Abstract
For years, bacillus Calmette-Guérin (BCG) has served as the unique vaccine against tuberculosis and has generally been regarded as safe. However, a clinical strain labeled 3281 that was isolated from a TB patient was identified to be BCG. Via the combination of next-generation sequencing (NGS) and comparative genomic analysis, unique 3281 genetic characteristics were revealed. A region containing the dnaA and dnaN genes that is closely related to the initial chromosome replication was found to repeat three times on the BCG Pasteur-specific tandem duplication region DU1. Due to the minimum number of epitopes in BCG strains, 3281 was inferred to have a high possibility for immune evasion. Additionally, variations in the virulence genes and predictions for potential virulence factors were analyzed. Overall, we report a pathogen that has never previously been thought to be pathogenic and initial insights that are focused on the genetic characteristics of virulent BCG.
Introduction
During the 20 years since the WHO declared that tuberculosis (TB) is a global public health emergency, great efforts have been made to control and eradicate this diseaseworldwide. Globally, the TB mortality rate has fallen by 45% since 1990. Although considerable progress has been made in these years,an estimated 8.6 million individuals stilldevelop TB, and 1.3 million die from the disease every year[1]. As one of the “three killers” of humans, TB remains a current major global health problem.Furthermore, one-third of the world population is latently infected with Mycobacterium tuberculosis (MTB), which makes the eradication of this this disease more difficult[2].
With the development of genomics and high-throughput sequencing technology, scientists have sought to disclose the “secret garden” of TB via the use of genomic methods[3,4]. H37Rv is a laboratoryvirulent MTB strain whose genome was the first to be completely sequenced, and it has typically been used as areference strain in comparative genomic research. The sole available TB vaccine, bacillus Calmette-Guérin (BCG), was derived from Mycobacterium bovis(M.bovis); the virulence of this mycobacterium was attenuated in the laboratory via cultivation on potatoglycerol medium, and this vaccine can only supply sufficient protection for children. However, this vaccine is incapable ofproviding the same efficacy for adolescents and adults[1,5]. Furthermore, the continual process of the subculturingof BCG in laboratories around the world has led to the generation of daughter strains, and the protective efficacies against these strains has been shown to varyacross laboratories and epidemiological investigations[6–8].To define the molecular basis of the attenuation of BCGs and the variation among daughter strains, comparative genomics research has been performed. Comparisons of BCG to M. bovisrevealed that several genes associated with virulence were lost[9].Further studies identified twotandem duplications, DU1 and DU2,which were shown to vary across all of the BCG vaccine strains[10–12]. In addition to these major mutations, it has been demonstrated that single nucleotide polymorphisms (SNPs) might also play significant roles in the attenuation and variation of BCGs[13,14].
In our study, a strain labeled 3281, which was derived from an adult TB patient who reported having never been inoculated with a TB vaccine and was determined to be free of HIV infection, was screened and identified to be BCG. Our interest was aroused by the question how BCG turned into a pathogen despitebeing regarded as safe for years. Thepresent research compared a virulent BCG isolate withBCG vaccines.
Results and Discussion
Case finding
The strain 3281 was isolated from a 33 year old male, who lived in Hebei province, which is a none-animal-husbandry regionlocated in northern China. The patient worked in a commercial company which was not involved with livestock. The patient had never previously been diagnosed with tuberculosis and there was no known tuberculosis case among his family members or friends. The patient reported a cough and expectorate for less than 3 weeks before he consulted a doctor. The chest X-ray and CT demonstrated sign of pneumonia. Three consecutive sputa were all Acid-Fast Bacilli (AFB) positive while the M. bovis BCG strain was cultured from all of the sputa. Given these reason, we suggested that the M. bovis BCG strain might be the pathogen of this pneumonia patient.
This isolate 3281 belonged to a predominant spoligotype (SB0120) which was frequently reported both among human bovine TB and among cattle[15]. This spoligotype is similar to the spoligotype of the vaccine strain BCG type, and four strains out of the 14 M. bovis strains isolated from cattle in China during 2007 and 2008 had the same spoligotype[16].
MIC(minimal inhibitory concentration)testing
Mycobacterium tuberculosis susceptibility to 12 first- and second-line drugs were performed using Trek Sensitre MYCOTB MIC plate (MYCOTB; Trek Diagnostic Systemes, Cleveland, OH), with incubation at 37°C for 30 days. The MIC was recorded as the lowest antibiotic concentration that reduced visible growth (Table 1). The result showed that BCG 3281 showed a higher resistance to Ethionamide (5μg/ml) than BCG Pasteur (2.5μg/ml), M.bovis(1.2μg/ml) and H37Rv (0.6μg/ml). Meanwhile, BCG 3281 showed similar resistance to Isoniazid as M.bovis (0.12μg/ml), twice that of BCG Pasteur and H37Rv (0.06μg/ml). In addition, the resistance to Para-aminosalicylic acid, Kanamycin, Ofloxacin and Moxifloxacin of BCG 3281 was different with BCG Pasteur, indicating that BCG 3281 was not a traditional BCG strain.
Table 1. MIC testing results.
| Antibiotic | Concentration Range (μg/ml) | MIC (μg/ml) | |||
|---|---|---|---|---|---|
| BCG 3281 | BCG Pasteur | M. bovis | H37Rv | ||
| Cycloserine | 2–256 | 16 | 16 | 16 | 8 |
| Ethambutol | 0.5–32 | 1 | 1 | 4 | 2 |
| Ethionamide | 0.3–40 | 5 | 2.5 | 1.2 | 0.6 |
| Isoniazid | 0.03–4 | 0.12 | 0.06 | 0.12 | 0.06 |
| Para-aminosalicylic acid | 0.5–64 | 0.5 | 1 | 64 | 0.5 |
| Rifabutin | 0.12–16 | 0.12 | 0.12 | 0.12 | 0.12 |
| Rifampicin | 0.12–16 | 0.12 | 0.12 | 0.5 | 0.25 |
| Kanamycin | 0.6–40 | 1.2 | 0.6 | 1.2 | 2.5 |
| Ofloxacin | 0.25–32 | 0.25 | 1 | 2 | 1 |
| Moxifloxacin | 0.06–8 | 0.06 | 0.25 | 0.5 | 0.5 |
| streptomycin | 0.25–32 | 0.25 | 0.25 | 1 | 0.5 |
| Amikacin | 0.12–16 | 0.12 | 0.12 | 0.5 | 0.5 |
General genomic features
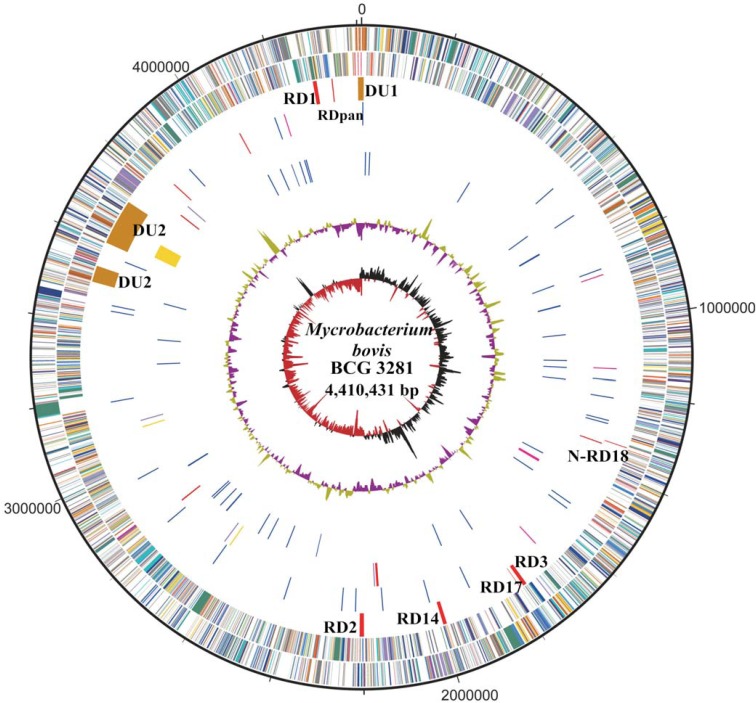
The size of the BCG 3281genome was 4,410,431 bp (Fig 1), and the sequencing error was less than 1/Mb. Thus far, BCG 3281 has the largest genome size in terms of the genomes of BCG that have been completed. The genome of 3281 is 135,909 bps larger than that of BCG Pasteur (Table 2). A total of 4,186 CDSs were identified by glimmer-prediction and reference gene-alignment[17]. Among these CDSs, 3,079 might be COG categories with e-values 1e-5. No credible prophage was found, despite the finding that prophage genes produced four hits in the BCG 3281 genomevia phage-finder[18]. Due to the polymorphic G+C-rich sequences (PGRSs), most of which consist of enzymes involved in lipogenesis and lipolysisandthe Pro-Glu(PE) motif-Pro-Pro-Glu(PPE) motifgene family, BCG 3281’s GC-content was as much as 65.6%, which is similar to the GC contents of MTB and M. bovis[3]. Forty-fivetRNA operons were predicted by tRNAscan-SE,and one rRNAoperon was located by RNAmmer[19,20].
Fig 1. Circular representation of the M. bovis BCG 3281 chromosome.
The outer black circle shows the coordinate. Moving inward, the next two circles show forward and reverse strand CDS, respectively, with colors representing the functional classification, the next circle shows RD(red) and DU (orange), followed by the 3281 unique SNP with nonsynonymous blue and synonymous red, then is the tRNA (blue) and rRNA (purple), final two are GC-content and GC-skew by using a 10-kb window.
Table 2. Genome messages of strains used in this paper.
| Strain | Length(bp) | GC | CDSs | rRNA Operons | tRNA Operons |
|---|---|---|---|---|---|
| M. tuberculosis H37Rv | 4,411,708 | 65.62% | 4111 | 1 | 45 |
| M. bovis AF2122 | 4,345,492 | 65.63% | 3918 | 1 | 45 |
| M. bovis BCG Mexico | 4,350,386 | 65.66% | 3951 | 1 | 45 |
| M. bovis BCG Tokyo | 4,371,711 | 65.64% | 3944 | 1 | 45 |
| M. bovis BCG Pasteur | 4,374,522 | 65.64% | 3949 | 1 | 47 |
| M. bovis BCG Korea | 4,376,711 | 65.64% | 4139 | 1 | 45 |
| M. bovis BCG 3281 | 4,410,431 | 65.65% | 4186 | 1 | 45 |
Genomic comparison with M.bovis and the four BCG strains revealed that the regions of difference (RDs)that contain virulence genes that were lost in the BCGswere also absent in 3281. Compared to the other BCGs and M. bovis, 35 BCG 3281-specific single nucleotide polymorphisms (SNPs) were identified (Fig 1), and 23of these SNPs produced nonsynonymous variations. Additionally, nineindels (threeinsertions and sixdeletions) were found to be exclusive to BCG 3281, and fourother deletions were shared only with M. bovis only. A total of 20 genes were affected by the 23 nonsynonymous variations (S1 Table), and 50 genes were affected by the 13 indels (Table 3 and S2 Table).
Table 3. CDSs involved in indels between M. bovis and BCGs.
| CDSs | Function | CDSs | Function |
|---|---|---|---|
| GS11_3486 | TetR family transcriptional regulator | Mb3236c | hypothetical protein |
| GS11_3501 | acetyl- CoA carboxylase biotin carboxyl carrier protein subunit | Mb3237 | ATP-dependent RNA helicase RhlE |
| GS11_3519 | hypothetical protein Mb3266c | Mb3238 | hypothetical protein |
| BCG_1955c | PPE family protein | Mb3239c | SOJ/PARA-like protein |
| BCG_2407 | hypothetical protein | Mb3240 | acid phosphatase |
| BCG_3228 | hypothetical protein | Mb3241 | isochorismate synthase |
| BCG_3354c | L-lysine-epsilon aminotransferase lat' | Mb3242 | acetyltransferase |
| Mb1951c | malto-oligosyltrehalose synthase | Mb3243c | hypothetical protein |
| Mb2572 | lipoprotein LppA | Mb3244 | hypothetical protein |
| Mb2573 | lipoprotein LprR | Mb3245 | transcriptional regulator WhiB |
| Mb3220 | ABC transporter ATP-binding protein | Mb3246c | two component sensor kinase |
| Mb3222c | DNA helicase II | Mb3247c | acetyl- CoA carboxylase biotin carboxyl carrier protein subunit |
| Mb3223 | glutaredoxin protein | Mb3248c | anti-sigma factor |
| Mb3224c | NADH pyrophosphatase | Mb3249c | hypothetical protein |
| Mb3225c | transmembrane cation transporter | Mb3250c | RNA polymerase sigma factor RpoE |
| Mb3226c | ATP-dependent DNA helicase | Mb3251 | short chain dehydrogenase |
| Mb3227c | ATP-dependent DNA helicase | Mb3254c | hypothetical protein |
| Mb3228 | lipase LipV | Mb3255c | hypothetical protein |
| Mb3229 | DNA-methyltransferase | Mb3256 | transferase |
| Mb3230c | hypothetical protein | Mb3257 | hypothetical protein |
| Mb3231c | molybdopterin biosynthesis-like protein MoeZ | Mb3258c | 3-phosphoshikimate 1-carboxyvinyltransferase |
| Mb3232c | hypothetical protein | Mb3259c | hypothetical protein |
| Mb3233 | TetR family transcriptional regulator | Mb3319c | AsnC family transcriptional regulator |
| Mb3234c | hypothetical protein | Mb3320 | hypothetical protein |
| Mb3235 | hypothetical protein | Mb3321 | piperideine-6-carboxilic acid dehydrogenase |
The GS11 is the official locus of BCG 3281 given by Genbank.
Unique genomic features of the BCG strains
Thirteen years of laboratory cultivation have caused great differences in virulence between the progeny and the original strainand resulted in the attenuated virulence and sufficient reserved antigenicity for protection against TB. Comparative genomic analyses have revealed massive discrepancies between BCG and M. bovis. The most significant two events were the loss of the RD1 regions that contain a specialized secretion system that is strongly associated with pathogenic ability[11,21]and the two tandem duplications, DU1 and DU2. DU1 is restricted to BCG Pasteur 1173P2, and DU2 is present in four different types in different BCGs[10,22].
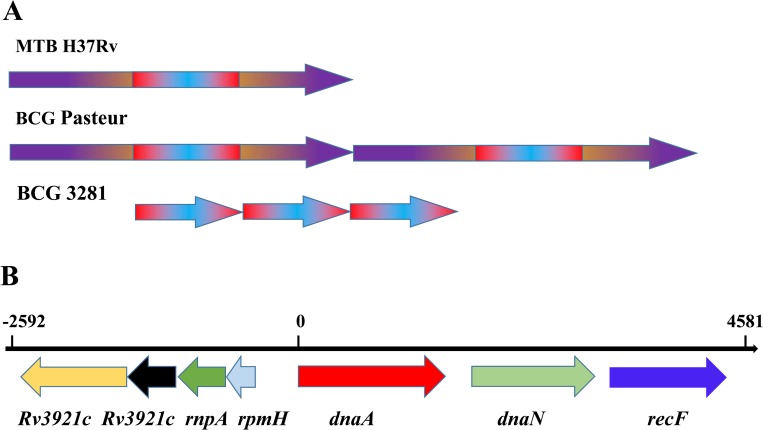
In the genome sequence of BCG 3281, a loss of RD1and duplications in the DU1 and DU2 regions were observed, which validates this stain as BCG. In the DU1 region, a 7 kb unit that covered six genes and crossed the oriC was found to be repeated three times (Fig 2); this duplication is specific to BCG 3281 and has never been reported before (Table 4 and 5). The DU1 in BCG Pasteur is 29.7 Kb, encompassing the region from Rv3910 to Rv0013, while the DU1 in BCG 3281 is only 7.2 Kb, including the region from Rv3921c to Rv0003. BCG 3281 has three copies of dnaA-dnaN region with functional oriC.Protein DnaA initiates chromosome replication when accumulated to the ‘initiation’ level[23], and multiple copies of dnaA in BCG 3281 might help the strain increase growth rate [24]and activate some gene expression [25].Thusthe triploidfor DNA replication elements might partly contribute to the pathogenic of BCG 3281.
Fig 2. Scheme showing the DU1 region of BCG 3281 and BCG Pasteur 1173p2.
(A). The color schemes means duplicated regions. (B). Details of genes involved in the BCG 3281 duplicated units (using H37Rv coordinate).
Table 4. Summary of DU1 regions within M. bovis BCG Pasteur, Mexico, Tokyo, Korea and 3281.
| Strain | H37Rv Coordinate | Unit Length | Repeat Times | Total Length |
|---|---|---|---|---|
| BCG Pasteur | 4398772..16733 | 29.7kb | 2 | 59.4kb |
| BCG Mexico | NA | |||
| BCG Tokyo | NA | |||
| BCG Korea | NA | |||
| BCG 3281 | 4409117..4581 | 7.2kb | 3 | 21.6kb |
“NA” means not present.
Table 5. Genes in the duplication unit that located at DU1 region of M. bovis BCG 3281.
| Genes | Length | Function |
|---|---|---|
| GS11_4181 | 507 | dnaA, chromosomal replication initiation protein |
| GS11_4182 | 402 | dnaN, DNA polymerase III subunit beta |
| GS11_4183 | 385 | recF, recombination protein F |
| GS11_4184 | 366 | oxaA, inner membrane protein translocase component YidC |
| GS11_4185 | 62 | rnpA, ribonuclease P protein component |
| GS11_4186 | 47 | rpmH, 50S ribosomal protein L34 |
The DU2 zone of BCG 3281 belongs to the DU2-Ⅳ type, which consists of two repeat units (41 kb and 37.5 kb) that correspondto regions 3,567,459–3,608,472 and 3,671,536–3,709,097 of M. tuberculosisH37Rv that are separate and repeattwice (Table 6).
Table 6. Summary of DU2 reagions with in M. bovis BCG Pasteur, Mexico, Tokyo, Korea and 3281.
| Strain | Type | H37Rv Coordinate | Unit Length | Repeat Times | Total Length |
|---|---|---|---|---|---|
| BCG Pasteur | DU2-Ⅳ | 3590899..3608474 | 17.5kb | 2 | 72kb |
| 3671533..3690125 | 18.5kb | 2 | |||
| BCG Mexico | DU2-Ⅳ | 3590899..3608474 | 17.5kb | 2 | 72kb |
| 3671533..3690125 | 18.5kb | 2 | |||
| BCG Tokyo | DU2-Ⅰ | 3684226..3705104 | 20.8kb | 3 | 62.4kb |
| BCG Korea | DU2-Ⅳ | 3590899..3608473 | 17.5kb | 3 | 106.5kb |
| 3671533..3690125 | 18kb | 3 | |||
| BCG 3281 | DU2-Ⅲ | 3567459..3608472 | 41kb | 2 | 157kb |
| 3671536..3709097 | 37.5kb | 2 |
The loss of RD1 and the two identified tandem duplications in BCG 3281 confirmed that the strain is a BCG. This result is completely contrasted with our expectation that BCG 3281 would be an M. bovis. Furthermore, the RD17 and RDpan, which are specific to BCGsand lost in M.bovisAF2122, were also found in BCG 3281[26].
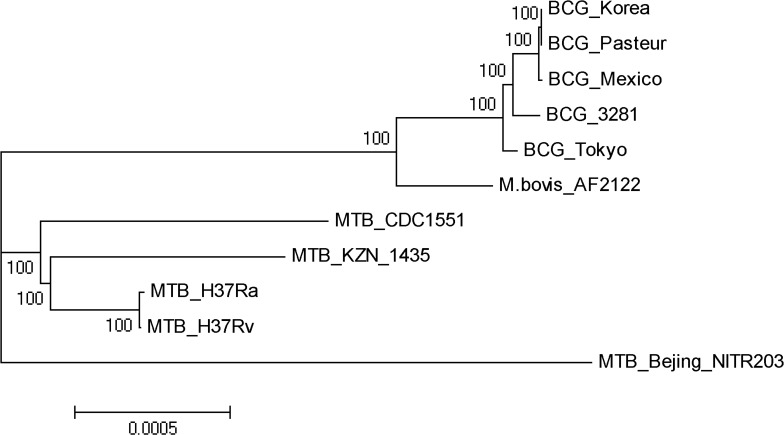
To ensure the accuracy of the strain identification, a SNP-based NJ phylogenetic tree was constructed (Fig 3). The phylogenetic position of BCG 3281was located near BCG Tokyo and far from the clinic strains, which validated 3281 as a BCG. For years, people have acknowledged that BCG strains are safe for vaccination and have notransmissibility. Nevertheless, thestrain 3281, which was isolated from an adult patient who had not been vaccinated with a BCG, was identified to be a BCG. We believe that the source of pathogen in this case was from the vaccine and had mutated to acquire the ability for horizontal transmission.
Fig 3. Phylogenetic tree of M. tuberculosis, M. Bovis and BCGS.
The tree was constructed employing Neighbor-joining method. It is based on the SNPs within 2263 core genes of the strains.
Antigen epitopevariations
Epitopes are the parts of antigens that are recognized by T-cell receptors (TCRs) and B-cell receptor (BCRs) and play the core role in the immune response. We believed similarities between the epitopes of BCG 3281 and M. Bovis or MTB would exist because all of these strains are pathogenic.
To identify the variations in the epitopes of these strains, 2,667 epitopes complied from the Immune Epitope Database(IEDB)[27], including 2,055 T-cell epitopes and 612 B-cell epitopes, were selected and renamed (S3 Table).These epitopes were subsequently positively experimentally identified by IEDB. Four complete genome BCG vaccines (i.e., BCG Pasteur 1173P2, BCG Tokyo 172, BCG Mexico and BCG Korea 1168P) were acquired from the National Center of Biotechnology Information (NCBI).
Only 100% identical match results were considered as the same epitopes because recent studies have shown that human T cell epitopes of Mycobacteriumtuberculosis are evolutionarily hyper-conserved[28]. For comparison, 1,600 epitopes, including 1,213 T-cell epitopes and 387 B-cell epitopes, were identified in all seven strains (BCG 3281, BCG Pasteur 1173P2, BCG Tokyo 172, BCG Mexico, BCG Korea1168P, M.bovisAF2122 and M. tuberculosis H37Rv). In contrast, 531 epitopes, including 404 T-cell epitopes and 127 B-cell epitopes, were absent in all seven strains. Moreover, 329 epitopes, including 290 T-cell epitopes and 39 B-cell epitopes, were found to be lost in only BCG 3281 and other BCGs. Additionally, 44 epitopes, including 33 T-cell epitopes and 11 B-cell epitopes, located in 22 geneswere found to be missing in only BCG 3281. When these 22 genes were examined, frameshiftswere found to have occurred in the coding regions of 19genes and 3 genes were lost (S4 Table).
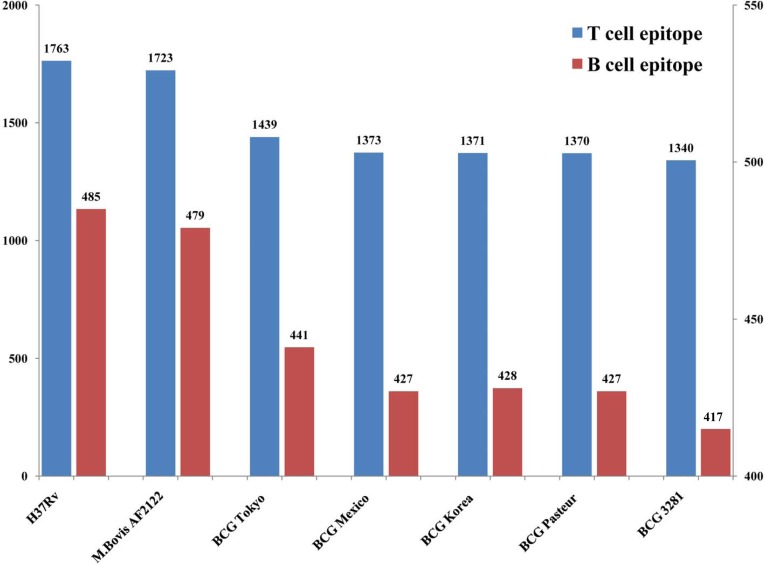
Despite sharing majorities of both T-cell and B-cell epitopes with H37Rv and M. bovis, the BCGs obviously possess fewer epitopes (Fig 4), whichmight result in reduced protection againstTB. In other words, fewer epitopes indicate poorer recognition of alien invadersby the human body. Moreover, BCG 3281 had the fewest number of epitopes among the BCGs, which amplifies the possibility for immune escape. Wen et al. found that BCG Tokyo possess the greatest number of both T-cell and B-cell epitopes among the BCGs and thus might be the vaccine that confers the best immune protection[29]. We found that 62 unique epitopes of BCG Tokyothat are locatedin two BCG Tokyo genes,JTY1991 and JTY1996, that were also present in M.bovis and H37Rvbut absent in other BCGs. The efficiency of BCG protection might be improved by the transduction of two genes into other BCG vaccines. No epitopes unique to 3281 among the other BCGs were identified. In one aspect, this might hint that BCG 3281 did not obtain exogenous genetic element through lateral gene transfer, emphasizing the possibility that pathogenic BCG 3281 might be formed through mutation from BCG vaccine. On the other hand, epitopes that had not been experimentally identified might existed in BCG 3281 unique genes.
Fig 4. Epitopes in M. tuberculosis H37Rv, M. bovis AF2122 and genome finished BCGs.
Duplicate epitopes were removed and only epitopes with 100% identical matches were considered present in the strain.
Virulence factors in BCG 3281
Variation in known virulence factors
BecauseBCG 3281 was considered to be a pathogenic bacterium, we expected that BCG 3281would share extensive similarities with MTB and M. bovisand possess distinct genetic differences from other BCGs, particularly with respect to virulence genes. To detect the variationsin the virulence factors, 88 virulence genes that were identified from the Virulence Factors Database (VFDB) were selected[30]. Blastpresults (Table 7) revealed that 51 virulence genes were 100% identical with M. bovis and the five CG strains,threegenes (located at RD5) were lost in both M. bovis and all of the BCGs, and sevengenes were M. bovis-specific; the latter genes were located at RD1 and were lost in all of the BCGs. A copy number variation (CNV) of one gene (VFG1412) was found and was located in the DU2 region. Additionally, a frameshiftin one virulence gene (VFG2388) was found in both M. bovis and the BCGs. Moreover, plentiful nonsynonymousmutations were identified. To our surprise, no virulence genes were found to be specific to BCG 3281 with respect to M. bovis and the other BCGs. Although the differences between M. bovisand BCG 3281were enormous, these differences were found to be common characteristics of BCGs.
Table 7. Comparison of mutative virulence factors within M. bovis AF2122 and M. bovis BCG 3281, Pasteur, Tokyo, Mexico and Korea.
| Virulence Factors | M. bovis | BCG 3281 | BCG Pasteur | BCG Tokyo | BCG Mexico | BCG Korea |
|---|---|---|---|---|---|---|
| VFG1382 | + | + | + | 1 | + | + |
| VFG1384 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1385 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1386 | + | 1 | + | + | + | + |
| VFG1390 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1391 | 2 | 2 | 2 | 2 | 2 | 2 |
| VFG1396 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1400 | - | - | - | - | - | - |
| VFG1401 | - | - | - | - | - | - |
| VFG1402 | - | - | - | - | - | - |
| VFG1407 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1408 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1409 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1412 | 1 copy | 2 copies | 2 copies | 1 copy | 2 copies | 3 copies |
| VFG1421 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1422 | + | - | - | - | - | - |
| VFG1423 | + | - | - | - | - | - |
| VFG1812 | + | 1 | 1 | 1 | 1 | 1 |
| VFG1815 | + | + | 1 | + | + | 1 |
| VFG1816 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1818 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG1820 | 2 | 2 | 2 | 2 | 2 | 2 |
| VFG1825 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG2378 | + | - | - | - | - | - |
| VFG2379 | + | - | - | - | - | - |
| VFG2380 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG2383 | + | trancated | trancated | trancated | trancated | trancated |
| VFG2384 | + | - | - | - | - | - |
| VFG2385 | + | - | - | - | - | - |
| VFG2387 | 2 | 2 | 2 | 2 | 2 | 2 |
| VFG2388 | frameshift | frameshift | frameshift | frameshift | frameshift | frameshift |
| VFG2389 | trancated | - | - | - | - | - |
| VFG2391 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG2394 | 3 | 3 | 3 | 3 | 3 | 3 |
| VFG2395 | 1 | 2 | 2 | 2 | 2 | 2 |
| VFG2397 | 1 | 1 | 1 | 1 | 1 | 1 |
| VFG2398 | 2 | 2 | 1 | 1 | 1 | 1 |
“+” means 100% in identit, “-” stands for lost, the number shows nonsynonymous mutations number or copy number.
Possible virulence genes
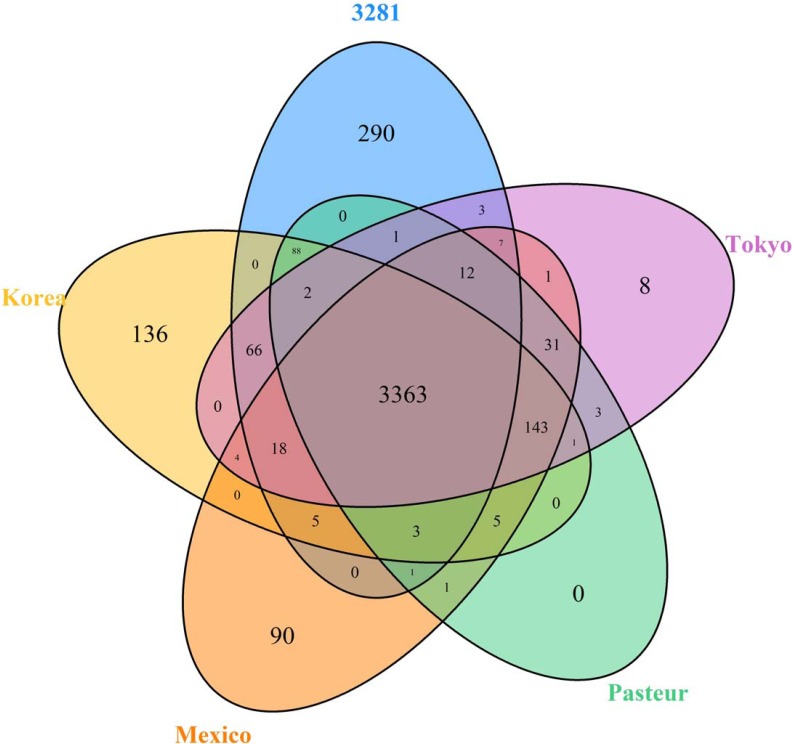
Because no large variations in confirmed virulence genes were detected within BCG 3281, a pan-genome analysis was performed to identify possiblenew virulence factors[31]. Via the use of the pan-genome analysis pipeline (PGAP),orthologous clusters within the 5 BCGs were grouped (Fig 5)[32]. The pan-genome clusters consisted of 4,282 orthologs and had a core of 3,363 orthologs.Two hundred and ninety orthologclusters contained 294 CDSs that were likely to be unique to 3281 and might have conferred additional virulence to BCG 3281.
Fig 5. The Venn diagram showing core orthologs of five genome finished BCGs.
Genes overlapping at least 50% length and 50% (by PGAP) of similarity were considered orthologs.
Considering the prediction discrepancyand the restrictions of the software, we searched these 294 CDSs within the genome and re-predicted the CDS libraries of the other fourBCGs. Ultimately, fourCDSs were proven to be 3281-specific, and all of these CDSs were generated by indels (Table 8).
Table 8. CDSs inferred with potential virulence in M. bovis BCG 3281.
| CDS | Length | Variation | Former Function |
|---|---|---|---|
| GS11_0276 | 1056 | a single nucleotide deletion | succinate dehydrogenase flavoprotein subunit |
| GS11_0578 | 1737 | 9 nucleotides deletion | PE-PGRS family protein |
| GS11_1865 | 2145 | a single nucleotide insertion | WAG22 antigen |
| GS11_3751 | 5442 | 9 nucleotides insertion | PE-PGRS family protein |
The GS11 is the official locus of BCG3281 given by Genbank.
Conclusions
Although BCG, which is an attenuated derivativeof M. bovis, has served for nearly 90 years as the sole vaccine that provides protection against tuberculosis, the clinical strain 3281 was proven to be a BCG and was found to be morbigenous. In an effort to determine the genetic structure of BCG 3281 and determine whether a BCG could be pathogenic, we sequenced the complete genome of BCG 3281 and compared its entire genome to four complete BCG genomesand the genome of M. bovis AF2122. First, to demonstrate the accuracy of the physiological and biochemical identification results, we examined the tandem duplicationsDU1 and DU2, which are significant characteristics of BCGs. Simultaneously, a genetic evolution analysis of the complete BCG genomes and the genome of M. bovis was constructed. The results of both analyses verified that strain 3281 is a BCG.
Examinations of all of the BCG genomes, includingthose of BCG Pasteur, Tokyo, Mexico, Korea,Frappier, Glaxo, Moreau, Phipps, Prague, Sweden, China, ATCC35733, ATCC35740 and ATCC35743,revealed that none contained the 7 kb duplication in the DU1 region.The presence of the dnaA and dnaNgenes is strongly associated with the initiation and regulation of chromosomal replication; thus, we inferred that BCG 3281would likely be capable of enduring greater burdens in replication[24].
To determine whether any identified virulence factors were unique to 3281 relative to the other BCGs, 88 virulence genes located at H37Rv were examined; 3281-unique indels anda single amino acid polymorphismwere located, but 3281-unique virulence factors were not found. We believe that these variations might influence the virulence of BCG 3281 to some extent but not so much as to convert an attenuated vaccine into a pathogenic bacterium. To identify the possible virulence factors, a pan-genome method was applied and four BCG 3281-unique CDSs were identified as puativevirulence genes since no other large variations in genome structure were found.Additionally, we detected antigen epitope variationsin BCG 3281. Compared to the other BCGs, BCG 3281 has lost more epitopes, which might intensify this strain’s potential for immune escape and increase the risk of secondary infection.Overall, this study provides initial insight into the characteristics of a pathogenic BCGthat should have significant effects on TB vaccine research.
Materials and Methods
Strain Information
The mycobacterial strain used in this study was acquired from the Beijing Bio-Bankof clinical resourceson Tuberculosis (D09050704640000)". This strain was originally isolated from an adult male patientwho was not infected with HIV.
Genome sequencing, assembly and annotation
Through a combination of next-generation sequencing (NGS) techniques, thegenome was sequenced with both a 454 GS-FLX system and a Hiseq2500. The 454 data were assembled with Newbler 2.5 withcoverage of 29.6.Using Soap 1.05, the Hiseq reads were assembled with a 108.9-fold coverage[33]. Gap closure was performed using the PCR method with the help of ContigScapeusing the 454 assembly results[34]. The low value dots were verified by the Hiseq assembly results. ORFs were predicted with Glimmer 3.0.2 and replenished by reference annotation[35].
SNP and Phylogenetic analyse
All SNPs were identified with Mauve 2.3.1, and they were localized to CDSsvia an in-house Perl script[36].The pangenomemethod was employed for the phylogenetic analysis. A core of 2,263 geneslengths of at least 0.8 and similarities of at least 0.8 was generated. The neighbor-joining tree was generated by MEGA with a bootstrap value of 1,000[37].
Supporting Information
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (973 program, No. 2012CB518800), the National Natural Science Foundation of China (Nos. 31201920, 31272538), Bank of clinical resources on Tuberculosis (D09050704640000), and the Transmission Mode of Tuberculosis project of the National Key Program of Mega Infectious Diseases (2013ZX10003006-002).
Data Availability
The complete genome sequence of BCB 3281 has been uploaded to the database of Genbank with the accession number CP008744 and the other genomes are avaliable at PATRIC: www.patricbrc.org. The epitopes employed in this manuscript were acquired from IEDB: www.iedb.org and the detailed dates were attached in the supporting materials.
Funding Statement
This work was supported by the Major State Basic Research Development Program of China (973 program, No. 2012CB518800), the National Natural Science Foundation of China (Nos. 31201920, 31272538), Bank of clinical resourceson Tuberculosis (D09050704640000), and the Transmission Mode of Tuberculosis project of the National Key Program of Mega Infectious Diseases (2013ZX10003006-002).
References
- 1. Organization WH. Global tuberculosis report 2013: World Health Organization; 2013. [Google Scholar]
- 2. Brennan MJ, Thole J. Tuberculosis vaccines: a strategic blueprint for the next decade. Tuberculosis. 2012;92:S6–S13. 10.1016/S1472-9792(12)70005-7 [DOI] [PubMed] [Google Scholar]
- 3. Cole S, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. [DOI] [PubMed] [Google Scholar]
- 4. Fleischmann R, Alland D, Eisen J, Carpenter L, White O, Peterson J, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. Journal of bacteriology. 2002;184(19):5479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu J, Tran V, Leung AS, Alexander DC, Zhu B. BCG vaccines: their mechanisms of attenuation and impact on safety and protective efficacy. Human vaccines. 2009;5(2):70–8. [DOI] [PubMed] [Google Scholar]
- 6. Davids V, Hanekom WA, Mansoor N, Gamieldien H, Sebastian JG, Hawkridge A, et al. The effect of bacille Calmette-Guerin vaccine strain and route of administration on induced immune responses in vaccinated infants. Journal of Infectious Diseases. 2006;193(4):531–6. [DOI] [PubMed] [Google Scholar]
- 7. Behr MA, Small PM. Has BCG attenuated to impotence? Nature. 1997;389(6647):133–4. [DOI] [PubMed] [Google Scholar]
- 8. Lagranderie M, Balazuc A-M, Deriaud E, Leclerc CD, Gheorghiu M. Comparison of immune responses of mice immunized with five different Mycobacterium bovis BCG vaccine strains. Infection and immunity. 1996;64(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garnier T, Eiglmeier K, Camus J-C, Medina N, Mansoor H, Pryor M, et al. The complete genome sequence of Mycobacterium bovis . Proceedings of the National Academy of Sciences. 2003;100(13):7877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proceedings of the National Academy of Sciences. 2007;104(13):5596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti . Molecular microbiology. 2002;46(3):709–17. [DOI] [PubMed] [Google Scholar]
- 12. Mostowy S, Tsolaki AG, Small PM, Behr MA. The in vitro evolution of BCG vaccines. Vaccine. 2003;21(27):4270–4. [DOI] [PubMed] [Google Scholar]
- 13. Collins DM, Kawakami RP, Buddle BM, Wards BJ, de Lisle GW. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis identifies phoT as having roles in tuberculosis virulence and phosphate transport. Microbiology. 2003;149(11):3203–12. [DOI] [PubMed] [Google Scholar]
- 14. Pelayo MCG, Uplekar S, Keniry A, Lopez PM, Garnier T, Garcia JN, et al. A comprehensive survey of single nucleotide polymorphisms (SNPs) across Mycobacterium bovis strains and M. bovis BCG vaccine strains refines the genealogy and defines a minimal set of SNPs that separate virulent M. bovis strains and M. bovis BCG strains. Infection and immunity. 2009;77(5):2230–8. 10.1128/IAI.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duarte EL, Domingos M, Amado A, Botelho A. Spoligotype diversity of Mycobacterium bovis and Mycobacterium caprae animal isolates. Veterinary microbiology. 2008;130(3–4):415–21. 10.1016/j.vetmic.2008.02.012 [DOI] [PubMed] [Google Scholar]
- 16. Du Y, Qi Y, Yu L, Lin J, Liu S, Ni H, et al. Molecular characterization of Mycobacterium tuberculosis complex (MTBC) isolated from cattle in northeast and northwest China. Research in veterinary science. 2011;90(3):385–91. 10.1016/j.rvsc.2010.07.020 [DOI] [PubMed] [Google Scholar]
- 17. Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic acids research. 2006;34(20):5839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178(5):1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast. 2000;17(2):111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q, Shi H. Coupling chromosomal replication to cell growth by the initiator protein DnaA in Escherichia coli . Journal of theoretical biology. 2012;314:164–72. 10.1016/j.jtbi.2012.08.045 [DOI] [PubMed] [Google Scholar]
- 24. Shuvaev A. DnaA dynamics could be linked with fitness cost in bacteria. Cell biochemistry and biophysics. 2014;70(1):295–9. 10.1007/s12013-014-9908-5 [DOI] [PubMed] [Google Scholar]
- 25. Hoover SE, Xu W, Xiao W, Burkholder WF. Changes in DnaA-dependent gene expression contribute to the transcriptional and developmental response of Bacillus subtilis to manganese limitation in Luria-Bertani medium. J Bacteriol. 2010;192(15):3915–24. 10.1128/JB.00210-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mostowy S, Inwald J, Gordon S, Martin C, Warren R, Kremer K, et al. Revisiting the evolution of Mycobacterium bovis . Journal of bacteriology. 2005;187(18):6386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughan K, Seymour E, Peters B, Sette A. Substantial gaps in knowledge of Bordetella pertussis antibody and T cell epitopes relevant for natural immunity and vaccine efficacy. Human immunology. 2014. [DOI] [PMC free article] [PubMed]
- 28. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nature genetics. 2010;42(6):498–503. 10.1038/ng.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang W, Zhang Y, Zheng H, Pan Y, Liu H, Du P, et al. Genome sequencing and analysis of BCG vaccine strains. PloS one. 2013;8(8):e71243 10.1371/journal.pone.0071243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, et al. VFDB: a reference database for bacterial virulence factors. Nucleic acids research. 2005;33(suppl 1):D325–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. D'Auria G, Jiménez-Hernández N, Peris-Bondia F, Moya A, Latorre A. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC genomics. 2010;11(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28(3):416–8. 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–4. 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- 34. Tang B, Wang Q, Yang M, Xie F, Zhu Y, Zhuo Y, et al. ContigScape: a Cytoscape plugin facilitating microbial genome gap closing. BMC genomics. 2013;14(1):289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hall BG. Building phylogenetic trees from molecular data with MEGA. Molecular biology and evolution. 2013;30(5):1229–35. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
The complete genome sequence of BCB 3281 has been uploaded to the database of Genbank with the accession number CP008744 and the other genomes are avaliable at PATRIC: www.patricbrc.org. The epitopes employed in this manuscript were acquired from IEDB: www.iedb.org and the detailed dates were attached in the supporting materials.