaussian GLMMs because the most reliable method for estimating P-values and CIs, parametric bootstrapping (Faraway 2005), is prohibitively slow for multiple simulations. We have therefore taken the approach of using fast, approximate methods to estimate P-values and CIs, while monitoring performance to identify scenarios where precision and power estimates are unacceptably inaccurate. In such cases, the more accurate methods should be used despite their slowness (see Appendix S1). The definition of ‘unacceptably inaccurate’ is subjective, but would cover, for example, a 95% CI that included the true value with only 85% probability. Small biases are acceptable because power analysis is an inherently approximate procedure, with results being strongly dependent on uncertain assumptions. However, in an analysis of real data, where computation time is much shorter because the analysis need be run only once, researchers should use the most accurate methods available, such as parametric bootstrapping (Faraway 2005).
We analysed each simulated data set by fitting the GLMM from which it was simulated using the lme4 package (Bates et al. 2014) for R. Wald z CIs around tick burden estimates were calculated as 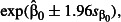 where
where 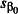 is the estimated standard error of
is the estimated standard error of 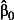 and 1·96 is the 97·5th percentile of the standard normal distribution. The null hypothesis of equal mortality between the standard (C) and experimental (E1) net types was tested using a Wald χ2-test with one degree of freedom for the β1 parameter, with the null hypothesis being rejected when P < 0·05. In GLMMs with overdispersion, Wald z CIs and χ2-tests are expected to give overconfident 95% CI coverage (i.e. true confidence being <95%) and inflated type I error rate (Bolker et al. 2009), leading to overestimation of precision and power. We therefore monitored type I error rate (the proportion of null hypotheses rejected when the null hypothesis is true) and 95% CI coverage (the proportion of 95% CIs that include the true effect size). We also monitored bias in parameter estimation.
and 1·96 is the 97·5th percentile of the standard normal distribution. The null hypothesis of equal mortality between the standard (C) and experimental (E1) net types was tested using a Wald χ2-test with one degree of freedom for the β1 parameter, with the null hypothesis being rejected when P < 0·05. In GLMMs with overdispersion, Wald z CIs and χ2-tests are expected to give overconfident 95% CI coverage (i.e. true confidence being <95%) and inflated type I error rate (Bolker et al. 2009), leading to overestimation of precision and power. We therefore monitored type I error rate (the proportion of null hypotheses rejected when the null hypothesis is true) and 95% CI coverage (the proportion of 95% CIs that include the true effect size). We also monitored bias in parameter estimation.
Power to detect the difference in mosquito mortality between the two LLIN types was estimated as the proportion of the 1000 simulated data sets in which the null hypothesis was rejected. Margin of error in the tick burden example was averaged over the 1000 simulated data sets. Mean computation time per 1000 simulations was 8·8 min for the binomial example and 2·0 min for the Poisson data example using a 2·7 GHz Intel Core i7 processor, parallelizing across 8 processor cores.
Results
Count Response Example: Estimating Tick Burden on Grouse Chicks
The precision of the estimates was greatly reduced by allowing counts of ticks on chicks to be non-independent within broods and locations (Fig.1). The analysis assuming independence among chicks (random effect and overdispersion variances = 0; black circles and solid lines in Fig.1) suggests that sampling only 20 locations will be sufficient to keep the expected margin of error within ± 25% (below the grey line in Fig.1) even at the lowest mean tick burden. Under more realistic assumptions that allow tick burdens to be correlated at the chick, brood and location levels (red triangles and dashed lines in Fig.1), the margin of error doubles. The impact on the study design of considering random effects is severe. A fivefold increase in sampling effort to 100 locations is required to achieve the desired level of precision, regardless of mean tick burden. In contrast to the binomial example (see below), random effects at all levels, not just overdispersion, contribute to reducing precision.
Figure 1.
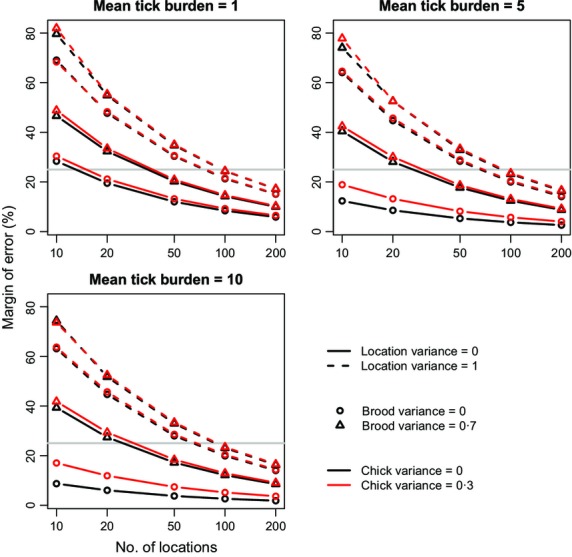
The relationship between margin of error in tick burden estimates and number of locations sampled. Margin of error was averaged over 1000 data sets simulated under scenarios that varied in mean tick burden and the degree of variation in mean tick burden at the location, brood and individual chick levels. The grey line shows the target margin of error of ± 25%.
Of the three random effects, the location effect had the largest impact on precision. The brood effect also substantially reduced precision, while the effect of the chick (or overdispersion) random effect was generally small. We confirmed that this effect was not simply due to location having the strongest random effect (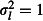 ) by repeating the simulations with random effects variances at all levels set to 0·5 (data not shown).
) by repeating the simulations with random effects variances at all levels set to 0·5 (data not shown).
Binomial Response Example: Comparing Mortality of Malaria Mosquitoes Exposed To Long-Lasting Insecticidal Nets
Standard power calculators ignore sources of non-independence such as the hut, week and overdispersion effects considered here, producing power estimates equivalent to the black circles in Fig.2 (i.e. variance for each random effect was set to 0). This was verified by estimating power assuming independent observations using g*power (Faul et al. 2007) and the power.prop.test function in R. Ignoring these effects and assuming that on average only five A. gambiae will enter each hut each night, 56% power is achieved after one rotation, 89% after two and 96% after three. Under the high abundance scenario of 25 A. gambiae per hut per night on average, a single rotation of the Latin square is sufficient to give 100% power. Thus, a cautious researcher might plan to run the experiment over two 6-week rotations to insure against low A. gambiae abundance. How does consideration of the random effects change this conclusion? Power is decreased slightly by the hut and week random effects (triangles in Fig.2), but is markedly reduced by overdispersion (red dashed and blue dotted lines). When mosquito abundance is low, realistic overdispersion (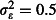 ; red dashed lines) necessitates three rotations to achieve at least 80% power, and strong overdispersion (
; red dashed lines) necessitates three rotations to achieve at least 80% power, and strong overdispersion (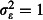 ; blue dotted lines) requires three to four rotations. At higher abundance, and assuming realistic overdispersion (red dashed lines), power falls just short of the 80% threshold after one rotation so that two rotations are required to exceed it. Two rotations are also just sufficient to achieve 80% power assuming strong overdispersion (blue dotted lines). Thus, consideration of non-independence, and overdispersion in particular, would motivate extending the duration of the trial by 50–100%, with important implications for planning and resourcing the study.
; blue dotted lines) requires three to four rotations. At higher abundance, and assuming realistic overdispersion (red dashed lines), power falls just short of the 80% threshold after one rotation so that two rotations are required to exceed it. Two rotations are also just sufficient to achieve 80% power assuming strong overdispersion (blue dotted lines). Thus, consideration of non-independence, and overdispersion in particular, would motivate extending the duration of the trial by 50–100%, with important implications for planning and resourcing the study.
Figure 2.
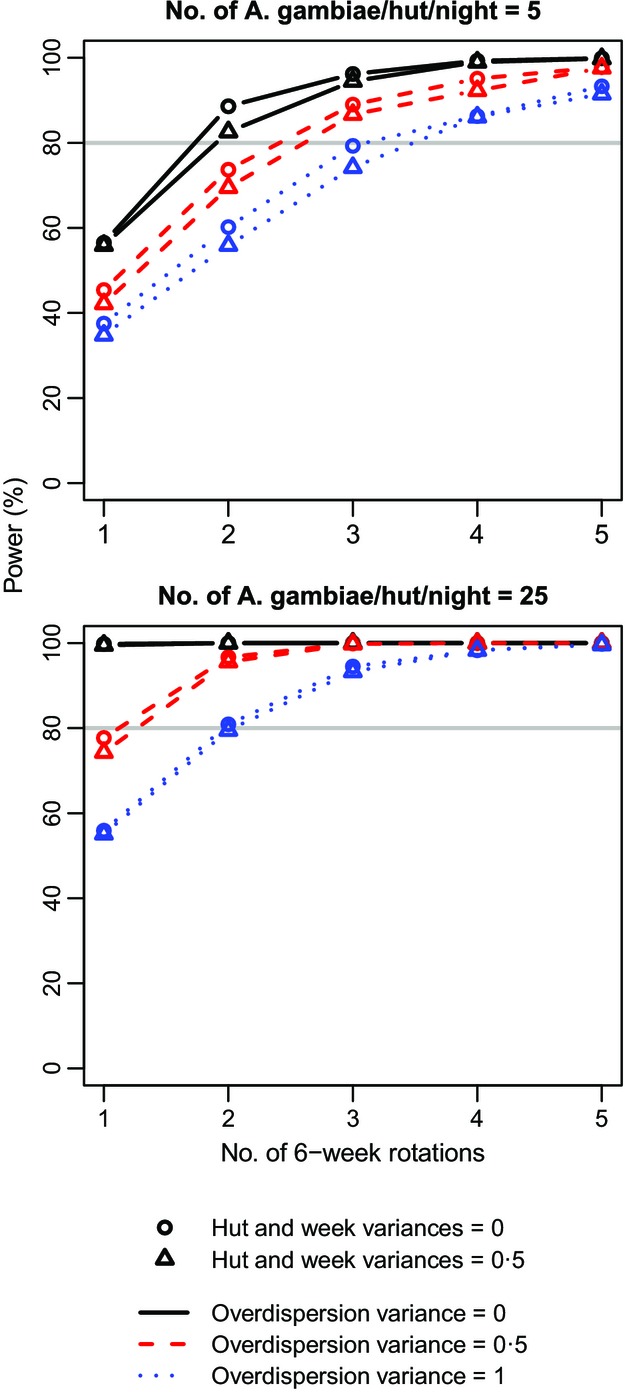
The relationship between the power of the long-lasting insecticidal net trial to detect a difference between nets causing 70% and 80% mortality and its duration in number of 6-week rotations of the Latin square. Each power estimate was derived from 1000 simulated data sets, generated under scenarios that varied in Anopheles gambiae abundance, degree of variation in mortality between huts and weeks, and strength of overdispersion. The horizontal grey line shows the target power of 80%.
An alternative to extending the duration of the trial would be to focus effort on the two net types of primary interest, the control net and the primary experimental net, by reducing the number of net types trialed. For example, if only the control net and two experimental nets were trialed, the number of observations per net type could be doubled. We found that this gives equivalent power to doubling the duration of the trial (data not shown).
Bias and Confidence Interval Coverage
Bias in the tick burden estimates ranged from −3·9% to 7·9% (Fig. S1), although the effect of bias was small relative to sampling error, with absolute bias always at least 4·3-fold smaller than the margin of error (median 28-fold), so that, relative to the overall error, bias was always small and usually negligible. In the LLIN trial example, bias in the odds ratio estimates was slightly lower, ranging from −0·2% to 7·7% (Fig. S2). In both binomial and count examples, bias was highest in scenarios where low sample size led to inadequate power or precision.
On average, CI coverage tended to be slightly overconfident (i.e. too narrow) in the count example and accurate in the binomial example (Fig. S3 and S4). Mean 95% CI coverage across all scenarios was 93·4% for the tick burden estimates and 94·4% for the mortality odds ratio estimates. Coverage of the odds ratio estimate under the null hypothesis of no difference between the nets (odds ratio = 1), which is equivalent to one minus the type I error rate, was similar to coverage under the alternative hypothesis (Fig. S5), with mean coverage of 94·6%, or, equivalently, a type I error rate of 5·4%, close to the nominal value of 5%. Under both the null and alternative hypotheses, scenarios that gave significantly low coverage (< 93%) tended to be those where A. gambiae abundance was low and overdispersion was at realistic (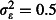 ) or strong (
) or strong (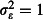 ) levels. CI coverage in the tick abundance example followed a similar pattern to that shown by bias, tending to be more problematic when sampling effort was low, with coverage typically around 90% in scenarios where the total number of ticks was low, either due to low tick abundance or low sample size.
) levels. CI coverage in the tick abundance example followed a similar pattern to that shown by bias, tending to be more problematic when sampling effort was low, with coverage typically around 90% in scenarios where the total number of ticks was low, either due to low tick abundance or low sample size.
Discussion
The key question to answer when assessing the utility of the power analysis methods presented here is: will they help researchers in ecology and evolution to design better studies? To do this, they must be (i) substantially more accurate than conventional power analysis methods to justify the extra time and effort required and (ii) reasonably straightforward to use. On the first point, consideration of random effects had a major impact on study design in both of the examples, motivating an increase in sampling effort of up to twofold in the binomial example and fivefold in the count example. On the second point, although our experience is that the perceived complexity of simulation from GLMMs can be intimidating, we argue that the methods presented here are no more complex conceptually, and only slightly more challenging technically, than fitting and interpreting a GLMM.
The methods presented here are flexible because GLMMs are flexible. Their flexibility is illustrated by the contrast between the two example studies: binomial vs. count responses; crossed vs. nested random effects; inference via NHST vs. confidence interval estimation; experimental vs. observational study design. This flexibility encompasses many common simpler analyses which can also be simulated as GLMMs, including linear mixed effects regression, GLMs, linear regression, anova and t-test. More complex models can also be simulated. For example, zero-inflated count models (Zuur, Saveliev & Ieno 2012) could be simulated as the product of binary and Poisson or negative binomial responses.
What do these results tell us about the impact of random effects and overdispersion on power and precision? Although we cannot generalize beyond the two examples, the results show that the effects of random variation on power and precision can depend on the specific study scenario. In the binomial example, overdispersion was the principal drain on power, while the higher level random effects (hut and week) had little impact. An implication of this observation is that when overdispersion is present, power can depend not just on the quantity of data (here total mosquito numbers), but also on how it is collected. For example, running the trial for one rotation catching 25 mosquitoes per hut per night yields exactly the same number of mosquitoes per treatment arm, 150, as running five rotations with an abundance of 5 mosquitoes per hut per night, yet in the presence of overdispersion, the latter scenario gives considerably more power (Fig.2). In the count example, by contrast, overdispersion had relatively little impact compared with the higher level random effects. The critical factor in determining precision was the variance at the highest level, among locations. This effect has been observed in multilevel LMMs, where power is limited principally by the highest level sample size (Maas & Hox 2005; Snijders 2005). The contrasting patterns between the two examples presented emphasize the unpredictability of the results of power analysis when there are multiple sources of variation, and the necessity of tailoring power analysis to apply to specific study designs and study systems.
Although we have shown that ignoring the influence of random effects and overdispersion on power analysis can grossly mislead study design, we are not suggesting that many researchers would be so naïve. EE researchers are generally alert to the impact of non-independence and overdispersion on inference, as evidenced by the widespread awareness of pseudoreplication (Hurlbert 1984) and the popularity of GLMMs (Bolker et al. 2009). It is more likely that they would not do a power analysis at all. Why not? Our experience is that there is simply not a culture of performing power analysis. This supposition is supported by the fact that few EE journals recommend that authors use power analysis. Together the Ecological Society of America (ESA; n = 3 journals), the British Ecological Society (BES; n = 4), the European Society for Evolutionary Biology (ESEB; n = 1) and the Society for the Study of Evolution (SSE; n = 1) publish nine of the most prominent primary research journals in EE. Only the ESA mentions power analysis in its guidance for authors, in tentatively supportive terms [‘Power analyses … occasionally can be very useful’ (Ecological Society of America Statistical Ecology Section 2012)], while none of the other journals mention the topic at all in their guidance. If journals are committed to raising the quality of the science they publish, journal editors should encourage and, where appropriate, require authors to include an a priori power analysis, or at least a justification of its omission. Although we have no objective evidence identifying other barriers to power analysis, in our experience these include a belief that power analysis cannot be extended beyond NHST inference; a lack of available power analysis methods for complex analyses; a lack of technical knowledge of even simple power analyses; and the perceived difficulty of defining a biologically meaningful effect size.
We argue that the last of these obstacles arises from a misunderstanding of the concept of a biologically meaningful effect size. This obstacle is (in our experience) frequently expressed as a question: ‘How can I power my study to detect an effect size of which I have no knowledge?’ The answer is that the study should be powered to detect not the actual effect (which cannot anyway be known before collecting the data) but the smallest effect that would be considered biologically meaningful. In other words, the study should be sufficiently sensitive to detect the smallest effect that, in the judgement of the researcher, is worth detecting. In the LLIN hut trial example, we chose to power the trial to detect a difference between a mortality of 80% with the experimental net and 70% with the control net. This choice implies that effect sizes that equate to experimental net mortalities in the range 80–100% are worth detecting, while those in the range 70–80% are not, because the study would be underpowered to detect them. Whichever combination of reasons explains the under-use of power analysis in EE, alleviation of this problem will require greater availability of methods and guidance on conducting power analysis for GLMMs such as those presented here, and greater recognition of the importance of power analysis in EE curricula.
Simulation-based power analysis for GLMMs has disadvantages. First, it is relatively slow. Secondly, while power analysis formulae can be rearranged to output any parameter, including sample size, sample size can only be an input when using simulations. It is therefore necessary to run simulations across a range of sample sizes in order to locate the one that gives the desired power, although an efficient algorithm to automate this procedure has been developed (Hooper 2013). Nevertheless, these disadvantages are easily outweighed by the much greater flexibility of simulation-based power analysis. Researchers should, however, resist the temptation to abuse this flexibility by overcomplicating power analysis. Most power analyses rest on strong assumptions with considerable inherent uncertainty whose impact is likely to dwarf the effect of fine adjustment to the simulation model. The simplifying assumptions made in both examples highlight an important distinction between retrospectively fitting a model to data and prospectively choosing a simulation model for power analysis. In the former, all available information is used to maximize the efficiency of the model, while in the latter, the absence of information and the practical necessity of limiting the number of scenarios to be explored necessitate simplification, while erring on the side of conservatism (i.e. we would rather underestimate than overestimate power).
In conclusion, power analysis should be much more widely used. Failing to include power analysis as a key element of study design misses an opportunity to increase the probability of a study being successful. However, power analysis is challenging for the dominant analysis framework – GLMMs – and hindered by lack of guidance and software. The guidance and methods presented here are intended to make power analysis for GLMMs more accessible to researchers and ultimately improve the standard of study design in ecology and evolution.
Acknowledgments
We acknowledge funding from the European Union Seventh Framework Programme FP7 (2007–2013) under grant agreement no 265660 AvecNet. We thank Mirko Winkler and coauthors for providing the hut trial data, and David Elston, Robert Moss and Xavier Lambin for providing the tick burden data. We are grateful to two anonymous reviewers whose comments on earlier drafts greatly improved this article.
Data accessibility
The data sets used in this paper have been published previously (Elston et al. 2001; Winkler et al. 2012).
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Tutorial showing examples of simulation-based power analysis for GLMMs using R.
R code for Appendix S1.
Fig. S1.The relationship between bias in estimating tick abundance and number of locations sampled.
Fig. S2. The relationship between bias in estimating the mortality odds ratio of 1·7 and trial duration in number of 6-week rotations of the Latin square.
Fig. S3. Variation in 95% confidence interval (CI) coverage for estimation of tick abundance by number of locations sampled.
Fig. S4. Variation in 95% confidence interval (CI) coverage for estimation of the mortality odds ratio of 1·7 by trial duration in number of 6-week rotations of the Latin square.
Fig. S5. Variation in 95% confidence interval (CI) coverage for estimation of the mortality odds ratio of 1 (i.e. under the null hypothesis) by trial duration in number of 6-week rotations of the Latin square.
References
- Barr DJ, Levy R, Scheepers C. Tily HJ. Random effects structure for confirmatory hypothesis testing: keep it maximal. Journal of Memory and Language. 2013;68:255–278. doi: 10.1016/j.jml.2012.11.001. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B. Walker S. 2014. & lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7. Retrieved July 19, 2014, from http://cran.r-project.org/package=lme4.
- Bliss CI. Fisher RA. Fitting the negative binomial distribution to biological data. Biometrics. 1953;9:176–200. &. [Google Scholar]
- Bolker BM. Ecological Models and Data in R. Princeton & Oxford: Princeton University Press; 2008. [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH. White J-SS. Generalized linear mixed models: a practical guide for ecology and evolution. Trends in Ecology and Evolution. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. &. [DOI] [PubMed] [Google Scholar]
- Burnham K. Anderson D. Kullback-Leibler information as a basis for strong inference in ecological studies. Wildlife Research. 2001;28:111–119. &. [Google Scholar]
- Crowder MJ. Beta-binomial anova for proportions. Journal of the Royal Statistical Society Series C (Applied Statistics) 1978;27:34–37. [Google Scholar]
- Cumming G. Understanding The New Statistics: Effect Sizes, Confidence Intervals, and Meta-Analysis. New York: Routledge; 2013. [Google Scholar]
- Di Stefano J. How much power is enough? Against the development of an arbitrary convention for statistical power calculations. Functional Ecology. 2003;17:707–709. [Google Scholar]
- Eberhardt L. Appraising variability in population studies. The Journal of Wildlife Management. 1978;42:207–238. [Google Scholar]
- Ecological Society of America Statistical Ecology Section. 2012. Guidelines for Statistical Analysis and Data Presentation. Retrieved August 13, 2013, from http://esapubs.org/esapubs/statistics.htm.
- Elston DA, Moss R, Boulinier T, Arrowsmith C. Lambin X. Analysis of aggregation, a worked example: numbers of ticks on red grouse chicks. Parasitology. 2001;122:563–569. doi: 10.1017/s0031182001007740. &. [DOI] [PubMed] [Google Scholar]
- Faraway JJ. Extending the Linear Model with R: Generalized Linear, Mixed Effects and Nonparametric Regression Models. Boca Raton: Chapman & Hall/CRC; 2005. [Google Scholar]
- Faul F, Erdfelder E, Lang A-G. Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Research Methods. 2007;39:175–191. doi: 10.3758/bf03193146. &. [DOI] [PubMed] [Google Scholar]
- Hooper R. Versatile sample-size calculation using simulation. The Stata Journal. 2013;13:21–38. [Google Scholar]
- Hurlbert SH. Pseudoreplication and the design of ecological field experiments. Ecological Monographs. 1984;54:187–211. [Google Scholar]
- Ioannidis JPA. Why most published research findings are false. PLoS Medicine. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennions M. Møller A. A survey of the statistical power of research in behavioral ecology and animal behavior. Behavioral Ecology. 2003;14:438–445. &. [Google Scholar]
- Lengeler C. 2004. The Cochrane Database of Systematic ReviewsInsecticide-treated bed nets and curtains for preventing malaria, CD000363.
- Maas CJM. Hox JJ. Sufficient sample sizes for multilevel modeling. Methodology. 2005;1:86–92. &. [Google Scholar]
- Malima RC, Magesa SM, Tungu PK, Mwingira V, Magogo FS, Sudi W, et al. An experimental hut evaluation of Olyset nets against anopheline mosquitoes after seven years use in Tanzanian villages. Malaria Journal. 2008;7:38. doi: 10.1186/1475-2875-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JGA, Nussey DH, Wilson AJ. Réale D. Measuring individual differences in reaction norms in field and experimental studies: a power analysis of random regression models. Methods in Ecology and Evolution. 2011;2:362–374. &. [Google Scholar]
- Ngufor C, Tchicaya E, Koudou B, N'Fale S, Dabire R, Johnson P, Ranson H. Rowland M. Combining organophosphate treated wall linings and long-lasting insecticidal nets for improved control of pyrethroid resistant Anopheles gambiae. PLoS ONE. 2014;9:e83897. doi: 10.1371/journal.pone.0083897. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey DH, Wilson AJ. Brommer JE. The evolutionary ecology of individual phenotypic plasticity in wild populations. Journal of Evolutionary Biology. 2007;20:831–844. doi: 10.1111/j.1420-9101.2007.01300.x. &. [DOI] [PubMed] [Google Scholar]
- O'Hara RB. Kotze DJ. Do not log-transform count data. Methods in Ecology and Evolution. 2010;1:118–122. &. [Google Scholar]
- R Core Team. 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved April 10, 2014, from http://www.r-project.org/
- Roll Back Malaria Partnership. 2008. The Global Malaria Action Plan: For a malaria-free world. Geneva, Switzerland.
- Schielzeth H. Forstmeier W. Conclusions beyond support: overconfident estimates in mixed models. Behavioral Ecology. 2009;20:416–420. doi: 10.1093/beheco/arn145. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hardy ICW. Gammell MP. Power rangers: no improvement in the statistical power of analyses published in Animal Behaviour. Animal Behaviour. 2011;81:347–352. &. [Google Scholar]
- Snijders TAB. Power and sample size in multilevel linear models. In: Howell DC, editor; Everitt BS, editor. Encyclopedia of Statistics in Behavioral Science Volume 3. Chichester: Wiley; 2005. pp. 1570–1573. & ) [Google Scholar]
- Snijders T. Bosker R. Standard errors and sample sizes for two-level research. Journal of Educational Statistics. 1993;18:237–259. &. [Google Scholar]
- Taborsky M. Sample size in the study of behaviour. Ethology. 2010;116:185–202. [Google Scholar]
- Thomas L. Juanes F. The importance of statistical power analysis: an example from Animal Behaviour. Animal Behaviour. 1996;52:856–859. &. [Google Scholar]
- Warton DI. Hui FKC. The arcsine is asinine: the analysis of proportions in ecology. Ecology. 2011;92:3–10. doi: 10.1890/10-0340.1. &. [DOI] [PubMed] [Google Scholar]
- Williams EJ. Experimental designs balanced for the estimation of residual effects of treatments. Australian Journal of Chemistry. 1949;2:149–168. [Google Scholar]
- Winkler MS, Tchicaya E, Koudou BG, Donzé J, Nsanzabana C, Müller P, Adja AM. Utzinger J. Efficacy of ICON® Maxx in the laboratory and against insecticide-resistant Anopheles gambiae in central Côte d'Ivoire. Malaria Journal. 2012;11:167. doi: 10.1186/1475-2875-11-167. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2013a. Guidelines for laboratory and field testing of long-lasting insecticidal nets. WHO/HTM/NTD/WHOPES/2013.1.
- World Health Organization. 2013b. World Malaria Report 2013. World Health Organization, Geneva, Switzerland.
- Zuur AF, Hilbe JM. Leno EN. A Beginner's Guide to GLM and GLMM with R: A Frequentist and Bayesian Perspective for Ecologists. Newburgh, UK: Highland Statistics Ltd; 2013. &. [Google Scholar]
- Zuur AF, Saveliev AA. Ieno EN. Zero Inflated Models and Generalized Linear Mixed Models with R. Newburgh, UK: Highland Statistics Ltd; 2012. &. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tutorial showing examples of simulation-based power analysis for GLMMs using R.
R code for Appendix S1.
Fig. S1.The relationship between bias in estimating tick abundance and number of locations sampled.
Fig. S2. The relationship between bias in estimating the mortality odds ratio of 1·7 and trial duration in number of 6-week rotations of the Latin square.
Fig. S3. Variation in 95% confidence interval (CI) coverage for estimation of tick abundance by number of locations sampled.
Fig. S4. Variation in 95% confidence interval (CI) coverage for estimation of the mortality odds ratio of 1·7 by trial duration in number of 6-week rotations of the Latin square.
Fig. S5. Variation in 95% confidence interval (CI) coverage for estimation of the mortality odds ratio of 1 (i.e. under the null hypothesis) by trial duration in number of 6-week rotations of the Latin square.
Data Availability Statement
The data sets used in this paper have been published previously (Elston et al. 2001; Winkler et al. 2012).


