Abstract
In treating hepatitis B virus (HBV) and human immunodeficiency virus (HIV) infections, the rapid reselection of resistance-associated variants (RAVs) is well known in patients with repeated exposure to the same class of antiviral agents. For chronic hepatitis C patients who have experienced virologic failure with direct-acting antiviral drugs, the potential for the reselection of persistent RAVs is unknown. Nine patients who received 14 days of telaprevir monotherapy were retreated with telaprevir-based triple therapy 4.3 to 5.7 years later. In four patients with virologic failure with both telaprevir-containing regimens, population-based and deep sequencing (454 GS-FLX) of the NS3 protease gene were performed before and at treatment failure (median coverage, 4,651 reads). Using deep sequencing, with a threshold of 1.0% for variant calling, no isolates were found harboring RAVs at the baseline time points. While population-based sequencing uncovered similar resistance patterns (V36M plus R155K for subtype 1a and V36A for subtype 1b) in all four patients after the first and second telaprevir treatments, deep sequencing analysis revealed a median of 7 (range, 4 to 23) nucleotide substitutions on the NS3 backbone of the resistant strains, together with large phylogenetic differences between viral quasispecies, making the survival of resistant isolates highly unlikely. In contrast, in a comparison of the two baseline time points, the median number of nucleotide exchanges in the wild-type isolates was only 3 (range, 2 to 8), reflecting the natural evolution of the NS3 gene. In patients with repeated direct antiviral treatment, a continuous evolution of HCV quasispecies was observed, with no clear evidence of persistence and reselection but strong signs of independent de novo generation of resistance. Antiviral therapy for chronic viral infections, like HIV, hepatitis B virus (HBV), or hepatitis C virus (HCV), faces several challenges. These viruses have evolved survival strategies and proliferate by escaping the host's immune system. The development of direct-acting antiviral agents is an important achievement in fighting these infections. Viral variants conferring resistance to direct antiviral drugs lead to treatment failure. For HIV/HBV, it is well known that viral variants associated with treatment failure will be archived and reselected rapidly during retreatment with the same drug/class of drugs. We explored the mechanisms and rules of how resistant variants are selected and potentially reselected during repeated direct antiviral therapies in chronically HCV-infected patients. Interestingly, in contrast to HIV and HBV, we could not prove long-term persistence and reselection of resistant variants in HCV patients who failed protease inhibitor-based therapy. This may have important implications for the potential to reuse direct-acting antivirals in patients who failed the initial direct antiviral treatment. (The phase IIIb study described in this paper is registered at ClinicalTrials.gov under registration number NCT01054573.)
INTRODUCTION
With an estimated 150 million people chronically infected worldwide and 3 to 4 million new infections each year (1), hepatitis C virus (HCV) infection is one of the major causes of liver cirrhosis and the subsequent development of hepatocellular carcinoma.
A large number of direct-acting antiviral agents (DAAs) are under investigation in clinical studies directed toward improving treatment response and tolerability, reducing treatment duration, and establishing interferon-free treatment options (2). Combination therapies of DAAs, including nonstructural 3 (NS3)/4A protease inhibitors, NS5A inhibitors, and NS5B polymerase inhibitors, with and without the addition of peginterferon alfa (PEG-IFN) and/or ribavirin (RBV), have led to higher sustained virologic response (SVR) rates and shorter treatment durations (3).
Telaprevir (TVR) is an effective and selective inhibitor of the HCV NS3/4A protease and is approved in combination with PEG-IFN and RBV for the treatment of genotype 1 chronic HCV infection (4, 5). Despite increased SVR rates using this combination compared to those with PEG-IFN–RBV without TVR, an SVR still led to a failure to eradicate HCV RNA permanently in numerous patients. The selection of resistance-associated variants (RAVs) as dominant strains was observed in most patients who failed TVR-based therapies (6, 7). For all NS3 protease inhibitors currently approved for antiviral combination therapies (TVR, boceprevir, and simeprevir), a continuous decline in resistant variants to undetectability within approximately 1 month to 1.5 years in patients with treatment failure has been described in long-term follow-up studies (8, 9). However, the persistence of resistant variants at low levels within the HCV quasispecies has been described using clonal and deep sequencing analyses in individual patients (10, 11). The potential for these variants to rapidly reemerge during reexposure to the same compound or the same class of drugs is largely unknown (12, 13).
In the present study, four patients were investigated who experienced virologic failure with TVR–PEG-IFN–RBV triple therapy in a phase IIIb, single-arm, and open-label study after short-term TVR monotherapy for 14 days in phase I studies (14, 15). The aim of the present study was to explore the presence and evolution of the RAVs selected during the first and second treatments with TVR. Full-length 454 deep sequencing of the NS3 protease gene was conducted before and at time of virologic failure with the first and second TVR-based therapies to differentiate the potential long-term persistence of RAVs from the de novo selection of identical resistance variants on different viral backbones. In addition, the phylogenetic relationships within and between viral quasispecies were examined in order to gauge the molecular evolution of the NS3 protease gene. We were able to describe the evolution of HCV genomes and show differences compared to those of other viral species, such as HBV or human immunodeficiency virus (HIV), in which the mechanism of archiving viral genomes, by integration into the host genome, contributes to a rapid reselection of resistant variants.
MATERIALS AND METHODS
Patients and study design.
Nine patients enrolled in the randomized, double-blind, placebo-controlled, 14-day, multiple-dose, and phase Ib TVR studies 101 (n = 8) and 103 (n = 1) (VX04-950-101 and VX05-950-103, respectively) (14, 15) were retreated with TVR and PEG-IFN plus RBV. All nine patients initially received TVR monotherapy for 14 days.
The single patient (HCV genotype 1a) who underwent study 103 received 750 mg of TVR every 8 h (q8h). The patients from study 101 were treated with 450 mg (n = 2) or 750 mg of TVR q8h (n = 3) or 1,250 mg of TVR q12h (n = 3). All patients suffered from chronic infection with HCV subtype 1a (n = 6) or 1b (n = 3), with plasma HCV RNA levels of ≥800,000 IU/ml, as well as negative hepatitis B surface antigen and antibodies to HIV-1 and -2. Written informed consent was obtained from each patient in accordance with the 1975 Declaration of Helsinki. None of these patients achieved an SVR after TVR monotherapy. These nine patients were then enrolled in the phase IIIb, single-arm, open-label, and rollover study C219 (VX-950-TiDP24-C219, registered at ClinicalTrials.gov under registration no. NCT01054573), in which they were retreated with TVR (750 mg q8h for 12 weeks) combined with PEG-IFN (180 μg/week) plus RBV (1,000 to 1,200 mg/day), for a total of 48 weeks. Futility rules were applied to all patients; i.e., TVR was stopped at week 4 or 8 if the HCV RNA level was >100 IU/ml (PEG-IFN–RBV were continued at standard doses), and all study drugs were discontinued if the HCV RNA level was >100 IU/ml at week 12 or ≥25 IU/ml at week 24 or 36.
Plasma samples were obtained before, during, and after treatment. HCV RNA levels were measured using the High Pure System (HPS)/Cobas TaqMan assay version 2.0 (Roche Molecular Diagnostics, Pleasanton, CA, USA; lower limit of quantification, 25 IU/ml). The primary study endpoint was SVR24, defined as HCV RNA levels of <25 IU/ml, with the target not detected, 24 weeks after the last actual dose of study drugs. On-treatment virologic failure was defined as having met a virologic stopping rule or having experienced viral breakthrough (a confirmed increase of >1 log in the HCV RNA level from the lowest level reached, or HCV RNA level of >100 IU/ml in patients whose HCV RNA level had previously become <25 IU/ml during treatment). Relapse was defined as having detectable HCV RNA during the follow-up period after a previous HCV RNA level of <25 IU/ml, with the target not detected, at the actual end of therapy (EOT).
HCV RNA extraction and reverse transcription.
Viral HCV RNA extraction (QIAamp viral RNA minikit; Qiagen, Hilden, Germany) from patient serum samples was followed by cDNA synthesis using the Life Technologies SuperScript III first strand synthesis system for reverse transcription-PCR (RT-PCR). All steps were performed according to the manufacturer's instructions.
RNA quality was assessed by calculating the absorbance ratio (optical density at 260 nm [OD260]/OD280) using NanoDrop model ND-1000 (Peqlab, Erlangen, Germany).
Amplification of the HCV NS3 protease gene and population-based sequencing.
In four patients with virologic failure with the first and second treatments, sequence analysis was performed at 4 time points: before (baseline 1 [BL1]) and at the end of (EOT1) the first TVR therapy course, as well as before the initiation (BL2) and at virologic failure (breakthrough or relapse) (EOT2) of the second TVR-containing regimen. In addition, for one patient (patient 2), a plasma sample from a second time point during follow-up of the second TVR treatment was available for sequence analysis (EOT3).
The region encoding the complete NS3 protease was amplified as one amplicon by nested-reverse transcription-PCR, using the following primer pairs (MID, multiplex identifiers): NS3-1a-in-f ([MID]-GTACGCCAGCAGACAAGGGGCCTCC) and NS3-1a-in-r ([MID]-CCGTGAACACCGGGGACCTCATGG), NS3-1b-in-f ([MID]-GACGCGAGGCCTACTTGGCTGCATC) and NS3-1b-in-r ([MID]-CGAGTTGTCCGTGAAGACCGGAGACC), NS3-1a-out-f (ATGGAGACCAAGCTCATCACGTGGG) and NS3-1a-out-r (ACCCGCCGTCGGCAAGGAACTTGCCGTA), and NS3-1b-out-f (ATGGAGACCAAGATCATCACCTGGG) and NS3-1b-out-r (CCGTCGGCAAGGAACTTGCCATAGGTGGA). The sequence adaptors and MIDs (barcode sequences) for the subsequent 454 deep sequencing were attached to the primers. To minimize bias due to PCR shift and reach the maximum number of different templates going into the deep sequencing process, three PCR replicates were pooled for each sample.
Population-based sequencing was performed as described previously (16). HCV RNA was isolated using a standard commercial silica gel membrane-binding method (QIAamp viral RNA minikit; Qiagen, Hilden, Germany), and the complete region encoding the NS3 protease catalytic domain was amplified by seminested-reverse transcription-PCR from stored plasma samples. A 912-bp region of genotype 1b NS3 was amplified using primers flanking the NS3 region (NS3-1b-1s [GGCGTGTGGGGACATCATC], NS3-1b-3a [GGTGGAGTACGTGATGGGGC], and NS3-1b-4a, [CATATACGCTCCAAAGCCCA]).
In the genotype 1a samples, a 620-bp fragment was amplified using the following primers: NS3-1a-1s (CCGGGAGATACTGCTCGGAC), NS3-1a-2s (CCGATGGAATGGTCTCCAAGG), NS3-1a-1a (GCTCTGGGGCACTGCTG), and NS3-1a-2a (GAGAGGAGTTGTCCGTGAACAC. The purified PCR products were subjected to sequence PCR, according to the manufacturer's instructions, using the appropriate forward or reverse primers of the second PCR (BigDye Deoxy Terminator; Applied Biosystems, Foster City, CA). Sequencing was performed by the 3130 DNA sequencer (Applied Biosystems, Foster City, CA).
454 deep sequencing analysis.
The viral quasispecies were analyzed using the 454-FLX+ system. An SFF file containing the nucleotide reads was generated for each sample by the 454 sequencing software (GS Run Processor; 454 Life Sciences, Branford, CT, USA) provided with the instrument. Subsequently, Phred quality scores were extracted from the reads, and primer sequences were removed from the start of the reads. Bases succeeding the first base call with a Phred quality score of <10 were trimmed, and only reads with no N base calls and a length of >25 bases were retained. The resulting high-quality reads were mapped to the HCV subgenotype-specific reference sequences H77 (1a) and HCV-J (1b) using SMALT version 0.7.5 (Wellcome Trust Sanger Institute, Cambridge, United Kingdom). The median coverage of the NS3 protease gene was 4,651 reads (range 1,034 to 35,916 reads) per sample. Tsibris et al. (17) conceded an error rate of 0.1 to 0.2% per nucleotide due to amplification and deep sequencing protocols after filtering out low-quality sequences, while Dietz et al. (18) found 0.507% erroneous substitutions per sequenced nucleotide when applying 454 deep sequencing. Since like Tsibris et al. (17), the deep sequencing workflow of our study contained both an RT-PCR and a second (nested) PCR, we settled for a conservative cutoff of 1.0% for the phylogenetic analyses. Like Dietz et al. (18), we used a 0.5% cutoff for calling potential, though unconfirmed minor resistance variants were below the 1.0% cutoff (Table 1).
TABLE 1.
Resistance-associated variants revealed by 454 deep sequencing with frequency cutoff set at 0.5%
| Patient | Resistance-associated variants (%) ina: |
|||
|---|---|---|---|---|
| BL1 | EOT1 | BL2 | EOT2/3 | |
| 1 | V36A (0.9) | V36A (2.8) | V36A (0.6) | |
| V36M (22.6) | V36M (4.4) | |||
| T54A (1.8) | ||||
| R155K (31.0) | R155K (51.1) | |||
| R155T (3.9) | ||||
| V36A + R155K (1.6) | ||||
| V36M + R155K (15.3) | V36M + R155K (38.1) | |||
| V36M + R155T (1.1) | ||||
| T54A + R155K (1.0) | ||||
| 2 | V36A (0.8) | V36A (32.3) | V36A (0.8) | V36A (0.8)/V36A (25.8) |
| T54A (57.5) | ||||
| V36A+ T54A (4.5) | ||||
| 3 | V36M (6.7) | V36M (3.5) | ||
| R155K (5.6) | ||||
| R155G (0.6) | R155G (0.6) | |||
| V36M + R155K (0.7) | V36M + R155K (85.2) | V36M + R155K (92.4) | ||
| V36M + T54A + R155K (0.7) | ||||
| 4 | V36A (0.6) | |||
| V36M (25.6) | V36M (2.9) | |||
| T54A (0.6) | ||||
| T54S (4.6) | ||||
| R155K (5.7) | R155K (1.9) | |||
| R155T (1.9) | ||||
| V36A + R155K (0.6) | ||||
| V36M + T54S (1.1) | ||||
| V36M + R155K (37.7) | V36M + R155K (94.8) | |||
| V36M + R155T (4.4) | ||||
| T54S + R155K (10.9) | ||||
| T54S + R155T (0.7) | ||||
| V36M + T54S + R155K (1.7) | ||||
Variants in italics carry the same linked neutral mutation within a patient.
IL28B genotyping.
The IL28B rs12979860 genotype was determined from patient blood samples using a custom TaqMan single nucleotide polymorphism (SNP) genotyping assay (Life Technologies, Darmstadt, Germany). The method was run on a StepOnePlus real-time PCR system (Applied Biosystems, Darmstadt, Germany) and analyzed using StepOne software version 2.0 (Applied Biosystems).
Bioinformatics region of interest.
Our region of interest comprises NS3 nucleotide positions 15 to 556 (NS3 amino acid positions 6 to 185) in genotype 1a, and NS3 nucleotide positions 28 to 563 (NS3 amino acid positions 10 to 187) in genotype 1b. This genomic region contains NS3 codons 36, 54, 155, and 156 associated with resistance to TVR and was the longest contiguous region whose coverage was >80% after quality control.
Consensus sequences.
Whole-population consensus sequences were derived from an alignment using the position-wise mode (i.e., taking the most prevalent base at each position).
Distance matrices.
For each patient, a 4 by 4 upper triangular matrix was constructed (5 by 5 in the case of patient 2; see the supplemental material) of pairwise Hamming distances between the whole-population consensus sequences.
Origin of resistance.
A preliminary insight into the origin of resistance at the EOT2 was gained by comparing the whole-population consensus pairwise distances BL2 to EOT2 and EOT1 to EOT2.
A more detailed picture was obtained by replacing the whole-population consensus sequences with subpopulation consensuses of reads carrying specific resistance variants.
In practice, the resistance variants 36M and 155K predominated in the data set, accounting for >80% of all resistance variants observed between them (correcting for variable numbers of reads per sample by normalizing coverage). In addition, the resistance variants at positions 36A, 54A, 54S, 155G, and 155T were also detected at various abundances of >0.5%.
Our third approach employed methods of unsupervised learning: a form of hierarchical clustering was implemented using neighbor joining, with complete linkage, and a variable threshold designed to ensure robust clusters containing no less than 5.0% of the reads. Complete linkage was chosen to favor dense clusters, tightly distributed around their mode, and thus were well represented by their consensus sequence.
The dynamic thresholding was introduced in order to reduce variance in the results introduced by experimental noise (particularly sequencing errors and outliers in sequence space) by excluding small clusters (containing <5.0% of the reads) that were deemed unreliable.
Clustering 3: resistance signature.
The subpopulation consensus sequences derived from the supervised and unsupervised methods described above (partitioning according to resistance variants and neighbor joining, respectively) were found to yield broadly consistent phylogenies. Therefore, the unsupervised approach was abandoned on the grounds of interpretability, as it lacked the meaningful labeling of clusters in terms of the presence/absence of resistance-associated variants in the supervised approach. In the final analysis, the samples were partitioned according to the specific combinations of resistance variants, or “resistance signature,” that they carried.
Choice of detection threshold.
An exploratory data analysis employed a frequency cutoff of 0.5%. For the present study, the detection threshold was raised to a conservative 1.0% cutoff to exclude potential false variants associated with the amplification and sequencing steps. Moreover, it was noted that BL resistance in the range of 0.5 to 1.0% was not selected at the EOT. For additional information, variants between frequencies of 0.5 and 1.0% are also presented in Table 1.
Linked neutral mutations.
In order to discern the BL population structure at the coarser frequency cutoff of 1.0% and better understand the origins of resistance at the EOT2, we identified synonymous (and presumed silent) single nucleotide polymorphisms significantly associated with resistance variants at the EOT. An association was confirmed using Fisher's exact test and significance using Bonferroni's method for multiple testing. A single linked neutral mutation was selected for plotting based on its observed associations and frequencies. The wild-type sequences at BL were partitioned in exactly the same manner as resistant sequences at the EOT, with the presence/absence of the chosen linked neutral mutation replacing the resistance signature as the criterion for clustering.
Tree construction.
The Hamming distances among the consensus sequences were again compared, paying particular attention to the nearest predecessors of the clusters at the EOT2.
Justification for excluding resistance-associated variants.
When calculating the Hamming distance between two haplotypes, care was taken to discount the nucleotide substitutions within codons 36, 54, 155, and 156 in order to reduce bias due to selection by TVR.
Method for choosing the nearest predecessor.
The rules for selecting the nearest predecessor of a given haplotype are as follows: (i) minimize genetic distance—among all past haplotypes, select that (those) with the smallest number of nucleotide substitutions, excluding positions associated with resistance (codons 36, 54, 155, and 156), since these variants arose under selection pressure; (ii) minimize time difference—in case of a tie, choose the most recent haplotype (later time); (iii) maximize probability—if a tie remains, choose the most abundant haplotype (higher frequency).
Phylogenetic tree.
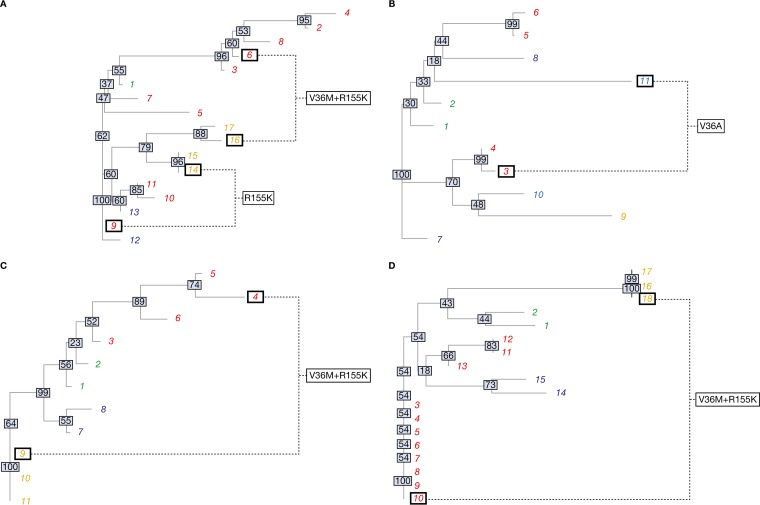
Neighbor-joining trees (Fig. 1) of the cluster consensus sequences, excluding those at codon positions 36, 54, 155, and 156, associated with resistance to TVR, were constructed using the Analyses of Phylogenetics and Evolution (APE) package in R (19, 20). The numbering of viral strains matches that of the corresponding evolutionary network (Fig. 2). Bootstrap values are commonly interpreted as a convenient proxy for the probability of the predicted phylogenetic relationships being correct. For our purposes, the bootstrap confidences and the tree itself are better thought of as representing a practical visualization of the matrices of pairwise genetic distances among viral strains (see the supplemental material).
FIG 1.
Neighbor-joining phylogenetic trees of the calculated distance matrices. Shown are data for patients 1 (A), 2 (B), 3 (C), and 4 (D). The different font colors represent different time points (green, BL1; red, EOT1; dark blue, BL2; orange, EOT2; light blue, EOT3). The numbering of leaves corresponds to the cluster numbering in the evolutionary networks (Fig. 2) and distance matrices (see the supplemental material). Clusters displaying identical resistance patterns at the EOT1 and EOT2/3 are highlighted by black squares to facilitate a direct comparison. The gray boxes superimposed on each internal node display the percentage bootstrap confidence with 10,000 replicates.
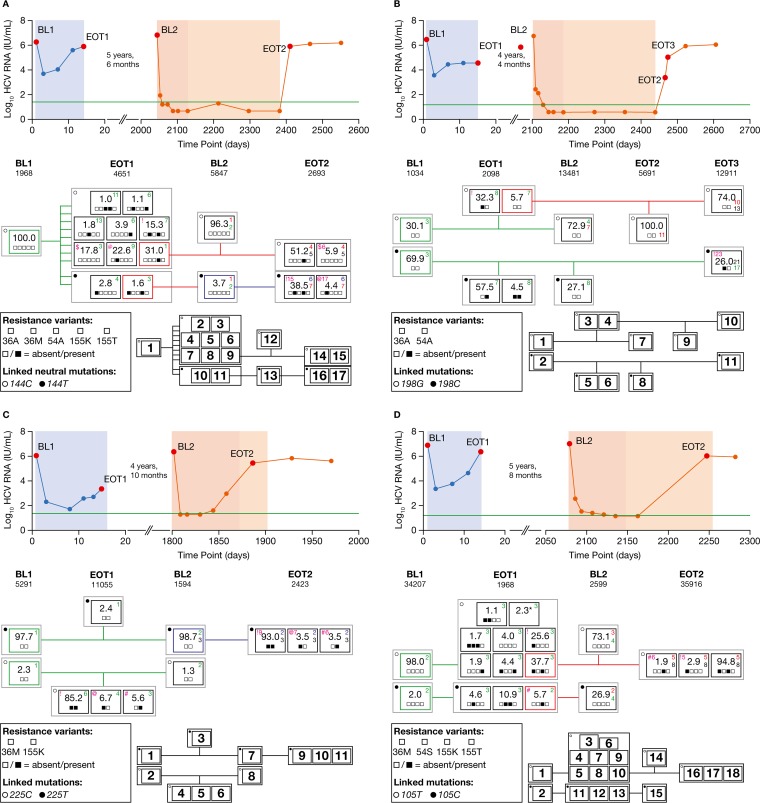
FIG 2.
Viral load profile, deep sequencing data, and phylogenetic relationships of clustered isolates at different time points. Shown are data for patients 1 (A), 2 (B), 3 (C), and 4 (D). Viral load profile: blue, TVR monotherapy; red, TVR–PEG-IFN–RBV triple therapy; orange, PEG-IFN–RBV dual therapy. The horizontal green line is the lower limit of quantification, and the red points show the times of sample acquisition. BL and EOT reads are shown, grouped (% of total quasispecies) by consensus sequence analysis, according to the presence/absence of resistance variants. The number of deep sequencing reads (coverage) used for analysis at the corresponding time points is given below each time point label (BL1, BL2, EOT1, EOT2, and EOT3). The clusters imply subpopulation consensuses of reads, either wild type or carrying specific resistance variants. The gray boxes collect clusters with the same linked neutral signature mutation. The colored boxes (green, red, and blue) represent the nearest predecessor population clusters. Each small square within the black or colored boxes shows the absence or presence of a particular resistance variant given in the legends within the figure. The colored numbers (green, red, and blue) give information about the number of nucleotide exchanges compared with that of the cluster consensus of the matching color; black numbers, included for rapid reference, give the distance to the next-nearest predecessor (see the supplemental material). The pairs of isolates at the EOT1 and EOT2/3 harboring identical resistance patterns are highlighted with magenta symbols ($, !, @, and #), accompanied by the number of nucleotide exchanges, also in magenta. The nearest predecessor clusters are connected by lines in corresponding colors. A table showing the numbers of nucleotide differences between all clusters can be found in the supplemental material. (D) At the EOT1, all clusters with a frequency of <1.0% were merged into a single cluster (cluster 6; 2.3%) and are marked with an asterisk. The numbering of the clusters in the black thumbnail plot next to each legend corresponds to the numbering of the leaves in the phylogenetic trees (Fig. 1A to D) and distance matrices (see the supplemental material).
Data accession number.
The data were deposited in the Sequence Read Archive (SRA) under accession no. SRP044323.
RESULTS
Clinical course of four patients with treatment failure.
While five patients achieved an SVR after the second full-course treatment with TVR plus PEG-IFN–RBV, four patients failed retreatment with TVR. These four patients were selected for deep sequence analysis to explore the evolution of wild-type sequences and resistant variants that potentially survived at low levels following the first exposure to TVR.
The treatment of these four patients consisted of TVR monotherapy for 14 days with 1,250 mg of TVR every 12 h (q12h) (patients 1 and 4), 450 mg of TVR every 8 h (q8h) (patient 2), or 750 mg of TVR q8h (patient 3). Patients 1, 3, and 4 were infected with HCV genotype 1a, while patient 2 was infected with HCV genotype 1b. All four patients experienced on-treatment virologic failure during initial TVR monotherapy. Three patients were nonresponders (patients 1, 2, and 4), and one patient experienced a relapse (patient 3) during treatment with PEG-IFN–RBV prior to the initiation of TVR triple therapy.
The IL28B rs12979860 genotypes were TT for patient 1 and CT for the other three patients. Two patients experienced relapse after TVR triple therapy, while two patients experienced on-treatment virologic failure (Table 2).
TABLE 2.
Clinical and virologic details of antiviral therapies and sequence analyses in patients with failure to both TVR therapies
| Measurement by TVR therapy typea | Patient 1 | Patient 2b | Patient 3 | Patient 4 |
|---|---|---|---|---|
| HCV genotype infection | 1a | 1b | 1a | 1a |
| IL28B genotype | TT | CT | CT | CT |
| TVR monotherapy | ||||
| VL BL1 (IU/ml) | 1,795,000 | 2,390,000 | 1,470,000 | 6,780,000 |
| RAVs at BL1 (direct + 454) | None | None | None | None |
| TVR dose (mg) | 1,250 q12h | 450 q8h | 750 q8h | 1,250 q12h |
| Duration (wk) | 2 | 2 | 2 | 2 |
| Treatment outcomec | On-treatment virologic failure | On-treatment virologic failure | On-treatment virologic failure | On-treatment virologic failure |
| VL EOT1 (IU/ml) | 889,000 | 38,800 | 2,620 | 2,440,000 |
| RAV(s) at EOT1 (direct) | V36M, R155K | V36A | V36M, R155K | V36M, R155K |
| RAV(s) at EOT1 (454) | V36A/M, T54A, R155K/T | V36A, T54A | V36M, R155K | V36M, T54S, R155K/T |
| Between-study measuresd | ||||
| Prior treatment response | Nonresponse | Nonresponse | Relapse | Null response |
| Time between studies | 5 yr 6 mo | 4 yr 4 mo | 4 yr 10 mo | 5 yr 8 mo |
| TVR triple therapy | ||||
| VL BL2 (IU/ml) | 9,290,000 | 1,200,000 | 2,440,000 | 19,900,000 |
| RAVs at BL2 (direct + 454) | None | None | None | None |
| TVR dose (mg q8h) | 750 | 750 | 750 | 750 |
| Duration (wk) | 52 | 52 | 12 | 24 |
| Treatment outcome | Relapse | Relapse | On-treatment virologic failuree | On-treatment virologic failuree |
| VL EOT2 (IU/ml) | 1,110,000 | 130,000 | 294,000 | 1,610,000 |
| RAV(s) at EOT2 (direct) | V36M, R155K | V36A | V36M, R155K | V36M, R155K |
| RAV(s) at EOT2 (454) | V36M, R155K | V36A | V36M, R155K | V36M, R155K |
454, detected by 454 ultradeep sequencing with 1.0% cutoff; direct, detected by population-based sequencing; VL, viral load.
V36A was detected at both EOT2 and EOT3 in patient 2.
In study 101, on-treatment virologic failure was defined as a >0.75-log10 increase in plasma HCV RNA level from the lowest measured HCV RNA level. In study 103, on-treatment virologic failure was defined as an increase from the lowest measured HCV RNA level during the 2-week study drug dosing period with a day 15 HCV RNA level of >50 IU/ml.
Patients received PEG-IFN–RBV between the end of the phase I study and the start of study C219. One of the four patients (patient 3) was PEG-IFN–RBV naive at initiation of the phase I study.
In study C219, on-treatment virologic failure was defined as having met a virologic stopping rule or experienced viral breakthrough.
Deep sequencing analysis was performed at baseline 1 (BL1) and the end of treatment 1 (EOT1) in the phase I studies and at BL (BL2) and time of virologic failure (EOT2) in the TVR triple-therapy study. The median number of sequence reads per position (coverage) for the HCV NS3 region over all patients and time points was 4,651 reads (range, 1,034 to 35,916 reads).
At both time points of treatment failure (EOT1 and EOT2), TVR-resistant variants were detected by 454 deep sequencing analysis in all patients as major quasispecies (Table 2), while at BL1/BL2, different minor resistant variants were detected at a frequency of only <1.0%, with uncertain significance (Table 1).
Viral kinetics, resistance, and phylogenetic analysis. (i) Patient 1 (subtype 1a).
Population-based sequencing revealed V36M plus R155K as dominant resistance pattern after both the first and second TVR-containing regimens, while no RAVs were observed at either BL time point. Using a 1.0% frequency cutoff to detect genuine variants within our deep sequencing assay, only wild-type virus was detected before the first exposure to TVR (BL1, Fig. 2A). However, the V36A variant, conferring low-level resistance to TVR, was observed at a frequency of 0.9%, being in the gray zone of potentially genuine variants (Table 1).
During the 2 weeks of TVR monotherapy (1,250 mg q12h), the HCV RNA levels initially declined, followed by an increase to near-BL levels by the EOT (Fig. 2A). At this time point (EOT1), 10 different isolates (clusters 2 to 11) containing the resistance-associated variants with the V36A, V36M, T54A, R155K, and R155T substitutions, alone or in combination, became detectable in the HCV quasispecies at frequencies ranging from 1.0% to 31.0%. Further grouping of these 10 clusters was made possible based on the detection of a linked neutral signature mutation at nucleotide position 144. Eight clusters (2–9) held the common signature 144C, while two (10 and 11, sole instances of variant V36A) contained 144T. The evolution of the different clusters from BL1 to the EOT1 was associated with a relatively large number of nucleotide substitutions (median, 6 substitutions; range, 1 to 13 substitutions). Among the variants present at the EOT1, the variant carrying R155K alone was present at the highest frequency (31.0%), suggesting its association with maximal replicational fitness within the viral quasispecies at that time point (Fig. 2A).
At BL2, 5 years and 6 months after the first exposure to TVR, 96.3% of the viruses were wild type (cluster 12) and most likely derived from the fittest R155K-containing cluster at the EOT1 at a distance of only one nucleotide exchange. Evolution from the surviving wild-type strain at the EOT1 (cluster 7) seemed less likely based on a longer distance of three nucleotide exchanges. Whereas the dominant cluster at BL2 (cluster 12) was associated with the signature variant 144C, the surviving variants with the 144T signature were also detectable at BL2 (cluster 13), albeit at a low frequency (3.7%), and their prevalences were comparable at the EOT1 (2.8% and 1.6%) (Fig. 2A). Interestingly, cluster 13 also contained traces of V36A below the 1.0% cutoff (0.6%; Table 1).
Patient 1 had an undetectable HCV RNA level at the end of the 48 weeks of TVR-based triple therapy. However, virologic relapse occurred 4 weeks after the EOT (Fig. 2A). Despite dosing of TVR for the first 12 weeks only, the majority of the variants observed at the time of relapse (EOT2) contained RAVs. Exactly as at the EOT1, the dominant resistant variant (cluster 14, 51.2%) harbored the R155K resistance variant alone. Here, the distance to the corresponding strain at the EOT1 (cluster 9) was 4 nucleotide exchanges versus 5 in the wild-type cluster 12 at BL2 (Fig. 2A). Thus, the survival of the resistant variant from the EOT1 to EOT2 seemed possible although it was deemed unlikely based on one nucleotide exchange less in comparison with the preceding wild-type cluster at BL2. Moreover, the survival of two other recurring mutational patterns (clusters 6 and 16 with V36M plus R155K, and clusters 8 and 17 with V36M alone) appeared highly implausible, due to the large differences in their NS3 backbones (15 and 17 nucleotide substitutions, respectively) (Fig. 2A).
(ii) Patient 2 (subtype 1b).
Population-based sequencing uncovered V36A following the first round of TVR therapy, and V36A again 4 weeks after the end of the second round, while only wild-type virus was observed at the respective BL time points. Deep sequencing at BL identified two distinct wild-type strains with an average separation of three nucleotide exchanges, including a 198C or 198G signature single nucleotide polymorphism (Fig. 2B). The V36A resistance variant was observed at a frequency of <1.0% within the BL 198C strains (Table 1). Patient 2 experienced viral breakthrough during TVR monotherapy (450 mg of TVR q8h) (Fig. 2B). Substantial evolution of the viral quasispecies was observed over the course of 14 days of TVR monotherapy, with the appearance of RAVs at positions V36 and T54 accompanied by an average of seven to eight additional nucleotide exchanges. Four years and 4 months later, again, only wild-type variants were detected, with further evolution of the NS3 nucleotide backbone (Fig. 2B). The minor resistance variant V36A was now observed at a frequency of 0.8% in the 198G strain (Table 1). Like patient 1, patient 2 also experienced a confirmed virologic response during the 48 weeks of the treatment period, followed by relapse 4 weeks after the EOT (Fig. 2B). An additional sample obtained a few days after the EOT2 was available for sequence analysis, displaying the further evolution of the wild-type virus together with the development of a cluster at 26.0% (cluster 11) containing the V36A resistant variant within the 198C lineage. The resistance signature of this cluster (V36A) was identical to that of cluster 3 (32.2%) at the EOT1. However, in view of an average 23 nucleotide exchanges between clusters 3 and 11, the survival and reselection of this variant from the EOT1 seemed highly improbable (Fig. 2B).
(iii) Patient3 (subtype 1a).
As for patient 1, population-based sequencing found wild-type virus at both BL time points and V36M plus R155K as the dominant resistance pattern at time of failure with both TVR therapies. Major and minor wild-type strains differing only at the signature nucleotide position 225 (255T or 255C, respectively) were observed by deep sequencing at BL1 before the initiation of TVR monotherapy (at 750 mg q8h) (Fig. 2C). After 14 days of TVR monotherapy, the patient experienced viral breakthrough (Fig. 2C), and deep sequencing detected a minor wild-type cluster (cluster 3; frequency, 2.4%) together with three clusters harboring the resistance variants V36M and/or R155K (clusters 4 to 6; total frequency, 97.5%). Interestingly, the signature variant at 225C suggests the evolution of resistance (clusters 4 to 6) from the minor BL quasispecies (cluster 2) (Fig. 2C). Four years and 10 months later, the distribution of wild-type strains at BL2 was similar to that at BL1 (Fig. 2C). Viral breakthrough (at EOT2) occurred at week 12 of TVR triple therapy, and deep sequencing analysis at this time point revealed patterns of resistance identical to those at the EOT1: V36M plus R155K, V36M only, and R155K only (clusters 9 to 11). However, a consensus distance between the corresponding resistant strains (Fig. 2C, magenta symbols and numerals) of 6 to 9 nucleotide exchanges on the NS3 backbone, including the signature variant at nucleotide position 225, made the persistence and direct evolution of these clusters from each other highly unlikely (Fig. 2C). Furthermore, despite the detection of V36M plus R155K (0.7%) and R155G (0.6%) at <1.0% within the dominant strain at BL1 (Table 1), these variants apparently did not give rise to dominant resistant strains at the EOT1, whose resistance-associated strains were characterized by the 225C signature single nucleotide polymorphism.
(iv) Patient 4 (subtype 1a).
Again, population-based sequencing uncovered wild-type virus at the BL time points, while at both times of treatment failure, V36M plus R155K was the predominant pattern of resistance. Based on the signature variant 105C or 105T, two viral clusters were observed at BL1, before the initiation of TVR monotherapy (at 1,250 mg q12h) (Fig. 2D). After viral breakthrough at the end of 14 days of TVR exposure (EOT1), a large number of different clusters was observed, including wild-type clusters and a variety of clusters containing resistance variants (Fig. 2D). Interestingly, despite the wide variety of resistance variants at the EOT1, the within-sample genetic diversity remained low across all different clusters, with a consensus variation in the NS3 backbone of at most three nucleotide substitutions. For both the 105T and 105C groups (clusters 3 to 10 and 11 to 13, respectively), the clusters with the highest prevalence and the smallest genetic distance were regarded as the origin for the evolution of wild-type clusters at BL2 (clusters 14 and 15). However, due to the very subtle genetic differences involved, no definite evolutionary pathway was established. The failure of TVR triple therapy in this patient occurred at week 24 of treatment with PEG-IFN–RBV. Once again, the EOT2 saw the recurrence of several resistance patterns already observed at the EOT1: V36M, R155K, and V36M plus R155K (clusters 16 to 18). The number of backbone nucleotide differences between the EOT2 and EOT1 clusters with matching patterns of resistance was lower (five to six substitutions) than that with the wild-type clusters at BL2 (eight substitutions). However, the overall number of nucleotide exchanges was nonetheless relatively high, precluding the possibility of unaltered long-term survival. Furthermore, for one strain (clusters 16 and 13), a change in the nucleotide signature made the persistence of resistance and reselection difficult to affirm.
Phylogenetic analysis.
The phylogenetic trees generated for patients 1 to 4 are shown in Fig. 1A to D, respectively. The trees illustrate the genetic relationships among the individual cluster consensus sequences. The major clusters carrying matching resistance patterns at the EOT1/2 were compared to assess the likelihood of direct evolution, i.e., reselection. In patient 1, it became apparent that none of the corresponding clusters (9 and 14, or 6 and 16) were located in the same clade; thus, a narrow relationship cannot be assumed. The phylogenetic distances between clusters 3 and 11 in patient 2 (both carrying the V36A RAV), clusters 4 and 9 in patient 3 (V36M plus R155K), and clusters 10 and 18 in patient 4 (V36M plus R155K) were all quite large. Therefore, these findings do not support the assumption of reselection of RAVs but provide an indication of de novo selection.
DISCUSSION
Since 2011, the standard treatment of genotype 1 HCV-infected patients has included the administration of HCV NS3/4A protease inhibitors in combination with PEG-IFN–RBV. A large number of patients worldwide received TVR- and boceprevir-based triple therapies, which exhibit improved SVR rates in comparison with those with the former standard treatment without protease inhibitors (4, 5, 21, 22). Subject to several negative predictors, such as previous response to PEG-IFN–RBV dual combination therapy and the stage of fibrosis, 25 to 69% of the patients failed to achieve viral eradication (4, 5, 21, 22). RAVs have been detected in 53 to 77% of the patients with treatment failure (8, 9). Recently, an all-oral combination therapy with the protease inhibitor simeprevir in combination with the nucleoside analogue polymerase inhibitor sofosbuvir has been approved in several countries. For patients failing TVR-boceprevir therapy, retreatment with a simeprevir-based all-oral regimen might be an attractive and effective treatment option. However, cross-resistance between TVR-boceprevir and simeprevir, together with the persistence of RAVs after treatment failure, may lead to an impaired virologic response. Diminished or depleted efficacies of direct antiviral drugs in hepatitis B virus (HBV) and human immunodeficiency virus (HIV) infection are well known, even if RAVs are no longer detectable by population-based sequence analysis. Therefore, in the present study, the potential of TVR-induced RAVs for long-term persistence and reselection of isolates containing these RAVs during a second exposure to the same drug was investigated by deep sequencing analysis. Hence, answering the question of whether a resistant variant once selected during initial exposure to a DAA is capable of surviving at low levels until retreatment with a second direct antiviral therapy, leading to rapid reselection and virologic treatment failure, is important.
In the present study, we analyzed the HCV quasispecies of four members of a unique cohort of patients treated twice with TVR-based regimens.
Deep sequencing analysis was performed at 4 to 5 time points before the initiation and after failure with the first and second exposures to TVR. To ensure reliable inference of viral haplotypes, we opted for 454 sequencing with a mean read length of 550 nucleotides. At both BL time points, neither population nor deep (1.0% cutoff) sequencing analysis detected RAVs.
While population-based sequencing showed similar resistance patterns after the first and second failures with TVR in individual patients, the results of the deep sequencing analysis made direct evolution from isolates persisting at low levels appear highly unlikely. In fact, a large number of nucleotide exchanges (median, 7 exchanges; range, 4 to 23 exchanges) on the NS3 backbone of isolates harboring identical RAVs at the first and second virologic failure time points (EOT1 versus EOT2/3) was detected. Moreover, large phylogenetic distances between pairs of isolates with identical patterns of resistance in individual patients supported independent evolution rather than reselection after long-term survival at very low frequencies.
Two other studies investigated the persistence of variants conferring resistance to NS3 protease inhibitors and retreatment with the same drug or class of drugs (12, 13). Lenz et al. (12) reported five patients who underwent repeated treatment with the NS3 protease inhibitor simeprevir. Deep sequencing analysis concluded that low-level persistence of RAVs was responsible for repeat treatment failure in two patients. However, in contrast with the present study, 454 deep sequencing did not cover the entire NS3 protease gene, which limited conclusions regarding persistence in the context of identical NS3 backbone sequences. Furthermore, Lenz et al. (12) observed divergent HCV RNA kinetics within the first days of the first and second courses of exposure to the NS3 protease inhibitor simeprevir. Specifically, the viral decline was slower during the second treatment with simeprevir, suggesting that low-frequency resistant variants within the HCV quasispecies may be responsible for partial nonresponsiveness. In our study, no differences in viral kinetics were observed between the first and second courses of treatment with TVR. However, HCV RNA sampling was performed only 8 to 9 days after the initiation of the second exposure to TVR. Thus, differences in very early viral kinetics were not detected in our patients. In the second study by Vermehren et al. (13) in HCV genotype 1-infected nonresponders who had been sequentially treated with boceprevir, clonal sequencing analysis of the NS3 resistance variants found mostly different mutational patterns after the first and second exposures to boceprevir. Here, boceprevir was administered at lower-than-normal doses (600 to 1,200 mg/day instead of 2,400 mg/day), and the majority of the HCV quasispecies isolates presented as the wild type. However, the intervals between different boceprevir exposures were very short (2 to 6 weeks) compared to those in both Lenz et al. (12) (approximately 1.5 years) and the present study (4.3 to 5.7 years).
The patients analyzed in this study received two treatments distinguished by the absence or presence of PEG-IFN and RBV. For patients receiving monotherapy with PEG-IFN or RBV, a very small number of nucleotide exchanges were observed in a recent study (18). Furthermore, other studies also described no significant changes in HCV quasispecies heterogeneity for virologic nonresponder patients before and after treatment with PEG-IFN and RBV together (23). Therefore, we considered a potential additional mutagenic effect of PEG-IFN and/or RBV as limited and most likely negligible.
While a fixed detection threshold of 1.0% was used throughout the present study, the 454 deep sequencing methods employed are capable of detecting low-level minority variants down to a prevalence of 0.5%, which represents another commonly used cutoff in other studies, although this may be associated with a concomitant increase in the risk of spurious variants arising during reverse transcription and amplification. The analysis of this gray area between 0.5% and 1.0%, in which variants might be real or false, may provide some interesting information about the potential evolution of RAVs and therefore is present as additional information. Yet, the overall assessment of this study is not affected by generally applying the 0.5% threshold. In line with previous studies, low-level (<1.0%) resistant variants were indeed observed in BL samples prior to TVR therapy (11). However, haplotype analysis found no evidence of selection of these variants at either the first or second EOT time points, making it difficult to decide whether or not variants below a 1.0% cutoff represent genuine virologically meaningful strains. Interestingly, in all patients, relationships were inferred between the strains at noncontiguous time points, pointing to the possibility that even deep sequencing is insufficiently sensitive to completely reconstitute the development of viral variants. Alternatively, the de novo generation of variants in a restricted sequence space constrained by drug and host immune system pressure is another possible explanation. The sequence analysis of additional HCV genes, especially of NS5B, may be of importance to further elucidate the question of persistence of RAVs.
To conclude, in patients with repeated TVR treatment, the continuous evolution of the NS3 quasispecies was observed with no clear evidence of persistence and reselection but strong signs of independent de novo generation of resistance. This finding might have important implications for the potential retreatment of patients with failure to direct antiviral therapies with direct antiviral agents binding to the same target, like the currently used protease inhibitor-based regimens involving simeprevir or ABT-450 (24–29).
Supplementary Material
ACKNOWLEDGMENTS
The present work was supported in part by a scientific grant from Janssen Infectious Diseases to S.S., S.Z., and C.S. and by the Deutsches Zentrum für Infektionsforschung, TTU Hepatitis, to C.S.
M.F. and G.L. declare no conflicts of interest. H.W.R. received research funding and consultancy fees from Abbvie, Astex, BMS, Boehringer Ingelheim, Gilead, GlaxoSmithKline, Janssen-Cilag, Merck/MSD, PRA International, Roche, Santaris, Regulus, R-Pharm, Korean Green Cross, and Replicor. S.Z. and C.S. received research funding and consultancy and speaker's fees from Janssen-Cilag, Merck/MSD, Roche, and Vertex. A.G., V.V.E., and S.D.M. are employees of Janssen Infectious Diseases. J.W. is an employee of Janssen Research & Development.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04911-14.
REFERENCES
- 1.World Health Organization. 2014. Hepatitis C: fact sheet no. 164. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs164/en/index.html. [Google Scholar]
- 2.Sarrazin C, Hézode C, Zeuzem S, Pawlotsky JM. 2012. Antiviral strategies in hepatitis C virus infection. J Hepatol 56:S88–S100. doi: 10.1016/S0168-8278(12)60010-5. [DOI] [PubMed] [Google Scholar]
- 3.Pawlotsky JM. 2014. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology 146:1176–1192. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S, ADVANCE Study Team. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med 364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 5.Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, Mullhaupt B, Pockros P, Terg R, Shouval D, van Hoek B, Weiland O, Van Heeswijk R, De Meyer S, Luo D, Boogaerts G, Polo R, Picchio G, Beumont M, REALIZE Study Team. 2011. Telaprevir for retreatment of HCV infection. N Engl J Med 364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 6.De Meyer S, Dierynck I, Ghys A, Beumont M, Daems B, Van Baelen B, Sullivan JC, Bartels DJ, Kieffer TL, Zeuzem S, Picchio G. 2012. Characterization of telaprevir treatment outcomes and resistance in patients with prior treatment failure: results from the REALIZE trial. Hepatology 56:2106–2115. doi: 10.1002/hep.25962. [DOI] [PubMed] [Google Scholar]
- 7.Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462. doi: 10.1053/j.gastro.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 8.Howe AY, Long J, Thompson S, Barnard RJ, Alves K, Howe JA, Wahl J. 2013. Analysis of the durability of response and persistence of resistance associated variants during long term follow up after boceprevir + pegylated interferon/ribavirin therapy—3 year analysis. Hepatology 58:1095A. [Google Scholar]
- 9.Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N, Martin EC, Beumont M, Jacobson IM, Sherman KE, Zeuzem S, Picchio G, Kieffer TL. 2013. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin Infect Dis 57:221–229. doi: 10.1093/cid/cit226. [DOI] [PubMed] [Google Scholar]
- 10.Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Fuller C, Perner D, Zeuzem S, Sarrazin C. 2011. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol 52:321–327. doi: 10.1016/j.jcv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Thomas XV, de Bruijne J, Sullivan JC, Kieffer TL, Ho CK, Rebers SP, de Vries M, Reesink HW, Weegink CJ, Molenkamp R, Schinkel J. 2012. Evaluation of persistence of resistant variants with ultra-deep pyrosequencing in chronic hepatitis C patients treated with telaprevir. PLoS One 7:e41191. doi: 10.1371/journal.pone.0041191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenz O, de Bruijne J, Vijgen L, Verbinnen T, Weegink C, Van Marck H, Vandenbroucke I, Peeters M, Simmen K, Fanning G, Verloes R, Picchio G, Reesink H. 2012. Efficacy of re-treatment with TMC435 as combination therapy in hepatitis C virus-infected patients following TMC435 monotherapy. Gastroenterology 143:1176–1178. doi: 10.1053/j.gastro.2012.07.117. [DOI] [PubMed] [Google Scholar]
- 13.Vermehren J, Susser S, Lange CM, Forestier N, Karey U, Hughes E, Ralston R, Tong X, Zeuzem S, Sarrazin C. 2012. Mutations selected in the hepatitis C virus NS3 protease domain during sequential treatment with boceprevir with and without pegylated interferon alfa-2b. J Viral Hepat 19:120–127. doi: 10.1111/j.1365-2893.2011.01449.x. [DOI] [PubMed] [Google Scholar]
- 14.Forestier N, Reesink HW, Weegink CJ, McNair L, Kieffer TL, Chu HM, Purdy S, Jansen PL, Zeuzem S. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640–648. doi: 10.1002/hep.21774. [DOI] [PubMed] [Google Scholar]
- 15.Reesink HW, Zeuzem S, Weegink CJ, Forestier N, van Vliet A, van de Wetering de Rooij J, McNair L, Purdy S, Kauffman R, Alam J, Jansen PL. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 17.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Parades R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietz J, Schelhorn SE, Fitting D, Mihm U, Susser S, Welker MW, Fuller C, Daumer M, Teuber G, Wedemeyer H, Berg T, Lengauer T, Zeuzem S, Herrmann E, Sarrazin C. 2013. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J Virol 87:6172–6181. doi: 10.1128/JVI.02778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 20.R Core Team. 2014. The R project for statistical computing. http://www.R-project.org.
- 21.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, Burroughs M, Brass CA, Albrecht JK, Esteban R, HCV RESPOND-2 Investigators. 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med 364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poordad F, McCone J Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP, SPRINT-2 Investigators. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med 364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasu A, Marusawa H, Ueda Y, Nishijima N, Takahashi K, Osaki Y, Yamashita Y, Inokuma T, Tamada T, Fujiwara T, Sato F, Shimizu K, Chiba T. 2011. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One 6:e24907. doi: 10.1371/journal.pone.0024907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r–ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 25.Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR, PEARL-III Study, PEARL-IV Study. 2014. ABT-450/r–ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med 370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 26.Lawitz E, Ghalib R, Rodriguez-Torres M, Younossi ZM, Corregidor A, Sulkowski MS, DeJesus E, Pearlman B, Rabinovitz M, Gitlin N, Lim JK, Pockros PJ, Fevery B, Lambrecht T, Ouwerkerk-Mahadevan S, Callewaert K, Symonds WT, Picchio G, Lindsay K, Beumont-Mauviel M, Jacobson IM. 2014. Simeprevir plus sofosbuvir with/without ribavirin in HCV genotype 1 prior null-responder/treatment-naive patients (COSMOS study): primary endpoint (SVR12) results in patients with Metavir F3-4 (cohort 2). J Hepatol 60:S524. doi: 10.1016/S0168-8278(14)61460-4. [DOI] [Google Scholar]
- 27.Poordad F, Hézode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM, Forns X, Lovell SS, Da Silva-Tillmann B, Collins CA, Campbell AL, Podsadecki T, Bernstein B. 2014. ABT-450/r–ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med 370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
- 28.Sulkowski MS, Jacobson IM, Ghalib R, Rodriguez-Torres M, Younossi Z, Corregidor A, Fevery B, Callewaert K, Symonds W, De La Rosa G, Picchio G, Ouwerkerk-Mahadevan S, Lambrecht T, Lawitz E. 2014. Once-daily simeprevir (TMC435) plus sofosbuvir (GS-7977) with or without ribavirin in HCV genotype 1 prior null responders with METAVIR F0-2: COSMOS study subgroup analysis. J Hepatol 60:S4. doi: 10.1016/S0168-8278(14)60009-X. [DOI] [Google Scholar]
- 29.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, Sulkowski MS, Wedemeyer H, Tam E, Desmond P, Jensen DM, Di Bisceglie AM, Varunok P, Hassanein T, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.