Abstract
The BRAF inhibitor vemurafenib is currently used for treating patients with BRAF V600E mutant melanoma. However, the responses to vemurafenib are generally partial and of relatively short duration. Recent evidence suggests that activation of the epidermal growth factor receptor (EGFR)/erbB signaling pathway may be responsible for the development of BRAF inhibitor resistance in melanoma patients. In this study, we characterized the erbB family of receptors and ligands in melanoma cell lines and examined whether targeting both BRAF and erbB provided enhanced antitumor activity in BRAF mutant melanoma. Variable levels of erbB2, erbB3, and truncated erbB4 were expressed in both BRAF wildtype and mutant melanoma cells with no significant differences between wildtype and mutant lines. EGFR was rarely expressed. Neuregulin 3 and neuregulin 4 were the major erbB ligands released by melanoma cells. Multi-erbB targeting with the irreversible tyrosine kinase inhibitor canertinib exerted a more effective growth inhibitory effect in both BRAF wildtype and mutant melanoma cells compared with the single-erbB or dual-erbB targeting inhibitors, gefitinib, erlotinib, and lapatinib. Canertinib inhibited both EGF-induced and neuregulin 1-induced erbB downstream signaling in both mutant and wildtype cell lines. However, canertinib induced apoptosis and sub-G1 arrest only in mutant cells. Canertinib statistically increased the antiproliferative effects of vemurafenib in the BRAF mutant melanoma cell lines while little or no enhanced effect was observed with the combination treatment in the wildtype cell lines. A combined inhibition strategy targeting BRAF together with multiple erbB family kinases is potentially beneficial for treating BRAF V600E mutant melanoma. Wildtype BRAF melanoma may also benefit from a multi-erbB kinase inhibitor.
Keywords: amphiregulin, canertinib, HB-EGF, neuregulin, transforming growth factor-α, vemurafenib
Introduction
Melanoma is curable when diagnosed at an early stage. However, despite many recent advances in novel immuno-therapies and targeted therapies for advanced melanoma, the prognosis for patients with advanced disease remains poor. Lack of effective treatment options is one of the major clinical challenges for management in this disease. The recent development of the BRAF inhibitor vemurafenib (PLX4032/Zelboraf) is a significant breakthrough in the development of molecularly-targeted therapy for melanoma patients carrying a particular BRAF activating mutation: valine substitution to glutamate at amino acid position 600 (V600E) [1]. However, the majority of the melanoma patients harboring the BRAF V600E mutation are only partially responsive to vemurafenib with relatively short response durations, most likely because of the emergence of acquired resistance to BRAF inhibitors [2]. Thus novel approaches are needed to treat this disease.
A recent phosphoproteomic screen in melanoma cells indicates that epidermal growth factor receptor (EGFR), erbB3, and erbB4 are promising targets for new molecular drug development [3]. EGFR expression has been detected, although in varying degrees, among the different stages of malignant melanoma [4]. It has also been shown that EGFR gene amplification along with chromosome 7 polysomy is detected in a subset of primary melanoma patients with poor prognosis [5,6]. Cetuximab, a recombinant human/mouse chimeric monoclonal antibody against the extracellular domain of EGFR can suppress the metastasis of M24met melanoma in SCID mice [7] and the growth and invasion of melanoma cells in vitro [6]. However, phase II clinical trials have indicated that the EGFR small molecule tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib, show only minimal clinical benefits towards melanoma patients [8,9]. EGFR inhibitors are ineffective in inhibiting the growth of tumor cells with high erbB2 expression levels [10]. However, gene amplification and overexpression of erbB2 are generally not found in malignant melanoma [11–13]. In contrast, high expression levels of other erbB family members like erbB3 and erbB4 are found in malignant melanoma [14,15]. Emerging data indicate that activation of the erbB receptor tyrosine kinase signaling by neuregulin (NRG) 1 is able to rescue the in-vitro growth inhibitory effect of vemurafenib in BRAF mutant melanoma [2,16,17]. Hence, a concomitant inhibition on erbB signaling may be beneficial to BRAF inhibitor treatment in BRAF mutant melanoma.
In this study, we show that melanoma cell lines, both BRAF mutant and wildtype (WT), express multiple erbB receptor family members and erbB ligands. Growth inhibition of melanoma cells is more effective with the pan-erbB targeting inhibitor canertinib than other single/dual-erbB targeting inhibitors. Canertinib also exerts stronger antitumor effects in the presence of vemurafenib in the BRAF mutant melanoma cells compared with this combination in WT cell lines. A combined inhibition strategy targeting BRAF together with multiple erbB family kinases is potentially beneficial for treating BRAF V600E mutant melanoma. WT BRAF melanoma may also benefit from a multi-erbB kinase inhibitor.
Methods
Chemicals and reagents
Recombinant human NRG1 (EGF domain), NRG4 (EGF domain), and EGF were obtained from Reprokine (Valley Cottage, New York, USA). Vemurafenib, canertinib, lapatinib, gefitinib, and erlotinib were purchased from ChemieTek (Indianapolis, Indiana, USA). General chemicals were purchased from Sigma-Aldrich (St Louis, Missouri, USA). Cell culture media, antibiotics, and fetal bovine serum (FBS) were obtained from Life Technologies (Grand Island, New York, USA).
Cell culture
SK-MEL147, SK-MEL19, SK-MEL94, SK-MEL100 were a generous gift from Paul Chapman and originally established at Sloan-Kettering Institute (New York, New York, USA) and routinely cultured in DMEM + 10% FBS. A375 was available from ATCC (Manassas, Virginia, USA) and also cultured routinely in DMEM + 10% FBS. IgR3, FEMX, M14, MEL526, 08-196-64, TPF-11-743 were obtained from the UPCI Melanoma Program (University of Pittsburgh Cancer Institute, Pittsburgh, Pennsylvania, USA) and cultured in RPMI1640 + 10% FBS. All cell lines had been verified within 2 months before use and routinely maintained in media supplemented with 1 × Pen/Strep antibiotic solution at 37°C in humidified CO2 incubator.
Cell viability assay
Melanoma cells were plated on 96-well plates with 6000 cells per well. The following day, EGFR TKIs and/or vemurafenib were added in each well at the concentrations indicated in the figures and incubated with the cells for 3 days at 37°C in humidified CO2 incubator. Cell viability was assessed by the MTT assay. Dose–response curves and IC50 were determined by the nonlinear regression function of GraphPad Prism version 4.03 for Windows, (GraphPad Software, San Diego, California, USA, http://www.graphpad.com).
Western blotting
Cultured cells were collected and lysed in lysis buffer [1% (v/v) NP-40, 150 mmol/l sodium chloride, 1 mmol/l EDTA, 10 mmol/l sodium phosphate buffer, pH 7.2, 1 mmol/l dithithreitol, 1 mmol/l phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 3 μg/ml aprotinin, 3 μg/ml pepstatin A, 1 mmol/l sodium orthovanadate, 0.05 mol/l sodium fluoride]. Lysate aliquots containing 50 μg proteins were resolved by 10% precast SDS-PAGE (Bio-Rad, Hercules, California, USA) and transferred to nitrocellulose membranes. Primary antibodies diluted in the blocking solution [2% (w/v) non-fat milk or BSA in 0.05% (v/v) Tween-20, 25 mmol/l Tris, 140 mmol/l sodium chloride, 2.7 mmol/l potassium chloride, pH 7.5] were incubated with the membrane at 4°C overnight. The following primary antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, California, USA) and used at the indicated dilutions: EGFR (1005; 1 : 500), erbB2 (C-18; 1 : 800), erbB3 (C-17; 1 : 500), erbB4 (C-18; 1 : 800), p-erbB2 (p-Neu Tyr1248; 1 : 500), p-erbB3 (Tyr1328; 1 : 800), and p-erbB4 (Tyr1056; 1 : 500). Antibodies against Akt (9272; 1 : 1000), pAkt (4051; 1 : 1000), MAPK (9102; 1 : 1000), pMAPK (9101; 1 : 2000), STAT3 (9139; 1 : 1000), pSTAT3 (9131; 1 : 1000), cleaved PARP (9541; 1 : 1000), and pEGFR (2234; 1 : 800) were from Cell Signaling Technology (Danvers, Massachusetts, USA). Actin antibody was purchased from Millipore (Billerica, Massachusetts, USA) and used at a 1 : 10 000 dilution. After washing, the membrane was incubated with diluted HRP conjugated secondary goat anti-rabbit/anti-mouse antibody from Life Technologies (Grand Island, New York, USA) at room temperature for 1 h. The membrane was developed with ECL 2 Reagents (Pierce, Rockford, Illinois, USA) and exposed on BioMax XAR films (Eastman Kodak, Rochester, New York, USA). Immunoreactive bands were quantitated using Un-Scan-It gel densitometry software (Silk Scientific, Orem, Utah, USA).
ELISA assays
Conditioned medium from each sample was harvested after 48 h of incubation at 37°C. The confluence of each cell line was 90–100% at the time of collection. Each sample was concentrated from 4 ml to 500 μl with the Amicon Ultra-4 Centrifugal Filter Devices (Millipore) by centrifuging at 4000 g for 6–7 min at room temperature. Each enzyme-linked immunosorbent assay (ELISA) experiment was performed using the following kits: Human amphiregulin DuoSet ELISA Development kit (R&D Systems, Minneapolis, Minnesota, USA), Human HBEGF DuoSet ELISA Development kit (R&D Systems), Quantikine Human TGF-α Immunoassay Kit (R&D Systems), Enzyme-linked Immunosorbent Assay Kit for NRG1 (Antibodies-Online Inc., Atlanta, Georgia, USA), Enzyme-linked Immunosorbent Assay Kit for NRG3 (Antibodies-Online Inc.), Enzyme-linked Immunosorbent Assay Kit for NRG4 (Antibodies-Online Inc.). Each cell line was harvested in triplicate and each assay was performed three times for each sample.
Immunohistochemistry
A selection of deidentified paraffin-embedded tumor blocks for patients diagnosed with malignant melanoma between 1990 and 1999 in Los Angeles County were used for immunohistochemical analysis. After antigen-retrieval and blocking, paraffin-embedded patient melanoma section slides were incubated for 1 h with diluted primary antibodies against EGFR (1 : 7500, E3138; Sigma-Aldrich), erbB2 (1 : 400, MS730-P; Thermo Scientific, Waltham, Massachusetts, USA), erbB3 (1 : 50, SC-285; Santa Cruz Biotechnology), or erbB4 (1 : 200, SC-238; Santa-Cruz Biotechnology). Afterwards, the sections were developed with MACH 4 Universal AP Link and Polymer and Warp Red chromagen (Biocare Medical, Concord, California, USA). All developed sections were counter-stained with hematoxylin (Dako, Carpinteria, California, USA). Immunohistochemical staining was evaluated by a dermatopathologist (J.H.). Tumor cells with greater than 10% specific (cytoplasmic/nuclear/both) staining in five high powered fields were considered to be positive. The percentage of positively stained cells was calculated by dividing the number of positive cells with the total number of cells in the field.
Cell cycle analysis
Cells were seeded on six-well plates at a density of 5 × 105 cells/well and allowed to adhere overnight. After treating the cells with DMSO, 10 μmol/l gefitinib, erlotinib, lapatinib, or canertinib (ChemieTek) for 24 h, cells were harvested, washed twice with PBS, and fixed with ice-cold 70% ethanol at 4°C for 24 h. Cells were centrifuged, washed once with PBS, and treated with 0.5 ml phosphate-citric acid buffer (192 ml 0.2 mol/l Na2HPO4 and 8 ml 0.1 mol/l citric acid, pH~7.8) at room temperature for 5 min to obtain a better defined sub-G1 peak. Cells were then centrifuged and resus-pended in 300 μl propidium iodide solution (15 μg propidium iodide and 3 Kunitz Units of RNase A in 1 × PBS) and incubated in the dark at room temperature for 30 min. Cell cycle distribution was measured with BD Accuri C6 flow cytometer (BD Biosciences, San Jose, California, USA) and analyzed with FlowJo software (Tree Star Inc., Ashland, Oregon, USA). Each sample was run in triplicate and the experiment was repeated one time.
Statistical analysis
Data were expressed as mean±SE. Statistical analyses were performed with GraphPad Prism version 4.03 for Windows (GraphPad Software). The statistical significance of the sample mean difference in the experiments was determined by unpaired t-test (for canertinib treatment with/without vemurafenib), or by one-way analysis of variance followed with Dunnett's test (for vemurafenib treatments with different concentrations of canertinib), with a 95% confidence level (P < 0.05). Effect of in-vitro drug combinations was tested for synergy using the method of Chou and Talalay [18].
Results
Multiple erbB family members and receptor ligands are expressed in melanoma cells
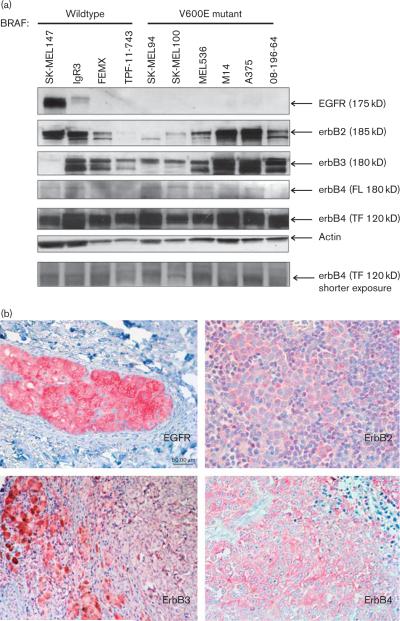
We examined the erbB receptor family (EGFR, erbB2, erbB3, and erbB4) expression profiles in a panel of four BRAF WT and six V600E mutant melanoma cell lines (Fig. 1a). All cell lines used in this analysis were NRAS WT. Previously it has been shown that EGFR is commonly overexpressed in melanoma [4]. However, in our chosen panel of cell lines, EGFR was undetectable in BRAF mutant cells while only the SK-MEL147 and IgR3 BRAF WT cells had detectable EGFR expression (Fig. 1a). Both erbB2 and erbB3 expression could be detected at differential levels among BRAF WT and mutant cells. In the BRAF WT cells, high levels of erbB2 were found in the SK-MEL147 and IgR3, whereas high levels of erbB3 were observed in IgR3, FEMX, and TPF-11-743 cells. All five BRAF mutant cell lines examined expressed both erbB2 and erbB3, in which the highest expression levels could be found in MEL526, M14, A375, and 08-196-64. Minor bands, which are possibly the proteolytic products of the erbB receptors (145 and 160 kD in the erbB2 and erbB3 blots, respectively), have been reported previously with the primary antibodies used [19]. In regard to erbB4 expression, the full-length erbB4 protein (180 kD) was present in very low levels in both BRAF WTand mutant cells, whereas a truncated (120 kD) form was the dominantly expressed form in melanoma cells. This truncated form (120 kD) of erbB4 has been previously reported to be highly expressed in human melanoma cells [20].
Fig. 1.
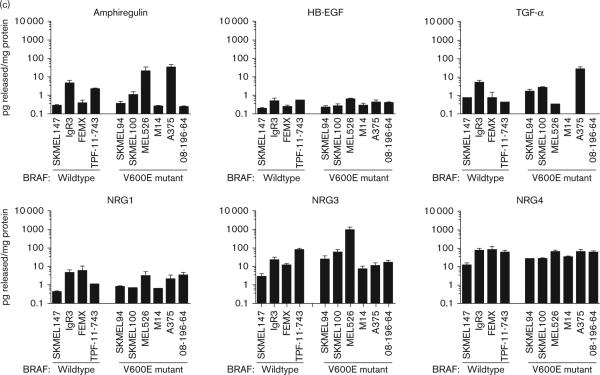
Multiple erbB receptors and ligands are expressed in BRAF V600E mutant and wild-type melanoma. (a) Representative western blot analyses showing the protein levels of different erbB family members (EGFR, erbB2, erbB3, and erbB4) in BRAF V600E mutants and wild-type cell lines. The major bands are indicated by arrows. Actin levels shown serve as the loading control. FL, full-length; TF, truncated form. Similar results were obtained in three independent experiments. (b) Representative positive immunohistochemical staining for EGFR, erbB2, erbB3, and erbB4 from patient tumor specimens. Photos were taken from different individual patient samples. (c) Release of various erbB ligands were detected from conditioned media of melanoma cell lines using ELISA. The release levels were normalized with the total protein content in the lysates of the cultured cells and results are presented as the mean pg released/mg protein±SE. EGFR, epidermal growth factor receptor; ELISA, enzyme-linked immunosorbent assay; NRG, neuregulin; TGF-α, transforming growth factor-α.
We confirmed expression of these receptors in human melanoma patient tissue samples. Expression of EGFR, erbB2, erbB3, and erbB4 was also detectable in primary melanomas (Clarks level: II–IV) using immunohistochemistry (Fig. 1b). Although the majority of the cell lines analyzed did not show detectable EGFR expression, cytoplasmic EGFR staining was observed in 8/13 patient samples (61.5%). ErbB4 staining was also observed in 8/13 samples, whereas erbB2 was detected in 11/13 samples (84.6%). Cytoplasmic erbB3 staining was detected in 7/8 samples (87.5%). BRAF mutation testing on the patient tumor tissues used in this analysis was inconclusive because of the limited availability of patient tissue material for genomic analyses.
After confirming the expression of multiple erbB receptors in melanoma cell lines and patients samples, we then measured the release of erbB ligands by ELISA in the same melanoma BRAF mutant and WT cell lines analyzed in Fig. 1a (Fig. 1c). Among the erbB ligands tested, we found that heparin-binding EGF-like growth factor was released at extremely low levels, less than 0.6 pg/mg protein by melanoma cells. Low levels were also detected for transforming growth factor-α (TGF-α) (0.4–4.8 pg/mg protein), and amphiregulin (0.3–4.4 pg/mg protein) in most of the cell lines. However, the BRAF mutant cell lines, A375 and MEL526, released high levels of amphiregulin (31.5±13.9 and 20.9±13.4 pg/mg protein, respectively). High levels of TGF-α (26.7±10.0 pg/mg protein) were also found in A375 cells. As both TGF-α and amphiregulin preferably bind to EGFR, which was absent in A375 and MEL526, it was unexpected to see such a high level of release of these ligands in these cells. However, previously, it has been reported that TGF-α and amphiregulin can alternatively activate erbB2/erbB4 and erbB3/ erbB4 heterodimers, respectively, in other cell types without EGFR expression [21,22]. The specific ligand for the erbB2/erbB3 heterodimer, NRG1, was released at 0.4–5.9 pg/mg protein. NRG3, the major erbB ligand for erbB4, was detected in the range 3.0–79.7 pg/mg protein in BRAF WT cells and 7.4–924.6 pg/mg protein in BRAF mutant cells. Another erbB4 ligand, NRG4, was released at moderate levels (12.3–80.8 pg/mg protein) in both BRAF WT and mutant cells. No significant differences were observed in ligand release between the BRAF WT and mutant cell lines. There were some differences in ligand release within the BRAF WT and mutant lines; however, these differences in ligand release did not correspond to receptor expression. Hence, the erbB receptor expression and ligand release profiles in the melanoma cells suggest the existence of an autocrine signaling loop between erbB2, erbB3, and erbB4 in both BRAF WTand mutant melanoma and the EGFR family expression profile does not play a role in differentiating BRAF mutant versus WT cells.
Growth of both BRAF wildtype and mutant melanoma cells is maximally inhibited by canertinib
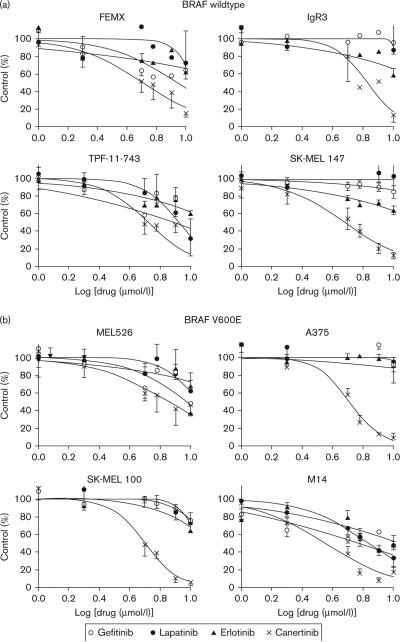
The expression of multiple erbB receptors and ligands among the melanoma cell lines suggested that TKIs targeting multiple erbB family members could be a feasible and effective erbB targeted molecular therapeutic approach for malignant melanoma. To explore this idea, we first compared the in-vitro antitumor activities of single EGFR targeting (gefitinib and erlotinib) versus multi-erbB targeting (lapatinib and canertinib) on human melanoma cell lines (Fig. 2). Most BRAF WT (Fig. 2a) and mutant cell lines (Fig. 2b) were weakly responsive to nonresponsive towards the EGFR-specific TKIs, gefitinib, and erlotinib, consistent with little evidence for autocrine signaling through EGFR. BRAF WT SK-MEL147 and IgR3 cells, which have detectable EGFR expression, were also among the unresponsive cell lines towards gefitinib, suggesting little dependence on the EGFR pathway. Although the panel of cell lines showed little variation in effectiveness of erlotinib (the most selective EGFR TKI assayed), gefitinib and lapatinib showed differential sensitivity (Table 1). Three cell lines (TPF-11-743, M14, and 08-196-64) showed greater sensitivity towards both gefitinib and lapatinib compared with the other cell lines. Both gefitinib and lapatinib are less selective towards EGFR than erlotinib. Gefitinib has some ability to inhibit both erbB2 and erbB4, whereas lapatinib also inhibits erbB2 [23]. In general, both BRAF WT and mutant cells, which express high levels of truncated erbB4 and often express erbB2, were more sensitive to gefitinib and lapatinib than erlotinib (Table 1). There was no direct relationship between the IC50 for the cell lines toward gefitinib or lapatinib and the amount of erbB2, erbB3, or erbB4 protein detected or in the amount of NRG released (Supplementary Table 1). This suggests that dependence on erbB pathways for growth or survival is not determined by the quantity of receptor or ligand detected alone.
Fig. 2.
Both BRAF wildtype and mutant melanoma cells are more sensitive to irreversible multi-erbB inhibitors canertinib than single-erbB or dual-erbB targeting agents. Dose-dependent response curves of growth inhibition in BRAF WT (a) and V600E mutant (b) cells by 3-day treatment of gefitinib, lapatinib, erlotinib, and canertinib. Cell viability after each drug treatment was assessed by MTT assay compared with vehicle control treatment and expressed as percent vehicle control (% control). n = 3–4 experiments. WT, wildtype.
Table 1.
IC50 of BRAF wildtype and mutant melanoma cells towards different small molecular erbB kinase Inhibitors
| Melanoma cell line | BRAF status | Gefitinib IC50 (μmol/l) | Erlotinib IC50 (μmol/l) | Lapatinib IC50 (μmol/l) | Canertinib IC50 (μmol/l) |
|---|---|---|---|---|---|
| SK-MEL147 | Wildtype | Unresponsive | 17.62±1.09 | Unresponsive | 4.37±1.19 |
| IgR3 | Wildtype | Unresponsive | 19.23±1.20 | Unresponsive | 6.73±1.12 |
| FEMX | Wildtype | 9.04±1.34 | 19.54±1.23 | 11.59±1.22 | 5.12±1.13 |
| TPF-11-743 | Wildtype | 8.46±1.07 | 15.00±1.10 | 6.18±1.05 | 5.59±1.08 |
| SK-MEL100 | V600E | 12.97±1.06 | 14.06±1.14 | 10.09±1.05 | 3.98±1.13 |
| MEL526 | V600E | 9.44±1.15 | 26.12±1.17 | 11.22±1.12 | 7.21±1.14 |
| M14 | V600E | 5.74±1.15 | 11.04±1.25 | 6.87±1.08 | 3.52±1.09 |
| A375 | V600E | Unresponsive | Unresponsive | Unresponsive | 5.15±1.05 |
| 08-196-64 | V600E | 6.89±1.07 | 17.74±1.21 | 9.38±1.07 | 4.16±1.10 |
The irreversible multi-erbB TKI canertinib exhibited the most effective decrease in cell proliferation of the TKIs tested. The IC50 of canertinib was in the range of 4.37–6.37 μmol/l in BRAF WT cells and 3.98–7.21 μmol/l in the BRAF mutant cells and was not significantly different based on mutation status (Table 1). The IC50 values in the melanoma cell lines analyzed in this study are much higher compared with EGFR driven cell lines such as A431 epidermoid carcinoma cells, which express very high levels of EGFR and are extremely sensitive to canertinib with a reported IC50 of 7.4 nmol/l [24]. On the basis of this in-vitro TKI sensitivity comparison, more potent growth inhibitory effects were always observed in melanoma cells whenever multiple erbB receptors (EGFR, erbB2, and erbB4) were targeted, regardless of BRAF mutation status. Besides the need to block several erbB members, the irreversible nature of canertinib's binding may also play a role in the enhanced growth inhibition observed.
Canertinib effectively inhibits multiple erbB family members and downstream signaling
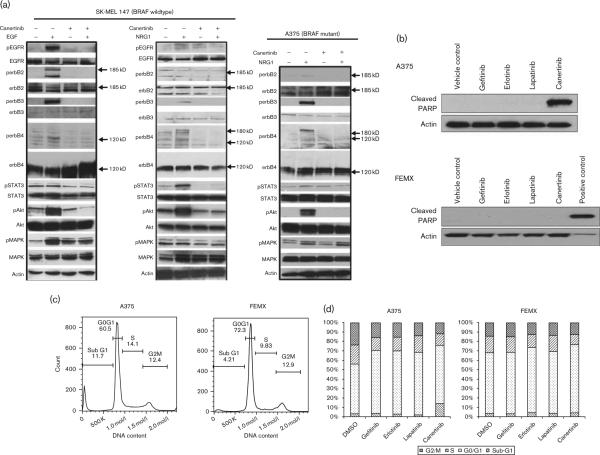
As canertinib was the most potent TKI against melanoma cells, its ability to block EGF-induced and NRG1-induced phosphorylation of erbB family members and downstream signaling pathways was examined. EGF was chosen to activate EGFR selectively, whereas NRG1 was chosen to activate ebB3/4 selectively. In BRAF WT/EGFR expressing SK-MEL147 cells, EGF stimulated the phosphorylation of EGFR at Tyr1068, erbB2 at Tyr1248, erbB3 at Tyr1328, and erbB4 at Tyr1056, whereas NRG1 induced the phosphorylation of EGFR, erbB3, and erbB4 (Fig. 3a). Given that erbB2 does not have any ligand binding activity and erbB3 retains no detectable tyrosine kinase activity, the costimulation of erbB2 and erbB3 by EGF suggests the existence of ligand-induced heterodimerization between EGFR and both erbB2 and erbB3. Similarly, heterodimerization between EGFR and truncated erbB4 may also be induced upon EGF stimulation. Phosphorylation of EGFR, erbB3, and erbB4 upon stimulation by NRG1 also is consistent with heterodimerization between EGFR and erbB3 or erbB4. Phosphorylation of erbB proteins in response to EGF or NRG1 was completely abolished after treatment with 5 μmol/l canertinib (Fig. 3a). Downstream Akt and MAPK phosphorylation was induced by EGF in SK-MEL147 cells, whereas STAT3 phosphorylation was not. NRG1 induced phosphorylation of Akt and STAT3 but not MAPK. This suggests some selectivity of MAPK signaling for the activated EGFR containing homo-dimers or heterodimers that contain several grb2 sites, whereas STAT3 activation may be initiated more effectively by erbB3/4 heterodimers. Although canertinib effectively inhibited EGF-induced Akt activation, MAPK activation was only partially inhibited. Canertinib reduced pSTAT3 levels even in the absence of EGF. Canertinib treatment abolished the NRG1-induced phosphorylation of EGFR, erbB3, and erbB4 as well as downstream pAkt and pSTAT3 in SKMEL147 WT cells. pMAPK expression was not modified in response to NRG1 suggesting that EGFR and/or erbB2 are more involved in this signaling in SK-MEL147 cells.
Fig. 3.
Canertinib inhibits ligand-induced coactivation between different erbB receptors and downstream Akt signaling and induces apoptosis and sub-G1 arrest in melanoma cells. (a) SK-MEL147 (BRAF WT) and A375 (BRAF mutant) cells were serum deprived for 48 h. Cells were pretreated with canertinib (5 μmol/l) for 1 h before stimulation with EGF (20 nmol/l) or NRG1 (20 nmol/l) for 15 min. Representative western blots are shown from three individual experiments. (b) A375 BRAF mutant cells or FEMX WT cells were treated for 16 h with gefitinib, erlotinib, lapatinib, or canertinib at 10 μmol/l. Cell lysates were prepared and analyzed for cleaved PARP expression by western analysis. β-Actin was used as a control. Representative western blots are shown from three individual experiments. (c) Representative histogram of cell cycle analysis in A375 and FEMX cells following treatment with 10 μmol/l canertinib for 16 h. The percentage of cells in each phase is shown. (d) Quantitative representation of cell cycle analysis in A375 and FEMX treated with gefitinib, erlotinib, lapatinib, or canertinib. Data are the mean of two independent experiments each of which had three samples per treatment. DMSO, dimethyl sulfoxide; EGF, epidermal growth factor; NRG, neuregulin; WT, wildtype.
In A375 BRAF mutant cells, which do not express EGFR as shown in Fig. 1a, NRG1 effectively induced p-erbB3 and p-erbB4 and canertinib completely inhibited this phosphorylation. perbB2 was not detected in these cells. Canertinib also effectively inhibited the NRG1 induction of the downstream signaling molecules pSTAT3 and pAKT, but not pMAPK, confirming what was observed with NRG1 in SK-MEL147 cells. Similar results of NRG1 stimulation and canertinib treatment were observed in IgR3 BRAF WT cells and M14 BRAF mutant cells (Supplementary Fig. 1).
Djerf Severinsson et al. [25] previously reported that canertinib at concentrations higher than 5 μmol/l was able to induce apoptosis in melanoma cells. Consistent with this observation, we also found that activation of caspase cleavage (indicated by the cleavage of endogenous caspase substrate PARP) was induced by overnight treatment of canertinib at 10 μmol/l, but not by gefitinib, erlotinib, nor lapatinib, in A375 BRAF mutant cells, but not BRAF WT cells (Fig. 3b). This is consistent with the ability of canertinib to cause complete reduction of pAkt in A375 cells (Fig. 3a). Cell cycle analysis confirms a significant 4.4-fold increase (P = 0.002) of cells in the sub-G1 phase in A375 cells treated with canertinib (13.9±2.38% average) compared with control-treated cells (3.2±1.1% average), whereas this was not observed in FEMX cells (canertinib 3.5±0.6% compared with control 4.2±0.3%). This was accompanied by a 50% decrease in cells in G2/M phase in canertinib treated compared with control-treated A375 cells. A representative histogram is shown in Fig. 3c. Quantitative analysis for all experiments is shown in Fig. 3d for all inhibitors tested. These results suggest that canertinib induces sub-G1 arrest in A375 BRAF mutant cells, but not FEMX cells, consistent with the cleaved PARP results shown in Fig. 3b.
Potent antitumor activity by multi-erbB and BRAF coinhibition in BRAF mutant melanoma
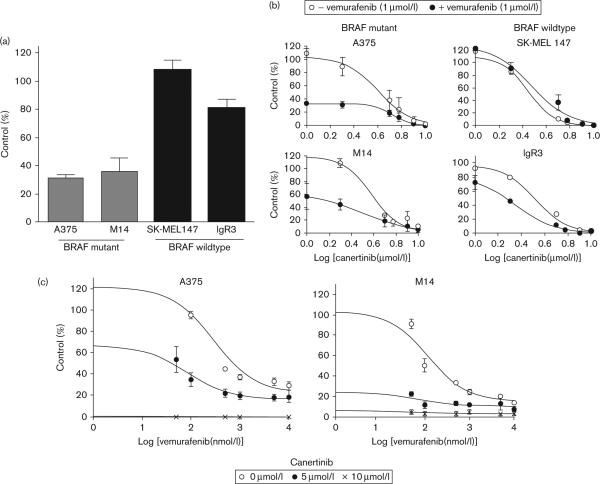
A recent report showed that NRG1 stimulation is able to potently rescue BRAF V600E melanoma cells from vemurafenib inhibition [26]. This suggested that melanoma cells, particularly the BRAF mutant cells, might retain intrinsic BRAF inhibitor resistance by the activation of multiple erbB/ligand autocrine loops. Therefore, we examined whether treatment of canertinib could exert a more effective growth inhibitory effect on BRAF mutant melanoma in the presence of BRAF inhibitors. We first verified that vemurafenib at 1 μmol/l concentration effectively caused a growth inhibition in BRAF mutant A375 and M14, but not in the BRAF WT cells (Fig. 4a). A 63–67% inhibition of cell growth with 1 μmol/l vemurafenib was observed in the BRAF mutant cells, whereas no inhibition was observed in the SK-MEL147 WT cells and only a slight 15% decrease was found in the IgR3 WT cells. Supplementary Table 2 shows the IC50 for vemurafenib for all of the cell lines tested.
Fig. 4.
In-vitro growth inhibitory effect of multi-erbB inhibitors canertinib is enhanced in BRAF V600E cells in the presence of BRAF inhibitor vemurafenib. (a) BRAF mutant and WT cells were treated with 1 μmol/l vemurafenib for 3 days and cell proliferation was monitored using the MTT assay. Effective growth inhibitory effect of vemurafenib could only be seen in BRAF mutant cells A375 and M14, but not in BRAF WT SK-MEL147 and IgR3. (b) The in-vitro antitumor effect of canertinib was greatly increased by vemurafenib (1 μmo/l) only in BRAF mutant cells. Cells were treated with increasing concentrations of canertinib in the presence or absence of 1 μmol/l vemurafenib for 3 days followed by MTT assay. (c) The dose-dependent responses of BRAF mutant cells A375 and M14 towards vemurafenib was enhanced by increasing concentrations of canertinib. A375 and M14 cells were treated with increasing concentrations of vemurafenib in the presence or absence of canertinib (5 or 10 μmol/l) for 3 days followed by MTT assay. WT, wildtype.
We next tested the effect of increasing concentrations of canertinib in the presence or absence of vemurafenib. In the presence of 1 μmol/l vemurafenib, the growth inhibitory effect of canertinib was significantly enhanced in BRAF mutant cells with little to no enhancement in the WT cells (Fig. 4b and Table 2). For example, a 63% inhibition with vemurafenib compared with only a 10% inhibition without vemurafenib at 2 μmol/l canertinib was observed in A375 cells. In contrast, the dose–response curves of canertinib in the BRAF WT cells SK-MEL147 were not significantly altered by vemurafenib. At this same dose of canertinib (2 μmol/l) in SK-MEL147 cells, no inhibition was observed in the absence of vemurafenib and only a slight 8.5% inhibition was observed in the presence of vemurafenib. Combination index dose–response experiments show that the effect of vemurafenib and canertinib in the BRAF mutant cells was synergistic at all concentrations evaluated with combination index values of less than 1 (data not shown). Additive effects of the two drugs at some concentrations were observed in BRAF WT cell lines suggesting that the combination of the two drugs is most effective in BRAF mutant cell lines. Consistent with these results, we also demonstrated that the dose-dependent antigrowth activity of vemurafenib could be significantly enhanced in the presence of canertinib (Fig. 4c). In the presence of 5 μmol/l canertinib, the IC50 for vemurafenib decreased to 104±31.9 nmol/l from 288.7±15.3 nmol/l in A375 cells (P = 0.002) and 66.6±7.4 from 133.5± 21.2 nmol/l in M14 cells (P = 0.041). Canertinib (10 μmol/l) completely inhibited cell growth in both of these cell lines. Together, these results suggest that canertinib can enhance the effects of vemurafenib in BRAF mutant cell lines.
Table 2.
Effect of 1 mmol/l vemurafenib on the IC50 of BRAF wildtype and mutant melanoma cells towards canertinib
| IC50 canertinib (mean±SEM, μmol/l) |
||||
|---|---|---|---|---|
| Cell line | BRAF status | –Vemurafenib | +Vemurafenib | P-value |
| SK-MEL147 | Wildtype | 2.92±0.08 | 3.92±0.63 | 0.189 |
| IgR3 | Wildtype | 3.19±0.17 | 1.67±0.21 | <0.001 |
| FEMX | Wildtype | 5.02±0.85 | 4.29±0.37 | 0.244 |
| A375 | V600E | 4.16±1.02 | 0.64±0.07 | <0.05 |
| M14 | V600E | 4.43±0.19 | 1.93±0.28 | <0.0001 |
| SK-MEL100 | V600E | 4.22±1.01 | 1.85±0.76 | <0.05 |
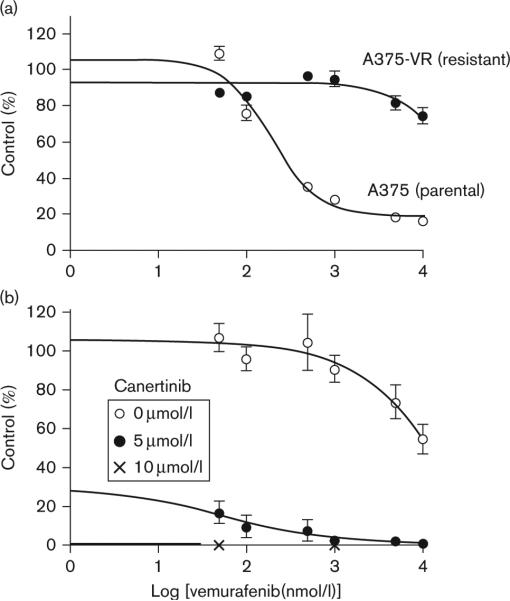
To demonstrate whether canertinib could overcome resistance to vemurafenib, we generated vemurafenib resistant A375 cells (A375-VR) by culturing A375 cells in the presence of increasing vemurafenib concentrations from 1 to 3 μmol/l sequentially. Figure 5a shows the vemurafenib dose–response between the parental A375 and A375-VR vemurafenib resistant cells. The IC50 for vemurafenib is not reached in the A375-VR cells with vemurafenib concentrations up to 10 μmol/l. The vemurafenib IC50 in A375 parental cells was 240 nmol/l. Addition of 5 or 10 μmol/l canertinib was able to completely overcome the vemurafenib resistance in the A375-VR with 83.6–92.5% decreases in cell growth (P < 0.0001) (Fig. 5b).
Fig. 5.
Canertinib can overcome vemurafenib resistance in A375-VR cells. (a) A375 BRAF mutant cells were treated with increasing concentrations of vemurafenib to generate a resistant cell line, A375-VR. A375-VR and A375 parental cells were treated with increasing concentrations of vemurafenib for 3 days and analyzed by MTT assay. (b) Vemurafenib treated cells were treated with or without 5 μmol/l canertinib for 3 days and analyzed by MTT. Percentage growth inhibition was calculated for each treatment group.
Discussion
BRAF inhibitors have shown great potential for melanoma patients; however, both intrinsic and acquired resistance remains a clinical problem. Thus, further understanding of BRAF mutant melanoma will be necessary to develop new treatments for this disease. In this study, we have shown that, rather than one dominant erbB family member, multiple erbB receptors were expressed in BRAF mutant and WT melanoma cell lines. Among the different erbB family members, EGFR was not frequently detected in BRAF V600E melanoma cells whereas high expression levels of EGFR were occasionally present in BRAF WT cells. Furthermore, we also detected very low or even undetectable release of EGFR preferential ligands (heparin-binding EGF-like growth factor, TGF-α, amphiregulin) by melanoma cells. Hence, in melanoma cells, the EGFR signaling axis may not be a common major contributor in most cases of melanoma development. However, previous clinical evidence did show that EGFR up-regulation and gene amplification are associated with malignancy progression, metastasis, and poor prognosis in a subset of melanoma cases [4,5].
erbB3 is commonly expressed in primary melanoma and metastases with high expression levels associated with poor patient prognosis [15]. erbB3, as well as erbB2, was often detected in high levels in both BRAF WT and mutant cells. We also detected the release of NRG1, the specific ligand of erbB3. When binding with NRG1, erbB3 can form heterodimers with other erbB partners for activating downstream signaling. We detected coactivation of erbB2 and erbB3 upon NRG1 stimulation in several melanoma cell lines, as well as coactivation of erbB3 and erbB4. Although the full-length form of erbB4 was not observed in immunoblots of unstimulated cells, phosphorylated erbB4 at 180 kD was observed after NRG1 stimulation, suggesting there is some full-length erbB4 present in these melanoma cells. This was seen in both BRAF WT and mutant cell lines. We also detected high levels of erbB4 ligands, NRG3, and NRG4, released by melanoma cells. Interestingly, we detected high expression levels of a truncated form of erbB4 in both BRAF WT and mutant cells. It has been shown that a truncated form (120 kD), rather than the full-length protein, is the dominant erbB4 species detected in human melanoma cells [20]. In breast cancer cells, erbB4 proteins undergo proteolysis in response to ligand stimulation. In return, the 80 kD intracellular domain is generated and then enters into the nucleus to carry out gene transcription activation functions [27]. At present, any transcription modulation activity exerted by truncated erbB4 in melanoma has not yet been described. However, our data suggested that the truncated erbB4 receptors may play an important role in melanoma biology. Recent evidence suggests that activating mutations in the ERBB4 gene are present in both primary melanomas as well as melanoma cell lines although the frequency of this mutation is variable ranging from 2 to 20% [28–31]. These mutations can coexist with BRAF or NRAS mutations. Whether or not these mutations exist in these cell lines is not known.
Although the involvement of the erbB family in melanoma development has long been known, the use of specific EGFR targeting agents for melanoma treatment has to date appeared to be mostly ineffective for clinical use. With limited knowledge on the expression profile of various erbB receptors in melanoma, previous phase II clinical trials involving the TKIs specifically targeting EGFR (gefitinib and erlotinib) in melanoma patients showed very limited therapeutic benefit [8,9]. In this study, our findings on multiple human melanoma cell lines indicate that (a) EGFR is not commonly present, and (b) multiple non-EGFR preferential erbB ligands, and erbB2/3/4 receptors are highly expressed in melanoma cells. Hence, it is not surprising that a single erbB targeting approach is not ideal for treating malignant melanoma patients.
It has been reported that human melanoma cell lines (RaH3 and RaH5) with high basal activated EGFR, erbB2, erbB3, and erbB4, both gefitinib and canertinib showed very effective growth inhibitory effects [25,32]. However, the therapeutic efficacy of TKIs targeting single versus multiple erbB proteins have never been compared for melanoma treatment. In this study, we compared the antigrowth effect of single-erbB and multi-erbB targeting inhibitors in BRAF WT and mutant melanoma cells. Our data strongly support the idea that a pan-erbB targeting approach, rather than the single/dual-erbB specific inhibition, is a more useful approach for treating melanoma in both BRAF WT and mutant tumors.
Previous clinical trials with canertinib show that thrombocytopenia and skin rash are the common toxicities associated with the drugs. The development of skin rash during canertinib treatment, a potential problem for melanoma patients, can be strategically solved through the optimization of treatment schedule as shown in other reports [33]. Another way to circumvent the toxicity of canertinib is by using a lower effective dose of canertinib by combining with other therapeutic agents. Our results indicate that, in BRAF mutant melanoma cells, the antigrowth effect of canertinib could be enhanced in the presence of BRAF inhibitor vemurafenib. Recent reports have already showed that NRG1 activation of erbB signaling can greatly attenuate BRAF inhibition by vemurafenib in BRAF mutant melanoma [2,16,17]. As NRG1 activates erbB3/4, an agent like canertinib that can block not only erbB3/4 but also their heterodimer binding partners would have the best potential for overcoming this effect. Furthermore, Prahallad et al. [26] have already described that inhibition of EGFR signaling renders BRAF mutant colon cancers subsequently sensitive to vemurafenib, although these are intrinsically resistant to BRAF inhibitors alone. Their data also show that ectopic EGFR expression in melanoma carrying BRAF V600E mutation is sufficient to induce BRAF inhibitor resistance.
Current use of vemurafenib in melanoma patients is often associated with the induction of secondary cutaneous squamous cell carcinoma development upon prolonged vemurafenib exposure. This is mainly because of the activation of MEK/MAPK/ERK pathway in BRAF normal cells by vemurafenib. Our data, however, indicate that canertinib is also effective in inhibiting the growth of BRAF WT tumor cells. Hence, the combined treatment of vemurafenib and canertinib may likely reduce the occurrence of secondary cutaneous squamous cell carcinoma development.
Conclusion
We have demonstrated that individual melanoma cell lines express more than one dominant member of the erbB receptor family, as well as ligands for these receptors, and that the different erbB family members signal together in response to a selective ligand for EGFR or erb3/4. Growth inhibition of melanoma cells is more effective with the pan-erbB targeting inhibitor canertinib than other single/dual erbB targeting agents, and canertinib can block the activation of multiple erbB receptors. In addition, we showed that multi-erbB inhibition with canertinib could enhance the antitumor efficacy of BRAF inhibitor vemurafenib in BRAF mutant melanoma. Further work to determine whether this enhanced effect in BRAF mutant melanoma can prevent or overcome vemurafenib resistance is warranted, along with exploration of the clinical applications of these concepts.
Supplementary Material
Acknowledgements
This study is supported by NIH SPORE in Skin Cancer Career Development Award (Y.K.N., L.P.S., P50CA121973), NIGMS Institutional Research and Academic Career Development Award (S.T., 1K12GM088021), and an Endowment Grant from the University of Pittsburgh Medical Center (UPMC) to J.M.S. This project used the UPCI TARPS Facility and Cytometry Facility that is supported by award P30CA047904. The Los Angeles Residual Tissue Repository is supported by grants and contracts P30CA014089 and HHSN261201000035C from the National Cancer Institute awarded to the University of Southern California. In addition, M.B. and S.T. were supported by K05 CA13675 and R01CA112524.
Footnotes
All supplementary data is available directly from the corresponding author.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, Muthusamy V, et al. Phosphoproteomic screen identifies potential therapeutic targets in melanoma. Mol Cancer Res. 2011;9:801–812. doi: 10.1158/1541-7786.MCR-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Wit PEJ, Moretti S, Koenders PG, Weterman MAJ, van Muijen GNP, Gianotti B, et al. Increasing epidermal growth factor receptor expression in human melanocytic tumor progression. J Invest Dermatol. 1992;99:168–173. doi: 10.1111/1523-1747.ep12616793. [DOI] [PubMed] [Google Scholar]
- 5.Rákosy Z, Vízkeleti L, Ecsedi S, Vokó Z, Bégány Á , Barok M, et al. EGFR gene copy number alterations in primary cutaneous malignant melanomas are associated with poor prognosis. Int J Cancer. 2007;121:1729–1737. doi: 10.1002/ijc.22928. [DOI] [PubMed] [Google Scholar]
- 6.Boone B, Jacobs K, Ferdinande L, Taildeman J, Lambert J, Peeters M, et al. EGFR in melanoma: clinical significance and potential therapeutic target. J Cutan Pathol. 2011;38:492–502. doi: 10.1111/j.1600-0560.2011.01673.x. [DOI] [PubMed] [Google Scholar]
- 7.Naramura M, Gillies S, Mendelsohn J, Reisfeld R, Mueller B. Therapeutic potential of chimeric and murine anti-(epidermal growth factor receptor) antibodies in a metastasis model for human melanoma. Cancer Immunol Immunother. 1993;37:343–349. doi: 10.1007/BF01518458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel SP, Kim KB, Papadopoulos NE, Hwu W-J, Hwu P, Prieto VG, et al. A phase II study of gefitinib in patients with metastatic melanoma. Melanoma Res. 2011;21:357–363. doi: 10.1097/CMR.0b013e3283471073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyman K, Kelley M, Puzanov I, Sanders K, Hubbard F, Krozely P, et al. Phase II study of erlotinib given daily for patients with metastatic melanoma (MM). J Clin Oncol. 2006;24:18S. [Google Scholar]
- 10.Christensen JG, Schreck RE, Chan E, Wang X, Yang C, Liu L, et al. High levels of HER-2 expression alter the ability of epidermal growth factor receptor (EGFR) family tyrosine kinase inhibitors to inhibit EGFR phosphorylation in vivo. Clin Cancer Res. 2001;7:4230–4238. [PubMed] [Google Scholar]
- 11.Inman JL, Kute T, White W, Pettenati M, Levine EA. Absence of HER2 overexpression in metastatic malignant melanoma. J Surg Oncol. 2003;84:82–88. doi: 10.1002/jso.10297. [DOI] [PubMed] [Google Scholar]
- 12.Kluger HM, DiVito K, Berger AJ, Halaban R, Ariyan S, Camp RL, et al. Her2/neu is not a commonly expressed therapeutic target in melanoma – a large cohort tissue microarray study. Melanoma Res. 2004;14:207–210. doi: 10.1097/01.cmr.0000130874.33504.2f. [DOI] [PubMed] [Google Scholar]
- 13.Persons DL, Arber DA, Sosman JA, Borelli KA, Slovak ML. Amplification and overexpression of HER-2/neu are uncommon in advanced stage melanoma. Anticancer Res. 2000;20:1965–1968. [PubMed] [Google Scholar]
- 14.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reschke M, Mihic-Probst D, van der Horst EH, Knyazev P, Wild PJ, Hutterer M, et al. HER3 is a determinant for poor prognosis in melanoma. Clin Cancer Res. 2008;14:5188–5197. doi: 10.1158/1078-0432.CCR-08-0186. [DOI] [PubMed] [Google Scholar]
- 16.Abel EV, Basile KJ, Kugal CH, Witkiewicz AK, Ke K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhbitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutterbach B, Ware C, Davis L, Zhang Q, Zawel L, Reilly J, et al. Growth factor-mediated resistance to BRAF/MEK inhibitors in BRAF mutant melanoma [abstract].. Proceedings of the conference growth factor-mediated resistance to BRAF/MEK inhibitors in BRAF mutant melanoma; Chicago, Illinois. Philadelphia (PA). 1–4 April 2012; AACR; 2012. [Google Scholar]
- 18.Chou TC, Talalay P. Quantitative analysis of dose–effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 19.Darcy KM, Zangani D, Wohlhueter AL, Huang R-Y, Vaughan MM, Russell JA, et al. Changes in ErbB2 (her-2/neu), ErbB3, and ErbB4 during growth, differentiation, and apoptosis of normal rat mammary epithelial cells. J Histochem Cytochem. 2000;48:63–80. doi: 10.1177/002215540004800107. [DOI] [PubMed] [Google Scholar]
- 20.Gordon-Thomson C, Jones J, Mason RS, Moore GP. ErbB receptors mediate both migratory and proliferative activities in human melanocytes and melanoma cells. Melanoma Res. 2005;15:21–28. doi: 10.1097/00008390-200502000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Mahtouk K, Hose D, Reme T, De Vos J, Jourdan M, Moreaux J, et al. Expression of EGF-family receptors and amphiregulin in multiple myeloma. Amphiregulin is a growth factor for myeloma cells. Oncogene. 2005;24:3512–3524. doi: 10.1038/sj.onc.1208536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang LM, Kuo A, Alimandi M, Veri MC, Lee CC, Kapoor V, et al. ErbB2 expression increases the spectrum and potency of ligand-mediated signal transduction through ErbB4. Proc Natl Acad Sci USA. 1998;95:6809–6814. doi: 10.1073/pnas.95.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa D, Yokota K, Gouda M, Narumi Y, Ohmoto H, Nishiwaki E, et al. Activity-based kinase profiling of approved tyrosine kinase inhibitors. Genes Cells. 2013;18:110–122. doi: 10.1111/gtc.12022. [DOI] [PubMed] [Google Scholar]
- 24.Slichenmyer WJ, Elliot WL, Fry DW. CI-1033, a pan-erbB tyrosine kinase inhibitor. Semin Oncol. 2001;28:80–85. doi: 10.1016/s0093-7754(01)90285-4. [DOI] [PubMed] [Google Scholar]
- 25.Djerf Severinsson EA, Trinks C, Green H, Abdiu A, Hallbeck AL, Stal O, et al. The pan-ErbB receptor tyrosine kinase inhibitor canertinib promotes apoptosis of malignant melanoma in vitro and displays anti-tumor activity in vivo. Biochem Biophys Res Commun. 2011;414:563–568. doi: 10.1016/j.bbrc.2011.09.118. [DOI] [PubMed] [Google Scholar]
- 26.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 27.Sundvall M, Peri L, Maatta JA, Tvorogov D, Paatero I, Savisalo M, et al. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene. 2007;26:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- 28.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutton-Regester K, Irwin D, Hunt P, Aoude LG, Tembe V, Pupo GM, et al. A high-throughput panel for identifying clinically relevant mutation profiles in melanoma. Mol Cancer Ther. 2012;11:888–897. doi: 10.1158/1535-7163.MCT-11-0676. [DOI] [PubMed] [Google Scholar]
- 30.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djerf EA, Trinks C, Abdiu A, Thunell LK, Hallbeck AL, Walz TM. ErbB receptor tyrosine kinases contribute to proliferation of malignant melanoma cells: inhibition by gefitinib (ZD1839). Melanoma Res. 2009;19:156–166. doi: 10.1097/CMR.0b013e32832c6339. [DOI] [PubMed] [Google Scholar]
- 33.Calvo E, Tolcher AW, Hammond LA, Patnaik A, de Bono JS, Elseman IA, et al. Administration of Cl-1033, an irreversible pan-erbB tyrosine kinase inhibitor, is feasible on a 7-day on, 7-day off schedule: a phase I pharmacokinetic and food effect study. Clin Cancer Res. 2004;10:7112–7120. doi: 10.1158/1078-0432.CCR-04-1187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.