SUMMARY
The dramatic rise in the incidence of antibiotic resistance demands that new therapeutic options will have to be developed. One potentially interesting class of antimicrobials are the modified bacteriocins termed lantibiotics, which are bacterially produced, posttranslationally modified, lanthionine/methyllanthionine-containing peptides. It is interesting that low levels of resistance have been reported for lantibiotics compared with commercial antibiotics. Given that there are very few examples of naturally occurring lantibiotic resistance, attempts have been made to deliberately induce resistance phenotypes in order to investigate this phenomenon. Mechanisms that hinder the action of lantibiotics are often innate systems that react to the presence of any cationic peptides/proteins or ones which result from cell well damage, rather than being lantibiotic specific. Such resistance mechanisms often arise due to altered gene regulation following detection of antimicrobials/cell wall damage by sensory proteins at the membrane. This facilitates alterations to the cell wall or changes in the composition of the membrane. Other general forms of resistance include the formation of spores or biofilms, which are a common mechanistic response to many classes of antimicrobials. In rare cases, bacteria have been shown to possess specific antilantibiotic mechanisms. These are often species specific and include the nisin lytic protein nisinase and the phenomenon of immune mimicry.
INTRODUCTION
Lantibiotics are ribosomally synthesized antimicrobial peptides produced by Gram-positive bacteria. They undergo posttranslational modifications which result in the creation of unusual amino acids, such as dehydroalanine (Dha) and dehydrobutyrine (Dhb), as well as the eponymous lanthionine/methyllanthionine residues. Lantibiotics are subclassified on the basis of their biosynthetic machinery and the amino acid sequence of the structural peptide (1). The most characterized lantibiotic is nisin A (subclass 1), which has been used in the dairy and food industries for decades. It is a 3,353-Da, cationic, linear peptide of 34 amino acids, produced by Lactococcus lactis subsp. lactis, that contains five intramolecular ring structures. It has a dual mode of action, in that it prevents cell wall biosynthesis and also forms pores in the cell membranes of susceptible cells (2). Lantibiotics are readily used as food preservatives and have frequently been suggested to have the potential to be utilized in a wide range of medical applications (for reviews, see references 3, 4, and 5). With these existing and prospective applications in mind, the incidence of lantibiotic resistance is a real consideration, especially in view of the prevalence of resistance to commercial antibiotics following their extensive utilization. To date, little is known of lantibiotic resistance in comparison to resistance to commercial antibiotics. However, lantibiotic resistance has been induced in laboratory settings, and this resistance, in addition to the innate resistance characteristics of the bacterial cell, is the subject of this review.
MODE OF ACTION
In order to appreciate the various mechanisms by which bacteria are or become resistant to lantibiotics, it is first necessary to understand the mode of action of these compounds. In the case of many cationic lantibiotics, or lantibiotics with cationic domains, there is an initial attraction to the anionic cell membrane of target microbes (6–8). Notably, changes in lantibiotic charge as a result of genetic engineering alter the efficacy of such lantibiotics, presumably due to a reduced attraction/interaction with the cell membrane (8, 9). In addition, it is believed that lantibiotics most probably recognize a specific receptor molecule on sensitive bacterial cells. To date, however, only one receptor-like protein has been discovered, with the identification in Streptococcus pyogenes of a gene encoding a protein named LsrS, which exhibits a receptor-like function for the lantibiotic Smb (10). LsrS is a membrane protein that belongs to the CAAX protease family and is well conserved across streptococcal species. Overexpression of LsrS in Streptococcus mutans increased cellular susceptibility to Smb. In addition, nisin also requires LsrS for optimum inhibitory activity. However, it is not required for the antimicrobial actions of haloduracin and galolacticin. The mechanism of action of this receptor-like function is as yet unknown; however, it has been shown that the CAAX protease domain is not essential for function.
Once at the membrane, lantibiotics, such as nisin, mersacidin, epidermin, plantaricin C, and lacticin 3147 (Ltnα; one of two components of a dual-peptide bacteriocin), bind to and form a complex with a docking molecule, lipid II (11–15). Lipid II is a precursor of cell wall peptidoglycan and is found at the outer leaflet of the bacterial membrane. The resulting lantibiotic-lipid II complex inhibits peptidoglycan synthesis. Nisin binds the pyrophosphate linker of lipid II (16), as does plantaricin C and lacticin 3147 (Ltnα) (13, 14). Mersacidin and members of the mersacidin group of lantibiotics target the N-acetylglucosamine (GlcNAc) moiety and most probably the sugar and phosphate residues of lipid II (17). Lipid I has also been identified as a target of nisin and epidermin (11); lipid I is essentially lipid II before the acquisition of the GlcNAc moiety. However, binding to lipid II has a more pronounced effect. It should be noted that lipid I or lipid II is not the target of all lantibiotics; for example, targets have yet to be found for Pep5 and epilancin K7 (18). For some lantibiotics that bind lipid II, including nisin, this binding not only inhibits cell wall biosynthesis but also facilitates pore formation and leads to the release of ions and small molecules from the target bacteria (17, 19). Two-component lantibiotics, such as lacticin 3147, are thought to function through the formation of a three-member complex, with the Ltnα peptide targeting lipid II and then sequestering the Ltnβ peptide for pore formation (13). There are a few models which describe how the structural conformation of lantibiotics within the membrane leads to pore formation. This includes the barrel-stave mechanism described by Sahl (20), the wedge model (21), and a model described by Chikindas et al. (22). However, not all lantibiotics form pores. Mersacidin, actagardine, and cinnamycin (all members of the globular subclass II lantibiotics) only block cell wall/membrane synthesis by binding to receptors such as lipid II or phosphatidylethanolamine (23, 24). Gallidermin (and related peptides) is unusual in that, while it possesses the same putative lipid II-binding motif as nisin, it is considerably shorter (22 amino acids, compared to the 34 amino acids of nisin). As a result, the ability of gallidermin to form pores depends on the membrane thickness of the target cell; thus, against some target cells, it is the interaction with lipids I and II alone, not pore formation, that is responsible for inhibiting these targets. The superior activity of gallidermin over that of nisin in a cell wall biosynthesis assay would seem to explain its high killing potency even in situations where it does not form pores (25).
Another activity that has been attributed to a number of lantibiotics, including nisin, subtilin, and sublancin, is the ability to prevent spore outgrowth by Bacillus and Clostridium species (26, 27). An in-depth study into the mechanism of action in Bacillus anthracis identified that the initiation of germination is essential for the action of nisin. The action of nisin is governed by two events during the outgrowth of spores: the establishment of an active metabolism and the shedding of the external spore structures (28). In L. lactis, truncated nisin A mutants lacking rings D and E were unable to form pores in the membranes or to cause a disruption of the membrane potential but were still able to inhibit the outgrowth of Bacillus subtilis spores (29). In contrast, mutants in the hinge region of nisin (N20P/M21P and M21P/K22P), which retained the ability to bind lipid II but were unable to form pores, had antimicrobial activity against vegetative cells of B. anthracis but did not inhibit spore outgrowth. Therefore, at least in B. anthracis, pore formation is essential for limiting spore outgrowth, which suggests that nisin utilizes lipid II as the germinated spore target during outgrowth inhibition and that nisin-mediated membrane disruption is essential for inhibiting spore development into vegetative cells (30).
It should be noted that there are a number of specific antimicrobial activities associated with individual lantibiotics (6). In addition, related peptides, referred to as lanthipeptides, have functions other than those of antimicrobial agents (31). The morphogenetic peptides SapB and SapT from Streptomycetes act in such a manner (32). These peptides are believed to function as biosurfactants during the formation of aerial hyphae (33).
ROLE OF CELL WALL MODIFICATIONS IN RESISTANCE
Lipid II
In addition to many lantibiotics, a number of other antibiotics also interact with lipid II (e.g., vancomycin), albeit by binding to different parts of the target molecule. Lipid II is the precursor of cell wall biosynthesis found at the outer leaflet of the bacterial membrane. The main structural component of the bacterial cell wall consists of peptidoglycan, which is a polymer of a repeating N-acetylmuramic acid (MurNAc) and GlcNAc motif, cross-linked via peptides attached to MurNAc. The final steps in maturation are catalyzed by a penicillin-binding protein (PBP), a bifunctional enzyme which catalyzes polymerization of the sugar units (glycosyltransfer) as well as peptide cross-linking (transpeptidation), utilizing lipid II as the substrate (34). Lipid II, as the corresponding monomer, consists of a single disaccharide pentapeptide bound by a pyrophosphate linker to a polyisoprenoid anchor called undecaprenyl. Binding of antimicrobial substances to lipid II interferes with peptidoglycan biosynthesis by physically sequestering the compound and preventing its utilization by transpeptidase and transglycosylase enzymes that install the cross-linked network of the bacterial cell wall (35).
Recently, it was shown that nisin and gallidermin also bind to other intermediates of the wall teichoic acid (WTA) biosynthesis pathway, including lipid III (undecaprenol-pyrophosphate-N-acetylglucosamine) and lipid IV (undecaprenol-pyrophosphate-N-acetylglucosamine-N-acetylmannosamine), and it has also been shown that the specific interaction with WTA precursors promotes pore formation in artificial lipid bilayers (36).
Despite the importance of lipid II to the activity of many lantibiotics, quantification of the levels of lipid II in nisin-sensitive Micrococcus flavus, nisin-sensitive Listeria monocytogenes, and their nisin-resistant variants indicated that nisin resistance/sensitivity is independent of lipid II levels (37).
DltA
Bacterial membranes usually have an overall negative net charge. Bacteriocins, including the lantibiotics, resemble a considerable proportion of the antimicrobial molecules associated with human, vertebrate, invertebrate, and plant host defense systems in that they have cationic properties that confer a high affinity for the anionic bacterial cell envelope (38). The Gram-positive bacterial cell wall comprises a thick peptidoglycan fabric as well as polymers of alternating phosphate and alditol groups, called teichoic acids. Teichoic acids are classified into two groups: WTA (as mentioned previously), which are phosphodiester linked via a linkage unit to muramic acid residues of peptidoglycan (39, 40); and lipoteichoic acids (LTA), which are macroamphiphiles anchored hydrophobically through the fatty acid residues of their glycolipid component in the outer layer of the cytoplasmic membrane (41). The dlt (d-alanyl-lipoteichoic acid) operon is responsible for d-alanine esterification of both LTA and WTA (Fig. 1A) (42). The dlt operon has been characterized across many species and contains four genes required for functionality: dltABCD (43). The role of DltA is to act as a d-alanine-d-alanyl carrier protein ligase (Dcl), which activates d-alanine by hydrolysis of ATP and transfers it to the phosphopantetheine cofactor of a specified alanine carrier protein (Dcp), encoded by dltC (44). The hydrophobic DltB protein, which may have a transmembrane location, is required for d-alanine incorporation into teichoic acids, possibly through the transfer of activated d-alanine across the cytoplasmic membrane (42). It is thought that DltD transfers d-alanine from the membrane carrier to teichoic acids, as indicated by the apparent presence of an N-terminal signal peptide. The dltABCD gene sequence and organization are highly conserved across a range of species (45). In Streptococcus agalactiae, two regulatory genes of the dlt operon, designated dltR and dltS, are located upstream of dltA (46). In Clostridium difficile, a putative regulatory protein (CD2850) is thought to negatively regulate the dlt operon (47).
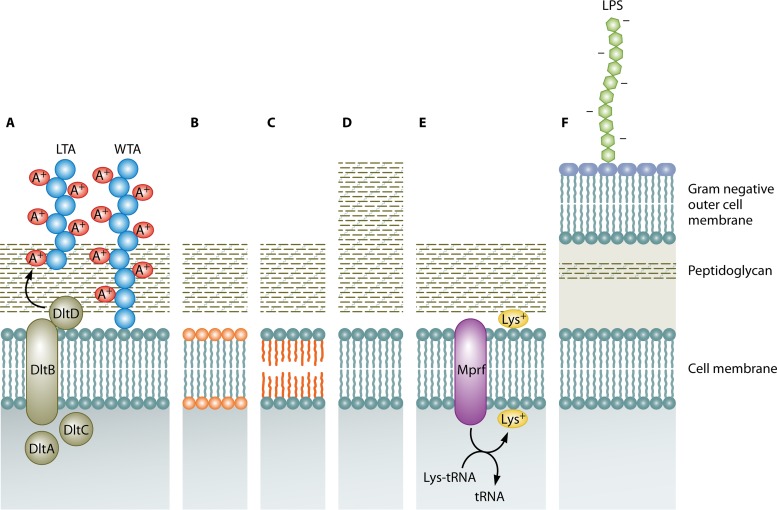
FIG 1.
Mechanisms of lantibiotic resistance which relate to the cell wall and membrane. (A) d-Alanylation of lipoteichoic acids (LTA) and wall teichoic acids (WTA) by the dltABCD operon, which confers a positive charge. (B) Changes in phospholipid composition. (C) Changes in membrane fatty acid composition. (D) Cell wall thickening. (E) Lysine esterification of one of the two hydroxyl groups of phosphatidylglycerol (PG) by Mprf. (F) Gram-negative outer cell membrane containing lipopolysaccharide (LPS).
As a result of the d-alanylation of teichoic acids, positive charges are incorporated into the cell wall. Hence, cationic antimicrobial peptides (CAMPs), such as nisin and gallidermin, are repelled from the cell envelope of target microorganisms, such as L. monocytogenes, Bacillus cereus, Clostridium difficile, Streptococcus pneumoniae, and Staphylococcus aureus (45, 47–50). This is a form of innate lantibiotic resistance and becomes particularly evident in the presence of antimicrobial peptides that trigger a signaling pathway that upregulates the process (45). This phenomenon was first reported by Peschel et al. (45), who determined that a dltA mutant of S. aureus and dltA, dltB, and dltD mutants of Staphylococcus xylosus did not produce d-alanine esters in their teichoic acids and that these conditions increased their sensitivity to CAMPs, including the lantibiotic gallidermin. Additionally, this study showed that wild-type strains of Staphylococcus bearing additional copies of the dlt operon contained larger amounts of d-alanylated teichoic acids and hence repelled cationic proteins more effectively and were less sensitive to gallidermin (45). It was also notable that spontaneously nisin-resistant mutants of L. lactis IL1403 expressed the dlt and gal operons at higher levels (51). The relevance of the gal operon, associated with the Leloir pathway (52), is that galE encodes a UDP-glucose 4-epimerase that is responsible for the synthesis of α-galactose, which is transported across the membrane to become a substituent of LTA. This observation led to further studies of L. lactis MG1363, which established that L. lactis MG1363 ΔgalAMK was twice as sensitive to nisin as the wild type, suggesting that α-galactose incorporation has an effect on LTA structure and thus is important in nisin resistance. In addition, the LTA of the resistant strain contained twice as much α-galactose as the wild type, which may indicate a more densely packed LTA, ultimately making the cell wall barrier less negatively charged due to the action of DltA (51). Nisin-resistant cells also appeared to have more lipoteichoic acid than nisin-sensitive cells. In addition, deesterified lipoteichoic acids from nisin-resistant cells migrated more slowly through a polyacrylamide gel than those from nisin-sensitive cells. These results indicated that lipoteichoic acids could be modified to increase the resistance of Streptococcus bovis to nisin (53). LTA alanylation also seems to affect the susceptibility of Streptococcus pneumoniae to nisin (49).
Abachin et al. (48) discovered that in the absence of dltA, L. monocytogenes was severely impaired in a mouse infection model (4-log increase in 50% lethal dose [LD50]), and also that, in vitro, the adherence of the mutant to various cell lines was impaired, although the amounts of surface proteins associated with virulence (ActA, InlA, and InlB) remained unaffected. These results show that the d-alanylation of LTA contributes to the virulence of the intracellular pathogen L. monocytogenes (48). However, to date, the opposite case, where increased levels of DltA contribute to virulence, has not been identified. Thus, whether such a mechanism enhances virulence due to sensing increased levels of DltA as well as enhancing the lantibiotic resistance of a strain has yet to be determined.
Penicillin-Binding Proteins
Multimodular penicillin-binding proteins (PBPs) are responsible for peptidoglycan assembly. They are subcategorized as class A or B PBPs, depending on the structure and catalytic activity of the N-terminal module. The C-terminal ends of the penicillin-binding protein modules of both classes catalyze peptide cross-linking between two adjacent glycan chains through their transpeptidase activity (54). The N-terminal module of PBPs belonging to class A is responsible for the glycosyltransferase activity leading to glycan chain elongation (55), whereas in class B PBPs, this domain is responsible for interactions with other proteins during septation and also regulates the shape of bacterial cells (56).
There have been a number of instances in which a relationship between lantibiotic resistance and PBPs has been noted. First, the expression level of a putative PBP was found to be increased significantly in a spontaneously nisin-resistant strain of L. monocytogenes compared to the wild-type, nonresistant strain (57). This strain was also slightly more resistant to mersacidin but was sensitive to a variety of beta-lactam antibiotics. Similarly, expression of the gene encoding PBP2A (a class B PBP) was higher in a nisin-resistant mutant of L. lactis IL1403 than in the corresponding wild-type strain, leading to speculation that resistance was provided by a thicker and more densely packed cell wall (51).
The class A penicillin-binding protein PBP4 (encoded by lmo2229 [also known as pbp2229]) of L. monocytogenes has been implicated in nisin resistance, as disruption of the encoding gene enhanced nisin sensitivity, with a 1.66-fold decrease in the MIC (58). It was also observed that increased expression of LiaS, a histidine kinase element which regulates PBP2229 (PBP4), resulted in increased levels of PBP and nisin resistance in Listeria (59). Gravesen et al. suggested that PBP2229 mediates enhanced nisin resistance by shielding lipid II and, in addition, reduces the extracellular lipid II concentration (59). This hypothesis is in accordance with previous studies suggesting that nisin sensitivity is affected by the accessibility of lipid II (12, 57). Curiously, mutation of another histidine kinase element, LisK, resulted in enhanced nisin resistance despite reducing expression of both pbp2229 and liaS (60).
IrpT/RmlD
Another novel gene involved in nisin resistance is irpT, whose disruption leads to an increased level of resistance. While IrpT is not directly involved in nisin resistance, it has a role in gene regulation. In the absence of IrpT, the rmlD gene and four others are upregulated in Lactococcus lactis subsp. lactis N8 (61). RmlD is involved in the synthesis of dTDP-l-rhamnose, which is a precursor involved in cell wall polysaccharide backbone production (62). The importance of RmlD was confirmed when it was established that overexpression of the corresponding gene in L. lactis MG1363 enhanced nisin resistance (61).
Resistance of Spores to Lantibiotics
Bacterial spores are extremely resistant to biocides, thanks in particular to their thickly layered proteinaceous spore coat (for a review, see reference 63). Although they do not kill spores, lantibiotics such as nisin can be sporostatic, preventing spore outgrowth. An investigation of nisin-resistant mutants of Clostridium botulinum created through continuous exposure to the lantibiotic resulted in resistant mutants (both spores and vegetative cells) which had the ability to germinate in levels of nisin that reduced the parental strain by 7 to 8 log (64). Although the mechanism by which the action of nisin is circumvented in these resistant mutants is unknown, it appears that it is not a nisin-specific phenomenon, as resistance was observed to a variety of bacteriocins across a range of classes.
CELL MEMBRANE MODIFICATIONS
Lipopolysaccharide
Lantibiotics are generally produced by Gram-positive bacteria and are most active against other Gram-positive strains. Gram-negative target microorganisms are usually very resistant to lantibiotics, as the architecture of their outer membrane prevents the penetration of the lantibiotics (with sizes of approximately 1,800 to 4,600 Da) to the cytoplasmic membrane (Fig. 1F) (65–67). The outer layer of the outer membrane serves as the site of attachment for lipid A, an anionic dimer of glucosamine linked to fatty acid chains and flanked by polar phosphate groups. Lipid A is covalently bound to a core polysaccharide and a specific O chain consisting of repeating oligosaccharide units. This complex is referred to as lipopolysaccharide (LPS), unless it lacks the O chain, in which case it is named lipooligosaccharide (LOS) (68). Chelating agents such as EDTA, as well as the application of sublethal stresses, such as heating or freezing, can disrupt the LPS barrier, leading to an increased sensitivity of enterobacteria to lantibiotics (66, 69, 70). Even without such treatments, some lantibiotics possess limited activity against Gram-negative targets. Microbisporicin, which selectively blocks peptidoglycan biosynthesis, causing cytoplasmic UDP-linked precursor accumulation, has some activity against Gram-negative species (such as Escherichia coli, Moraxella catarrhalis, Neisseria spp., and Haemophilus influenzae), which is unseen for other lantibiotics (71). However, the mechanistic basis for this enhanced activity has yet to be elucidated. Some bioengineered nisin mutants, altered at serine residue 29, are also exceptional by virtue of their enhanced activity against Gram-negative organisms, such as Cronobacter sakazakii, E. coli, and Salmonella enterica serovar Typhimurium (72).
Lipid Composition
The lipid composition of the bacterial cell membrane also has impacts on the levels of resistance. Membranes contain phospholipids whose composition and relative abundance vary profoundly, both between species and under various environmental conditions and growth phases (73, 74). The most common bacterial phospholipids are phosphatidylglycerol (PG) and diphosphatidylglycerol (cardiolipin), whose head groups are negatively charged (75). Others, such as phosphatidylethanolamine (PE) (a zwitterion), are found in Enterobacteriaceae (76) and bacilli (77) but not in Staphylococcus (78) or Listeria (79) species. In a nisin-resistant variant of L. monocytogenes, Scott A, increased levels of PG over cardiolipin were identified as the mechanism of resistance (80). There are corroborating reports that nisin has a greater propensity to penetrate membranes composed of cardiolipin over PG, PE, phosphatidylcholine (PC), monogalactisyldiacylglycerol (MGDG), and digalactisyldiacylglycerol (DGDG) (81). Decreased levels of the anionic phospholipids PG and cardiolipin and increased levels of the neutrally charged phospholipid PE were also associated with nisin resistance in L. monocytogenes ATCC 700302 (82). Such a phospholipid composition results in a decrease in membrane net negative charge and hence potentially diminishes the ability of nisin to interact with the membrane (Fig. 1B). Nisin-resistant cells have been reported to have decreased levels of phospholipids and to have a higher ratio of straight-chain/branched-chain fatty acids than that of parental cells (Fig. 1C) (83). Reduced membrane fluidity has also been associated with nisin resistance, with increased long-chain fatty acids and reduced ratios of C15/C17 fatty acids contributing to a more rigid membrane conformation (84–86).
MprF
Lysine esterification of one of the two hydroxyl groups of PG results in the free amino groups imparting a net positive charge on PG, creating lysyl-phosphatidylglycerol (L-PG) (73, 87). The presence of L-PG and the process of lysinylation have been described for several bacterial pathogens, including S. aureus (78, 87), L. monocytogenes (79, 88), and Mycobacterium tuberculosis (89), as well as some soil organisms, such as Bacillus species (90, 91). This lysinylation process is performed by MprF (multiple peptide resistance factor), which catalyzes the transfer of lysine residues from lysyl-tRNAs to PG (92–94) and which appears to have a major role in resistance to antimicrobial peptides (Fig. 1E). Identification of the protein responsible led to the corresponding gene being detected in a considerable number of Gram-positive and Gram-negative genomes (38). In fact, sequence comparisons with known PG synthase sequences allowed the identification of mprF homologues in 347 microorganisms, including 31 genera of Gram-positive bacteria (mostly Firmicutes and Actinobacteria), 59 genera of Gram-negative bacteria (mostly Proteobacteria), and 3 species of Archaea, within the genus Methanosarcina (95).
MprF proteins are integral membrane proteins with two separable functional domains: a well-conserved hydrophilic cytoplasmic domain at the C terminus that synthesizes L-PG and a large hydrophobic domain at the N terminus that facilitates the flipping of L-PG (96). Both domains of MprF are necessary for the resistance phenotype. The C-terminal domain and 6 of the 14 proposed transmembrane segments of MprF are sufficient for the full synthesis of L-PG, but they do not lead to an efficient resistance phenotype, since most of the L-PG remains in the inner layer of the cytoplasmic membrane. However, full resistance is achieved when the N-terminal domain is coexpressed with the L-PG synthase domain and the L-PG is translocated to the outer layer of the membrane (96). L-PG-deficient mutants exhibit increased susceptibilities to many cationic peptides. This was first noted when a mutant of S. aureus devoid of L-PG was found to be more susceptible to gallidermin and nisin (87). Comparable effects due to a loss of L-PG synthesis were observed for L. monocytogenes, with the corresponding deletion mutant being significantly more susceptible to gallidermin and the α-defensins HNP-1 and HNP-2 (88).
Some Gram-positive bacteria, such as Clostridium perfringens SM101, possess two mprF-homologous genes (mprF1 and mprF2). While mprF2 encodes a lysyl-phosphatidylglycerol synthase (L-PGS), mprF1 encodes an alanyl phosphatidylglycerol synthase (A-PGS). The formation of alanyl phosphatidylglycerol (A-PG) and L-PG was shown to be tRNA dependent, using Ala-tRNAs and Lys-tRNAs, respectively (94, 97). In Enterococcus faecalis, mprF1 and mprF2 homologues have been identified, although only MprF2 is involved in aminoacylation and formation of L-PG, A-PG, and R-PG (98), a task also carried out by a single MprF protein found in Enterococcus faecium (99). Aminoacylation of PG with glycine and ornithine has also been described (93, 100). By producing the MprF A-PGS protein of C. perfringens in an S. aureus mprF deletion mutant, it was established that the production of A-PG rather than L-PG did not affect susceptibility to antimicrobials, such as nisin and gallidermin, or to the antibiotic daptomycin (101). This indicates that the zwitterion A-PG is just as effective as the cationic L-PG phospholipid in protecting the cell and maintaining resistance. A-PG contributes up to 6% of the overall lipid content of Pseudomonas aeruginosa. A-PG deficiency (due to deletion of an mprf homologue) leads to increased susceptibility to β-lactam antibiotics (ampicillin, oxacillin, and cefsulodin), the lipopeptide antibiotic daptomycin, and other antimicrobials (protamine sulfate, poly-l-lysine, and polymyxin E), as well as chromium ions (102, 103), indicating that MprF contributes to reducing the antimicrobial susceptibility of both Gram-positive and Gram-negative microorganisms.
A similar protective mechanism has been identified in M. tuberculosis. In this bacterium, the lysX gene, encoding the two-domain lysyl-transferase (MprF)–lysyl-tRNA synthetase (LysU) protein, is responsible for L-PG production. A lysX deletion mutant shows sensitivity to cationic antibiotics and antimicrobial peptides. This mutant also shows altered membrane potential compared to the wild-type strain and an increased association with lysosome-associated membrane protein-positive vesicles. This indicates that the lysX mutant strain is not as adept at preventing fusion of phagosomes with lysosomes, a process which is partly required to allow intramacrophage replication (89). It is interesting that the expression of the mprF fragment of lysX alone does not lead to the production of L-PG. This is notable because two lysU genes are expressed by M. tuberculosis, one as a cytosolic essential protein and the other as a domain of LysX. It therefore appears that unlike the case in other bacteria, cytosolic LysU and membrane-bound MprF do not cooperate to produce L-PG (89).
In conclusion, the presence of these aminoacylated PGs provides a way for bacteria to shield themselves from the action of lantibiotics and the action of CAMPs in general. The extent of this protection can be considerable, ranging from a 1.5-fold increase in the amount of gallidermin required to inhibit L. monocytogenes (88) to 7- and 28-fold increases in resistance to gallidermin and nisin, respectively, in S. aureus (87).
fab Operon
In a nisin-resistant L. lactis strain, the fabDG1G2Z1Z2 operon is expressed to a lesser extent than in the sensitive wild type (51). This operon is involved in membrane synthesis via saturation and elongation of phospholipids (104). The decreased expression of the fab operon might lead to a reduced amount of saturated fatty acids and fewer elongated fatty acids in the membrane, making it less densely packed. Such alterations in cytoplasmic membrane composition might influence the ability of nisin to interact with the membrane and thus to increase resistance. However, further investigations are required to definitively establish the contribution of this operon to providing protection against lantibiotics.
TWO-COMPONENT SYSTEMS ASSOCIATED WITH LANTIBIOTIC RESISTANCE
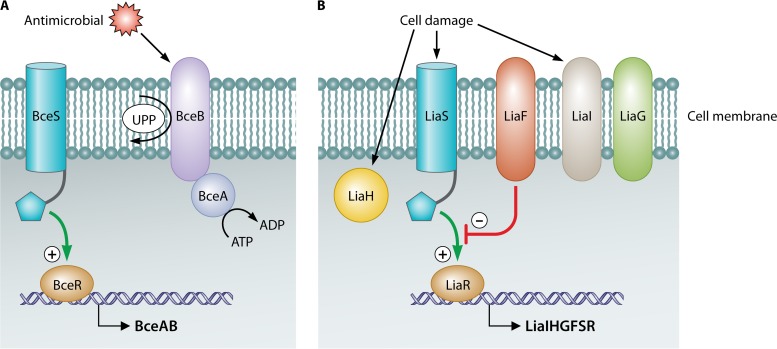
Two-component systems (TCSs) are thought to function as monitors that allow the cell to sense and respond to specific environmental conditions. They contain a sensor that encodes a sensory histidine kinase (HK) and a regulator that encodes a cognate response regulator. In addition, an intramembrane-sensing HK (IM-HK) lacks an extracytoplasmic domain, indicative of a sensing process at or within the membrane. Two major groups are found in Firmicutes, linked to maintaining cell envelope integrity, to mediating antibiotic resistance, or to detoxification processes, and can be differentiated based on sequence similarity and genomic context: (i) BceS-like IM-HK that are functionally and genetically linked to ABC transporters and (ii) LiaS-like IM-HK as part of three-component systems (Fig. 2).
FIG 2.
Two main types of two-component systems (TCSs) are responsible for lantibiotic resistance: the Bce-like TCSs (A) and the Lia-like TCSs (B). The presence of antimicrobials, such as lantibiotics, or the cell damage incurred as a result of their presence causes these TCSs to mediate the transcription of genes whose products confer a resistance phenotype.
BceRS-Like Two-Component Systems
BceRS of B. subtilis.
BceS-like IM-HK possess two transmembrane helices but lack any extracellular sensory domains. They are functionally and genetically linked to ABC transporters named BceAB (105, 106). They are found almost exclusively in Firmicutes, with 80% of the transporters associated with a BceRS-like TCS (105). It appears that the presence of antimicrobial peptides cannot be detected by these histidine kinases alone but rather that this resistance module relies on the transporters for stimulus recognition, i.e., in the presence of an antimicrobial peptide, the transporter somehow communicates with the sensor kinase. It is believed that this involves direct contact between the transporter and the histidine kinase (105, 107). However, the mechanism by which signaling occurs is as yet unknown. This communication leads to activation of the cognate response regulator and, subsequently, an induction of transporter gene expression. Importantly, ATP hydrolysis by the transporter, and therefore active transport, is required for the signaling process (Fig. 2) (108, 109).
Experimental evidence from a number of homologous systems, from B. subtilis, Staphylococcus aureus, Streptococcus mutans, Enterococcus faecalis, and Lactobacillus casei, confirms that this signaling pathway is a general characteristic of the Bce-type modules, and on a number of occasions, this pathway has been associated with lantibiotic resistance (107–113). For example, in L. lactis, a homologue named LlrG/KinG is found encoded to either side of the gene for a VanZ-like protein. VanZ is found in E. faecium and confers resistance to teicoplanin (114). Downstream from llrG/kinG is an ABC transporter gene, ysaBC. The expression of this transporter is induced by nisin and consequently confers resistance to this lantibiotic (51).
The BceAB-type transporters are well characterized and have been classified within the peptide 7 exporter (Pep7E) family in the Transport Classification Database (TCDB) (115). These transporters are composed of an ATPase (BceA) and a permease (BceB) component with 10 transmembrane helices and a characteristic, large, extracellular domain of approximately 200 amino acids between helices VII and VIII. These transporters contribute to B. subtilis bacitracin and lantibiotic resistance, with the observed MIC of bacitracin for the bceAB mutant reduced ∼30-fold and resistance to actagardine and mersacidin reduced ∼2- to 4-fold (109, 110, 116, 117). Through random mutagenesis, the specific functions of the transporter were found to be associated primarily with the C-terminal region of the permease, BceB, particularly in the eighth transmembrane helix. Furthermore, although signaling and resistance are functionally interconnected, several identified mutations had stronger effects over one than the other (118). The cooccurrence and coevolution of Pep7E transporters and the BceRS-like TCS led to the hypothesis that a functional link always exists between these systems, in which they cooperate in both signaling and detoxification (105). The exact means by which BceAB detoxification occurs has not been fully elucidated. A recent study on bacitracin resistance, however, revealed that rather than acting as an efflux pump for the antimicrobial peptide, BceAB may function by transfer of the bacitracin target undecaprenyl pyrophosphate (UPP) from the external to the internal leaflet of the inner membrane, where it is no longer susceptible to binding by bacitracin or, indeed, lantibiotics that utilize the same lipid II target (Fig. 2) (119).
In addition to BceRS-AB, there are two further paralogous peptide-sensing and detoxification (ABC transporter) modules encoded within the B. subtilis genome, i.e., YxdJK-LM and PsdRS-AB (formerly YvcPQ-RS). The YxdJK-LM system responds to the human antimicrobial peptide LL-37 (120), and the PsdRS-AB system responds primarily to lipid II-binding lantibiotics, such as nisin, gallidermin, subtilin, and actagardine (strongest inducer), as well as the lipid II-binding lipopeptide enduracidin. Other lantibiotics, including Pep5, mersacidin, sublancin, and duramycin, as well as the lipid II-binding antibiotics vancomycin and ramoplanin, are not inducers (110). On the basis of these findings and the different lipid II-associated motifs that these peptides target, it seems that PsdRS-induced expression is linked to antimicrobials with an N-terminal lipid II-binding motif, referred to as a pyrophosphate cage. A range of TCSs were characterized for L. casei BL23, and insertional mutations identified that among these, TC09 and TC12, which are homologous to the three paralogous TCSs of B. subtilis, BceRS, PsdRS, and YxdJK, are responsible for nisin resistance. Of the remaining mutated TCSs identified, some showed more sensitivity to nisin and one (T04) showed more resistance (121).
BraRS of S. aureus.
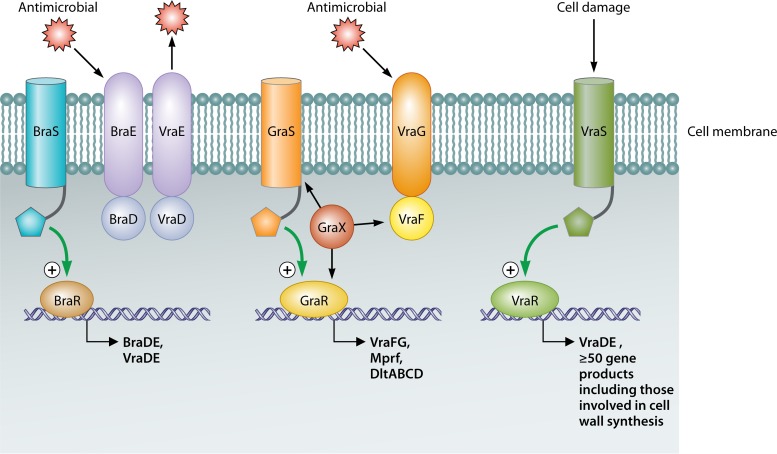
A BceRS-like (IM-HK) TCS associated with S. aureus, BraRS, has been shown to be essential for resistance to bacitracin, nisin, and nukacin ISK-1 (108, 122–124). As a consequence of having been identified in three separate studies, this TCS is also referred to as BceRS (123) and NsaRS (125). The name BraRS (bacitracin resistance associated) is representative of its characteristics and thus has been chosen for further referral (108). Through use of a lacZ reporter-gene fusion, it was demonstrated that braRS expression is upregulated by a variety of cell-envelope-damaging antibiotics, including phosphomycin, ampicillin, nisin, gramicidin, carbonyl cyanide m-chlorophenylhydrazone, and penicillin G (122). Immediately downstream of braRS are the genes which encode the ABC transporter BraDE (an example of the BceAB-type transporters discussed above), which is also of key importance to bacitracin and nisin resistance through sensing and signaling through BraRS. BraRS also activates transcription of the detoxification ABC transporter encoded by vraDE (108) (Fig. 3). Highly conserved imperfect palindromic sequences, essential for transcriptional activation by BraRS, have been identified upstream from the braDE and vraDE transcription start sites (108). Microarray analysis revealed that the transcription of 245 genes is altered in a ΔbraS mutant (with the majority being downregulated). These 245 genes include several that are involved in transport, drug resistance, cell envelope synthesis, transcriptional regulation, amino acid metabolism, and virulence (126). Thus, BraRS has an important resistance role in S. aureus and functions to reprogram gene expression to modify cell envelope architecture, facilitating adaptation and survival.
FIG 3.
In S. aureus, three main TCSs are responsible for lantibiotic/antimicrobial resistance. These include two Bce-like TCSs, BraRS and GraRS, and an Lia-like TCS, VraSR. A coordinated resistance effort results from the actions of these TCSs, causing upregulation of genes whose products alter the composition of the cell wall and membrane and also of genes encoding ABC transporters which expel antimicrobials from the cell.
The extent of the contribution of BraRS to antimicrobial resistance relative to those of other TCSs has been assessed through the creation and analysis of a number of mutants. When an S. aureus ΔbraRS mutant was cocultured with a nukacin ISK-1 or nisin A producer, a significant decrease in numbers was observed, which was not apparent for the parental S. aureus strain. In contrast, ΔvraSR and ΔgraRS (GraRS is yet another TCS) mutants were more moderately affected, thus establishing the greater importance of BraRS and its associated transporters (124). Interestingly, through screening of a bank of nisin-resistant mutants, a single mutation was identified in braS (A20E) that conferred an 8- to 16-fold increase in nisin resistance (125). It is thus apparent that changes to BraRS can provide a means via which S. aureus can attain enhanced resistance to lantibiotics.
GraRS of S. aureus.
Overexpression of a second IM-HK identified in S. aureus and its associated response regulator, named GraRS, has been linked with the vancomycin-intermediate S. aureus (VISA) phenotype. This TCS is also referred to as vraCA (127) and closely resembles bceRS of B. subtilis (128). GraRS induces expression of the adjacently located genes vraFG, which encode an ABC transporter (129) known to be overexpressed in strains exhibiting increased vancomycin resistance (130). This transporter was revealed to be involved in antimicrobial sensing and signaling through GraRS (107). A recent study identified an upstream gene whose product, GraX, appears to be a cytosolic accessory protein which acts by signaling through the GraS kinase and also effectively interacts with VraF and VraR. GraXRS-VraFG has been defined as a five-component system involved in cationic antimicrobial peptide sensing and signal transduction to promote resistance to these peptides in S. aureus (Fig. 3) (107). It has also been suggested that GraRS may regulate more than just vraFG. Analysis of graR and graS mutants highlighted alterations in the overall surface charge, with the charge becoming more negative than that of wild-type cells (129). It has also been discovered that these cell surface changes are due to the regulation of dlt and mprF by GraRS (127, 131). GraRS is also thought to contribute to tolerance of high temperatures, survival upon exposure to oxidative stress, and regulation of pathogenicity (132).
With respect to lantibiotic resistance/sensitivity, it has been noted that a graRS-negative mutant exhibits a greater susceptibility to nukacin ISK-1 than to nisin A, while no susceptibility to bacitracin has been observed (124). A previous study found that a graR mutant had increased susceptibility to vancomycin and polymyxin B. Interestingly, single or multiple amino acid changes within GraR, including an S197N mutation, have been associated with enhanced resistance (128, 129). Expression of graR transcripts was not found to increase upon exposure of the cell to nukacin ISK-1 or nisin (124). This supports the model proposed in the same study, which details the coordinated effort achieved by GraRS and BraRS upon exposure of S. aureus to a relatively high level of nisin A, nukacin ISK-1, or such cationic antimicrobials. That is, GraRS is seen as important for resistance until a significant induction of VraDE expression by BraRS occurs. In conclusion, S. aureus possesses three distinct class I bacteriocin resistance systems, implemented through the independent action of VraSR (an LiaRS-like TCS) and the coordinated function of the two TCSs mentioned above (Fig. 3) (124). Staphylococcus epidermidis also harbors a GraRS equivalent, named ApsRS, that has been found to be induced by nisin and other antimicrobials (133).
AnrAB transporter of L. monocytogenes.
AnrAB was named on the basis of its role, i.e., it is an ABC transporter involved in nisin resistance. The permease component of the transporter was discovered as a result of screening of a mariner transposon bank of L. monocytogenes EGD-e mutants for nisin sensitivity and was found to be a BceAB-like transporter. Examination of a subsequently generated ΔanrB deletion mutant also revealed associated increased sensitivities to the lantibiotic gallidermin, to bacitracin, and to a large number of β-lactam antibiotics (134). Based on these results, it was proposed that AnrAB is a multidrug resistance (MDR) transporter that contributes to the innate antimicrobial resistance of L. monocytogenes. The TCS VirRS and RpoN (σ54) are responsible for regulation of anrAB (135, 136). It has been proposed that Lmo1746-Lmo1747 (a BceAB-like ABC transporter encoded downstream from VirR), along with AnrAB and VirRS, may form an antimicrobial sensing and detoxification module similar to the VraDE-BraRS-BraDE network in S. aureus (137).
Interestingly, AnrAB, which is a BceAB-like transporter, is most similar to an ABC transporter in Streptococcus pneumoniae D39 (Sp0912-Sp0913) which was found to contribute to the innate resistance of this strain to nisin, bacitracin, gramicidin (all cell envelope-active antimicrobials), and lincomycin (a protein synthesis inhibitor). Furthermore, expression of sp0912 was induced by up to 13-fold upon treatment with nisin or bacitracin (138) and, more recently, was found to be regulated by a TCS designated Rr01-Hk01 (105).
LiaRS and LiaRS-Like Two-Component Systems
Most members of the phylum Firmicutes harbor a TCS, generally referred to as LiaRS (formerly known as YvqEC), which is involved in the response to cell envelope stress elicited by inhibitors of the lipid II cycle (Fig. 2B). Upon the addition of vancomycin or bacitracin, LiaRS autoregulates the genes liaIHGFSR, which are organized across two operons, liaIH and lia(G)FSR. Across the Firmicutes, liaG is present only in Bacillus spp. closely related to B. subtilis (139). Systematic deletion of the operon revealed the functions of each gene product. LiaF is a potent negative regulator of LiaR-dependent gene expression. As the sequence and genomic location of the liaF gene are conserved in the Firmicutes, it seems that LiaFRS is a common three-component system, with both positive- and negative-feedback loops that sense and respond to cell envelope stress signals. LiaH, a homologue of E. coli phage shock protein A, responds through damage sensing. In silico analysis could not predict the role of liaI or liaG, though the fact that liaI harbors two transmembrane domains and liaG harbors one transmembrane domain is indicative of membrane localization. However, LiaI seems to be involved in sensing and counteracting membrane damage (139, 140). In addition to characterization of the genes within the operon, the promoter of the lia operon and the cis-acting sequences necessary for antibiotic-inducible gene expression were also identified (141). This promoter responded strongly to a subset of cell wall-active antibiotics that interfere with the lipid II cycle, such as bacitracin, ramoplanin, vancomycin, and nisin (141). A detailed analysis strongly suggests that the stimulus sensed by the LiaRS system is some aspect of interference with cell wall biosynthesis by such antimicrobials, rather than the system directly sensing their presence. In that instance, LiaRS acts as a damage-sensing and signal-transducing system and thus contrasts with BceRS-AB, which is thought to function through direct drug sensing and signal transduction (142).
The role of LiaRS in L. monocytogenes has been investigated (59, 143, 144). LiaRS-mediated signal transduction is also negatively regulated by LiaF, in a fashion similar to that described for B. subtilis (143). LiaFRS in B. subtilis is seen to regulate expression of itself and liaIH (139), and as seen in L. monocytogenes sv1/2a EGD, this occurs in response to exposure to the cell wall-active antibiotics vancomycin and bacitracin. In L. monocytogenes, however, the expression of 27 genes (143), including a PBP gene (57) and telA, which encodes a protein involved in toxic ion resistance (145), is also regulated. Collins et al. (144) reported that a ΔliaS strain grew more successfully than the corresponding wild type in the presence of nisin. A mutant lacking another histidine kinase, the ΔlisK mutant, which also has a nisin resistance phenotype, was previously reported to alter liaS and PBP expression. With this in mind, the nisin resistances of 3 mutants, the ΔliaS, ΔliaS ΔlisK, and ΔlisK mutants, were compared, and it was discovered that the ΔlisK mutant was 2-fold more resistant than the wild type and that the ΔliaS and ΔliaS ΔlisK mutants were another 2-fold more resistant. On the basis of the nisin resistance phenotypes of these mutants, it was postulated that these phenotypes were attributable to the increased production of the corresponding PBP, whose expression was also increased in the ΔliaS mutant (144). In contrast, mutation of the corresponding response regulator (ΔliaR mutation) in seven wild-type strains of L. monocytogenes resulted in an average decrease in cell density of 5.2 ± 0.7 log10 CFU ml−1 after 24 h for the 7 ΔliaR strains compared to the wild type on exposure to nisin (146). Furthermore, this study established that lmo1746, a gene downstream from virR (encoding a response regulator) that encodes an ABC transporter permease, contributed to LiaR-mediated nisin resistance (146). A group B Streptococcus ΔliaR mutant has also been found to be more susceptible to cell wall-active antibiotics (vancomycin and bacitracin) and to antimicrobial peptides (polymyxin B, colistin, and nisin) than the parent strain. In the absence of LiaR, this strain also became avirulent, no longer causing sepsis and pneumonia in mouse models (147). The different consequences of mutating LiaS and LiaR most likely reflect the inactivation of different members of the three-component system, with deletion of LiaS potentially resulting in the cross-activation of LiaR by other histidine kinases.
The LiaFSR system of S. mutans has also been characterized and is known to regulate genes encoding membrane- and cell wall-associated proteases and chaperones, as well as genes encoding proteins involved in cell envelope biogenesis and remodeling (148). LiaRS has also been identified as upregulating dltA expression in Streptococcus gordonii (149). CesSR (also referred to as TCS-D) (141, 150) is an LiaRS homologue found in L. lactis. Upstream of cesSR is llmg1650, an liaF homologue. An L. lactis ΔcesR mutant was found to be approximately 2-fold more sensitive to nisin, plantaricin C, and bacitracin than the corresponding parent strain. It has also been established that the bacteriocins lactococcin 972 and plantaricin C, as well as bacitracin and vancomycin, act as inducers of the llmg1650 promoter, in a concentration-dependent manner, but that nisin does not induce this promoter (151).
VraSR of S. aureus.
To date, four two-component systems involved in both cell wall synthesis and drug resistance have been identified in S. aureus: WalKR, BraRS, GraRS, and VraSR. While WalKR is more specifically involved in cell wall metabolism and its regulation, the other three TCSs contribute significantly to antimicrobial resistance (106, 152). The vraSR genes are homologous to liaRS of B. subtilis. VraSR, a vancomycin resistance-associated sensor/regulator, contributes to resistance to cell wall-targeting agents, such as lantibiotics, and has been shown to protect against cell damage by modulating components of the peptidoglycan biosynthesis pathway. Through microarray analysis of S. aureus strain N315 and its vraSR null mutant, Kuroda et al. discovered that of 139 transcripts induced by exposure of the parent strain to vancomycin, 46 were no longer transcribed in the mutant strain (153). Among the genes regulated by VraSR are those encoding SgtB (a monofunctional glycosyltransferase involved in the polymerization of peptidoglycan), PBP2 (penicillin-binding protein 2; also involved in peptidoglycan polymerization), and MurZ (UDP-N-acetylglucosamine enolpyruvyl transferase; required for murein monomer precursor synthesis), all involved in cell wall biosynthesis. A number of genes previously associated with β-lactam as well as glycopeptide resistance in S. aureus have also been identified to be under the control of VraSR. Among them are the vraDE genes, which encode an ABC transporter that is linked with bacitracin and nisin resistance. This transporter is also regulated by another S. aureus TCS, BraRS (108). It has been demonstrated that overproduction of PBP2 significantly increases resistance to the lipid II cycle-disrupting antibiotic teicoplanin (154). Therefore, the significant reduction in teicoplanin resistance observed in vraSR null mutants agrees well with the loss of PBP2 induction. Kuroda et al. also revealed that overexpression of vraSR via a high-copy-number plasmid alone did not increase transcription of pbp2 and sgtB (153). Induction thus requires exposure to cell wall synthesis inhibitors in the presence of VraSR. In other words, resistance is dependent on attack in the form of inhibition of cell wall synthesis, which in turn activates the VraS sensor kinase. In addition, VraSR is independently activated upon inhibition of cell wall biosynthesis, such as that observed through the action of mersacidin, daptomycin, and vancomycin (124, 155). However, a study by Muthaiyan et al. revealed that unlike daptomcin and vancomycin in the previous study, nisin treatment did not induce vraSR expression (156). This was confirmed by a second study that demonstrated that the lantibiotic nukacin ISK-1 had such induction properties; this difference was attributed to their differing structures and modes of action (124).
In the absence of VraSR, S. aureus shows a significant increase in susceptibility to various cell wall synthesis inhibitors and antimicrobials, including nukacin ISK-1, oxacillin, cefoxitin, tunicamycin, cycloserine, daptomycin, vancomycin, and bacitracin (124, 153, 157). The vraSR genes have been found to be more highly expressed in some vancomycin-intermediate S. aureus (VISA) strains than in vancomycin-susceptible S. aureus (VSSA). In fact, VraSR has been proposed to be involved in the VISA-type resistance mechanism via a contribution to cell wall thickening which prevents antimicrobials from reaching their target molecules, as is the case for vancomycin and lipid II (Fig. 1D). In the case of the thickened cell wall, increased free amounts of false target sites are present, leading to a reduced diffusion velocity of vancomycin through the cell wall. Although mersacidin also has the same target molecule, it does not bind to the d-alanyl–d-alanine terminus. Resistance to mersacidin is not hindered in such a manner but is still observed to induce the VraSR TCS (130, 155, 158). However, a recent study by Berscheid et al. identified two mutations in VraS (L114S and D242G) which contribute to an enhanced thickening of the cell wall and lead to increased resistance to mersacidin (159).
Other Two-Component Systems
LisRK of L. monocytogenes.
LisRK is a TCS associated with a significant role in the virulence potential of L. monocytogenes. Originally identified in strain LO28, it has since been revealed that in addition to its major contribution in responding to ethanol, pH, and hydrogen peroxide stresses, LisRK is involved in the cell's tolerance to antimicrobials, including nisin and the cephalosporin family of antibiotics (160, 161). A ΔlisK mutant showed a large reduction in the expression of three genes. One is thought to encode a penicillin-binding protein that shows homology to PBP2a of Streptococcus pneumoniae and PBP1a of B. subtilis, both of which are high-molecular-weight PBPs possessing glycosyltransferase and transpeptidase domains (60). The other LisK-regulated genes encode another histidine kinase that is homologous to LiaR of B. subtilis and a protein of unknown function with homology to B. subtilis YvlB (60).
VirRS of L. monocytogenes.
In almost all cases, genes encoding two-component systems are contiguous on the chromosome. This is not the case for virR (regulator gene) and virS (HK gene), which are separated by three genes. L. monocytogenes transcriptomic analysis of both the virR::Tn917 and ΔvirS mutants revealed that VirR positively controlled 12 genes and that, while VirS also regulated these genes, it also regulated another 108 genes, in both a positive and a negative manner. The simplest explanation for this difference is that the VirS histidine kinase is able to interact with one or several other response regulators, leading to the activation/repression of the transcription of these genes (135). The putative VirR DNA-binding site, a palindromic region of 16 bases, was found upstream of transcriptional units commonly regulated by VirR and VirS. The palindrome was highly conserved between the different promoter regions and showed a common organization (135).
The VirR/VirS system was originally identified as being involved in L. monocytogenes virulence, with in vivo upregulation of VirR mediated by the major virulence regulator PrfA (162). With respect to genes known to contribute to innate nisin resistance, it is notable that dltA, mprF, and anrB have been shown to be regulated by VirRS in L. monocytogenes (134, 135). This strongly suggests that a significant role of the VirR/VirS system is to regulate resistance of L. monocytogenes to cationic peptides.
CprRK of C. difficile.
The CprK sensor kinase and an orphan response regulator, CprR, encoded elsewhere on the chromosome, are responsible for the resistance of C. difficile to cationic antimicrobial peptides, including lantibiotics such as nisin and gallidermin (163). The lantibiotics nisin, gallidermin, subtilin, mutacin 1140, and cinnamycin were found to activate the CprK-CprR system, while sublancin, cytolysin, and actagardine did not. It was proposed by Suarez et al. (163) that a combination of the (methyl)lanthionine groups along with proline and glycine residues forms a motif that interacts with CprK and leads to activation of the CprK-CprR two-component system. CprK and CprR are both expressed at low levels prior to lantibiotic exposure, but CprK expression is markedly induced upon activation, while CprR expression is not (163). The separate locations of the cprR and cprK transcripts may explain the fact that they are not coordinately regulated. The genes encoding the ABC transporter CprABC are regulated by CprK-CprR and found adjacent to cprK in the genome. Insertional disruption of one of the transporter genes resulted in significant decreases in resistance to both nisin and gallidermin (164). Hence, this TCS and ABC transporter pair contributes to the resistance of C. difficile to many lantibiotics.
NsrRS and LcrRS of S. mutans.
In S. mutans, two novel two-component systems, NsrRS and LcrRS, are associated with resistance to nisin A, lacticin 481, and nukacin ISK-1. NsrRS regulates the expression of NsrX, a protein which shares homology with several acetyltransferases, including those of the TraX family, which is associated with F pilin acetylation in E. coli. Nisin-binding studies revealed that more nisin bound to cells that had a complemented ΔnsrX mutation than to the mutant. Thus, it was determined that NsrX (or an as yet unidentified factor modified through NsrX) is involved in binding nisin A and preventing its interaction with lipid II. LcrRS regulates the expression of the ABC transporter LctFEG. Mutation of the lcrRS and lctFEG genes results in increased susceptibility to nukacin ISK-1 (and lacticin 481 with respect to the ΔlcrRS strain) (165).
OTHER RESISTANCE ELEMENTS
Biofilms
The resistance of biofilms to lantibiotics has been investigated, and it is clear that, like many other antimicrobials, lantibiotics can be effective at preventing the formation of biofilms but are less successful at killing microorganisms present in established biofilms. In one instance, the lantibiotic gallidermin and the well-studied biofilm-forming S. aureus SA113 and S. epidermidis O47 strains were the subjects of investigation (166). When gallidermin was used at MICs determined with planktonic cells, cells treated with gallidermin were completely unable to form a biofilm. In the case of S. aureus, a biofilm-inhibiting effect was evident even in the sub-MIC range. It was particularly notable that the transcription levels of a gene involved in primary adhesion (the major autolysin gene atl) and another involved in exopolysaccharide production (the intercellular adhesin gene ica) were significantly decreased in the presence of gallidermin. In contrast, when biofilm-associated staphylococci were treated with 8× MIC of gallidermin, the number of CFU was decreased 3 log in 24-h biofilms and only 1 to 2 log in 5-day biofilms (166). The activity of the two-peptide lantibiotic lacticin 3147 against S. mutans in biofilms has also been tested. At 2× MIC (6.25 μmol liter−1), lacticin 3147 reduced S. mutans biofilm formation by ∼90%. However, when 50 μmol liter−1 lacticin 3147 was tested against 1-day-old biofilms of S. mutans, only 24 to 50% reductions were apparent (167). It is worth noting that many S. mutans strains are capable of producing lantibiotics and/or other bacteriocins, known as mutacins. The production of these antimicrobials may allow these strains to compete within the dental biofilm and to persist when nutrients become limited in the dental plaque (168). Similarly, nisin has been shown to prevent biofilm formation of 25 S. aureus strains at the MICs; however, at subinhibitory concentrations, biofilm formation was not prevented (169).
A recent study compared the potentials of lantibiotics nisin A, nukacin ISK-1, and the class II bacteriocin lacticin Q to inhibit 24-h established methicillin-resistant S. aureus (MRSA) biofilms (170). Despite targeting lipid II, nukacin ISK-1 has a bacteriostatic mode of action only, inhibiting cell wall synthesis but not forming pores (171). Lacticin Q functions by forming toroidal pores in target membranes (172). From this study, it appears that pore formation leading to ATP efflux is important for bactericidal activity against biofilm cells. Of the antimicrobials tested, nisin A most closely fits this requirement, with ATP efflux observed through the action of nisin A at 0.25× MIC (0.625 μM), whereas 1× MIC (5.0 μM) lacticin Q was required. Nukacin ISK-1 had no effect on ATP efflux even at 8× MIC (96 μM) (170).
Nisin has been encapsulated within liposomes (nisin-liposomes) in order to prolong the inhibition of S. mutans strain 10449b glucan biofilms on microplates. While unencapsulated nisin lost its potency within 6 h, only 76% of the nisin within nisin-liposomes was released within this time, thus prolonging the inhibitory effects (173). Nisin has also been combined with sodium fluoride to provide a stronger bactericidal effect on S. mutans. The observed synergy revealed through checkerboard and survival assays was also more effective at preventing biofilm formation over 4 h and 16 h than the action of sodium fluoride alone (174). Nisin also functions synergistically with daptomycin and ciprofloxacin with respect to inhibiting established (24 h) MRSA biofilms. Time-kill assays using the minimum biofilm eradication concentrations of these antimicrobials demonstrated that a 3-log reduction in CFU of an MRSA control and two clinically obtained MRSA biofilms occurred within 4 h when the antimicrobials were used in combination, while the individual antimicrobials never obtained such kill levels even at 24 h postapplication (175).
The immobilization of nisin via a polyethylene glycol 1000 (PEG 1000) linker to multiwalled carbon nanotubes significantly enhanced the antimicrobial and antibiofilm properties of the nanotubes, up to 7-fold (176). Similarly, nisin has been covalently bound to stainless steel surfaces, with the same effect, reducing adhesion of bacteria and thus biofilm formation (177). Incorporating different concentrations (0.1%, 0.5%, and 1%) of nisin into polyethylene-covinyl acetate (EVA) films reduced biofilm formation on these surfaces, with the beneficial effects being more evident against a strain of S. epidermidis than against representative L. monocytogenes or S. aureus strains (178). A very recent breakthrough involves acyldepsipeptides, which have been shown to effectively assist in the killing of biofilm persister cells as a result of uncontrolled activation of a subunit (ClpP) of the protease enzyme Clp. This activation causes a dramatic increase in the size of the central pore in ClpP, allowing access of peptides and proteins to the proteolytic chamber, resulting in an increase in protein degradation and, ultimately, cell death (179).
Nisin Resistance Protein
A 35-kDa nisin resistance protein (NSR) has been found to be present in some strains of L. lactis. Originally nsr was observed as a specific nisin resistance gene located on a 60-kb plasmid in the nisin nonproducer L. lactis subsp. diacetylactis DRC3 (180). Since then, several groups have isolated nisin-resistant lactococcal strains containing nsr on a plasmid (181–183). The mechanism by which NSR confers resistance is proposed to involve the proteolytic degradation of nisin, mediated by a C-terminally conserved, tail-specific protease (TSPc) domain (184, 185). TSPc-containing enzymes efficiently cleave substrates that have apolar residues and a free alpha-carboxylate at the C terminus and have been identified and characterized in a range of species, including E. coli (186). Such proteases contain a conserved PDZ domain adjacent to the TSPc domain, which is indispensable for binding of the TSPc domain to nonpolar C termini of its peptide substrates and thus for the catalytic activity (187, 188). However, NSR does not possess a PDZ domain. NSR also differs from other TSPc-containing proteins in that it harbors charged and polar residues (Lys, His, and Ser) at the C terminus (189). More detailed analyses have revealed that NSR and purified NSRSD (a version of NSR without the predicted N-terminal signal peptide sequence) proteolytically inactivate nisin by cleaving the peptide bond between MeLan28 and Ser29. This truncated nisin (nisin1–28) shows a noticeably reduced affinity for the lactococcal membrane, a significantly reduced effectiveness in pore formation in the target membrane and a 100-fold decrease in bactericidal activity against L. lactis MG1363 compared to whole nisin (183). An nsr gene has also been identified in S. agalactiae ATCC 13813; the gene product is referred to as SaNSR. A plasmid expressing this resistance gene was shown to confer a 20-fold increase in resistance to the host strain. In silico screening for nsr-like genes revealed their presence across a range of species, including Corynebacterium casei and Corynebacterium ammoniagenes and various strains of Leuconostoc mesenteroides, Leuconostoc carnosum, Enterococcus faecium, Staphylococcus epidermidis, Streptococcus ictaluri, Streptococcus sanguinis, Streptococcus dysgalactiae, and Streptococcus agalactiae. These genes were found within operons that also contained genes encoding an ABC transporter, named NsrFP, and a TCS, NsrRK. It is proposed that these genes confer nisin resistance in these strains (190).
Nisinase
In a study dating back to 1967, antinisin activity was detected in extracts of bacilli and endospores of organisms that had a lytic mechanism for rupture of the spore coat (type L), which was not detectable in spores which ruptured the spore coat mechanically (type M). Further investigation of cell extracts of Bacillus cereus and Bacillus polymyxa revealed that the antinisin phenotype was not a result of proteolytic activity and was somewhat specific, as it did not affect polymyxin, gramicidin, or bacitracin but did inactivate nisin and subtilin (191). Nisinase was isolated from several Bacillus sp. and was shown to be a dehydropeptide reductase, as it specifically reduced the C-terminal dehydroalanyl-lysine of nisin to alanyl-lysine (192). Nisinase activity has also been associated with Lactobacillus plantarum (193), Streptococcus thermophilus (194), C. botulinum (195), L. lactis subsp. cremoris, E. faecalis, and S. aureus (196).
Gad System
In L. monocytogenes, several systems help the organism to withstand low-pH stress, but the glutamate decarboxylase (GAD) system is probably the most important (197–199). Of the five gad genes, gadD1, gadD2, and gadD3 encode decarboxylases that catalyze the conversion of glutamate to γ-aminobutyrate (GABA) and carbon dioxide (CO2). gadT1 and gadT2 encode antiporters which import glutamate and export GABA. Deletion of gadD1 impairs the ability of a strain to tolerate exposure to both sublethal and lethal levels of nisin (200). It was discovered that the intracellular ATP levels were reduced in the ΔgadD1 mutant, being only approximately 60% of those of the parent, suggesting that GadD1 contributes significantly to ATP pools and hence to tolerance of nisin (200).
arc Operon
The arc genes are involved in the breakdown of arginine via the arginine deiminase pathway. This pathway is responsible for the breakdown of arginine into ornithine, ammonium, and carbon dioxide. Arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC) are the three enzymes responsible for this degradation process (201). The arc operon is thought to contribute to acquired nisin resistance, as a 4-fold overexpression of the arcAC1C2DT2 genes was observed in a nisin-resistant L. lactis strain compared to the corresponding wild-type strain. The conversion of arginine to ammonium might result in a locally less acidic pH at the outer side of the cytoplasmic membrane, preventing nisin from attaching to the lipid II molecule (51).
TelA
The tellurite resistance gene, telA, was identified from the screening of a mariner random mutant bank for nisin-sensitive L. monocytogenes mutants. The telA gene encodes a toxic ion resistance transporter that also plays a role in resistance to cell wall-acting antibiotics. LiaR has been reported to regulate telA expression under salt-induced nisin resistance conditions (146). The telA gene also forms a two-gene operon with xpaC, which encodes a putative member of the halogen hydrol superfamily. The same genetic organization is conserved across many members of the Firmicutes and Proteobacteria. The creation of a nonpolar deletion mutant of telA confirmed its involvement in resistance, as the strain was found to be 4-fold more sensitive to nisin, and the ΔtelA strain was also 8-fold more susceptible to gallidermin and 2-fold more susceptible to bacitracin, cefuroxime, cefotaxime, and tellurite (145).
PurR
Three mutations within the purR gene (encoding a purine operon repressor) were identified in a nisin-resistant mutant of S. aureus SH1000. However, this gene does not seem to be essential for resistance, as it is intact in other cases of spontaneous nisin resistance in S. aureus (125). However, the fact that the three mutations (G228E, V229G, and E184STOP) were retrieved from independently selected nisin-resistant mutants of SH1000 strongly suggests that this locus is involved in resistance in this strain.
IreK-IreP-IreB
One of the mechanisms employed by E. faecalis to survive in the presence of therapeutic concentrations of cephalosporins may also be involved in determining resistance to lantibiotics. This means of resistance involves a signaling system comprised of a eukaryotic-like serine/threonine kinase (IreK) and phosphatase (IreP) pair that antagonistically regulates cephalosporin resistance (202). A third protein, IreB, is modulated by IreK-dependent phosphorylation, and although its role is unknown, it has been found to contribute to negative regulation of resistance and to be an endogenous substrate of both IreK and IreP (203). Homologues of these three proteins are found in the genomes of nearly all Gram-positive bacteria. A strain bearing a deletion in ireK exhibited reduced resistance to sodium cholate (a detergent found in bile) and to nisin (202, 203). In addition, a double mutant lacking ireK and ireB showed enhanced resistance to both these bactericidal agents (202, 203).
Sigma Factors
One important mediator of the stress response in many Gram-positive bacteria is the alternative sigma factor SigB. In B. subtilis, SigB activity is regulated by a complex network of protein-protein interactions governed by a variety of environmental or metabolic stresses, such as heat shock, osmotic shock, ethanol treatment, or entry into stationary growth phase (204). The growth of an L. monocytogenes EGDe sigB mutant was noticeably reduced in the presence of nisin or lacticin 3147 relative to that of its parental strain (205). It was hypothesized that SigB regulates general stress proteins or proteins involved in extrusion of antimicrobials out of the cell. Indeed, SigB-binding sites are located upstream of htrA (lmo0292), which encodes a putative molecular chaperone shown to be involved in tolerance of penicillin G (206). Another SigB-binding site is upstream of mdrL (lmo1409), encoding an antibiotic efflux pump (207).
Another alternative sigma factor, SigL, also contributes to antimicrobial resistance. Nisin survival assays showed that both SigB and SigL affect the sensitivity of L. monocytogenes 10403S to nisin in broth survival assays. In contrast to the findings of Begley et al. (205), the ΔsigB mutant was seen to be more resistant to nisin. The differences observed were attributed to the class of peptide, the strain, the initial number of bacteria, the growth phase, and the assay used for evaluation (208). Interestingly, a sigB null mutation was revealed to sensitize the cell in a ΔsigL background, leading to reduced nisin resistance (208). In addition, Butcher and Helmann proposed that the extracytoplasmic function (ECF) sigma factors σM, σX, and σW all contribute to antibiotic resistance in B. subtilis (209). A B. subtilis sigX deletion mutant has been found to be more sensitive to nisin than the wild type. Furthermore, most genes under σX control processes contribute to the biosynthesis or metabolism of the cell envelope, including the dlt operon and pbpX (penicillin-binding protein gene), among many others (210). σM was found to contribute to bacitracin resistance via regulation of the bcrC (ywoA) gene, encoding a putative bacitracin transport permease (211, 212). With respect to nisin resistance, the major contribution of σM is expression of ltaSa, encoding a stress-activated lipoteichoic acid synthase. Lipoteichoic acids (LTA) adsorb cations from the environment, including antimicrobials such as nisin. Binding of such cations to LTA may inhibit their transit through the cell wall and thereby increase resistance (213). Together, σM and σX regulate cell envelope structure to decrease access of nisin to its lipid II target. SigW has been linked with increased rigidity and decreased fluidity of the membrane (214, 215) and has been linked directly with lantibiotic resistance. The σW-dependent operons, such as the B. subtilis resistance module yqeZ-yqfAB, are seen as critical for sublancin resistance. The membrane-integrated protease encoded within this operon, YqeZ, is thought to degrade lantibiotics that integrate into the membrane (209). In terms of nisin resistance, the σW-associated SppA protein appears to be the only signal peptide peptidase in B. subtilis that makes a contribution. In addition, σW regulates expression of the phage shock proteins PspA and YvlC, which enhance membrane stability and thus provide resistance specifically against the membrane-perturbing and pore-forming activities of nisin (213). Tellurite resistance-related proteins (YceGHI) are also regulated by σW, with the protein YceG, homologous to TelA, found in L. monocytogenes (145, 213). These self-protective mechanisms also provide resistance to other lantibiotics, such as mersacidin, gallidermin, and subtilin (213).
Immune Mimicry
Lantibiotic self-immunity mechanisms are the means by which lantibiotic producers protect themselves against their own bactericidal agents. Immunity-specific proteins (LanI) or specific self-protection ABC transporters [LanFE(G)] are found alone or in combination, providing specific resistance to lantibiotic self-producers. Cross immunity is extremely rare between lantibiotic producers, and these immunity mechanisms are specific to the particular lantibiotic produced by the cell (for a review, see reference 216). An unusual means of resistance was recently revealed, in which non-lantibiotic-producing bacteria were shown to possess genes homologous to the lantibiotic immunity genes. With respect to lacticin 3147, functional immunity homologues were found in E. faecium DO and Bacillus licheniformis DSM 13. The B. licheniformis BliI protein has 37% identity with the lacticin 3147 immunity protein LtnI, and when bliI was expressed constitutively in a lacticin 3147-sensitive strain, L. lactis MG1363, the resistance of the strain was increased 20-fold (217). A similar result was seen when the genes encoding the LtnFE ABC transporter homologue from E. faecium DO, i.e., EfdFE, were also expressed in the MG1363 background. The protection provided was specific in that no resistance to the distantly related one-peptide lantibiotic nisin was apparent (217). This phenomenon appears to extend beyond lacticin 3147 resistance, as genes encoding immunity homologues have been found in Streptococcus infantarius subsp. infantarius BAA-102. These genes, spiFEG, encode an ABC transporter with >50% identity to that encoded in the nisin U operon and associated with immunity. Interestingly, genes homologous to the individual components of the nisin two-component system known as NisRK (or NsuRK in the case of nisin U), referred to as spiRR′K, are colocated with the spiFEG genes. While these regulatory genes did not confer a nisin U resistance phenotype when coexpressed with spiFEG, protection was provided when spiFEG was expressed constitutively in the nisin U-sensitive strain Lactococcus lactis subsp. cremoris HP (218).
CONCLUSIONS
The emergence of antimicrobial resistance represents a grand challenge to humanity; an understanding of how and by what means bacteria become resistant facilitates the development of new therapeutic treatments. In tandem, it allows for the reassessment of current antimicrobials and the potential to improve current therapeutic regimens. With low levels of naturally occurring lantibiotic resistance, these modified peptides serve as a credible alternative to commercial antibiotics. As some lantibiotics have a dual mode of action, forming pores and also inhibiting cell wall biosynthesis, they represent a significant challenge to the target cell. A detailed classification of the known means of lantibiotic resistance is summarized in Table 1. Such a future characterization may allow for further tailoring of genetic variants of lantibiotic peptides that may circumvent resistance issues and inform the development of appropriate therapeutic regimens to overcome such issues if and when lantibiotics are deployed in clinical settings.
TABLE 1.
Summary of lantibiotic resistance methodsa
| Resistance mechanism and relevant protein(s) or operon | Species | Regulator(s) | Lantibiotic resistance(s) |
|---|---|---|---|
| Cell wall/membrane-altering mechanisms | |||
| Mprf | S. aureus, L. monocytogenes | VirRS/GraRS | Nisin, gallidermin |
| DltA | Streptococcus spp., Staphylococcus spp., L. monocytogenes, L. lactis, B. subtilis, C. difficile | VirRS/sigma factors | Nisin, gallidermin |
| gal operon | L. lactis | Nisin | |
| Penicillin-binding protein | L. monocytogenes, L. lactis, S. aureus | LiaRS, LisRK, VraSR, sigma factors | Nisin |
| Lipid composition, including the fab operon | L. monocytogenes, L. lactis | Nisin | |
| Miscellaneous resistance mechanisms | |||
| Nisin resistance protein* | L. lactis, Corynebacterium spp., Leuconostoc spp., Enterococcus spp., Streptococcus spp. | Nisin | |
| Nisinase* | S. thermophilus, L. plantarum, C. botulinum, L. lactis, E. faecalis, S. aureus, Bacillus spp. | Nisin, subtilin | |
| Gad | L. monocytogenes | Nisin | |
| NsrX | S. mutans | NsrRS | Nisin |
| arc operon | L. lactis | Nisin | |
| PurR | S. aureus | Nisin | |
| IreK-IreP-IreB | E. faecalis | Nisin | |
| Sigma factors | L. monocytogenes, B. subtilis | Nisin, sublancin, mersacidin, subtilin gallidermin | |
| VraFG | S. aureus | GraXRS | Nisin, nukacin ISK-1 |
| CprABC | C. difficile | CprRK | Nisin, gallidermin |
| AnrAB | L. monocytogenes | VirRS | Nisin |
| VraDE | S. aureus | BraRS, VraSR | Nisin |
| YsaBC | L. lactis | LlrG/KinG | Nisin |
| BraDE | S. aureus | BraRS | Nisin |
| Lmo1746-Lmo1747 | L. monocytogenes | VirRS, LiaRS | Nisin |
| BceAB | B. subtilis | BceRS | Actagardine, mersacidin |
| PsdAB | B. subtilis | PsdRS | Nisin, gallidermin, subtilin, actagardine |
| Sp0912-Sp0913 | S. pneumoniae | Rr01-Hk01 | Nisin |
| LctFEG | S. mutans | LcrRS | Nukacin ISK-1, lacticin 481 |
| NsrFP | Corynebacterium spp., Leuconostoc spp., Enterococcus spp., Streptococcus spp. | NsrRK | Nisin |
| TelA | L. monocytogenes | LiaRS | Nisin, gallidermin |
| EfdFE* | E. faecium | Lacticin 3147 | |
| SpiFEG* | S. infantarius | SpiRR′K? | Nisin |
| Two-component systems | |||
| TC09, TC12 | L. casei | Nisin | |
| GrsRS | S. aureus | Nisin, nukacin ISK-1 | |
| BraRS | S. aureus | Nisin, nukacin ISK-1 | |
| PsdRS | B. subtilis | Nisin, gallidermin, subtilin, actagardine | |
| LlrG/KinG | L. lactis | Nisin | |
| ApsRS | S. epidermidis | Nisin | |
| BceRS | Bacillus spp. | Actagardine, mersacidin | |
| LiaRS | Bacillus spp., Streptococcus spp., L. monocytogenes | Nisin | |
| NsrRS | S. mutans | Nisin, lacticin 481, nukacin ISK-1 | |
| LcrRS | S. mutans | Nukacin ISK-1, lacticin 481 | |
| VraSR | S. aureus | Nisin, mersacidin, nukacin ISK-1 | |
| LisRK | L. monocytogenes | Nisin | |
| CesSR | L. lactis | Nisin, plantaricin C | |
| VirRS | L. monocytogenes | Nisin | |
| CprRK | C. difficile | Nisin, gallidermin, subtilin, mutacin 1140, cinnamycin |
The table lists the strains in which lantibiotic resistance has been observed and the lantibiotics to which this resistance is targeted. Not included in the table are the general resistance mechanism inferred by the Gram-negative cell wall and that of spore or biofilm formation. Lantibiotic-specific resistance methods are indicated with asterisks.
ACKNOWLEDGMENT
We were supported in part by research grants from Science Foundation Ireland (SFI) (grants 10/IN.1/B3027, SFI/12/RC/2273, and 12/RC/2273).
Biographies

Lorraine A. Draper obtained her M.Sc. (2007) and Ph.D. (2014) in the area of molecular microbiology at University College Cork, Ireland. She is now a Postdoctoral Researcher with the Alimentary Pharmabiotic Centre (APC), Cork, Ireland, a multidisciplinary research center focusing on the role of gut microbiota in health and disease. Dr. Draper's research focuses on investigating food-grade antimicrobials and how they can be employed in prevention and treatment of human-pathogenic organisms.

Paul D. Cotter obtained his Ph.D. (2001) in molecular microbiology at University College Cork, Ireland. He is currently a Principal Research Officer at Teagasc Food Research Centre, Moorepark, Ireland, and a Principal Investigator with the APC. His research focuses on the microbiology of the gut (with a view to establishing and maintaining health) and foods, the investigation of ribosomally synthesized peptide antibiotics and toxins produced by Gram-positive bacteria and the mechanisms employed by bacteria to survive exposure to these and other stresses.

Colin Hill has a Ph.D. in molecular microbiology and is Professor of Microbial Food Safety in the School of Microbiology at University College Cork, Ireland. He is a Principal Investigator in the APC. His main interests are in infectious disease, particularly in defining the mechanisms of virulence of foodborne pathogens and in developing strategies to prevent and limit the consequences of microbial infections in the gastrointestinal tract.

R. Paul Ross graduated from UCC with a B.Sc. in Biochemistry and Microbiology and a Ph.D. in Microbiology. Following postdoctoral research and an Assistant Professorship at Wake Forest University, NC, USA, he moved to Teagasc’s Moorepark Food Research Centre to lead the Food Biotechnology Programme, which encompasses aspects ranging from gut microbiota to novel antimicrobials, including bacteriocins and bacteriophage. Until recently he was Head of Food Research at Teagasc and Adjunct Professor at UCC. Paul is now Head of College of Science Engineering & Food Science at UCC and is a Principal Investigator in the Alimentary Pharmabiotic Centre (APC). He was awarded a D.Sc. in 2009 based on published work and received the William C. Haines Dairy Science in 2007 and the Enterprise Ireland Commercialization award in 2008.
REFERENCES
- 1.Rea MC, Ross RP, Cotter PD, Hill C. 2011. Classification of bacteriocins from Gram-positive bacteria, p 29–53. In Drider D, Rebuffat S (ed), Prokaryotic antimicrobial peptides. Springer, New York, NY. [Google Scholar]
- 2.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Hill C, Ross RP. 2005. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci 6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 4.Piper C, Cotter PD, Ross RP, Hill C. 2009. Discovery of medically significant lantibiotics. Curr Drug Discov Technol 6:1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- 5.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 6.Asaduzzaman SM, Nagao J, Aso Y, Nakayama J, Sonomoto K. 2006. Lysine-oriented charges trigger the membrane binding and activity of nukacin ISK-1. Appl Environ Microbiol 72:6012–6017. doi: 10.1128/AEM.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moll GN, Roberts GC, Konings WN, Driessen AJ. 1996. Mechanism of lantibiotic-induced pore-formation. Antonie Van Leeuwenhoek 69:185–191. doi: 10.1007/BF00399423. [DOI] [PubMed] [Google Scholar]
- 8.Deegan LH, Suda S, Lawton EM, Draper LA, Hugenholtz F, Peschel A, Hill C, Cotter PD, Ross RP. 2010. Manipulation of charged residues within the two-peptide lantibiotic lacticin 3147. Microb Biotechnol 3:222–234. doi: 10.1111/j.1751-7915.2009.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Kraaij C, Breukink E, Rollema HS, Siezen RJ, Demel RA, Kruijff B, Kuipers OP. 1997. Influence of charge differences in the C-terminal part of nisin on antimicrobial activity and signaling capacity. Eur J Biochem 247:114–120. doi: 10.1111/j.1432-1033.1997.00114.x. [DOI] [PubMed] [Google Scholar]
- 10.Biswas S, Biswas I. 2014. A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J Bacteriol 196:1578–1587. doi: 10.1128/JB.00028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brotz H, Josten M, Wiedemann I, Schneider U, Gotz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 12.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 13.Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, Sahl HG. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol 61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- 14.Wiedemann I, Bottiger T, Bonelli RR, Schneider T, Sahl HG, Martinez B. 2006. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl Environ Microbiol 72:2809–2814. doi: 10.1128/AEM.72.4.2809-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brotz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob Agents Chemother 42:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 17.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 18.Pag U, Heidrich C, Bierbaum G, Sahl HG. 1999. Molecular analysis of expression of the lantibiotic pep5 immunity phenotype. Appl Environ Microbiol 65:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benz R, Jung G, Sahl HG. 1991. Mechanism of channel-formation by lantibiotics in black lipid membranes, p 359–372. In Jung G, Sahl HG (ed), Nisin and novel lantibiotics: proceedings of the First International Workshop on Lantibiotics. Escom, Leiden, The Netherlands. [Google Scholar]
- 20.Sahl HG. 1991. Pore formation in bacterial membranes by cationic lantibiotics, p 347–358. In Jung G, Sahl HG (ed), Nisin and novel lantibiotics: proceedings of the First International Workshop on Lantibiotics. Escom, Leiden, The Netherlands. [Google Scholar]
- 21.Driessen AJ, van den Hooven HW, Kuiper W, van de Kamp M, Sahl HG, Konings RN, Konings WN. 1995. Mechanistic studies of lantibiotic-induced permeabilization of phospholipid vesicles. Biochemistry 34:1606–1614. doi: 10.1021/bi00005a017. [DOI] [PubMed] [Google Scholar]
- 22.Chikindas ML, Novak J, Driessen AJ, Konings WN, Schilling KM, Caufield PW. 1995. Mutacin II, a bactericidal antibiotic from Streptococcus mutans. Antimicrob Agents Chemother 39:2656–2660. doi: 10.1128/AAC.39.12.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brotz H, Bierbaum G, Markus A, Molitor E, Sahl HG. 1995. Mode of action of the lantibiotic mersacidin: inhibition of peptidoglycan biosynthesis via a novel mechanism? Antimicrob Agents Chemother 39:714–719. doi: 10.1128/AAC.39.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machaidze G, Seelig J. 2003. Specific binding of cinnamycin (Ro 09-0198) to phosphatidylethanolamine. Comparison between micellar and membrane environments. Biochemistry 42:12570–12576. doi: 10.1021/bi035225b. [DOI] [PubMed] [Google Scholar]
- 25.Bonelli RR, Schneider T, Sahl HG, Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob Agents Chemother 50:1449–1457. doi: 10.1128/AAC.50.4.1449-1457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst A. 1981. Nisin. Adv Appl Microbiol 27:85–123. doi: 10.1016/S0065-2164(08)70342-3. [DOI] [Google Scholar]
- 27.Paik SH, Chakicherla A, Hansen JN. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem 273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 28.Gut IM, Prouty AM, Ballard JD, van der Donk WA, Blanke SR. 2008. Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrob Agents Chemother 52:4281–4288. doi: 10.1128/AAC.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJ, Kuipers OP, Moll GN. 2007. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl Environ Microbiol 73:5809–5816. doi: 10.1128/AEM.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gut IM, Blanke SR, van der Donk WA. 2011. Mechanism of inhibition of Bacillus anthracis spore outgrowth by the lantibiotic nisin. ACS Chem Biol 6:744–752. doi: 10.1021/cb1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. 2005. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol Microbiol 58:1368–1380. doi: 10.1111/j.1365-2958.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- 33.Willey JM, Willems A, Kodani S, Nodwell JR. 2006. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol Microbiol 59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz B, Markwalder JA, Wang Y. 2001. Lipid II: total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Escherichia coli. J Am Chem Soc 123:11638–11643. doi: 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- 35.Lazar K, Walker S. 2002. Substrate analogues to study cell-wall biosynthesis and its inhibition. Curr Opin Chem Biol 6:786–793. doi: 10.1016/S1367-5931(02)00355-1. [DOI] [PubMed] [Google Scholar]
- 36.Müller A, Ulm H, Reder-Christ K, Sahl HG, Schneider T. 2012. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb Drug Resist 18:261–270. doi: 10.1089/mdr.2011.0242. [DOI] [PubMed] [Google Scholar]
- 37.Kramer NE, Smid EJ, Kok J, Kruijff B, Kuipers OP, Breukink E. 2004. Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol Lett 239:157–161. doi: 10.1016/j.femsle.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 38.Weidenmaier C, Kristian SA, Peschel A. 2003. Bacterial resistance to antimicrobial host defenses—an emerging target for novel antiinfective strategies? Curr Drug Targets 4:643–649. doi: 10.2174/1389450033490731. [DOI] [PubMed] [Google Scholar]
- 39.Ward J. 1981. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol Rev 45:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hancock IC, Baddiley J. 1985. Biosynthesis of the bacterial envelope polymers teichoic acid and teichuronic acid, p 279–307. In Martonosi A. (ed), The enzymes of biological membranes. Springer, New York, NY. [Google Scholar]
- 41.Fischer W. 1988. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol 29:233–302. doi: 10.1016/S0065-2911(08)60349-5. [DOI] [PubMed] [Google Scholar]
- 42.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270:15598–15606. [DOI] [PubMed] [Google Scholar]
- 43.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heaton MP, Neuhaus FC. 1992. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J Bacteriol 174:4707–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 46.Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol 183:6324–6334. doi: 10.1128/JB.183.21.6324-6334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride SM, Sonenshein AL. 2011. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 49.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khattar ZA, Rejasse A, Destoumieux-Garzón D, Escoubas J, Sanchis V, Lereclus D, Givaudan A, Kallassy M, Nielsen-Leroux C, Gaudriault S. 2009. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol 191:7063–7073. doi: 10.1128/JB.00892-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kramer NE, van Hijum SA, Knol J, Kok J, Kuipers OP. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob Agents Chemother 50:1753–1761. doi: 10.1128/AAC.50.5.1753-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grossiord BP, Luesink EJ, Vaughan EE, Arnaud A, de Vos WM. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J Bacteriol 185:870–878. doi: 10.1128/JB.185.3.870-878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantovani HC, Russell JB. 2001. Nisin resistance of Streptococcus bovis. Appl Environ Microbiol 67:808–813. doi: 10.1128/AEM.67.2.808-813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goffin C, Ghuysen JM. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev 62:1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala JA, Ghuysen JM, Nguyen-Disteche M. 1999. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol 34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 56.Marrec-Fairley M, Piette A, Gallet X, Brasseur R, Hara H, Fraipont C, Ghuysen JM, Nguyen-Disteche M. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol Microbiol 37:1019–1031. doi: 10.1046/j.1365-2958.2000.02054.x. [DOI] [PubMed] [Google Scholar]
- 57.Gravesen A, Sorensen K, Aarestrup FM, Knochel S. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb Drug Resist 7:127–135. doi: 10.1089/10766290152045002. [DOI] [PubMed] [Google Scholar]
- 58.Guinane CM, Cotter PD, Ross RP, Hill C. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob Agents Chemother 50:2824–2828. doi: 10.1128/AAC.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gravesen A, Kallipolitis B, Holmstrom K, Hoiby PE, Ramnath M, Knochel S. 2004. pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl Environ Microbiol 70:1669–1679. doi: 10.1128/AEM.70.3.1669-1679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cotter PD, Guinane CM, Hill C. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob Agents Chemother 46:2784–2790. doi: 10.1128/AAC.46.9.2784-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xuanyuan Z, Wu Z, Li R, Jiang D, Su J, Xu H, Bai Y, Zhang X, Saris PE, Qiao M. 2010. Loss of IrpT function in Lactococcus lactis subsp. lactis N8 results in increased nisin resistance. Curr Microbiol 61:329–334. doi: 10.1007/s00284-010-9615-4. [DOI] [PubMed] [Google Scholar]
- 62.Tsukioka Y, Yamashita Y, Nakano Y, Oho T, Koga T. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol 179:4411–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. 2012. Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol 113:485–498. doi: 10.1111/j.1365-2672.2012.05336.x. [DOI] [PubMed] [Google Scholar]
- 64.Mazzotta AS, Crandall AD, Montville TJ. 1997. Nisin resistance in Clostridium botulinum spores and vegetative cells. Appl Environ Microbiol 63:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schved F, Henis Y, Juven BJ. 1994. Response of spheroplasts and chelator-permeabilized cells of Gram-negative bacteria to the action of the bacteriocins pediocin SJ-1 and nisin. Int J Food Microbiol 21:305–314. doi: 10.1016/0168-1605(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 66.Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR. 1991. Nisin treatment for inactivation of Salmonella species and other Gram-negative bacteria. Appl Environ Microbiol 57:3613–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brotz H, Sahl HG. 2000. New insights into the mechanism of action of lantibiotics—diverse biological effects by binding to the same molecular target. J Antimicrob Chemother 46:1–6. doi: 10.1093/jac/46.1.1. [DOI] [PubMed] [Google Scholar]
- 68.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zahringer U, Seydel U, Di Padova F, Schreir M, Brade H. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J 8:217–225. [DOI] [PubMed] [Google Scholar]
- 69.Kalchayanand N, Hanlin MB, Ray B. 1992. Sublethal injury makes Gram-negative and resistant Gram-positive bacteria sensitive to the bacteriocins, pediocin AcH and nisin. Lett Appl Microbiol 15:239–243. doi: 10.1111/j.1472-765X.1992.tb00773.x. [DOI] [Google Scholar]
- 70.Boziaris IS, Adams MR. 2000. Transient sensitivity to nisin in cold-shocked Gram negatives. Lett Appl Microbiol 31:233–237. doi: 10.1046/j.1365-2672.2000.00804.x. [DOI] [PubMed] [Google Scholar]
- 71.Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, Parenti F. 2008. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15:22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 72.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. 2012. Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLoS One 7:e46884. doi: 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ratledge C, Wilkinson SG. 1988. Microbial lipids. Academic Press, London, United Kingdom. [Google Scholar]
- 74.DiRusso CC, Nystrom T. 1998. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol Microbiol 27:1–8. doi: 10.1046/j.1365-2958.1998.00645.x. [DOI] [PubMed] [Google Scholar]
- 75.Huijbregts RP, de Kroon AI, de Kruijff B. 2000. Topology and transport of membrane lipids in bacteria. Biochim Biophys Acta 1469:43–61. doi: 10.1016/S0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 76.Matsumoto K. 2001. Dispensable nature of phosphatidylglycerol in Escherichia coli: dual roles of anionic phospholipids. Mol Microbiol 39:1427–1433. doi: 10.1046/j.1365-2958.2001.02320.x. [DOI] [PubMed] [Google Scholar]
- 77.de Mendoza D, Grau R, Cronan JE. 1993. Biosynthesis and function of membrane lipids, p 411–421. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC. [Google Scholar]
- 78.Nahaie MR, Goodfellow M, Minnikin DE, Hajek V. 1984. Polar lipid and isoprenoid quinone composition in the classification of Staphylococcus. J Gen Microbiol 130:2427–2437. [DOI] [PubMed] [Google Scholar]
- 79.Fischer W, Leopold K. 1999. Polar lipids of four Listeria species containing l-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. Int J Syst Bacteriol 49:653–662. doi: 10.1099/00207713-49-2-653. [DOI] [PubMed] [Google Scholar]
- 80.Verheul A, Russell NJ, Hof RVT, Rombouts FM, Abee T. 1997. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl Environ Microbiol 63:3451–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Demel RA, Peelen T, Siezen RJ, Kruijff B, Kuipers OP. 1996. Nisin Z, mutant nisin Z and lacticin 481 interactions with anionic lipids correlate with antimicrobial activity. Eur J Biochem 235:267–274. doi: 10.1111/j.1432-1033.1996.00267.x. [DOI] [PubMed] [Google Scholar]
- 82.Crandall AD, Montville TJ. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol 64:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ming X, Daeschel MA. 1995. Correlation of cellular phospholipid content with nisin resistance of Listeria monocytogenes Scott A. J Food Prot 58:416–420. [DOI] [PubMed] [Google Scholar]
- 84.Mazzotta AS, Montville TJ. 1997. Nisin induces changes in membrane fatty acid composition of Listeria monocytogenes nisin-resistant strains at 10 degrees C and 30 degrees C. J Appl Microbiol 82:32–38. doi: 10.1111/j.1365-2672.1997.tb03294.x. [DOI] [PubMed] [Google Scholar]
- 85.Ming X, Daeschel MA. 1993. Nisin resistance of foodborne bacteria and the specific resistance responses of Listeria monocytogenes Scott A. J Food Prot 11:927–1005. [DOI] [PubMed] [Google Scholar]
- 86.Martinez B, Rodriguez A. 2005. Antimicrobial susceptibility of nisin resistant Listeria monocytogenes of dairy origin. FEMS Microbiol Lett 252:67–72. doi: 10.1016/j.femsle.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 87.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol 62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 89.Maloney E, Stankowska D, Zhang J, Fol M, Cheng QJ, Lun S, Bishai WR, Rajagopalan M, Chatterjee D, Madiraju MV. 2009. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog 5:e1000534. doi: 10.1371/journal.ppat.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Staubitz P, Peschel A. 2002. MprF-mediated lysinylation of phospholipids in Bacillus subtilis—protection against bacteriocins in terrestrial habitats? Microbiology 148:3331–3332. [DOI] [PubMed] [Google Scholar]
- 91.Samant S, Hsu FF, Neyfakh AA, Lee H. 2009. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J Bacteriol 191:1311–1319. doi: 10.1128/JB.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lennarz WJ, Nesbitt JA 3rd, Reiss J. 1966. The participation of sRNA in the enzymatic synthesis of O-l-lysyl phosphatidylglycerol in Staphylococcus aureus. Proc Natl Acad Sci U S A 55:934–941. doi: 10.1073/pnas.55.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gould RM, Lennarz WJ. 1967. Biosynthesis of aminoacyl derivatives of phosphatidylglycerol. Biochem Biophys Res Commun 26:512–515. [DOI] [PubMed] [Google Scholar]
- 94.Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. 2004. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol Lett 231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 95.Roy H. 2009. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life 61:940–953. doi: 10.1002/iub.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roy H, Ibba M. 2008. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A 105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, Hartke A, Huebner J. 2012. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 7:e38458. doi: 10.1371/journal.pone.0038458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roy H, Ibba M. 2009. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J Biol Chem 284:29677–29683. doi: 10.1074/jbc.M109.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Houtsmuller UM, van Deenen L. 1963. Identification of a bacterial phospholipid as an O-ornithine ester of phosphatidyl glycerol. Biochim Biophys Acta 70:211–213. doi: 10.1016/0006-3002(63)90743-1. [DOI] [PubMed] [Google Scholar]
- 101.Slavetinsky CJ, Peschel A, Ernst CM. 2012. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 56:3492–3497. doi: 10.1128/AAC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arendt W, Hebecker S, Jager S, Nimtz M, Moser J. 2012. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases. J Bacteriol 194:1401–1416. doi: 10.1128/JB.06576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J. 2009. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol 71:551–565. doi: 10.1111/j.1365-2958.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 104.Heath RJ, Rock CO. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem 270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 105.Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S. 2011. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J Bacteriol 193:3851–3862. doi: 10.1128/JB.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mascher T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 264:133–144. doi: 10.1111/j.1574-6968.2006.00444.x. [DOI] [PubMed] [Google Scholar]
- 107.Falord M, Karimova G, Hiron A, Msadek T. 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:1047–1058. doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol 81:602–622. doi: 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 109.Rietkotter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol Microbiol 68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- 110.Staron A, Finkeisen DE, Mascher T. 2011. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob Agents Chemother 55:515–525. doi: 10.1128/AAC.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ouyang J, Tian XL, Versey J, Wishart A, Li YH. 2010. The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob Agents Chemother 54:3895–3906. doi: 10.1128/AAC.01802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Revilla-Guarinos A, Gebhard S, Alcantara C, Staron A, Mascher T, Zuniga M. 2013. Characterization of a regulatory network of peptide antibiotic detoxification modules in Lactobacillus casei BL23. Appl Environ Microbiol 79:3160–3170. doi: 10.1128/AEM.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gebhard S, Fang C, Shaaly A, Leslie DJ, Weimar MR, Kalamorz F, Carne A, Cook GM. 2014. Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob Agents Chemother 58:1425–1433. doi: 10.1128/AAC.02111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87–92. doi: 10.1016/0378-1119(94)00851-I. [DOI] [PubMed] [Google Scholar]
- 115.Saier MH Jr, Yen MR, Noto K, Tamang DG, Elkan C. 2009. The Transporter Classification Database: recent advances. Nucleic Acids Res 37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 117.Ohki R, Giyanto, Tateno K, Masuyama W, Moriya S, Kobayashi K, Ogasawara N. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol 49:1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x. [DOI] [PubMed] [Google Scholar]
- 118.Kallenberg F, Dintner S, Schmitz R, Gebhard S. 2013. Identification of regions important for resistance and signalling within the antimicrobial peptide transporter BceAB of Bacillus subtilis. J Bacteriol 195:3287–3297. doi: 10.1128/JB.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kingston AW, Zhao H, Cook GM, Helmann JD. 2014. Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol Microbiol 93:37–49. doi: 10.1111/mmi.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pietiainen M, Gardemeister M, Mecklin M, Leskela S, Sarvas M, Kontinen VP. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577–1592. doi: 10.1099/mic.0.27761-0. [DOI] [PubMed] [Google Scholar]
- 121.Alcantara C, Revilla-Guarinos A, Zuniga M. 2011. Influence of two-component signal transduction systems of Lactobacillus casei BL23 on tolerance to stress conditions. Appl Environ Microbiol 77:1516–1519. doi: 10.1128/AEM.02176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J, Elasri MO, Shaw LN. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157:2206–2219. doi: 10.1099/mic.0.049692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yoshida Y, Matsuo M, Oogai Y, Kato F, Nakamura N, Sugai M, Komatsuzawa H. 2011. Bacitracin sensing and resistance in Staphylococcus aureus. FEMS Microbiol Lett 320:33–39. doi: 10.1111/j.1574-6968.2011.02291.x. [DOI] [PubMed] [Google Scholar]
- 124.Kawada-Matsuo M, Yoshida Y, Zendo T, Nagao J, Oogai Y, Nakamura Y, Sonomoto K, Nakamura N, Komatsuzawa H. 2013. Three distinct two-component systems are involved in resistance to the class I bacteriocins, nukacin ISK-1 and nisin A, in Staphylococcus aureus. PLoS One 8:e69455. doi: 10.1371/journal.pone.0069455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Blake KL, Randall CP, O'Neill AJ. 2011. In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob Agents Chemother 55:2362–2368. doi: 10.1128/AAC.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kohler T, Weidenmaier C, Peschel A. 2009. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J Bacteriol 191:4482–4484. doi: 10.1128/JB.00221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 128.Cui L, Lian JQ, Neoh HM, Reyes E, Hiramatsu K. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 49:3404–3413. doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meehl M, Herbert S, Gotz F, Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 51:2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuroda M, Kuwahara-Arai K, Hiramatsu K. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun 269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 131.Matsuo M, Oogai Y, Kato F, Sugai M, Komatsuzawa H. 2011. Growth-phase dependence of susceptibility to antimicrobial peptides in Staphylococcus aureus. Microbiology 157:1786–1797. doi: 10.1099/mic.0.044727-0. [DOI] [PubMed] [Google Scholar]
- 132.Falord M, Mader U, Hiron A, Debarbouille M, Msadek T. 2011. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6:e21323. doi: 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob Agents Chemother 54:4416–4423. doi: 10.1128/AAC.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol 57:1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 136.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hebraud M, Hechard Y. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581–1590. doi: 10.1099/mic.0.26860-0. [DOI] [PubMed] [Google Scholar]
- 137.Gebhard S, Mascher T. 2011. Antimicrobial peptide sensing and detoxification modules: unravelling the regulatory circuitry of Staphylococcus aureus. Mol Microbiol 81:581–587. doi: 10.1111/j.1365-2958.2011.07747.x. [DOI] [PubMed] [Google Scholar]
- 138.Majchrzykiewicz JA, Kuipers OP, Bijlsma JJ. 2010. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob Agents Chemother 54:440–451. doi: 10.1128/AAC.00769-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wolf D, Kalamorz F, Wecke T, Juszczak A, Mäder U, Homuth G, Jordan S, Kirstein J, Hoppert M, Voigt B. 2010. In-depth profiling of the LiaR response of Bacillus subtilis. J Bacteriol 192:4680–4693. doi: 10.1128/JB.00543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol 188:5153–5166. doi: 10.1128/JB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolf D, Dominguez-Cuevas P, Daniel RA, Mascher T. 2012. Cell envelope stress response in cell wall-deficient L-forms of Bacillus subtilis. Antimicrob Agents Chemother 56:5907–5915. doi: 10.1128/AAC.00770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fritsch F, Mauder N, Williams T, Weiser J, Oberle M, Beier D. 2011. The cell envelope stress response mediated by the LiaFSRLm three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaSLm. Microbiology 157:373–386. doi: 10.1099/mic.0.044776-0. [DOI] [PubMed] [Google Scholar]
- 144.Collins B, Guinane CM, Cotter PD, Hill C, Ross RP. 2012. Assessing the contributions of the LiaS histidine kinase to the innate resistance of Listeria monocytogenes to nisin, cephalosporins, and disinfectants. Appl Environ Microbiol 78:2923–2929. doi: 10.1128/AEM.07402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Collins B, Joyce S, Hill C, Cotter PD, Ross RP. 2010. TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob Agents Chemother 54:4658–4663. doi: 10.1128/AAC.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bergholz TM, Tang S, Wiedmann M, Boor KJ. 2013. Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl Environ Microbiol 79:5682–5688. doi: 10.1128/AEM.01797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Klinzing DC, Ishmael N, Dunning Hotopp JC, Tettelin H, Shields KR, Madoff LC, Puopolo KM. 2013. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology 159:1521–1534. doi: 10.1099/mic.0.064444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McCormick NE, Halperin SA, Song F. 2011. Regulation of d-alanylation of lipoteichoic acid in Streptococcus gordonii. Microbiology 157:2248–2256. doi: 10.1099/mic.0.048140-0. [DOI] [PubMed] [Google Scholar]
- 150.O'Connell-Motherway M, van Sinderen D, Morel-Deville F, Fitzgerald GF, Ehrlich SD, Morel P. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935–947. [DOI] [PubMed] [Google Scholar]
- 151.Martinez B, Zomer AL, Rodriguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol Microbiol 64:473–486. doi: 10.1111/j.1365-2958.2007.05668.x. [DOI] [PubMed] [Google Scholar]
- 152.Dubrac S, Boneca IG, Poupel O, Msadek T. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 154.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum RS, Labischinski H, Hiramatsu K. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother 42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 155.Sass P, Jansen A, Szekat C, Sass V, Sahl HG, Bierbaum G. 2008. The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol 8:186. doi: 10.1186/1471-2180-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother 52:980–990. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McCallum N, Meier PS, Heusser R, Berger-Bachi B. 2011. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother 55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, Projan S, Tomasz A. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol 188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Berscheid A, François P, Strittmatter A, Gottschalk G, Schrenzel J, Sass P, Bierbaum G. 2014. Generation of a vancomycin-intermediate Staphylococcus aureus (VISA) strain by two amino acid exchanges in VraS. J Antimicrob Chemother 69:3190–3198. doi: 10.1093/jac/dku297. [DOI] [PubMed] [Google Scholar]
- 160.Cotter PD, Emerson N, Gahan CG, Hill C. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol 181:6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kallipolitis BH, Ingmer H. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol Lett 204:111–115. doi: 10.1111/j.1574-6968.2001.tb10872.x. [DOI] [PubMed] [Google Scholar]
- 162.Camejo A, Buchrieser C, Couve E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Suarez JM, Edwards AN, McBride SM. 2013. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J Bacteriol 195:2621–2631. doi: 10.1128/JB.00166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kawada-Matsuo M, Oogai Y, Zendo T, Nagao J, Shibata Y, Yamashita Y, Ogura Y, Hayashi T, Sonomoto K, Komatsuzawa H. 2013. Involvement of the novel two-component NsrRS and LcrRS systems in distinct resistance pathways against nisin A and nukacin ISK-1 in Streptococcus mutans. Appl Environ Microbiol 79:4751–4755. doi: 10.1128/AEM.00780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Saising J, Dube L, Ziebandt AK, Voravuthikunchai SP, Nega M, Gotz F. 2012. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 56:5804–5810. doi: 10.1128/AAC.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dobson A, O'Connor PM, Cotter PD, Ross RP, Hill C. 2011. Impact of the broad-spectrum antimicrobial peptide, lacticin 3147, on Streptococcus mutans growing in a biofilm and in human saliva. J Appl Microbiol 111:1515–1523. doi: 10.1111/j.1365-2672.2011.05153.x. [DOI] [PubMed] [Google Scholar]
- 168.Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, Qi F. 2009. Genes involved in the repression of mutacin I production in Streptococcus mutans. Microbiology 155:551–556. doi: 10.1099/mic.0.021303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sudagidan M, Yemenicioglu A. 2012. Effects of nisin and lysozyme on growth inhibition and biofilm formation capacity of Staphylococcus aureus strains isolated from raw milk and cheese samples. J Food Prot 75:1627–1633. doi: 10.4315/0362-028X.JFP-12-001. [DOI] [PubMed] [Google Scholar]
- 170.Okuda KI, Zendo T, Sugimoto S, Iwase T, Tajima A, Yamada S, Sonomoto K, Mizunoe Y. 2013. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob Agents Chemother 57:5572–5579. doi: 10.1128/AAC.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Asaduzzaman SM, Nagao J, Iida H, Zendo T, Nakayama J, Sonomoto K. 2009. Nukacin ISK-1, a bacteriostatic lantibiotic. Antimicrob Agents Chemother 53:3595–3598. doi: 10.1128/AAC.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yoneyama F, Imura Y, Ohno K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob Agents Chemother 53:3211–3217. doi: 10.1128/AAC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yamakami K, Tsumori H, Sakurai Y, Shimizu Y, Nagatoshi K, Sonomoto K. 2013. Sustainable inhibition efficacy of liposome-encapsulated nisin on insoluble glucan-biofilm synthesis by Streptococcus mutans. Pharm Biol 51:267–270. doi: 10.3109/13880209.2012.717227. [DOI] [PubMed] [Google Scholar]
- 174.Tong Z, Zhou L, Jiang W, Kuang R, Li J, Tao R, Ni L. 2011. An in vitro synergetic evaluation of the use of nisin and sodium fluoride or chlorhexidine against Streptococcus mutans. Peptides 32:2021–2026. doi: 10.1016/j.peptides.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 175.Dosler S, Mataraci E. 2013. In vitro pharmacokinetics of antimicrobial cationic peptides alone and in combination with antibiotics against methicillin resistant Staphylococcus aureus biofilms. Peptides 49:53–58. doi: 10.1016/j.peptides.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 176.Qi X, Poernomo G, Wang K, Chen Y, Chan-Park MB, Xu R, Chang MW. 2011. Covalent immobilization of nisin on multi-walled carbon nanotubes: superior antimicrobial and anti-biofilm properties. Nanoscale 3:1874–1880. doi: 10.1039/c1nr10024f. [DOI] [PubMed] [Google Scholar]
- 177.Hequet A, Humblot V, Berjeaud JM, Pradier CM. 2011. Optimized grafting of antimicrobial peptides on stainless steel surface and biofilm resistance tests. Colloids Surf B Biointerfaces 84:301–309. doi: 10.1016/j.colsurfb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 178.Nostro A, Scaffaro R, Ginestra G, D'Arrigo M, Botta L, Marino A, Bisignano G. 2010. Control of biofilm formation by poly-ethylene-co-vinyl acetate films incorporating nisin. Appl Microbiol Biotechnol 87:729–737. doi: 10.1007/s00253-010-2598-z. [DOI] [PubMed] [Google Scholar]
- 179.Conlon B, Nakayasu E, Fleck L, LaFleur M, Isabella V, Coleman K, Leonard S, Smith R, Adkins J, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Froseth BR, McKay LL. 1991. Molecular characterization of the nisin resistance region of Lactococcus lactis subsp. lactis biovar diacetylactis DRC3. Appl Environ Microbiol 57:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu CQ, Harvey ML, Dunn NW. 1997. Cloning of a gene encoding nisin resistance from Lactococcus lactis subsp. lactis M189 which is transcribed from an extended 210 promoter. J Gen Appl Microbiol 43:67–73. doi: 10.2323/jgam.43.67. [DOI] [PubMed] [Google Scholar]
- 182.Yun XX, Hu J, Chen Q. 2006. Screening, isolation and identification of nisin resistance determinant gene in strains of Lactococcus lactis. Nan Fang Yi Ke Da Xue Xue Bao 26:839–842. [PubMed] [Google Scholar]
- 183.Sun Z, Zhong J, Liang X, Liu J, Chen X, Huan L. 2009. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob Agents Chemother 53:1964–1973. doi: 10.1128/AAC.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem Rev 105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 185.O'Driscoll J, Glynn F, Fitzgerald GF, van Sinderen D. 2006. Sequence analysis of the lactococcal plasmid pNP40: a mobile replicon for coping with environmental hazards. J Bacteriol 188:6629–6639. doi: 10.1128/JB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Keiler KC, Silber KR, Downard KM, Papayannopoulos IA, Biemann K, Sauer RT. 1995. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci 4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 188.Spiers A, Lamb HK, Cocklin S, Wheeler KA, Budworth J, Dodds AL, Pallen MJ, Maskell DJ, Charles IG, Hawkins AR. 2002. PDZ domains facilitate binding of high temperature requirement protease A (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of the small stable RNA A (ssrA)-encoded peptide. J Biol Chem 277:39443–39449. doi: 10.1074/jbc.M202790200. [DOI] [PubMed] [Google Scholar]
- 189.Keiler KC, Sauer RT. 1996. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem 271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 190.Khosa S, Alkhatib Z, Smits SH. 2013. NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol Chem 394:1543–1549. doi: 10.1515/hsz-2013-0167. [DOI] [PubMed] [Google Scholar]
- 191.Jarvis B. 1967. Resistance to nisin and production of nisin-inactivating enzymes by several Bacillus species. J Gen Microbiol 47:33–48. doi: 10.1099/00221287-47-1-33. [DOI] [PubMed] [Google Scholar]
- 192.Jarvis B. 1970. Enzymic reduction of the C-terminal dehydroalanyl-lysine sequence in nisin. Biochem J 119:56P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kooy J. 1952. Strains of Lactobacillus plantarum which inhibit the activity of the antibiotics produced by Streptococcus lactis. Neth Milk Dairy J 6:323. [Google Scholar]
- 194.Alifax R, Chevalier R. 1962. Study of the nisinase produced by Streptococcus thermophilus. J Dairy Res 29:233–240. [Google Scholar]
- 195.Rayman K, Malik N, Hurst A. 1983. Failure of nisin to inhibit outgrowth of Clostridium botulinum in a model cured meat system. Appl Environ Microbiol 46:1450–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Carlson S, Bauer H. 1957. A study of problems associated with resistance to nisin. Arch Hyg Bakteriol 141:445–460. [PubMed] [Google Scholar]
- 197.Cotter PD, O'Reilly K, Hill C. 2001. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J Food Prot 64:1362–1368. [DOI] [PubMed] [Google Scholar]
- 198.Cotter PD, Ryan S, Gahan CG, Hill C. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol 71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cotter PD, Gahan CG, Hill C. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 40:465–475. doi: 10.1046/j.1365-2958.2001.02398.x. [DOI] [PubMed] [Google Scholar]
- 200.Begley M, Cotter PD, Hill C, Ross RP. 2010. Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes. Appl Environ Microbiol 76:6541–6546. doi: 10.1128/AEM.00203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Poolman B, Driessen A, Konings W. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol 169:5597–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kristich CJ, Wells CL, Dunny GM. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci U S A 104:3508–3513. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hall CL, Tschannen M, Worthey EA, Kristich CJ. 2013. IreB, a Ser/Thr kinase substrate, influences antimicrobial resistance in Enterococcus faecalis. Antimicrob Agents Chemother 57:6179–6186. doi: 10.1128/AAC.01472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Boylan SA, Redfield AR, Brody MS, Price CW. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J Bacteriol 175:7931–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Begley M, Hill C, Ross RP. 2006. Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl Environ Microbiol 72:2231–2234. doi: 10.1128/AEM.72.3.2231-2234.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stack HM, Sleator RD, Bowers M, Hill C, Gahan CG. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl Environ Microbiol 71:4241–4247. doi: 10.1128/AEM.71.8.4241-4247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mata MT, Baquero F, Perez-Diaz JC. 2000. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol Lett 187:185–188. doi: 10.1111/j.1574-6968.2000.tb09158.x. [DOI] [PubMed] [Google Scholar]
- 208.Palmer ME, Wiedmann M, Boor KJ. 2009. Sigma(B) and sigma(L) contribute to Listeria monocytogenes 10403S response to the antimicrobial peptides SdpC and nisin. Foodborne Pathog Dis 6:1057–1065. doi: 10.1089/fpd.2009.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Butcher BG, Helmann JD. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol Microbiol 60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 210.Cao M, Helmann JD. 2004. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol 186:1136–1146. doi: 10.1128/JB.186.4.1136-1146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cao M, Helmann JD. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J Bacteriol 184:6123–6129. doi: 10.1128/JB.184.22.6123-6129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bernard R, El Ghachi M, Mengin-Lecreulx D, Chippaux M, Denizot F. 2005. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J Biol Chem 280:28852–28857. doi: 10.1074/jbc.M413750200. [DOI] [PubMed] [Google Scholar]
- 213.Kingston AW, Liao X, Helmann JD. 2013. Contributions of the σW, σM and σX regulons to the lantibiotic resistome of Bacillus subtilis. Mol Microbiol 90:502–518. doi: 10.1111/mmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee YH, Kingston AW, Helmann JD. 2012. Glutamate dehydrogenase affects resistance to cell wall antibiotics in Bacillus subtilis. J Bacteriol 194:993–1001. doi: 10.1128/JB.06547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kingston AW, Subramanian C, Rock CO, Helmann JD. 2011. A sigmaW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol Microbiol 81:69–79. doi: 10.1111/j.1365-2958.2011.07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Draper LA, Ross RP, Hill C, Cotter PD. 2008. Lantibiotic immunity. Curr Protein Pept Sci 9:39–49. doi: 10.2174/138920308783565750. [DOI] [PubMed] [Google Scholar]
- 217.Draper LA, Grainger K, Deegan LH, Cotter PD, Hill C, Ross RP. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol Microbiol 71:1043–1054. doi: 10.1111/j.1365-2958.2008.06590.x. [DOI] [PubMed] [Google Scholar]
- 218.Draper LA, Tagg JR, Hill C, Cotter PD, Ross RP. 2012. The spiFEG locus in Streptococcus infantarius subsp. infantarius BAA-102 confers protection against nisin U. Antimicrob Agents Chemother 56:573–578. doi: 10.1128/AAC.05778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]