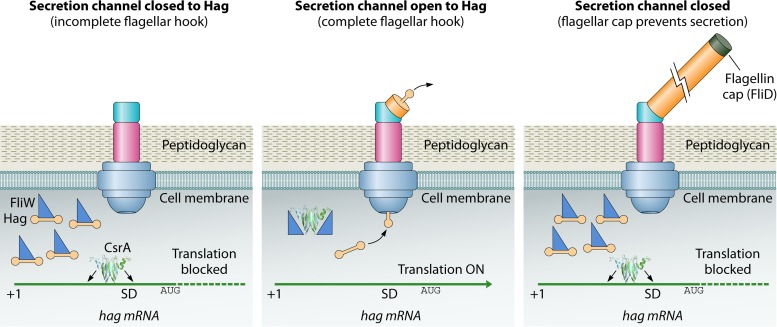
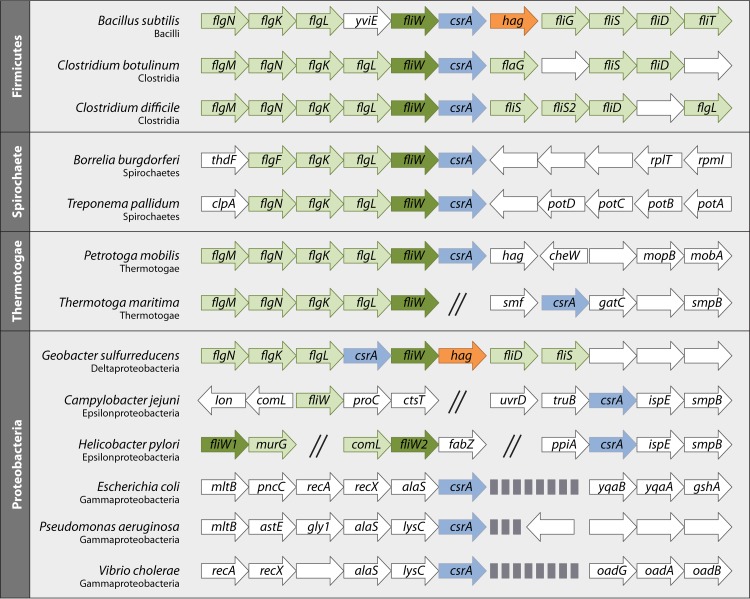
SUMMARY
Most bacterial pathogens have the remarkable ability to flourish in the external environment and in specialized host niches. This ability requires their metabolism, physiology, and virulence factors to be responsive to changes in their surroundings. It is no surprise that the underlying genetic circuitry that supports this adaptability is multilayered and exceedingly complex. Studies over the past 2 decades have established that the CsrA/RsmA proteins, global regulators of posttranscriptional gene expression, play important roles in the expression of virulence factors of numerous proteobacterial pathogens. To accomplish these tasks, CsrA binds to the 5′ untranslated and/or early coding regions of mRNAs and alters translation, mRNA turnover, and/or transcript elongation. CsrA activity is regulated by noncoding small RNAs (sRNAs) that contain multiple CsrA binding sites, which permit them to sequester multiple CsrA homodimers away from mRNA targets. Environmental cues sensed by two-component signal transduction systems and other regulatory factors govern the expression of the CsrA-binding sRNAs and, ultimately, the effects of CsrA on secretion systems, surface molecules and biofilm formation, quorum sensing, motility, pigmentation, siderophore production, and phagocytic avoidance. This review presents the workings of the Csr system, the paradigm shift that it generated for understanding posttranscriptional regulation, and its roles in virulence networks of animal and plant pathogens.
INTRODUCTION
Prior to the discovery of CsrA in the early 1990s, bacterial gene expression was understood to be regulated at the level of transcription initiation and to a lesser extent by transcription termination. At that time, examples of posttranscriptional regulation included the role of Hfq (then called host factor 1) in bacteriophage Qβ replication and limited roles of ribosomal proteins in translation control (1, 2). Hfq is now recognized as an RNA chaperone that promotes the interaction of numerous base-pairing small RNAs (sRNAs) with their mRNA targets and CsrA as a sequence-specific RNA binding protein, both of which globally influence gene expression and virulence (3–8). Other posttranscriptional regulators also participate in bacterial virulence networks, including RNA helicases, ribonucleases, and the Crc protein of pseudomonads (9–15).
The csrA (carbon storage regulator A) gene was originally uncovered by a transposon mutagenesis screen that was designed to identify regulators of gene expression in the stationary phase of growth. The csrA mutation had pleiotropic effects on genes involved in carbon flux pathways and phenotypes, including cell morphology and surface adhesion (16, 17). A CsrA homolog (95% identity) called RsmA (repressor of secondary metabolites) was later identified in Pectobacterium carotovorum and shown to repress pathogenicity in plant hosts through effects on extracellular lytic enzymes and quorum sensing (18, 19). CsrA homologs have since been studied in a variety of animal and plant pathogens. In addition to controlling metabolism and fundamental physiological properties, CsrA is critical for the regulation of dedicated virulence systems required for host infection.
This review addresses the workings of the Csr system, including the structure and function of the CsrA protein, the Csr sRNAs that introduced the concept of molecular mimicry as a strategy for riboregulation, and the global role of this system in regulating gene expression. A discussion of the Csr regulatory circuits is provided for a number of important pathogens, including Escherichia coli, Pseudomonas spp., Legionella pneumophila, and Vibrio cholerae. These sections will highlight (i) important regulatory targets of CsrA/RsmA pertinent to virulence, (ii) environmental factors and regulatory circuits that control CsrA/RsmA activity, and (iii) features of CsrA/RsmA regulatory circuitry that are unique for each pathogen. The contributions of CsrA to virulence in Salmonella spp., Yersinia spp., pathogenic E. coli strains, and plant pathogens, as well as its regulatory role in beneficial biocontrol species, will also be discussed. Finally, we address recent studies on the regulation of CsrA by an anti-CsrA protein (FliW) in the Gram-positive bacterium Bacillus subtilis and the implications of these findings for bacterial pathogens. A summary of Csr (Rsm) systems is presented in Table 1.
TABLE 1.
Overview of Csr (Rsm) systems
| Organism | Homolog | TCS(s) | Inhibitory RNA(s) | CsrD (MshH) | Direct target(s) (reference)a | Regulated phenotype(s)c |
|---|---|---|---|---|---|---|
| E. coli | CsrA | BarA/UvrY | CsrB, CsrC, McaS | + | glgCAP (21), sdiA (33), pgaABCD (29), nhaR (34), cstA (28), csrA (32), flhDC (46, 47), LEE4 locus (312), grlRA (312) | Motility, biofilm formation, glycogen metabolism, T3SS |
| S. Typhimurium | CsrA | BarA/SirA | CsrB, CsrC, fimAICDHF | + | hilD (269) fimAICDHF (285), pefACDEF (285), STM1987, STM3375, STM3611, STM1344, STM1697 (84) | Motility, SPI1 T3SS, biofilm formation |
| P. aeruginosa | RsmA | GacS/GacA | RsmY, RsmZ | − | psl (40), tssA1 (80), fha1 (80), phz2 (45), rahU, algU, pqsR, PA1300 (56), magA (80) | T3SS, T6SS, quorum sensing, biofilm formation, type IV pili |
| RsmF (RsmN) | ? | ? | − | tssA1 (157) | T6SS, biofilm formation | |
| P. fluorescens | RsmA | GacS/GacA | RsmX, RsmY, RsmZ | − | hcnA (167) | Motility, secreted biocontrol factors |
| RsmE | GacS/GacA | RsmX, RsmY, RsmZ | − | hcnA (167) | Motility, secreted biocontrol factors | |
| L. pneumophila | CsrA | LetS/LetA | RsmX, RsmY, RsmZ | − | Motility, T4SS, pigment production | |
| V. cholerae | CsrA | VarS/VarA | CsrB, CsrC, CsrD | + | Quorum sensing, biofilm formation | |
| Y. pseudotuberculosis | CsrA | BarA/UvrY, PhoQ/PhoP | CsrB, CsrC | + | flhDC (333) | Invasin production |
| P. carotovorum | RsmA | GacS (ExpS)/GacA (ExpA) | RsmB | + | Cell wall-degrading exoenzymes | |
| Xanthomonas spp. | RsmA | ? | ? | − | hrpG, hrpD (100) | Motility and T3SS, exoenzymes, EPS |
| P. syringae | RsmA | GacS/GacA | RsmB, RsmX, RsmY, RsmZ | − | Motility, secreted virulence factors | |
| B. subtilis | CsrA | ? | ?b | − | hag (381) | Motility |
Direct targets, discussed in this review, were demonstrated by in vitro binding assays.
The CsrB gene was predicted via bioinformatics approaches, but it has not been tested (55).
The list of regulated phenotypes is not all-inclusive and is meant to suggest the conservation as well as breadth of regulated processes.
CsrA/RsmA: Structure and Function
Because the amino acid sequence of CsrA was unrelated to known proteins, its regulatory mechanism was originally unclear (17, 20). Studies on its regulatory target glgC, which encodes the glycogen biosynthetic enzyme ADP-glucose pyrophosphorylase, allowed its mode of action to be determined. Surprisingly, CsrA regulation of a glgC′-′lacZ fusion did not depend on the presence of a native glgC promoter and required a region that overlapped the glgC ribosome binding site (RBS) (20). These results suggested that CsrA may control gene expression at the posttranscriptional level. Consistent with this hypothesis, glgC mRNA was destabilized by CsrA (20). It was later determined that CsrA binding to the glgC mRNA leader blocks ribosome access, suggesting that its effect on transcript stability in this case may be secondary to its effect on translation (21).
Subsequent to those seminal discoveries, the understanding of CsrA structure and function has advanced dramatically. Structural studies of E. coli CsrA and its homolog, Yersinia enterocolitica RsmA, revealed a dimer of identical subunits, each containing 5 tandem β-strands, a short α-helix, and a flexible C terminus (Fig. 1) (22, 23). The β-strands of the two polypeptide monomers are interwoven to form a hydrophobic core, and two C-terminal wing-like α-helices extend away from the protein. A comprehensive alanine-scanning mutagenesis study by Mercante et al. revealed that two identical surfaces of E. coli CsrA mediate RNA binding and regulation, which are formed primarily by the parallel β1 and β5 strands of opposite polypeptides (24). Structural analyses of CsrA-RNA complexes later confirmed that the amino acid residues that are critical for regulation interact directly with bound RNA (25). In addition, these structural studies revealed contacts between the polypeptide backbone and RNA that were not identified by alanine-scanning mutagenesis (24, 25).
FIG 1.
Structure of CsrA/RsmA/RsmE orthologs and the RsmE-hcnA RNA complex. (A) Protein secondary structure is shown at the top, with β-strands and α-helices depicted as arrows and cylinders, respectively, and amino acid sequence comparisons are shown immediately below. Sequence alignments from the Gammaproteobacteria (top) and from species containing the csrA gene in proximity to fliW (bottom) are depicted. Red boxes indicate the locations of conserved residues that are important for RNA binding and in vivo regulation in E. coli (24). (B) Ribbon diagram of the RsmE dimer (P. fluorescens) in a 1:2 complex with a 20-nucleotide segment of hcnA RNA. Individual RsmE polypeptides are colored with green or cyan, and the hcnA ribose-phosphate backbone is shown in orange. The critical R44 and L4 (red boxes) residues of RsmE and the GGA (blue boxes) recognition motif present on each hcnA molecule are indicated. The PDB structure file for RsmE-hcnA (2JPP) was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/), and the protein structure was rendered using PyMOL. (Adapted from reference 25 by permission from Macmillan Publishers Ltd., copyright 2007.) (C) Two-dimensional interpretation of the interaction of RsmE with hcnA RNA. Amino acid residues contributed by an individual RsmE polypeptide are shown in green or cyan. Hydrogen bonds and hydrophobic interactions are indicated with dashed blue lines and orange highlights, respectively. The effects of alanine substitution on in vivo regulatory activities (% of wild type [WT], top) and in vitro binding affinities (Kd, bottom) for the E. coli CsrA protein were determined by Mercante et al. (24). Asterisks indicate amino acid positions that differ between E. coli CsrA and P. fluorescens RsmE. (Adapted from reference 25 by permission from Macmillan Publishers Ltd., copyright 2007.)
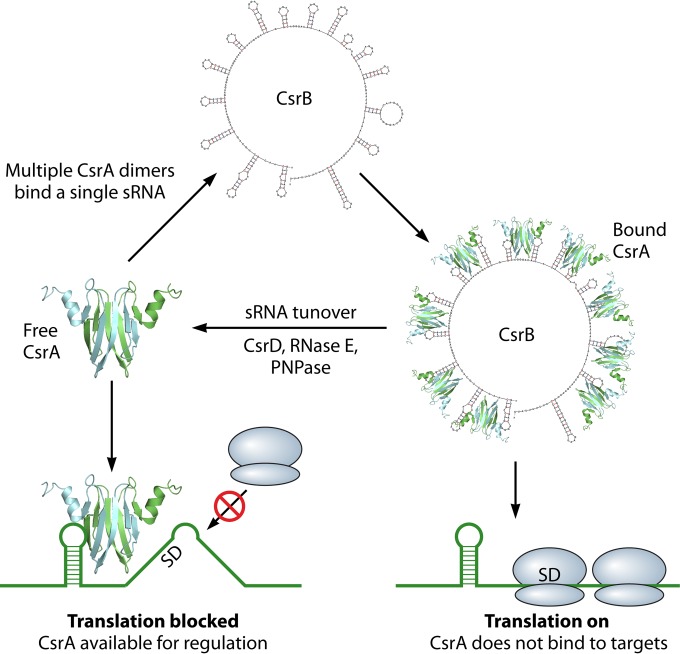
The features of RNA that are recognized by CsrA were first suggested following the discovery of the CsrA-inhibitory sRNA CsrB. CsrB unexpectedly copurified in a large globular complex with CsrA and was found to compete with lower-affinity mRNA targets for CsrA occupancy (26, 27). Finding CsrB was fortuitous not only because of its importance as a regulator of the Csr system (discussed below) but also because it suggested the RNA sequence and structure elements of the CsrA binding site. A stochastic RNA folding algorithm predicted that CsrB contains 15 RNA hairpin structures, most of which possess a GGA motif located in a single-stranded loop with conserved flanking sequences, predominantly CAGGA(U/A/C)G (26). Single-stranded GGA sequence motifs also typify the CsrA binding sites of mRNA untranslated leader segments (21, 28–35). This conserved sequence resembles the Shine-Dalgarno (SD) sequence of ribosome binding sites, to which CsrA often binds in translation repression mechanisms.
To further define the CsrA binding site, systematic evolution of ligands by exponential enrichment (SELEX) experiments were performed to identify RNA ligands that contain high-affinity CsrA binding sites from a pool of random RNAs (36). A total of 55 selected RNA ligands were analyzed, 100% of which contained either one (50 RNAs) or two (5 RNAs) GGA motifs. A majority (92%) of the RNAs (51 RNAs) were predicted to contain GGA motifs within the loop of an RNA hairpin, and in fact mutations that disrupted the hairpin reduced CsrA binding affinity. The SELEX-derived consensus was determined to be RUACARGGAUGU, with the ACA and GGA motifs being 100% conserved. Recently, solution nuclear magnetic resonance (NMR) and other studies of the CsrA homolog RsmE binding to the sRNA RsmZ or short oligonucleotides derived from RsmZ have provided additional evidence that the sequence and structural context in which the GGA motif resides determine the binding affinity (37).
Since CsrA exists as a symmetrical dimer with two RNA binding surfaces, it was predicted to be able to bridge two sites on a single RNA molecule. Mutant CsrA dimers containing an alteration in one or both RNA binding surfaces were tested for binding to a series of model RNAs, revealing that the homodimer is able to bridge two binding sites separated by 10 to 63 nucleotides (nt) (38). This intersite bridging mechanism plays a regulatory role in glgC expression, where binding of a CsrA homodimer to a high-affinity site upstream of the RBS apparently allows CsrA to bridge to a lower-affinity site that overlaps the RBS, thus inhibiting translation (Fig. 2) (38). Other examples of CsrA function may not involve bridged CsrA binding sites (30). Recent structural studies confirmed dual-site binding and further revealed that CsrA/RsmA dimers assemble sequentially onto the binding sites of regulatory sRNAs, as opposed to binding to them randomly (37, 39).
FIG 2.
Models for repression and activation by CsrA/RsmA. (A) E. coli CsrA represses glgC translation by competing with ribosome (30S) binding. (Top) CsrA homodimer first binds to a high-affinity site present in the single-stranded region of an RNA hairpin, located in the 5′ untranslated leader of the glgC transcript (20, 21, 38). The tethered CsrA homodimer can then bind via its available RNA binding surface to a low-affinity site that overlaps the SD sequence, thus blocking ribosome binding. (Bottom) In the absence of free CsrA, the ribosome can bind to the SD sequence, and translation can proceed. (B) P. aeruginosa RsmA represses translation of psl by stabilizing a stem-loop structure that sequesters the RBS. (Top) RsmA can bind to a single site (GGA) present in the 5′ untranslated leader of the psl transcript (40). RsmA binding stabilizes an RNA hairpin formed between the SD and anti-SD sequences, thus blocking ribosome access. (Bottom) In the absence of RsmA, the predicted hairpin structure is unstable, and ribosome binding and translation can proceed. (C) E. coli CsrA binding promotes Rho-dependent transcription termination of pgaA. (Top) CsrA binds to six sites in the pgaA mRNA (29), two of which are located in a segment that forms a hairpin in the absence of CsrA (53). CsrA binding prevents hairpin formation and exposes rut sites for entry of Rho transcription termination protein. Rho binding leads to premature termination (dashed line) of pgaA transcription. (Bottom) In the absence of CsrA, rut sites are shielded by RNA base pairing, Rho is unable to bind pgaA mRNA, and transcription can proceed. (D) P. aeruginosa RsmA binding to the phz2 untranslated leader prevents the formation of secondary structure that blocks translation (45). (E) E. coli activates translation of moaA by altering RNA structure. (Top) CsrA binds to two sites (GGA) present within the predicted moaA riboswitch aptamer that overlaps the SD (35). CsrA binding alters the aptamer structure, which reveals the SD for ribosome binding. (Bottom) In the absence of CsrA, the riboswitch aptamer sequesters the SD, thus blocking ribosome access. (F) E. coli CsrA stabilizes flhDC by preventing endonuclease cleavage by RNase E. (Top) CsrA binds to sites (GGA) present in the single-stranded region of two RNA hairpins located at the 5′ end of flhDC (47). CsrA binding prevents 5′ end-dependent cleavage by RNase E. (Bottom) In the absence of CsrA, RNase E binds to the 5′ monophosphorylated end of flhDC and performs several cleavages that initiate turnover of the transcript (47).
Dynamic Effects of CsrA on Translation, RNA Turnover, and Transcription Termination
As discussed above, CsrA represses translation of glgC by directly occluding the RBS (Fig. 2), and similar translation repression mechanisms have been described for sdiA, nhaR, cstA, and other genes of E. coli and other organisms (25, 33, 34). Studies by Irie et al. revealed that CsrA can also repress translation by stabilizing the formation of RNA secondary structure that blocks translation (40) (Fig. 2). The following discussion addresses recently described mechanisms by which CsrA regulates translation, RNA stability, or Rho-mediated transcription termination.
Riboswitches are cis-acting RNA elements in which the binding of a small molecule ligand within a folded RNA aptamer regulates gene expression (41, 42). A class of riboswitches was recently proposed to bind the molybdenum cofactor (Moco), which serves as a redox center for many metabolic enzymes (43). E. coli moaABCDE mRNA is required for Moco biosynthesis and contains a prototypical riboswitch element from this family, which is negatively regulated by Moco. Studies by Patterson-Fortin et al. showed that CsrA binds to the 5′ untranslated leader of moaA at two sites within its proposed aptamer domain and activates translation, likely by increasing ribosome accessibility (Fig. 2) (35). These studies introduced a new model for riboswitch function in which two different factors interact with an RNA aptamer to regulate expression.
Pseudomonads synthesize phenazine compounds that serve as precursors of secreted factors that affect biocontrol and virulence pathways (44). P. aeruginosa contains two phenazine biosynthetic gene clusters referred to as phz1 and phz2. Ren et al. proposed a model in which RsmA posttranscriptionally activates expression of phz2 by binding to the leader transcript and disrupting an RNA structure that would otherwise block the RBS (Fig. 2) (45). Though this model has yet to be demonstrated in vitro, it is supported by RNA structure predictions and in vivo expression studies with wild-type and mutant phz2 leaders. Similar to CsrA regulation of moaA expression in E. coli, this is an example of translational activation and is related to repression mechanisms wherein CsrA mediates accessibility of the RBS by modifying RNA structure (40).
Several regulatory circuits converge on the master operon for flagellum biosynthesis, flhDC, to regulate motility in E. coli. Wei et al. demonstrated that CsrA activates expression of flhDC by binding to this mRNA and stabilizing it (46). Yakhnin et al. later identified the molecular mechanism of CsrA-mediated activation; CsrA binds to two sites within the flhDC RNA leader, which prevents 5′ end-dependent cleavage of this transcript by RNase E (Fig. 2) (47). While effects on translation often cause secondary effects on transcript stability (48–50), in this case, regulation of flhDC stability is a primary consequence of CsrA binding (Fig. 2).
The pgaABCD operon of E. coli is required for the synthesis and secretion of a biofilm polysaccharide adhesin (51, 52), and CsrA binding to pgaA mRNA represses ribosome binding to and translation of this transcript (29). In addition to this translation repression mechanism, Figueroa-Bossi et al. recently demonstrated that CsrA binding to the 5′ untranslated region (5′ UTR) of the pgaA transcript remodels its structure, unveiling binding sites for Rho protein, which causes premature termination of the elongating transcript (53, 54) (Fig. 2). This is the first example of CsrA/RsmA directly controlling transcription. How typical this mechanism is for CsrA function remains to be seen. Nevertheless, it is notable that the pgaABCD mRNA contains a long untranslated leader, with 6 CsrA binding sites, the largest number presently known for an mRNA and consistent with its complex regulation by CsrA (29).
Regulation of the Csr/Rsm System
sRNAs that sequester CsrA/RsmA proteins.
The molecular mimicry paradigm for Csr (Rsm) sRNA function appears to be conserved throughout the Gammaproteobacteria, and many bacterial species have multiple Csr sRNAs (3, 55, 56). The inhibitory sRNAs likely represent the primary means of controlling CsrA activity in the species that produce them, and in a few cases, their synthesis and stability have been studied (57–59). Under the growth conditions that have been tested, CsrB appears to be the principle antagonist of CsrA activity in E. coli (26, 60). Mutation of csrB affects the expression of downstream CsrA targets, and its absence causes pleiotropic effects on E. coli physiology (Fig. 3). E. coli also produces additional sRNAs that control CsrA activity, CsrC and McaS, and these sRNAs substantially antagonize CsrA activity when overproduced, or in the case of CsrC, its effects are evident in a ΔcsrB strain background (60, 61). Nevertheless, all three sRNAs bind to CsrA with high affinity in vitro. In principle, the presence of multiple negative regulators of CsrA might imply regulatory redundancy, designed to decrease intrinsic noise and increase the robustness of the Csr genetic circuitry (62), or it may also allow the cell to fine-tune CsrA activity in response to different environmental stresses and/or stimuli. Because growth conditions (carbon source) and regulatory factors (Crp, integration host factor [IHF], Hfq, etc.) differentially influence csrB, csrC, and mcaS expression (63–65), these sRNAs apparently have distinct regulatory roles.
FIG 3.
Outline of the Csr system in E. coli. CsrA binding to target mRNAs can have several regulatory outcomes: blocking translation initiation (as shown), stabilizing or destabilizing mRNA, or resulting in premature transcriptional termination. The concentration of free CsrA and therefore its regulatory activity depends on the levels of inhibitory small RNAs (CsrB is shown here). These sRNAs can bind to multiple CsrA dimers with high affinity and prevent them from binding target mRNAs. sRNA levels are regulated at the level of transcription (not shown) and turnover. Ribosomes are depicted in blue.
The BarA/UvrY TCS activates transcription of CsrA-inhibitory sRNAs.
The BarA/UvrY two-component system (TCS) (also referred to as GacS/GacA, VarS/VarA, ExpS/ExpA, LetS/LetA, and BarA/SirA in various species) is the primary regulator of csrB and csrC expression and is highly conserved throughout the Gammaproteobacteria (Table 1) (58, 66). BarA is a membrane-bound, tripartite histidine sensor kinase that uses an unusual phosphorelay mechanism to phosphorylate its cognate response regulator, UvrY (66). BarA activity is stimulated by short-chain fatty acids (SCFAs), the potency of which is negatively correlated with the aliphatic chain length (67, 68). The mechanistic basis of this stimulation remains to be determined. The SCFA acetate is abundant in the intestinal tracts of mammalian hosts, where it may serve as a potent activator of BarA during enteric colonization or infection (68). Alternatively, BarA-independent phosphorylation of UvrY has been observed to occur (both in vitro and in vivo) via acetyl-phosphate, a high-energy intermediate in the synthesis of acetate (68, 69). Though phosphorylation of UvrY and other response regulators by acetyl-phosphate has been observed in various bacterial species, the physiological relevance/significance of this observation is poorly understood (70–72).
UvrY is a member of the FixJ family of DNA binding proteins, which contain amino-terminal transceiver domains and carboxy-terminal DNA binding domains. UvrY directly binds to the csrB and csrC promoters in vitro (63, 73), yet the details of its transcription activation mechanism have not been determined. Though Csr/Rsm-inhibitory sRNAs of various species may bear little sequence identity outside the CsrA/RsmA binding sequences, their expression in most cases is highly dependent on the UvrY ortholog (73–79). Whether UvrY and its various orthologs have additional regulatory roles remains to be determined, although the GacA protein of P. aeruginosa was proposed only to regulate Rsm sRNA transcription (80).
Stability of CsrA-inhibitory sRNAs is tightly controlled in E. coli.
In a genetic screen for novel regulators of the Csr system, Suzuki et al. discovered CsrD, a protein that specifically targets CsrB and CsrC for RNase E-dependent turnover (57). CsrD-mediated turnover of CsrB/C indirectly regulates the expression of all CsrA target genes and processes that have been examined, including motility, glycogen biosynthesis, and biofilm production. The molecular mechanism by which CsrD facilitates RNase E-dependent cleavage of CsrB/C is presently unclear. CsrD is a membrane-bound protein (C. A. Vakulskas and T. Romeo, unpublished data) that contains GGDEF and EAL domains, which are normally associated with the synthesis and breakdown of the bacterial secondary messenger cyclic di-GMP (c-di-GMP), respectively. However, CsrD contains degenerate or, more aptly stated, evolved domains and does not synthesize or degrade this nucleotide. Other GGDEF or EAL domains of bacterial proteins have also evolved novel roles (81, 82). A CsrD homolog in Vibrio cholerae, MshH, was found to interact with the glucose-specific enzyme IIA (EIIAGlc) of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS), which is a central regulator of carbon metabolism (83). Whether carbon flux regulates CsrB/C RNA turnover through this interaction and which CsrD domain(s) is responsible for binding to EIIAGlc remain to be determined. In E. coli and Salmonella enterica serovar Typhimurium, CsrA also represses the expression of csrD, suggestive of a negative feedback loop wherein CsrA indirectly stabilizes its sRNA antagonists, although the effect of CsrA on CsrB turnover seems to be negligible (57, 84).
CsrA apparently controls the stability of CsrA/RsmA-inhibitory sRNAs in Pseudomonas fluorescens through a mechanism that involves RsmA-dependent protection from RNase cleavage (59). Though the details of this mechanism are uncertain, a csrA mutation had little or no effect on the stability of CsrB/C sRNAs in E. coli (57). Perhaps this discrepancy is due to the presence of the decay specificity factor (CsrD) in E. coli, which is absent in the pseudomonads. A comparison of Csr/Rsm sRNA stability and the presence or absence of a csrD homolog in several bacterial genera is needed to further explore these relationships.
CsrA regulates its own expression.
Autoregulation is common for global regulatory proteins, and CsrA is no exception. CsrA binds to the untranslated leader of its mRNA and represses translation by competing with ribosome binding (32). CsrA concomitantly activates its transcription indirectly through a mechanism that involves the σS-dependent P3 promoter, one of five csrA promoters that use either σ70 or σS for transcription. The finding that CsrA simultaneously activates its own transcription and represses its translation highlights the exquisite level of control that is maintained in this system. The fact that translational repression can be enacted more rapidly than transcriptional activation may suggest the former as a mechanism to rapidly shut off CsrA synthesis when its activity reaches a critical level (32). Thus, the autoregulation of CsrA activity is extremely complex, involving positive and negative feedback loops in CsrA synthesis and negative feedback loops that control the levels of its sRNA antagonists.
Multiple regulatory circuits feed into the Csr system.
Other regulatory components besides BarA/UvrY control the levels of CsrB/C sRNAs, many of which function through or in conjunction with the BarA/UvrY TCS (Fig. 4). For example, CsrA indirectly activates the expression of its sRNA antagonists, CsrB/C, indicative of negative feedback regulation (32, 58, 60), and RsmA from Pseudomonas spp. similarly regulates expression of the sRNAs RsmX, RsmY, and RsmZ (85, 86). Epistasis analyses and other experiments indicate that these pathways involve UvrY (GacA). Though the available genetic data suggest that CsrA activates CsrB/C expression through the BarA/UvrY TCS, CsrA-dependent regulation of BarA/UvrY protein levels and/or phosphorylation state remains to be thoroughly examined. CsrA/RsmA activation of its antagonists may serve as a homeostatic mechanism that has evolved to minimize dramatic fluctuations in CsrA/RsmA activity under a given condition (3, 87).
FIG 4.
Csr regulatory circuitry in E. coli. The inner and outer membranes are indicated. Solid lines indicate regulatory connections with molecular mechanisms supported by experimental evidence, whereas dashed lines show regulatory effects for which a mechanism is lacking. Activation or repression is indicated by a black arrowhead or red T-bar, respectively. A phosphoryl group is indicated by “P.” See the accompanying text for details.
Integration host factor (IHF) binds to the promoter of the csrB gene of Salmonella enterica serovar Typhimurium and the rsmZ promoter from P. fluorescens (63, 74). Deletion of ihfA in S. Typhimurium severely decreases csrB expression, similar to the case for a uvrY deletion, although it does not affect csrC expression. IHF typically modifies DNA architecture, affecting DNA supercoiling, duplex stabilization, and global gene expression (88). It commonly regulates transcription from promoters that are directly regulated by multiple transcription factors. Perhaps it should be no surprise that UvrY and IHF function in concert to activate csrB expression or that csrB transcription is tightly controlled by specific (UvrY) and global (IHF) regulatory factors. Whether IHF activates csrB expression by a typical DNA bending mechanism and the physiological role of this regulation are open questions.
Recently, Vakulskas et al. determined that the DEAD box RNA helicases DeaD and SrmB activate CsrB/C expression in the exponential phase of growth (10). These bacterial proteins traditionally have been associated with rRNA maturation activities, particularly at low temperatures (89). DeaD was found to activate uvrY translation by overcoming the effect of a long-range inhibitory RNA structure that forms between the uvrY RNA leader and proximal coding sequence. Though the mechanism of SrmB function is not understood, it did not affect the levels or phosphorylation state of UvrY (10). Despite their established roles as cold shock proteins, DeaD and SrmB activated CsrB expression over a broad temperature range, indicative of wider physiological roles for these proteins than was previously appreciated. DeaD also regulated sirA (uvrY) expression in S. Typhimurium, and bioinformatics analysis suggested that this regulatory mechanism is conserved in most of the Enterobacteriaceae. The physiological purpose of this kind of regulation remains to be determined, but it may help to maintain adequate levels of uvrY expression as the capacity for protein synthesis in the cell declines (10), similar to the biological role of (p)ppGpp in CsrB/C expression (31).
Components of the stringent response, including the alarmone (p)ppGpp and the (p)ppGpp-responsive transcription regulator DksA, strongly activate CsrB and CsrC expression (31). Interestingly, CsrA represses the relA gene, which encodes (p)ppGpp synthase I, and (p)ppGpp levels are elevated during the stringent response in a csrA mutant background. These findings depict a reciprocal regulatory circuit wherein amino acid starvation activates CsrB/C expression, and the resulting reduction in CsrA activity increases (p)ppGpp production, thus strengthening the stringent response. The Csr system also reinforces effects of the stringent response in another way: genes that are known to be posttranscriptionally repressed (glgC) or activated (flhDC) by CsrA are transcriptionally activated or repressed by (p)ppGpp, respectively. Thus, when the stringent response is invoked, the direct regulation of these genes by (p)ppGpp is heightened by the decrease in CsrA activity that is caused by (p)ppGpp-mediated induction of CsrB and CsrC expression (31).
The RNA binding protein Hfq positively affects CsrB levels under certain growth conditions (57). Hfq typically mediates pairing of sRNA and mRNA molecules that share weak complementarity, affecting the stability of one or both RNAs and/or translation of the mRNA (90). Because Hfq did not affect stability of the sRNA CsrB, it likely activates csrB gene expression indirectly (57).
The Csr system captures the outputs of stress response systems and converges them into global regulation.
In various species, CsrA/RsmA regulates environmental stress responses and regulatory factors that mediate stress responses, including the stringent response (31), osmotic resistance (91), heat resistance (91), oxidative stress (92–94), the global stress sigma factors RpoS (94–96) and AlgU (97), and the RNA chaperone Hfq (30). Global studies of CsrA/RsmA target RNAs and transcriptome effects suggest that CsrA/RsmA regulates the expression of many additional stress response genes that have not been studied in detail (31, 56, 98–101). Furthermore, the levels of the CsrA/RsmA-inhibitory sRNAs are controlled by several regulators that mediate stress responses, including DeaD/SrmB (10), Hfq (57, 102), RpoS (32), AlgU (86), Crp (103), and (p)ppGpp/DksA (104–106). These diverse connections of the Csr system to stress responses, in which CsrA/RsmA activity is regulated by the effects of stress responses on Csr/Rsm sRNA levels and CsrA/RsmA regulates stress response genes, imply that Csr/Rsm systems are a centerpiece of global stress response circuitry. Apparently CsrA/RsmA, similar to RpoS, governs its regulon in response to diverse environmental stresses (3, 107). Just how extensively the Csr system serves to capture and channel the effects of stress responses into global posttranscriptional regulation is an important question that is worthy of additional study.
The Repertoire of Direct and Indirect CsrA Targets: the Csr Regulon
The effects of csrA deletion and overexpression have been studied in several species using microarray experiments, revealing a profound influence of CsrA/RsmA on the transcriptome. For example, Lawhon et al. demonstrated that in S. Typhimurium, a csrA::kan mutation causes ≥2-fold activation or repression of the steady-state levels of 365 different transcripts, including mRNAs critical for virulence factors such as motility and type III secretion systems (T3SS) (98). Microarray analysis of the P. aeruginosa transcriptome revealed that 9% of genes present on the array (506 of 5,570) were significantly affected by rsmA deletion, including those associated with virulence systems for iron acquisition, quorum-sensing, motility, and antibiotic resistance (99). Similar experiments have been performed in plant pathogens, Xanthomonas campestris pv. campestris, and Pectobacterium wasabiae, all of which demonstrated CsrA/RsmA-dependent effects on the expression of a large and diverse set of genes (100, 101). These studies greatly expanded the potential roles of the Csr/Rsm system but were unable to distinguish between direct and indirect regulatory targets.
In an attempt to identify the direct targets of CsrA, Jonas et al. pulse-induced CsrA expression in E. coli and monitored changes in mRNA levels as a function of time (108). The rationale was that the direct targets of CsrA would show changes in gene expression more rapidly than indirect targets, which was confirmed using known direct (pgaA) (29) and indirect (csrB) (58, 87) targets. Though this method was limited by the fact that CsrA is capable of regulating translation independently of effects on mRNA transcription and/or stability, this set of experiments revealed interesting new direct targets of CsrA, including the c-di-GMP signaling proteins YcdT and YdeH (108).
In an approach designed to identify binding targets of P. aeruginosa RsmA, Brencic and Lory coprecipitated RNA with RsmA, cloned cDNAs derived from the resulting RNA pool, and sequenced a number of these clones (80). Altogether, 40 likely direct targets of RsmA were identified, including genes involved in T6SS, fatty acid metabolism, cell division, and proteolysis. More recently, Edwards et al. introduced the use of high-throughput RNA sequencing (RNA-seq) to analyze RNA that copurified with E. coli CsrA (31). This study identified 721 transcripts that copurified with CsrA, representing genes with extremely diverse functions. These studies to identify the direct RNA targets of CsrA, in concert with observations that CsrA/RsmA serves as a regulator of many other regulators and has remarkable effects on bacterial physiology and virulence, confirm the critical role that the Csr system plays in bacterial gene expression. Current state-of-the-art approaches for studying RNA binding proteins can be used to identify the complete set of bound RNAs as well as the minimum region that is protected by a bound protein (10, 109–111).
Csr/Rsm REGULATE VIRULENCE LIFESTYLES OF GAMMAPROTEOBACTERIAL PATHOGENS
The Pseudomonads
Opportunistic pathogens of the Pseudomonadaceae reside primarily in soil and aquatic environments and are known to infect a broad range of hosts using a vast arsenal of secreted factors, including protein toxins and secondary metabolites with antihost properties. P. aeruginosa is the most prevalent human pathogen of this group and a major cause of morbidity and mortality in immunocompromised, burned, or wounded patients, as well as debilitating and difficult-to-treat chronic lung infections of cystic fibrosis (CF) patients (112–114). Although P. fluorescens is a biocontrol agent that protects plants against fungal pathogens, it is included here because it has served as an important model for studies of Pseudomonas Csr/Rsm systems (115–117). Studies have focused on regulation of the virulence and biocontrol systems of each organism. In P. fluorescens, this includes genes involved in the production and secretion of the antimicrobial factors alkaline metalloprotease A, hydrogen cyanide, and 2,4-diacetylphloroglucinol (59, 118, 119). In P. aeruginosa, RsmA studies have focused on genes involved in T3SS, T6SS, motility, quorum sensing, and biofilm formation (80, 85, 112, 120, 121). Pseudomonas syringae strains cause many important diseases of higher plants and will be discussed below along with the other plant pathogens.
P. aeruginosa: RsmA regulates the switch between acute and chronic infection.
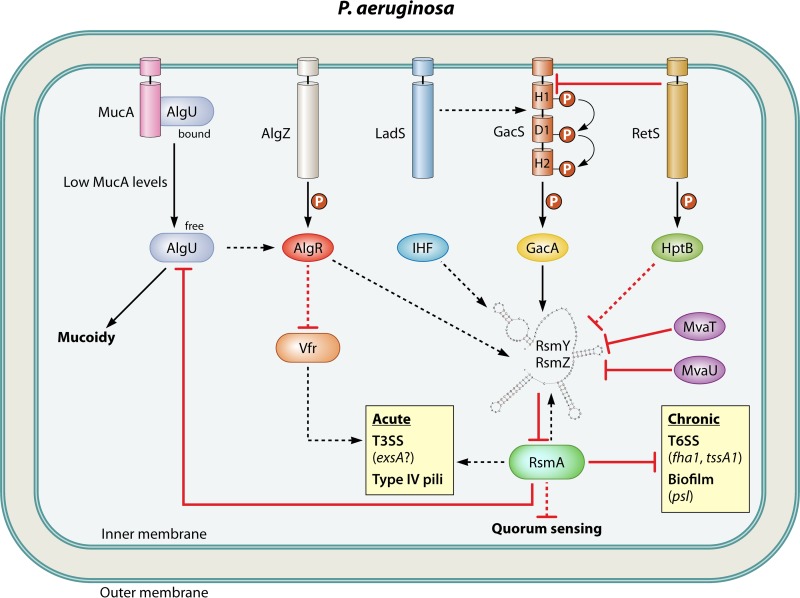
P. aeruginosa causes acute respiratory infections that frequently become chronic in patients with the genetic disorder cystic fibrosis (CF) (122, 123). Patients suffer recurring episodes of acute P. aeruginosa infection that can become septic and are nearly impossible to eradicate with antibiotic therapy (124). The switch between acute and chronic infection is characterized by major changes in virulence factor gene expression, which RsmA mediates by positively regulating factors associated with acute virulence, including T3SS and type IV pili, and negatively regulating factors associated with chronic virulence, including biofilm formation and a T6SS (Fig. 5) (113, 125). In a mouse model of acute P. aeruginosa infection, rsmA mutation significantly reduced colonization during the initial stages of infection, but the loss of rsmA ultimately favored persistent infection (112). RsmA also represses the expression of the acyl homoserine lactone (AHL)-producing quorum-sensing systems las and rhl in P. aeruginosa; however, quorum-sensing signals have complex and seemingly conflicting roles in the regulation of acute and chronic virulence phenotypes (126).
FIG 5.
Rsm regulatory circuitry in P. aeruginosa, depicted as in Fig. 4.
Expression of T3SS genes of P. aeruginosa is tightly controlled by a partner-switching mechanism involving a cascade of regulatory proteins called ExsA, ExsC, ExsD, and ExsE (127). Transcription of T3SS-associated genes is intimately coupled to the state of the T3SS secretory apparatus by the small, T3SS-secreted regulatory factor ExsE (128). In the absence of inducing conditions (high Ca2+ or host cell contact) the secretion channel is closed, and ExsE remains cytoplasmic and bound to its chaperone, ExsC. Cytoplasmic ExsE sequesters ExsC away from its other binding partner, ExsD. ExsD is then free to bind ExsA and inhibit its DNA binding ability. ExsA is the master transcriptional activator of all T3SS genes. In the presence of induction, the secretory apparatus is open and ExsE is secreted or translocated into host cells, ExsC binds to and sequesters ExsD, and ExsA is then free to bind T3SS-associated promoters. ExsA activates transcription by recruiting RNA polymerase and promoting open complex formation through a direct interaction with σ70 (129–134).
Microarray experiments by Brencic et al. determined that rsmA mutation reduces the expression of 22 T3SS-associated genes, including representatives from genes that facilitate secretion, translocation, regulation, and every exotoxin secreted by P. aeruginosa strain PAK (80, 133). These results implied that a T3SS-associated transcription factor was likely a target for RsmA regulation. Consistent with this hypothesis, RsmA regulated the T3SS by activating exsA expression (80, 86). Whether RsmA directly binds to exsA mRNA and how it controls ExsA protein levels is presently unknown. While this RsmA activation pathway serves as a regulatory output for the GacA/GacS TCS to control the T3SS, in the absence of a known environmental stimulus for GacS, the physiological importance of this regulation remains unclear. Other studies by Intile et al. (discussed below) suggested that P. aeruginosa isolates from chronic CF patients may lack a functional T3SS in part due to disruption of RsmA-ExsA regulation (86).
Type IV pili are fiber-like protein structures present on the surfaces of a wide variety of bacteria and archaea that are utilized for activities ranging from the uptake of DNA to electron transport (134). In P. aeruginosa, type IV pili have known roles in adhesion to host cells, biofilm formation, twitching motility, host immune system evasion, and phage transduction and are critical for establishing acute infection (80, 125, 135, 136). Studies by Brencic and Lory showed that RsmA activates expression of 9 genes involved in type IV pilus biogenesis (80). RsmA-dependent activation of the pilMNOPQ operon was observed using transcriptional fusions to lacZ, but no effect was observed using translational fusions to lacZ driven by a constitutive promoter (also called a leader fusion) (80). These results indicated that most of these effects likely occur indirectly through other factors. AlgR is a known regulator of type IV pilus gene expression; however, it was not identified in the microarray studies (80, 137). The magnitude of RsmA's effect on type IV pilus gene expression (∼2 to 4-fold) pales in comparison to its effects on T3SS gene expression (∼3- to 25-fold), suggesting that RsmA plays a more nuanced role in controlling type IV pilus production. Additional experiments to determine the effects of RsmA on type IV pilus-dependent phenotypes, such as twitching motility, adhesion, biofilm formation, and pathogenicity, are still needed.
Patients with chronic P. aeruginosa infections preferentially express virulence factors that increase persistence and immune system evasion (138–142). Examples of RsmA-regulated virulence factors associated with chronic disease include the ability to form biofilm and the type VI secretion system (T6SS), described below (80, 143, 144). P. aeruginosa strains secrete one or more polysaccharides that contribute to the biofilm extracellular matrix, namely, alginate, Pel, and/or Psl (145). Whereas alginate is a uronic acid polymer that is produced by mucoid strains commonly found in patients with late-stage CF infections (146, 147), Pel and Psl are complex biofilm polysaccharides produced by nonmucoid strains from early- to mid-stage CF infections (148, 149). The Psl polysaccharide is a repeating pentamer of d-mannose, l-rhamnose, and d-glucose. Psl surrounds the cell in a helical distribution and is thought to play a role in facilitating cell-cell and cell-surface interactions during biofilm formation (145, 150). Irie et al. demonstrated that deletion of rsmA resulted in high biofilm formation and increased levels of psl mRNA (40). RsmA binds to the 5′ UTR of psl mRNA and facilitates the formation of a stem-loop structure that blocks the ribosome binding site. The Pel polysaccharide forms a fabric-like matrix that connects cells at the air-liquid interface (145, 148). Microarray experiments indicate that RsmA represses the expression of genes involved in Pel polysaccharide biosynthesis (80), though it is presently unclear whether this regulation occurs directly or indirectly.
The T6SS is a specialized secretion system and was originally thought to secrete/translocate only toxic products into host eukaryotic host cells (151–155). However, it is now known to also mediate interbacterial interactions by directly injecting proteins into the cytoplasm of neighboring bacterial cells (156). RsmA-dependent control of the T6SS in P. aeruginosa was suggested by results from microarray studies comparing gene expression in wild-type and rsmA mutant strains (80). Furthermore, RsmA repressed the expression of at least 12 T6SS-associated genes and bound directly to tssA1 and fha1 mRNAs (80, 157). Interestingly, both RsmA and its paralog RsmF (described below) bind the tssA1 mRNA in vitro, and both proteins repress tssA1 expression in vivo (157). RsmF binding to fha1 was not examined in these studies. Both tssA1 and fha1 encode structural components of the T6SS (158). Further studies are needed to better define the mechanism(s) by which RsmA and RsmF repress T6SS and the role(s) played by T6SS in chronic infection.
P. fluorescens: RsmA regulates the expression of secreted biocontrol factors.
P. fluorescens is a soilborne bacterium that protects plant roots from fungal and nematode diseases. P. fluorescens induces systemic resistance phenotypes in plants and secretes lytic enzymes (phospholipase C, tryptophan side chain oxidase, and exoprotease) and antimicrobial compounds (pyoluteorin and 2,4-diacetylphloroglucinol) (118, 159–164). The GacS/GacA (BarA/UvrY) TCS controls the expression of genes involved in the production/secretion of these factors, including the genes for 2,4-diacetylphloroglucinol biosynthesis, phlACBDE, the major exoprotease, aprA, and hydrogen cyanide synthase, hcnABC (119, 160, 163–165). In the closely related pathogen P. aeruginosa, GacA activates the expression of RsmY and RsmZ sRNAs, and GacA from P. fluorescens follows a similar trend via RsmX, RsmY, and RsmZ (74, 76). Therefore, some or all of the GacA effects on secreted biocontrol factors probably occur through activation of RsmX/Y/Z expression and, hence, inhibition of RsmA activity. Consistent with this hypothesis, deletion of either gacA or the rsmXYZ sRNA genes reduces hydrogen cyanide secretion, 2,4-diacetylphloroglucinol secretion, exoprotease activity, and swarming motility to a similar extent (166). Lapouge et al. demonstrated that RsmA directly bound to the hcnA mRNA at three sites within the untranslated leader using RNA footprinting and toeprinting techniques (167).
RsmA negatively regulates the production of quorum-sensing AHLs in Pseudomonas spp.
P. aeruginosa encodes at least two different AHL pathways, the rhl and las pathways, which collectively affect the expression of approximately 10% of the genome (168). The production of AHL-dependent exoproducts in P. aeruginosa, including exoenzymes, pyocyanin, and hydrogen cyanide, is repressed by RsmA (169). Consistent with this finding, RsmA negatively affected the production of both las and rhl AHLs. Nevertheless, the addition of exogenous AHLs did not fully restore exoproduct production in a strain that overproduces RsmA, which suggested that RsmA may affect exoproduct production at multiple levels. For example, RsmA regulates the P. aeruginosa hydrogen cyanide synthesis transcript hcnABC directly (169). It was later confirmed that RsmY and RsmZ positively affect the production of quorum-sensing AHLs and exoproducts, consistent with their roles as negative regulators of RsmA (85). The generally accepted role for RsmA in P. aeruginosa pathogenesis is that it activates gene expression of virulence factors associated with acute infection (T3SS and motility) and that it represses the expression of virulence factors associated with chronic infection (T6SS and biofilm). Quorum-sensing systems are important for both acute and chronic infections, where they regulate secreted bacterial virulence factors and the host immune response (170). Additional studies are needed to fully understand how RsmA-mediated repression of quorum-sensing systems fits into the chronic/acute paradigm for RsmA regulation or if this model is an oversimplification of its regulatory responsibilities.
In P. fluorescens strain CHA0, which lacks quorum-sensing mechanisms controlled by AHLs and other known autoinducers (85, 171), a solvent-extractable, cell population density-dependent signal activates RsmX/Y/Z expression. The chemical nature of this signal is not known; however, it did not appear to be related to AHLs or other known signaling molecules (172). Carboxylate compounds with short aliphatic tails stimulate BarA activity in E. coli, and Takeuchi et al. recently demonstrated that increased cellular pools of the carboxylate compounds succinate, malate, and 2-oxoglutarate correlated with an increase in RsmXYZ sRNA levels in P. fluorescens (67, 173). While an uncharacterized quorum-sensing chemical could certainly be influencing GacS/GacA function, these findings might also be explained by changes in carboxylate-containing metabolites that fluctuate throughout growth (174).
CsrA paralogs of Pseudomonas spp.: RsmA, RsmE, and RsmF.
Pseudomonads contain RsmA paralogs that appear to have redundant regulatory roles in P. fluorescens (RsmA and RsmE) and both redundant and unique regulatory roles in P. aeruginosa (RsmA and RsmF). The RsmA and RsmE proteins from P. fluorescens share 71% amino acid identity, and mutations in the rsmA and rsmE genes have similar effects on repression of the target mRNAs hcnA, aprA, and phlA and activation of the inhibitory sRNAs rsmY and rsmZ (59). Both RsmA and RsmE bind to the RsmY/RsmZ sRNAs in vitro with similar affinity/specificity and antagonize the regulatory effects of these proteins similarly in vivo. Though the functions of these two proteins are remarkably similar, their regulation appears to be different. RsmA levels modestly increase throughout growth, while RsmE levels peak sharply in the stationary phase. Regulation of rsmE expression appears to occur at least in part due to repression by both RsmA and RsmE (59). At present, regulators that control RsmA levels in P. fluorescens have yet to be reported. While future experiments might reveal regulatory targets exclusive to RsmA or RsmE, the available data suggest that the role of RsmE is to reinforce RsmA function in the stationary phase of growth.
In contrast, RsmA and RsmF (RsmN) in P. aeruginosa share only 31% amino acid identity and have both shared and exclusive RNA targets (157, 175). The rsmF deletion strain appears phenotypically virtually indistinguishable from wild-type cells when comparing the expression of known RsmA targets, including genes involved in biofilm, T3SS, and T6SS. However, deletion of both rsmA and rsmF greatly exacerbated these effects of an rsmA deletion, implying a supportive role for the RsmF protein. Unlike RsmA/RsmE from P. fluorescens, however, RsmF binds to the RsmY and RsmZ sRNAs with 245- and 58-fold-lower affinity (Keq) than RsmA, and predictably, neither sRNA appeared to regulate RsmF activity in vivo. Furthermore, RsmA but not RsmF binds to the pslA mRNA with high affinity and specificity. How these differences in the Csr/Rsm paralogs favor the unique lifestyles of these two species remains to be seen. Studies are needed to determine whether RsmF and RsmA are expressed at different phases of growth or under different growth conditions.
The structure of RsmF has been solved by X-ray crystallography, and although the RNA binding surfaces are similar to those of CsrA and RsmE, the overall dimer structure is distinctly different (Fig. 6). For example, RsmF is a homodimer that contains antiparallel β-sheets formed by β1, β3, and β4 from one polypeptide and β2 and β5 from the opposite polypeptide (157, 175). This is in contrast to the case for CsrA/RsmA/RsmE, wherein β-sheets are formed by β2, β3, and β4 from one polypeptide and β1 and β5 from the opposite polypeptide. Furthermore, whereas the C-terminal α-helices of CsrA/RsmA/RsmE form wing-like structures that extend away from the body of the protein, the α-helices encoded by RsmF are located between β-strands β2 and β3 and form a solvent-exposed, unique interaction surface.
FIG 6.
Structural comparison between RsmE of P. fluorescens and RsmF (RsmN) of P. aeruginosa. (A and C) Cartoons depicting the NMR solution structure of RsmE and crystal structure of RsmF (157, 175). Individual polypeptides of the RsmE homodimer are shown in green or cyan, RsmF polypeptides are shown in blue or maroon, and secondary structural elements are labeled with subscript “A” or “B,” as shown. (B and D) Structures of RsmE (top) and RsmF (RsmN) (bottom) rotated 90°. The β-strands that form the RNA interaction surfaces are highlighted using the coloring scheme from panels A and C. Protein structures were rendered using PyMOL. The PDB structure files for RsmE-hcnA (2JPP) and RsmF (4K59) were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/).
The crystal structure of RsmF bound to a hairpin from the RsmZ sRNA has been solved, revealing that, in contrast to the case for CsrA and RsmE (25), where each RNA binding surface of the dimer is comprised of two β-strands contributed by opposite polypeptides, the RsmF RNA binding surface is composed of two β-strands originating from the same polypeptide (Fig. 6) (175). How do these topological differences in the RsmE and RsmF proteins explain how both proteins bind to a single RsmZ hairpin with similar affinities (175), yet RsmA binds the complete RsmZ RNA with 58-fold-higher affinity than does RsmF (157)? Perhaps the difference in the proximity of RNA binding surfaces in RsmF constrains the ability of this protein to bridge two binding sites within an RNA target relative to CsrA/RsmE (38), especially a target such as RsmZ that contains multiple closely spaced binding sites (39).
The RsmA/RsmE-inhibitory sRNAs.
P. aeruginosa and P. fluorescens have different pairs of CsrA-like proteins, as well as a different set of inhibitory sRNAs. Both organisms produce the sRNAs RsmY and RsmZ, and P. fluorescens also produces a third sRNA called RsmX. All of these sRNAs antagonize the respective RsmA protein in vivo (59, 85, 166). It is important to note that RsmA-inhibitory sRNAs from pseudomonads differ greatly from those of E. coli and Salmonella spp. in length, number of predicted CsrA/RsmA binding sites, and stability (57, 59, 176). The CsrB RNA from E. coli, for example, is 369 nt in length, possesses 18 to 22 predicted CsrA binding sites, and has a half-life of 1.7 min (26, 57). In contrast, the Rsm sRNAs of the pseudomonads are on average half as long, possess one-half to one-third as many RsmA binding sites, and are much more stable (20 to 60 min) (59, 177). The greater stability of the latter sRNAs may be related to the absence of an apparent CsrD homolog in Pseudomonas spp., which is essential for turnover of E. coli CsrB/C sRNAs (57). Rapid synthesis and turnover of CsrB/C suggest that CsrA activity can be rapidly altered in response to changes in environmental conditions in E. coli, an idea that is supported by mathematical modeling studies (178). The kinetic response of RsmA activity to conditions that affect the Rsm sRNAs remains to be investigated.
Complex regulatory networks of Pseudomonas Rsm systems.
The genetic circuitry that controls the expression of Rsm sRNAs in Pseudomonas spp. is complex, involving several regulatory proteins and presumably multiple environmental stimuli. Regulation is mediated through the BarA/UvrY homologs GacS/GacA. GacS is a membrane-bound sensor kinase that phosphorylates GacA, which binds to the rsmX, rsmY, and rsmZ promoters and activates transcription (59). Though a specific stimulus has yet to be conclusively demonstrated for any BarA homolog, mutations in Krebs cycle genes affect GacA-dependent activation of RsmZ expression, suggesting the involvement of Krebs cycle intermediate(s) or derivatives thereof in GacS activation (173). The Krebs cycle involves 9 intermediates that possess one or more carboxylate groups, and carboxylate compounds with short aliphatic tails stimulate BarA activity in E. coli (67).
Two unusual hybrid sensor kinase-response regulator proteins called RetS (LemA) and LadS regulate GacS through repression and activation mechanisms, respectively (Fig. 5). RetS is a membrane-bound protein that binds to GacS and prevents autophosphorylation, which reduces GacA phosphorylation and ultimately Rsm sRNA expression (179). LadS is a membrane-bound protein that binds to GacS and stimulates its activity through an unknown mechanism (180, 181). Though the stimuli for RetS and LadS regulation are unclear, these proteins act antagonistically to control T3SS, motility, T6SS, quorum sensing, and biofilm formation through GacS/GacA and RsmA (180). Recent studies demonstrated that a novel phosphotransfer protein called HptB interacts with and is phosphorylated by RetS and that HptB, similar to RetS, represses RsmY expression (182, 183). Nevertheless, RetS and HptB regulate RsmY expression through separate pathways. Interestingly, whereas RetS repressed both RsmY and RsmZ expression, HptB appeared to control only RsmY expression. As in E. coli, these findings imply that the presence of multiple Csr/Rsm-inhibitory sRNAs may serve to provide multiple regulatory inputs to control the Csr/Rsm regulon(s) in response to different environmental stimuli.
Strains of P. aeruginosa isolated from chronic CF patients typically lack a functional T3SS. One reason for this is the frequent occurrence of mutations affecting the MucA/AlgU and AlgZ/AlgR signal transduction systems (141, 142). MucA is a membrane-bound anti-sigma factor that sequesters AlgU (sigma 22), preventing it from activating the expression of ∼300 genes, including genes for the T3SS (184–187). AlgU activates transcription of the response regulator AlgR, which inhibits T3SS gene expression via two independent pathways (86). The first involves AlgR activation of RsmY and RsmZ expression through an unknown mechanism. The second pathway involves AlgR-dependent repression of Vfr, a cyclic AMP (cAMP)-activated global transcription factor that is homologous to E. coli cAMP receptor protein (188). Though the mechanism has yet to be established, it is believed that Vfr activates transcription of T3SS genes through activation of the exsCEBA operon (188). Given the shear number of TCSs in P. aeruginosa (approximately 64 response regulators and 63 sensor histidine kinases) and the diverse repertoire of potential hosts/environments it can inhabit, it is perhaps no surprise that multiple signal transduction networks should control the Rsm system.
A number of cytoplasmic global regulators also control the levels of RsmA-inhibitory sRNAs in Pseudomonas species. For example, IHF (discussed above) binds to and activates transcription from the rsmZ promoter in P. fluorescens. IHF serves to maintain DNA architecture (supercoiling and duplex stabilization). The mechanism for IHF effects on rsmZ expression is presently unknown. Interestingly, the H-NS (histone-like nucleoid structuring protein) orthologs MvaT and MvaU bound to the rsmZ promoter region in vivo and repressed its expression (76). Similar to IHF, H-NS also affects DNA architecture, and it is known to repress gene expression under conditions of low temperature or low osmolarity (189). While the conditions that control MvaT/MvaU activity are unknown, IHF antagonizes H-NS-dependent silencing of gene expression in bacterial pathogens, including E. coli, V. cholerae, and S. enterica (190–192). Further studies are needed to determine whether IHF antagonizes MvaT/MvaU-dependent repression of RsmZ.
Posttranscriptional regulators of gene expression also influence RsmA activity in Pseudomonas species. For example, Hfq appears to modestly stabilize RsmY in P. aeruginosa through a direct interaction that prevents RNase E-dependent cleavage (102, 177). Deletions of hfq and rsmY similarly reduced expression of the quorum-sensing AHL synthetase rhlI. It has been proposed that Hfq activates rhlI expression indirectly via RsmA, though this hypothesis has not been experimentally tested. Hfq also enhances CsrB expression in E. coli, apparently through indirect effects on its synthesis (57).
Legionella pneumophila
Legionella pneumophila is the causative agent of a respiratory pneumonia called Legionnaires' disease. This bacterium is found primarily in aquatic environments, where it infects and replicates within numerous species of protozoa (193). Humans are infected with L. pneumophila by inhaling contaminated water droplets, which are present in both natural and man-made sources (194). The infectious life cycle of L. pneumophila can be broken down into two phases: replication and transmission. In the replicative phase, abundant nutrients cause intracellular replication to proceed rapidly and transmission traits to be repressed. When nutrients become limiting, transmission traits are expressed, including motility, resistance to heat and osmotic pressure, sodium sensitivity, and the ability to avoid phagosome-lysosome fusion (195, 196). The postexponential growth phase of L. pneumophila is strongly correlated with virulence phenotypes, and bacteria from the exponential or stationary phase of growth are used to study the replicative or transmission phase, respectively. The leading hypothesis to explain these findings is that nutrient depletion perturbs fatty acid biosynthesis, which in turn stimulates production of ppGpp by SpoT. When ppGpp levels are high, both the stationary-phase sigma factor RpoS and the LetS/LetA TCS cooperate to induce the expression of transmission genes (197–200).
CsrA controls the switch from replicative to transmissive virulence phase of infection.
CsrA in L. pneumophila was originally studied in plasmid overexpression experiments, where it was determined that CsrA represses transmission traits, including pigmentation and motility, and promotes morphological changes leading to filamentation (201). Molofsky and Swanson demonstrated that CsrA-dependent repression of transmission traits is alleviated by the LetS/LetA (BarA/UvrY) TCS (91). The RsmY and RsmZ sRNAs were identified by bioinformatics analyses (55), and Sahr et al. demonstrated that RsmY/Z link the LetS/LetA TCS and CsrA to the control of replication versus transmission phases (202). The function of RsmY/Z in L. pneumophila is consistent with the Gammaproteobacteria model in which LetS/LetA (BarA/UvrY) modulates CsrA activity by activating transcription of the RsmY/Z sRNAs (3).
Though the majority of LetS/LetA-regulatory effects depended on RsmY/Z, regulation of several motility genes did not (202). This may suggest that L. pneumophila encodes additional CsrA-inhibitory sRNAs or that LetA directly controls the expression of these genes. A third Csr sRNA, RsmX, is present in many (but not all) L. pneumophila strains. Its expression is also dependent on LetS/LetA (203). The regulatory contributions of this sRNA, however, did not account for LetA-dependent control of motility gene expression. Further studies are needed to determine how the LetS/LetA TCS affects motility gene expression and whether this regulatory circuit involves CsrA. Whether RsmX, RsmY, and RsmZ of L. pneumophila function in a redundant fashion or whether they are independently expressed in response to different environmental parameters is an open question.
The Dot/Icm type IV secretion system.
When humans inhale aerosolized L. pneumophila, the bacterium either immediately triggers contact-dependent apoptosis or rapidly multiplies within the engulfing alveolar macrophage by establishing the Legionella-containing vacuole (LCV) (204). Either fate depends on effectors that are secreted by the Dot/Icm type IV secretion system (T4SS), which is important for transporting L. pneumophila virulence components into the host. The T4SS structural genes are homologous to the T-DNA transfer system of Agrobacterium tumefaciens and the Tra transfer genes of the IncI ColIb-P9 plasmid of Shigella flexneri (205). The Dot/Icm complex includes approximately 27 protein components that are assembled into one of three subcomplexes: the inner membrane substrate receptor, the transmembrane bridge connecting the inner and outer core subcomplexes, and the outer membrane core containing the secretory components for host cell penetration (206). This T4SS secretes approximately 300 effectors (207–210) and is important for intracellular replication, organelle trafficking, and effector-dependent egress from the host cell (204, 211–213). CsrA regulates the expression of at least 26 of 44 tested Dot/Icm effector genes and at least three genes encoding regulatory proteins that influence Dot/Icm expression (78, 214). Though many of these putative CsrA targets appear to contain multiple CsrA binding sites, in vitro CsrA binding experiments in L. pneumophila have yet to be reported.
Complex regulation of CsrA and the L. pneumophila life cycle.
Three TCSs in L. pneumophila apparently regulate CsrA activity: LetS/LetA, PmrB/PmrA, and LqsS/LqsT/LqsR (Fig. 7). Although it is known that rsmY/Z expression is stimulated as growth approaches stationary phase, the exact nature of the inducing signal is unknown. In other Gram-negative bacteria, the signal for the homologous sensor kinase (BarA, GacA, etc.) seems to be tied to carbon metabolism intermediates, including short-chain fatty acids such as acetate or intermediates of the tricarboxylic acid cycle (67, 173). Likewise, L. pneumophila requires its LetS/LetA TCS to induce transmission traits in response to SCFAs, as well as to other metabolic stresses and ppGpp (199, 200). However, a direct stimulatory factor has never been definitively demonstrated in this or any other bacterium.
FIG 7.
Csr regulatory circuitry in in L. pneumophila, depicted as in Fig. 4. Pentagons depict the autoinducer LAI-1.
The PmrB/PmrA TCS modulates the composition of lipopolysaccharide (LPS) in response to signals in host and nonhost environments (215) in a variety of pathogens, (216–222). The PmrB sensor kinase is activated by high Fe3+ (∼100 μm), high Al3+ (∼100 μM), and mildly acidic pH (pH 5.8) (215). Surprisingly, L. pneumophila PmrA did not regulate LPS composition (223) and instead activated transcription of several Dot/Icm T4SS genes (224). PmrA also activated transcription of the csrA gene in both exponential and stationary growth phases (78). The net result of these effects was PmrA-dependent activation of Dot/Icm gene expression at the transcriptional level and CsrA-dependent repression of Dot/Icm genes at the posttranscriptional level. An explanation for this somewhat paradoxical finding may be that PmrA and CsrA work in concert to fine-tune the levels of Dot/Icm products as the infection cycle progresses.
L. pneumophila produces the quorum-sensing autoinducer molecule 3-hydroxypentadecane-4-one (LAI-1 or Legionella autoinducer 1). LAI-1 is produced by the LAI-1 synthase LqsA and is sensed by sensor kinase proteins LqsS/LqsT that stimulates phosphorylation of the response regulator LqsR (225, 226). LqsA and LqsS are homologs of V. cholerae CqsA and CqsS, respectively (227). L. pneumophila lqsR mutants are poorly phagocytosed by amoeba and macrophage hosts and are defective in intracellular replication (226). Mutations in letA lead to reduced LqsR levels, suggesting that LqsR is repressed by CsrA. Perhaps the Lqs quorum-sensing system facilitates the transition from transmission-phase gene expression to the replication phase, a role complementary to that of Csr, which facilitates the switch from replication to transmission phase.
In many bacterial species, the regulatory activities of CsrA/RsmA and RpoS appear to oppose one another (3), yet in L. pneumophila, the LetS/LetA TCS destabilizes the rpoS transcript in stationary phase (228). Furthermore, while RpoS in other bacterial species promotes stationary-phase resistance to various environmental stresses (229), RpoS in L. pneumophila does not appear to perform this function (197, 230). Clearly, RpoS has evolved a unique role to accommodate the life cycle of L. pneumophila.
IHF in L. pneumophila is expressed in stationary phase and is required for full virulence in amoebae (231). IHF directly binds to and activates transcription of the rsmY and rsmZ promoters (232). Expression of the genes encoding IHF, ihfA and ihfB, is negatively regulated by LetA. This regulatory circuit, wherein LetA directly activates rsmY/Z and indirectly represses rsmY/Z through ihfA/B, appears to comprise an incoherent feedforward loop. Incoherent feedforward loops are known to have diverse regulatory effects, including the ability to alter the dynamics or the dynamic range of target regulation, to facilitate the generation of transient pulses in regulation, or to permit the integration of multiple signals and accelerate response times (233).
Salmonella Species
The top three food-borne infections causing hospitalization in the United States are Salmonella, norovirus, and Campylobacter, with Salmonella being the leading cause of death (234). Salmonella enterica can infect a remarkably broad range of host species, including a large number of different animals and even plants (235–238). In humans, the serovar S. Typhimurium causes an acute gastroenteritis characterized by an inflammatory diarrhea and fever (239–242). In rare cases this is followed by reactive arthritis (243, 244). Besides being a significant cause of human morbidity and mortality, S. Typhimurium was one of the first and most powerful models for bacterial genetics.
The primary virulence systems of S. Typhimurium are two distinct type III secretion systems (T3SS) both of which are regulated by CsrA. T3SS1 is encoded within SPI1 (Salmonella pathogenicity island 1) and is involved primarily in the intestinal phase of disease. T3SS1 effectors are directly injected into intestinal epithelial cells (245–247), which causes uptake of bacterial cells via macropinocytosis and an inflammatory response (247–263). T3SS2 (encoded within SPI2) is induced after Salmonella has invaded or is phagocytized by eukaryotic cells and is required for survival in macrophages and systemic disease (264–268). These observations led to a simple model in which T3SS1 is needed to invade intestinal cells but is not required during the subsequent phases of Salmonella pathogenesis, while T3SS2 is expressed only when the bacteria reside within eukaryotic cells. However, the regulation and function of the Salmonella T3SSs may be more complicated. SPI1 and SPI2, which contribute to inflammation, and the anaerobic respiration pathways that take advantage of inflammation are all regulated by CsrA, as detailed below (98, 269).
SPI1 is regulated by SirA/CsrA.
The uvrY ortholog in Salmonella, sirA (Salmonella invasion regulator A), was first discovered by screening for mutations that confer a defect in SPI1 gene regulation (270). Another study identified barA and csrB as regulators of SPI1, which led to further studies on the effects of csrA (271, 272). CsrC was identified by its homology with E. coli csrC (77, 273). Mutations in barA, sirA, csrB, and csrC all cause decreases in SPI1 gene expression. Interestingly, either mutation of csrA or overexpression of csrA causes a decrease in SPI1 gene expression (271).
CsrA regulates the dedicated regulators of SPI1. There are three large operons in SPI1 that encode regulatory, structural, and effector proteins. There are also at least five regulatory genes encoded within SPI1: hilA, hilC, hilD, sprB, and invF. HilC, HilD, and RtsA (not encoded within SPI1) are AraC/XylS family regulators which comprise a feedforward loop that regulates hilA (274–277). HilA is an OmpR/ToxR family protein that regulates a number of SPI1 genes by activating transcription from the invF and prgH promoters (278, 279). InvF is another AraC/XylS family regulator that, in association with a coactivating chaperone SicA, activates the transcription of numerous genes encoding T3SS effector proteins, encoded inside and outside SPI1, including sip/sspABCD, sopA, sopB/sigD, sopE, sopE2, sspH1, sptP, and slrP (280–283).
The effects of barA, sirA, csrB, and csrC on SPI1 are mediated through the binding of CsrA to the hilD transcript (269). BarA phosphorylates SirA (73, 269), which then binds as a dimer to an 18-bp inverted repeat sequence between positions −190 and −173 of csrB and −168 and −151 of csrC relative to their transcription start sites (63). CsrB and CsrC antagonize CsrA, preventing it from binding the hilD transcript. The net result of BarA/SirA activity is increased HilD expression, which has downstream effects on the remainder of SPI1 (269). HilD also regulates SPI2 under certain conditions (269, 284). CsrA affects invF and sipC, but not prgH, in the absence of hilA, suggesting that CsrA may also directly regulate other genes within SPI1, which lie above and below the hilA regulatory level (271).
Other genes regulated by CsrA.
SirA and CsrA also regulate the expression of type 1 and Pef fimbriae (285, 286). Under laboratory conditions, only type 1 fimbriae are expressed, unless the 5′ UTR of the fimAICDHF mRNA is deleted, which then allows expression of Pef fimbriae. The mechanism of this regulation is that CsrA binds the 5′ UTR of the fimAICDHF transcript, which has weak regulatory effects on the expression of type 1 fimbriae but has surprisingly large effects on the expression of Pef fimbriae (285). The 5′ UTR of the fimAICDHF mRNA is so abundant that it titrates CsrA from other transcripts, including the pefA transcript, for which CsrA is a positive regulator. Thus, the 5′ UTR of fimAICDHF acts as a third regulatory RNA in addition to csrB and csrC (285).
In a microarray study, mRNAs whose levels are affected by the csrA gene included those of SPI1, flagellum synthesis and chemotaxis, maltose, propanediol and ethanolamine utilization, tetrathionate reductase and hydrogen sulfide production, and cobalamin biosynthesis (98). As in E. coli, CsrA represses biofilm formation, and sirA or csrB csrC double mutants are defective in biofilm development (286). CsrA has been shown to bind to the mRNAs of 5 of the 20 GGDEF/EAL domain proteins in Salmonella, which are involved in switching between biofilm and motile phases of growth (84).
Csr and the response to short-chain carboxylate compounds.
The intestinal environment contains high concentrations of short-chain fatty acids (SCFAs), which regulate Salmonella invasion through Csr-dependent and -independent pathways. Acetate appears to activate Salmonella invasion independently of barA, but dependent upon sirA, through acetyl-phosphate (68). Formate activates Salmonella invasion, apparently independently of barA/sirA and csrA (287). Propionate represses invasion gene expression via the posttranslational control of HilD, independently of the Csr system (288). Because formate and acetate levels are elevated in the small intestine, while propionate is elevated in the colon, these SCFAs may serve as cues to prepare Salmonella for invasion of or exit from the host, respectively. In contrast to these findings, all of these SCFAs activated csrB expression via BarA signaling in E. coli (67).
Vibrio cholerae
V. cholerae primarily inhabits aquatic environments and causes the human disease known as cholera (290). Virulent serogroups of V. cholerae form robust biofilms and express the virulence factors cholera toxin (CT) and the toxin-coregulated pilus (TCP) (290, 291). The maturation and dispersal of V. cholerae biofilms, as well as the production of CT and TCP, are controlled by multiple independent quorum-sensing systems, whose effects converge on a single regulatory circuit (Fig. 8) (227, 292, 293). CsrA exerts its influence on virulence factor expression and quorum sensing in V. cholerae through this circuitry.
FIG 8.
Csr regulatory circuitry in V. cholerae, depicted as in Fig. 4. Autoinducers CAI-1 and AI-2 are shown as pentagons and triangles, respectively. (Adapted from reference 75 with permission of the publisher.)
There are two well-studied quorum-sensing circuits in V. cholerae that regulate gene expression in response to autoinducers that are referred to as CAI-1 [cholera autoinducer-1; (S)-3-hyroxytridecan-4-one] and AI-2 [autoinducer 2; (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate] (227, 289, 294, 295). These quorum-sensing systems coordinate gene expression so that virulence factors are produced at low cell population density. At low population density the concentration of CAI-1 and AI-2 is low, and the sensor kinases CqsS (CAI-1) and LuxQ (AI-2) facilitate phosphorylation of LuxU and ultimately the LuxO response regulator protein (296–298). LuxO-P, Fis, and σ54-RNA polymerase activate transcription of the Qrr1 to -4 sRNAs, which bind to and destabilize the hapR mRNA in an Hfq-dependent manner (299–301). HapR represses transcription of vpsR and vpsT, which encode transcription factors that activate biofilm formation genes. HapR also regulates the expression of 14 genes involved in the synthesis and degradation of c-di-GMP, whose net effect is to reduce c-di-GMP levels (302). The Qrr1 to -4 sRNAs also activate translation of the transcription factor AphA (303, 304). AphA indirectly activates toxT expression, and ToxT activates transcription of genes encoding the CT and TCP (227, 305, 306). At high cell population density, the autoinducers CAI-1 and AI-2 are produced in sufficient quantities to switch CqsS and LuxQ from kinases to phosphatases. This system is reinforced by mutual repression wherein HapR represses aphA transcription and AphA represses hapR transcription. High cell population density therefore triggers quorum-sensing systems to reprogram gene expression, which halts production of virulence factors and increases production of transmission factors such as motility.
Using a genetic screen to isolate genes that affect quorum-sensing gene expression, Lenz et al. determined that transposon insertion mutations in the barA/uvrY orthologs varS/varA exhibited reduced luxCDABE expression (75). luxCDABE is a bioluminescent reporter for quorum-sensing gene expression in Vibrio harveyi, and it was determined that this locus also faithfully reports effects on quorum-sensing gene expression in V. cholerae (75). These effects did not depend on the CAI-1 or AI-2 quorum-sensing system, and epistasis experiments subsequently demonstrated that the VarS/VarA TCS most likely functions through the response regulator LuxO. Genetic screens and epistasis experiments determined that VarS/VarA regulates quorum sensing indirectly through CsrA and three CsrA-inhibitory sRNAs called CsrB, CsrC, and CsrD. As is true for the other Gammaproteobacteria, transcription of the CsrA-inhibitory sRNAs depends on VarS/VarA and likely is activated directly by VarA.
Studies are needed to determine the pathway(s) by which CsrA (and VarS/VarA/CsrB/CsrC/CsrD) affects LuxO/HapR/AhpA and/or other components of the quorum-sensing circuitry. Because CsrA/RsmA is known to regulate the expression of virulence factors in other pathogens by both direct and indirect mechanisms, these pathways are likely to be complex. Perhaps intermediates in carbon pathways, such as acetate, other SCFAs, or tricarboxylic acid (TCA) cycle intermediates, stimulate VarS activity and therefore tie the metabolic status of V. cholerae to virulence factor gene expression. Toward this end, it is also of high interest to understand the effects of CsrA on the V. cholerae transcriptome.
Pathogenic Escherichia coli and Related Species
A variety of E. coli strains have gained and/or lost genetic information relative to commensal isolates and in so doing have become important sources of intestinal or extraintestinal infections (307–309). Pathotypes causing infections are classified according to the traits and genes that distinguish them. Nevertheless, sharing of genetic information among strains is common, and new hybrid forms can arise that complicate the classification schemes. A common core of roughly 1,700 genes is found in all strains. Along with the genes that vary among strains, they constitute a pangenome of over 16,000 genes, highlighting the remarkable plasticity of the E. coli genome (310, 311). Genes contributing specifically to virulence vary among the pathotypes and are often clustered together in horizontally transferred loci, present in the chromosome or in mobile elements such as plasmids or phage and referred to as pathogenicity islands. Although not yet demonstrated, it seems likely that the central role of the Csr system in controlling the expression of common genes for metabolism, biofilm formation, motility, and stress responses, such as the stringent response, broadly impacts the host interactions of all E. coli strains (3, 27, 31). In addition, however, the Csr system is integrated directly into virulence gene networks of E. coli pathogens.
EPEC.
A relatively detailed analysis of CsrA regulation in the enteropathogenic E. coli (EPEC) virulence gene network was conducted (312, 313). EPEC causes noninvasive diarrhea, characterized histopathologically by the formation of attaching and effacing (A/E) lesions in the small intestine, and is responsible for thousands of infant deaths each year in developing countries (307, 314, 315). The A/E lesions involve destruction of the intestinal microvilli and the formation of actin rearrangements in EPEC-attached host cells to form pathognomonic protrusions, referred to as pedestals (316). The main virulence factor of EPEC is a T3SS, encoded within the locus of enterocyte effacement (LEE) of the bacterial chromosome (317). This system transfers protein effectors into host cells, which interact with host signaling proteins and lead to pedestal formation (318). The EPEC LEE locus consists of five multigene operons, LEE1 to -5, along with grlRA and several monocistronic operons (313). Expression of LEE genes is coordinated by a large number of regulators that converge to govern expression of a DNA binding protein, referred to as the LEE expression regulator or Ler. Ler is encoded in LEE1 and acts as the master activator of the LEE gene expression, in large part by relieving repression caused by its paralog H-NS (319). CsrA does not directly regulate ler expression but directly activates expression of the Lee4 locus by binding to its mRNA leader and indirectly activates escD (312). The escD and LEE4 genes encode translocators that must be produced and assembled prior to the secretion of the effectors, suggesting that CsrA specifically activates this step in the secretion pathway. CsrA also appears to regulate LEE at a higher tier in the circuitry, based on ectopic expression studies showing that elevated CsrA levels repress grlRA expression by binding directly to grlRA mRNA (312). Because GrlA activates Ler expression (320), this causes extensive repression of LEE operons and genes. The ability of CsrA to switch from being an activator to a repressor of virulence properties as its levels vary was first observed in Salmonella (271). While the biological relevance of this kind of “seesaw” regulation needs to be established (3), its features hint that CsrA activity may be governed according to the particular niche environment within the host and in turn coordinates the workings of the T3SS and other processes. Interestingly, CsrA also activates expression of tryptophanase in EPEC, which produces indole and derivatives of indole that can be can be toxic to Caenorhabditis elegans (321) or can affect LEE expression and attenuate disease in a mouse model of infection (322).
UPEC.
Uropathogenic E. coli (UPEC) strains commonly cause urinary tract infections, where they replicate in intracellular and extracellular locations with the help of a battery of virulence factors that include fimbriae, toxins, and iron siderophores (323). The BarA/UvrY TCS and CsrA have been implicated in regulating UPEC virulence in animal models and virulence factors expressed in vitro, although the mechanisms for these effects have not been determined (324–326).
Shigella species.
Shigella species are closely related to E. coli and cause invasive, localized diarrheal disease. Infection by Shigella involves entry of the bacterium into host M cells of the colon and rectum, destruction of the phagosomal membrane and release into the cytoplasm, intracellular motility via the host actin system, and lysis of host cells and infection of neighboring cells during repeated cycles of reinfection (327). Pathogenicity is dependent upon the presence of a 220-kb virulence plasmid and the T3SS that it encodes (328). In S. flexneri, CsrA was found to regulate metabolic genes of the chromosome, which are important for causing infection, e.g., pfkA for phosphofructokinase, as well as the virF and virB genes of the virulence plasmid, which activate expression of secreted T3SS effectors (329). However, the mechanisms for these effects remain to be determined.
Yersinia species
The Csr system of Yersinia pseudotuberculosis is the best studied of this important genus. This species, along with the related Yersinia enterocolitica, is the causative agent of a variety of gut-associated diseases, including colitis, diarrhea, and mesenterial lymphadenitis, collectively referred to as yersiniosis (330). These pathogens live both in environmental reservoirs and in a variety of animal hosts and are spread by fecal-oral contamination. Infections in humans result mainly from consumption of contaminated food, most commonly pork (4).
Infection begins when the bacteria tightly associate with the mucosal surface of the intestine. This initial interaction occurs at M cells of Peyer's patches and is mediated primarily via the cell surface-associated virulence factor invasin (331). The bacteria are rapidly translocated across the intestinal barrier into underlying lymphoid tissue, where they replicate. Infection can then spread to mesenteric lymph nodes and, in later stages of infection, to the liver and spleen. Expression of invasin and other early-stage virulence factors of Y. pseudotuberculosis pathogenesis is regulated by RovA, a member of the MarR/SlyA family of transcription regulators (332, 333). A rovA mutant is deficient in its ability to invade and colonize Peyer's patches, in part due to reduced expression of invasin and ability to induce inflammation through interleukin-1 (IL-1) production. After the initial stages of infection, virulence factors important for immune evasion and dissemination of the infection are induced, while expression of early virulence factors is reduced. These virulence factors include the adhesins Ail and YadA as well as T3SSs and their associated effectors, including Yops (Yersinia outer proteins) (331).
RovA expression varies in response to nutrient availability, which is in part due to effects of another transcriptional regulator, RovM. This LysR family protein is induced during growth in minimal media and it acts in concert with H-NS to repress the expression of RovA (334). Studies to determine how expression of RovM and RovA is linked to nutrient availability led to the identification of a Csr system in Y. pseudotuberculosis (333). In minimal media, CsrA was found to indirectly activate RovM expression, leading to repression of RovA. As in many other species, two inhibitory small RNAs, CsrB and CsrC, regulate CsrA activity. Of these, CsrC was found to provide the most dramatic regulatory effect, likely as a result of the very low level of CsrB expression under the conditions tested. It appears that the nutrient-sensitive expression of RovM and thus RovA is due primarily to high expression of CsrC in rich media. Subsequent work has shown that Crp is also involved in linking nutrient availability to CsrA activity via indirect activation of CsrC and repression of CsrB (103). The relevance of these opposing effects of Crp is uncertain. Regulation of CsrA activity in response to nutrient availability may allow Y. pseudotuberculosis to express early virulence factors only in the appropriate host compartment and under growth conditions that might lead to a successful infection.
Further studies identified an interesting distinction in the Y. pseudotuberculosis Csr regulatory circuitry: the BarA/UvrY TCS directly activates only the expression of CsrB, and not that of CsrC (333), while another TCS, the PhoQ/PhoP system, directly activates CsrC but not CsrB expression (335). The latter TCS regulates a number of virulence determinates in response to low magnesium, acidic pH, and antimicrobial peptides and is important for replication in macrophages (336). Such differential expression of Csr sRNAs by distinct TCSs has yet to be identified in the other characterized Csr systems, and its physiological role remains uncertain.
Plant Pathogens
Pectobacterium carotovorum.
Pectobacterium carotovorum (previously known as Erwinia carotovora) is an important plant pathogen and is the etiological agent of soft-rot diseases in a variety of commercially important plant species (337, 338). As a close relative of E. coli (family Enterobacteriaceae), P. carotovorum is a genetically tractable organism in which some of the first studies on the structure and function of plant cell wall-degrading enzymes were conducted. These studies revealed that secreted proteases, cellulases, and pectinases play critical roles in P. carotovorum virulence (338–340). Further investigation of P. carotovorum virulence pathways led to the discovery of a quorum-sensing signal used to alter gene expression in response to changes in cell population density (338, 341–345). The quorum-sensing signal N-(3-oxohexanoyl)-l-homoserine lactone (acyl homoserine lactone [AHL]) is required for production of P. carotovorum virulence factors and related products, including the previously mentioned cell wall-degrading enzymes, antibiotics, hypersensitive responses, and the T3SS (346, 347).
Shortly after the discovery of CsrA in E. coli, its homolog RsmA in P. carotovorum was found to repress tissue maceration, the production of cell wall-degrading enzymes, and the AHL quorum-sensing signal (18, 19). Furthermore, Chatterjee et al. determined that AHL-dependent activation of exoenzyme production is regulated by rsmA (19). As found in many other species, P. carotovorum produces an RsmA-inhibitory sRNA called RsmB, which depends on the GacS/GacA TCS for its expression (348, 349). It was later shown that AHL activates the production of cell wall-degrading enzymes by repressing rsmA expression (350, 351). More specifically, AHL production inhibits the activity of the AHL receptor proteins ExpR1 and ExpR2, both of which activate rsmA expression in the absence of AHL (352–354). P. carotovorum subspecies may contain one or both ExpR receptor proteins.
RsmA and RsmB have been studied in other soft-rot disease-causing Pectobacterium species, including P. atrosepticum and P. wasabiae. RsmA also represses the expression of cell wall-degrading enzymes in these bacteria (101, 355). It is interesting to note that induction of the alarmone (p)ppGpp activates rsmB expression in P. atrosepticum, which is reminiscent of links between the Csr and the stringent response systems in E. coli and L. pneumophila (31, 198, 355, 356). Whether nutrient starvation through the stringent response system generally serves as a trigger for inhibiting CsrA activity in other bacteria remains to be seen.
Xanthomonas spp.
The genus Xanthomonas includes a diverse set of pathogens that infect over 200 different families of plants (357). Two Xanthomonas pathovars of great concern to agriculture are (i) X. campestris pathovar campestris, the causal agent of black rot, a disease of cruciferous plants, and (ii) X. axonopodis pathovar citri, the causal agent of citrus canker, a disease that reduces the vitality of susceptible citrus hosts. RsmA has been studied in the X. campestris pv. campestris and X. axonopodis pv. citri pathovars (100, 358). In both cases, RsmA promoted virulence through the activation of genes associated with a T3SS. Comparative genome analyses indicate that species of Xanthomonas encode every known type of bacterial secretion system (type I to type VI) and suggest that the T3SS is essential for pathogenicity of most Xanthomonas pathovars (359). Xanthomonas species typically translocate as many as 40 T3SS effectors that suppress host defenses and alter host gene expression to promote dissemination and tissue destruction (359).
Studies by Chao et al. revealed that rsmA is required for infection of host plants by X. campestris pv. campestris and significantly reduces the hypersensitive response in nonsusceptible hosts (358). Deletion of rsmA significantly reduced motility and the extracellular levels of virulence factors, including endoglucanase, amylase, and extracellular polysaccharide (EPS) (also called “xanthan gum”). Microarray analysis of wild-type cells versus rsmA deletion mutants revealed that rsmA affects the expression of at least 435 transcripts whose gene products are associated with a diverse variety of cellular processes. These studies did not differentiate between direct and indirect RsmA targets, which is needed to define the RsmA regulon.
Work by Andrade et al. similarly demonstrated that rsmA deletion in X. axonopodis pv. citri significantly affects its pathogenicity and ability to elicit a hypersensitive response in nonsusceptible plant hosts (100). Microarray experiments comparing an rsmA deletion mutant to the wild type revealed effects of rsmA on the levels of 170 transcripts. As seen in X. campestris pv. campestris, several T3SS regulatory and effector genes were affected by rsmA deletion. RsmA bound to the 5′ UTR of the hrpG and hrpD transcripts in vitro and stabilized these messages against degradation in vivo. HrpG is the master regulator of T3SS effector gene expression, and RsmA may regulate T3SS gene expression through HrpG. Consistent with this hypothesis, ectopic expression of hrpG from a plasmid restored pathogenicity and the hypersensitive response in an rsmA deletion strain. RsmA-dependent control of T3SS gene expression via master transcription regulatory factors, as seen in both animal and plant pathogens, is a conserved feature of virulence-associated secretion systems.
Pseudomonas syringae.
P. syringae is a remarkable species that includes over 50 pathovars that infect a wide variety of plant hosts. Economically important diseases caused by P. syringae pathovars include bacterial speck of the tomato (360), bleeding canker of horse chestnut (361), halo blight of bean (362), and various cankers and distortions of trees (338). As is the case for many other plant and animal pathogens, P. syringae secretes a variety of products, including quorum-sensing compounds, phytotoxins, antibiotics, exoproteases, fluorescent pigments, and alginate, which influence infection and disease processes (363). These secreted products as well as biofilm formation, host colonization, and lesion formation are all regulated by the GacS/GacA TCS (364–379). Furthermore, genetic analyses of various P. syringae pathovars have identified homologs of the RsmA-inhibitory sRNAs RsmX, RsmY, and RsmZ of P. fluorescens, as well as RsmB of P. carotovorum (55). Studies by Kong et al. demonstrated that rsmA overexpression from a plasmid negatively affects pathogenicity of several P. syringae pathovars and appears to do so by repressing the production of numerous secreted virulence factors, including phytotoxin, exoprotease, and pyoverdin (363). All tested P. syringae pathovars also showed reduced swarming motility in response to rsmA overexpression, while its impact on biofilm formation and alginate production varied by pathovar. An analysis of the phenotypic effects of deleting the rsmA and/or rsmXYZ genes has yet to be performed in P. syringae.
A PARTNER-SWITCHING MECHANISM REGULATES CsrA ACTIVITY IN BACILLUS SUBTILIS: IMPLICATIONS FOR BACTERIAL PATHOGENS
Although the Csr system has been extensively studied in a variety of Gram-negative bacteria, Bacillus subtilis is the only Gram-positive organism in which this system has been examined. In B. subtilis, csrA is the terminal gene of a large flagellum-biosynthetic operon. In addition to a σD-dependent promoter that controls expression of the entire operon, a σA-dependent promoter drives expression of the last two genes (fliW and csrA) (380). The gene encoding flagellin, hag, is located in a separate σD-dependent operon, immediately downstream from csrA. CsrA represses translation initiation of hag by binding to two sites in the hag leader transcript, one of which overlaps the hag SD sequence (380). This translation repression mechanism is similar to those controlling gene expression in Gram-negative species. Although a csrB gene has been predicted in the B. subtilis genome, experimental evidence of its role as an sRNA antagonist of CsrA is lacking (55).
A recent and unexpected discovery from studies of B. subtilis CsrA regulation was that a protein (FliW) functions as an antagonist of CsrA, and together these proteins constitute part of a morphogenic checkpoint in flagellum assembly (Fig. 9) (381). Prior to flagellum hook assembly, FliW is largely bound in a complex with cytoplasmic Hag (flagellin) protein, leaving CsrA free to repress hag translation. Once assembly of the flagellar hook structure is completed, Hag can be secreted and assembled to form the flagellum filament. The resulting unbound FliW protein in the cytoplasm is then capable of binding to CsrA, thereby relieving CsrA-mediated translational repression of hag. This leads to derepression of Hag synthesis at the precise time in which Hag is needed in large quantities (381). Thus, flagellin homeostatically restricts its own translation as a consequence of a partner-switching mechanism involving alternative FliW-Hag, FliW-CsrA, and CsrA-hag mRNA complexes. The finding of this regulatory mechanism suggests that CsrA in B. subtilis might play a more limited role than it does in the Gammaproteobacteria, in which it controls gene expression in coordination with flagellum morphogenesis (381).
FIG 9.
Model for CsrA-FliW-Hag control of flagellin homeostasis in B. subtilis. Flagellin homeostasis is controlled by a partner-switching mechanism involving FliW (dark blue triangles), CsrA, and Hag (orange barbells). The basal body (blue and pink), flagellar hook (cyan), filament (orange), cytoplasmic membrane, and peptidoglycan layer are indicated. (Adapted from reference 381 with permission of the publisher.)
Bioinformatics analysis of the phylogenetic occurrence of fliW and csrA together in a flagellum gene cluster further suggests that CsrA may have had an ancestral role in controlling flagellar assembly and motility, which subsequently evolved to include additional cellular processes in other bacteria (381). The csrA and fliW genes are encoded adjacent to each other in many Firmicutes, Spirochaetales, Thermotogales, and Delta- and Epsilonproteobacteria, including human pathogens (Fig. 10). Although largely absent from the Alpha- and Betaproteobacteria, csrA, but not fliW, reappears in the Gammaproteobacteria, along with its associated sRNA antagonists. These observations raise the possibility that early in proteobacterial evolution, the Gammaproteobacteria may have obtained csrA via horizontal gene transfer, followed by the emergence of a large number of CsrA-controlled genes and the sRNA antagonists that replace FliW (381). In view of the fact that motility is often required for pathogenesis, the CsrA-FliW partner-switching mechanism seems worthy of investigation in additional species.
FIG 10.
The csrA and fliW genes tend to cluster in the genomes of many taxonomically diverse species, with the notable exception of the Gammaproteobacteria, which lack FliW. Gene neighborhoods around csrA (blue) and/or fliW (dark green) in Clostridium botulinum A strain ATCC 19397, Clostridium difficile 630, Borrelia burgdorferi B31, Treponema pallidum subsp. pallidum strain Nichols, Petrotoga mobilis SJ95, Thermotoga maritima MSB8, Geobacter sulfurreducens PCA, Campylobacter jejuni NCTC 11168, Helicobacter pylori J99, Escherichia coli MG1655, Pseudomonas aeruginosa PAO1 and Vibrio cholerae MZO-2 are shown. The associated phylum and class taxonomic information is shown to the left of and immediately beneath the species names. Protein-coding genes are represented as arrows, and tRNA genes are represented with gray bars. Flagellin genes are colored orange, and other motility-associated genes are colored pale green. Slashes indicate that csrA and fliW are not within the same genomic neighborhood of a species. Annotation and gene position information was determined using MicrobesOnline (http://www.microbesonline.org/) and reference 381.
CONCLUDING REMARKS
The Csr system is extensively integrated into gene expression networks that govern bacterial physiology, metabolism, and virulence. Inhibitory sRNAs integrate signaling from the BarA/UvrY TCS and pathogen-specific regulatory factors into this system by sequestering the CsrA/RsmA protein away from its lower-affinity mRNA targets. Studies in E. coli K-12 revealed that CsrA often has opposite effects on opposing metabolic pathways, e.g., gluconeogenesis versus glycolysis, or physiological processes such as motility versus biofilm formation (3). Pathogens appear to use the Csr system to coordinate the expression of virulence factors according to alternate stages of infection or shifts in the host microenvironment. For example, CsrA activates L. pneumophila virulence factors associated with intracellular replication and represses genes for transmission traits (91, 201). RsmA activates P. aeruginosa genes associated with acute infection and represses genes associated with chronic infection (80). CsrA represses V. cholerae motility genes and activates genes for biofilm formation and cholera toxin (75). CsrA regulates genes necessary for the switch between early colonization and persistent infection in Y. pseudotuberculosis (4). Thus, the Csr system governs pivotal lifestyle decisions of Gammaproteobacteria, including the pathogens of this bacterial class.
Our understanding of the role of CsrA/RsmA in the physiology and virulence of pathogens is heavily influenced by the known virulence mechanisms, which tend to attract the greatest research attention. However, CsrA/RsmA protein is a global regulator of gene expression in every pathogen examined thus far, and its functions other than regulation of virulence factor expression are likely to be required for successful infection. High-throughput RNA sequencing and next-generation protein-RNA interaction studies that utilize the HITS-CLIP (high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation) (10, 109), PAR-CLIP (photoactivatable-ribonucleoside-enhanced CLIP) (110), or iCLIP-seq (individual-nucleotide-resolution CLIP) (111) method would be helpful for assessing the overall influence of CsrA/RsmA on gene expression in pathogens. Yet even with this information, it will be important to determine how global gene expression varies during the life cycle of a pathogen and the role played by CsrA/RsmA in this dynamic process. This is a goal that has not yet been achieved, even for a model pathogen.
Some questions arising from studies of proteobacterial pathogens are how and why the Csr system has evolved to become integrated into so many diverse virulence factor regulatory circuits. The virulence factors and their dedicated regulators typically are encoded within pathogenicity islands that are acquired by horizontal gene transfer (382). Immediately following acquisition, transcription of foreign genetic material tends to be silenced by the action of nucleoid-associated proteins, e.g., H-NS and Stp (383). If the newly acquired virulence genes are to confer a selective advantage for a pathogen, then evolution must occur to permit them to be expressed and appropriately regulated. While the details remain obscure, there must be strong selection for regulating virulence circuitry by CsrA. Perhaps the broad use of the Csr system in controlling metabolic and physiological functions makes it well suited for regulating virulence genes in response to metabolic shifts that occur during infection. Without a doubt, determining the environmental cues or physiological signals that control the BarA/UvrY-orthologous TCSs of various species will be necessary for addressing this hypothesis. It is worth noting that CsrA regulates gene expression by recognizing a relatively small and adaptable sequence motif, which should evolve rapidly. The striking similarity of the CsrA binding sequence to the Shine-Dalgarno sequence was recognized early (26) and no doubt allows translational regulation by CsrA to evolve with minimal mRNA sequence alteration. The Csr sRNAs may have evolved by duplication of a CsrA binding mRNA target, which subsequently accumulated additional CsrA binding sequences. The Salmonella fimAICDHF mRNA exemplifies how this might have occurred, as this abundant mRNA contains an untranslated leader that permits its own regulation by CsrA and competes with pefACDEF mRNA for CsrA binding (285). Finally, spontaneous mutations in the Csr system have been observed as colony morphotypes, although the significance of such mutations for pathogen evolution remains to be determined (40).
Given the wide distribution of the BarA/UvrY TCS, its strong effects on Csr/Rsm sRNA expression, and its critical roles in physiology and virulence pathways, it is remarkable that a direct stimulus for this TCS has yet to be identified. Some of the most compelling findings toward this goal originate from Chavez et al., who demonstrated that the addition of carboxylate compounds containing short aliphatic tails (such as acetate and formate) stimulates BarA/UvrY-dependent expression of E. coli csrB in vivo (67). Consistent with these observations, P. fluorescens strains lacking enzymes in the Krebs cycle accumulated carboxylate-containing compounds and stimulated RsmXYZ expression (173). These results suggest that the BarA/UvrY TCS and its homologs may serve as a barometer for the metabolic state of the bacterial cell. BarA autophosphorylation may occur when products of metabolism are abundant, for example, in the mammalian intestinal tract or in a localized nidus of infection. Furthermore, LetS/LetA-dependent differentiation of L. pneumophila occurs in response to perturbations in fatty acid biosynthesis, a stress that also activates SpoT to generate ppGpp (199, 200). Additional studies are needed to determine (i) whether carboxylate compounds bind directly to and stimulate BarA or GacS phosphorylation in E. coli or P. fluorescens, respectively, (ii) whether similar compounds serve as activators for other species of bacteria, and (iii) the way in which the metabolic environment of the host niche influences signaling through this TCS during an infection.
Recent studies have uncovered novel mechanisms of regulation by CsrA and new ways of regulating CsrA activity in the cell. The first detailed molecular mechanism of activation by CsrA was described for the E. coli flhDC operon, in which bound CsrA inhibits 5′ end-dependent cleavage of the transcript by RNase E (47). In addition, CsrA was recently shown to regulate termination of the E. coli biofilm transcript pgaABCD by altering its structure and making it accessible to Rho protein (53). This is the fourth mechanism by which CsrA regulates the expression of this transcript, along with effects on transcript stability, translation initiation, and transcription initiation (29, 34). An mRNA (fimA) that is naturally expressed at high levels and contains two CsrA binding sites in its untranslated leader antagonizes CsrA activity in S. Typhimurium, in a mechanism resembling regulation by sRNAs (285). This mechanism contributes to the hierarchical synthesis of Salmonella fimbriae. Finally, Mukherjee et al. demonstrated a new model for regulation in the B. subtilis Csr system (381), a partner-switching mechanism in which the FliW protein binds to CsrA and prevents it from repressing translation of the flagellin (hag) mRNA. Whether CsrA of B. subtilis is also regulated by sRNAs or regulates other genes besides hag is unclear. Bioinformatics analyses suggest that this partner-switching mechanism might operate in diverse Gram-positive and negative pathogens (Fig. 10). Without a doubt, many such surprises must be unearthed before the workings of these complex regulatory systems can be fully appreciated.
ACKNOWLEDGMENTS
This is not a comprehensive review of literature related to the Csr/Rsm regulatory system. Accordingly, we offer our sincere apologies to colleagues and collaborators whose work was not discussed.
Studies in our laboratories that contributed to this body of literature were funded in part by the National Institute of Allergy and Infectious Diseases (F32AI100322 and R01AI097116) and the National Institute of General Medical Sciences (R01GM059969 and R01GM066794) of the National Institutes of Health. This material is also based upon work supported by the National Science Foundation Graduate Research Fellowship Program awarded under grant no. DGE-1315138. Additional support was provided by the University of Florida CRIS project, FLA-004949.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Biographies

Christopher A. Vakulskas obtained his B.S. and Ph.D. degrees from the University of Iowa in biology and microbiology, respectively. He is presently a National Institutes of Health postdoctoral research fellow in the laboratory of Dr. Tony Romeo in the Microbiology and Cell Science Department at the University of Florida. His research focuses on the mechanisms by which bacteria utilize posttranscriptional gene regulation to adapt to changes in environmental conditions.

Anastasia H. Potts obtained her B.S. in microbiology and cell science at the University of Florida in 2011. She is currently a National Science Foundation Graduate Research Fellow in the laboratory of Dr. Tony Romeo at the University of Florida. Her Ph.D. research is focused on understanding the global regulatory role of CsrA in E. coli and S. enterica.

Paul Babitzke obtained his Ph.D. in genetics from the University of Georgia. He worked as a postdoctoral scientist for 3 years at Stanford University prior to joining the faculty at the Pennsylvania State University (Penn State) in 1994. He is currently Professor of Biochemistry and Molecular Biology and Director of the Center for RNA Molecular Biology at Penn State. His research interests lie in elucidating the roles that RNA structure and RNA binding proteins play in regulating transcription attenuation, translation initiation, and mRNA stability. He has an ongoing interest in mechanisms that control RNA polymerase pausing and transcription termination.

Brian M. M. Ahmer obtained a B.S. in microbiology from Colorado State University in 1990 and a Ph.D. in genetics and cell biology from Washington State University in 1994, working on E. coli iron acquisition. He did postdoctoral research with Dr. Fred Heffron at Oregon Health Sciences University, working on Salmonella. In 1999 he moved to the Ohio State University, where he is currently an Associate Professor in the Department of Microbial Infection and Immunity. His main interests are Salmonella interactions with the host and other microbes.

Tony Romeo (Ph.D., 1987) received his B.S., M.S., and Ph.D. degrees from the University of Florida in microbiology. Following postdoctoral studies at Michigan State University in biochemistry, he held faculty positions at the University of North Texas Health Science Center, Emory University, and the University of Florida, where he is currently Professor of Microbiology and Cell Science. He is interested in the molecular mechanisms and regulatory circuitry that bacteria use to govern their physiology and metabolism in response to diverse and ever-changing environmental conditions. The workings of the Csr system and the complex phenotypes that it controls have been a focus of the Romeo laboratory since the early 1990s.
REFERENCES
- 1.Carmichael GG, Weber K, Niveleau A, Wahba AJ. 1975. The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J Biol Chem 250:3607–3612. [PubMed] [Google Scholar]
- 2.Babitzke P, Baker CS, Romeo T. 2009. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol 63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heroven AK, Bohme K, Dersch P. 2012. The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol 9:379–391. doi: 10.4161/rna.19333. [DOI] [PubMed] [Google Scholar]
- 5.Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. 2008. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- 6.Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Harris JF, Micheva-Viteva S, Li N, Hong-Geller E. 2013. Small RNA-mediated regulation of host-pathogen interactions. Virulence 4:785–795. doi: 10.4161/viru.26119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steimer L, Klostermeier D. 2012. RNA helicases in infection and disease. RNA Biol 9:751–771. doi: 10.4161/rna.20090. [DOI] [PubMed] [Google Scholar]
- 10.Vakulskas CA, Pannuri A, Cortes-Selva D, Zere TR, Ahmer BM, Babitzke P, Romeo T. 2014. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol Microbiol 92:945–958. doi: 10.1111/mmi.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaberdin VR, Blasi U. 2013. Bacterial helicases in post-transcriptional control. Biochim Biophys Acta 1829:878–883. doi: 10.1016/j.bbagrm.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Gao Q, Chen W, Qin H, Hengzhuang W, Chen Y, Yang L, Zhang G. 2013. Regulation of pqs quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiology 159:1931–1936. doi: 10.1099/mic.0.066266-0. [DOI] [PubMed] [Google Scholar]
- 13.Linares JF, Moreno R, Fajardo A, Martinez-Solano L, Escalante R, Rojo F, Martinez JL. 2010. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol 12:3196–3212. doi: 10.1111/j.1462-2920.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- 14.Sonnleitner E, Blasi U. 2014. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet 10:e1004440. doi: 10.1371/journal.pgen.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F. 2 June 2014. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol doi: 10.1111/1462-2920.12499. [DOI] [PubMed] [Google Scholar]
- 16.Romeo T, Gong M. 1993. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J Bacteriol 175:5740–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui Y, Chatterjee A, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol 177:5108–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl Environ Microbiol 61:1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu MY, Yang H, Romeo T. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol 177:2663–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol 44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol 187:3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol 355:1026–1036. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem 281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 25.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 26.Liu MY, Gui G, Wei B, Preston JF 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 27.Romeo T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 28.Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol 185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 30.Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. 2007. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol 189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, Babitzke P. 2011. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ70 and EσS, and indirect transcriptional activation by CsrA. Mol Microbiol 81:689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, Babitzke P. 2011. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-l-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J Bacteriol 193:6162–6170. doi: 10.1128/JB.05975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannuri A, Yakhnin H, Vakulskas CA, Edwards AN, Babitzke P, Romeo T. 2012. Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J Bacteriol 194:79–89. doi: 10.1128/JB.06209-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson-Fortin LM, Vakulskas CA, Yakhnin H, Babitzke P, Romeo T. 2013. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J Mol Biol 425:3662–3677. doi: 10.1016/j.jmb.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duss O, Michel E, Diarra dit Konte N, Schubert M, Allain FH. 2014. Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res 42:5332–5346. doi: 10.1093/nar/gku141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. 2009. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol 392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FH. 2014. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509:588–592. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- 40.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serganov A, Patel DJ. 2012. Molecular recognition and function of riboswitches. Curr Opin Struct Biol 22:279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serganov A, Patel DJ. 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regulski EE, Breaker RR. 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 44.Huang J, Xu Y, Zhang H, Li Y, Huang X, Ren B, Zhang X. 2009. Temperature-dependent expression of phzM and its regulatory genes lasI and ptsP in rhizosphere isolate Pseudomonas sp. strain M18. Appl Environ Microbiol 75:6568–6580. doi: 10.1128/AEM.01148-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren B, Shen H, Lu ZJ, Liu H, Xu Y. 2014. The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLoS One 9:e89653. doi: 10.1371/journal.pone.0089653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 47.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. 2013. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87:851–866. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takyar S, Hickerson RP, Noller HF. 2005. mRNA helicase activity of the ribosome. Cell 120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 49.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. 2011. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deneke C, Lipowsky R, Valleriani A. 2013. Effect of ribosome shielding on mRNA stability. Phys Biol 10:046008. doi: 10.1088/1478-3975/10/4/046008. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Preston JF 3rd, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF III, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-d-glucosamine. J Bacteriol 190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Figueroa-Bossi N, Schwartz A, Guillemardet B, D'Heygere F, Bossi L, Boudvillain M. 2014. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev 28:1239–1251. doi: 10.1101/gad.240192.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson JP. 2003. Loading Rho to terminate transcription. Cell 114:157–159. doi: 10.1016/S0092-8674(03)00554-3. [DOI] [PubMed] [Google Scholar]
- 55.Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res 34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulkarni PR, Jia T, Kuehne SA, Kerkering TM, Morris ER, Searle MS, Heeb S, Rao J, Kulkarni RV. 2014. A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res 42:6811–6825. doi: 10.1093/nar/gku309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol 184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reimmann C, Valverde C, Kay E, Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 61.Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wall ME, Hlavacek WS, Savageau MA. 2004. Design of gene circuits: lessons from bacteria. Nat Rev Genet 5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- 63.Martinez LC, Martinez-Flores I, Salgado H, Fernandez-Mora M, Medina-Rivera A, Puente JL, Collado-Vides J, Bustamante VH. 2014. In silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC genes. J Bacteriol 196:325–336. doi: 10.1128/JB.00806-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jonas K, Melefors O. 2009. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol Lett 297:80–86. doi: 10.1111/j.1574-6968.2009.01661.x. [DOI] [PubMed] [Google Scholar]
- 65.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pernestig AK, Melefors O, Georgellis D. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J Biol Chem 276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 67.Chavez RG, Alvarez AF, Romeo T, Georgellis D. 2010. The physiological stimulus for the BarA sensor kinase. J Bacteriol 192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 69.Tomenius H, Pernestig AK, Mendez-Catala CF, Georgellis D, Normark S, Melefors O. 2005. Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J Bacteriol 187:7317–7324. doi: 10.1128/JB.187.21.7317-7324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem 269:31567–31572. [PubMed] [Google Scholar]
- 71.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A 89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teplitski M, Goodier RI, Ahmer BM. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol 185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humair B, Wackwitz B, Haas D. 2010. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl Environ Microbiol 76:1497–1506. doi: 10.1128/AEM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol 58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 76.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fortune DR, Suyemoto M, Altier C. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 79.Mukherjee A, Cui Y, Ma W, Liu Y, Chatterjee AK. 2000. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Environ Microbiol 2:203–215. doi: 10.1046/j.1462-2920.2000.00093.x. [DOI] [PubMed] [Google Scholar]
- 80.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pickering BS, Smith DR, Watnick PI. 2012. Glucose-specific enzyme IIA has unique binding partners in the Vibrio cholerae biofilm. mBio 3(6):e00228–12. doi: 10.1128/mBio.00228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. 2010. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol 12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. 2001. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol 183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vivas P, Velmurugu Y, Kuznetsov SV, Rice PA, Ansari A. 2012. Mapping the transition state for DNA bending by IHF. J Mol Biol 418:300–315. doi: 10.1016/j.jmb.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 89.Iost I, Bizebard T, Dreyfus M. 2013. Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. Biochim Biophys Acta 1829:866–877. doi: 10.1016/j.bbagrm.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 90.Sobrero P, Valverde C. 2012. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol 38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 91.Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 92.Barnard FM, Loughlin MF, Fainberg HP, Messenger MP, Ussery DW, Williams P, Jenks PJ. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol Microbiol 51:15–32. doi: 10.1046/j.1365-2958.2003.03788.x. [DOI] [PubMed] [Google Scholar]
- 93.Fields JA, Thompson SA. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J Bacteriol 190:3411–3416. doi: 10.1128/JB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heeb S, Valverde C, Gigot-Bonnefoy C, Haas D. 2005. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol Lett 243:251–258. doi: 10.1016/j.femsle.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 95.Ge Y, Yang S, Fang Y, Yang R, Mou D, Cui J, Wen L. 2007. RpoS as an intermediate in RsmA-dependent regulation of secondary antifungal metabolites biosynthesis in Pseudomonas sp. M18. FEMS Microbiol Lett 268:81–87. doi: 10.1111/j.1574-6968.2006.00562.x. [DOI] [PubMed] [Google Scholar]
- 96.Mukherjee A, Cui Y, Ma W, Liu Y, Ishihama A, Eisenstark A, Chatterjee AK. 1998. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J Bacteriol 180:3629–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R, Martin M. 2012. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One 7:e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol 48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- 99.Burrowes E, Baysse C, Adams C, O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 100.Andrade MO, Farah CS, Wang N. 2014. The Post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5′ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas. PLoS Pathog 10:e1003945. doi: 10.1371/journal.ppat.1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koiv V, Andresen L, Broberg M, Frolova J, Somervuo P, Auvinen P, Pirhonen M, Tenson T, Mae A. 2013. Lack of RsmA-mediated control results in constant hypervirulence, cell elongation, and hyperflagellation in Pectobacterium wasabiae. PLoS One 8:e54248. doi: 10.1371/journal.pone.0054248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U. 2007. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun 352:769–773. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 103.Heroven AK, Sest M, Pisano F, Scheb-Wetzel M, Steinmann R, Bohme K, Klein J, Munch R, Schomburg D, Dersch P. 2012. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front Cell Infect Microbiol 2:158. doi: 10.3389/fcimb.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Owttrim GW. 2013. RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol 10:96–110. doi: 10.4161/rna.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol 189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 107.Klauck E, Typas A, Hengge R. 2007. The σS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci Prog 90:103–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp 41:2034. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yao C, Weng L, Shi Y. 2014. Global protein-RNA interaction mapping at single nucleotide resolution by iCLIP-seq. Methods Mol Biol 1126:399–410. doi: 10.1007/978-1-62703-980-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun 76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Furukawa S, Kuchma SL, O'Toole GA. 2006. Keeping their options open: acute versus persistent infections. J Bacteriol 188:1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luzar MA, Montie TC. 1985. Avirulence and altered physiological properties of cystic fibrosis strains of Pseudomonas aeruginosa. Infect Immun 50:572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weller DM, Landa BB, Mavrodi OV, Schroeder KL, De La Fuente L, Blouin Bankhead S, Allende Molar R, Bonsall RF, Mavrodi DV, Thomashow LS. 2007. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol 9:4–20. doi: 10.1055/s-2006-924473. [DOI] [PubMed] [Google Scholar]
- 116.Haas D, Blumer C, Keel C. 2000. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Curr Opin Biotechnol 11:290–297. doi: 10.1016/S0958-1669(00)00098-7. [DOI] [PubMed] [Google Scholar]
- 117.Blumer C, Haas D. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol 173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 118.Heeb S, Blumer C, Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blumer C, Heeb S, Pessi G, Haas D. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci U S A 96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O'Grady EP, Mulcahy H, O'Callaghan J, Adams C, O'Gara F. 2006. Pseudomonas aeruginosa infection of airway epithelial cells modulates expression of Kruppel-like factors 2 and 6 via RsmA-mediated regulation of type III exoenzymes S and Y. Infect Immun 74:5893–5902. doi: 10.1128/IAI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mulcahy H, O'Callaghan J, O'Grady EP, Adams C, O'Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sadikot RT, Blackwell TS, Christman JW, Prince AS. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hoiby N, Johansen HK. 2007. Isolation measures for prevention of infection with respiratory pathogens in cystic fibrosis: a systematic review? J Hosp Infect 65:374–375. (Author reply, 65: 375–376.) doi: 10.1016/j.jhin.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 124.Garau J, Gomez L. 2003. Pseudomonas aeruginosa pneumonia. Curr Opin Infect Dis 16:135–143. doi: 10.1097/00001432-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 125.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 126.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brutinel ED, Yahr TL. 2008. Control of gene expression by type III secretory activity. Curr Opin Microbiol 11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Urbanowski ML, Lykken GL, Yahr TL. 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci U S A 102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vakulskas CA, Brutinel ED, Yahr TL. 2010. ExsA recruits RNA polymerase to an extended −10 promoter by contacting region 4.2 of sigma-70. J Bacteriol 192:3597–3607. doi: 10.1128/JB.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vakulskas CA, Brady KM, Yahr TL. 2009. Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J Bacteriol 191:6654–6664. doi: 10.1128/JB.00902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brutinel ED, Vakulskas CA, Yahr TL. 2010. ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol 192:1479–1486. doi: 10.1128/JB.01457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. 2008. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 68:657–671. doi: 10.1111/j.1365-2958.2008.06179.x. [DOI] [PubMed] [Google Scholar]
- 133.Yahr TL, Wolfgang MC. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 134.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 135.Woods DE, Straus DC, Johanson WG Jr, Berry VK, Bass JA. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun 29:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 137.Belete B, Lu H, Wozniak DJ. 2008. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J Bacteriol 190:2023–2030. doi: 10.1128/JB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 139.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jain M, Bar-Meir M, McColley S, Cullina J, Potter E, Powers C, Prickett M, Seshadri R, Jovanovic B, Petrocheilou A, King JD, Hauser AR. 2008. Evolution of Pseudomonas aeruginosa type III secretion in cystic fibrosis: a paradigm of chronic infection. Transl Res 152:257–264. doi: 10.1016/j.trsl.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, Bar-Meir M, Sullivan CL, McColley SA, Hauser AR. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wagner VE, Iglewski BH. 2008. P. aeruginosa biofilms in CF infection. Clin Rev Allergy Immunol 35:124–134. doi: 10.1007/s12016-008-8079-9. [DOI] [PubMed] [Google Scholar]
- 144.Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, Levesque RC. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5:1294–1308. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 145.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, Deshazer D. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol 64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 152.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 154.Ma AT, Mekalanos JJ. 2010. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A 107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bladergroen MR, Badelt K, Spaink HP. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- 156.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Silverman JM, Brunet YR, Cascales E, Mougous JD. 2012. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Laville J, Blumer C, Von Schroetter C, Gaia V, Defago G, Keel C, Haas D. 1998. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol 180:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laville J, Voisard C, Keel C, Maurhofer M, Defago G, Haas D. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci U S A 89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maurhofer M, Hase C, Meuwly P, Metraux JP, Defago G. 1994. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0—influence of the gacA gene and of pyoverdine production. Phytopathology 84:139–146. doi: 10.1094/Phyto-84-139. [DOI] [Google Scholar]
- 162.Oberhansli T, Dfago G, Haas D. 1991. Indole-3-acetic acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens: role of tryptophan side chain oxidase. J Gen Microbiol 137:2273–2279. doi: 10.1099/00221287-137-10-2273. [DOI] [PubMed] [Google Scholar]
- 163.Sacherer P, Defago G, Haas D. 1994. Extracellular protease and phospholipase-C Are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol Lett 116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 164.Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, Reimmann C, Notz R, Defago G, Haas D, Keel C. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol 182:1215–1225. doi: 10.1128/JB.182.5.1215-1225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Blumer C, Haas D. 2000. Multicopy suppression of a gacA mutation by the infC operon in Pseudomonas fluorescens CHA0: competition with the global translational regulator RsmA. FEMS Microbiol Lett 187:53–58. doi: 10.1111/j.1574-6968.2000.tb09136.x. [DOI] [PubMed] [Google Scholar]
- 166.Kay E, Dubuis C, Haas D. 2005. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A 102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol 66:341–356. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- 168.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 169.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Willcox MD, Zhu H, Conibear TC, Hume EB, Givskov M, Kjelleberg S, Rice SA. 2008. Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology 154:2184–2194. doi: 10.1099/mic.0.2008/019281-0. [DOI] [PubMed] [Google Scholar]
- 171.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol 24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 172.Dubuis C, Haas D. 2007. Cross-species GacA-controlled induction of antibiosis in pseudomonads. Appl Environ Microbiol 73:650–654. doi: 10.1128/AEM.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Takeuchi K, Kiefer P, Reimmann C, Keel C, Dubuis C, Rolli J, Vorholt JA, Haas D. 2009. Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem 284:34976–34985. doi: 10.1074/jbc.M109.052571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Camara M, Emsley J, Williams P, Searle MS. 2013. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21:1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res 35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol 59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 178.Adamson DN, Lim HN. 2013. Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. Proc Natl Acad Sci U S A 110:13120–13125. doi: 10.1073/pnas.1308476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Workentine ML, Chang L, Ceri H, Turner RJ. 2009. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol Lett 292:50–56. doi: 10.1111/j.1574-6968.2008.01445.x. [DOI] [PubMed] [Google Scholar]
- 182.Hsu JL, Chen HC, Peng HL, Chang HY. 2008. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J Biol Chem 283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, Filloux A. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol 76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Firoved AM, Deretic V. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol 185:7029–7029. doi: 10.1128/JB.185.23.7029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 187.Ohman DE, Chakrabarty AM. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun 33:142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lory S, Wolfgang M, Lee V, Smith R. 2004. The multi-talented bacterial adenylate cyclases. Int J Med Microbiol 293:479–482. doi: 10.1078/1438-4221-00297. [DOI] [PubMed] [Google Scholar]
- 189.Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 190.Queiroz MH, Madrid C, Paytubi S, Balsalobre C, Juarez A. 2011. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology 157:2504–2514. doi: 10.1099/mic.0.049197-0. [DOI] [PubMed] [Google Scholar]
- 191.Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. 2008. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J Bacteriol 190:4736–4748. doi: 10.1128/JB.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 193.Abu Kwaik Y, Gao LY, Stone BJ, Venkataraman C, Harb OS. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol 64:3127–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66:3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hammer BK, Swanson MS. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol 33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 197.Bachman MA, Swanson MS. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol 40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 198.Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol 44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 199.Edwards RL, Dalebroux ZD, Swanson MS. 2009. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol 71:1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- 200.Dalebroux ZD, Edwards RL, Swanson MS. 2009. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol 71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- 201.Fettes PS, Forsbach-Birk V, Lynch D, Marre R. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int J Med Microbiol 291:353–360. doi: 10.1078/1438-4221-00141. [DOI] [PubMed] [Google Scholar]
- 202.Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol 9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 204.Zink SD, Pedersen L, Cianciotto NP, Abu Kwaik Y. 2002. The Dot/Icm type IV secretion system of Legionella pneumophila is essential for the induction of apoptosis in human macrophages. Infect Immun 70:1657–1663. doi: 10.1128/IAI.70.3.1657-1663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Segal G, Shuman HA. 1999. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol Microbiol 33:669–670. doi: 10.1046/j.1365-2958.1999.01511.x. [DOI] [PubMed] [Google Scholar]
- 206.Qiu J, Luo ZQ. 2013. Effector translocation by the Legionella Dot/Icm type IV secretion system. Curr Top Microbiol Immunol 376:103–115. doi: 10.1007/82_2013_345. [DOI] [PubMed] [Google Scholar]
- 207.Ensminger AW, Isberg RR. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr Opin Microbiol 12:67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shin S, Roy CR. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol 10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 209.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. 2011. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol 2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Segal G, Shuman HA. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol 6:253–255. doi: 10.1016/S0966-842X(98)01308-0. [DOI] [PubMed] [Google Scholar]
- 212.Vogel JP, Isberg RR. 1999. Cell biology of Legionella pneumophila. Curr Opin Microbiol 2:30–34. doi: 10.1016/S1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 213.Kirby JE, Vogel JP, Andrews HL, Isberg RR. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol Microbiol 27:323–336. doi: 10.1046/j.1365-2958.1998.00680.x. [DOI] [PubMed] [Google Scholar]
- 214.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol 196:681–692. doi: 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hagiwara D, Yamashino T, Mizuno T. 2004. A genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem 68:1758–1767. doi: 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- 217.Winfield MD, Groisman EA. 2004. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A 101:17162–17167. doi: 10.1073/pnas.0406038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 219.Mitrophanov AY, Jewett MW, Hadley TJ, Groisman EA. 2008. Evolution and dynamics of regulatory architectures controlling polymyxin B resistance in enteric bacteria. PLoS Genet 4:e1000233. doi: 10.1371/journal.pgen.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Winfield MD, Latifi T, Groisman EA. 2005. Transcriptional regulation of the 4-amino-4-deoxy-l-arabinose biosynthetic genes in Yersinia pestis. J Biol Chem 280:14765–14772. doi: 10.1074/jbc.M413900200. [DOI] [PubMed] [Google Scholar]
- 221.Viau C, Le Sage V, Ting DK, Gross J, Le Moual H. 2011. Absence of PmrAB-mediated phosphoethanolamine modifications of Citrobacter rodentium lipopolysaccharide affects outer membrane integrity. J Bacteriol 193:2168–2176. doi: 10.1128/JB.01449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock RE. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol 188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Al-Khodor S, Kalachikov S, Morozova I, Price CT, Abu Kwaik Y. 2009. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect Immun 77:374–386. doi: 10.1128/IAI.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 225.Schell U, Kessler A, Hilbi H. 2014. Phosphorylation signalling through the Legionella quorum sensing histidine kinases LqsS and LqsT converges on the response regulator LqsR. Mol Microbiol 92:1039–1055. doi: 10.1111/mmi.12612. [DOI] [PubMed] [Google Scholar]
- 226.Tiaden A, Spirig T, Weber SS, Bruggemann H, Bosshard R, Buchrieser C, Hilbi H. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- 227.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bachman MA, Swanson MS. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect Immun 72:3284–3293. doi: 10.1128/IAI.72.6.3284-3293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hales LM, Shuman HA. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol 181:4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Morash MG, Brassinga AK, Warthan M, Gourabathini P, Garduno RA, Goodman SD, Hoffman PS. 2009. Reciprocal expression of integration host factor and HU in the developmental cycle and infectivity of Legionella pneumophila. Appl Environ Microbiol 75:1826–1837. doi: 10.1128/AEM.02756-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pitre CA, Tanner JR, Patel P, Brassinga AK. 2013. Regulatory control of temporally expressed integration host factor (IHF) in Legionella pneumophila. Microbiology 159:475–492. doi: 10.1099/mic.0.062117-0. [DOI] [PubMed] [Google Scholar]
- 233.Beisel CL, Storz G. 2010. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev 34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Swearingen MC, Porwollik S, Desai PT, McClelland M, Ahmer BM. 2012. Virulence of 32 Salmonella strains in mice. PLoS One 7:e36043. doi: 10.1371/journal.pone.0036043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Iniguez AL, Dong Y, Carter HD, Ahmer BM, Stone JM, Triplett EW. 2005. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact 18:169–178. doi: 10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- 237.Dong Y, Iniguez AL, Ahmer BM, Triplett EW. 2003. Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl Environ Microbiol 69:1783–1790. doi: 10.1128/AEM.69.3.1783-1790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, Adams LG, Konjufca V, Curtiss R III, Slauch JM, Ahmer BM. 2008. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nuccio SP, Baumler AJ. 2014. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5(2):e00929–914. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.van der Heijden J, Finlay BB. 2012. Type III effector-mediated processes in Salmonella infection. Future Microbiol 7:685–703. doi: 10.2217/fmb.12.49. [DOI] [PubMed] [Google Scholar]
- 241.Sanchez-Vargas FM, Abu-El-Haija MA, Gomez-Duarte OG. 2011. Salmonella infections: an update on epidemiology, management, and prevention. Travel Med Infect Dis 9:263–277. doi: 10.1016/j.tmaid.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 242.Godinez I, Keestra AM, Spees A, Baumler AJ. 2011. The IL-23 axis in Salmonella gastroenteritis. Cell Microbiol 13:1639–1647. doi: 10.1111/j.1462-5822.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- 243.Tuuminen T, Lounamo K, Leirisalo-Repo M. 2013. A review of serological tests to assist diagnosis of reactive arthritis: critical appraisal on methodologies. Front Immunol 4:418. doi: 10.3389/fimmu.2013.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ajene AN, Fischer Walker CL, Black RE. 2013. Enteric pathogens and reactive arthritis: a systematic review of Campylobacter, Salmonella and Shigella-associated reactive arthritis. J Health Popul Nutr 31:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Collazo CM, Galan JE. 1997. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene 192:51–59. doi: 10.1016/S0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 246.Collazo CM, Galan JE. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol 24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 247.Garcia-del Portillo F, Finlay BB. 1994. Salmonella invasion of nonphagocytic cells induces formation of macropinosomes in the host cell. Infect Immun 62:4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3(3):e00143–112. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sekirov I, Gill N, Jogova M, Tam N, Robertson M, de Llanos R, Li Y, Finlay BB. 2010. Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1:30–41. doi: 10.4161/gmic.1.1.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Valdez Y, Grassl GA, Guttman JA, Coburn B, Gros P, Vallance BA, Finlay BB. 2009. Nramp1 drives an accelerated inflammatory response during Salmonella-induced colitis in mice. Cell Microbiol 11:351–362. doi: 10.1111/j.1462-5822.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- 254.Woo H, Okamoto S, Guiney D, Gunn JS, Fierer J. 2008. A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS One 3:e1603. doi: 10.1371/journal.pone.0001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hapfelmeier S, Hardt WD. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol 13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 257.Stecher B, Macpherson AJ, Hapfelmeier S, Kremer M, Stallmach T, Hardt WD. 2005. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun 73:3228–3241. doi: 10.1128/IAI.73.6.3228-3241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Tsolis RM, Kingsley RA, Townsend SM, Ficht TA, Adams LG, Baumler AJ. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv Exp Med Biol 473:261–274. [PubMed] [Google Scholar]
- 259.Lee CA, Silva M, Siber AM, Kelly AJ, Galyov E, McCormick BA. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc Natl Acad Sci U S A 97:12283–12288. doi: 10.1073/pnas.97.22.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Baumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect Immun 71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Francis CL, Ryan TA, Jones BD, Smith SJ, Falkow S. 1993. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 262.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. 1994. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med 179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 265.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun 67:5690–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Chakravortty D, Hansen-Wester I, Hensel M. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med 195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun 67:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Johnston C, Pegues DA, Hueck CJ, Lee A, Miller SI. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol 22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 271.Altier C, Suyemoto M, Lawhon SD. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect Immun 68:6790–6797. doi: 10.1128/IAI.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol 35:635–646. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 273.Teplitski M, Goodier RI, Ahmer BM. 2006. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int J Med Microbiol 296:449–466. doi: 10.1016/j.ijmm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 274.Olekhnovich IN, Kadner RJ. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol 184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Schechter LM, Lee CA. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol 40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- 276.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 277.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol 185:5096–5108. doi: 10.1128/JB.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lostroh CP, Lee CA. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of PprgH from Salmonella pathogenicity island 1. J Bacteriol 183:4876–4885. doi: 10.1128/JB.183.16.4876-4885.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lostroh CP, Bajaj V, Lee CA. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol Microbiol 37:300–315. doi: 10.1046/j.1365-2958.2000.01991.x. [DOI] [PubMed] [Google Scholar]
- 280.Darwin KH, Miller VL. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J 20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Darwin KH, Miller VL. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol Microbiol 35:949–960. doi: 10.1046/j.1365-2958.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- 282.Eichelberg K, Galan JE. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun 67:4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Dieye Y, Dyszel JL, Kader R, Ahmer BM. 2007. Systematic analysis of the regulation of type three secreted effectors in Salmonella enterica serovar Typhimurium. BMC Microbiol 7:3. doi: 10.1186/1471-2180-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A 105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sterzenbach T, Nguyen KT, Nuccio SP, Winter MG, Vakulskas CA, Clegg S, Romeo T, Baumler AJ. 2013. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J 32:2872–2883. doi: 10.1038/emboj.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Teplitski M, Al-Agely A, Ahmer BM. 2006. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- 287.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol 190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. 2013. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87:1045–1060. doi: 10.1111/mmi.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 290.Lutz C, Erken M, Noorian P, Sun S, McDougald D. 2013. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol 26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 293.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303–314. doi: 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 295.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 296.Freeman JA, Lilley BN, Bassler BL. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol 35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- 297.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 298.Freeman JA, Bassler BL. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol 181:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 300.Lilley BN, Bassler BL. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 301.Lenz DH, Bassler BL. 2007. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol Microbiol 63:859–871. doi: 10.1111/j.1365-2958.2006.05545.x. [DOI] [PubMed] [Google Scholar]
- 302.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Shao Y, Bassler BL. 2012. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol 83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Withey JH, DiRita VJ. 2006. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol 59:1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 306.Kovacikova G, Skorupski K. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 307.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Leimbach A, Hacker J, Dobrindt U. 2013. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358:3–32. doi: 10.1007/82_2012_303. [DOI] [PubMed] [Google Scholar]
- 309.Manges AR, Johnson JR. 2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 55:712–719. doi: 10.1093/cid/cis502. [DOI] [PubMed] [Google Scholar]
- 310.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. 2009. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun 77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bhatt S, Romeo T, Kalman D. 2011. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol 19:217–224. doi: 10.1016/j.tim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Chen HD, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev 29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 316.Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun 41:1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.McDaniel TK, Kaper JB. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol 23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 318.Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol 12:101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. 2009. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol 17:361–370. doi: 10.1016/j.tim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 320.Jimenez R, Cruz-Migoni SB, Huerta-Saquero A, Bustamante VH, Puente JL. 2010. Molecular characterization of GrlA, a specific positive regulator of ler expression in enteropathogenic Escherichia coli. J Bacteriol 192:4627–4642. doi: 10.1128/JB.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Bhatt S, Anyanful A, Kalman D. 2011. CsrA and TnaB coregulate tryptophanase activity to promote exotoxin-induced killing of Caenorhabditis elegans by enteropathogenic Escherichia coli. J Bacteriol 193:4516–4522. doi: 10.1128/JB.05197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bommarius B, Anyanful A, Izrayelit Y, Bhatt S, Cartwright E, Wang W, Swimm AI, Benian GM, Schroeder FC, Kalman D. 2013. A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PLoS One 8:e54456. doi: 10.1371/journal.pone.0054456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, Schembri MA. 2013. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol 16:100–107. doi: 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 324.Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. 2013. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One 8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, Zhu X, Mukhopadhyay S. 2012. BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PLoS One 7:e31348. doi: 10.1371/journal.pone.0031348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Tomenius H, Pernestig AK, Jonas K, Georgellis D, Mollby R, Normark S, Melefors O. 2006. The Escherichia coli BarA-UvrY two-component system is a virulence determinant in the urinary tract. BMC Microbiol 6:27. doi: 10.1186/1471-2180-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Carayol N, Tran Van Nhieu G . 2013. The inside story of Shigella invasion of intestinal epithelial cells. Cold Spring Harb Perspect Med 3:a016717. doi: 10.1101/cshperspect.a016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ashida H, Ogawa M, Mimuro H, Sasakawa C. 2009. Shigella infection of intestinal epithelium and circumvention of the host innate defense system. Curr Top Microbiol Immunol 337:231–255. doi: 10.1007/978-3-642-01846-6_8. [DOI] [PubMed] [Google Scholar]
- 329.Gore AL, Payne SM. 2010. CsrA and Cra influence Shigella flexneri pathogenesis. Infect Immun 78:4674–4682. doi: 10.1128/IAI.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Bottone EJ. 1997. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev 10:257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Dube P. 2009. Interaction of Yersinia with the gut: mechanisms of pathogenesis and immune evasion. Curr Top Microbiol Immunol 337:61–91. doi: 10.1007/978-3-642-01846-6_3. [DOI] [PubMed] [Google Scholar]
- 332.Nagel G, Lahrz A, Dersch P. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol Microbiol 41:1249–1269. doi: 10.1046/j.1365-2958.2001.02522.x. [DOI] [PubMed] [Google Scholar]
- 333.Heroven AK, Bohme K, Rohde M, Dersch P. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol 68:1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- 334.Heroven AK, Dersch P. 2006. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol Microbiol 62:1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x. [DOI] [PubMed] [Google Scholar]
- 335.Nuss AM, Schuster F, Kathrin Heroven A, Heine W, Pisano F, Dersch P. 2014. A direct link between the global regulator PhoP and the Csr regulon in Y. pseudotuberculosis through the small regulatory RNA CsrC. RNA Biol 11:580–593. doi: 10.4161/rna.28676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Grabenstein JP, Marceau M, Pujol C, Simonet M, Bliska JB. 2004. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect Immun 72:4973–4984. doi: 10.1128/IAI.72.9.4973-4984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Perombelon MCM, Kelman A. 1980. Ecology of the soft rot Erwinias. Annu Rev Phytopathol 18:361–387. doi: 10.1146/annurev.py.18.090180.002045. [DOI] [Google Scholar]
- 338.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Hinton JC, Gill DR, Lalo D, Plastow GS, Salmond GP. 1990. Sequence of the peh gene of Erwinia carotovora: homology between Erwinia and plant enzymes. Mol Microbiol 4:1029–1036. doi: 10.1111/j.1365-2958.1990.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 340.Liu Y, Chatterjee A, Chatterjee AK. 1994. Nucleotide sequence, organization and expression of rdgA and rdgB genes that regulate pectin lyase production in the plant pathogenic bacterium Erwinia carotovora subsp. carotovora in response to DNA-damaging agents Mol Microbiol 14:999–1010. [DOI] [PubMed] [Google Scholar]
- 341.Barnard AM, Bowden SD, Burr T, Coulthurst SJ, Monson RE, Salmond GP. 2007. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philos Trans R Soc Lond B Biol Sci 362:1165–1183. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Jones S, Yu B, Bainton NJ, Birdsall M, Bycroft BW, Chhabra SR, Cox AJ, Golby P, Reeves PJ, Stephens S, et al. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J 12:2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR, Salmond GP, Toth IK. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog 4:e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Pirhonen M, Flego D, Heikinheimo R, Palva ET. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J 12:2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 346.Cui Y, Madi L, Mukherjee A, Dumenyo CK, Chatterjee AK. 1996. The RsmA-mutants of Erwinia carotovora subsp. carotovora strain Ecc71 overexpress hrpNEcc and elicit a hypersensitive reaction-like response in tobacco leaves. Mol Plant Microbe Interact 9:565–573. doi: 10.1094/MPMI-9-0565. [DOI] [PubMed] [Google Scholar]
- 347.Mukherjee A, Cui Y, Liu Y, Chatterjee AK. 1997. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol Plant Microbe Interact 10:462–471. doi: 10.1094/MPMI.1997.10.4.462. [DOI] [PubMed] [Google Scholar]
- 348.Liu Y, Cui Y, Mukherjee A, Chatterjee AK. 1998. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol 29:219–234. doi: 10.1046/j.1365-2958.1998.00924.x. [DOI] [PubMed] [Google Scholar]
- 349.Cui Y, Chatterjee A, Chatterjee AK. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol Plant Microbe Interact 14:516–526. doi: 10.1094/MPMI.2001.14.4.516. [DOI] [PubMed] [Google Scholar]
- 350.Chatterjee A, Cui Y, Chatterjee AK. 2002. RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-l-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. J Bacteriol 184:4089–4095. doi: 10.1128/JB.184.15.4089-4095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Koiv V, Mae A. 2001. Quorum sensing controls the synthesis of virulence factors by modulating rsmA gene expression in Erwinia carotovora subsp. carotovora. Mol Genet Genomics 265:287–292. doi: 10.1007/s004380000413. [DOI] [PubMed] [Google Scholar]
- 352.Cui Y, Chatterjee A, Hasegawa H, Chatterjee AK. 2006. Erwinia carotovora subspecies produce duplicate variants of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N-acylhomoserine lactone signals. J Bacteriol 188:4715–4726. doi: 10.1128/JB.00351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Chatterjee A, Cui Y, Hasegawa H, Leigh N, Dixit V, Chatterjee AK. 2005. Comparative analysis of two classes of quorum-sensing signaling systems that control production of extracellular proteins and secondary metabolites in Erwinia carotovora subspecies. J Bacteriol 187:8026–8038. doi: 10.1128/JB.187.23.8026-8038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Cui Y, Chatterjee A, Hasegawa H, Dixit V, Leigh N, Chatterjee AK. 2005. ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA-binding protein. J Bacteriol 187:4792–4803. doi: 10.1128/JB.187.14.4792-4803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Bowden SD, Eyres A, Chung JC, Monson RE, Thompson A, Salmond GP, Spring DR, Welch M. 2013. Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p) ppGpp. Mol Microbiol 90:457–471. doi: 10.1111/mmi.12369. [DOI] [PubMed] [Google Scholar]
- 356.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 357.Boch J, Bonas U. 2010. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 358.Chao NX, Wei K, Chen C, Meng QL, Tang DJ, He YO, Lu GT, Jiang BL, Liang XX, Feng JX, Chen B, Tang JL. 2008. The rsmA-like gene rsmAXcc of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol Plant Microbe Interact 21:411–423. doi: 10.1094/MPMI-21-4-0411. [DOI] [PubMed] [Google Scholar]
- 359.Buttner D, Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 360.Shenge KC, Mabagala RB, Mortensen CN, Stephan D, Wydra K. 2007. First report of bacterial speck of tomato caused by Pseudomonas syringae pv. tomato in Tanzania. Plant Dis 91:462–462. doi: 10.1094/PDIS-91-4-0462C. [DOI] [PubMed] [Google Scholar]
- 361.Steele H, Laue BE, MacAskill GA, Hendry SJ, Green S. 2010. Analysis of the natural infection of European horse chestnut (Aesculus hippocastanum) by Pseudomonas syringae pv. aesculi. Plant Pathol 59:1005–1013. doi: 10.1111/j.1365-3059.2010.02354.x. [DOI] [Google Scholar]
- 362.Romantschuk M, Bamford DH. 1986. The causal agent of halo blight in bean, Pseudomonas syringae pv. phaseolicola, attaches to stomata via its pili. Microb Pathog 1:139–148. doi: 10.1016/0882-4010(86)90016-1. [DOI] [PubMed] [Google Scholar]
- 363.Kong HS, Roberts DP, Patterson CD, Kuehne SA, Heeb S, Lakshman DK, Lydon J. 2012. Effect of overexpressing rsmA from Pseudomonas aeruginosa on virulence of select phytotoxin-producing strains of P. syringae. Phytopathology 102:575–587. doi: 10.1094/PHYTO-09-11-0267. [DOI] [PubMed] [Google Scholar]
- 364.Barta TM, Kinscherf TG, Willis DK. 1992. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J Bacteriol 174:3021–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Chatterjee A, Cui Y, Yang H, Collmer A, Alfano JR, Chatterjee AK. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol Plant Microbe Interact 16:1106–1117. doi: 10.1094/MPMI.2003.16.12.1106. [DOI] [PubMed] [Google Scholar]
- 366.De la Torre-Zavala S, Aguilera S, Ibarra-Laclette E, Hernandez-Flores JL, Hernandez-Morales A, Murillo J, Alvarez-Morales A. 2011. Gene expression of Pht cluster genes and a putative non-ribosomal peptide synthetase required for phaseolotoxin production is regulated by GacS/GacA in Pseudomonas syringae pv. phaseolicola. Res Microbiol 162:488–498. doi: 10.1016/j.resmic.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 367.Dumenyo CK, Mukherjee A, Chun W, Chatterjee AK. 1998. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur J Plant Pathol 104:569–582. doi: 10.1023/A:1008651300599. [DOI] [Google Scholar]
- 368.Hirano SS, Charkowski AO, Collmer A, Willis DK, Upper CD. 1999. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc Natl Acad Sci U S A 96:9851–9856. doi: 10.1073/pnas.96.17.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Hrabak EM, Willis DK. 1993. Involvement of the IemA gene in production of syringomycin and protease by Pseudomonas syringae pv syringae. Mol Plant Microbe Interact 6:368–375. doi: 10.1094/MPMI-6-368. [DOI] [Google Scholar]
- 370.Hrabak EM, Willis DK. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv syringae on bean is a member of a family of 2-component regulators. J Bacteriol 174:3011–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Kinscherf TG, Willis DK. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J Bacteriol 181:4133–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Kitten T, Kinscherf TG, McEvoy JL, Willis DK. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol 28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 373.Marutani M, Taguchi F, Ogawa Y, Hossain MM, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2008. Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol Genet Genomics 279:313–322. doi: 10.1007/s00438-007-0309-y. [DOI] [PubMed] [Google Scholar]
- 374.Quinones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol Plant Microbe Interact 18:682–693. doi: 10.1094/MPMI-18-0682. [DOI] [PubMed] [Google Scholar]
- 375.Records AR, Gross DC. 2010. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol 192:3584–3596. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Rich JJ, Kinscherf TG, Kitten T, Willis DK. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol 176:7468–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Wang N, Lu SE, Wang JL, Chen ZJ, Gross DC. 2006. The expression of genes encoding lipodepsipeptide phytotoxins by Pseudomonas syringae pv. syringae is coordinated in response to plant signal molecules. Mol Plant Microbe Interact 19:257–269. doi: 10.1094/MPMI-19-0257. [DOI] [PubMed] [Google Scholar]
- 378.Willis DK, Holmstadt JJ, Kinscherf TG. 2001. Genetic evidence that loss of virulence associated with gacS or gacA mutations in Pseudomonas syringae B728a does not result from effects on alginate production. Appl Environ Microbiol 67:1400–1403. doi: 10.1128/AEM.67.3.1400-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Willis DK, Hrabak EM, Rich JJ, Barta TM, Lindow SE, Panopoulos NJ. 1990. Isolation and characterization of a Pseudomonas syringae pv syringae mutant deficient in lesion formation on bean. Mol Plant Microbe Interact 3:149–156. doi: 10.1094/MPMI-3-149. [DOI] [Google Scholar]
- 380.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol 64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 381.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol 82:447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Dorman CJ. 2009. Regulatory integration of horizontally-transferred genes in bacteria. Front Biosci 14:4103–4112. [DOI] [PubMed] [Google Scholar]
- 383.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]